Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common cancer worldwide and the third most common cause of
cancer-related mortality (1).
Despite improvements in surgical techniques and other treatments
for HCC, the prognosis of HCC patients remains unsatisfactory. It
is imperative to understand the molecular mechanisms of HCC and
find optimized strategies to improve HCC treatment (2).
A growing number of studies have revealed that
microRNAs (miRNAs) are highly conserved non-coding RNA
oligonucleotides (18–25 nt long) that play essential roles in
regulating diverse biological processes, such as cell
proliferation, invasion, migration, apoptosis and the cell cycle
(3–5). miRNAs have been indicated in the
regulation of cancer development. Wang et al revealed that
downregulation of miR-122 promoted proliferation, migration and
invasion of HCC by activating epithelial-mesenchymal transition
(6). miR-383 was reported to
inhibit HCC cell proliferation via targeting a
proliferation-inducing ligand (APRIL) (7). Furthermore, Zhang et al
indicated that miR-381 inhibited cell growth and invasion by
targeting the liver receptor homolog-1 in HCC (8). However, the biological function of
miR-922 and the related mechanisms in HCC remain unclear.
Cylindromatosis (CYLD), a deubiquitination enzyme,
is a tumor-suppressor gene in different types of cancer (9,10).
Herein, we indicated that expression of miR-922 was upregulated in
HCC cells and clinical tissues. Overexpression of miR-922 promoted
HCC cell proliferation. Bioinformatic analyses revealed that
miR-922 is able to bind to the 3 untranslated region (3UTR) of CYLD
mRNA to prevent its translation, which was confirmed by luciferase
reporter assay. Further experiments indicated that CYLD
downregulation counteracted suppress of HCC cell proliferation by
the inhibitor of miR-922, miR-922-in. Together, our data suggest
that miR-922 overexpression promotes HCC cell growth through the
regulation of CYLD expression.
Materials and methods
Cell culture
Human HCC cell lines Huh7, MHCC97H, HepG2, QGY-7703,
Hep3B, MHCC97L, HCCC-9810 and BEL-7402 were obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA), and
were cultured in Dulbecco's modified Eagle's medium (Gibco, Grand
Island, NY, USA) supplemented with 10% fetal bovine serum (FBS;
Sigma, St. Louis, MO, USA), and 100 U/ml of penicillin-streptomycin
(Invitrogen, Carlsbad, CA, USA). A human hepatic cell line THLE3
was purchased from the China Center for Type Culture Collection
(CCTCC; Wuhan, China), and was cultured in McCoy's 5A modified
medium (Invitrogen, Life Technologies, Carlsbad, CA, USA)
supplemented with 10% FBS and 100 U/ml of penicillin-streptomycin.
All cells were maintained at 37°C in a humidified incubator
containing 5% CO2.
Clinical specimens
Eight pairs of primary human HCC tissue samples and
adjacent normal tissue (ANT) samples were obtained from HCC
patients at the Department of Hepato-Biliary-Pancreatic Surgery,
Sun Yat-sen Memorial Hospital, Sun Yat-sen University (Guangzhou,
China), following surgical dissections. Written informed consent
was obtained from each participant. The present study was approved
by the Ethics Committee of Sun Yat-sen Memorial Hospital, Sun
Yat-sen University. Tissues were snap-frozen in liquid
nitrogen.
Plasmids, small interfering RNA and
transfection
miR-922 mimic, miR-922 inhibitor (miR-922-in) and
negative controls were purchased from GeneCopoeia Co., Ltd.
(Guangzhou, China) and transfected into OS cells using
Lipofectamine 2000 reagent (Invitrogen) according to the
manufacturer's protocol. The CYLD ORF was amplified from the Hep3B
cell cDNA and subcloned into pEGFP-N3 (Invitrogen). The wild-type
3′UTR of CYLD was synthesized and subcloned into the firefly
luciferase reporter (RiboBio, Guangzhou, China), and then
transfected into HCC cells using Lipofectamine 2000 reagent
(Invitrogen) according to the manufacturer's instructions. shRNA
lentiviruses against CYLD were purchased from GeneCopoeia (Co.,
Ltd.) and transfected into Hep3B cells in 24-well plates using
Lipofectamine 2000 reagent (Invitrogen) according to the
manufacturer's instructions.
RNA extraction and real-time
quantitative PCR
Total RNA was extracted using TRIzol reagent
(Invitrogen), and then cDNA was synthesized from 5 ng of total RNA
using the miRNA First-Strand cDNA Synthesis SuperMix (TransGen
Biotech, Beijing, China). The expression of miR-922 was detected by
TransScript® Green miRNA Two-Step qRT-PCR SuperMix
(TransGen Biotech). In addition, the relative miR-922 expression
levels after normalization to U6 were calculated. The cyclin D1 and
MYC levels were quantified using qRT-PCR with the
TaqMan®. We assessed the RNA expression according to
relative quantification using the 2−ΔΔCt method to
determine the fold-change in the expression. Each sample was
analyzed in triplicate and the mean expression level was
calculated.
MTT and colony formation assays
For the MTT assay, at four different time points
(24, 48, 72 and 96 h) 20 µl of 5 mg/ml MTT solution (Sigma-Aldrich,
St. Louis, MO, USA) was added to each well, followed by incubation
for 4 h at 37°C and the absorbance was read at 490 nm by a Thermo
Scientific Multiskan (Thermo Fisher Scientific, USA). For the
colony formation assays, Hep3B cells after transfection were
harvested and seeded into 6-well plates at 1×103
cells/well. Two weeks after seeding, the cells were stained with
0.1% crystal violet for 1 min and fixation with 10% formaldehyde
for 10 min. Images were captured and the colonies were counted.
Each time point was repeated in three wells and the experiment was
independently performed for three times.
Anchorage-independent growth
assay
One thousand HepG2 cells were trypsinized and
resuspended in 2 ml complete medium plus 0.3% agar (Sigma). The
agar-cell mixture was plated on top of a bottom layer comprising
0.66% complete medium agar mixture. The plates were incubated at
37°C for >2 weeks until the visible colonies had formed, and
colonies >0.1 mm in diameter were counted. The experiment was
performed independently three times for each cell line.
Luciferase assays
Cells were transferred with miR-922, or miR-922-in,
or miR-922-mut into 24-well plates at a density of 30,000
cells/well. After 24 h, the cells were co-transfected with the
pRL-TK plasmid (Promega, Madison, WI, USA) containing the
Renilla luciferase gene, and various constructs containing
the seed sequence of CYLD 3′UTR. Luciferase reporter assays were
performed using the Dual-Luciferase reporter assay system according
to the manufacturer's instructions.
Western blotting
The cultured cells were washed with 1X
phosphate-buffered saline (PBS) and lysed with RIPA buffer. Equal
quantities of protein (30 µg) were used for the detection of CYLD,
c-Myc, cyclin D1, Rb and p-Rb protein (1:1,000) and α-tubulin
(1:1,000) (both from Cell Signaling Technology, Danvers, MA, USA)
was used as the loading control. In addition, the membranes were
washed with PBS and incubated with the respective goat anti-rabbit
secondary antibodies (Beyotime Biotechnology, Shanghai, China). The
bound antibodies were detected using an ECL kit. Quantity One
software was used to quantify protein band intensities.
Statistical analysis
All results in the present study were statistically
presented as mean ± standard error of the mean using SPSS 19.0
(IBM). P<0.05 was considered to indicate a statistically
significant result by Student's t-test.
Results
miR-922 expression is upregulated in
HCC cell lines and HCC tissues
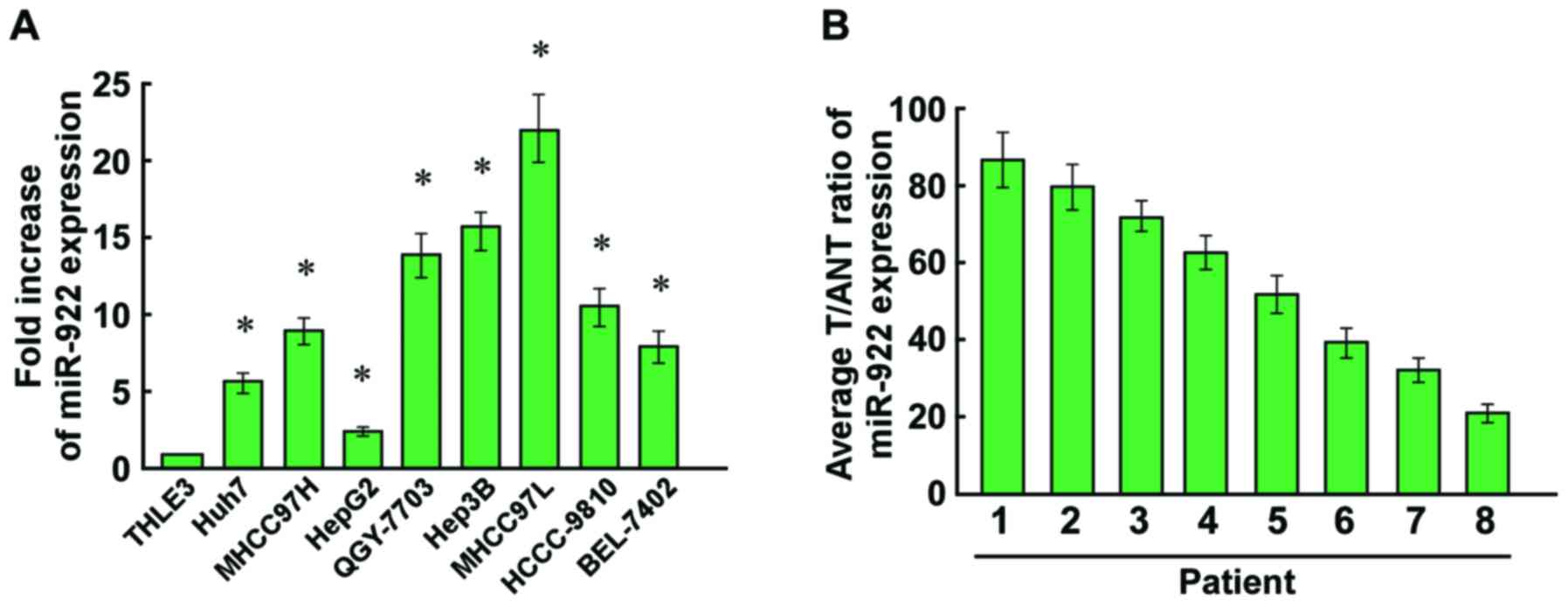
To detect the expression of miR-922 in HCC cells,
qRT-PCR was used to analyze miR-922 expression in HCC cells and
human hepatic cell line THLE3. Expression of miR-922 was markedly
upregulated in the HCC cell lines compared with that in the THLE3
cells (Fig. 1A). We also evaluated
the expression of miR-922 in HCC specimens. As showed in Fig. 1B, upregulation of miR-922 was
observed in the HCC tissues compared with adjacent non-tumor
tissues (ANT). Collectively, miR-992 may play a tumor-promoting
role in the progression of HCC.
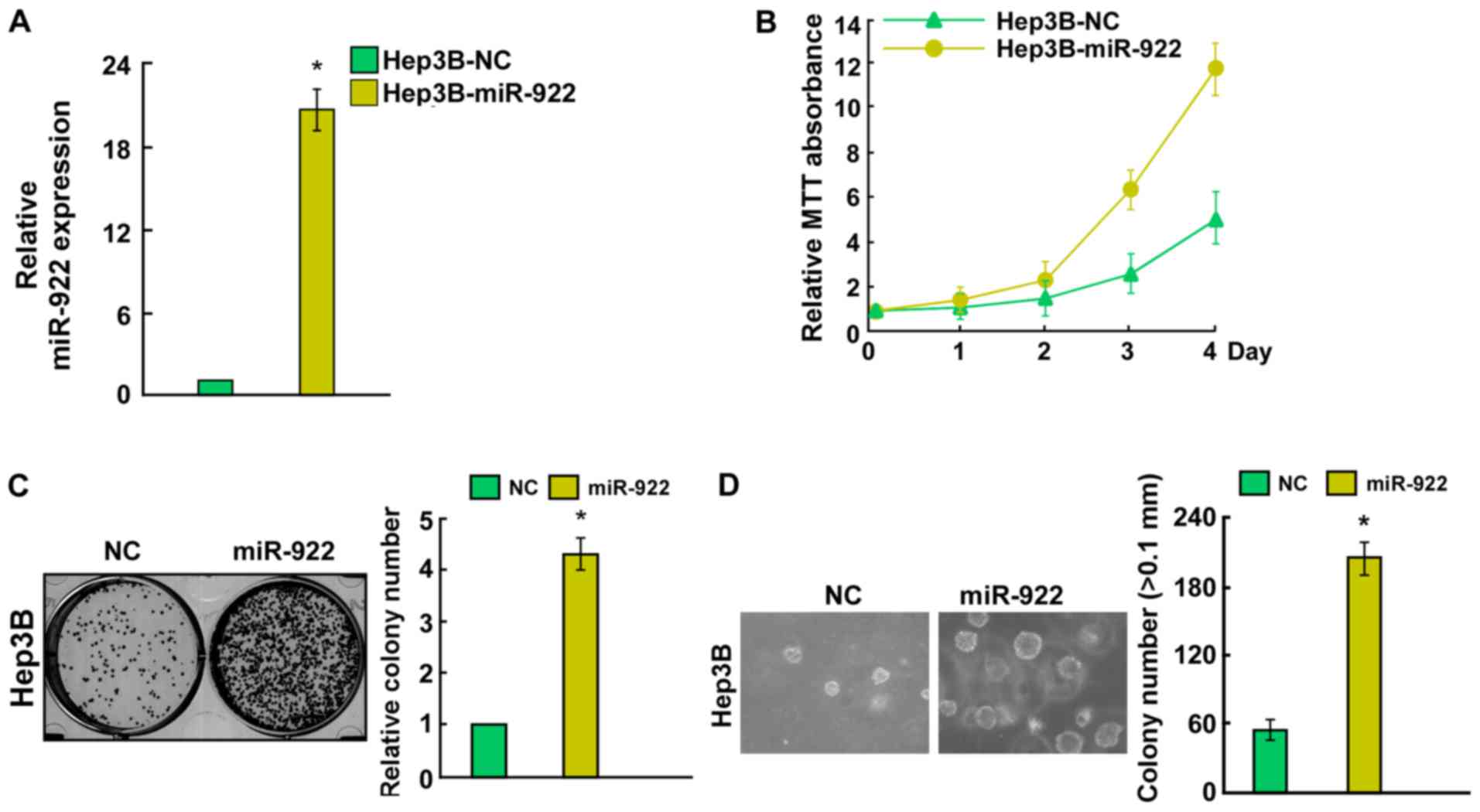
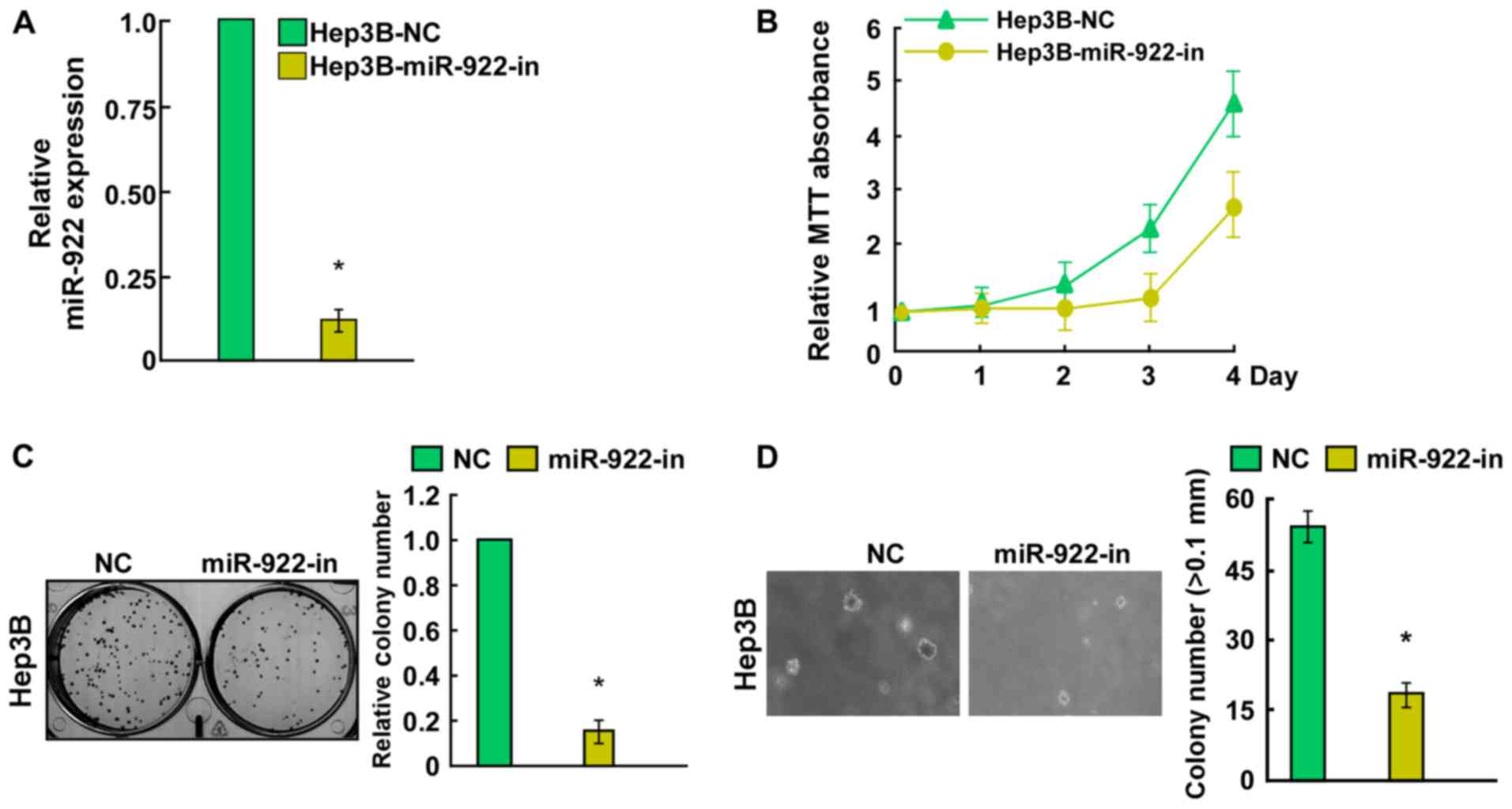
miR-922 promotes, while its inhibitor
miR-922-in suppresses HCC cell proliferation
To investigate the biological function of miR-922 in
HCC, Hep3B cells were transfected with miR-922 mimic, miR-922-in or
negative controls. Transfection efficiency was confirmed by qRT-PCR
(Figs. 2A and 3A). Cell growth assay showed that
upregulation of miR-922 efficiently promoted the proliferation of
Hep3B cells (Fig. 2B). Conversely,
a significant decrease in the number of Hep3B cells was observed in
the miR-922-in group (Fig. 3B).
Notably, colony formation assays indicated that miR-922
significantly promoted, while miR-922-in markedly inhibited the
ability of colony formation in the Hep3B cells (Figs. 2C and 3C). Results of anchorage-independent
growth showed that miR-922-transfected Hep3B cells formed more and
larger colonies (Fig. 2D). In
contrast, Hep3B cells after transfection with miR-922-in showed a
significant reduction in the ability to form colonies on soft agar
(Fig. 3D). Collectively, miR-922
promoted the proliferation of HCC cells in vitro.
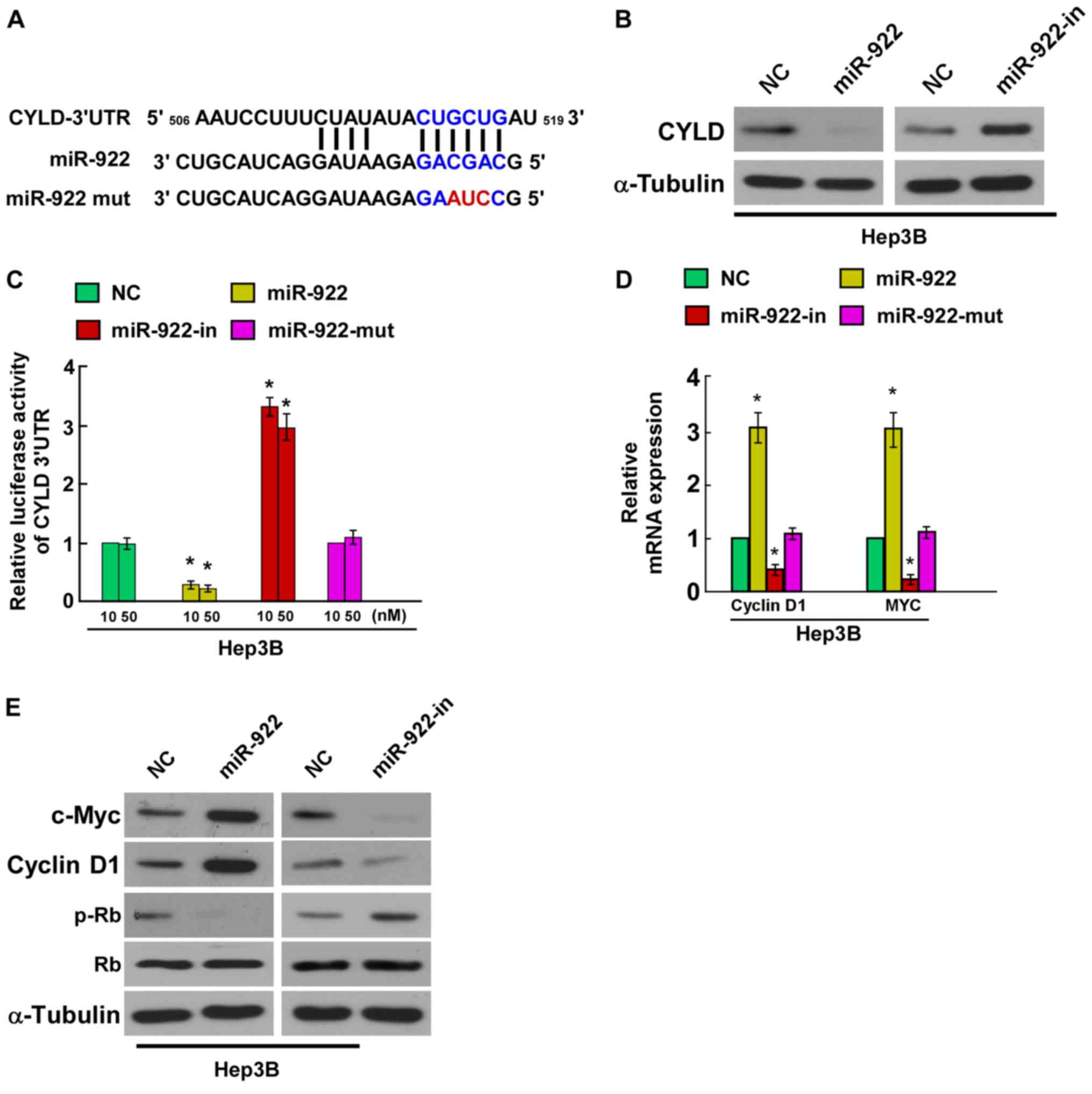
miR-922 directly targets CYLD by
binding to its 3′UTR and alters levels of proteins related to cell
proliferation in Hep3B cells
In order to confirm whether CYLD is the direct
target of miR-4262, we identified CYLD as a potential target of
miR-922 using bioinformatic analysis (TargetScan). We found that
the 3′UTR of CYLD was complementary to the miR-922 seed region
(Fig. 4A). Hep3B cells were
transfected with miR-922 mimic, miR-922-in or negative controls,
and the effect of miR-922 or miR-922-in on the expression of CYLD
was detected by western blotting. The results indicated that CYLD
protein expression was significantly decreased in the Hep3B cells
stably expressing miR-922, whereas CYLD protein expression was
markedly increased by miR-922-in (Fig.
4B). In addition, we tested whether miR-922 directly targets
CYLD by luciferase reporter assay. The results showed that
overexpression of miR-922 markedly reduced the luciferase activity
of the CYLD wild-type 3′UTR, whereas miR-922-in increased this
activity, and the luciferase activity of the CYLD wild-type 3′UTR
was not affected by the miR-922-mut (Fig. 4C), indicating that miR-922 can bind
to the CYLD 3′UTR. Collectively, these results demonstrated that
CYLD is a direct target of miR-922.
CYLD was reported to negatively
regulate NF-κB activity, and Jun N-terminal kinase and
Wnt/β-catenin signaling pathways (11–13)
In the present study, we evaluated the mRNA of the
CYLD downstream genes (cyclin D1, c-Myc, Rb and p-Rb). Results of
quantitative PCR revealed a significant increase in the mRNA levels
of cyclin D1 and c-Myc, while miR-922-in showed the opposite effect
in the Hep3B cells (Fig. 4D).
Western blot analysis showed that the levels of cyclin D1 and c-Myc
were increased while the level of p-Rb was decreased in the miR-922
group compared with the miR-NC group, while miR-922-in showed the
opposite function (Fig. 4E). Taken
together, our results demonstrated that miR-922 functionally
modulated CYLD, and then regulated cellular proliferation
regulators, cyclin D1, c-Myc and p-Rb, thus relevant to cell
proliferation. These results demonstrated that miR-922 could
combine with CYLD 3′UTR and play a role in suppressing the
expression of the CYLD gene.
CYLD downregulation counteracts the
cell proliferation arrest by miR-922-in
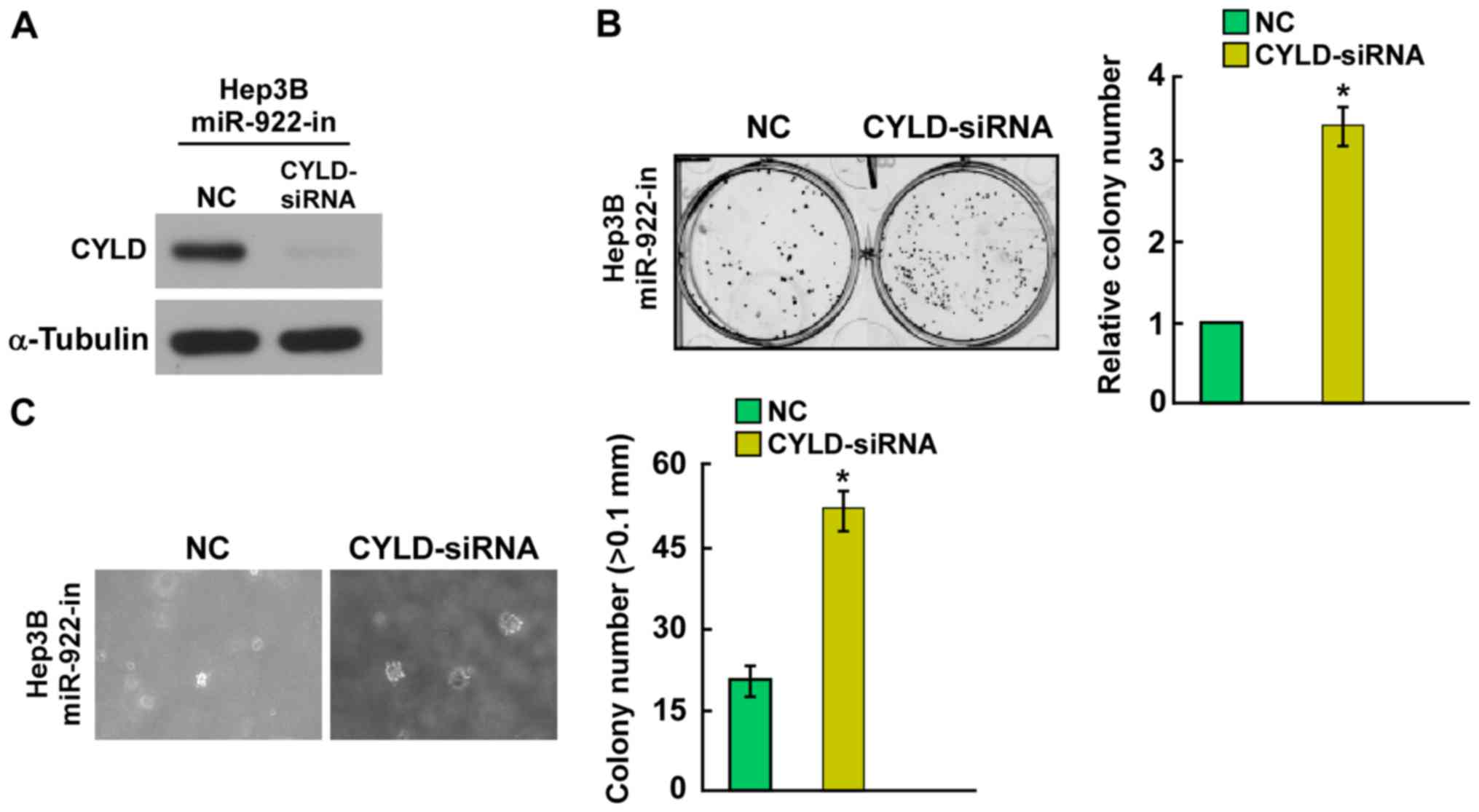
CYLD protein expression was suppressed in the
miR-922-in transfected Hep3B cells using CYLD-siRNA (Fig. 5A). Knockdown of CYLD enhanced cell
colony formation and anchorage-independent growth ability of
miR-922-in-transfected Hep3B cells (Fig. 5B and C). Taken together, these
results suggest that the tumor-suppressive effect of miR-922-in is
mediated by regulation of CYLD in HCC.
Discussion
In the present study, we characterized the
functional significance of miR-922 in hepatocellular carcinoma
(HCC). We demonstrated that miR-922 expression was upregulated in
HCC cell lines and clinical tissues. Moreover, ectopic miR-922
expression promoted cell proliferation of the Hep3B cells.
Furthermore, miR-922 overexpression increased Hep3B cell colony
formation and anchorage-independent growth ability. Additionally,
we identified cylindromatosis (CYLD) as a direct target of miR-922.
Knockdown of CYLD counteracted the cell proliferation arrest by
miR-922-in in the HCC Hep3B cells. These results indicated that
miR-922 may function as an oncogene in HCC.
An increasing number of studies indicated that
dysregulation of miRNAs plays essential roles in multiple
biological processes, such as cell invasion, migration,
proliferation, apoptosis, cell cycle and angiogenesis (14–16).
miR-765 was found to promote HCC cell proliferation (17). miR-1180 was reported to be
significantly increased and to promote the proliferation of HCC
(18). Furthermore, miR-379-5p
acted as a tumor suppressor and inhibited cell invasion and
metastasis in HCC (19). Zhou et
al indicated that miR-761 was upregulated and regulated
tumorigenesis in HCC (20).
However, to date, the biological function and regulatory mechanisms
of miR-922 in HCC tumorigenesis have not been elucidated.
Currently, we demonstrated that miR-922 acts as a tumor promoter
and promotes the cell proliferation of HCC.
The CYLD gene encoding a deubiquitinating enzyme,
has been identified as a tumor-suppressor gene and is downregulated
in many types of cancer, including HCCs, gliomas, melanoma and
colon carcinomas (21–24). CYLD is a direct target of miR-454,
the increased expression of which resulted in CYLD reduction and
promoted cell proliferation of human colorectal cancer cells
(25). miR-501-5p was reported to
regulate CYLD expression and promote cell proliferation in human
HCC (26). However, whether miR-922
can directly target CYLD in HCC remains unknown. In the present
study, the results of the bioinformatic analysis indicated sequence
complementarity between miR-922 and the 3′UTR of CYLD and was
confirmed by luciferase assays. Further experiment revealed that
knockdown of CYLD by siRNA counteracted the cell proliferation
arrest by miR-922-in. These results suggest that miR-922 directly
regulates CYLD expression in HCC.
In summary, the present study indicated that miR-922
promoted HCC cell proliferation by suppressing CYLD expression and
offers a new insight into the mechanisms of miR-922 in HCC,
suggesting that miR-922 could be a potential therapeutic target in
HCC.
Acknowledgements
The present study was supported by the Guangzhou
Major Projects on Collaborative Innovation of Industry
(1561000198).
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie W, Qin W, Kang Y, Zhou Z and Qin A:
MicroRNA-340 inhibits tumor cell proliferation and induces
apoptosis in endometrial carcinoma cell line RL 95–2. Med Sci
Monit. 22:1540–1546. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang M, Zhuang Q and Cui L: MiR-194
inhibits cell proliferation and invasion via repression of RAP2B in
bladder cancer. Biomed Pharmacother. 80:268–275. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin L, Li Y, Liu J, Yang S, Gui Y, Mao X,
Nie G and Lai Y: Tumor suppressor miR-149-5p is associated with
cellular migration, proliferation and apoptosis in renal cell
carcinoma. Mol Med Rep. 13:5386–5392. 2016.PubMed/NCBI
|
|
6
|
Wang N, Wang Q, Shen D, Sun X, Cao X and
Wu D: Downregulation of microRNA-122 promotes proliferation,
migration, and invasion of human hepatocellular carcinoma cells by
activating epithelial-mesenchymal transition. Onco Targets Ther.
9:2035–2047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen L, Guan H, Gu C, Cao Y, Shao J and
Wang F: miR-383 inhibits hepatocellular carcinoma cell
proliferation via targeting APRIL. Tumour Biol. 37:2497–2507. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Q, Zhao S, Pang X and Chi B:
MicroRNA-381 suppresses cell growth and invasion by targeting the
liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep.
35:1831–1840. 2016.PubMed/NCBI
|
|
9
|
Massoumi R: CYLD: A deubiquitination
enzyme with multiple roles in cancer. Future Oncol. 7:285–297.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Massoumi R, Chmielarska K, Hennecke K,
Pfeifer A and Fässler R: Cyld inhibits tumor cell proliferation by
blocking Bcl-3-dependent NF-kappaB signaling. Cell. 125:665–677.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ni F, Gui Z, Guo Q, Hu Z, Wang X, Chen D
and Wang S: Downregulation of miR-362-5p inhibits proliferation,
migration and invasion of human breast cancer MCF7 cells. Oncol
Lett. 11:1155–1160. 2016.PubMed/NCBI
|
|
12
|
Pannem RR, Dorn C, Ahlqvist K, Bosserhoff
AK, Hellerbrand C and Massoumi R: CYLD controls c-MYC expression
through the JNK-dependent signaling pathway in hepatocellular
carcinoma. Carcinogenesis. 35:461–468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tauriello DV, Haegebarth A, Kuper I,
Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H
and Maurice MM: Loss of the tumor suppressor CYLD enhances
Wnt/beta-catenin signaling through K63-linked ubiquitination of
Dvl. Mol Cell. 37:607–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang G, Zhang X and Shi J: MiR-98 inhibits
cell proliferation and invasion of non-small cell carcinoma lung
cancer by targeting PAK1. Int J Clin Exp Med. 8:20135–20145.
2015.PubMed/NCBI
|
|
15
|
Lv H, Zhang Z, Wang Y, Li C, Gong W and
Wang X: MicroRNA-92a promotes colorectal cancer cell growth and
migration by inhibiting KLF4. Oncol Res. 23:283–290. 2016.
View Article : Google Scholar
|
|
16
|
Shen F, Cai WS, Feng Z, Li JL, Chen JW,
Cao J and Xu B: MiR-492 contributes to cell proliferation and cell
cycle of human breast cancer cells by suppressing SOX7 expression.
Tumour Biol. 36:1913–1921. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie BH, He X, Hua RX, Zhang B, Tan GS,
Xiong SQ, Liu LS, Chen W, Yang JY, Wang XN, et al: Mir-765 promotes
cell proliferation by downregulating INPP4B expression in human
hepatocellular carcinoma. Cancer Biomark. 16:405–413. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou X, Zhu HQ, Ma CQ, Li HG, Liu FF,
Chang H and Lu J: MiR-1180 promoted the proliferation of
hepatocellular carcinoma cells by repressing TNIP2 expression.
Biomed Pharmacother. 79:315–320. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen JS, Li HS, Huang JQ, Dong SH, Huang
ZJ, Yi W, Zhan GF, Feng JT, Sun JC and Huang XH: MicroRNA-379-5p
inhibits tumor invasion and metastasis by targeting FAK/AKT
signaling in hepatocellular carcinoma. Cancer Lett. 375:73–83.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Zhang L, Zheng B, Yan Y, Zhang Y,
Xie H, Zhou L, Zheng S and Wang W: MicroRNA-761 is upregulated in
hepatocellular carcinoma and regulates tumorigenesis by targeting
Mitofusin-2. Cancer Sci. 107:424–432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Urbanik T, Köhler BC, Boger RJ, Wörns MA,
Heeger S, Otto G, Hövelmeyer N, Galle PR, Schuchmann M, Waisman A,
et al: Down-regulation of CYLD as a trigger for NF-κB activation
and a mechanism of apoptotic resistance in hepatocellular carcinoma
cells. Int J Oncol. 38:121–131. 2011.PubMed/NCBI
|
|
22
|
Guo J, Shinriki S, Su Y, Nakamura T,
Hayashi M, Tsuda Y, Murakami Y, Tasaki M, Hide T, Takezaki T, et
al: Hypoxia suppresses cylindromatosis (CYLD) expression to promote
inflammation in glioblastoma: Possible link to acquired resistance
to anti-VEGF therapy. Oncotarget. 5:6353–6364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ke H, Augustine CK, Gandham VD, Jin JY,
Tyler DS, Akiyama SK, Hall RP and Zhang JY: CYLD inhibits melanoma
growth and progression through suppression of the JNK/AP-1 and
β1-integrin signaling pathways. J Invest Dermatol. 133:221–229.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hellerbrand C, Bumes E, Bataille F,
Dietmaier W, Massoumi R and Bosserhoff AK: Reduced expression of
CYLD in human colon and hepatocellular carcinomas. Carcinogenesis.
28:21–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang HL, Hu AP, Li SL, Xie JP, Ma QZ and
Liu JY: MiR-454 prompts cell proliferation of human colorectal
cancer cells by repressing CYLD expression. Asian Pac J Cancer
Prev. 16:2397–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang DH, Wang GY, Zhang JW, Li Y, Zeng XC
and Jiang N: MiR-501-5p regulates CYLD expression and promotes cell
proliferation in human hepatocellular carcinoma. Jpn J Clin Oncol.
45:738–744. 2015. View Article : Google Scholar : PubMed/NCBI
|