Introduction
Prostate cancer (PCa) continues to be the most
common lethal malignancy diagnosed in men and metastatic
progression of PCa is a major cause of death. During progression to
metastasis, PCa cells are thought to acquire a mesenchymal
phenotype, which allows them to dissociate from the primary tumor,
invade, and migrate to distant organs (1,2). Cell
metastasis involves the accumulation of sequential disrupted
expression of genes, which in turn promotes cancer progression
(3,4). MicroRNAs (miRNAs), a class of small
non-coding RNAs, are single-stranded RNAs consisting of ~22
nucleotides that negatively regulate expression of their target
genes at the 3-untranslated regions (3UTRs) (5–7). A
growing number of miRNAs have been described as candidate oncogenes
or tumor-suppressor genes through respective binding to target
tumor-suppressor genes or oncogenic genes (8–11).
Over the last two decades, distinct expression profiles of miRNAs
have been linked to tumor development and progression in PCa
(12–15). Nevertheless, the exact mechanism of
multiple molecular events in PCa initiation, growth, invasion and
metastasis remains unclear.
The miRNA-194 family is comprised of miR-215,
miR-194-1, miR-192 and miR-194-2, which are clustered and expressed
as two separate polycistronic pri-miRNA transcripts. The miR-192
and miR-194-2 cluster on chromosome 11q13.1 and the miR-215 and
miR-194-1 cluster on chromosome 1q41. miR-194-1 and miR-194-2 have
the identical mature sequence. miR-192 and miR-215 are closely
related with similar seed sequence. Like many other miRNAs, miR-194
may contribute to the development and progression of different
types of cancers, implicating a tumor suppressor function. Emerging
evidence suggests that miR-194 is downregulated in a number of
different malignancies, such as colorectal cancer (16), renal childhood neoplasms (17), liver (18) and endometrial cancer (19). Conversely, increased miR-194
expression is found in a range of cancer cellss, including those of
gastric (20), colorectal (21) and endometrial cancer (22) and is associated with reduced cancer
metastasis. However, the biological functions of the deregulation
of miR-194 in tumor progression have not yet been completely
defined.
The mammalian NudC nuclear distribution protein
(NUDC) is expressed in a broad range of tissues and is increasingly
being recognized as a multifunctional protein that affects various
cellular responses, such as cell division (23), proliferation (24), migration (25,26),
and cytokinesis (27,28). Aberrant regulation of hNUDC
expression is correlated with a variety of pathologies. For
example, the expression levels of hNUDC are much higher in
erythroid precursor cells compared to other human tissues (29). In addition, hNUDC protein expression
was found to be significantly upregulated in patients with acute
myelogenous and acute lymphoblastic leukemia compared to aspirates
from normal controls (30,31). Furthermore, hNUDC was found
expressed in neuroectodermal tumors, but not in non-neoplastic
brain tissue (32). There is also
an inverse correlation between hNUDC expression and nodal
metastasis in esophageal cancer (33). Downregulation of human hNUDC mRNA,
including the use of the antisense oligonucleotides and small
interfering (si)RNAs resulted in impairment of both cell
proliferation in various types of cancer in vitro (27,34).
As a consequence, the cells exhibited nuclear enlargement and
multiple nuclei, possibly due to a failure to complete the
inhibition of both mitosis and cytokinesis. As may be expected, a
protein involved in mitotic cell division is also found to play a
role in cancer. According to previous studies in the literature,
adenovirus-mediated overexpression of hNUDC in LNCaP, DU-145 and
PC-3 cells significantly attenuatedthe rate of cell proliferation
through G2/M phase arrest. This has been interpreted as a potential
requirement for hNUDC to accomplish the first steps of metastasis
(35). Previous results have
demonstrated that both upregulation and downregulation of hNUDC
play an important role in anticancer intervention.
In the present study, the regulation of biological
functions by miR-194 was identified for the first time in PCa by
the targeting of the hNUDC gene. Transfection of DU-145 and PC-3
cell lines with miR-194 decreased hNUDC mRNA and protein expression
and reduced cell migration and invasion. In vitro and in
vivo experiments indicated that the overexpression of miR-194
suppressed colony formation and tumorigenicity of PCa cells in nude
mice. These findings suggest that miR-194 may act as a tumor
suppressor in PCa, which is consistent with its role in other human
cancers.
Materials and methods
Cell lines and culture conditions
Human prostate cancer cell lines, PC-3 and DU-145,
were obtained from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were grown in Dulbeccos modified Eagles
medium (DMEM; Gibco® by Life Technologies™, Grand
Island, NY, USA) supplemented with 10% v/v inactivated fetal bovine
serum (FBS; Biological Industries Israel Beit Haemek, Ltd., Kibbutz
Beit-Haemek, Israel) and 100 U/ml penicillin + 100 µg/ml
streptomycin. Cells were maintained in a humidified incubator at
37°C with 5% CO2.
Transfection procedures
Cells were plated at 1×105 in a 6-well
plate. Twelve hours later, cells were transfected with 50 nM of
scrambled control, miR-194, miR-194 inhibitor, inhibitor control or
siRNA for hNUDC (Suzhou GenePharma Co., Ltd., Suzhou, China) with
the use of Lipofectamine® 2000 reagent (Invitrogen by
Life Technologies, Carlsbad, CA, USA). siRNA for hNUDC used here
has been previously described (27). After transfection, the cells were
processed for western blot analysis, quantitative real-time PCR
(RT-qPCR), migration and invasion assays.
RNA isolation and RT-qPCR
Total RNA was isolated from the prostate cell lines
using TRIzol reagent, and quantified spectrophotometrically. The
reverse transcription of miRNAs from total RNA (1 µg) was performed
with miR-194-specific stem-loop primer
(5-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCACATG-3) using a Bestar™
qPCR RT kit (DBI® Bioscience, Ludwigshafen, Germany). To
measure hNUDC mRNA expression levels, the first strand of cDNA was
synthesized using oligo (dT) primers by Bestar™ qPCR RT kit and the
reverse transcription product was amplified using Bestar™ SybrGreen
qPCR Mastermix (DBI® Bioscience). The sequences of the
primers specific for hNUDC were 5-CAGTGGGGTCTTGCTGTCATCT-3′
(forward) and 5-CTAACCCTTGCCTTTCAACTCA-3′ (reverse). Quantitative
PCR was performed using the StepOnePlus™ Real-Time PCR System
(Applied Biosystems). The PCR reaction consisted in a denaturation
step at 95°C for 10 min, followed by 40 amplification cycles at
95°C for 10 sec, and annealing at 60°C for 35 sec and extension at
72°C for 30 sec. Gene expression was normalized using endogenous
β-actin or U6 as a control. Densitometric quantification was
perfomed with the comparative cycle threshold method
(2−ΔΔCt) (36).
Western blot analysis
Total lysates from DU-145 and PC-3 cells (10 µg)
were applied to 10% SDS-PAGE and transferred onto polyvinylidene
difluoride (PVDF) membranes. Membranes were blocked with 5% BSA in
1X Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at
room temperature. Western blot analysis was performed by incubating
the membranes with primary antibodies for hNUDC (Abcam, Cambridge,
MA, USA) at a dilution of 1:3,000 in 5% BSA in 1X TBST. After
overnight incubation at 4°C, the membranes were incubated with goat
anti-mouse IgG alkaline phosphatase (AP) conjugated secondary
antibodies (Cell Signaling Technology, Inc.) at 1:1,000 for 1 h at
room temperature. Signals were visualized using an enhanced
chemiluminescence (ECL) detection system (Pierce, Rockford, IL,
USA). Densitometric quantification was performed using ImageJ
software.
Dual-Luciferase assay
3′UTR mRNA sequence of hNUDC containing a miR-194
binding site was amplified by the following primers:
5′-ATTACGCGTCCCCTGTTTTTTCCTCCCTG-3′ (forward) and
5′-CGGAAGCTTAGCTGGGCAAATCGTTTTAA-3′ (reverse). The PCR product was
then cloned downstream to the luciferase gene in plasmid
pMIR-Report. A mutation construct was made by deletion of several
bases within the binding site and cloned into the same vector. The
control vector was used for normalization of cell number and
transfection efficiency. Co-transfection of synthetic miR-194 and
luciferase report constructs were transfected in HEK-293T cells by
using Lipofectamine 2000 reagent. Luciferase assays were performed
using the Dual-Luciferase® Reporter Assay System kit
(Promega Corp., Madison, WI, USA) and luminescence was measured on
a GloMax® 96 microplate luminometer (Promega).
In vitro invasion and migration
assays
Cellular invasion and migration were assessed using
the Cultrex® Basement Membrane Extract (BME),
PathClear® (Trevigen, Gaithersburg, MD, USA) according
to the manufacturers protocol. Cells at a density of
5×104 were seeded onto Transwell® Permeable
Supports (Corning Incorporated, Corning, NY, USA). For the
migration assay, the plates were incubated for 12 h at 37°C. The
cells that migrated through the pores without the matrix to the
lower surface of the membrane were fixed in 70% ethanol and stained
with crystal violet. For the invasion assay, the plates were
incubated for 48 h and the cells that invaded through the pores
covered with the matrix to the lower surface of the membrane were
fixed and stained. The cells were counted under a light microscope
in five separate fields. Each blue point indicated an individual
cell and all the images were scanned and counted at ×40
magnification.
Determination of nuclear
morphology
Cells were transfected for 48 h and fixed with
methanol prior to being stained with May-Grünwald-Giemsa staining
solution for 15 min at room temperature. The stained slides were
examined by a phase contrast microscope at ×40 magnification.
Generation of stable cell lines
The packaged lentivirus expressing miR-194,
anti-miR-194-sponge and GFP-control were purchased directly from
ViGene Biosciences (Shandong, China). Stable expression of miR-194,
GFP-control and anti-miR-194-sponge in PC-3 and DU-145 cells was
achieved through selection with 2 µg/ml puromycin (Amresco, Solon,
OH, USA).
Clonogenic cell growth assay
The clonogenic cell growth of PC-3 and DU-145 cell
lines was examined on soft agar using Cell Transformation Detection
Assay (Millipore, Temecula, CA, USA). Briefly, each well of a
24-well plate was first layered with 0.8% agarose in growth medium
(DMEM supplemented with 10% FBS). The cell lines to be tested were
trypsinized, and 1×104 cells were resuspended in a
growth medium containing 0.4% agarose and then they were poured as
a top layer in the 24-well plates. The plates were incubated at
37°C for 28 days. Colony formation was observed using Cell Stain
Solution overnight and the diameter of colonies was scored under a
microscope at ×40 magnification. Cells were counted with the Cell
Quantification Solution after a 4-h incubation at 37°C, followed by
a spectrophotometer reading at OD490.
Nude mouse xenograft assay
The female BALB/c nude mice (18 g, aged 6 weeks)
were obtained from Guangdong Medical Laboratory Animal Center
(permit no. SCXK 2013-0002). Animals were maintained and cared for
in the Traditional Chinese Medicine and Marine Drugs Laboratory of
Sun Yat-Sen University (permit no. SYXK 2014-0020) which conformed
to the Guide for the Care and Use of Laboratory Animals. In
vivo experiments were performed in accordance with a protocol
approved by the Animal Ethics and Welfare Committee of Sun Yat-Sen
University. PC-3 and DU-145 cell lines stably expressing miR-194,
anti-miR-194-sponge or GFP-control were suspended in a PBS buffer
at a concentration of 1×107 cells/ml. Cell suspension
(2×106 cells/mouse) were injected subcutaneously below
the right scapula of the BALB/c-nude mice. Seven mice from each of
the three groups were used. The experiment ended when the tumor
volume reached 2,000 mm3. The tumors were measured with
microcalipers three times a week and the volumes were calculated as
V (mm3) = length (mm) × (width (mm))2 × 0.5.
The mice were euthanized on day 30 and the tumors were harvested
and weighed.
Significance testing
Statistical relevant differences were assigned to a
p-value of at least ≤0.05 using an unpaired Students t-test in all
functional assays and densitometric analyses of western blot
analysis assays. All data represent at least three independent
experiments. Results are expressed as the mean ± standard error
(SE).
Results
hNUDC is a direct target of
miR-194
To determine whether hNUDC can be regulated by
miRNAs, we used TargetScan (37,38)
(http://www.targetscan.org/) and miRanda
(39–41) (http://www.microrna.org/) to predict miRNAs that could
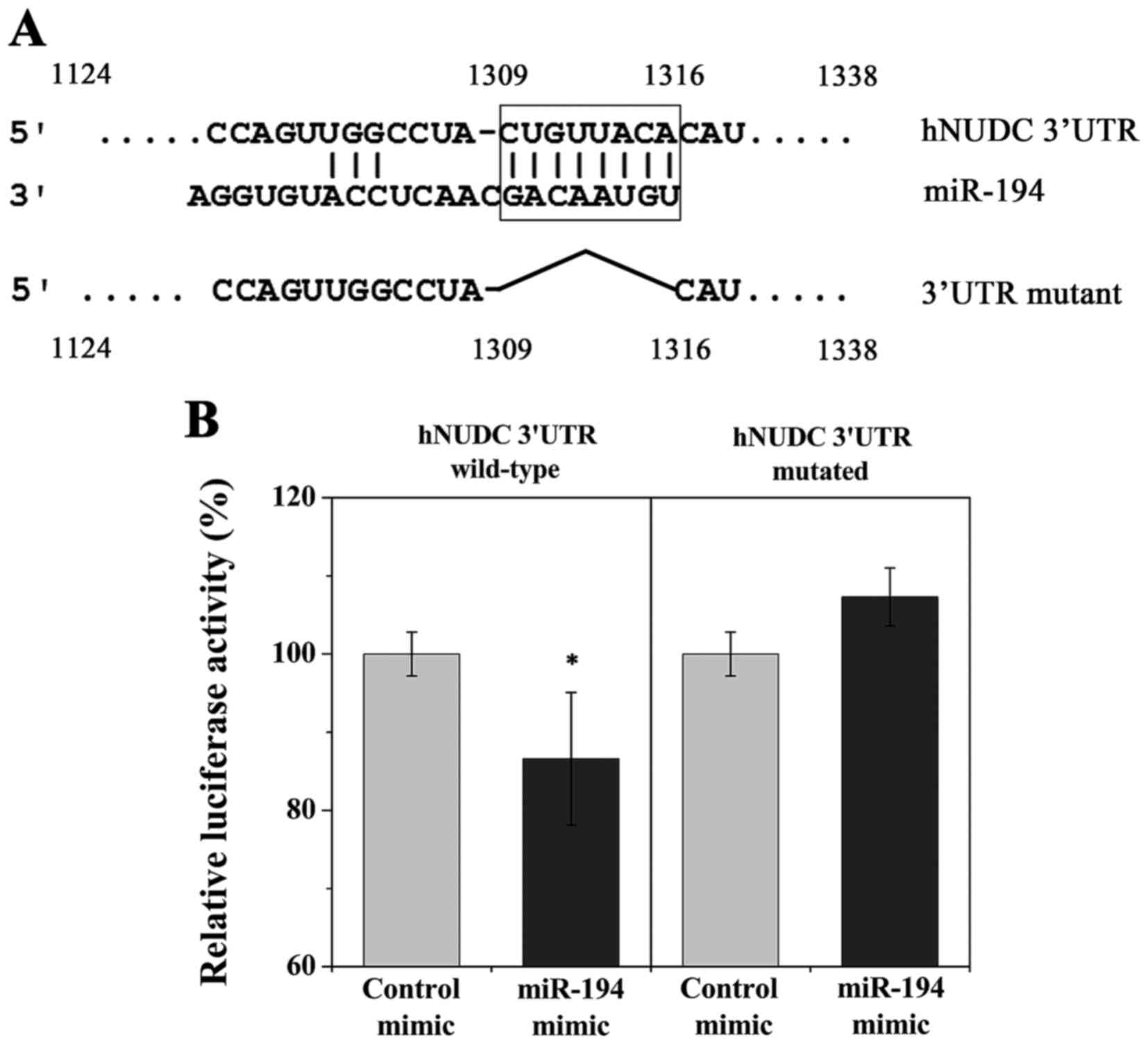
potentially target the hNUDC 3UTR. A potential binding site for
miR-194 was identified at the hNUDC 3UTR from nucleotides 1309–1316
(Fig. 1A). To validate hNUDC as a
direct target of miR-194, both wild-type and mutant hNUDC 3UTRs
were cloned downstream of the firefly luciferase ORF in a
pMIR-Report vector (Fig. 1A). The
wild-type and mutant hNUDC 3UTRs luciferase expression vectors were
co-transfected with scrambled control or miR-194 mimic into
HEK-293T cells. Relative luciferase activity was significantly
reduced for wild-type hNUDC 3UTR, indicating that hNUDC is a
potential direct target of miR-194 (Fig. 1B). Mutation in the predicted miR-194
target site abrogated inhibition by miR-194 mimic, confirming the
functionality of this target site (Fig.
1B).
hNUDC expression levels are regulated
by miR-194 in prostate cancer cells
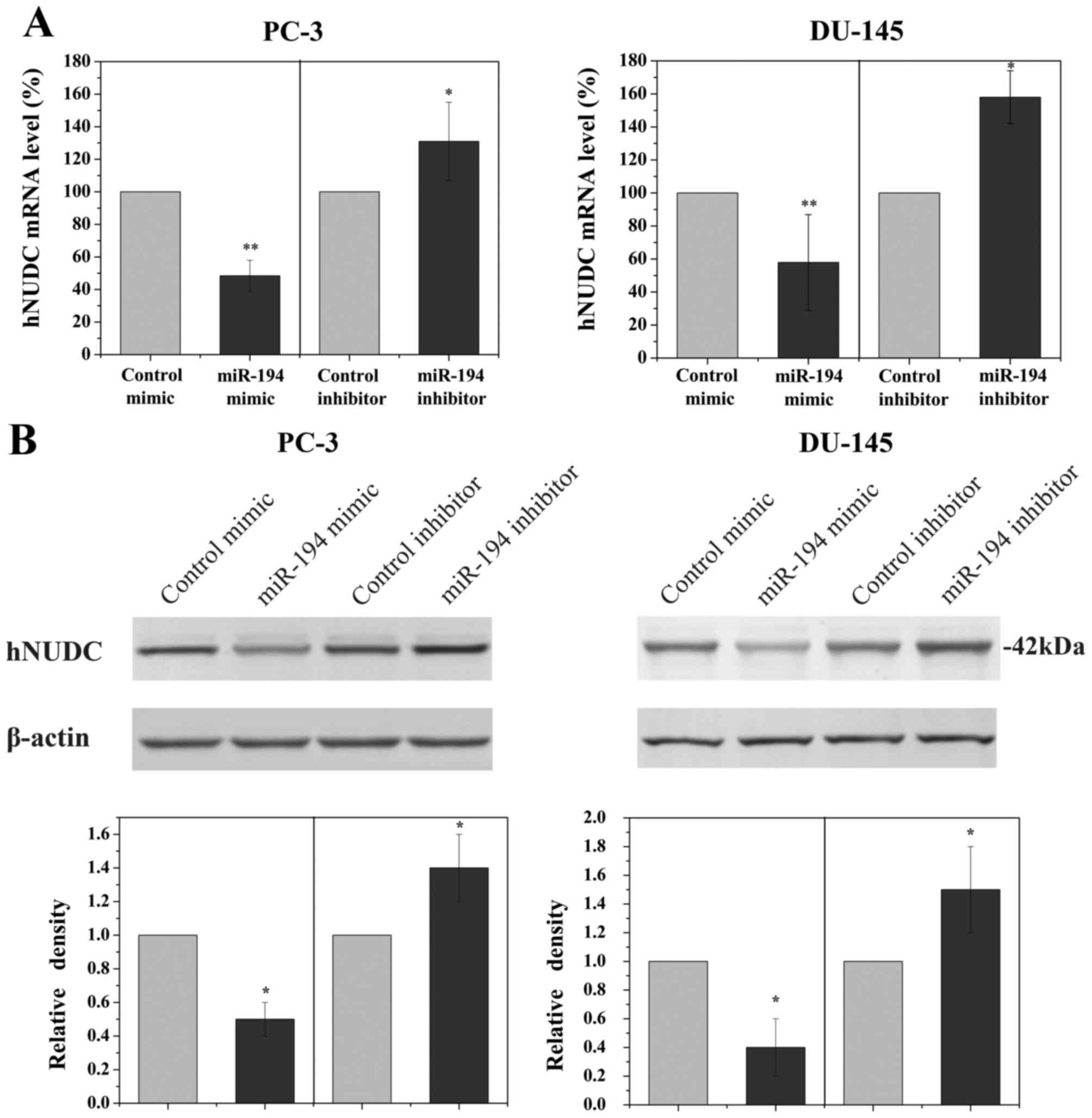
To determine if the hNUDC gene was a biologically
relevant target of miR-194 in PCa cells, PC-3 and DU-145 cell lines
were transfected with miR-194 mimic or miR-194 inhibitor. To
demonstrate the specificity of miR-194 in order to evaluate the
magnitude of the changes in hNUDC mRNA levels, non-targeting miRNA
inhibitor and non-targeting scrambled miRNA were used as controls.
qRT-PCR analyses revealed a significant reduction in the RNA
expression of hNUDC following transfection with miR-194 mimic,
whereas miR-194 inhibitor increased hNUDC levels (Fig. 2A). Western blot analysis was also
employed to examine the hNUDC protein, showing a prominent decrease
in the expression of hNUDC when PC-3 and DU-145 cells were
transfected with miR-194 mimic. Conversely, inhibiting the
endogenous miR-194 in PC-3 and DU-145 cells using miR-194 inhibitor
increased the hNUDC protein level (Fig.
2B).
miR-194 attenuates the migration and
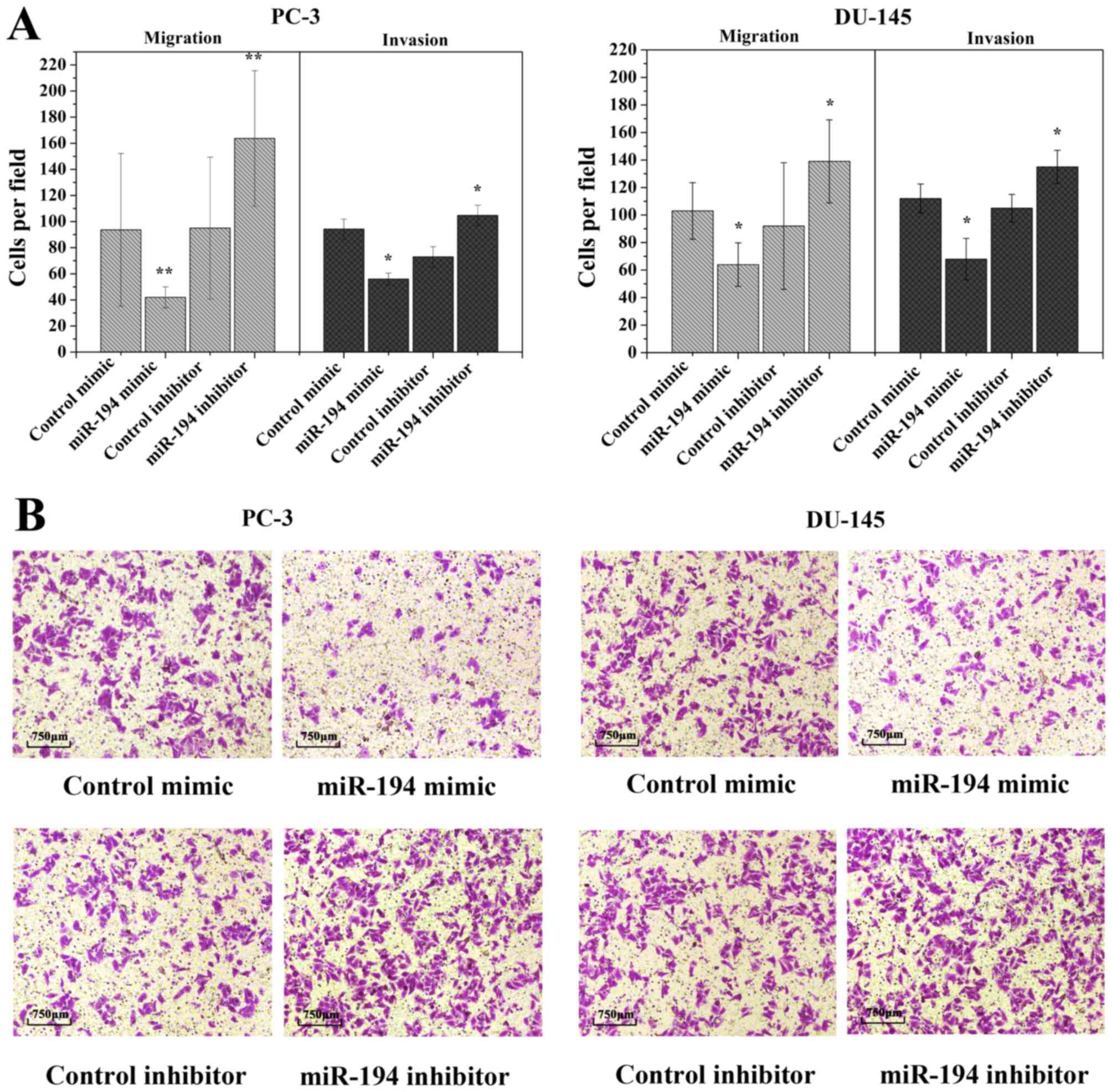
invasion of prostate cancer cells
To date, migration and invasion are known as the key
processes in many cancers. In order to investigate whether the
ectopic overexpression of miR-194 is involved in these processes,
we assessed the in vitro migratory and invasive capacities
by Transwell migration assay. The ectopic expression of miR-194
decreased the invasiveness of PC-3 and DU-145 cells compared to the
control mimic-transfected cells (Fig.
3A). Upon inhibition of endogenous miR-194 by miR-194
inhibitor, a visible opposite result was seen than in the control
inhibitor transfected cells (Fig.
3A). Microscopy results revealed that upon transfection of
miR-194 mimic, the loss of migratory capability was accompanied by
a loss of cell elongation (Fig.
3B). These findings indicate that upregulation of miR-194
inhibits cell invasion and migration.
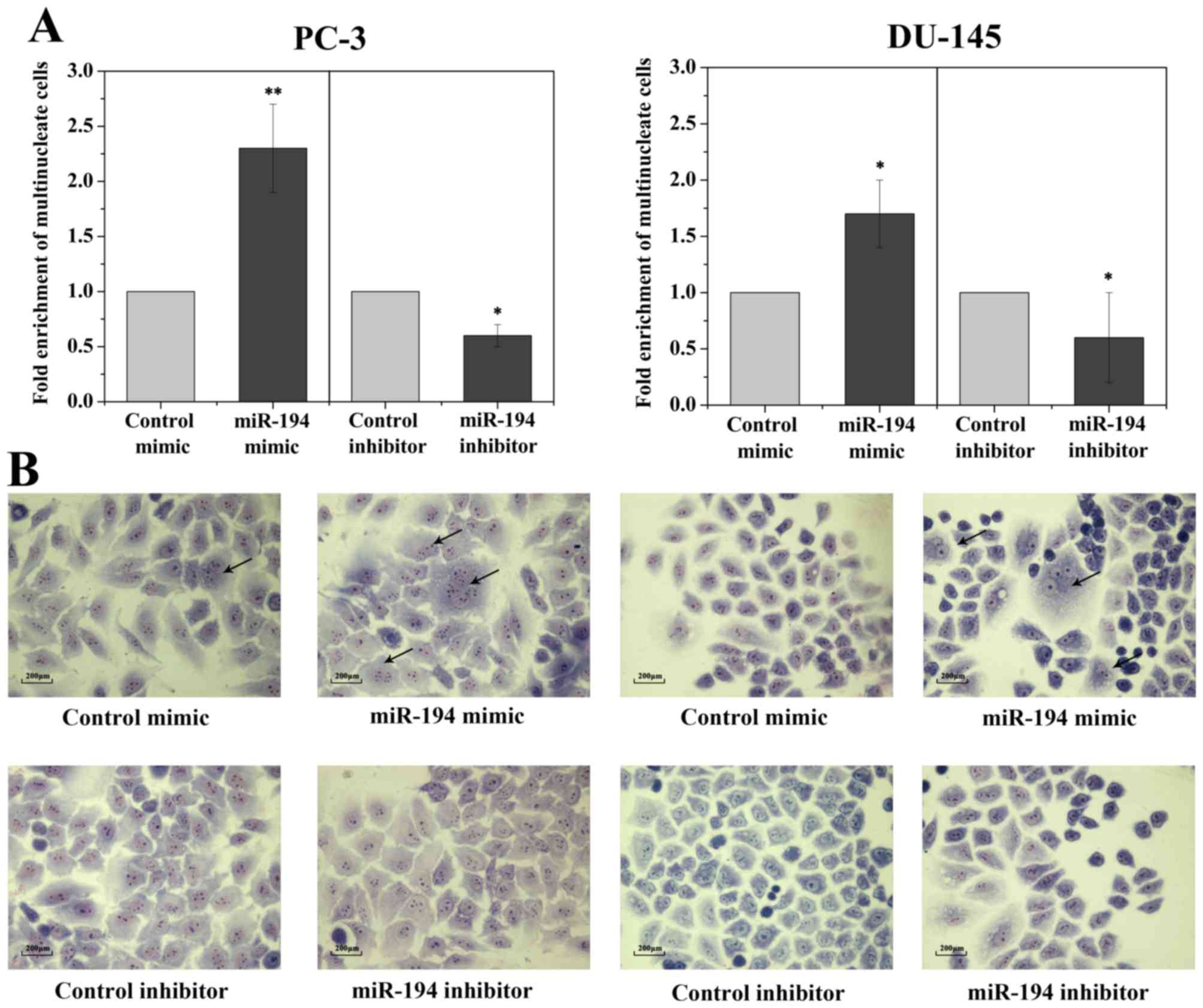
Overexpression of miR-194 induces
multinucleated cells
Since hNUDC is considered an important mediator of
cell proliferation and cytokinesis, thus, it is expected that the
inhibition of hNUDC would result in the failure of cell growth and
give rise to cells with multiple nuclei. However, our cell
proliferation and cell cycle analyses revealed that transfection of
the miR-194 mimics into two PC-3 and DU-145 cell lines showed no
influence on cell proliferation and cell cycle compared to the
scrambled control (data not shown). Morphologically, transfection
of cells with the miR-194 resulted in an increase in multinucleated
cells coupled with abnormally large, flattened and accumulated
multiple nuclei, whereas cells transfected with the mimic and
inhibitor controls had normal nuclear morphology (Fig. 4A and B).
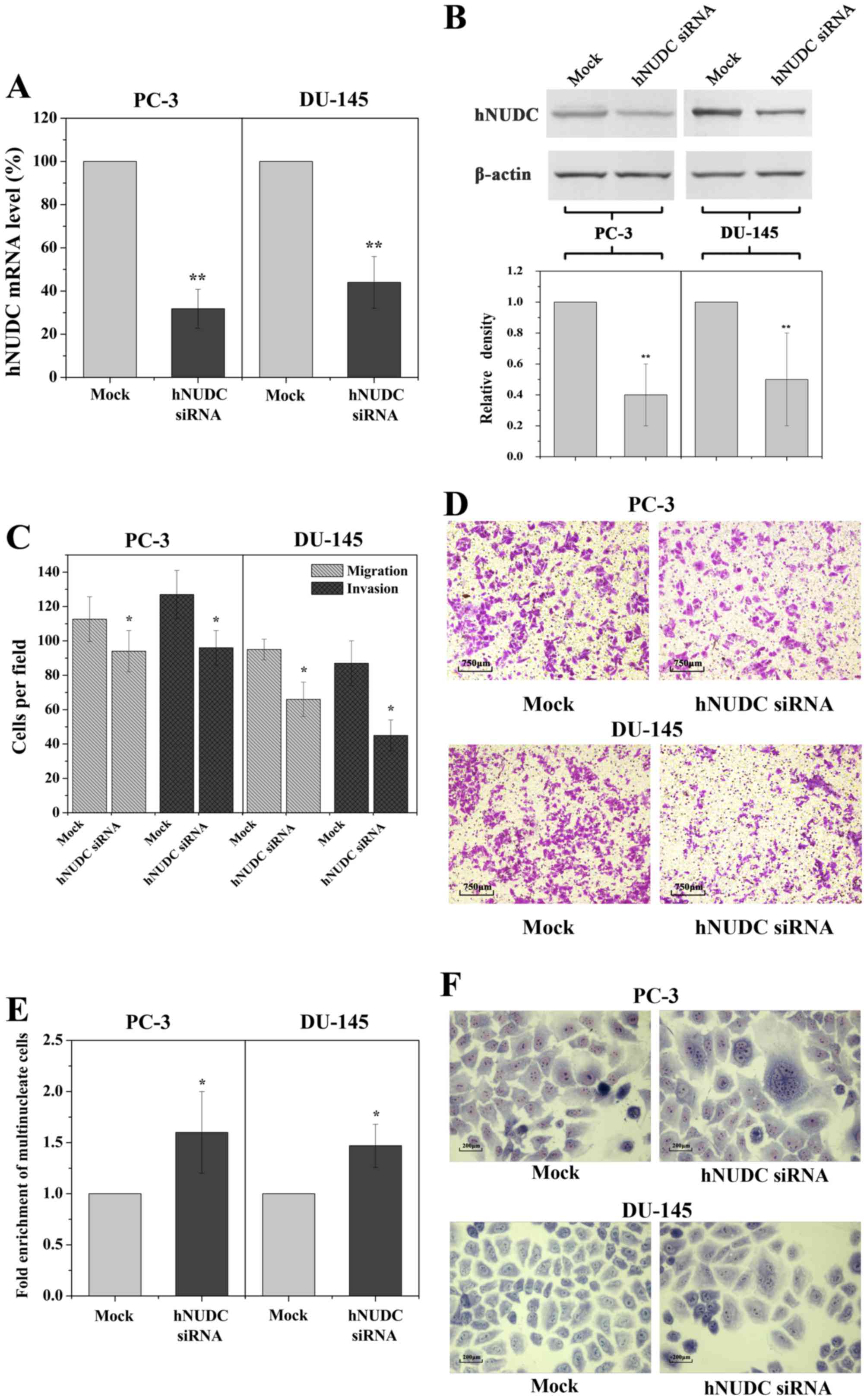
siRNA-mediated inhibition of hNUDC
mimics the phenotypes induced by miR-194
The aforementioned results demonstrated that miR-194
is a potent suppressor of cell migration and invasion through the
targeting of hNUDC mRNA. Thus, we reasoned that siRNA-mediated
knockdown of hNUDC should result in a cell phenotype similar to
that of miR-194 overexpression. Indeed, by using hNUDC siRNA we
were able to obtain a decrease in the expression of the hNUDC gene,
reducing mRNA levels by 3.3-fold in the PC-3 cells and by 2.5-fold
in the DU-145 cells, as determined by qRT-PCR analysis 48 h
post-transfection (Fig. 5A). This
result was confirmed by a significant reduction in hNUDC at the
protein level (Fig. 5B). Similarly,
miR-194 overexpression, significantly reduced the migration and
invasion potential following knockdown of hNUDC expression
(Fig. 5C and D). Following
transfection with hNUDC siRNA, we observed that the multinucleated
cells were enhanced following inhibition of endogenous hNUDC
(Fig. 5E and F). These results
strongly suggest that downregulation of hNUDC expression promotes
invasion and migration of prostate cancer cells.
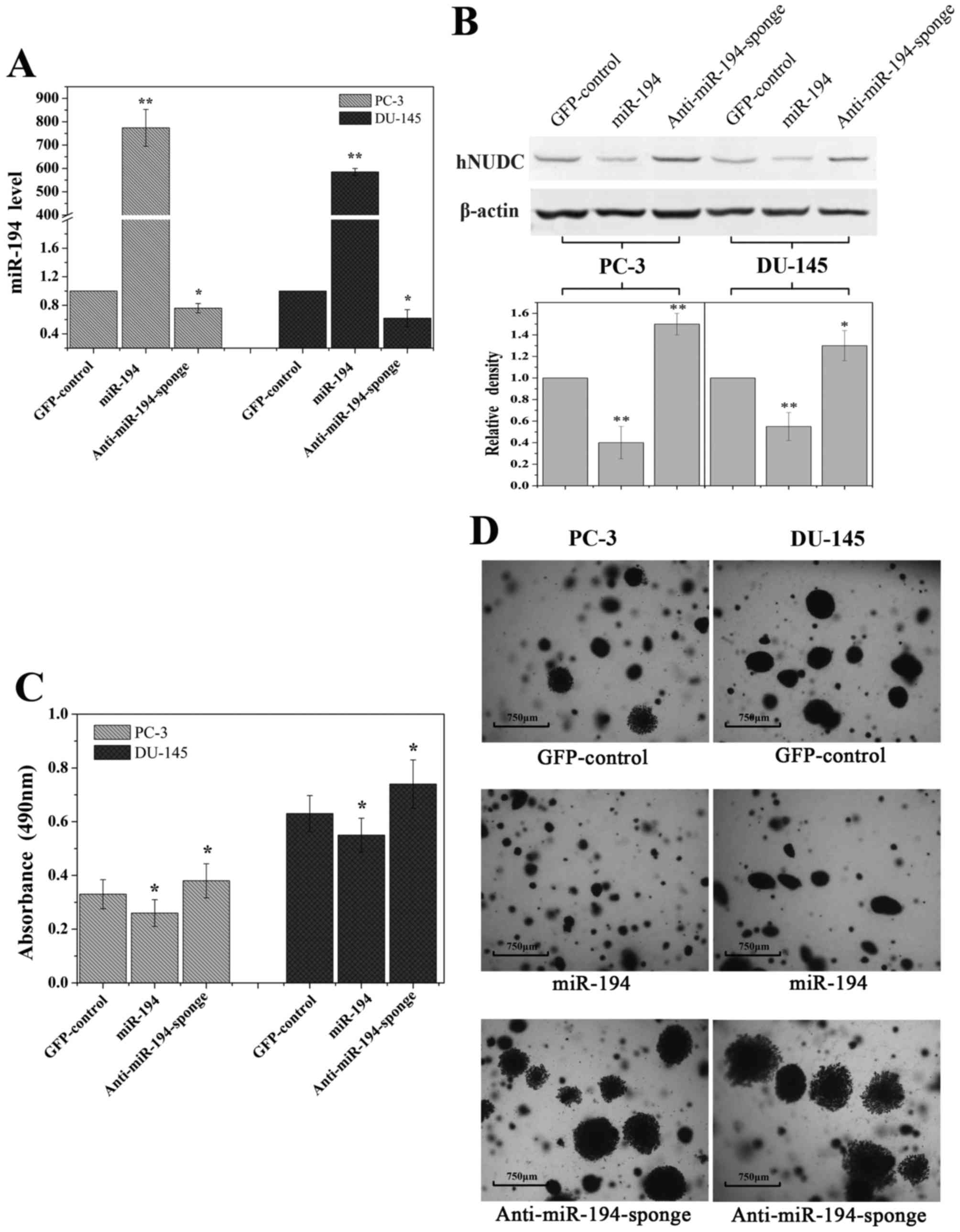
Overexpression of miR-194 inhibits
cell growth potential
To examine whether miR-194 regulates primary tumor
growth, a standard colony formation assay was used for these
studies. We overexpressed miR-194 in PC-3 and DU-145 cells by
transduction with high-titer lentivirus expressing miR-194 or
anti-miR194-sponge using an empty vector as a control. An ~700-fold
increase of miR-194 expression was detected by qRT-PCR in the
transduced cells, in comparison to cells that were transduced with
a control vector expressing GFP (Fig.
6A). As expected, a small decrease in miR-194 in the lentiviral
expressing anti-miR-194-sponge was observed (Fig. 6A). The expression level of hNUDC
protein was significantly decreased in response to ectopic miR-194
expression (Fig. 6B).
Overexpression of miR-194 decreased the total number of cells
observed when compared with the control (Fig. 6C). In contrast, anti-miR-194-sponge
did not suppress cell growth in soft agar (Fig. 6C). Moreover PC-3 and DU-145 cells
transfected with miR-194 formed smaller colonies compared to larger
disseminated colonies formed by the GFP-control and
anti-miR-194-sponge transduced cells after 30 days (Fig. 6D). The significant reduction of
colonies suggests that miR-194 exhibits a tumor-suppressive
function in cancer cells.
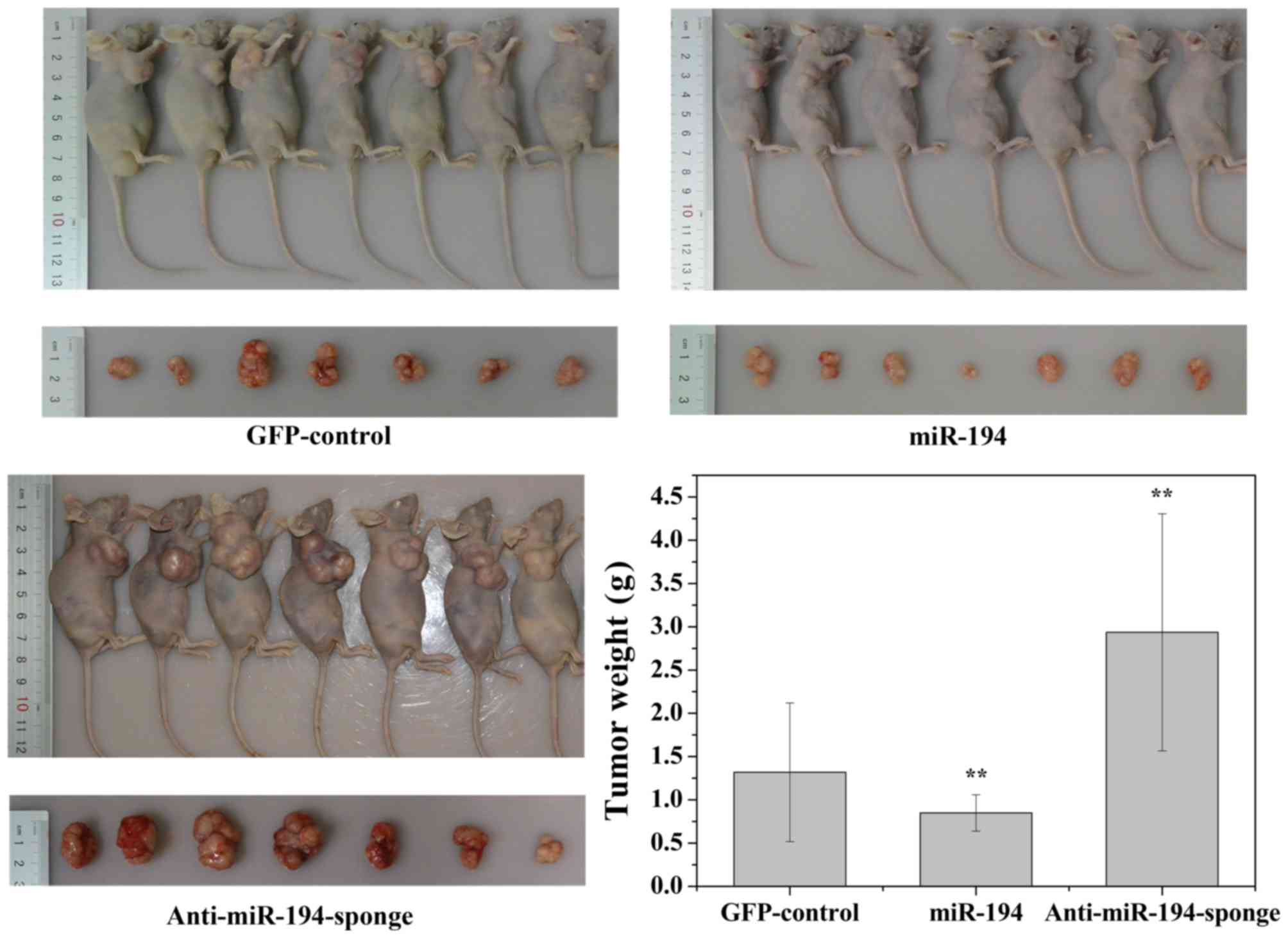
Overexpression of miR-194 suppresses
the growth of PCa xenografts
We next examined the effect of ectopic miR-194
expression on prostate cancer tumorigenesis in vivo. DU-145
cells stably expressing either miR-194 or anti-miR-194-sponge was
subcutaneously injected into 6-week-old nude mice. The control
group was injected with DU-145 stably expressing GFP. Tumor growth
was monitored twice weekly for 30 days. As shown in Fig. 7, tumors derived from cells
expressing anti-miR-194-sponge grew more rapidly. By the end of the
study, the tumors were larger (tumor volume) and heavier (tumor
weight) than the GFP-control. In contrast, tumors derived from the
cells with miR-194 overexpression appeared significantly smaller
than the tumors formed from the cells transduced with the
GFP-control.
Discussion
Little is known regarding the role of hNUDC in
cancer. Previous results have demonstrated that hNUDC is
ubiquitously expressed in normal human tissues. However, its
protein levels are markedly overexpressed in almost all types of
cancer cells, including cutaneous T-cell lymphoma and
neuroectodermal tumors (29–33).
Human NUDC has been identified in various molecular contexts
accounting for its numerous aliases: a dynein-associated nuclear
movement protein (27), chaperone
protein (42), neuronal migration
protein (43). Hence, hNUDC is a
multifunctional protein and is involved in diverse biological
events. Our laboratory has recently reported that hNUDC acts as a
secondary ligand for thrombopoietin receptor (Mpl) involved in
regulating proliferation and differentiation of different types of
megakaryocyte cells (44–46). However, a complete understanding of
all of the hNUDC functions remains unclear since this protein
interacts with a number of other proteins with multi-functional
effects. The present study is the second study concerning hNUDC in
prostate cell lines. Our study focused on ectopic expression of
miR-194 and its effects on prostate cancer by suppressing hNUDC
expression. The preliminary data indicated that downregulation of
hNUDC by miR-194 or si-hNUDC in DU-145 and PC-3 cells did not
induce cell proliferation or cell cycle progression (data not
shown) but rather resulted in morphological changes usually
associated with an enhanced multinucleated phenotype. However,
suppression of colony formation in the soft agar assay in the PC-3
and DU-145 cells suggests that hNUDC may play a role in the
inhibition of tumor cell growth. We also observed that
downregulation of endogenous hNUDC impaired prostate tumor cell
migration and invasion with either miR-194 or siRNA strategies,
although the mechanism by which this effect is mediated remains to
be elucidated. To demonstrate that miR-194 contributes to PCa
development in vivo, we employed a xenograft nude mouse
model and found that primary tumor growth was reduced
subcutaneously. Thus, it is possible that miR-194 may play a more
general role in suppressing oncogenic processes in prostate cancer.
This conclusion is further supported by the overexpression of
miR-194 whereby it reduced protein bone morphogenetic protein 1
(BMP1) levels in PC-3 cells causing a significant decrease of
invasion in Matrigel-coated Transwell chambers (47). During prostate cancer progression,
most deaths from prostate cancer are not due to the primary tumor
but rather to secondary metastases to distant organs. For this
reason it is of fundamental importance to study the mechanisms that
drive prostate cancer invasion and metastases. These data support
the biological relevance of model systems implicating miR-194 and
hNUDC expression as potential clinical markers.
Although it goes beyond the scope of this study, we
initially tested five cell lines, RWPE-1 and WPMY-1 which are
classically defined as non-tumoral, and the cancer cell lines
LNCaP, DU-145 and PC-3. We found that RWPE-1, WPMY-1 and LNCaP
inherently display a relative higher expression of miR-194 compared
to the DU-145 and PC-3 cells. However, the inverse relationship
between the hNUDC and miR-194 expression levels was observed in all
the normal and prostate tumor cell lines (data not shown). Both the
DU-145 and PC-3 cell lines, derived from secondary metastatic
prostate tumors, were used in our study as it remains possible that
low expression of miR-194 is associated with acquisition of a more
metastatic phenotype. We did not find any correlation between hNUDC
expression and miR-194 in clinical parameters used to assess the
poor prognosis of prostate cancer. In a future study, we will
address this issue in more depth, since this issue remains to be
investigated in a larger series of cases.
Recently, miR-194 expression profiles have been
detected in a variety of tumor entities and it has been suggested
that miRNAs are mostly downregulated in these cancer cells,
although some are overexpressed, playing a critical role in tumor
initiation and progression (16–19).
One of the largest qRT-PCR analyses of tissue cohorts reported by
Selth et al, indicated that miR-194 was robustly expressed
in malignant prostate tissue and its expression in primary tumors
was associated with a poor prognosis (48). One question that arises from this
result is the mechanism that is involved in the increased
upregulation in malignant prostate tissues compared to the matched
poor prognosis tissues. As prostate tumors are very heterogeneous,
the relative higher levels of expression of the miRNAs may be
masked by the contribution of the stromal levels of miR-194 that
may remain elevated. In addition, the miR-194 family consists of
four members whose clusters are located on two separate
chromosomes, and we believe that an explanation for this
discrepancy is likely due to the fact that their spatial and
temporal expression is tightly regulated by two genomic loci.
Although hNUDC is identified as a target of miR-194,
it is a well accepted fact that an miRNA can target many genes.
Database mining revealed that a single miR-194 may have multiple
targets in tumorigenesis. One study implicated ectopic
overexpression of miR-194 in the regulation of colon cancer
angiogenesis in vivo, by suppressing its target,
thrombospondin-1 (49). Moreover,
upregulation of miR-194 induced downregulation of RBX1 showing
significant inhibition of tumor size, invasion and tumor node
metastasis (50). Similarly, BMI-1
knockdown inhibited cell proliferation and clone growth and BMI-1
was recently proposed as a biologically relevant miR-194 target in
endometrial cancer cells (22).
Furthermore, miR-194 was found to inhibit chondrogenic
differentiation of human adipose-derived stem cells by targeting
Sox5 and suppressed osteosarcoma cell proliferation and metastasis
in vitro (51) and in
vivo by targeting CDH2 and IGF1R (52). Therefore, we cannot exclude the
possibility that these candidate targets for miR-194 other than
hNUDC may be involved in tumor suppression. The activity of hNUDC
as a cancer candidate target for miR-194 needs to be confirmed in
future studies.
In conclusion, our study is the first to indicate
that miR-194 interacts directly with hNUDC and regulates its
expression and activity. More research is required to examine the
effect of miR-194 on cellular growth, shape and function,
especially in clinical cancerous cells, in order to determine its
therapeutic potential in regulating aberrantly expressed hNUDC,
especially in downregulated hNUDC-related tumors.
Acknowledgments
The authors would like to acknowledge funding
attributed to this study, namely by the National Natural Science
Foundation of China (grant no. 31271230).
References
|
1
|
Koutsilieris M: Osteoblastic metastasis in
advanced prostate cancer. Anticancer Res. 13:443–449.
1993.PubMed/NCBI
|
|
2
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bagnyukova TV, Pogribny IP and Chekhun VF:
MicroRNAs in normal and cancer cells: A new class of gene
expression regulators. Exp Oncol. 28:263–269. 2006.PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hurst DR, Edmonds MD and Welch DR:
Metastamir: The field of metastasis-regulatory microRNA is
spreading. Cancer Res. 69:7495–7498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jerónimo C, Bastian PJ, Bjartell A,
Carbone GM, Catto JW, Clark SJ, Henrique R, Nelson WG and Shariat
SF: Epigenetics in prostate cancer: Biologic and clinical
relevance. Eur Urol. 60:753–766. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shen MM and Abate-Shen C: Molecular
genetics of prostate cancer: New prospects for old challenges.
Genes Dev. 24:1967–2000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Coppola V, De Maria R and Bonci D:
MicroRNAs and prostate cancer. Endocr Relat Cancer. 17:F1–F17.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chiang Y, Song Y, Wang Z, Liu Z, Gao P,
Liang J, Zhu J, Xing C and Xu H: microRNA-192, −194 and −215 are
frequently downregulated in colorectal cancer. Exp Ther Med.
3:560–566. 2012.PubMed/NCBI
|
|
17
|
Senanayake U, Das S, Vesely P, Alzoughbi
W, Fröhlich LF, Chowdhury P, Leuschner I, Hoefler G and Guertl B:
miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated
and their common target ACVR2B is strongly expressed in renal
childhood neoplasms. Carcinogenesis. 33:1014–1021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove
R, Xu R and Huang W: miR-194 is a marker of hepatic epithelial
cells and suppresses metastasis of liver cancer cells in mice.
Hepatology. 52:2148–2157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhai H, Karaayvaz M, Dong P, Sakuragi N
and Ju J: Prognostic significance of miR-194 in endometrial cancer.
Biomark Res. 1:12013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song Y, Zhao F, Wang Z, Liu Z, Chiang Y,
Xu Y, Gao P and Xu H: Inverse association between miR-194
expression and tumor invasion in gastric cancer. Ann Surg Oncol.
19:(Suppl 3). S509–S517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang B, Shen ZL, Gao ZD, Zhao G, Wang CY,
Yang Y, Zhang JZ, Yan YC, Shen C, Jiang KW, et al: MiR-194,
commonly repressed in colorectal cancer, suppresses tumor growth by
regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle.
14:1046–1058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong P, Kaneuchi M, Watari H, Hamada J,
Sudo S, Ju J and Sakuragi N: MicroRNA-194 inhibits epithelial to
mesenchymal transition of endometrial cancer cells by targeting
oncogene BMI-1. Mol Cancer. 10:992011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gocke CD, Osmani SA and Miller BA: The
human homologue of the Aspergillus nuclear migration gene nudC is
preferentially expressed in dividing cells and ciliated epithelia.
Histochem Cell Biol. 114:293–301. 2000.PubMed/NCBI
|
|
24
|
Zhang MY, Huang NN, Clawson GA, Osmani SA,
Pan W, Xin P, Razzaque MS and Miller BA: Involvement of the fungal
nuclear migration gene nudC human homolog in cell proliferation and
mitotic spindle formation. Exp Cell Res. 273:73–84. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morris SM, Albrecht U, Reiner O, Eichele G
and Yu-Lee LY: The lissencephaly gene product Lis1, a protein
involved in neuronal migration, interacts with a nuclear movement
protein, NudC. Curr Biol. 8:603–606. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aumais JP, Tunstead JR, McNeil RS, Schaar
BT, McConnell SK, Lin SH, Clark GD and Yu-Lee LY: NudC associates
with Lis1 and the dynein motor at the leading pole of neurons. J
Neurosci. 21:RC1872001.PubMed/NCBI
|
|
27
|
Aumais JP, Williams SN, Luo W, Nishino M,
Caldwell KA, Caldwell GA, Lin SH and Yu-Lee LY: Role for NudC, a
dynein-associated nuclear movement protein, in mitosis and
cytokinesis. J Cell Sci. 116:1991–2003. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou T, Aumais JP, Liu X, Yu-Lee LY and
Erikson RL: A role for Plk1 phosphorylation of NudC in cytokinesis.
Dev Cell. 5:127–138. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Miller BA, Zhang MY, Gocke CD, De Souza C,
Osmani AH, Lynch C, Davies J, Bell L and Osmani SA: A homolog of
the fungal nuclear migration gene nudC is involved in normal and
malignant human hematopoiesis. Exp Hematol. 27:742–750. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gocke CD, Reaman GH, Stine C, Zhang MY,
Osmani SA and Miller BA: The nuclear migration gene NudC and human
hematopoiesis. Leuk Lymphoma. 39:447–454. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hartmann TB, Mattern E, Wiedemann N, van
Doorn R, Willemze R, Niikura T, Hildenbrand R, Schadendorf D and
Eichmüller SB: Identification of selectively expressed genes and
antigens in CTCL. Exp Dermatol. 17:324–334. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Suzuki SO, McKenney RJ, Mawatari SY,
Mizuguchi M, Mikami A, Iwaki T, Goldman JE, Canoll P and Vallee RB:
Expression patterns of LIS1, dynein and their interaction partners
dynactin, NudE, NudEL and NudC in human gliomas suggest roles in
invasion and proliferation. Acta Neuropathol. 113:591–599. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hatakeyama H, Kondo T, Fujii K, Nakanishi
Y, Kato H, Fukuda S and Hirohashi S: Protein clusters associated
with carcinogenesis, histological differentiation and nodal
metastasis in esophageal cancer. Proteomics. 6:6300–6316. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin SH, Nishino M, Luo W, Aumais JP,
Galfione M, Kuang J and Yu-Lee LY: Inhibition of prostate tumor
growth by overexpression of NudC, a microtubule motor-associated
protein. Oncogene. 23:2499–2506. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Faircloth LM, Churchill PF, Caldwell GA
and Caldwell KA: The microtubule-associated protein, NUD-1,
exhibits chaperone activity in vitro. Cell Stress Chaperones.
14:95–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xiang X, Osmani AH, Osmani SA, Xin M and
Morris NR: NudF, a nuclear migration gene in Aspergillus nidulans,
is similar to the human LIS-1 gene required for neuronal migration.
Mol Biol Cell. 6:297–310. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pan RM, Yang Y, Wei MX, Yu XB, Ge YC and
Xu P: A microtubule associated protein (hNUDC) binds to the
extracellular domain of thrombopoietin receptor (Mpl). J Cell
Biochem. 96:741–750. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wei MX, Yang Y, Ge YC and Xu P: Functional
characterization of hNUDC as a novel accumulator that specifically
acts on in vitro megakaryocytopoiesis and in vivo platelet
production. J Cell Biochem. 98:429–439. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tang YS, Zhang YP and Xu P: hNUDC promotes
the cell proliferation and differentiation in a leukemic cell line
via activation of the thrombopoietin receptor (Mpl). Leukemia.
22:1018–1025. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang C, Shu L, Kim H, Khor TO, Wu R, Li W
and Kong AN: Phenethyl isothiocyanate (PEITC) suppresses prostate
cancer cell invasion epigenetically through regulating
microRNA-194. Mol Nutr Food Res. 60:1427–1436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Selth LA, Townley SL, Bert AG, Stricker
PD, Sutherland PD, Horvath LG, Goodall GJ, Butler LM and Tilley WD:
Circulating microRNAs predict biochemical recurrence in prostate
cancer patients. Br J Cancer. 109:641–650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sundaram P, Hultine S, Smith LM, Dews M,
Fox JL, Biyashev D, Schelter JM, Huang Q, Cleary MA, Volpert OV, et
al: p53-responsive miR-194 inhibits thrombospondin-1 and promotes
angiogenesis in colon cancers. Cancer Res. 71:7490–7501. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen X, Wang Y, Zang W, Du Y, Li M and
Zhao G: miR-194 targets RBX1 gene to modulate proliferation and
migration of gastric cancer cells. Tumour Biol. 36:2393–2401. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu J, Kang Y, Liao WM and Yu L: MiR-194
regulates chondrogenic differentiation of human adipose-derived
stem cells by targeting Sox5. PLoS One. 7:e318612012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Han K, Zhao T, Chen X, Bian N, Yang T, Ma
Q, Cai C, Fan Q, Zhou Y and Ma B: microRNA-194 suppresses
osteosarcoma cell proliferation and metastasis in vitro and in vivo
by targeting CDH2 and IGF1R. Int J Oncol. 45:1437–1449.
2014.PubMed/NCBI
|