Introduction
Panax ginseng C.A. Meyer (P. ginseng)
has been used worldwide as a traditional medicine for the treatment
of cancer, including lung cancer and other diseases. Among the
varieties of ginseng used in medical treatment, P. ginseng
(Korean ginseng) and P. quinquefolius (American ginseng)
have been the ones most studied to date (1,2). We
found that the anticancer effect of ginseng increased after
enzymatic processing of ginseng saponin (3), which may be the result of the
accumulation of minor saponins, such as Rh1, Rg3, compound K and
protopanaxatriol type (PPT) constituents in ginseng saponin. The
modified regular ginseng extract (MRGX), which reinforces various
ginsenosides from regular ginseng butanol-extract (GBX) by treating
them with the enzymes laminarinase and pectinase to produce
fortified anticancer ginsenosides, is reported to have anticancer
effects against lung cancer cells (3–5). In
addition, the chemical profiles of enzymatically modified ginseng
extract were addressed by high-performance liquid chromatography
(HPLC) compared to butanol extract as previously reported by us.
Five ginsenosides, including Rh1, Rg3(S), Rg3(R), compound K and
Rh2, were exclusively detected (6).
Lung cancer, which consists mostly of non-small cell
lung cancer (NSCLC) in up to 85% of the total cases, is the second
leading cause of cancer-related death worldwide (7), and is considered to be an extremely
lethal form of cancer (8). Over the
past several decades, surgical and combined therapies have
contributed to reducing the mortality and the morbidity associated
with lung cancer; however, these approaches still have limitations.
Complementary and alternative therapy using herbal medicine,
particularly using gensenosides have recently received increased
attention in the Western medical society (2,9,10).
Yet, studies concerning their mechanism of action at the molecular
biological level are lacking.
Autophagy, a catabolic process for the degradation
and recycling of proteins and organelles, is one type of cell death
(11,12). It is initiated by the formation of
autophagosomes, which are double-membrane vesicles that fuse to
lysosomes to form autolysosomes, where the sequestered cellular
components are digested by lysosomal enzymes. The autophagic
process is fundamental in ridding the cell of aged and aggregated
proteins and is the only known mechanism for the disposal of
damaged organelles under normal conditions and for disposal of
cells under stress. This process is regulated by autophagy factors
such as ULK1, ATG13, ATG7, ATG5 and Beclin-1 (13,14).
In addition, autophagy occurs as a result of inappropriate cellular
signal transduction through the AMPK-linked mammalian target of
rapamycin (mTOR). Generally, mTOR is considered a therapeutic
target for lung cancer since mTOR is a major pathway that
coordinates proper cell growth and proliferation by regulating
ribosomal biogenesis and protein translation (15).
In the present study, MRGX was investigated to
determine its potential for use as a therapeutic agent for the
treatment of lung cancer. The antiproliferative effect of MRGX was
assessed in human non-small cell lung adenocarcinoma cells.
Additionally, the molecular mechanisms underlying the anticancer
activity of MRGX were studied, with a special focus on the
autophagy-related multiple signaling pathways in this particular
lung cancer type.
Materials and methods
Preparation of the MRGX extract
The root of regular ginseng (4 years old) was
purchased from the National Agricultural Cooperative Foundation
(Chuncheongnam-do, Korea). A total of 20 g of pulverized ginseng
root powder was suspended in 380 ml of distilled water and
sterilized at 121°C for 15 min. The extract was fractionated via
extraction with various solvents, including water, methanol and
butanol. Ginseng butanolic extract (GBX) was used as a control for
regular ginseng. An aliquot of sterilized laminarinase and
pectinase (1:1, specific activity units) was added to the
suspension, which was then incubated at 40°C for 2 days, after
which it was evaporated to powder at 60°C. This enzyme-modified
ginseng powder was suspended in 400 ml of 80% (v/v) methanol. The
suspension was sonicated and filtered through Whatman no. 2 filter
paper (Whatman International Ltd., Maidstone, UK). The filtrates
were combined and evaporated to dryness at 50°C. The extract was
dissolved in 200 ml distilled water, then separated by a funnel and
extracted with 200 ml of butanol. The extract was dissolved to a
concentration of 10% (w/v) in 70% ethanol. A total of 7
ginsenosides, including Rg1, Re, Rf, Rb1, Rc, Rb2 and F1, were
present in the GBX. By contrast, 5 ginsenosides, including Rh1,
Rg3(S), Rg3(R), compound K and Rh2, were exclusively detected in
MRGX (6,10). The final obtained sample source was
named MRGX in the present study. This sample was deposited
(BP1234034, http://biorp.kribb.re.kr) in the
Biological Resource Center (BRC) in Korea Research Institute of
Bioscience and Biotechnology (KRIBB).
Cell lines
Human non-small cell lung cancer cell lines (A549,
H596 and H1299) were obtained from the American Type Culture
Collection (ATCC; Rockville, MD, USA). Cells were grown in
Dulbeccos modified Eagles medium (DMEM), supplemented with 10%
(v/v) fetal bovine serum (FBS) and 1% (w/v) penicillin-streptomycin
(both from Gibco, Grand Island, NY, USA), at 37°C with 5% (v/v)
CO2.
Cell viability assay
Cells (5×103/well) were placed in a
96-well plate. After a 24-h incubation, the cells were treated with
0, 25, 50 or 100 µg/ml of MRGX for 72 h. Cell viability assays were
performed following the procedures published in a previous study
(16). In brief, at the end of the
treatment, 10 µl of Cell Counting Kit-8 solution (Dojindo,
Kumamoto, Japan) was added to the cell solution and incubated for 1
h at 36°C. In addition, the cells were pretreated with 100 nM of
the autophagy inhibitor bafilomycin A1 (BafA1; Sigma-Aldrich, St.
Louis, MO, USA), and treated with MRGX at the same concentrations.
Cell viability was determined using a microplate reader (Sunrise,
Tecan, Switzerland) to measure the absorbance at 450 nm. The assays
were performed in triplicate.
Cell cycle analysis with propidium
iodide staining
The cell cycle was analyzed using propidium iodide
(PI) staining kits (Roche Diagnosis GmbH, Mannheim, Germany)
following the instructions provided by the manufacturer. In brief,
the cells were incubated with the extract for 48 h, trypsinized,
washed twice with phosphate-buffered solution (PBS), and
centrifuged at 2,000 rpm for 5 min at 4°C. Then, the cells were
incubated with 1.4 mg/ml of PI for 15 min at room temperature.
Measurements were carried out on a flow cytometer (MoFlo Astrios
flow cytometer; Beckman-Coulter, Indianapolis, IN, USA) with a
670-nm high-pass filter for PI detection. The data were analyzed
using Summit V6.0 software.
Microarray analysis
Total RNA was extracted from vehicle- or 100 µg/ml
MRGX-treated A549 lung cancer cells. The total RNA from each sample
was extracted using TRIzol reagent (Gibco-BRL, Rockville, MD, USA)
according to the manufacturer's instructions. For microarray
analysis of the MRGX-treated lung cancer cells, the Human Twin
Chip™ Human 44K (Genocheck, Seoul, Korea) was used for the
transcription profiling analysis. The microarray analysis was
performed following the procedures published in a previous study
(3). In brief, the Cy3- (vehicle)
and Cy5-labeled (MRGX-treated) cRNAs were hybridized with the Human
44K microarray, and the hybridization images were analyzed with an
Agilent DNA Microarray Scanner. All data were normalization and
selection of upregulated and downregulated genes was performed
using GeneSpring GX 7.3 (Agilent Technologies, Inc., Santa Clara,
CA, USA).
Ontology-related network analysis
To study the biological functions of
ontology-related regulated genes and proteins through their
interaction network, we conducted a bioinformatic network analysis
using an Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com). The IPA identifies a gene
interaction network on the basis of a regularly updated ‘Ingenuity
Pathways Knowledge-base’. The updatable database was retrieved from
the biological literature. Network generation was optimized from
the inputted expression profile when possible and was aimed at the
production of highly connected networks.
Acridine orange staining for
autophagy
Autophagy is characterized by increased formation of
acidic vesicular organelles (AVOs; lysosomes and
autophagolysosomes), which can be quantified using flow cytometry
after staining the cells with acridine orange (AO). AO is a weak
base that accumulates in AVOs. The AVOs can be quantified based on
the fact that the increase in the intensity of the red fluorescence
is proportional to the degree of acidity (17). For the FACS analysis, A549 cells
(5×106) were placed in a 100-mm culture dish and treated
with 0, 50 or 100 µg/ml of MRGX. After a 48-h incubation, 1 µg/ml
of AO was added, and the cells were incubated for 15 min at 37°C,
and then subjected to flow cytometry examination.
Identification of autophagy using
immunofluorescence and confocal microscopy
Human full-length LC3 (microtubule-associated
protein 1 light chain 3α), Beclin-1 and ATG5 (autophagy-related
gene 5, a marker of autophagy), and complementary DNA cloned into
pCMV-SPORT6 vectors (LC3, hMU000295; Beclin-1, hMU003282; and ATG5,
KU032585) were purchased from the 21 Century Frontier Human Gene
Bank (Daejeon, Korea), along with a cloning green fluorescent
protein (GFP) sequence. A549 cells were seeded at 5×104
cells/well on the coverslip of an 8-well plate. After 24 h, the
cells were transfected using the HilyMax transfection reagent
(Dojindo, Kumamoto, Japan) with GFP-LC3, GFP-Beclin-1 and GFP-ATG5
vector for 48 h. After MRGX treatment, cells were washed twice with
PBS, fixed with 4% (v/v) formaldehyde for 15 min, and then
permeabilized with cell permeabilization reagents (FIX & PERM
Cell Permeabilization reagents; Invitrogen, Carlsbad, CA, USA).
They were further blocked with 10% (w/v) goat serum for 1 h,
followed by staining with 4,6-diamidine-2-phenylindole
dihydrochloride (DAPI) solution. For immunostaining, cells were
incubated in primary antibody (LC3, Beclin-1, ATG5; Abcam, Eugane,
CA, USA) diluted to 1:100 in 10% (w/v) goat serum in PBS overnight
at 4°C. After the cells had been washed 3 times for 5 min each with
PBS, they were incubated in the dark for 1 h with a 1:1,000
dilution of Alexa Fluor 488-conjugated secondary antibody
(Invitrogen). Finally, the slides were washed 3 times with PBS and
mounted in mounting medium (Vector, Burlingame, CA, USA). Images
were captured using a laser-scanning confocal microscope (LSM 710;
Carl Zeiss, Jena, Germany) equipped with a C-Apochromat 40x/1.2
water immersion lens (48-nm argon laser/505- to 550-nm detection
range). Final image data were analyzed using ZEN 2009 Light Edition
software.
Western blot analysis
The expression levels of autophagy-related signaling
proteins in the cells treated with MRGX and compound C (5 µmol/l;
CAS 866405-64-3; Sigma-Aldrich) were examined using western
blotting, as previously described (18). In brief, 30 µg of the denatured
protein was separated using 12% polyacrylamide gel electrophoresis
and transferred onto a nitrocellulose membrane. The nitrocellulose
membrane was then stained with Ponceau S to position the proteins.
The blotted membrane was blocked for 1 h with 5% (w/v) skimmed milk
in Tween-20 and Tris-buffered saline (TTBS), followed by incubation
with a dilution of primary antibodies, LC3-I, p62, Akt, p-Akt,
AMPK, p-AMPK, mTOR, p-mTOR, 4EBP1, p-4EBP1, Ulk1, p-Ulk1 (S317) and
GAPDH (Cell Signaling Technology, Danvers, MA, USA), at room
temperature for 2 h or at 4°C overnight. The membrane was washed 3
times for 5 min each time with 0.1% (v/v) Tween-20 in TBS before
incubation with horseradish-peroxidase (HRP)-conjugated goat
anti-mouse IgG or HRP-conjugated rabbit anti-goat IgG with a
1:2,000 dilution in TBS containing 5% (w/v) skimmed milk at room
temperature for 1 h. The membrane was rinsed 3 times with TBS-0.1%
(v/v) Tween-20 for 5 min each time and an enhanced
chemiluminescence system (Thermo Scientific, San Jose, CA, USA) was
used to visualize the bands on a ChemiDoc MP system (Bio-Rad,
Hercules, CA, USA). The expression levels of the proteins were
quantitatively analyzed by comparison with GAPDH as an internal
control.
Statistical analyses
GraphPad Prism software (GraphPad, San Diego, CA,
USA) was used for the statistical analyses. The Student's t-test
was used to assess the statistical difference between the control
and the MRGX-treated groups. P-values <0.05 were considered to
indicate a statistically significant result.
Results
Inhibition of the growth of lung
cancer cells by MRGX
In order to investigate the effect of MRGX on the
growth of various lung cancer cell lines, we directly treated A549,
H596 and H1299 cells with MRGX, and evaluated the growth inhibition
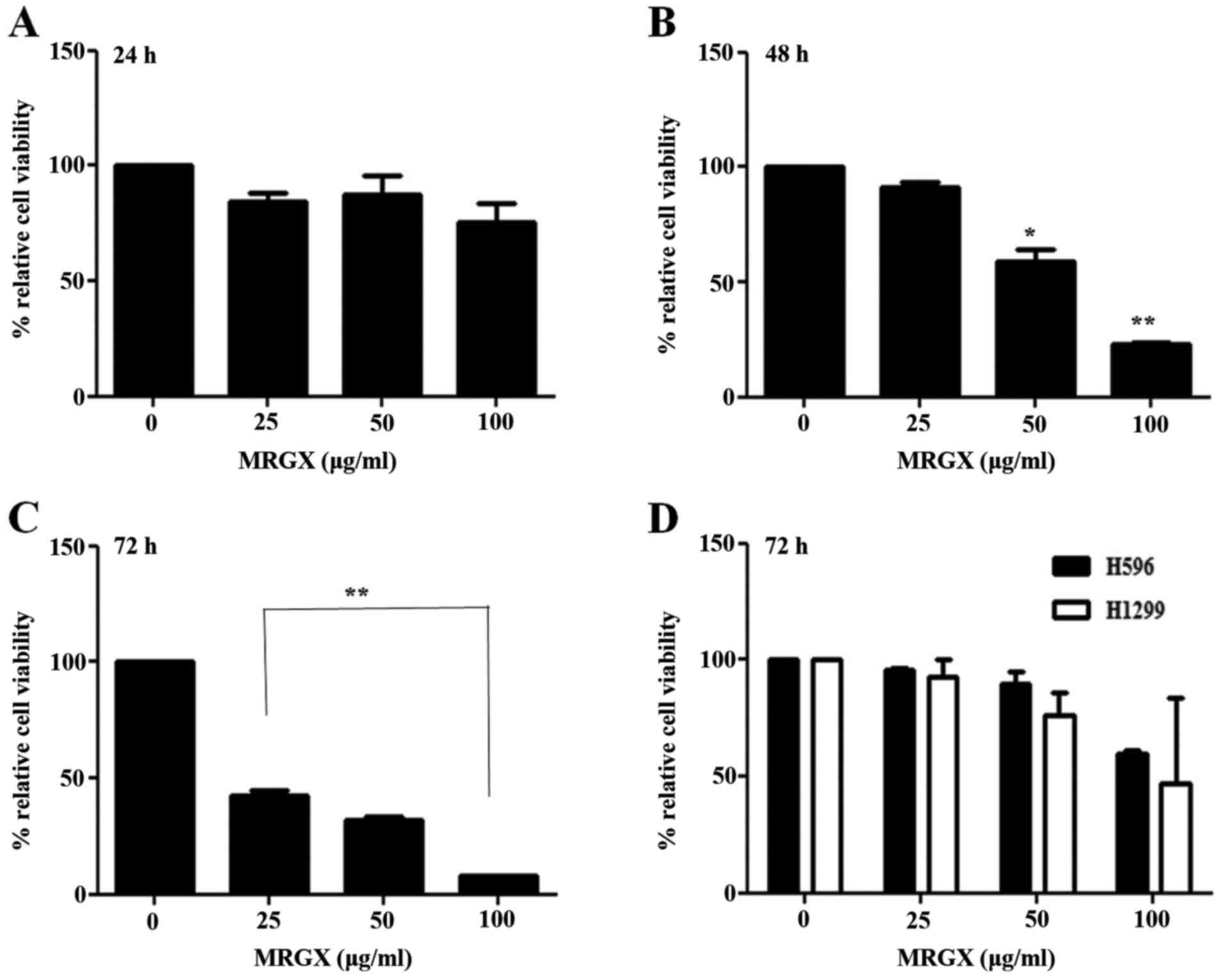
using cell viability assays. As shown in Fig. 1A, MRGX inhibited the growth of A549
cells after 24 h of incubation with no statistical significance
(Fig. 1A). Treatment with MRGX for
48 and 72 h markedly and significantly inhibited the proliferation
of the A549 cells (Fig. 1B and C).
The inhibitory effect of MRGX on the growth of A549 cells was
concentration-dependent. Intriguingly, MRGX only exerted moderate
inhibitory effect on the growth of H596 and H1299 cells even after
the cells had been treated for 72 h (Fig. 1D).
Cell cycle analysis of A549 cells
following treatment with MRGX
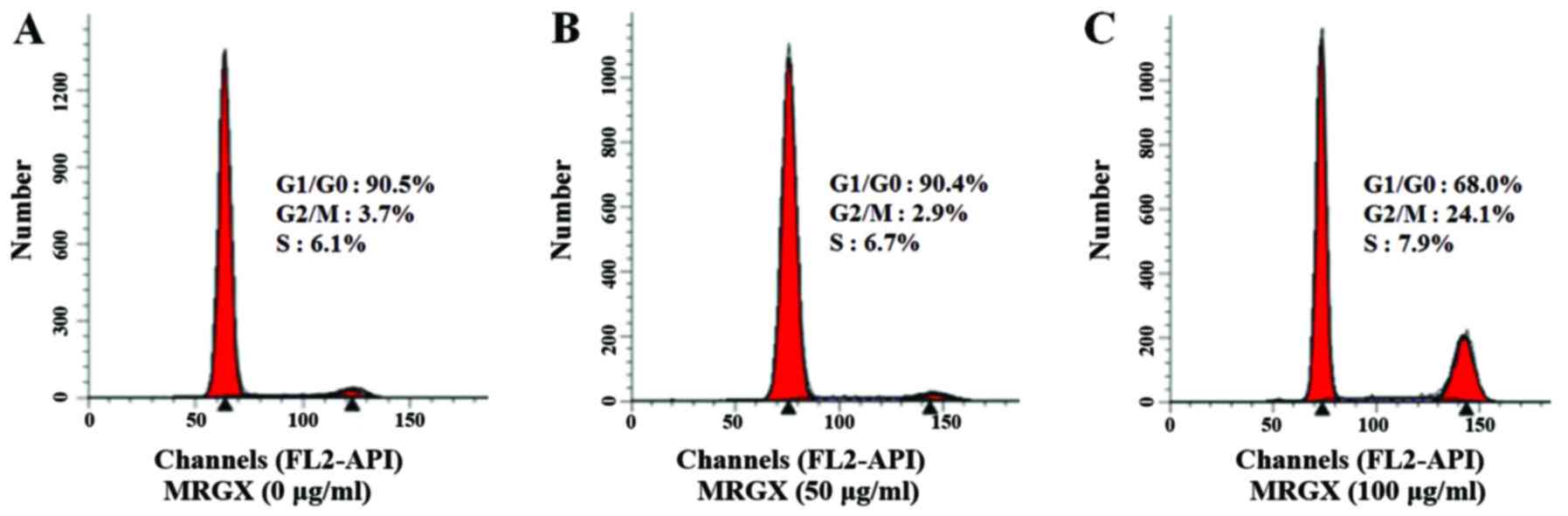
The effect of MRGX on the cell cycle distribution of
A549 cells was evaluated using PI staining and flow cytometry
(Fig. 2). The flow cytometric
analysis of A549 cells showed an apparent shift from the G1 to the
G2/M phase. In A549 cells treated with a vehicle control for 48 h,
90.5% of the cells were in the G1 phase, and 3.7% were in the G2/M
phase (Fig. 2A). In contrast, in
the A549 cells treated with 100 µg/ml of MRGX, 68.0% were in the G1
phase, and 24.1% were in the G2/M phase (Fig. 2C), suggesting that MRGX treatment
caused the cells to undergo G2/M phase arrest.
Analysis of MRGX-related gene
expression in lung cancer cells
To analyze the MRGX-related gene expression in lung
cancer cells, we used a cDNA microarray analysis approach.
Microarray data were filtered and combined using gene symbols; and
then network analyses were performed. The clustered microarray data
showed that groups of genes in the untreated and MRGX-treated (100
µg/ml) lung cancer cells were differentially regulated. Genes were
considered to be differentially expressed when the global M-value,
log2 (R/G fluorescence), exceeded 1.0 (2-fold-change)
with a P-value <0.05 after the significance analysis of the
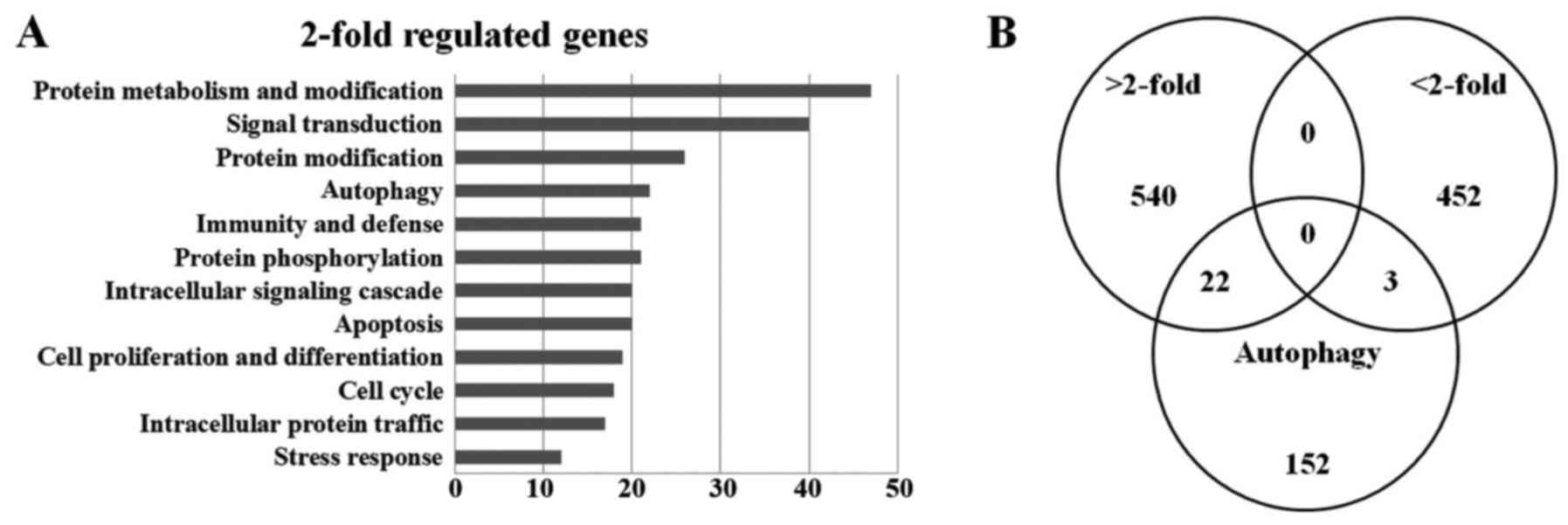
microarray (SAM). A total of 1,169 genes were ultimately identified
and categorized in terms of their cellular functions using Gene
ontology annotation (Fig. 3A). The
Venn diagram for autophagy-related increases (22 genes) and
decreases (3 genes) is shown (Fig.
3B). The relative abundances of the regulated genes between the
untreated and MRGX-treated lung cancer cells are listed in Table I.
 | Table I.Autophagy-related genes affected by
MRGX. |
Table I.
Autophagy-related genes affected by
MRGX.
|
| Signal
intensity |
|
|
|---|
|
|
|
|
|
|---|
| Ratio | MRGX | Ctl | Gene symbol | GenBank acc.
no. |
|---|
| 10.66 | 26 |
2 | HSPA9 | NM_004134 |
| 9.82 | 19 |
2 | SMAD2 | NM_001003652 |
| 8.85 | 54 |
6 | BECN1 | NM_003766 |
| 8.21 | 32 |
4 | CTNNB1 | NM_001098209 |
| 7.84 | 60 |
8 | AIFM1 | NM_001130846 |
| 4.53 | 23 |
5 | CLTC | NM_004859 |
| 4.51 | 17 |
4 | CD46 | NM_002389 |
| 4.30 | 23 |
5 | ATG12 | NM_004707 |
| 4.24 | 254 | 60 | SUMO1 | NM_001005781 |
| 4.10 | 38 |
9 | PIK3C3 | NM_002647 |
| 4.05 | 14 |
3 | HIF1A | NM_001530 |
| 3.81 | 27 |
7 | MFN1 | NM_033540 |
| 3.42 | 13 |
4 | XIAP | NM_001167 |
| 3.29 | 266 | 81 | SQSTM1 | NM_001142298 |
| 3.16 | 20 |
6 | PPARG | NM_005037 |
| 3.01 | 75 | 25 | ITGB1 | NM_002211 |
| 2.87 | 280 | 98 | ANXA7 | NM_001156 |
| 2.84 | 420 | 148 | RRAGA | NM_006570 |
| 2.77 | 13 |
5 | PDCD6IP | NM_001162429 |
| 2.62 | 36 | 14 | AURKA | NM_003600 |
| 2.37 | 66 | 28 | ATG3 | NM_022488 |
| 2.22 | 98 | 44 | MAPRE1 | NM_012325 |
| 1.25 | 15 | 12 | ULK1 | NM_003565 |
| 0.43 | 137 | 319 | GSK3A | NM_019884 |
| 0.40 | 141 | 354 | SCRIB | NM_015356 |
| 0.28 | 66 | 239 | TGM2 | NM_004613 |
Network analysis based on Gene
Ontology analysis
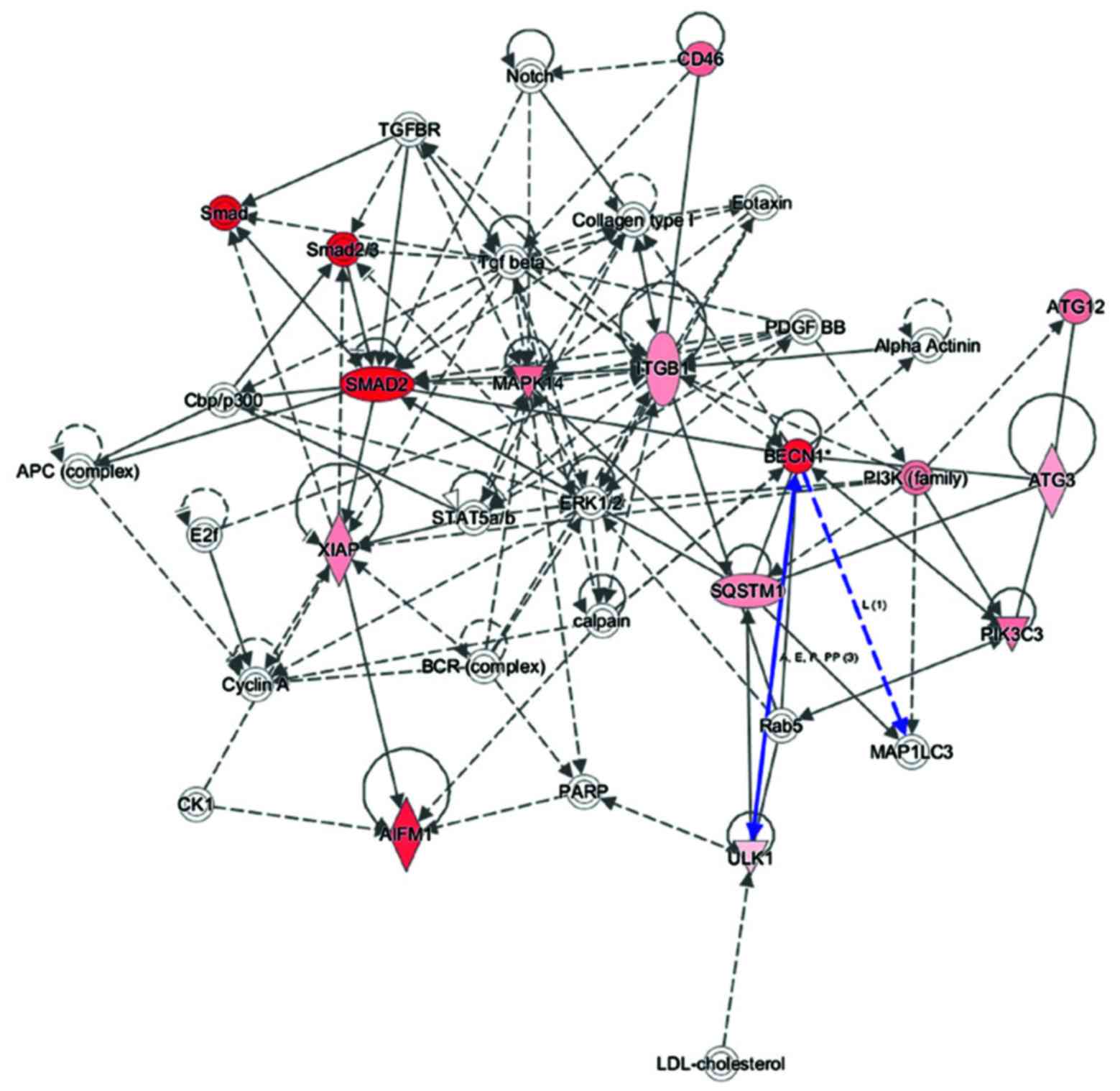
To explore key autophagy proteins related to MRGX
treatment in the Gene Ontology analysis of gene functions, we used
IPA to query 25 autophagy-related proteins belonging to the
proteins upregulated and downregulated by MRGX, resulting in a
distinct interconnected network (Fig.
4). Quantitative and qualitative alterations were observed in
the MRGX-treated lung cancer cells between the regulation groups.
Among them, LC3, ATG5, Beclin-1 and Ulk1 were identified as centers
of the MRGX-treated autophagy-related protein network in the lung
cancer cells. LC3, a mammalian homolog of yeast Atg8, is a reliable
marker of autophagosomes. Tracking the conversion of LC3-I to
LC3-II is indicative of autophagic activity (19). ATG5 is a protein encoded by the ATG5
gene (20). It is an E3 ubiquitin
ligase, which is necessary for autophagy due to its role in
autophagosome elongation. Beclin-1 is a Bcl-2-homology (BH)-3
domain only protein. It is expressed in many human tissues and is
localized in cytoplasmic structures. Beclin-1 is important for
localization of autophagic proteins to a pre-autophagosomal
structure (PAS), depending on interactions with the class-III-type
phosphoinositide 3-kinase (PI3KC3) (21).
In the present study, Beclin-1 was markedly
increased in the MRGX-treated lung cancer cells. The
protein-protein network analysis suggests that Beclin-1 is a major
protein that interacts with multiple proteins and that it is
directly or indirectly downregulated or upregulated in MRGX-treated
lung cancer cells. Proteins linked to Beclin-1 include ATG3 and
Ulk1. Ulk1 plays a specific role in autophagy in lung cancer
(22). However, the detailed
mechanism has not yet been studied.
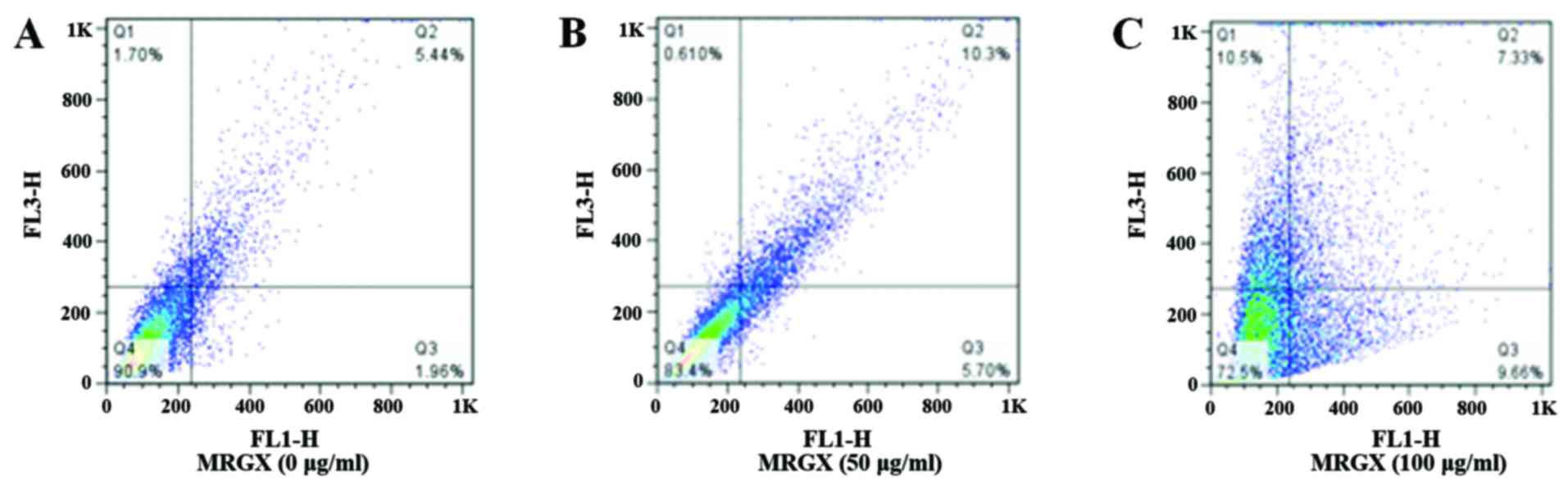
AO staining for autophagy
AO is a versatile fluorescence dye used to stain
acidic vacuoles (lysosomes, endosomes and autophagosomes), RNA and
DNA in living cells. AVOs were quantified using AO staining and
flow cytometry. In AO-stained cells, the cytoplasm and the
nucleolus fluoresce bright green and dark red, respectively,
whereas the acidic compartment fluoresces bright red (23). The proportion of AVOs shifted from
1.96 to 9.66% (Quadrant 3 of Fig. 5A
and C) and from 5.44 to 7.33% (Quadrant 2 of Fig. 5A and C) in the cells treated with
100 µg/ml of MRGX compared with 5.70% (Quadrant 3 of Fig. 5B) and 10.3% (Quadrant 2 of Fig. 5B) for cells treated with 50 µg/ml of
MRGX. AO-stained A549 cells showed AVOs that had mostly accumulated
in the cytoplasm of the cells after having been exposed to 100
µg/ml of MRGX (Fig. 5C).
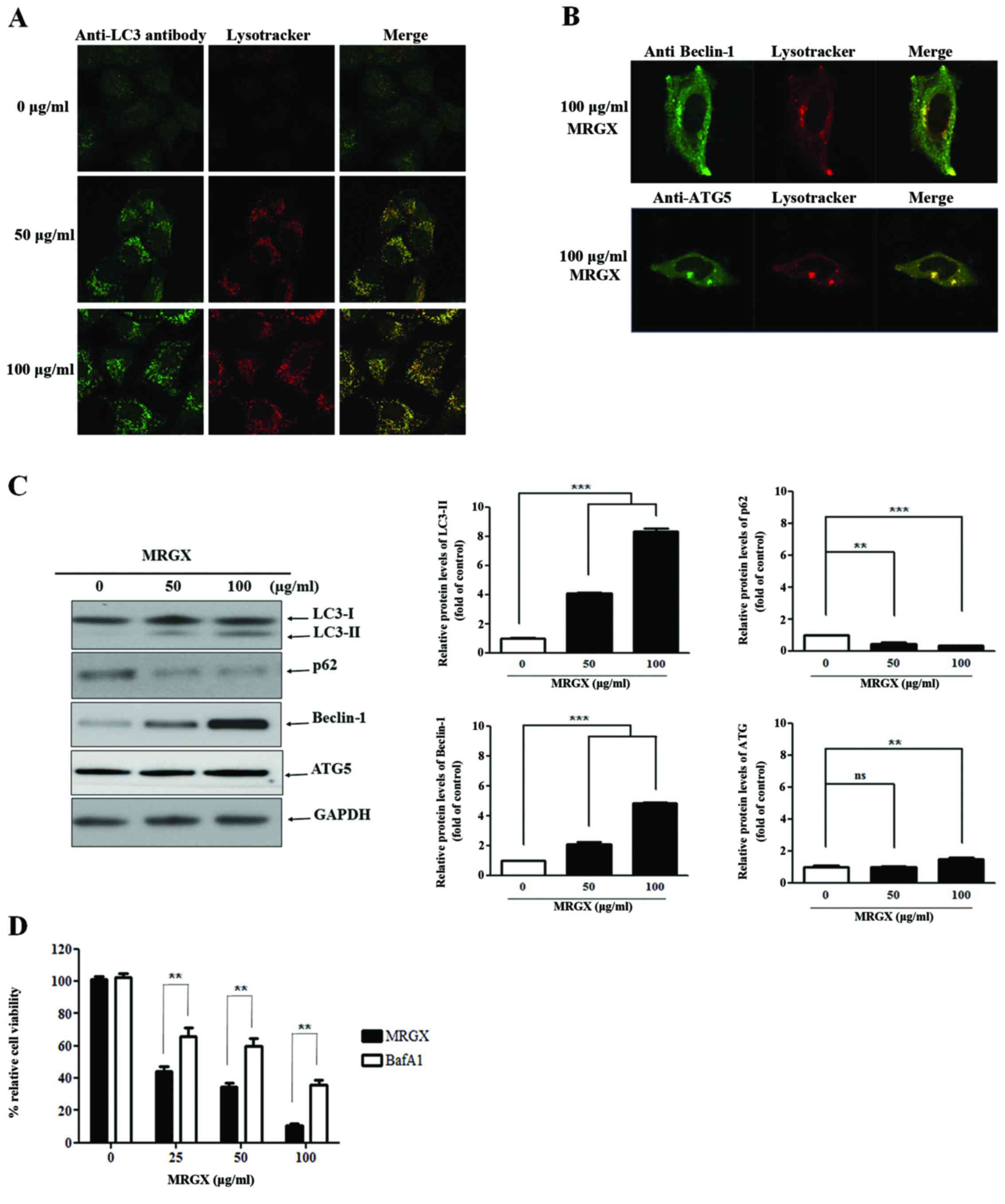
MRGX treatment induces the autophagy
of A549 cells
We screened the A549 cells for autophagy-inducing
compounds that stably expressed GFP-LC3, GFP-Beclin-1 and GFP-ATG5
(24–26). When autophagy is induced, GFP-LC3,
Beclin-1 and ATG5 are distinctive compared to the control. We
treated transfected A549 cells with MRGX and used fluorescent
microscopy to examine LC-3, Beclin-1 and ATG5 staining. MRGX (50
and 100 µg/ml) induced autophagy, as evidenced by the increased and
distinctive GFP-LC3 staining in the cytoplasm of the cells treated
with MRGX at a concentration of 50 µg/ml, suggesting that MRGX
induces autophagy in a dose-dependent manner (Fig. 6A). In addition, Beclin-1 and ATG5
were stained in the cytoplasm of the cells treated with 50 µg/ml of
MRGX (Fig. 6B). The induction of
autophagy by MRGX in A549 cells was further tested by examining the
expression levels of the autophagy marker proteins LC3, p62,
Beclin-1 and ATG5. Similarly, the levels of the LC3-II, Beclin-1
and ATG5 proteins were increased and the level of p62 was decreased
in a dose-dependent manner, collectively indicating that MRGX
actively triggers autophagy (Fig.
6C). To confirm that MRGX induces autophagy, we analyzed the
effect of MRGX-induced autophagy in the A549 cells cultured for 72
h in the presence of BafA1 (an autophagy inhibitor). The inhibitory
effect of BafA1 on MRGX-induced autophagy in the A549 cells was
concentration-dependent (Fig. 6D).
These data therefore indicated that MRGX induced autophagy of the
A549 cells.
MRGX regulates the Ulk-dependent
autophagy pathway by AMPK
Autophagy in A549 cells is involved in the
activation of Akt-mediated mTOR-AMPK-Ulk1 pathways (27,28).
To investigate whether MRGX-induced autophagy is mediated through
the AMPK-Ulk1 pathway, we compared the expression levels of Akt,
mTOR, 4EBP1, AMPK and Ulk1 in A549 cells with and without MRGX
treatment using western blot analysis. The expression level of
phosphorylated AMPK (T172) was increased in the A549 cells treated
with 100 µg/ml of MRGX, as was that of p-Akt. p-mTOR and p-4EBP1
expression levels were decreased in the A549 cells treated with 50
and 100 µg/ml of MRGX, suggesting that mTOR signaling was inhibited
by MRGX through AMPK-Akt signaling. Ulk1 is a large protein that
can be phosphorylated in a serine/threonine-rich region of a
molecule such as S317. MRGX increased the phosphorylation of Ulk1
(S317), but not p-Ulk1 (S757) (Fig.
7A). Compound C (dorsomorphin) is an available agent that is
used as a cell-permeable AMPK inhibitor. When A549 cells were
treated with compound C together with MRGX, p-AKT and p-AMPK were
increased (Fig. 7B). Even though
compound C induces protective autophagy in cancer cells, MRGX
induced autophagy via an AMPK-Akt pathway.
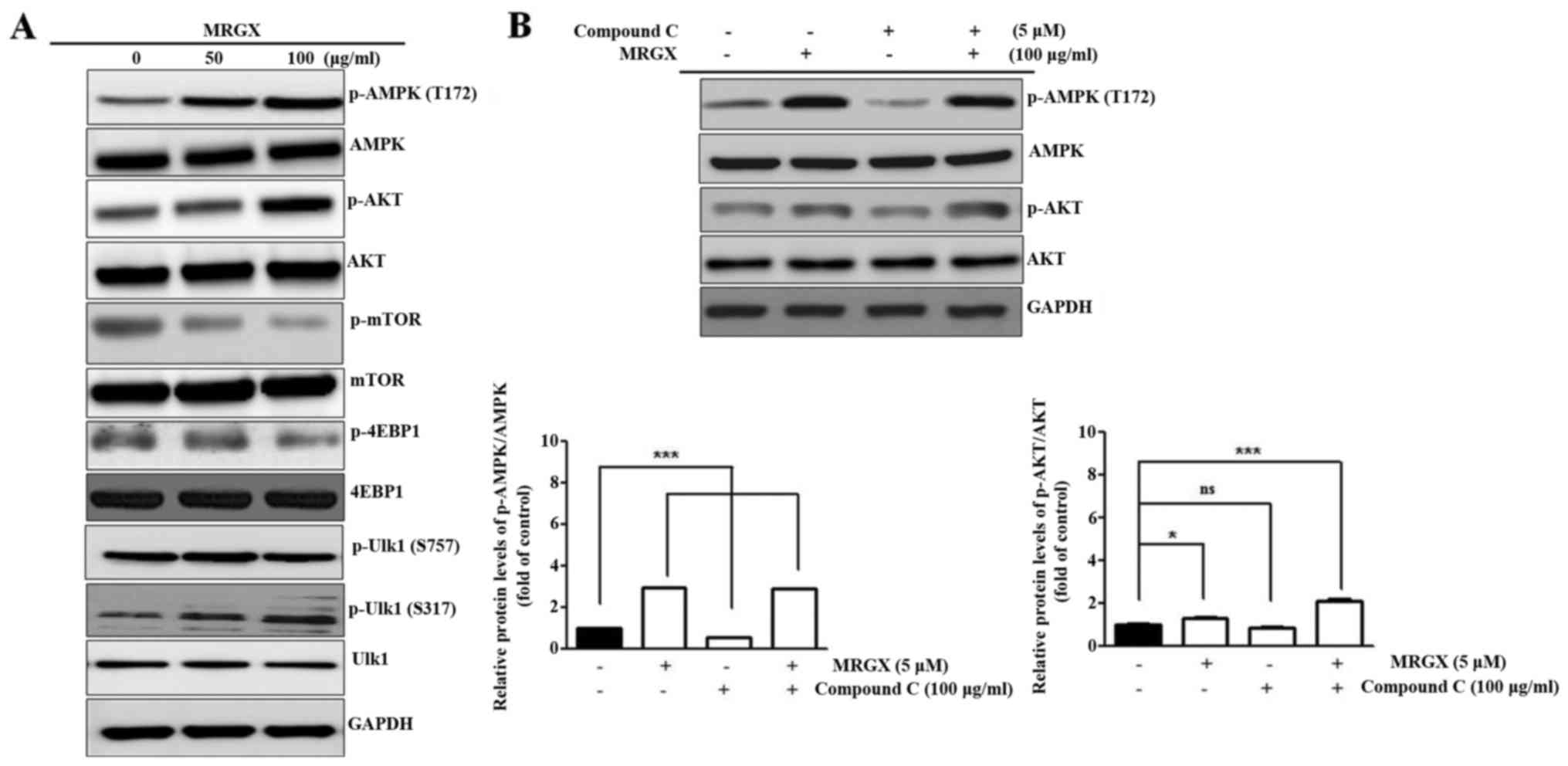 | Figure 7.MRGX regulates the Ulk-dependent
autophagy pathway by AMPK. A549 cells were incubated with MRGX for
48 h. (A) Insoluble cellular fractions were lysed directly with SDS
buffer and equal amounts of cell lysate were subjected to western
blot analysis with AMPK, p-AMPK (T172), Akt, p-Akt, mTOR, p-mTOR,
4EBP1, p-4EBP1, Ulk1 and p-Ulk1 antibodies. (B) MRGX activated AMPK
and Akt phosphorylation compared to compound C-treated A549 cells.
GAPDH was used as an internal standard. n.s., not significant. Data
are expressed as means ± SD. *P<0.05, ***P<0.001. |
Discussion
The pharmacological actions of ginseng are
attributed to ginsenosides (29).
More than 60 ginsenosides have been isolated from American ginseng,
and novel ginsenosides are still being discovered (30,31).
Steaming or heating American ginseng alters the ginsenoside
profiles and increases the anticancer activity. The boiled water
and butanol extracts of mountain ginseng exhibit strong anti-lung
cancer activity, and ginsenoside profiling of these extracts has
identified Rg3, Rh2 and compound K as being more prevalent than
they are in regular ginseng (32).
In the present study, we evaluated the anticancer activity of
enzyme-treated regular ginseng root in lung cancer cells via
autophagy activation. Treatment with MRGX at concentrations of
50–100 µg/ml for 48 and 72 h markedly and significantly inhibited
the proliferation of A549 cells (Fig.
1). The inhibitory effect of MRGX was mediated through G2/M
phase arrest (Fig. 2) and induction
of autophagic cell death. To analyze MRGX treatment-mediated
expression of autophagy-related genes in lung cancer cells, we used
a cDNA microarray approach. The clustered microarray data
identified groups of genes that were regulated differentially in
the control and MRGX-treated cells. The genes whose expression
levels were altered by at least 2-fold are shown in Fig. 3A. Three groups of genes (2-fold
increased, 2-fold decreased and autophagy-related genes) are shown.
Among these, 22 genes were increased by >2-fold, and 3 were
decreased by >2-fold; autophagy-related genes are shown
individually using Venn diagrams (Fig.
3B). The autophagy-related genes are listed in Table I. To explore the major
autophagy-related proteins identified using the Gene Ontology
analyses, we used IPA to query 25 proteins that were upregulated or
downregulated by MRGX. These analyses resulted in a distinct
interconnected network of 25 proteins (Fig. 4). Among these, Beclin-1, ATG5 and
Ulk1 were the centers of the MRGX-related autophagy protein
network.
Autophagy is a conserved catabolic process that
depends not only on cellular stress response, but also on the
recycling of protein and cellular organelles for cell death.
Autophagy is initiated by the formation of a double-membrane
vesicle called an autophagosome, which upon formation sequesters
cellular components for delivery to the lysosome. Although it is
not an ideal autophagy marker, acridine orange staining can be used
to assess the autophagy status. In A549 cells treated with 0–100
µg/ml of MRGX, AVOs were quantified using acridine orange staining
and flow cytometry. The number of AVOs were found to increase
gradually in an MRGX concentration-dependent manner (Fig. 5). The autophagy process requires
several different AuTophaGy (ATG) factors. Ulk1-ATG13 complex
controls initiation, Beclin-1-class III PI3K is needed for
nucleation, and microtubule-associated protein light chain 3 (LC3)
regulates elongation. Autophagy can be monitored using microscopic
and bio-chemical methods, and the LC3, Beclin-1 and ATG5 proteins
are useful autophagy markers since they associate with the
isolation membrane and spherical autophagosomes (33). The cellular localization of LC3,
Beclin-1 and ATG5 can be easily visualized using a recombinant
protein fused with a green fluorescent protein (GFP) (34). Punctate GFP-LC3, GFP-Beclin-1 and
GFP-ATG5 spots in the cytoplasm reveal autophagosome formation
(Fig. 6A and B), and the numbers of
GFP-LC3, GFP-Beclin-1 and GFP-ATG5 spots represent the level of
autophagy induction. Cytosolic LC3-I and autophagosome-bound LC3-II
levels are established indicators of autophagy (35). Since transient overexpression of
GFP-LC3 often results in false-positive outcomes, we used a GFP-LC3
stable cell line for testing. MRGX treatment resulted in
cytoplasmic LC3 foci in the A549 cells and altered the expression
of autophagy markers, specifically increasing the expression of
LC3-II. The linker protein p62/SQSTM1 (p62) includes a
ubiquitin-associated binding domain and an LC3-interacting region.
In this way, p62 scavenges ubiquitinated proteins for degradation
and binds LC3 to anchor these proteins into the forming
autophagosome. Beclin-1 and ATG5 were also increasingly expressed
cytoplasmically with lysosomes (Fig.
6C). Together, these data suggest that MRGX is, indeed, an
autophagy inducer.
The mTOR pathway is an integral cell growth
regulator involving TORC1 and TORC2, which have been defined by
both their association with Raptor or Rictor, respectively, and
their sensitivity to short-term rapamycin inhibition. AMPK is a
ubiquitously expressed kinase that plays a key role in energy
homeostasis. Stimulation of AMPK activates autophagy directly
through phosphorylation of Ulk1, a key initiator of the autophagic
process. The Akt pathway to mTOR signaling is a feature of control
in mammalian autophagy caused by AMPK. In Fig. 7A, AMPK phosphorylation (T172)
expression markedly increased the level of p-Akt. The increase in
the Akt activity in A549 cells treated with a 100 µg/ml
concentration of MRGX was correlated with a decrease in mTOR
activity, as determined from 4EBP1 phosphorylation. 4EBP1 plays a
prominent role in mediating the effects of these pathways in cancer
cells. AMPK promoted autophagy by directly activating Ulk1 through
phosphorylation of S317, but not S757. There are over 20
phosphorylation sites on Ser, and residue phosphorylation is
essential for controlling its activity (36). Mechanistically, the level of p-AKT
was increased, but that of p-mTOR was decreased, in the A549 cells
treated with MRGX at a concentration of 100 µg/ml compared to the
control. The downregulation of mTOR due to MRGX treatment depended
on the levels of p-4EBP1 and p-Ulk (S317), which depend on the
autophagy pathway caused by AMPK (T172). Active AMPK inhibits mTOR
and 4EBP1 to ensure that cells maintain essential nutrients and
energy during metabolic crises. Despite the importance of kinase,
no specific chemical inhibitors are available to examine its
function. However, compound C (dorsomorphin) has been widely used
in cellular, biochemical, and in vivo assays as a selective
AMPK inhibitor. When cells were treated with compound C,
phosphorylation of AMPK and Akt was decreased. However, following
treatment with compound C and MRGX treatment, the p-AKT and the
p-AMPK levels were increased (Fig.
7B), and Ulk1 phosphorylation induced autophagy. The ability of
MRGX to inhibit mTOR signaling in an Akt-dependent manner and to
induce autophagy signaling in an AMPK-Ulk1-dependent manner
suggests the existence of a unique signaling pathway in these
cells. Akt inhibited 4EBP1 phosphorylation through mTOR, which led
to the activation of the TORC1 complex pathway whereas AMPK
upregulated by MRGX activated LC3, Beclin-1 and ATG5. These results
suggest that MRGX induces autophagy signaling by modulating
AMPK-Ulk1 in A549 lung cancer cells.
In the present study, we showed activation of
autophagy as a result of MRGX treatment. As excessive autophagy has
potential to induce tumor-cell death, further study will be
required to reveal the role of MRGX-mediated autophagy induction in
lung cancer cell death.
Acknowledgements
The present study was supported by the Creative
Fusion Research Program through the Creative Allied Project funded
by the Korean Research Council for Fundamental Science and
Technology (CAP-12-1-KIST), the Korea Basic Science Institute
Research Program (C36955) and Korea Institute of Oriental Medicine
(K16060).
References
|
1
|
Ang-Lee MK, Moss J and Yuan CS: Herbal
medicines and perioperative care. JAMA. 286:208–216. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nag SA, Qin JJ, Wang W, Wang MH, Wang H
and Zhang R: Ginsenosides as anticancer agents: In vitro and in
vivo activities, structure-activity relationships, and molecular
mechanisms of action. Front Pharmacol. 3:252012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kwon YK, Lee SY, Kang HS, Sung JS, Cho CK,
Yoo HS, Shin S, Choi JS, Lee YW and Jang IS: Differential
expression of gene profiles in MRGX-treated lung cancer. J
Pharmacopuncture. 16:30–38. 2013. View Article : Google Scholar
|
|
4
|
Kim KH, Choi I, Lee YW, Cho CK, Yoo HS,
Lee SB, Ho Choi S, Kwon KR and Jang JH: Target genes involved in
antiproliferative effect of modified ginseng extracts in lung
cancer A549 cells. Acta Biochim Biophys Sin. 46:441–449. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jang SI, Lee YW, Cho CK, Yoo HS and Jang
JH: Identification of target genes involved in the
antiproliferative effect of enzyme-modified ginseng extract in
HepG2 hepatocarcinoma cell. Evid Based Complement Alternat Med.
2013:5025682013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hwang JW, Baek YM, Jang IS, Yang KE, Lee
DG, Yoon SJ, Rho J, Cho CK, Lee YW, Kwon KR, et al: An
enzymatically fortified ginseng extract inhibits proliferation and
induces apoptosis of KATO3 human gastric cancer cells via
modulation of Bax, mTOR, PKB and IκBα. Mol Med Rep. 11:670–676.
2015.PubMed/NCBI
|
|
7
|
Stinchcombe TE, Lee CB and Socinski MA:
Current approaches to advanced-stage non-small-cell lung cancer:
First-line therapy in patients with a good functional status. Clin
Lung Cancer. 7 Suppl 4:S111–S117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yun TK: Panax ginseng - a
non-organ-specific cancer preventive? Lancet Oncol. 2:49–55. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee DG, Jang SI, Kim YR, Yang KE, Yoon SJ,
Lee ZW, An HJ, Jang IS, Choi JS and Yoo HS: Anti-proliferative
effects of ginsenosides extracted from mountain ginseng on lung
cancer. Chin J Integr Med. 22:344–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gustafsson AB and Gottlieb RA: Recycle or
die: The role of autophagy in cardioprotection. J Mol Cell Cardiol.
44:654–661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kundu M and Thompson CB: Autophagy: Basic
principles and relevance to disease. Annu Rev Pathol. 3:427–455.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie Z, Zhang J, Wu J, Viollet B and Zou
MH: Upregulation of mitochondrial uncoupling protein-2 by the
AMP-activated protein kinase in endothelial cells attenuates
oxidative stress in diabetes. Diabetes. 57:3222–3230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colombo SL and Moncada S: AMPKalpha1
regulates the antioxidant status of vascular endothelial cells.
Biochem J. 421:163–169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vellai T: Autophagy genes and ageing. Cell
Death Differ. 16:94–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stockert JC, Blázquez-Castro A, Cañete M,
Horobin RW and Villanueva A: MTT assay for cell viability:
Intracellular localization of the formazan product is in lipid
droplets. Acta Histochem. 114:785–796. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Choi IK, Cho YS, Jung HJ and Kwon HJ:
Autophagonizer, a novel synthetic small molecule, induces
autophagic cell death. Biochem Biophys Res Commun. 393:849–854.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hwang JW, Baek YM, Yang KE, Yoo HS, Cho
CK, Lee YW, Park J, Eom CY, Lee ZW, Choi JS, et al: Lactobacillus
casei extract induces apoptosis in gastric cancer by inhibiting
NF-κB and mTOR-mediated signaling. Integr Cancer Ther. 12:165–173.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mizushima N and Yoshimori T: How to
interpret LC3 immunoblotting. Autophagy. 3:542–545. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hammond EM, Brunet CL, Johnson GD,
Parkhill J, Milner AE, Brady G, Gregory CD and Grand RJ: Homology
between a human apoptosis specific protein and the product of APG5,
a gene involved in autophagy in yeast. FEBS Lett. 425:391–395.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kang R, Zeh HJ, Lotze MT and Tang D: The
Beclin 1 network regulates autophagy and apoptosis. Cell Death
Differ. 18:571–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheong H, Wu J, Gonzales LK, Guttentag SH,
Thompson CB and Lindsten T: Analysis of a lung defect in
autophagy-deficient mouse strains. Autophagy. 10:45–56. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Traganos F and Darzynkiewicz Z: Lysosomal
proton pump activity: Supravital cell staining with acridine orange
differentiates leukocyte subpopulations. Methods Cell Biol.
41:185–194. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoon JH, Her S, Kim M, Jang IS and Park J:
The expression of damage-regulated autophagy modulator 2 (DRAM2)
contributes to autophagy induction. Mol Biol Rep. 39:1087–1093.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L,
Cai Y, Norberg HV, Zhang T, Furuya T, et al: Beclin1 controls the
levels of p53 by regulating the deubiquitination activity of USP10
and USP13. Cell. 147:223–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guévin C, Manna D, Bélanger C, Konan KV,
Mak P and Labonté P: Autophagy protein ATG5 interacts transiently
with the hepatitis C virus RNA polymerase (NS5B) early during
infection. Virology. 405:1–7. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Egan D, Kim J, Shaw RJ and Guan KL: The
autophagy initiating kinase ULK1 is regulated via opposing
phosphorylation by AMPK and mTOR. Autophagy. 7:643–644. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alers S, Löffler AS, Wesselborg S and
Stork B: Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy:
Cross talk, shortcuts, and feedbacks. Mol Cell Biol. 32:2–11. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan CS, Wu JA and Osinski J: Ginsenoside
variability in American ginseng samples. Am J Clin Nutr.
75:600–601. 2002.PubMed/NCBI
|
|
30
|
Lei J, Li X, Gong XJ and Zheng YN:
Isolation, synthesis and structures of cytotoxic ginsenoside
derivatives. Molecules. 12:2140–2150. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han M, Hou JG, Dong CM, Li W, Yu HL, Zheng
YN and Chen L: Isolation, synthesis and structures of ginsenoside
derivatives and their anti-tumor bioactivity. Molecules.
15:399–406. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hwang JW, Oh JH, Yoo HS, Lee YW, Cho CK,
Kwon KR, Yoon JH, Park J, Her S, Lee ZW, et al: Mountain ginseng
extract exhibits anti-lung cancer activity by inhibiting the
nuclear translocation of NF-κB. Am J Chin Med. 40:187–202. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Levine B and Klionsky DJ: Development by
self-digestion: Molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu D, Wu J, Xu L, Zhang R and Chen L: A
method for the establishment of a cell line with stable expression
of the GFP-LC3 reporter protein. Mol Med Rep. 6:783–786.
2012.PubMed/NCBI
|
|
35
|
Ding Z, Wang X, Schnackenberg L, Khaidakov
M, Liu S, Singla S, Dai Y and Mehta JL: Regulation of autophagy and
apoptosis in response to ox-LDL in vascular smooth muscle cells,
and the modulatory effects of the microRNA hsa-let-7g. Int J
Cardiol. 168:1378–1385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Roach PJ: AMPK → ULK1 → autophagy. Mol
Cell Biol. 31:3082–3084. 2011. View Article : Google Scholar : PubMed/NCBI
|