Introduction
Thyroid cancer is a common endocrine malignancy and
ranked the ninth most frequent in the USA in 2014 with estimated
62,980 new patients with thyroid carcinoma in the USA in 2014. Over
time, the incidence and morbity rates were increased by 5.5% and
0.8%, respectively each year (1,2). The
incidence of thyroid cancer is the sixth highest in women and more
than three-fold in women compared to men (3). Thyroid cancer is classed into three
categories: MTC (medullary thyroid cancer), DTC (differentiated
thyroid cancer, include papillary thyroid cancer, follicular
thyroid cancer and poorly differentiated thyroid cancer) and ATC
(anaplastic thyroid cancer) (3,4). MTC
is originated from the parafollicular C-cells and the remaining
subtypes are derived from thyroid follicular cells (1,5,6).
Fine-needle aspiration (FNA) biopsy is a useful clinical tool to
examine thyroid cancer, but the accuracy of diagnosis still need
improvement (7). Radiotherapy and
chemotherapy are effective but the appropriate selection and dosage
are still controversial (6).
Currently, molecular analysis provides better understanding of the
development and progression of thyroid cancer.
Transcription is regulated through various
chromatin-remodeling complexes such as NuRD (nucleosome remodeling
and histone deacetylation) complex. GATAD2A (GATA zinc finger
domain containing 2A), is a subunit of NuRD complex, which
comprises HDAC1/2 (histone deacetylases), CHD3/4 (ATP-dependent
remodeling enzymes), RbAp46/48 (histone chaperones) and MTA1/2/3
(specific DNA-binding proteins) (8). The NuRD complex is a key factor in
various biological progresses, including tumor growth inhibition,
cellular differentiation, embryonic development and the general
repression of transcription (8,9).
GATAD2B is a paralog of GATAD2A (10). GATAD2A/B have two conserved regions
known as CR1 and CR2 which could bind to MBD3 (methyl-CpG binding
domain protein 3), a key protein of the NuRD (10,11).
It is demonstrated that the GATAD2A plays essential roles in early
mouse development (12). However,
little information is known regarding its biology in thyroid cancer
up to now.
In this study, thyroid cancer cell lines (Cal-62 and
8305C) were used to detect the expression of GATAD2A by qPCR and
western blotting. The effects of GATAD2A on Cal-62 and 8305C
proliferation, cell cycle and apoptosis were analyzed. Moreover, we
detected the expression of apoptosis-associated proteins, caspase-3
and PARP. These results might help to diagnose the thyroid cancer
and improve the targeted therapies.
Materials and methods
Cell culture
The cell lines (Cal-62, 8305C and 293T) were
obtained from Cell Bank of Chinese Academy of Science (Shanghai,
China). Cal-62, a thyroid cancer cell line, was cultured in DMEM
supplemented with 10% FBS (fetal bovine serum, Biowest, S1810).
8305C, a thyroid cancer cell line, was cultured in EMEM containing
10% FBS, 1% L-Glu (21127022, Gibco, Cambrex, MD, USA) and 1% NEAA.
293T cell line was also cultured in DMEM containing 10% FBS (fetal
bovine serum, Biowest, S1810). All cell lines were maintained at
37°C in an incubator (311, Thermo) with 5% CO2.
Construction of GATAD2A knockdown
lentivirus
According to the sequence of GATAD2A, two vectors
shRNA S1 and shRNA S2 were designed. The sequences are as follows:
shRNA S1:
5′-CGTGCTGAAGCAGGTCATAAACTCGAGTTTATGACCTGCTTCAGCACGTTTTT-3′; shRNA
S2: 5′-CAGAACCTACTGGAGACACAACTCGAGTTGTGTCTCCAGTAGGTTCTGTTTTT-3′;
control shRNA:
5′-GCGGAGGGTTTGAAAGAATATCTCGAGATATTCTTTCAAACCCTCCGCTTTTTT-3′. The
three shRNAs were cloned into the plasmid pFH-L (Shanghai Hollybio,
China). Recombinant lentiviruses were generated by transfection of
80% 293T cells with lentivirus (including controls) and the helper
plasmids including pVSVG-I and pCMV∆R8.92 (Shanghai Hollybio,
China). After filtered and determined the viral titers, 10 and 8
MOI (multiplicities of infection) were achieved in Cal-62 and 8305C
cell lines, respectively. After infected for 96 and 72 h, the
achieved infection efficiency was 80%, and the expression of
GATAD2A was analyzed by RT-qPCR and western blotting.
Quantitative real-time PCR
(RT-qPCR)
After infected with lentivirus for 5 days, total
RNAs of cultured cells (Cal-62 and 8305C) were extracted using
TRIzol (15596–018, Invitrogen, Carlsbad, CA, USA). The mRNA level
of GATAD2A was evaluated by RT-qPCR using the iQ™ SYBR Green
Supermix kit (1708880, Bio-Rad, Hercules, CA, USA) on Bio-Rad
Connet Real-time PCR platform. The 20 µl mixture reaction system
contained 2X SYBR Premix Ex Taq 10 µl, cDNA 5 µl, forward and
reverse primers (2.5 µM) 0.5 µl, and ddH2O 4.5 µl. Actin
was applied as the input reference. Primers were designed based on
cDNA to amplify GATAD2A (NM_017660.3) and actin. The primers used
were as follows: GATAD2A: Forward, 5′-TGCTCGCCTCCTGCCTGTAG-3′;
Reverse, 5′-GACACAAAGCCAAGCCAGACC-3′; actin: Forward,
5′-GTGGACATCCGCAAAGAC-3′; Reverse, 5′-AAAGGGTGTAACGCAACTA-3′. The
mRNA was determined by using the 2−∆∆CT method.
Western blotting
After infection for 6 days, cells were collected,
washed with PBS and lysed in protein lysate (2X SDS Sample Buffer
(100 mMTris-HCl (pH 6.8), 10 mM EDTA (Sangon, Shanghai, China), 4%
SDS (SB0485-500 g, Sangon), 10% glycine). Total protein was
measured through BCA method and separated by 10% SDS-PAGE (45
µg/lane). Then proteins were transferred to PVDF (polyvinylidene
difluoride) membranes and blocked in 40–50 ml TBST (Tris-buffered
saline containing 0.1% Tween-20) including 5% skim milk for 1 h at
room temperature. The membranes were then incubated with primary
antibodies (rabbit anti-GATAD2A, Abgent, #20258a, 1:500; rabbit
anti-PARP, Cell Signaling, #9542, 1:1000; rabbit anti-caspase-3,
Cell Signaling, #9661, 1:500; rabbit anti-GAPDH antibody,
Proteintech, 10484-1-AP, 1:500000) at 4°C overnight. The membranes
were then washed three times with TBST for 10 min and probed with
secondary antibody (goat anti-rabbit HRP, Santa Cruz Biotechnology,
SC-2054, dilution: 1:5000) for 2 h at room temperature. After three
washes with TBST, the membranes were visualized using the ECL Plus
Western Blotting System kit (Amersham, RPN2132), and the
chemiluminescence was detected using X-ray film (Kodak).
MTT proliferation and colony formation
assays
Cal-62 and 8305C cell lines were infected for 96 and
120 h, respectively, the inoculation density of cell lines was
adjusted to 2×103 per well of Cal-62 and
5×103 per well of 8305C in 96-well plates, respectively.
After cell adhesion, 20 µl MTT (5 mg/ml, #M2128, Sigma, St. Louis,
MO, USA) was added and cultured for 4 h. Then, 100 µl acidified
isopropanol (10% SDS, 5% isopropanol and 0.01 mol/l HCl) was added
to stop the reaction and OD was determined on microplate
reader.
For evaluation of colony formation, the Cal-62 cell
lines were infected for 96 h and the density was achieved to 600
cells per well in 6-well plate. After cultured for 7 days, the
cells were fixed by 700 µl 4% paraformaldehyde per well. The plates
were washed with PBS (phosphate-buffered saline) twice and stained
with 700 µl crystal purple per well for 5 min. After washed with
PBS and air-dried, the number of colonies was manually counted
under the microscope.
Determination of cell cycle by flow
cytometry
After infection for 6 and 8 days, the Cal-62 and
8305c cell lines were seeded in 6-cm dishes at 2×105
cells and 8×104 cells per well, respectively. The CAL-62
cells were cultured for 6 days and harvested when the density
reached 80%. The collected cells were fixed by 75% cold ethanol
(10009269, Sinopharm Chemical Regant Co. Ltd., Shanghai, China) for
1 day at 4°C, the cells were washed with cold PBS and stained with
500 µl PI (Propidium Iodide) buffer (C1052, Beyotime Biotechnology,
Shanghai, China) for 1 h at 37°C in the dark. The cell cycles were
analyzed by flow cytometry at 24 h.
Determination of cell apoptosis by
flow cytometry
Apoptosis of Cal-62 and 8305C cell lines were
detected using Annexin V/7-AAD double staining kit (KGA1026,
Nanjing KeyGen Biotech. Co., Ltd.) after lentivirus infection for 6
and 8 days, respectively. When cells (5×105 cells per
dish) were cultured in 10-cm dishes to reach 80% confluency, they
were harvested, washed twice with PBS, and stained with 5 µl
Annexin V-APC and 5 µl 7-AAD for 5–15 min. Then cells were kept on
ice in the dark and subjected to apoptosis analysis on flow
cytometry.
Statistical analysis
All the experiments were repeated three times. The
SPSS 17.0 was used to analyze the results with t-test or one-way
ANOVA. The results were considered to be statistically significant
at P-value <0.05.
Results
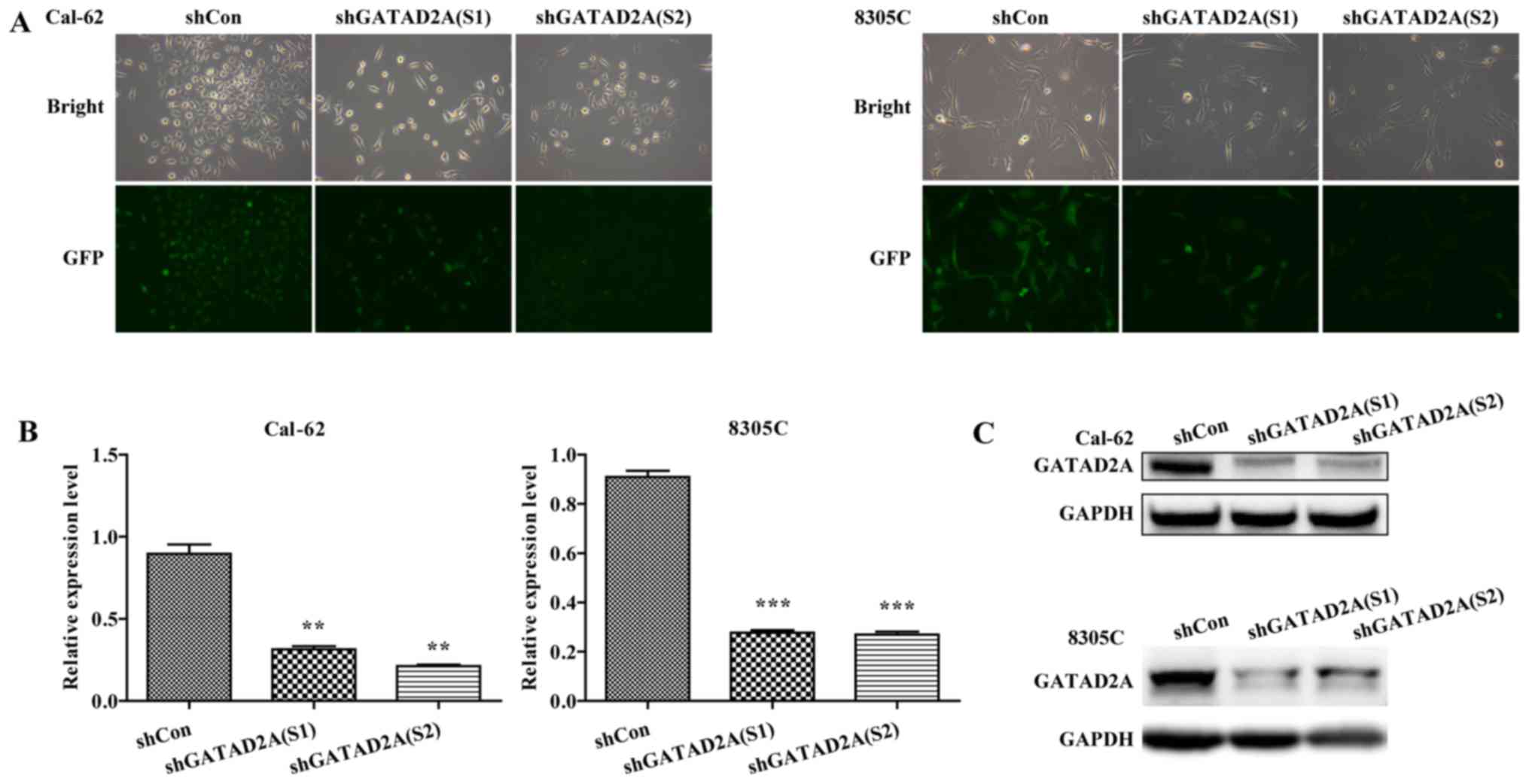
Inhibition of GATAD2A in Cal-62 and
8305C cell lines
To investigate the function of GATAD2A in thyroid
cancer, the expression of GATAD2A was inhibited by infection with
shGATAD2A (S1) or shGATAD2A (S2) in Cal-62 and 8305C cell lines. We
further examined the expression level of GATAD2A by RT-PCR and
western blotting in shGATAD2A and shCon infected cell lines. As
shown in Fig. 1A, most cells
presented GFP-positive expression under a microscope, suggesting
higher infection efficiency. Furthermore, the mRNA levels of
GATAD2A were significantly downregulated in shGATAD2A (S1 and S2)
compared to the shCon group in Cal-62 and 8305C cell lines
(Fig. 1B, P<0.01, P<0.001).
Consistently, the shGATAD2A (S1 and S2) obviously downregulated the
protein level of GATAD2A in Cal-62 and 8305C cells (Fig. 1C). There was a difference in the
expression levels of S1 and S2 in Cal-62 cells, but the difference
is not significant. Collectively, we successfully constructed
GATAD2A-silencing Cal-62 and 8305C cell lines.
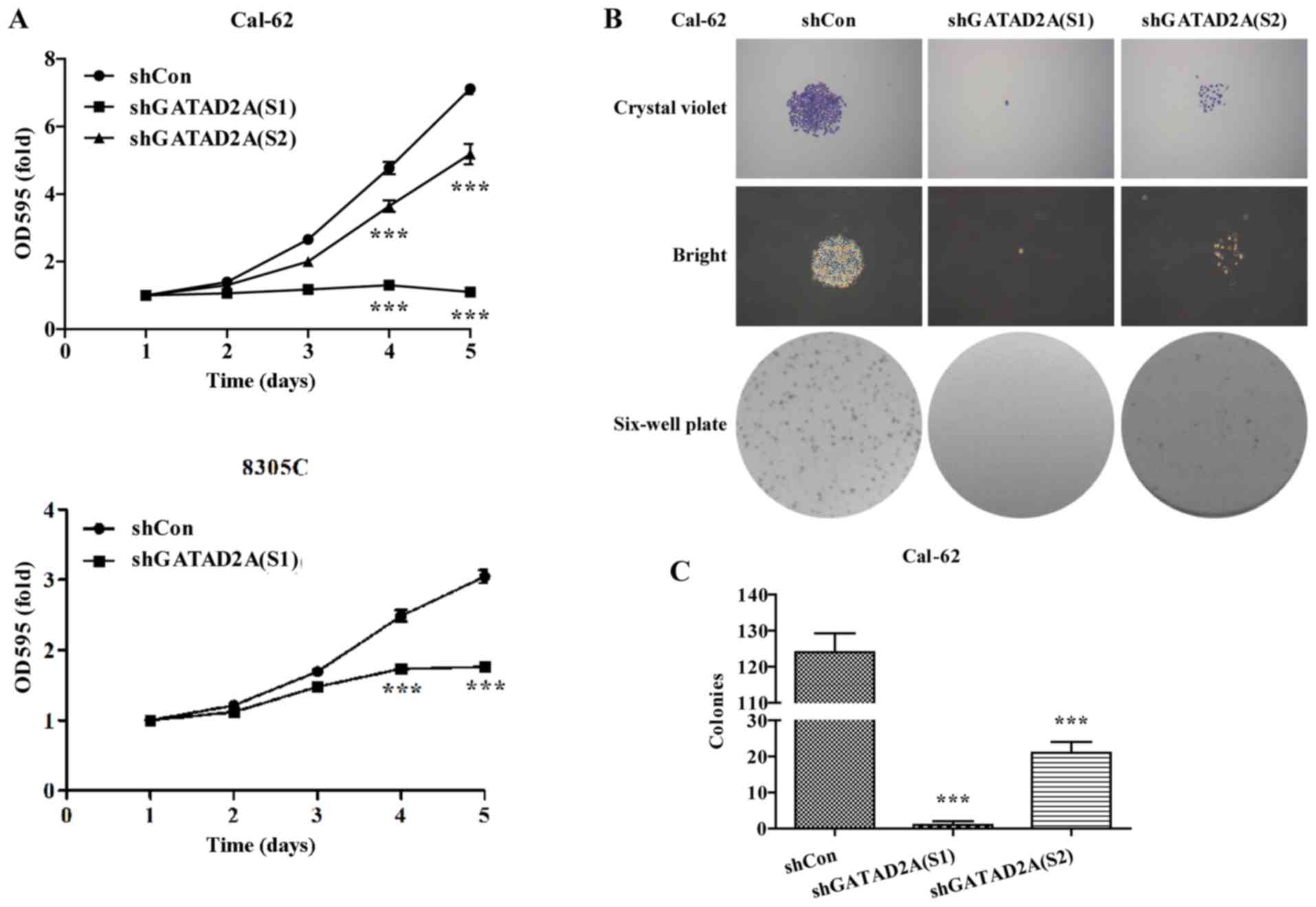
Effects of GATAD2A on cell
proliferation and colony formation
The potential effects of GATAD2A on cell
proliferation and colony formation were explored in Cal-62 and
8305C cell lines. The same number of CAL-62 and 8305C cells from
shGATAD2A and shCon group was subjected to MTT assay. The results
showed that Cal-62 and 8305C cells presented significantly slower
proliferative rate (P<0.001, Fig.
2A) after transfection with shGATAD2A (S1) or shTAGAD2A (S2).
Notably, the suppressive effects in Cal-62 cells were more obvious
than that in 8305C cells.
To further determine the effects of GATAD2A on cell
proliferation, we chose Cal-62 cells to perform colony formation
assay. As shown in Fig. 2B, The
size was relatively smaller and colonies were fewer in shGATAD2A
(S1) or shGATAD2A (S2) groups compared with shCon groups in Cal-62
cells. Statistical analysis further confirmed that knockdown of
GATAD2A significantly reduced colonies formed in Cal-62 cells
(Fig. 2C, P<0.001). Notably,
Cal-62 cells treated with shGATAD2A (S1) presented more obvious
growth inhibition than shGATAD2A (S2). The above results revealed
that GATAD2A might play an important role in thyroid cancer cell
growth and proliferation.
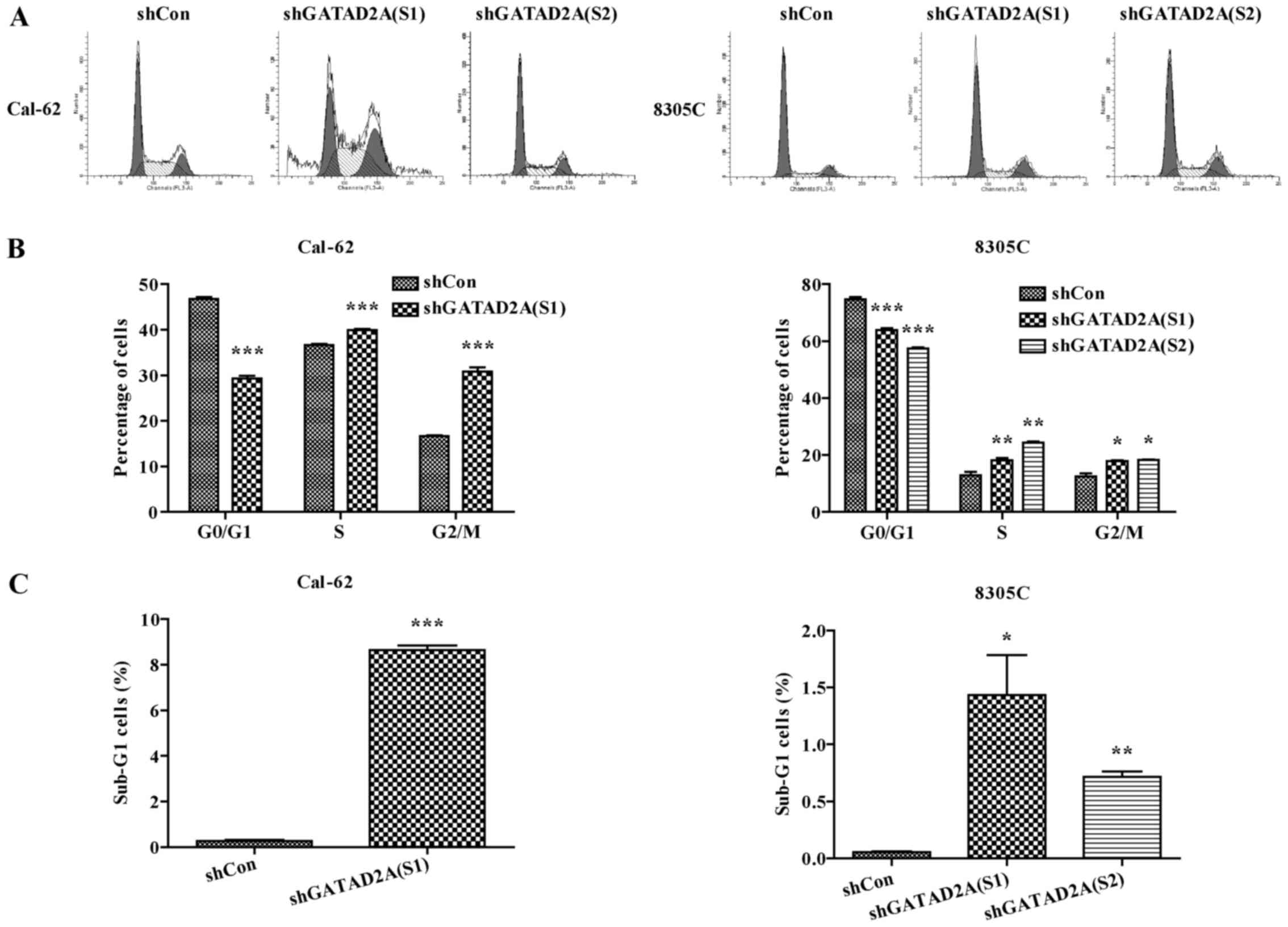
Effects of GATAD2A on cell cycle
progression
To determine the underlying mechanism of inhibition
of cell proliferation, cell cycle was analyzed by flow cytometry.
The percentage of cell distribution was observed as significantly
different in shGATAD2A and Con groups (Fig. 3A). Compared to shCon, the percentage
of cells in G0/G1 phase was significantly decreased, while the
cells in G2/M phase were remarkably increased in shGATAD2A infected
Cal-62 and 8305C cell lines (Fig.
3B, P<0.001), indicating cell cycle was arrested in G2/M
phase by GATAD2A knockdown. In addition, the sub-G1 cell number of
both cell lines infected with shGATAD2A was significantly increased
(Fig. 3C, P<0.05, P<0.01,
P<0.001). Taken together, these data suggested that knockdown of
GATAD2A-suppressed cell proliferation might be via cell cycle
arrest.
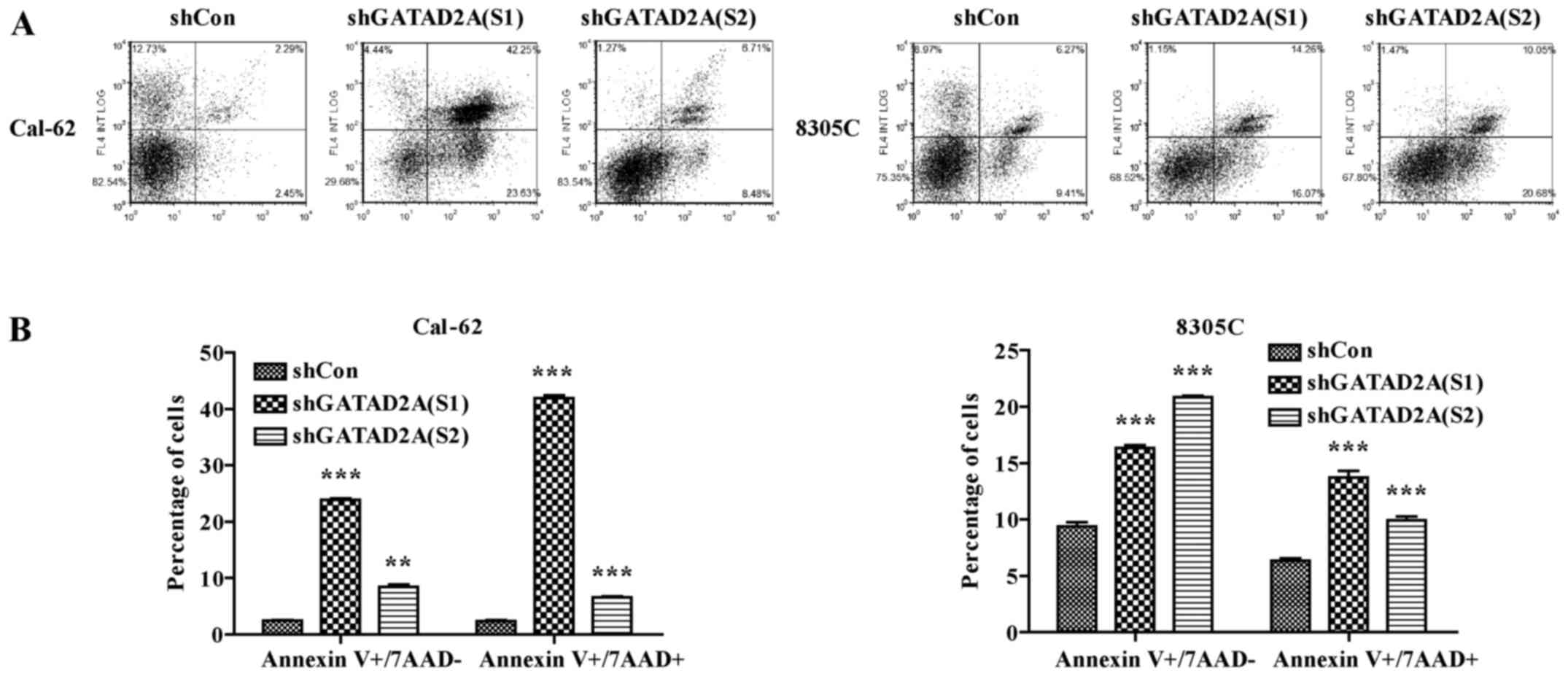
Effects of GATAD2A on cell
apoptosis
In addition, we further investigated the effects of
GATAD2A knockdown on cell apoptosis by flow cytometry (Fig. 4A). As shown in Fig. 4B, the number of apoptotic cells in
shGATAD2A infected Cal-62 and 8305C cell lines was significantly
higher than that in shCon group (P<0.001). Based on the above
data, we observed there existed differences in suppressing cell
proliferation and inducing cell cycle arrest and apoptosis between
S1 and S2. Overall, functional experiments revealed shGATAD2A (S1)
was more powerful than shGATAD2A (S2) in suppressing cell
proliferation, induced cell cycle arrest and apoptosis. Therefore,
shGATAD2A (S1) was chosen to study the effect of GATAD2A in
regulating the apoptosis-related proteins in Cal-62 cells. In line
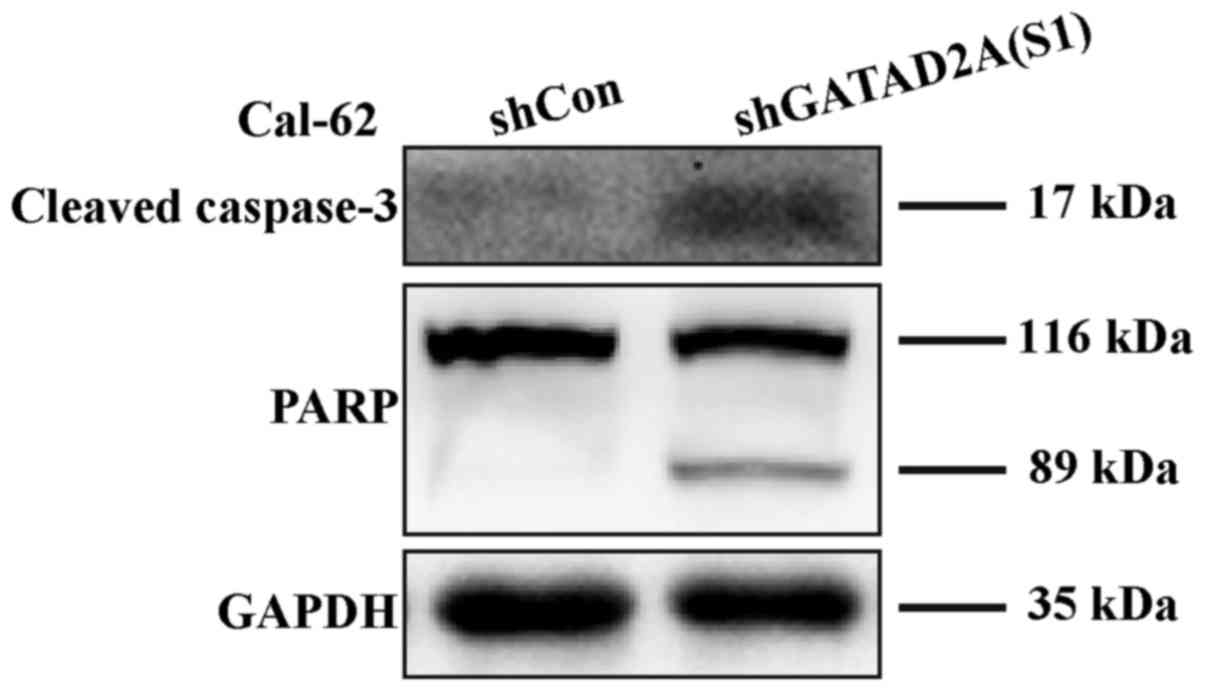
with flow cytometry, caspase-3 cleavage was higher in shGATAD2A
(S1) infected cells and its substrate PARP (poly ADP-ribose
polymerase), a marker of apoptosis and activation of caspase-3, was
also elevated compared to the control group (Fig. 5). The results suggested that GATAD2A
plays essential roles in cell proliferation of thyroid cancer.
Discussion
It has been implicated that NuRD participates in the
regulation of transcriptional events, chromatin assembly, cell
cycle progression and genetic instability (13). Depending on cell types, the NuRD
complex plays roles in both promoting and suppressing tumorigenesis
(13). It has been reported that
GATAD2A is an important structural subunit of the NuRD complex and
is directly associated with histone tails (11,14).
The amino-terminal conserved region of GATAD2A directly interacts
with MBD2 or MBD3 and the C-terminal conserved region can interact
with histone tails and is essential for targeting to the specific
genomic loci (11,14). This evidence suggests GATAD2A might
be closely associated with tumorigenesis. Thus, our study revealed
the close relationship between GATAD2A and thyroid cancer
progression. Functional analysis demonstrated that knockdown of
GATAD2A suppressed thyroid cancer cell growth and
proliferation.
To better understand the underlying mechanism of
cell growth inhibition, the distribution of cell cycle phases was
analyzed in thyroid cancer cells after GATAD2A knockdown. As
expected, knockdown of GATAD2A resulted in G2/M arrest in the cell
cycle. A previous study showed NuRD complex is an active regulator
of both the G1/S and the G2/M checkpoints and its defection affects
checkpoint control of G2/M through disruption in histone
modification (13). It might
indicate that GATAD2A is the key regulator in G2/M check point
through interaction with other partners within the NuRD
complex.
Moreover, the flow cytometry results revealed that
the abnormal cell group distributed in the sub-G1 phase. Sub-G1
peak was defined as the cells with deficit in DNA content and the
DNA fragments was stained (15–17).
DNA fragmentation is the hallmark of cell apoptosis (18,19).
Quantification of the percentage of apoptotic cells in sub-G1 phase
is commercially available to detect cell apoptosis. However, the
sub-G1 DNA content cannot be used as a sole marker of apoptotic
cells. Further, we detected cell apoptosis by flow cytometry and
the results revealed that knockdown of GATAD2A increased cell
apoptosis in both early and late stages compared to the control
group. These results kept in line with the results from the
previous assay, which indicated that GATAD2A played potential roles
in thyroid cancer cell proliferation.
In apoptosis progress, caspase-3 is a crucial
mediator of apoptosis and cleaved to activate (20). Cleaved caspase-3 displayed
upregulation in the GATAD2A knockdown group. Moreover, activation
of PARP is a response to DNA damaging compounds and acts as a
target of caspase-3 for degradation. The PARP was cleaved into two
polypeptide fragments and the subsequent loss in its activity
accompanies apoptosis (21–23). In addition, it has been shown that
the NuRD complex is rapidly recruited to the sites of DSBs
(double-strand breaks) through the activity of the poly(ADP ribose)
polymerase (PARP) (24,25). It might indicate that GATAD2A
knockdown is the executor response to DNA damage and active
downstream cascade of cell death.
Based on these data, we could conclude that
suppression of GATAD2A attenuated thyroid cancer cell proliferation
and colony formation and promoted apoptosis, through activating the
caspase-3 and PARP. To further explore its clinical significance,
more work is still required to validate the findings of the current
study in terms of a clinical application, due to the low number of
clinical specimens examined.
References
|
1
|
Weitzman SP and Cabanillas ME: The
treatment landscape in thyroid cancer: A focus on cabozantinib.
Cancer Manag Res. 7:265–278. 2015.PubMed/NCBI
|
|
2
|
Grande C and Brose MS: The evolving
treatment landscape for metastatic differentiated thyroid cancer. J
Adv Pract Oncol. 5:461–465. 2014.PubMed/NCBI
|
|
3
|
LiVolsi VA: The pathology of autoimmune
thyroid disease: a review. Thyroid. 4:333–339. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aschebrook-Kilfoy B, Ward MH, Sabra MM and
Devesa SS: Thyroid cancer incidence patterns in the United States
by histologic type, 1992–2006. Thyroid. 21:125–134. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moley JF: Medullary thyroid carcinoma.
Curr Treat Options Oncol. 4:339–347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carling T and Udelsman R: Thyroid cancer.
Annu Rev Med. 65:125–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Enewold L, Zhu K, Ron E, Marrogi AJ,
Stojadinovic A, Peoples GE and Devesa SS: Rising thyroid cancer
incidence in the United States by demographic and tumor
characteristics, 1980–2005. Cancer Epidemiol Biomarkers Prev.
18:784–791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Torchy MP, Hamiche A and Klaholz BP:
Structure and function insights into the NuRD chromatin remodeling
complex. Cell Mol Life Sci. 72:2491–2507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smits AH, Jansen PW, Poser I, Hyman AA and
Vermeulen M: Stoichiometry of chromatin-associated protein
complexes revealed by label-free quantitative mass
spectrometry-based proteomics. Nucleic Acids Res. 41:e282013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brackertz M, Boeke J, Zhang R and
Renkawitz R: Two highly related p66 proteins comprise a new family
of potent transcriptional repressors interacting with MBD2 and
MBD3. J Biol Chem. 277:40958–40966. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brackertz M, Gong Z, Leers J and Renkawitz
R: p66alpha and p66beta of the Mi-2/NuRD complex mediate MBD2 and
histone interaction. Nucleic Acids Res. 34:397–406. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marino S and Nusse R: Mutants in the mouse
NuRD/Mi2 component P66alpha are embryonic lethal. PLoS One.
2:e5192007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lai AY and Wade PA: Cancer biology and
NuRD: A multifaceted chromatin remodelling complex. Nat Rev Cancer.
11:588–596. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng Q, Cao R, Xia L, Erdjument-Bromage H,
Tempst P and Zhang Y: Identification and functional
characterization of the p66/p68 components of the MeCP1 complex.
Mol Cell Biol. 22:536–546. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Umansky SR, Korol' BA and Nelipovich PA:
In vivo DNA degradation in thymocytes of gamma-irradiated or
hydrocortisone-treated rats. Biochim Biophys Acta. 655:9–17. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gong J, Traganos F and Darzynkiewicz Z: A
selective procedure for DNA extraction from apoptotic cells
applicable for gel electrophoresis and flow cytometry. Anal
Biochem. 218:314–319. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Darzynkiewicz Z, Juan G, Li X, Gorczyca W,
Murakami T and Traganos F: Cytometry in cell necrobiology: Analysis
of apoptosis and accidental cell death (necrosis). Cytometry.
27:1–20. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagata S, Nagase H, Kawane K, Mukae N and
Fukuyama H: Degradation of chromosomal DNA during apoptosis. Cell
Death Differ. 10:108–116. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagata S: Apoptotic DNA fragmentation. Exp
Cell Res. 256:12–18. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Porter AG and Jänicke RU: Emerging roles
of caspase-3 in apoptosis. Cell Death Differ. 6:99–104. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heller B, Wang ZQ, Wagner EF, Radons J,
Bürkle A, Fehsel K, Burkart V and Kolb H: Inactivation of the
poly(ADP-ribose) polymerase gene affects oxygen radical and nitric
oxide toxicity in islet cells. J Biol Chem. 270:11176–11180. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Karabay AZ, Aktan F, Sunguroğlu A and
Buyukbingol Z: Methylsulfonylmethane modulates apoptosis of
LPS/IFN-γ-activated RAW 264.7 macrophage-like cells by targeting
p53, Bax, Bcl-2, cytochrome c and PARP proteins. Immunopharmacol
Immunotoxicol. 36:379–389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kaufmann SH: Induction of endonucleolytic
DNA cleavage in human acute myelogenous leukemia cells by
etoposide, camptothecin, and other cytotoxic anticancer drugs: A
cautionary note. Cancer Res. 49:5870–5878. 1989.PubMed/NCBI
|
|
24
|
Olsen JV, Vermeulen M, Santamaria A, Kumar
C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al:
Quantitative phosphoproteomics reveals widespread full
phosphorylation site occupancy during mitosis. Sci Signal.
3:ra32010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Polo SE, Kaidi A, Baskcomb L, Galanty Y
and Jackson SP: Regulation of DNA-damage responses and cell-cycle
progression by the chromatin remodelling factor CHD4. EMBO J.
29:3130–3139. 2010. View Article : Google Scholar : PubMed/NCBI
|