Introduction
Worldwide, approximately 4% of all oral squamous
cell carcinomas occur in men, with an increased frequency in males
over the age of 50; geographical variations also affect the
incidence of disease (1). Multiple
gene changes accumulate as cell transition from normal cells into
cancer cells during a multi-step process that includes changes in
oncogenes and tumor suppressor genes. Many carcinogens and other
factors are related to OSCC, including the use of tobacco and
alcohol, which are the most important risk factors for head and
neck cancers (2–4). Chewing betel quid is another primary
risk factor for OSCC (5), and 85%
of all patients with OSCC chew betel quid on a regular basis
(6). Betel quid contains artificial
supplements such as arecoline and other alkaloids, which are
carcinogenic.
New strategies to detect the early stages of OSCC
are an essential and emergent issue. Several studies have focused
on gene expression profiles, using cDNA microarrays to reveal
genetic alterations in OSCC patients. Comparing aberrant miRNA
expression profiles with matched normal controls in tissues and
cell lines are beginning to unveil the mechanisms of OSCC disease
progression (7). There is an
increasing number of studies that analyze miRNA expression profiles
in several cancers, and differentially expressed miRNAs are
involved in the development of many malignancies, including OSCC
(8,9).
MicroRNAs (miRNAs) are 21–23 nucleotides long and
inhibit protein synthesis (10) or
cause mRNA degradation. Recent evidence has demonstrated that there
are distinct microRNA expression signatures between tumor tissues
and their normal counterparts, and it seems that miRNAs function as
either oncogenes or tumor suppressors (11). Therefore, in the present study, we
survey miRNA expression profiles and identify specific miRNA
signatures by comparing normal and tumor tissues in patients with
OSCC using a combination of miRNA microarray data mining with
bioinformatics. These data can profoundly impact the development of
clinically relevant diagnostic tools for the treatment of oral
cancers.
Materials and methods
Chemicals and reagents
Pre-miR™ precursors, Anti-miR™ inhibitors, siPORT™
NeoFX™ reagents and the miRNA expression reporter vector were
purchased from Applied Biosystems (Foster City, CA, USA).
Patients and tissue samples
The present study was reviewed and approved by an
Institutional Review Board (IRB) at the China Medical University
Hospital (CMUH IRB no. DMR98-IRB-202), Taichung, Taiwan (CMUH).
After acquiring informed consent from each patient in the study at
the Department of Otolaryngology, China Medical University Hospital
(CMUH, Taichung, Taiwan), paired normal and tumor samples, mostly
obtained from the tongue and other areas of the mouth, were
collected from 39 patients who presented with primary OSCC. All
tissues were frozen in liquid nitrogen immediately after surgery
and stored at −80°C until the extraction of RNA. The control group
consisted of patients who obtained surgery for non-neoplastic
diseases of the head and neck. Histological studies were also
performed at the Department of Pathology in CMUH, and all tumors
were confirmed as squamous cell carcinoma.
MicroRNA arrays
RNA samples were extracted and isolated using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) and performed according to
the manufacturers instructions. One microgram of RNA was prepared
for microarray analysis. The expression profiles of 365 mature
miRNAs were assessed using TaqMan Human MicroRNA Arrays v2.0
(Applied Biosystems) according to the manufacturers
instructions.
Quantification of microRNA
expression
TaqMan miRNA assays (Applied Biosystems) quantified
the maturity of the miRNA samples from miR-375, miR-204 and
miR-196a. RNU6B was the reference gene control. Quantitative
polymerase chain reaction assays were performed according to the
manufacturer's protocol. Briefly, RNA samples were reverse
transcribed into specific cDNA; these specific cDNAs were
quantified according to the manufacturers instructions. All
amplification reactions were performed in triplicate. The threshold
cycle (Ct) values were obtained and analyzed using the ABI 7900HT
SDS 2.2 software.
Predicting microRNA target genes
Several computer software programs, including
miRBase (http://microrna.sanger.ac.uk/), TargetScan (http://www.targetscan.org/), and miRanda (http://www.microrna.org/), were used to analyze and
compare potential microRNA target genes.
Cell culture
Oral cancer cell lines including CAL27 and HSC-3
were cultured in Dulbeccos modified Eagles medium and Dulbecco's
modified Eagles medium/F-12 (Invitrogen), respectively. The media
were supplemented with 10% heat-inactivated fetal bovine serum
(FBS), 100 Units/ml penicillin and 100 µg/ml streptomycin
(Invitrogen) (12).
miRNA transfection
miRNA transfection was performed using the NeoFX™
reagent according to the manufacturers instructions (Applied
Biosystems). Briefly, CAL27 cells were seeded onto 6-well plates
and transfected with a 10 nM solution of pre-miR-375, pre-miR-204,
pre-miR-196a or pre-miR, which were used as control
oligonucleotides (Applied Biosystems) for 48 h.
Vector construction and luciferase
reporter gene assays
Oligonucleotides for the potential target genes,
miR-375, miR-204 and miR-196a, were cloned into a pMIR-Report
vector (Applied Biosystems). The putative miR-375, miR-204 and
miR-196a binding sites were cloned into the same vector and used as
controls. Using jetSI-ENDO reagents (PolyPlus Transfection,
Illkirch, France) in a luciferase reporter assay, CAL27 cells were
co-transfected with 1.5 mg of a pMIR-Report firefly luciferase
construct, and 500 ng of a pRL-CMV Renilla luciferase was
used as a normalised control (Promega, Madison, WI, USA). In the
presence of microRNA precursors and after 48 h of transfection, the
luciferase activities were analyzed using a Dual-Luciferase
reporter assay system (Promega, Madison, WI, USA) according to the
manufacturers instructions. All experiments were performed twice
and repeated in three independent experiments (13).
Western blot analysis
Western blot analysis was carried out as previously
described (14–16). Briefly, 30 µg of protein was
separated using a 10% SDS-polyacrylamide gel and transferred onto
polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica,
MA, USA). The membranes were incubated with a specific primary
antibody overnight and then incubated with secondary antibody for 1
h at room temperature. Bands were detected using a commercially
enhanced chemiluminescence system (GE Healthcare Biosciences,
Piscataway, NJ, USA). The band intensities were analyzed using
Adobe Photoshop software.
Stable miR-196a knockdown clones
HSC-3 human oral cancer cells
(3×105/well) were seeded onto 6-well plates overnight.
Transfection was performed using Arrest-In reagents following the
manufacturers protocol (Thermo Fisher Scientific-Open Biosystems,
Huntsville, AL, USA). Specific miR-196a knockdown stable clones
were selected using 2.5 µg/ml of puromycin and were identified by
western blot analysis.
Cell proliferation assay
HSC-3 cells or miR-196a knockdown HSC-3 cells were
seeded onto 6-well plates at a density of 2×105 cells.
Cells were trypsinized and counted every 24 h. Cell proliferation
assays were measured in triplicate using the Beckman Coulter Z1
Particle Counter (17,18).
Statistical analysis
All data were expressed as the mean ± standard
deviations (SD). Differences between the groups were examined using
the two-tailed unpaired Student's t-tests and an analysis of
variance for the repeated measurements. Statistical significance
between the groups was determined based on P-values set at
P<0.05 (19).
Results
Validation of miR-375, miR-204 and
miR-196a expression in 39 pairs of oral cancer patients
The 384-well TaqMan Human MicroRNA array screened
and analysed the miRNA expression profiles for 2 patients. The
miRNA expression profiles for OSCCs were compared with those in
normal tissue. We found that 29 miRNAs were significantly
upregulated (Table I), whereas 34
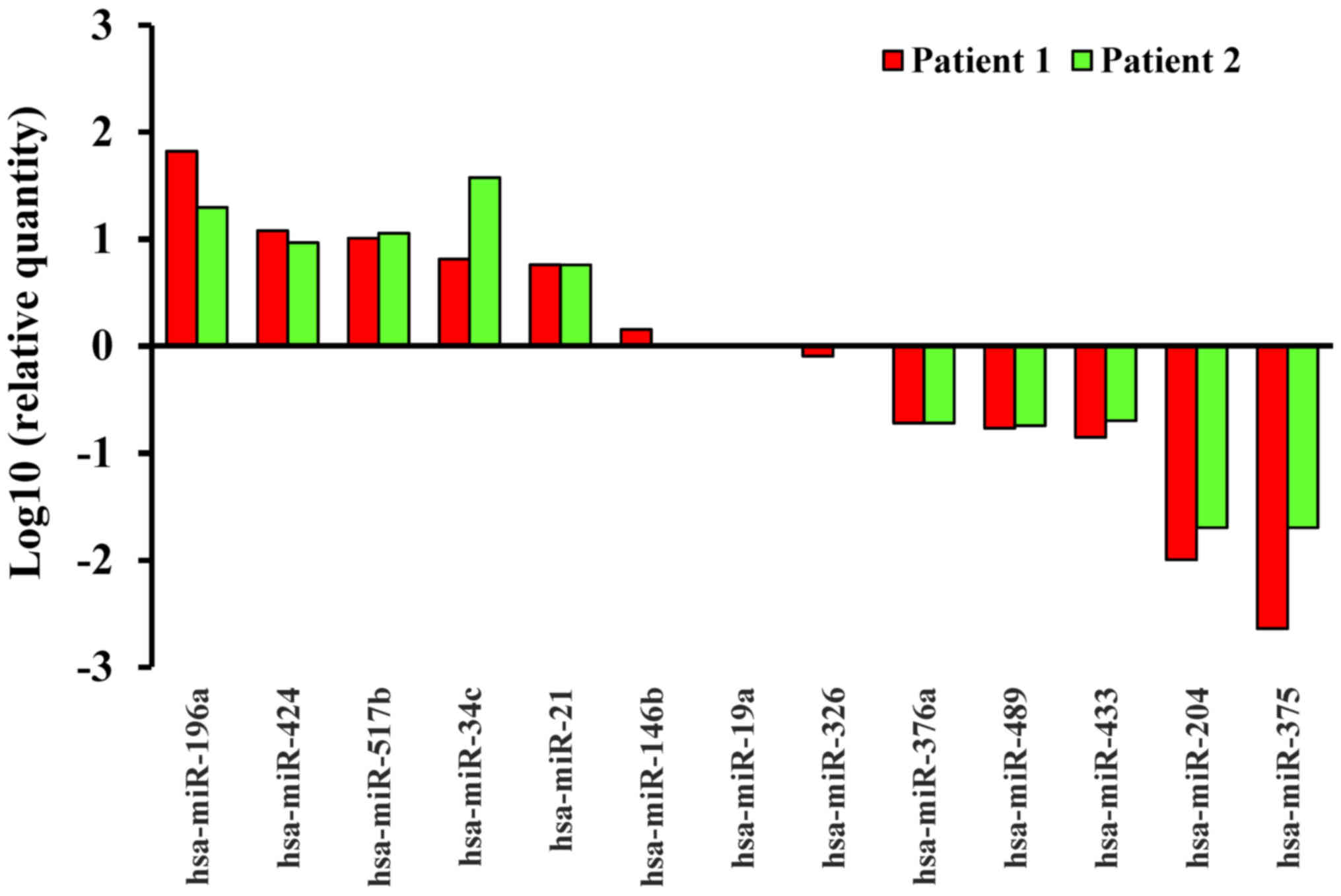
miRNAs were downregulated in these oral cancer patients (Table II). In Fig. 1, for example, the top 5 upregulated
miRNAs of 2 patients were miR-196a, miR-424, miR-517b, miR-34c and
miR-21, whereas the top 5 downregulated miRNAs were miR-375,
miR-204, miR-433, miR-489 and miR-376a. In addition, although 218
miRNAs remained the same, 72 miRNAs were differentially expressed
in oral cancer tissue.
 | Table I.Upregulated miRNAs in oral cancer
patients after analyzing by TaqMan® Human MicroRNA
array. |
Table I.
Upregulated miRNAs in oral cancer
patients after analyzing by TaqMan® Human MicroRNA
array.
|
| Patient 1 stage
IV | Patient 2 stage
IV |
|---|
|
|
|
|
|---|
| miRNA | Fold change (tumor
vs. normal) | Fold change (tumor
vs. normal) |
|---|
| hsa-miR-196a | 66.13 | 19.66 |
| hsa-miR-424 | 11.91 |
9.21 |
| hsa-miR-517b | 10.13 | 11.29 |
| hsa-miR-34c |
6.46 | 37.37 |
| hsa-miR-21 |
5.72 | 5.7 |
| hsa-miR-503 | 20.1 |
4.76 |
| hsa-miR-644 | 17.83 |
4.88 |
| hsa-miR-432 | 14.62 |
4.73 |
| hsa-miR-618 |
8.65 |
2.36 |
| hsa-miR-221 |
8.12 |
3.51 |
| hsa-miR-31 |
8.12 |
2.75 |
| hsa-miR-490 |
7.42 |
3.57 |
| hsa-miR-196b |
5.61 |
3.07 |
| hsa-miR-452 |
5.43 |
3.84 |
| hsa-miR-222 |
4.75 |
2.25 |
| hsa-miR-301 |
4.39 |
3.28 |
| hsa-miR-130b |
4.06 |
3.57 |
| hsa-miR-576 |
3.91 |
2.18 |
| hsa-miR-556 |
3.66 |
4.82 |
| hsa-miR-512-3p |
3.36 | 41.72 |
| hsa-miR-18a |
3.17 |
5.09 |
| hsa-miR-181d |
3.07 |
2.49 |
| hsa-miR-324-5p |
2.92 |
3.19 |
| hsa-miR-520h |
2.75 | 38.03 |
| hsa-miR-34b |
2.62 |
4.99 |
| hsa-miR-193b |
2.33 |
2.53 |
| hsa-miR-365 |
2.31 |
2.33 |
| hsa-miR-15b |
2.12 |
2.63 |
| hsa-miR-450 | 2 | 19.4 |
 | Table II.Downregulated miRNAs in oral cancer
patients after analyzing by TaqMan® Human MicroRNA
array. |
Table II.
Downregulated miRNAs in oral cancer
patients after analyzing by TaqMan® Human MicroRNA
array.
|
| Patient 1 stage
IV | Patient 2 stage
IV |
|---|
|
|
|
|
|---|
| miRNA | Fold change (tumor
vs. normal) | Fold change (tumor
vs. normal) |
|---|
| hsa-miR-379 | 0.47 | 0.44 |
| hsa-miR-656 | 0.44 | 0.07 |
| hsa-miR-432 | 0.39 | 0.23 |
| hsa-miR-554 | 0.39 | 0.06 |
| hsa-miR-506 | 0.39 | 0.03 |
| hsa-miR-100 | 0.37 | 0.12 |
| hsa-miR-20b | 0.35 | 0.34 |
| hsa-miR-195 | 0.34 | 0.28 |
| hsa-miR-27b | 0.33 | 0.38 |
| hsa-miR-125b | 0.33 | 0.22 |
| hsa-miR-410 | 0.32 | 0.36 |
| hsa-miR-99a | 0.32 | 0.09 |
| hsa-miR-26a | 0.28 | 0.43 |
| hsa-miR-218 | 0.27 | 0.15 |
| hsa-miR-127 | 0.26 | 0.27 |
| hsa-miR-369-5p | 0.26 | 0.25 |
| hsa-miR-485-5p | 0.25 | 0.24 |
| hsa-miR-328 | 0.23 | 0.29 |
| hsa-miR-411 | 0.23 | 0.25 |
| hsa-let-7c | 0.23 | 0.05 |
| hsa-miR-149 | 0.22 | 0.22 |
| hsa-miR-296 | 0.2 | 0.28 |
| hsa-miR-126 | 0.18 | 0.4 |
| hsa-miR-30a-3p | 0.17 | 0.4 |
| hsa-miR-139 | 0.17 | 0.23 |
| hsa-miR-487b | 0.13 | 0.42 |
| hsa-miR-95 | 0.09 | 0.32 |
| hsa-miR-486 | 0.09 | 0.27 |
| hsa-miR-9 | 0.04 | 0.33 |
| hsa-miR-376a | 0.19 | 0.19 |
| hsa-miR-489 | 0.17 | 0.18 |
| hsa-miR-433 | 0.14 | 0.2 |
| hsa-miR-204 | 0.01 | 0.02 |
| hsa-miR-375 | <0.01 | 0.02 |
Thirty-nine oral cancer patients were examined to
determine whether the observed miRNA expression profiling was
specific to the individual. Patients were classified into four
groups: group I patients possessed tumor sizes ≤4 cm without
metastasis; group II patients possessed tumor sizes >4 cm
without metastasis; patients with tumor sizes <4 cm who
presented with metastasis belonged to group III; and patients
possessing tumor sizes >4 cm who presented with metastasis
belonged to group IV (Table III).
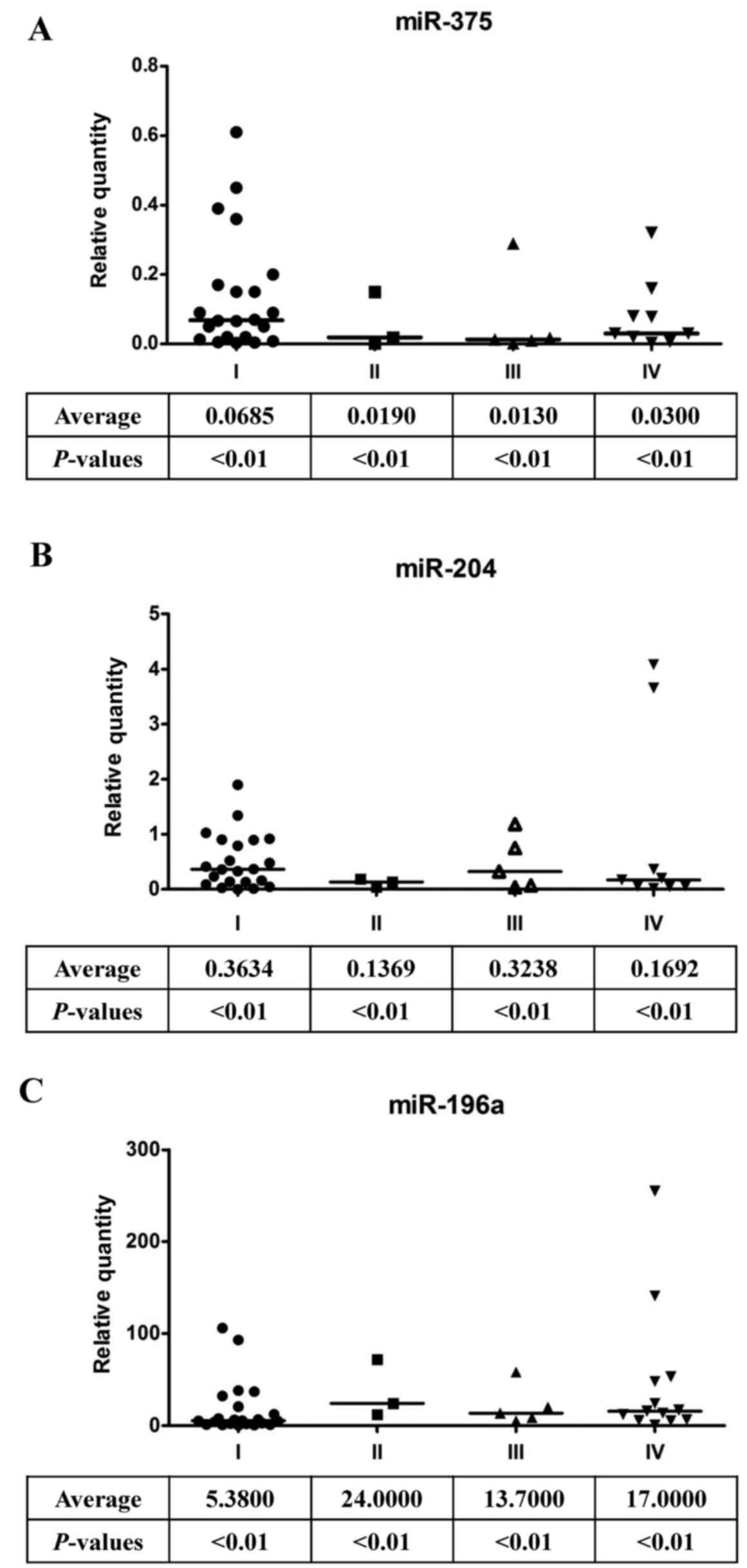
Indeed, miR-375 (Fig. 2A) and
miR-204 (Fig. 2B) expression levels
were significantly low in oral cancer patients. In contrast,
miR-196a expression was dramatically increased in oral cancer
patients (Fig. 2C). These data
suggest that the expression levels of miR-375, miR-204 and miR-196a
are good indicators of oral cancer progression. In addition, after
analysing the clinical pathological characteristics of 39 patients,
we found that >67% of the patients were habitual smokers and
chewed betel quid, which are both risk factors for OSCC in Taiwan
(Table III).
 | Table III.Patient demographics and
clinicopathological characteristics (N=39). |
Table III.
Patient demographics and
clinicopathological characteristics (N=39).
| Stage | Early (I, II) | Late (III, IV) | Early (I, II) and
metastasis | Late (III, IV) and
metastasis |
|---|
| Female | 1 | 0 | 0 | 2 |
| Male | 21 | 3 | 5 | 7 |
| Age (years) | 33–67 | 28–50 | 41–57 | 32–74 |
| Smoking | 18 (82%) | 2 (67%) | 4 (80%) | 7 (78%) |
| Chewing betel | 17 (77%) | 2 (67%) | 2 (40%) | 6 (67%) |
| Tumor site | SCC | SCC | SCC | SCC |
Targeted prediction and Gene Ontology
analysis for miR-375, miR-204 and miR-196a
The combined use of different computer software
programs helped predict the target genes of miRNAs (miR-375,
miR-204 and miR-196a) (Table IV).
The HSPA12A, INSM1, MTPN, PDK1, UBE2E and USP1 were potential
miR-375 gene targets and are associated with the development of
oral cancers (20). Specifically,
previous studies have demonstrated that PDPK1 and MTPN are miR-375
gene targets in pancreatic cancer (21,22).
Bcl-2, FJX1, HMGA2, MEIS1, RAB1A and SOX4 are potential target
genes for miR-204 and have been associated with cancer development
using similar approaches (23,24).
Lastly, the potential target genes in miR-196 were ANXA1, p27
(CDKN1B), HOXB8, HOXC8, ING5 and LRP1B, which are associated with
the development of oral cancers (25–27).
 | Table IV.Target gene predictions of miR-375,
miR-204 and miR-196a. |
Table IV.
Target gene predictions of miR-375,
miR-204 and miR-196a.
| miR-375 target
genes | miR-204 target
genes | miR-196a target
genes |
|---|
| MTPN
(myotrophin) | HMGA2 (high
mobility group AT-hook 2) | HOXB8 (homeo box
B8) |
| PDK1
(3-phosphoinositide dependent protein kinase-1) | SOX4 (SRY (sex
determining region Y)-box 4) | HOXC8 (homeo box
C8) |
| USP1 (ubiquitin
specific protease 1) | FJX1 (four jointed
box 1) | ANXA1 (Annexin
A1) |
| UBE2E2
(ubiquitin-conjugating enzyme E2E2) | RAB1A (RAS oncogene
family) | ING5 (inhibitor of
growth family, member 5) |
| HABP2 (hyaluronan
binding protein 2) | Bcl-2 (B-cell
CLL/lymphoma 2) | CDKN1B
(cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
| INSM1
(insulinoma-associated 1) | MEIS1 (myeloid
ecotropic integration site 1) | LRP1B (low density
lipoprotein-related protein 1B) |
Validation of miR-375, miR-204 and
miR-196a targeted genes using luciferase reporter assays
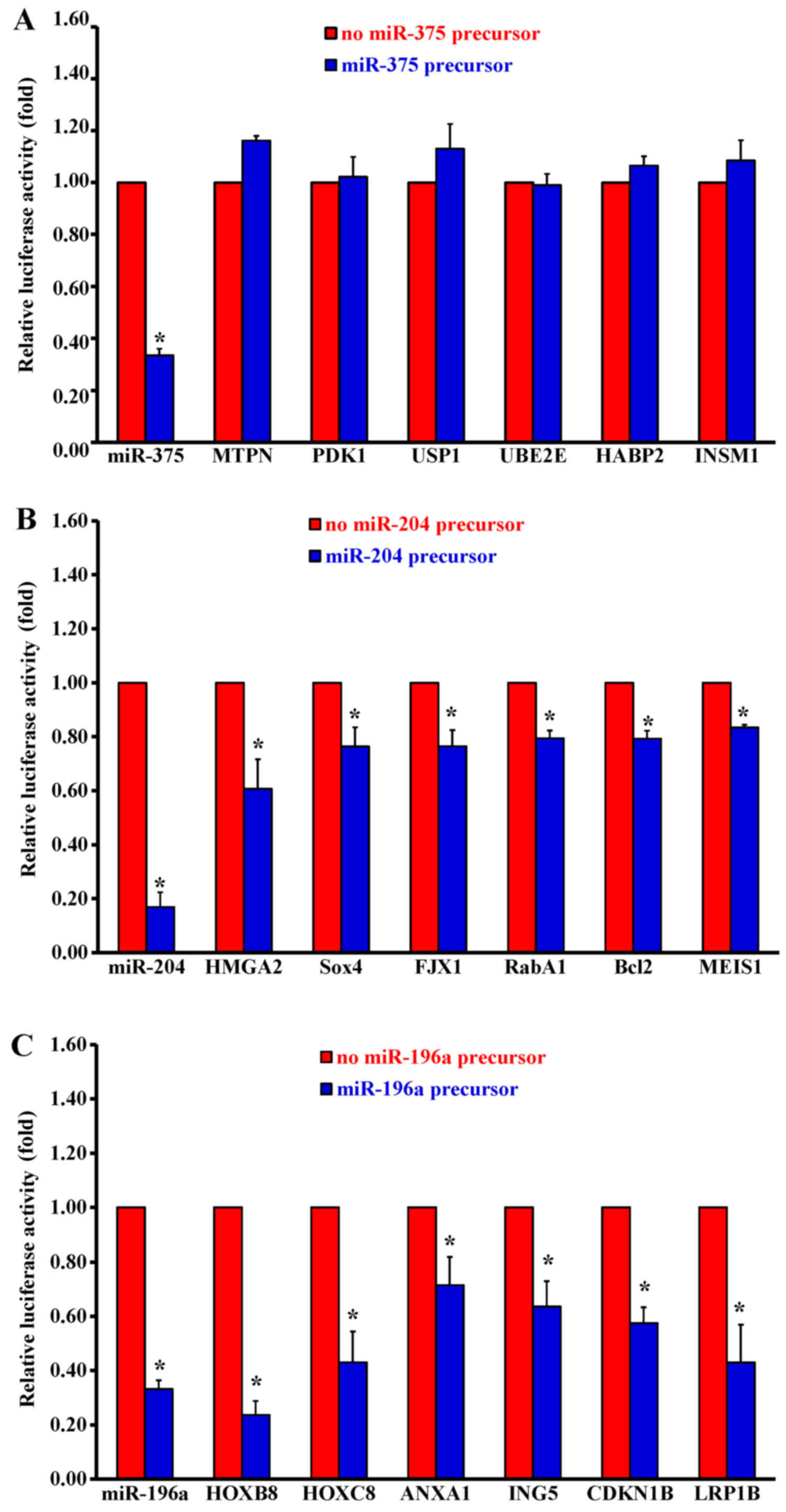
Constructs of miRNA target genes were introduced
into a 3′UTR miRNA luciferase gene in an expression reporter
vector. The CAL27 cells were transfected with the luciferase
reporter gene plasmid either in the presence or absence of a
miR-375 precursor. There was no change in inhibition for the
luciferase activity in 6 potential miR-375 target genes (Fig. 3A). Unexpectedly, MTPN and PDK1 are
target genes for miR-375 in pancreatic cancer (22,28),
but not in oral cancers. The luciferase activity from 6 potential
miR-204 target genes decreased dramatically, including HMGA2
(Fig. 3B) in CAL27 cells.
Furthermore, the luciferase activity from 6 potential miR-196a
target genes was dramatically inhibited, especially in HOXB8 and
p27 (CDKN1B) (Fig. 3C).
Characterisation of miR-375, miR-204
and miR-196a targeted genes using western blot analysis
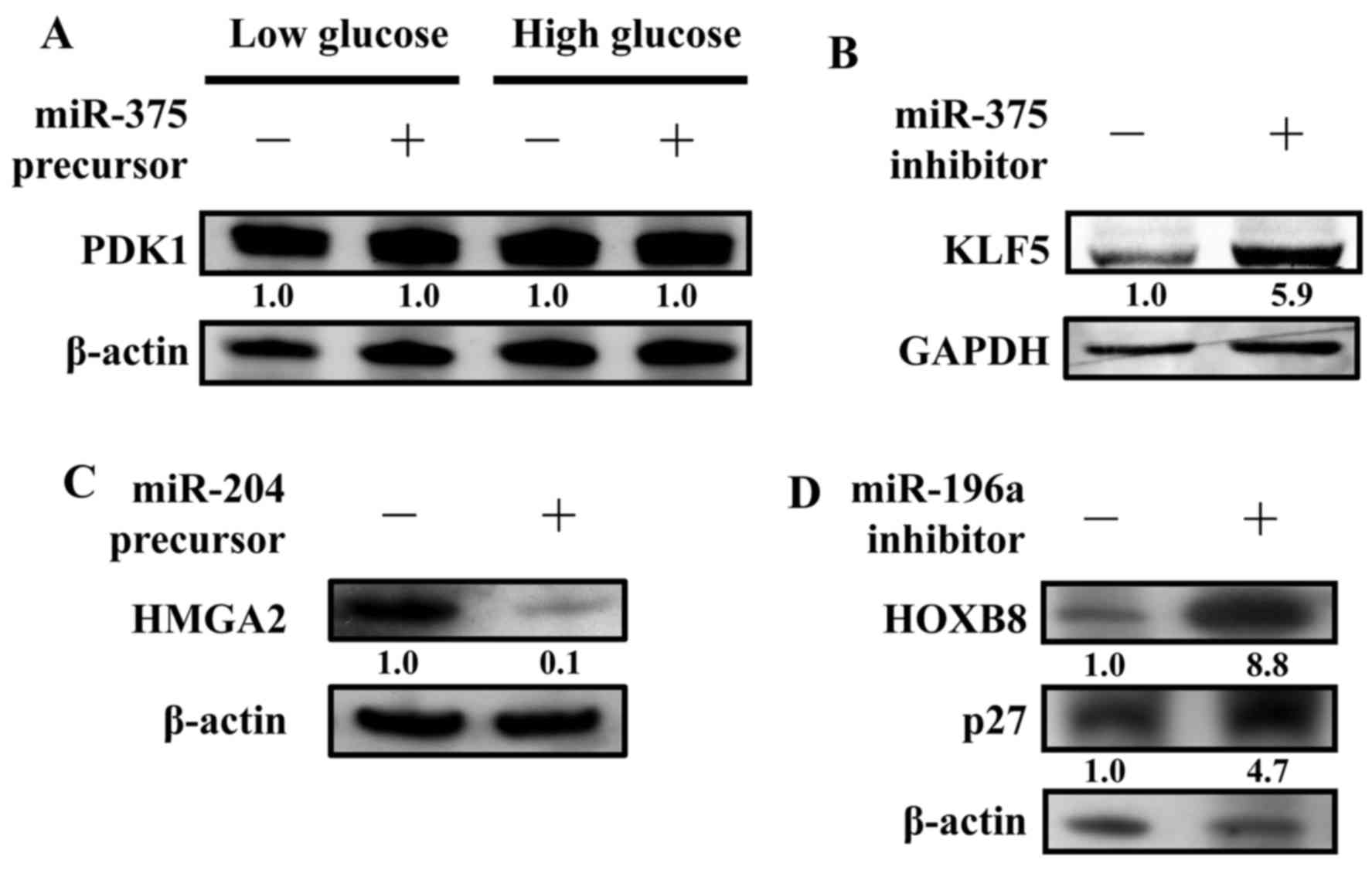
CAL27 cells were transfected with or without a
miR-375 precursor to detect PDK1 protein expression levels. In
previous studies, the concentration of glucose affected PDK1
protein expression levels (22).
Our results, however, indicate that the expression level remained
the same regardless of the glucose concentrations added to CAL27
(Fig. 4A). Thus, PDK1 may not be
the miR-375 targeted gene in oral cancers. Recently, KLF5 was found
to be regulated by miR-375 (29).
We examined the protein expression level of KLF5 in the presence of
a miR-375 inhibitor. Indeed, inhibition of miR-375 caused the
upregulation of KLF5 in CAL27 (Fig.
4B). Next, the protein expression level of HMGA2 was examined
in the presence of a miR-204 precursor. As predicted, the protein
expression levels of HMGA2 were significantly inhibited by the
miR-204 precursor. These results suggest that HMGA2 is one of the
miR-204 directly targeted genes (Fig.
4C). In addition, the protein expression levels of HOXB8 and
p27 (CDKN1B) were also examined in the presence of a miR-196a
inhibitor. As predicted, the protein expression levels of HOXB8 and
p27 (CDKN1B) were significantly increased by the miR-196a
inhibitor, suggesting that HOXB8 and p27 (CDKN1B) are two genes
that are directly targeted by miR-196a (Fig. 4D).
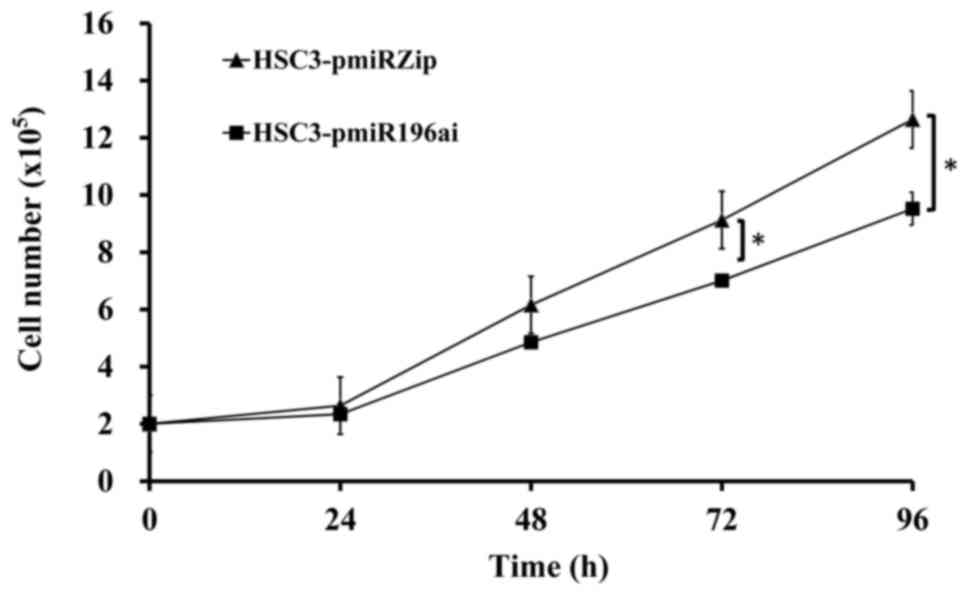
miR-196a knockdown cells reduces cell
proliferation
HSC3 cells were transfected with either the pmiRZIP
vector only or pmiRZIP-miR-196a. After 48-h transfection, cells
were cultured in the medium containing 2.5 µg/ml and medium was
changed every 3 days until a single clone formed. HSC3 knockdown
miR-196a (HSC3-pmiR-196ai) cells and the control HSC3-pmiRZIP cells
were seeded with 2×105/well into a 6-well plate. Cells
were counted every 24 h. Indeed, HSC3-pmiR-196ai cells grow slower
than control HSC3 cells (Fig.
5).
Discussion
To date, several published studies have addressed
the differential expression of miRNAs in oral cancers. However, a
comprehensive analysis of miRNA-targeted genes could lead to the
elucidation of pathways that could deregulate cancer cells and
subsequently identify therapeutic targets. In the present study,
the expression levels of 365 microRNAs were investigated in OSCC.
After validation by quantitative reverse transcription-PCR, we
discovered that miR-375 and miR-204 were downregulated, whereas
miR-196a was upregulated (Fig. 1
and Tables I and II). The downregulation of miR-375 in
HNSCC tumors is consistent with previous findings that miR-375 is
expressed at significantly greater levels in laryngeal tumors
compared with those in the oral cavity (Fig. 2A). The elevated expression of
miR-375 is significantly associated with alcohol consumption
(30). Notably,
3-phosphoinositide-dependent protein kinase 1 (PDK1) is a kinase
that activates anti-apoptotic AKT. However, our data did not show
any change in the protein expression levels of PDK1 (Fig. 4A).
Several lines of evidence have demonstrated that
miR-204 is responsible for different expression patterns in cancer.
Overexpression of miR-204, for example, is correlated with
insulinomas (31). In contrast, the
downregulation of miR-204 targets HOXA10 and MEIS1 in acute myeloid
leukaemia (32). The data presented
here show that miR-204 is downregulated in oral cancers and in the
high mobility group A2 (HMGA2) gene, which is one of its direct
targets (Figs. 2B, 3B and 4C).
In addition, miR-196a was highly expressed in oral cancers, and
HOXB8 (homeobox B8) is one of the potential target genes of
miR-196a (Figs. 2C, 3C and 4D).
Thus, our results are consistent with previous studies (33). HOXB8, located on chromosome 17, is
one of the homeobox gene family members and is involved in
development. Furthermore, several studies have shown that aberrant
HOXB8 expression is correlated with cancer formation. Elevated
expression of HOXB8, for example, is associated with colorectal
cancer (34). Additionally, HOXB8
has been identified as a cause of leukaemia, and it regulates
smooth muscle cell differentiation. It is speculated that abnormal
HOXB8 expression leads to tumorigenesis in OSCC (35). Here, we demonstrated that HOXB8
expression is reduced via miR-196a suppression. It is important to
understand the role that HOXB8-mediated molecular mechanisms play
in the development of oral cancers. Moreover, p27 (CDKN1B), a
cyclin-dependent kinase (Cdk) inhibitor, may be another potential
target of miR-196a, which functions as a negative cell cycle
regulator. It was demonstrated that p27 (CDKN1B) was downregulated
and is strongly associated with various cancers, including OSCC
(36) (Fig. 4D). HSC3 cells were either
individually transfected with the pmiRZIP vector or with
pmiRZIP-miR-196a. After a 48-h transfection, the cells were
cultured in a medium containing 2.5 µg/ml; the medium was changed
every 3 days until a single clone formed. An HSC3 knockdown of
miR-196a (HSC3-pmiR-196ai) cells and the control HSC3-pmiRZIP cells
were counted every 24 h. It was observed that the HSC3-pmiR-196ai
cells grew slower than the control HSC3 cells (data not shown).
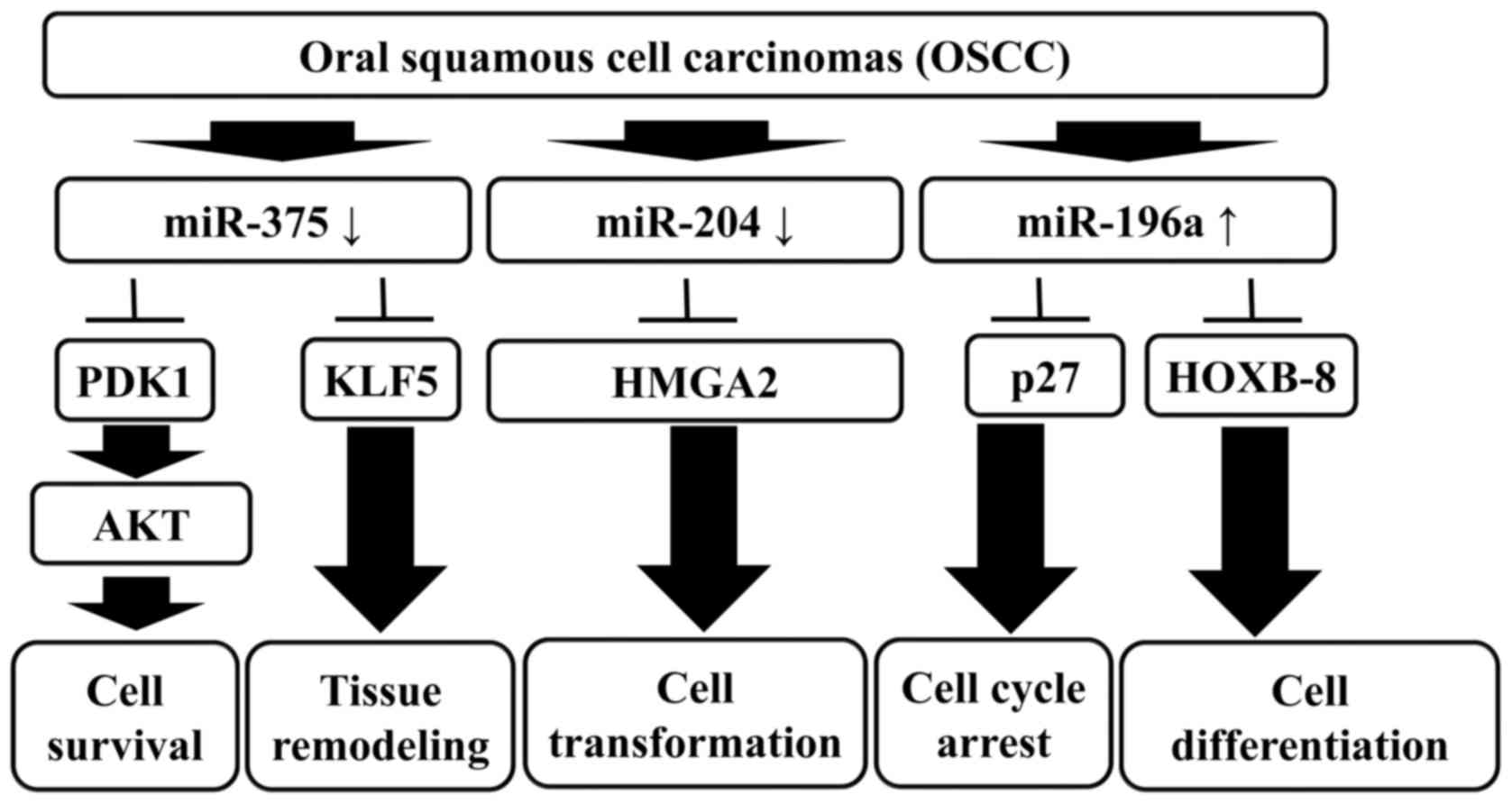
Overall, this study provides evidence that miRNA
signature profiling is a potential diagnostic tool and therapy to
target cancer. Furthermore, the characterization of miRNA profiling
provides new insights into the pathogenesis and progression of
OSCC. Although the underlying biological mechanisms of miRNAs
remain largely unknown, there is compelling evidence that miRNAs
will advance the management of OSCC in the near future (37). Our results suggest that miR-375,
miR-204 and miR-196a are differentially expressed in OSCC, and the
combined expression signatures of miR-375, miR-204 and miR-196a
provide promising biomarkers for the diagnosis, prognosis and
therapeutic utility of OSCC clinical treatment (Fig. 6).
Acknowledgements
The present study was supported in part by a grant
from the China Medical University (CMU95-305) and in part by a
grant from the National Science Council (NSC 98-2815-C-039-016-B).
We thank Jian-Chiao Wang and the lab members in Drs Tsai and Kao
for their suggestions during this study.
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Camisasca DR, Silami MA, Honorato J, Dias
FL, de Faria PA and Lourenço SQ: Oral squamous cell carcinoma:
Clinicopathological features in patients with and without
recurrence. ORL J Otorhinolaryngol Relat Spec. 73:170–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang CH, Lee CY, Lu CC, Tsai FJ, Hsu YM,
Tsao JW, Juan YN, Chiu HY, Yang JS and Wang CC: Resveratrol-induced
autophagy and apoptosis in cisplatin-resistant human oral cancer
CAR cells: A key role of AMPK and Akt/mTOR signaling. Int J Oncol.
50:873–882. 2017.PubMed/NCBI
|
|
4
|
Yuan CH, Horng CT, Lee CF, Chiang NN, Tsai
FJ, Lu CC, Chiang JH, Hsu YM, Yang JS and Chen FA: Epigallocatechin
gallate sensitizes cisplatin-resistant oral cancer CAR cell
apoptosis and autophagy through stimulating AKT/STAT3 pathway and
suppressing multidrug resistance 1 signaling. Environ Toxicol.
32:845–855. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tovosia S, Chen PH, Ko AM, Tu HP, Tsai PC
and Ko YC: Prevalence and associated factors of betel quid use in
the Solomon Islands: A hyperendemic area for oral and pharyngeal
cancer. Am J Trop Med Hyg. 77:586–590. 2007.PubMed/NCBI
|
|
6
|
Chiang SL, Jiang SS, Wang YJ, Chiang HC,
Chen PH, Tu HP, Ho KY, Tsai YS, Chang IS and Ko YC:
Characterization of arecoline-induced effects on cytotoxicity in
normal human gingival fibroblasts by global gene expression
profiling. Toxicol Sci. 100:66–74. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang SS, Jiang WW, Smith I, Poeta LM,
Begum S, Glazer C, Shan S, Westra W, Sidransky D and Califano JA:
MicroRNA alterations in head and neck squamous cell carcinoma. Int
J Cancer. 123:2791–2797. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scapoli L, Palmieri A, Lo Muzio L,
Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and
Carinci F: MicroRNA expression profiling of oral carcinoma
identifies new markers of tumor progression. Int J Immunopathol
Pharmacol. 23:1229–1234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gorenchtein M, Poh CF, Saini R and Garnis
C: MicroRNAs in an oral cancer context - from basic biology to
clinical utility. J Dent Res. 91:440–446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hammond SM: MicroRNAs as oncogenes. Curr
Opin Genet Dev. 16:4–9. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu FS, Yang JS, Yu CS, Lu CC, Chiang JH,
Lin CW and Chung JG: Safrole induces apoptosis in human oral cancer
HSC-3 cells. J Dent Res. 90:168–174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tsai SC and Seto E: Regulation of histone
deacetylase 2 by protein kinase CK2. J Biol Chem. 277:31826–31833.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsai SC, Valkov N, Yang WM, Gump J,
Sullivan D and Seto E: Histone deacetylase interacts directly with
DNA topoisomerase II. Nat Genet. 26:349–353. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang WW, Chiu YJ, Fan MJ, Lu HF, Yeh HF,
Li KH, Chen PY, Chung JG and Yang JS: Kaempferol induced apoptosis
via endoplasmic reticulum stress and mitochondria-dependent pathway
in human osteosarcoma U-2 OS cells. Mol Nutr Food Res.
54:1585–1595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang SH, Wu LW, Huang AC, Yu CC, Lien JC,
Huang YP, Yang JS, Yang JH, Hsiao YP, Wood WG, et al: Benzyl
isothiocyanate (BITC) induces G2/M phase arrest and apoptosis in
human melanoma A375.S2 cells through reactive oxygen species (ROS)
and both mitochondria-dependent and death receptor-mediated
multiple signaling pathways. J Agric Food Chem. 60:665–675. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai KC, Huang AC, Hsu SC, Kuo CL, Yang JS,
Wu SH and Chung JG: Benzyl isothiocyanate (BITC) inhibits migration
and invasion of human colon cancer HT29 cells by inhibiting matrix
metalloproteinase-2/−9 and urokinase plasminogen (uPA) through PKC
and MAPK signaling pathway. J Agric Food Chem. 58:2935–2942. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liao CL, Lai KC, Huang AC, Yang JS, Lin
JJ, Wu SH, Wood W Gibson, Lin JG and Chung JG: Gallic acid inhibits
migration and invasion in human osteosarcoma U-2 OS cells through
suppressing the matrix metalloproteinase-2/−9, protein kinase B
(PKB) and PKC signaling pathways. Food Chem Toxicol. 50:1734–1740.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma YS, Weng SW, Lin MW, Lu CC, Chiang JH,
Yang JS, Lai KC, Lin JP, Tang NY, Lin JG, et al: Antitumor effects
of emodin on LS1034 human colon cancer cells in vitro and in vivo:
Roles of apoptotic cell death and LS1034 tumor xenografts model.
Food Chem Toxicol. 50:1271–1278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kato H, Uzawa K, Onda T, Kato Y, Saito K,
Nakashima D, Ogawara K, Bukawa H, Yokoe H and Tanzawa H:
Down-regulation of 1D-myo-inositol 1,4,5-trisphosphate 3-kinase A
protein expression in oral squamous cell carcinoma. Int J Oncol.
28:873–881. 2006.PubMed/NCBI
|
|
21
|
Mello CC and Czech MP: Micromanaging
insulin secretion. Nat Med. 10:1297–1298. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
El Ouaamari A, Baroukh N, Martens GA,
Lebrun P, Pipeleers D and van Obberghen E: miR-375 targets
3-phosphoinositide-dependent protein kinase-1 and regulates
glucose-induced biological responses in pancreatic beta-cells.
Diabetes. 57:2708–2717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Conte I, Carrella S, Avellino R, Karali M,
Marco-Ferreres R, Bovolenta P and Banfi S: miR-204 is required for
lens and retinal development via Meis2 targeting. Proc Natl Acad
Sci USA. 107:15491–15496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu
Y, Li J, Hasina R, Cheng C, Lingen MW, et al: Network modeling
identifies molecular functions targeted by miR-204 to suppress head
and neck tumor metastasis. PLOS Comput Biol. 6:e10007302010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim YJ, Bae SW, Yu SS, Bae YC and Jung JS:
miR-196a regulates proliferation and osteogenic differentiation in
mesenchymal stem cells derived from human adipose tissue. J Bone
Miner Res. 24:816–825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luthra R, Singh RR, Luthra MG, Li YX,
Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, et al:
MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of
annexin A1 downregulation in cancers. Oncogene. 27:6667–6678. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schimanski CC, Frerichs K, Rahman F,
Berger M, Lang H, Galle PR, Moehler M and Gockel I: High miR-196a
levels promote the oncogenic phenotype of colorectal cancer cells.
World J Gastroenterol. 15:2089–2096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P,
et al: A pancreatic islet-specific microRNA regulates insulin
secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi W, Yang J, Li S, Shan X, Liu X, Hua H,
Zhao C, Feng Z, Cai Z, Zhang L, et al: Potential involvement of
miR-375 in the premalignant progression of oral squamous cell
carcinoma mediated via transcription factor KLF5. Oncotarget.
6:40172–40185. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Avissar M, McClean MD, Kelsey KT and
Marsit CJ: MicroRNA expression in head and neck cancer associates
with alcohol consumption and survival. Carcinogenesis.
30:2059–2063. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Roldo C, Missiaglia E, Hagan JP, Falconi
M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et
al: MicroRNA expression abnormalities in pancreatic endocrine and
acinar tumors are associated with distinctive pathologic features
and clinical behavior. J Clin Oncol. 24:4677–4684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garzon R, Garofalo M, Martelli MP,
Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG,
Schnittger S, Haferlach T, et al: Distinctive microRNA signature of
acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin.
Proc Natl Acad Sci USA. 105:3945–3950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yekta S, Shih IH and Bartel DP:
MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vider BZ, Zimber A, Hirsch D, Estlein D,
Chastre E, Prevot S, Gespach C, Yaniv A and Gazit A: Human
colorectal carcinogenesis is associated with deregulation of
homeobox gene expression. Biochem Biophys Res Commun. 232:742–748.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kudo Y, Kitajima S, Ogawa I, Miyauchi M
and Takata T: Down-regulation of Cdk inhibitor p27 in oral squamous
cell carcinoma. Oral Oncol. 41:105–116. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu BH, Xiong XP, Jia J and Zhang WF:
MicroRNAs: New actors in the oral cancer scene. Oral Oncol.
47:314–319. 2011. View Article : Google Scholar : PubMed/NCBI
|