Introduction
microRNAs (miRNAs) inhibit the expression of targets
by contributing to the degradation or translational inhibition of
target mRNAs (1). They have been
found to be actively involved in different cellular processes
(2,3) including proliferation, apoptosis,
differentiation and movement. Emerging studies showed that abnormal
expression and function of miRNAs play important roles in the
pathogenesis and tumorigenicity of human malignancies (4–6).
Otherwise, miRNAs have been demonstrated to be hopeful diagnostic
biomarkers and drug-targets of colorectal cancer (CRC) (7). Investigation of the expression and
biological function of miRNAs will facilitate the discovery of new
biomarkers and drug-targets for CRC patients.
miR-944 functions as one of prognostic microRNAs in
cancer tissue from patients operated for pancreatic cancer
(8). Furthermore, miR-944 is
identified as one potential driver miRNA in non-small cell lung
cancers (NSCLC) (9,10). Overexpression of miR-944 promotes
tumorigenesis of NSCLC by targeting suppressor of cytokine
signaling (SOCS4) (11). Increased
plasma circulation miR-944 acts as a potential diagnostic biomarker
of squamous cell carcinoma in lung cancer (12). miR-944 is significantly
overexpressed in cervical cancer and promotes proliferation as well
as migration and invasion in cancer cells (13). Upregulation of miR-944 is observed
in breast cancer patients' serum and tumor tissues and it promotes
the chemotherapy of breast cancer by targeting BCL2 interacting
protein 3 (BNIP3). While, miR-944 is identified to be prominently
downregulated in exosomes arising from adenocarcinoma of the
esophagus (14). Flores-Pérez et
al (15) reported that miR-944
expression was significantly silenced in clinical specimens and
breast cancer cell lines, and miR-944 promoted cell migration
through targeting of Siah E3 ubiquitin protein ligase 1 (SIAH1) and
protein tyrosine phosphatase type IVA, member 1 (PTP4A1). A recent
study reported that the levels of miR-944 in recurrent CRC patients
were evidently lower than that in non-recurrent cases, suggesting
that miR-944 may function as a tumor suppressive miRNA in CRC
(16). However, the clinical value
and biological role of miR-944 in CRC remain largely unknown.
In the present study, we confirmed that miR-944 was
underexpressed in CRC specimens and cells. The low level of miR-944
correlated with malignant clinical features of CRC patients and
reduced survival. Our data showed that miR-944 inhibited the
invasive ability of cancer cells in CRC. Moreover, the
metastasis-associated in colon cancer-1 (MACC1) was identified as a
downstream molecule of miR-944 and possibly mediated the biological
functions of miR-944 in CRC.
Materials and methods
Clinical samples
Clinical specimens were obtained from 86 patients
histologically diagnosed as CRC in the Department of
Gastrointestinal Surgery, Sun Yat-sen Memorial Hospital. Patients
who received immunotherapy, chemotherapy or radiotherapy before
surgical treatment were excluded. Informed consent were signed by
each patient before clinical specimens were collected and used. All
specimens were stored in liquid nitrogen or fixed with formalin for
further investigation. The present study was permitted by the
Research Ethics Committee of Sun Yat-sen University.
Cell culture and transfection
Human CRC cell lines including HCT116, Caco-2, HT29,
SW620 and SW480, and human intestinal epithelial cells (HIEC) as
well as HEK293 cells were obtained from the Institute of
Biochemistry and Cell Biology, Chinese Academy of Sciences
(Shanghai, China). All the cells were cultured in Dulbeccos
modified Eagles medium (DMEM; HyClone Laboratories, Inc., Logan,
UT, USA) along with fetal bovine serum (10%) (FBS; HyClone
Laboratories), penicillin (100 U/ml) and streptomycin (100 µg/ml).
Cell cultures were kept in an incubator containing of 5%
CO2 and humidified atmosphere at 37°C.
miR-944 mimic, miR-944 inhibitor and the
corresponding control vectors were from Genecopoeia (Guangzhou,
China) and were then tranduced into CRC cells with Lipofectamine
2000 following the manufacturers protocol. Retroviral vectors
pMMP-MACC1 were constructed by inserting the corresponding cDNA
into pMMP. The retroviruses were packaged and tranfected into CRC
cells as previously described (17).
Quantitative real-time RT-PCR
(qRT-PCR)
Total RNA from CRC cells was isolated by miRNeasy
Mini kit (Qiagen, Hilden, Germany) and total RNA from CRC tissues
were extracted with TRIzon reagent. miR-944 levels in these samples
were assayed using TaqMan MicroRNA assays based on the
manufacturers instructions (Applied Biosystems, Inc., Carlsbad, CA,
USA). PCR of MACC1 was performed using UltraSYBR Mixture (CW0957;
Cwbio, Beijing, China) and LightCycler 480 PCR System (Roche
Diagnostics, Indianapolis, IN, USA). The primers for miR-944 and
U6, MACC1 and GAPDH were from Genecopoeia. U6 was used as the
control gene for the relative level of miR-944 while GAPDH served
as internal control for MACC1.
Luciferase reporter assay
3′-UTR of MACC1 was amplified and cloned into
pmiR-RB-REPORT™ Luciferase. Mutant (mt) miR-944 was constructed by
performing mutation on the seed sequences. Then, the 3′-UTR of
MACC1 and corresponding miRNA vectors were co-transduced into
HEK293 cells, respectively. Forty-eight hours after
co-transduction, the cells were lysed and detected using a
Dual-Luciferase® reporter assay kit (Promega, Madison,
WI, USA) based on the manufacturers protocols.
Wound healing assay
CRC cells transfected with corresponding vectors
were seeded in 6-well plates to form a single confluent cell layer.
The wounds were made with 100 µl tips in the confluent cell layer.
After wound scratching at 0 and 24 h, the width of wound was
photographed with a phase-contrast microscope.
Proliferation assays
For cell proliferation, CRC cells that were treated
with miR-944 mimic or inhibitor were seeded into 96-well plates
(1.5×103 cells/well). Twenty-four, 48, 72 and 96 h after
transfection, the cell proliferation assay was performed by
addition of 10 µl Cell Counting kit-8 solution (CCK-8; Beyotime
Institute of Biotechnology, Shanghai, China) to each well, followed
by incubation at 37°C for 2 h. Absorbance was measured at a
wavelength of 490 nm using a microplate reader (Flexstation III ROM
V2.1.28; Molecular Devices, Sunnyvale, CA, USA).
Transwell migration and invasion
assay
The migratory and invasive ability of CRC cells were
evaluated with Transwell chambers (BD Biosciences, Franklin Lakes,
NJ, USA). CRC cells (5–10×104) suspended in 100 µl
medium without serum were seeded into the upper chamber, and lower
chamber was full of 20% FBS to induce CRC cell migration or
invasion through the membrane. Matrigel (1:6 dilution) was added on
the upper chamber for invasion assay. Twenty-four hours later,
cells with crystal violet staining that migrated or invaded across
the Transwell membrane were numbered under an optical
microscope.
Western blot analysis
Cell proteins were collected with RIPA lysis buffer,
and 40 µg protein was subjected to 4–20% SDS gel electrophoresis
and then transferred to PVDF membranes. Then, membranes were
blocked in 5% skimmed milk and incubated with MACC1 (Abcam,
Cambridge, MA, USA), Met (Cell Signaling Technology, Inc., Danvers,
MA, USA), AKT (Cell Signaling Technology), p-AKT (Ser473) (Cell
Signaling Technology), GSK3β (Cell Signaling Technology) or p-GSK3β
(Ser9) (Cell Signaling Technology) antibody and subsequently
incubated with matched secondary antibodies (Cell Signaling
Technology). Then, signals for each protein expression was detected
with the Bio-Rad Gel imaging system. GAPDH (G8140; US Biological,
Swampscott, MA, USA) was used as a loading control.
Immunohistochemistry (IHC)
Before IHC staining, CRC tissues were fixed with 4%
formalin and embedded with paraffin. Then, the paraffin-embedded
specimens were cut into 4 µm sections. IHC staining following
standard protocol was performed to evaluate the expression level of
MACC1 (Abcam) in CRC tissues. The percentage of positive tumor
cells was graded as per the following criteria: 0, <10%; 1,
10–30%; 2, 30–50%; 3, >50%.
Statistical analysis
All data were collected and showed as the mean ±
SEM. Statistical analyses including Pearson Chi-squared test, a
two-tailed Students t-test, ANOVA, Kaplan-Meier method, log-rank
test and Spearmans correlation analysis were performed with
GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA,
USA) used in this study to perform statistical analysis. P<0.05
was considered to be statistically different.
Results
miR-944 expression is downregulated in
CRC
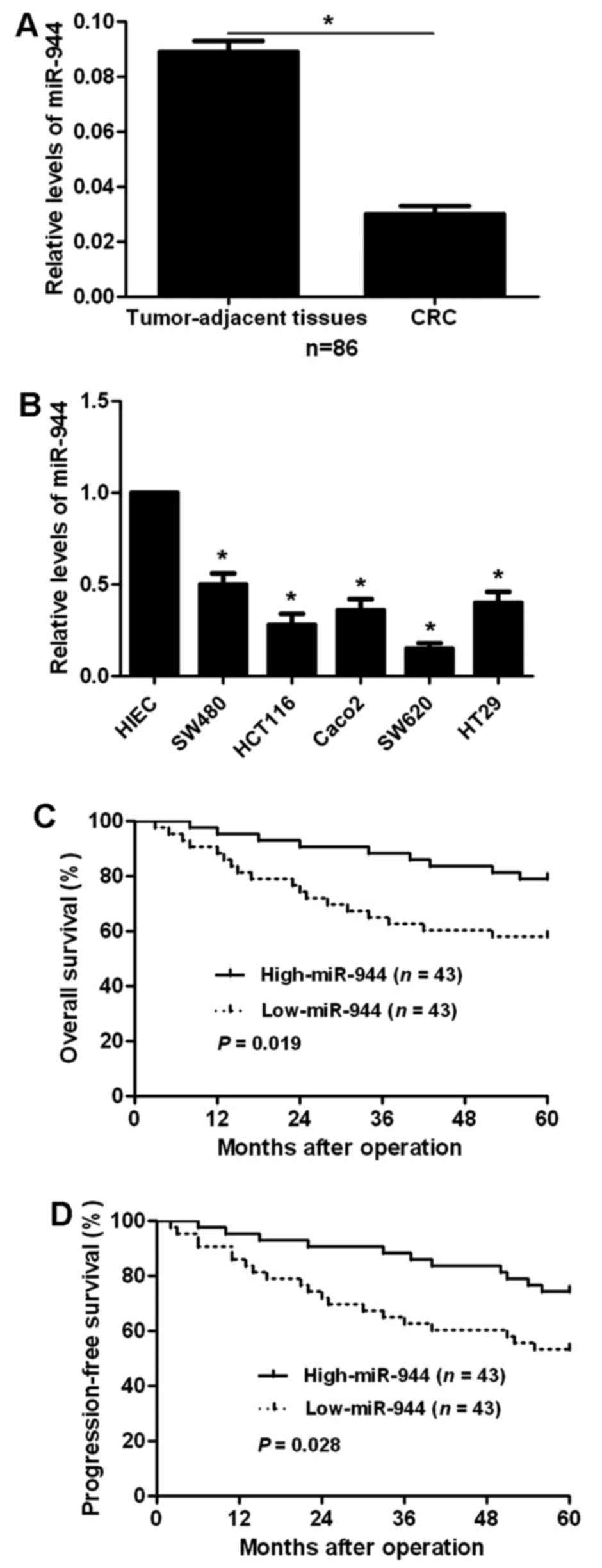
To examine the status of miR-944 in CRC, qRT-PCR was
performed for 86 CRC cases. Our data disclosed that CRC tissues had
significant decreased expression levels of miR-944 compared to
tumor-adjacent tissues (P<0.05; Fig.
1A). Next, we compared the expression levels of miR-944 between
CRC cell lines and HIEC cells. Compared with HIEC cells, the levels
of miR-944 in all CRC cells (SW480, HCT116, Caco2, SW620 and HT29)
were significantly reduced (P<0.05; Fig.1B). These data indicate that miR-944
probably plays a tumor suppressive role in CRC.
Decrease in tissue miR-944 predicts
malignant clinical parameters and poor prognosis of CRC
patients
To clarify the clinical value of miR-944 in CRC, all
patients were divided into miR-944 low and high group according to
the median expression of miR-944. As shown in Table I, CRC patients with low expression
of miR-944 had high tumor invasion stage (P=0.002), more lymph node
and distant metastasis (P=0.001 and P=0.019, respectively), and
advanced tumor-node-metastasis (TNM) stage (P=0.018). Furthermore,
survival analyses indicated that patients with low expression
showed significantly reduced 5-year overall and progression-free
survival (P=0.019 and P=0.028, respectively; Fig. 1C and D). We suggest that miR-944 is
a possible prognostic biomarker for CRC patients.
 | Table I.Clinicopathological findings and
correlation with miR-944 expression in CRC. |
Table I.
Clinicopathological findings and
correlation with miR-944 expression in CRC.
|
|
| miR-944
expression |
|
|---|
|
|
|
|
|
|---|
| Features | N (86) | Low | High | P-value |
|---|
| Age (years) |
|
<65 | 57 | 29 | 28 | 0.820 |
| ≥65 | 29 | 14 | 15 |
|
| Sex |
| Male | 46 | 25 | 21 | 0.387 |
|
Female | 40 | 18 | 22 |
|
| Tumor grade |
|
G1+G2 | 65 | 34 | 31 | 0.451 |
|
G3+G4 | 21 | 9 | 12 |
|
| Size (cm) |
|
<5 | 38 | 16 | 22 | 0.193 |
| ≥5 | 48 | 27 | 21 |
|
| Tumor invasion |
|
T1+T2 | 20 | 4 | 16 | 0.002a |
|
T3+T4 | 66 | 39 | 27 |
|
| Lymph node
status |
|
<1 | 46 | 15 | 31 | 0.001a |
| ≥1 | 40 | 28 | 12 |
|
| Distant
metastasis |
|
Absent | 67 | 29 | 38 | 0.019a |
|
Present | 19 | 14 | 5 |
|
| TNM stage |
|
I+II | 41 | 15 | 26 | 0.018a |
|
III+IV | 45 | 28 | 17 |
|
miR-944 inhibits the proliferation and
metastasis of CRC cells
Since increased cancer cell proliferation and
metastasis is an important reason for the metastasis and recurrence
of human cancer (18), we explored
whether miR-944 could modulate the proliferation, migration and
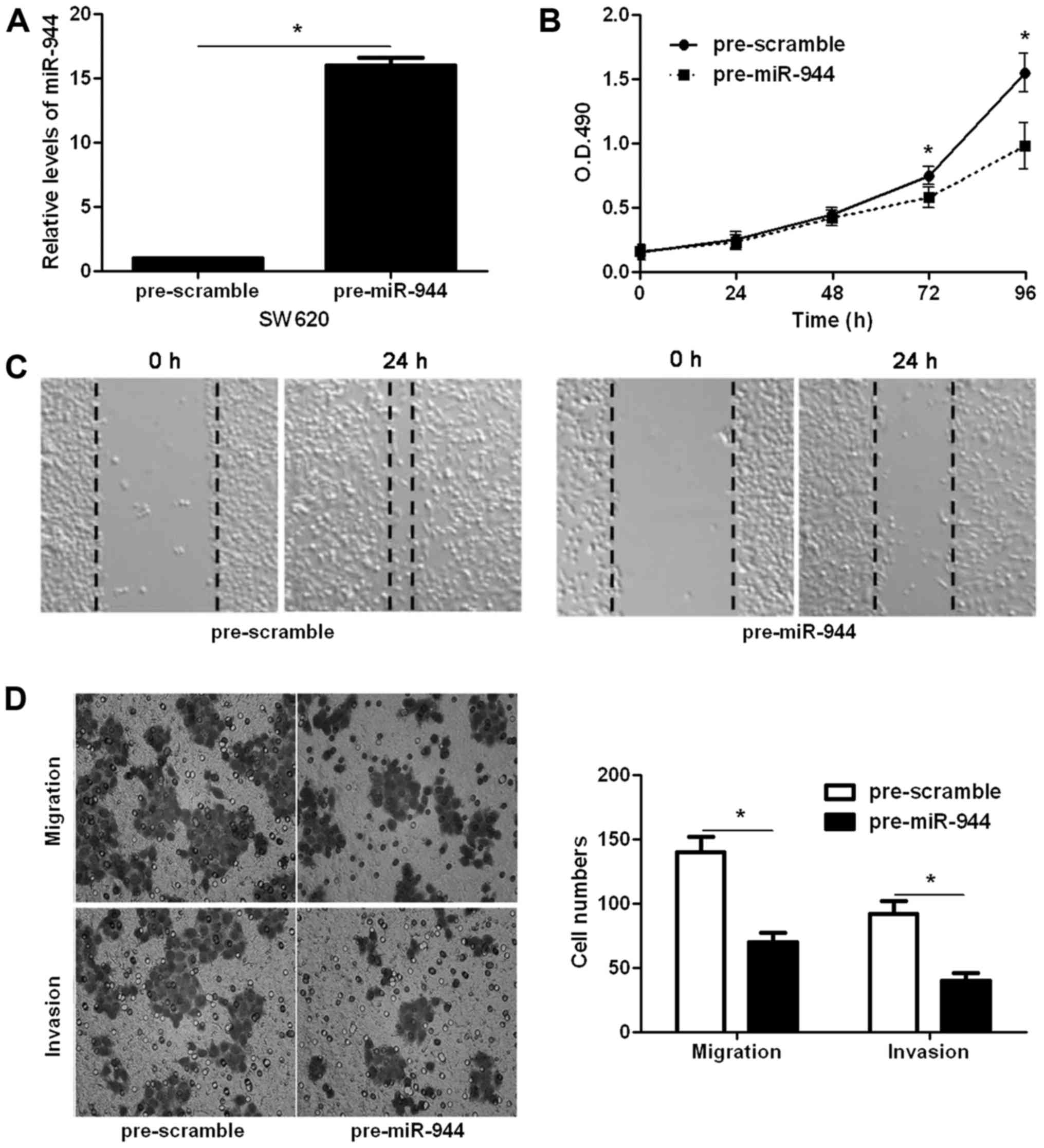
invasion of CRC cells. Transfection of miR-944 mimic obviously
upregulated the level of miR-944 in SW620 cells (P<0.05;
Fig. 2A). CCK-8 assays indicated
that miR-944 overexpression inhibited SW620 cell proliferation
(P<0.05; Fig. 2B). The wound
healing assays showed that miR-944 overexpression notably reduced
cell migration in SW620 cells (Fig.
2C). In addition, Transwell assays indicated that ectopic
expression of miR-944 significantly reduced the number of migrated
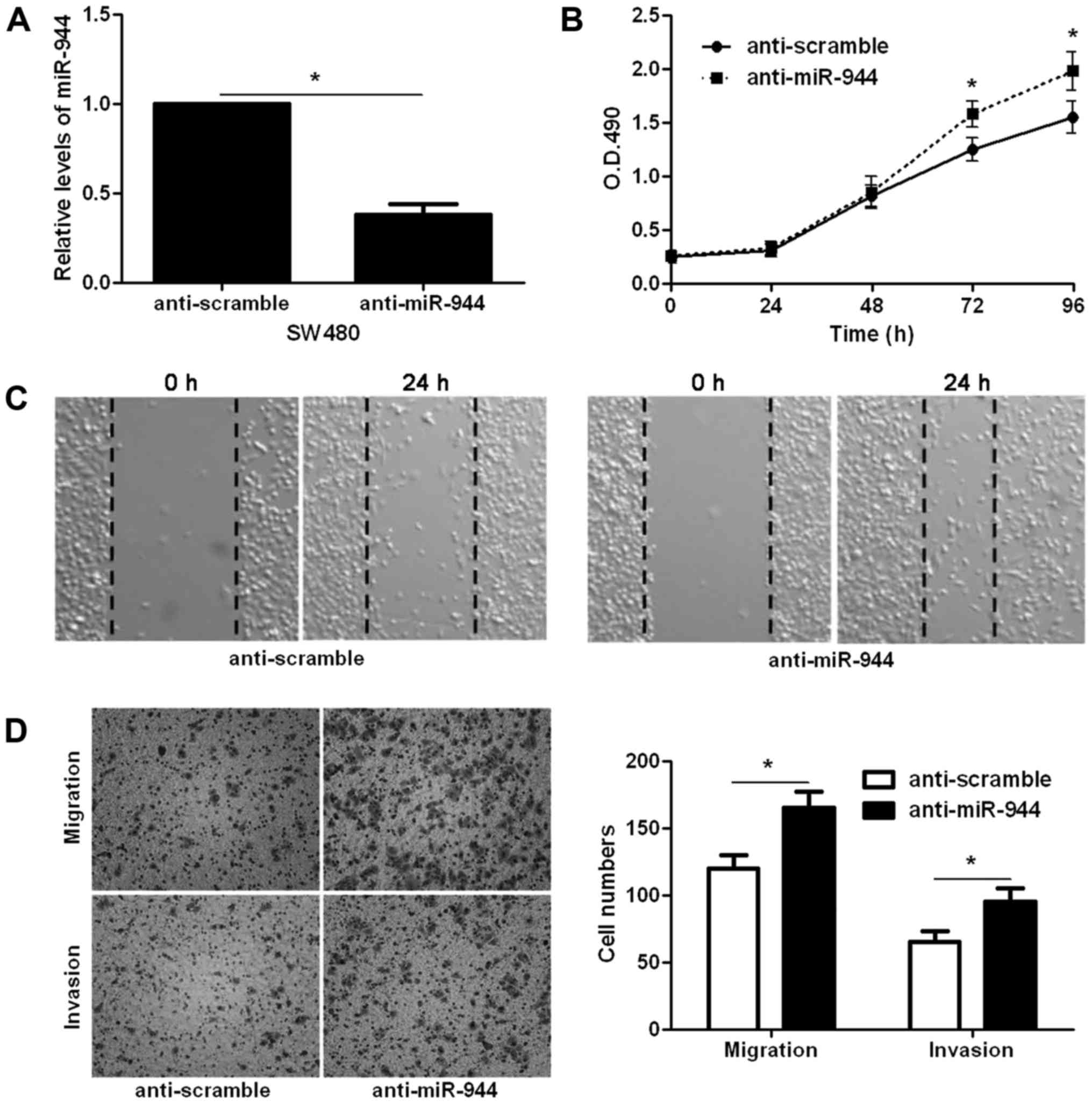
and invaded SW620 cells (P<0.05, respectively; Fig. 2D). In contrast, miR-944 inhibitor
significantly decreased the level of miR-944 in SW480 cells
(P<0.05; Fig. 3A). Subsequently,
miR-944 silencing notably facilitated SW480 cell proliferation,
migration and invasion (P<0.05, respectively; Fig. 3B-D). Thus, miR-944 exerts an
anticancer role in CRC cells.
miR-944 post-transcriptionally
regulates MACC1 expression
To disclose the underlying molecular mechanisms
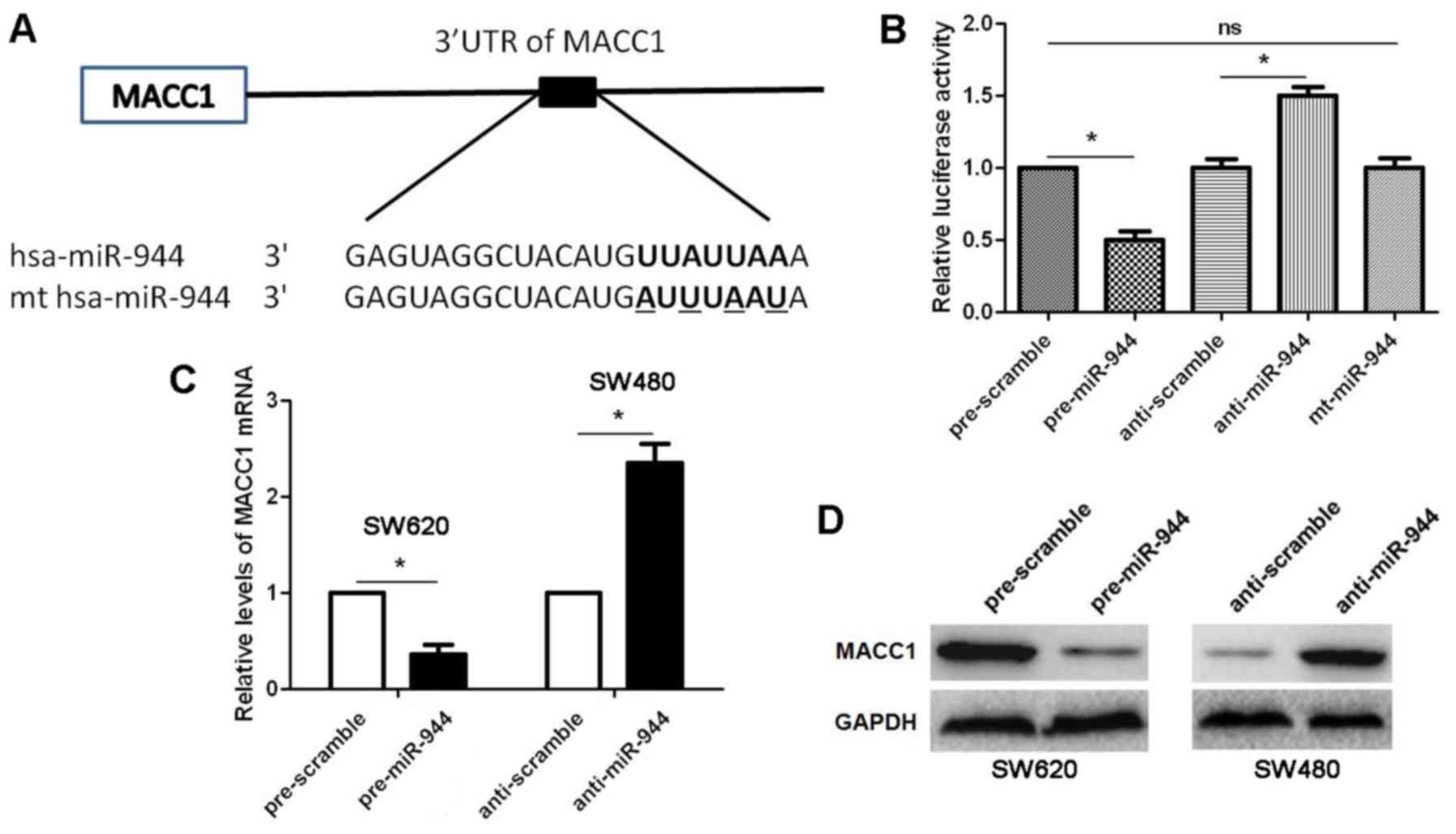
involved in the role of miR-944 in CRC cells, we searched for
candidate target genes of miR-944 using publicly available
databases, including TargetScanHuman 7.1 (http://www.targetscan.org/) and miRanda (microrna.org and miRbase). MACC1, a pro-metastatic
molecule in CRC (19), was
recognized as a potential target molecule of miR-944, because the
complementary sequence of miR-944 was identified in the 3′-UTR of
MACC1 mRNA by TargetScan analysis (Fig.
4A). Then, our data indicated that miR-944 overexpression
decreased while miR-944 knockdown increased the luciferase activity
of MACC1 3′-UTR (P<0.05, respectively; Fig. 4B), while mt miR-944 did not have any
influence on the luciferase activity of MACC1 3′-UTR in HEK293
cells (Fig. 4B). Further
experiments indicated that miR-944 overexpression reduced while
miR-944 silencing upregulated the expression of MACC1 mRNA and
protein (P<0.05, respectively; Fig.
4C and D). Next, qRT-PCR and IHC were performed to detected
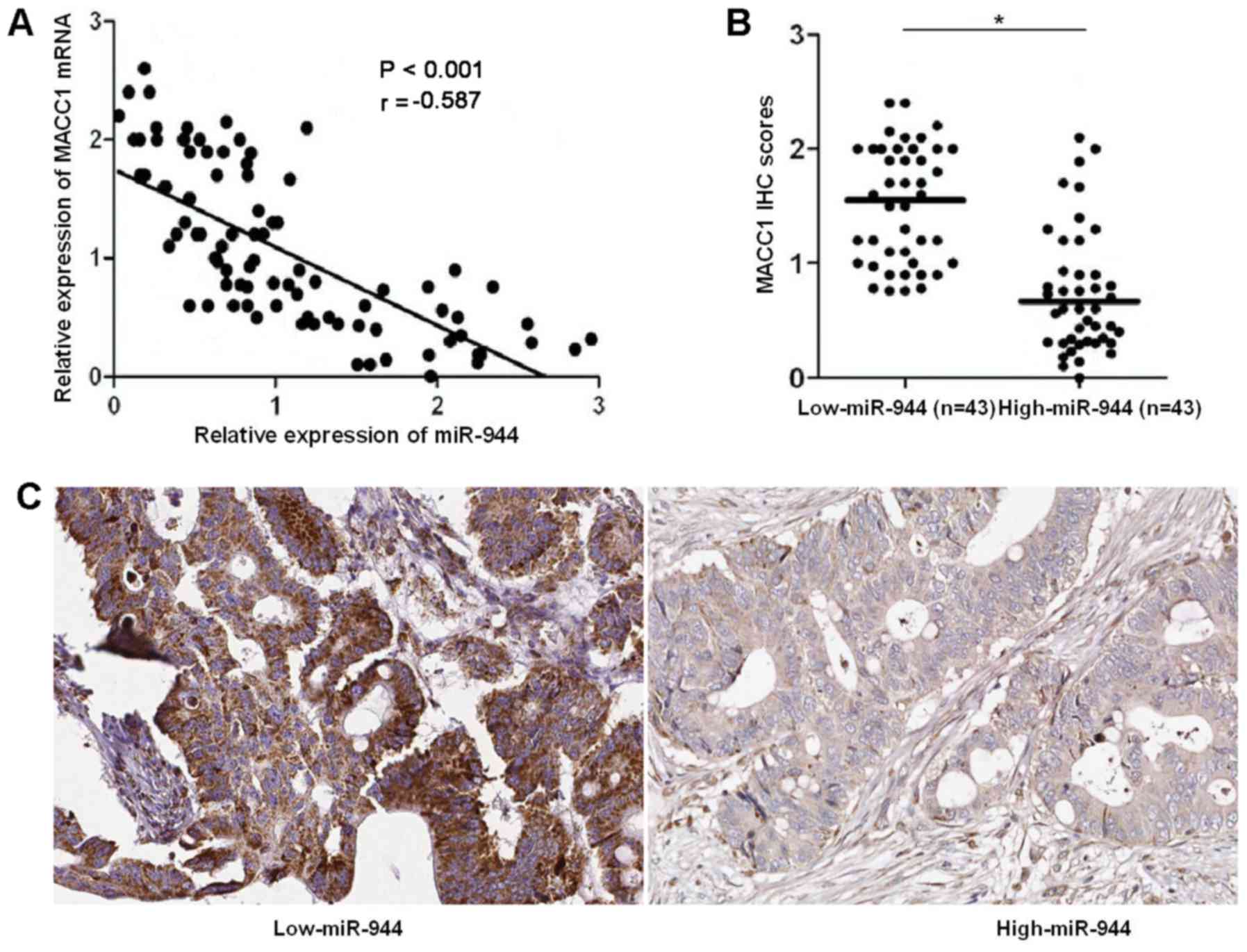
MACC1 in CRC tissues. Spearmans correlation analysis disclosed that
miR-944 was negatively correlated with MACC1 mRNA in CRC specimens
(r=−0.587, P<0.001; Fig. 5A).
Notably, IHC data suggested that the expressions of MACC1 in
miR-944 low expressing tumors were notably higher than those in
miR-944 high expressing cases (P<0.05; Fig. 5B and C). Altogether, miR-944
negatively regulates MACC1 abundance in CRC cells.
MACC1/Met/AKT signaling potentially
mediates the function of miR-944 in CRC
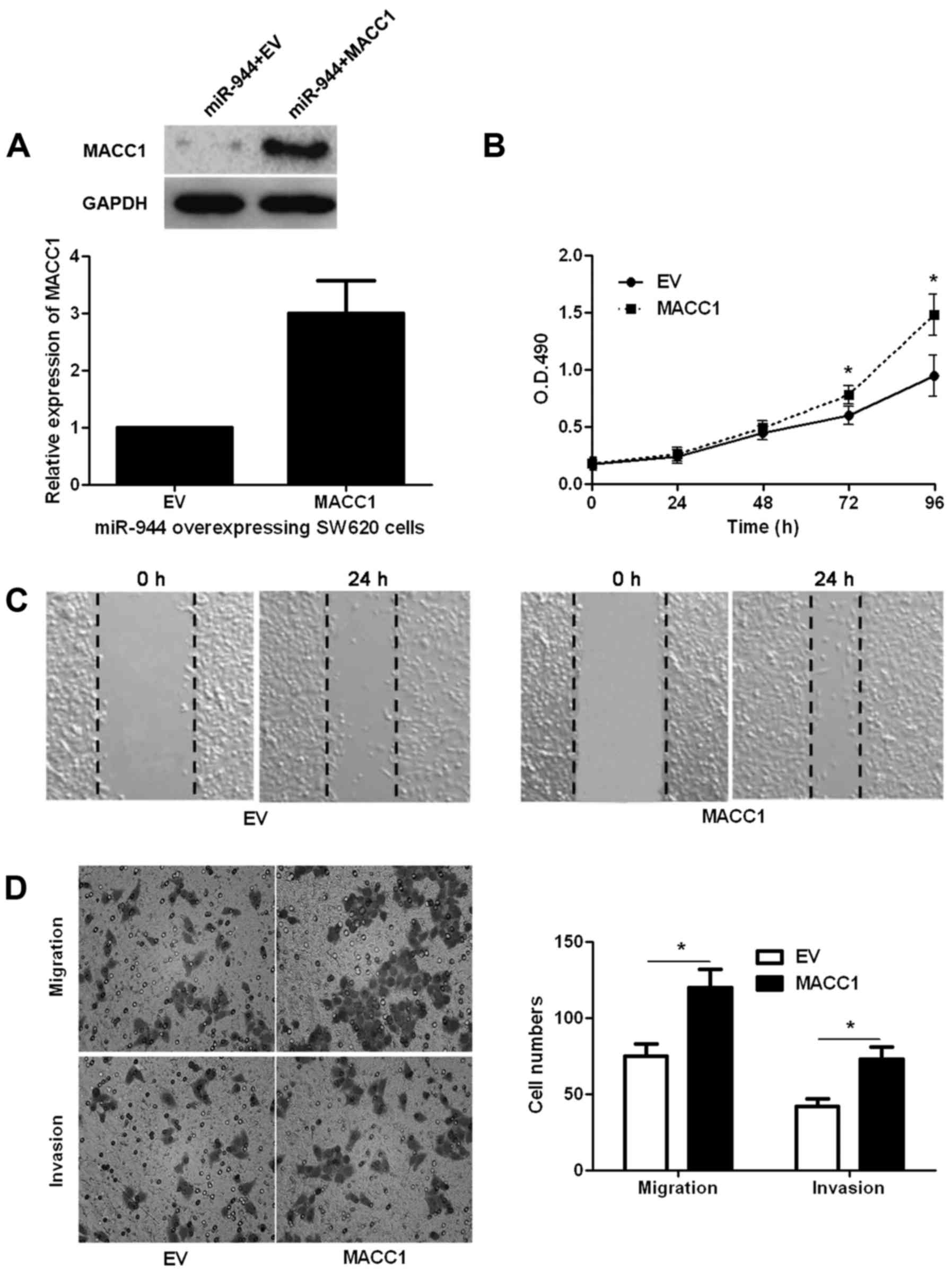
Since we confirmed that MACC1 was a target molecule
of miR-944, MACC1 retroviruses were employed to disclose whether
MACC1 restoration abolished the anti-metastatic role of miR-944 in
CRC cells. As shown in Fig. 6A,
MACC1 retroviruses infection significantly increased the level of
MACC1 in miR-944 overexpressing SW620 cells (P<0.05; Fig. 6A). Consequently, restoration of
MACC1 promoted the malignant behavior of miR-944 overexpressing
SW620 cells with enhanced cell proliferation, migration and
invasion (P<0.05, respectively; Fig.
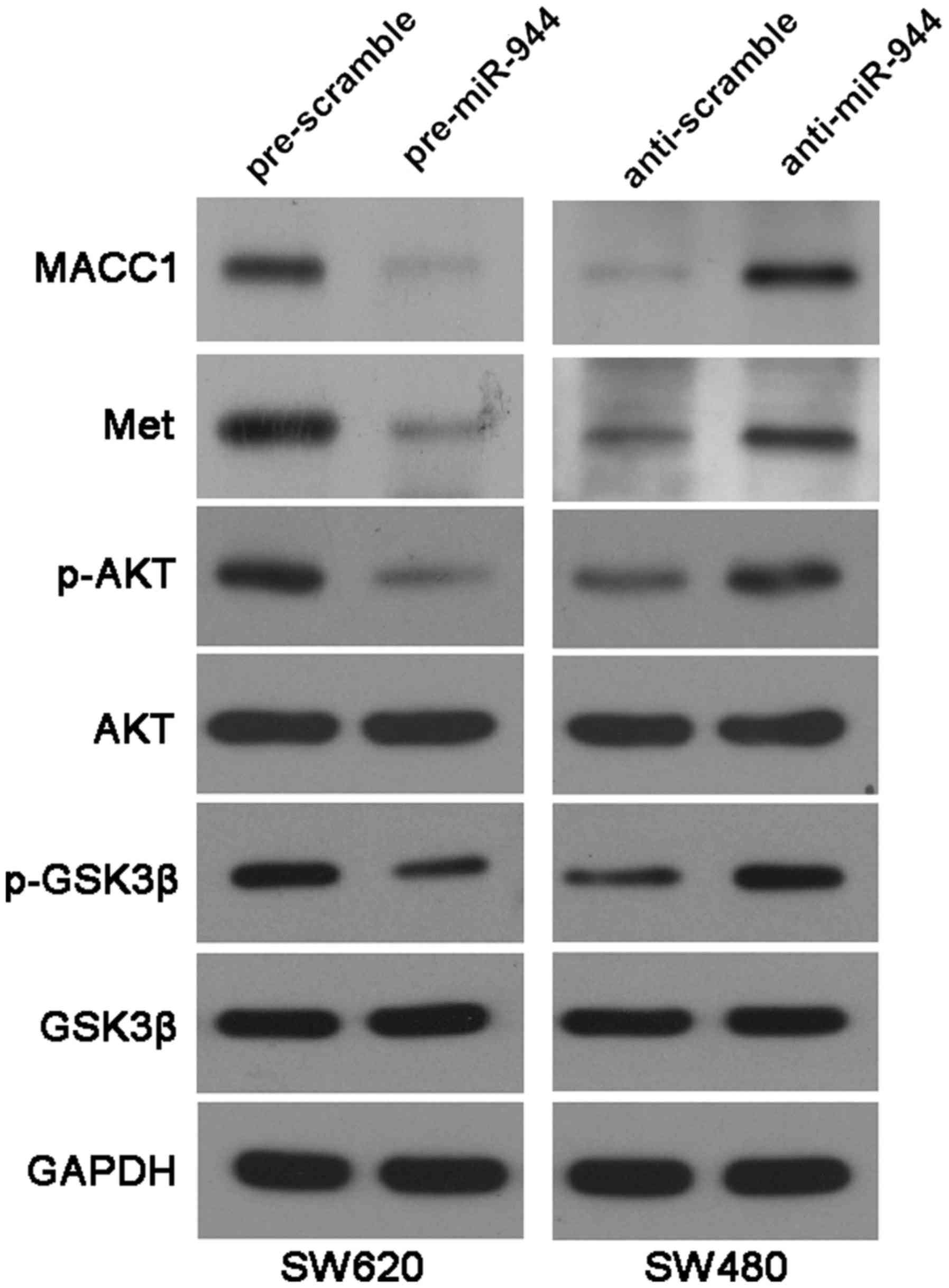
6B-D). Previous studies have reported that MACC1 regulates the
expression of Met (19), which
activates Akt and abrogates GSK-3β activity (20). As shown in Fig. 7, miR-944 overexpression reduced the
levels of MACC1, Met, p-AKT (Ser473) and p-GSK3β (Ser9) in SW620
cells. While, miR-944 knockdown increased the expression of MACC1,
Met, p-AKT (Ser473) and p-GSK3β (Ser9) in SW480 cells. These
experiments suggest that MACC1 is not only a downstream target but
also a possible mediator of miR-944 in CRC.
Discussion
Emerging evidence has confirmed that miRNAs are
actively involved in the pathogenic process of CRC (21). In addition, miRNAs have been
reported to be an important mediator of metastasis and epithelial
mesenchymal transition of CRC cells (22). According to the important function
of miRNAs in CRC, miRNAs have been considered as potential
diagnostic biomarkers and drug-targets of CRC (23). In this study, miR-944 was found to
be significantly downregulated in CRC. The low expression of
miR-944 conferred malignant clinical parameters of CRC patients
including high tumor invasion stage, more lymph node and distant
metastasis and advanced clinical stage. More importantly, the
decreased expression of miR-944 correlated with shortened 5-year
overall and progression-free survival. Therefore, miR-944 plays a
tumor suppressive role in CRC and potentially serves as a promising
biological target for the prognosis of patients.
Systemic metastasis is the important reason for the
unsatisfactory prognosis of CRC patients (24). Increased migratory and invasive
ability of cancer cells underlies the systemic metastasis of CRC.
Thus, it is fundamental to disclose the underlying mechanisms for
the metastasis of CRC cells. Here, we found that miR-944 inhibited
the proliferation, migration and invasion of CRC cells in
vitro. These data confirmed that miR-944 exerted an anticancer
role by inhibiting proliferation and metastasis in CRC cells. MACC1
was reported to be an independent prognostic marker for metastasis
and progression-free survival (19). Otherwise, MACC1 was found to
function as a pro-metastatic factor by promoting the migratory and
invasive ability of CRC cells (19). In the present study, we disclosed
that miR-944 suppressed the expression of MACC1 in CRC cells. The
levels of MACC1 mRNA in CRC tissues were negatively correlated with
the expression of miR-944. Furthermore, we found that miR-944 could
directly interact with the 3′-UTR of MACC1. These experiments
suggest that MACC1 is a downstream molecule of miR-944. We found
that restoration of MACC1 could abrogate the anticancer effects of
miR-944 on CRC cell proliferation, migration and invasion. Previous
studies have reported that MACC1 regulates the expression of Met
(19), which activates Akt and
abrogates GSK-3β activity (20).
Our data showed that miR-944 inversely regulated MACC1/Met/Akt
signaling in CRC cells. Our results suggest MACC1 is not only a
downstream target but also a possible mediator of miR-944 in
CRC.
Collectively, the present study demonstrates that
miR-944 expression is significantly decreased in CRC. The low level
of miR-944 correlates with malignant clinical parameters of CRC
patients and shortened survival. miR-944 inhibits the proliferation
and metastasis of CRC cells. Furthermore, MACC1 is a downstream
target of miR-944 in CRC. miR-944 exerts its inhibitory effects on
CRC proliferation and metastasis, at least in part, by targeting
MACC1/Met/AKT signaling.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81572925).
References
|
1
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang W, Dahlberg JE and Tam W: MicroRNAs
in tumorigenesis: A primer. Am J Pathol. 171:728–738. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomas J, Ohtsuka M, Pichler M and Ling H:
MicroRNAs: Clinical Relevance in Colorectal Cancer. Int J Mol Sci.
16:28063–28076. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schultz NA, Andersen KK, Roslind A,
Willenbrock H, Wøjdemann M and Johansen JS: Prognostic microRNAs in
cancer tissue from patients operated for pancreatic cancer-five
microRNAs in a prognostic index. World J Surg. 36:2699–2707. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lazar V, Suo C, Orear C, van den Oord J,
Balogh Z, Guegan J, Job B, Meurice G, Ripoche H, Calza S, et al:
Integrated molecular portrait of non-small cell lung cancers. BMC
Med Genomics. 6:532013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu M, Zhou K and Cao Y: MicroRNA-944
affects cell growth by targeting EPHA7 in non-small cell lung
cancer. Int J Mol Sci. 17:172016. View Article : Google Scholar
|
|
11
|
Ma J, Mannoor K, Gao L, Tan A, Guarnera
MA, Zhan M, Shetty A, Stass SA, Xing L and Jiang F:
Characterization of microRNA transcriptome in lung cancer by
next-generation deep sequencing. Mol Oncol. 8:1208–1219. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Powrózek T, Krawczyk P, Kowalski DM,
Winiarczyk K, Olszyna-Serementa M and Milanowski J: Plasma
circulating microRNA-944 and microRNA-3662 as potential histologic
type-specific early lung cancer biomarkers. Transl Res.
166:315–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie H, Lee L, Scicluna P, Kavak E, Larsson
C, Sandberg R and Lui WO: Novel functions and targets of miR-944 in
human cervical cancer cells. Int J Cancer. 136:E230–E241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Warnecke-Eberz U, Chon SH, Hölscher AH,
Drebber U and Bollschweiler E: Exosomal onco-miRs from serum of
patients with adenocarcinoma of the esophagus: Comparison of miRNA
profiles of exosomes and matching tumor. Tumour Biol. 36:4643–4653.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Flores-Pérez A, Marchat LA,
Rodríguez-Cuevas S, Bautista VP, Fuentes-Mera L, Romero-Zamora D,
Maciel-Dominguez A, de la Cruz OH, Fonseca-Sánchez M, Ruíz-García
E, et al: Suppression of cell migration is promoted by miR-944
through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC
Cancer. 16:3792016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Christensen LL, Tobiasen H, Holm A,
Schepeler T, Ostenfeld MS, Thorsen K, Rasmussen MH,
Birkenkamp-Demtroeder K, Sieber OM, Gibbs P, et al: COLOFOL
steering group: MiRNA-362-3p induces cell cycle arrest through
targeting of E2F1, USF2 and PTPN1 and is associated with recurrence
of colorectal cancer. Int J Cancer. 133:67–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tu K, Yang W, Li C, Zheng X, Lu Z, Guo C,
Yao Y and Liu Q: Fbxw7 is an independent prognostic marker and
induces apoptosis and growth arrest by regulating YAP abundance in
hepatocellular carcinoma. Mol Cancer. 13:1102014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arlt F and Stein U: Colon cancer
metastasis: MACC1 and Met as metastatic pacemakers. Int J Biochem
Cell Biol. 41:2356–2359. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stein U, Walther W, Arlt F, Schwabe H,
Smith J, Fichtner I, Birchmeier W and Schlag PM: MACC1, a newly
identified key regulator of HGF-MET signaling, predicts colon
cancer metastasis. Nat Med. 15:59–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Birchmeier C, Birchmeier W, Gherardi E and
Woude GF Vande: Met, metastasis, motility and more. Nat Rev Mol
Cell Biol. 4:915–925. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chi Y and Zhou D: MicroRNAs in colorectal
carcinoma - from pathogenesis to therapy. J Exp Clin Cancer Res.
35:432016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Muhammad S, Kaur K, Huang R, Zhang Q, Kaur
P, Yazdani HO, Bilal MU, Zheng J, Zheng L and Wang XS: MicroRNAs in
colorectal cancer: Role in metastasis and clinical perspectives.
World J Gastroenterol. 20:17011–17019. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan S, Cao Y and Mao A: MicroRNAs in
colorectal cancer: Potential biomarkers and therapeutic targets.
Front Biosci (Landmark Ed). 20:1092–1103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X, Ji Q, Fan Z and Li Q: Cellular
signaling pathways implicated in metastasis of colorectal cancer
and the associated targeted agents. Future Oncol. 11:2911–2922.
2015. View Article : Google Scholar : PubMed/NCBI
|