Introduction
Globally, colorectal cancer (CRC) is the second and
third most common causes of cancer in women (9.2% of diagnoses) and
men (10.0%), respectively (1).
Colorectal cancer death ranks fourth after lung, stomach, and liver
cancer, which is more common in developed countries (2). Treatments used for colorectal cancer
include rational combination of surgery, radiation therapy,
chemotherapy and targeted therapy.
In recent years, autophagy has become a topic of
debate in cancer (3). Autophagy is
a conserved lysosomal degradation process of intracellular
recycling and degraded metabolites, and can maintain the cellular
homeostasis and adaption to stress conditions. In cancer, autophagy
plays a complex role in tumor initiation and growth. It can either
induce cancer cell death by eliminating carcinogenic factors and
damaged organelles in the initial stage of cancer; or, promote
cancer cell survival by intracellular recycling of degraded
metabolites to cope with starvation or stress conditions in cancer
progression (4). In addition,
autophagy may contribute to drug resistance due to an adaptive
response to chemotherapy and radiation therapy (5).
The mammalian target of rapamycin (mTOR) signaling
plays a role in suppressing autophagy (6). The dysregulation of the mTOR activity
is found linking to the initiation and development of many human
tumors (7). The most established
mTOR inhibitors such as rapamycin have been evaluated as anticancer
agents, and used in the treatment of cancer in clinical trials
(8).
Rapamycin inhibits mTOR by binding the FK506 binding
protein 1A, 12 kDa (FKBP-12) (9).
However, the use of mTOR inhibitors is limited because of the
occurrence of cancer escape due to drug resistance. The mechanism
is related to oncogene Akt reactivation by negative mTOR-PI3K
feedback (10).
The use of natural plant-derived products in cancer
therapy has gained attention in recent years. Garlic, a plant
within the genus Allium, has been explored by focusing on
many organosulfur compounds (OSCs). S-allylmercaptocysteine (SAMC),
the water-soluble fraction, is one of the major part in OSCs
(11). SAMC has been studied in
various cancer cells for its antiproliferative effect (12,13),
and in vivo studies also showed tumor suppressive ability
(14,15). Moreover, SAMC is a good adjuvant in
combination with chemopreventive agents (16).
The relationship between autophagy and Nrf2 pathway
has been studied, promoting a series of antioxidant programs
(17). Also, garlic and garlic
extract has shown to facilitate Nrf2-HO-1 signaling in endothelial
(18) and B35 neural cells
(19). Nuclear factor
(erythroid-derived 2)-like 2, also known as Nrf2, is a
transcription factor that protects against oxidative damage
triggered by injury and inflammation. Nrf2 is a basic leucine
zipper (bZIP) protein that regulates a variety of detoxification
enzymes such as heme oxyegenase-1 (HO-1), NAD(P)H: quinone
oxidoreductase 1 (NQO1). In many cancer cell lines and tumor
tissues, high level of Nrf2 is detected, and Nrf2 is thought to
play dual roles in cancer cell growth and survival (20).
In this study, we used both rapamycin and SAMC as
anticancer reagents in human colon cancer cells and tumor xenograft
mice, and investigated the underlying mechanisms of tumor growth
inhibition, especially the relationship between autophagy and
Nrf2.
Materials and methods
Reagents
SAMC (purity of 99%) was synthesized and purified in
our laboratory with a modified procedure as previously reported
(21). SAMC was freshly prepared as
a stock solution in PBS for the in vitro assay and was
suspended in 10% (w/v) L-dextrose, 1% (w/v) gum Arabic
(Sigma-Aldrich, St. Louis, MO, USA) for application in mice.
Cell culture
The human colorectal cancer cell line HCT-116 was
purchased from American Type Culture Collection (ATCC, Manassas,
VA, USA). HCT-116 cells were cultured in RPMI-1640 (Hyclone, Logan,
UT, USA) supplemented with 10% fetal bovine serum (BI, Cromwell,
CT, USA), 100 U/ml of penicillin and 100 mg/ml streptomycin
(Solarbio, Beijing, China). All cells were maintained in a
humidified incubator at 37°C in 5% CO2/95% air.
Cell viability assay
A stock solution of SAMC (5 mM) was prepared fresh
in PBS. Rapamycin was dissolved in dimethyl sulfoxide (DMSO) as a
5-mM stock solution and dilutions were made in RPMI-1640. The total
DMSO concentrations were kept below 0.05% (v/v), which showed no
influence on cell growth. Cells were seeded in 96-well plates at a
concentration of 1.5×103 cells/well. After 24 h, cells
were categorized into five groups: control, rapamycin (Rapa) (0.5
µM), SAMC (200 and 400 µM) and Rapa+SAMC combination group. The two
doses of SAMC were selected based on our previous experiment (data
not shown). Cell viability was measured by the SRB method. Briefly,
the treated cells were then fixed with 10% TCA for 1 h at 4°C, the
96-well plates were washed three times with distilled water and
allowed to dry in the air. Each well was added with 100 µl of
sulphorhodamine (SRB) solution and the staining was completed at
room temperature for 15 min. The SRB stain solution was removed by
washing the plates quickly with 1% (v/v) acetic acid three times,
and the plates were dried in the air. The dried materials in each
well were solubilized by adding 200 µl of 10 mM unbuffered Tris
base (pH 10.5). The cell viability was detected by measuring the
absorbance at 540 nm on a plate reader (Safire2, Tecan, France).
All experiments were repeated at least three times.
DAPI staining
The HCT-116 cells were seeded into 24-well plates
for 24 h. Then Rapa and SAMC (200 and 400 µM) were directly added
to the well and incubated for 48 h. The treated cells were washed
with PBS and fixed with cold methanol/acetone (1:1, stored at
−20°C) for 5 min at room temperature. The solution was removed and
washed with PBS, and then incubated with the DAPI solution for 10
min at room temperature. Fluorescent cells were observed under a
fluorescence microscope (Olympus, Tokyo, Japan).
Animal experiments
Female BALB/c nude mice (16 g, aged 5–6 weeks) were
purchased from Institute of Laboratory Animal Sciences,
Cams&Pumc (Beijing, China, SCXK 2014–0004). Mice were housed
under standard conditions (12:12 h light/dark cycle at 25±2°C and
40–70% humidity) in specific pathogen-free (SPF) conditions, with
ad libitum access to food and water. The protocol of the
animal experiments were performed in accordance with the
institutional guidelines of the Animal Care and Use Committee of
Shandong University.
In vivo xenograft implantation and
tumor growth
The human colorectal adenocarcinoma HCT-116 cells
used for implantation were harvested during log phase growth and
resuspended in phosphate-buffered saline (PBS) at 5×107
cells/ml. Each Balb/c nu/nu mouse was injected s.c. in the right
flank with 1×107 cells (0.2 ml cell suspension). When
tumor volume reached ~100 mm3, mice were randomly
divided into four groups (n=6): the control, rapamycin (Rapa) (5
mg/kg/per two days), S-allylmercaptocysteine (SAMC) (300 mg/kg/d),
and Rapa + SAMC combination. All groups were treated for 28 days,
Rapa was given as intraperitoneal injection three times a week, and
SAMC was given by oral gavage daily.
Mice were carefully observed daily, and tumor
volumes and body weights were measured every four days. The
diameters of the tumors were measured with a caliper, and the tumor
volume was calculated using the formula: V = 1/2 × L ×
W2, where length (L) and width (W) were determined in
millimeters (mm). The inhibition rate (%) of tumor growth was
defined as the ratio of tumor weight to that of the control.
At the end of the experiment, all animals were
euthanized. The tumor weight and spleen, liver, kidney weight were
measured. Tumors were resected and snap-frozen in liquid nitrogen
or transferred at −80°C for western blotting and PCR.
Western blot analysis
Tumor tissues were homogenized with RIPA lysis
buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1%
sodium deoxycholate, 0.1% SDS] containing protease and phosphatase
inhibitor cocktails (Roche), at 4°C with vortexing. The total
protein concentration was determined by the BCA protein assay
reagent (Pierce Biomedical Co., Rockford, IL, USA). Tissue lysates
were separated using 12% SDS-PAGE and electro-transferred onto the
polyvinylidene difluoride (PVDF) membrane. The protein expression
levels were determined using primary antibodies with the
appropriate dilution. The PVDF membranes were washed in
Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBST) and
incubated with appropriate secondary antibodies. The immunoreactive
bands were visualized by an enhanced chemiluminescence reagent
(Millipore) using Alphalmager HP system (Cell Biosciences,
USA).
The density of each band was measured using
Image-Pro Plus, standardized by the density of β-actin. The primary
antibodies included those of anti-LC3 (L7543, Sigma-Aldrich),
anti-p62 (ab91526, Abcam), anti-Bcl-2 (ab32124, Abcam), anti-Bax
(ab32503, Abcam), anti-Nrf2 (C-20) (sc-722, Santa Cruz
Biotechnology), anti-NQO1 (ab34173, Abcam), anti-Akt (sc-8312,
Santa Cruz Biotechnology), anti-phospho-Akt/Sre-473 (sc-135651,
Santa Cruz Biotechnology), p53(ab28, Abcam) and β-actin (TA-09,
ZSGB-BIO).
Quantitative real-time (q-PCR)
Total RNA from the tumor was extracted using TRIzol
Reagent (Invitrogen Corp.). Reverse transcription reactions
(SuperScript III First-Strand Synthesis system for RT-PCR;
Invitrogen Corp., Carlsbad, CA, USA) were performed with 0.5 µg of
DNase I (Qiagen)-treated RNA. PCR and quantitative real-time PCR
(q-PCR) were carried out using the CFX96 Touch™ Real-Time PCR
Detection system (Bio-Rad, Laboratories, Inc.). Expression levels
of target genes were normalized by concurrent measurement of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels.
Primers for target gene amplification were as follows: Nrf2
(NM_010902.3): 5′-ATGATGGACTTGGAGTTGCC-3′,
5′-TCCTGTTCCTTCTGGAGTTG-3′; NQO1 (NM_008706.5):
5′-CGGTATTACGATCCTCCCTCAACA-3, 5′-AGCCTCTACAGCAGCCTCCTTCAT-3′;
GAPDH (NM_008084.2): 5′-ATGTTCCAGTATGACTCCACTCACG-3′,
5′-GAAGACACCAGTAGACTCCACGACA-3′.
Measurements of the oxidative stress
markers and antioxidant enzyme activities
A 10% liver tissue homogenate was prepared with the
phosphate buffer saline (50 mM, pH 7.4) using Teflon homogenizer. A
part of the homogenate was mixed with an equal volume of 10%
trichloroacetic acid (TCA) and centrifuged at 3,000 rpm for 15 min
at 4°C. The supernatant was used to determine the content of
glutathione (GSH-Px) and malondialdehyde (MDA) (oxidative stress
marker) enzymes. The remaining part of the homogenate was
centrifuged at 12,000 g for 45 min at 4°C and the supernatant was
used for estimation of antioxidant enzymes activities as superoxide
dismutase (SOD) and catalase (CAT) enzymes. All treatments were
conducted in an ice bath. The activities of GSH-Px, MDA, SOD and
CAT were then assayed using commercial reagent kits (Nanjing
Jiancheng Bioengineering Institute, Nanjing, China) according to
the manufacturer's instructions.
Statistical analysis
Data are shown as mean ± SD. Statistical differences
were analyzed by one-way ANOVA followed by Bonferroni test for
multiple comparisons using GraphPad Prism software. Differences
were considered significant at p<0.05. Statistical analysis was
performed with SPSS/Win 13.0 software (SPSS, Inc., Chicago, IL,
USA).
Results
Rapamycin and SAMC combination
inhibits colon cancer cell proliferation and induces apoptosis in
HCT-116 cells
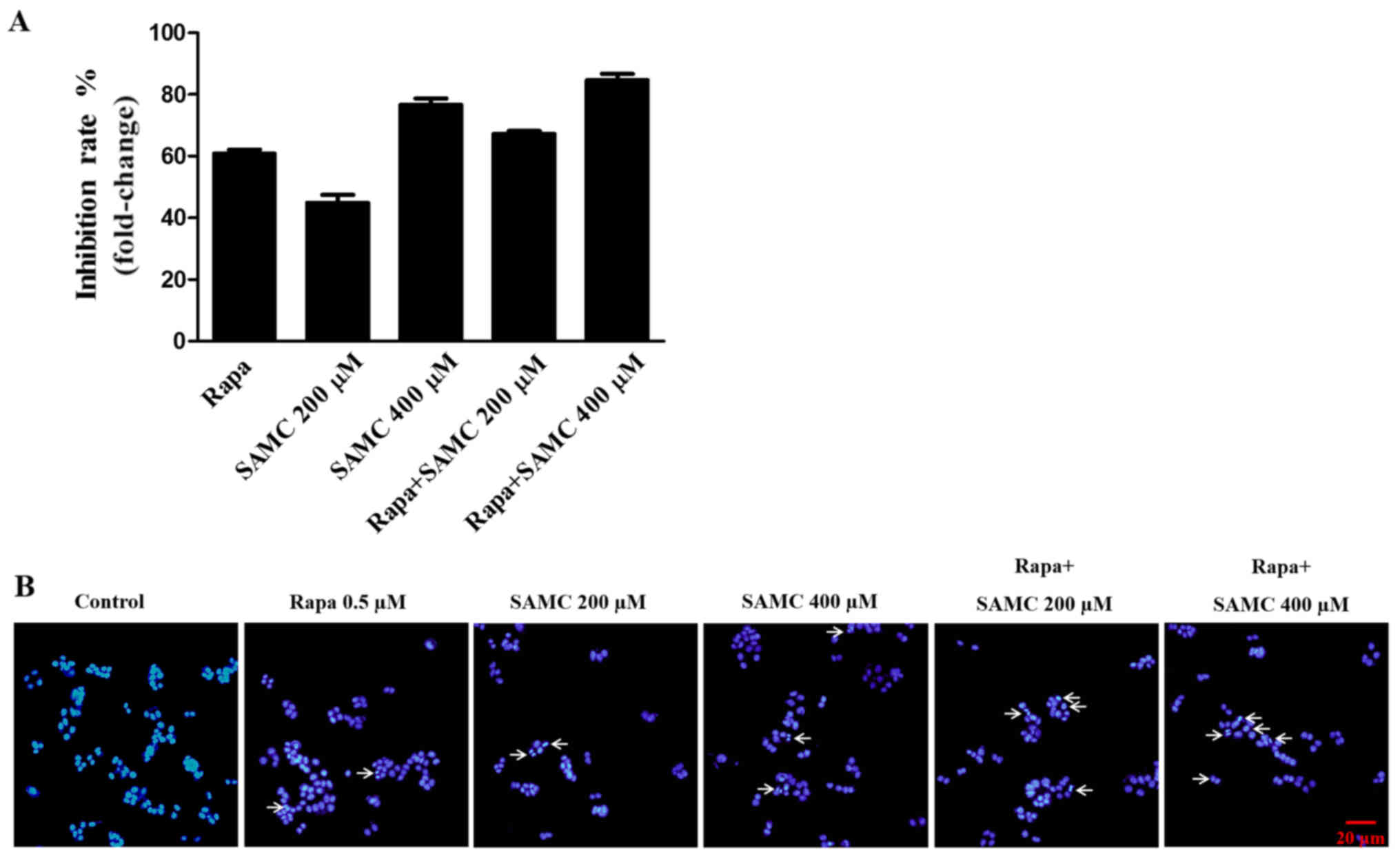
To assess the antiproliferative activity of
rapamycin, SAMC and co-treatment group, HCT-116 cells were exposed
to different concentrations (200 and 400 µM) of SAMC for 48 h and
assayed for proliferation using the SRB method. As shown in
Fig. 1A, the proliferation of
HCT-116 cells was significantly suppressed by Rapa, SAMC, and the
combination.
SAMC has been shown to induce colorectal carcinoma
cell apoptosis (22). To detect the
cell apoptosis induced by rapamycin and SAMC, the HCT-116 cells
were stained with DAPI to assess the morphological change in this
study. The cells exhibited typical morphological signs of
apoptosis, such as fragmented nuclei and apoptotic bodies, as
evidenced by the arrows in Fig.
1B.
SAMC potentiates antitumor effect of
rapamycin in mouse subcutaneous HCT-116 xenograft tumor models
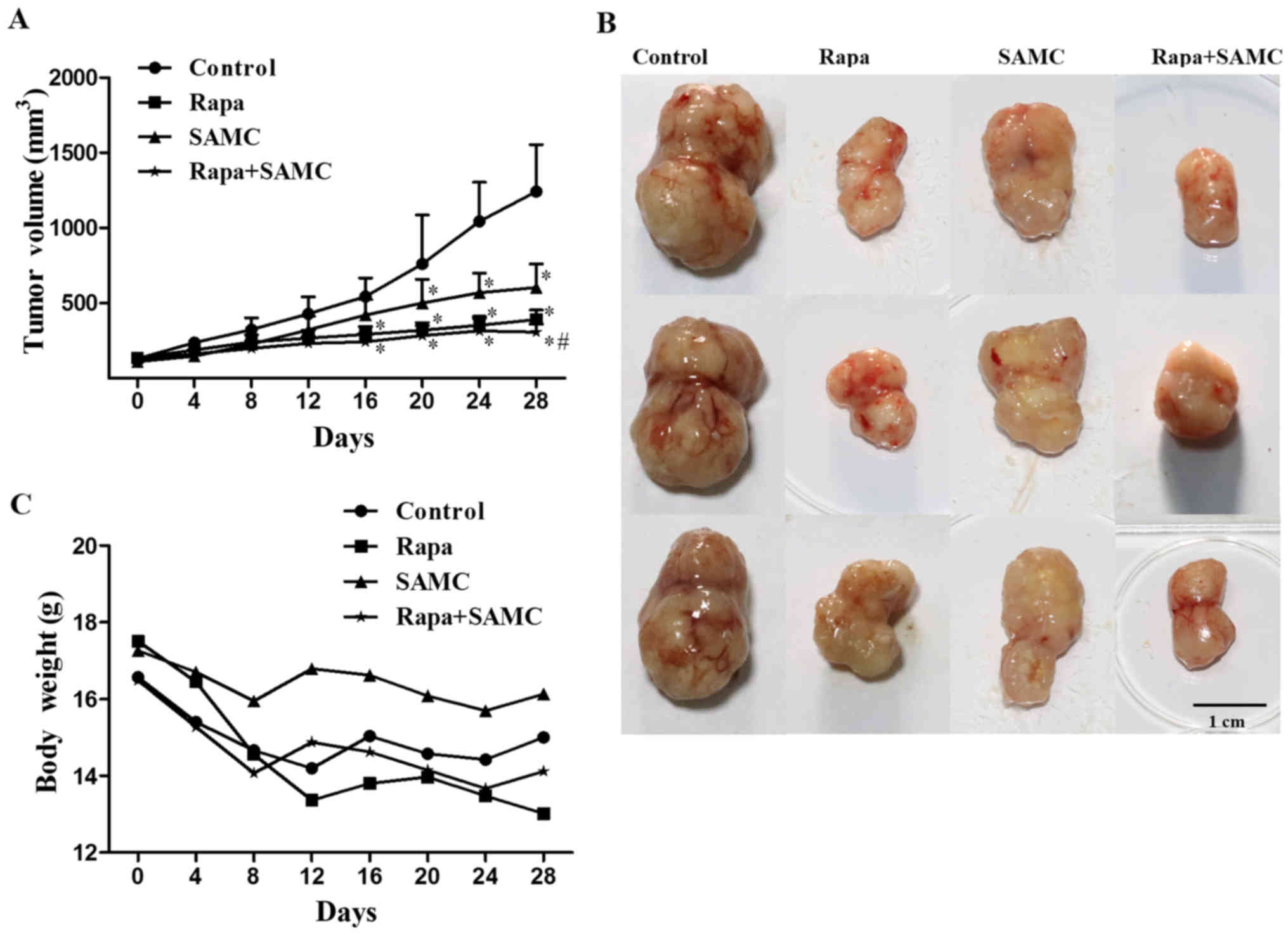
The antitumor effect of rapamycin and SAMC was
evaluated in the mouse model of HCT-116 xenografts. Compared with
the control group, Rapa treatment (10 mg/kg per day three times a
week) significantly inhibited tumor growth (Fig. 2A and B) by 63.64% (p<0.05), while
SAMC alone inhibited tumor growth by 59.09% (p<0.05),
respectively. However, the tumor growth inhibition rate of the
combination reached 80.17% (p<0.05) compared with the control.
The average tumor volumes on day 28 were 390.97±62.71,
603.42±157.42 and 307.84±53.36 mm3 in Rapa, SAMC and
combination groups, respectively, compared to 1,242.72±311.60
mm3 of control group (Table
I). The body weight of mice decreased in the control group, and
Rapa treatment showed a similar degree of body weight loss to the
control; but SAMC treatment showed no obvious weight loss.
Moreover, SAMC and Rapa combination partially reversed the weight
loss caused by Rapa (Fig. 2C).
 | Table I.Effect of rapamycin and SAMC on the
tumor growth of HCT-116 xenografts in mice. |
Table I.
Effect of rapamycin and SAMC on the
tumor growth of HCT-116 xenografts in mice.
|
| Tumor volume
(mm3) |
|
|
|---|
|
|
|
|
|
|---|
| Group | Beginning | At the end | Average tumor
weight (g) | Inhibition rate
(%) |
|---|
| Control |
130.93±25.34 |
1,242.72±311.60 |
2.42±0.59 |
|
| Rapa |
135.22±22.24 |
390.97±62.71a |
0.88±0.17a | 63.64 |
| SAMC |
110.47±17.64 |
603.42±157.42a,b |
0.99±0.14a | 59.09 |
| Rapa+SAMC |
129.79±20.92 |
307.84±53.36a,b |
0.48±0.11a,b | 80.17 |
Rapamycin and SAMC combination
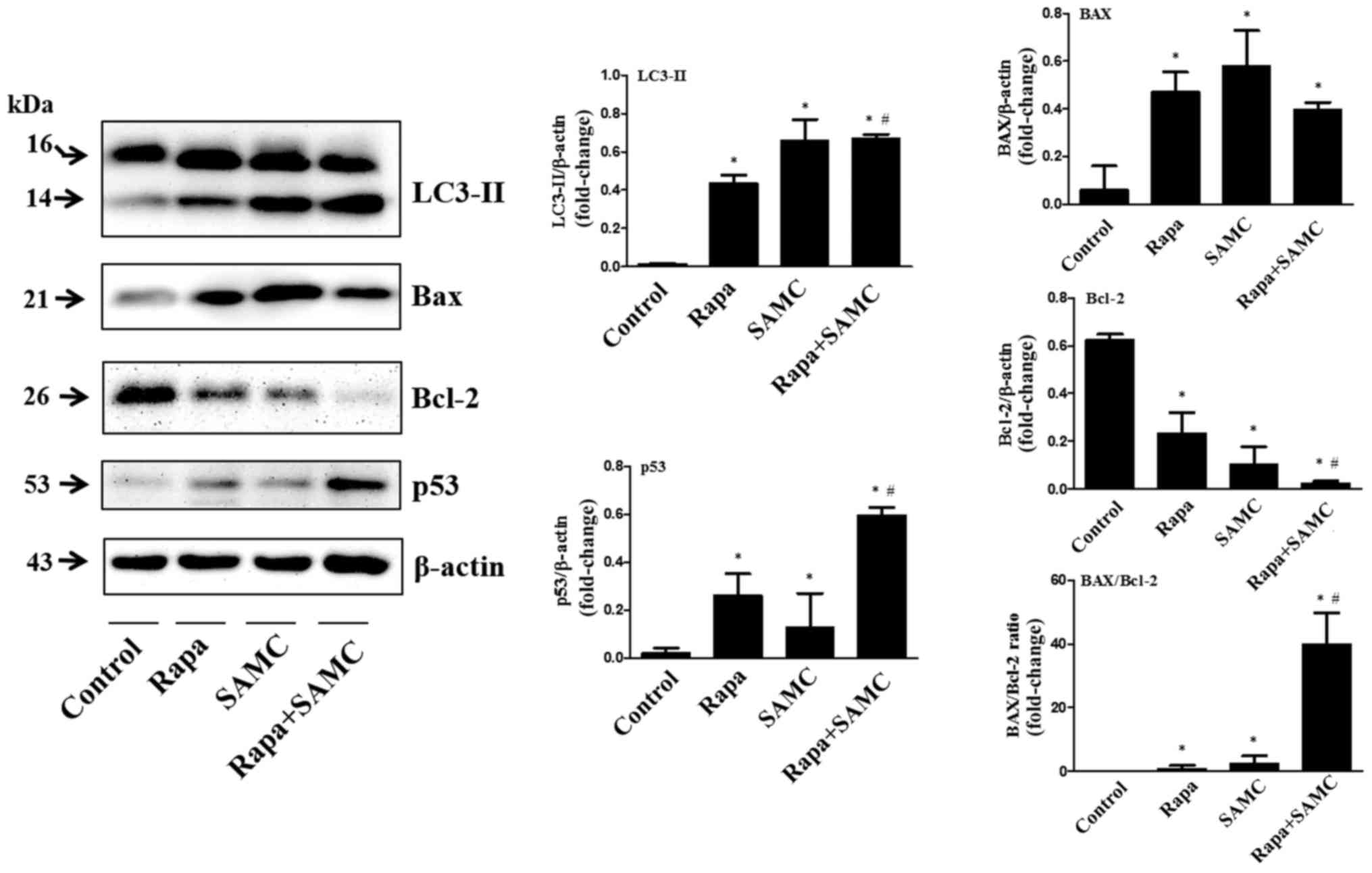
regulates autophagy process and induces apoptosis
In order to examine whether autophagy inhibition can
enhance the antitumor ability of rapamycin on colorectal cancer, we
tested the tumor tissues of HCT-116 xenografts treated with both
Rapa and SAMC. The autophagy-related protein, LC3-II, represent the
autophagic activity (23). Western
blot analysis showed that the LC3-II expression level was increased
by Rapa alone and further increased when combined with SAMC
(Fig. 3).
Apoptotic protein Bax and Bcl-2 expression levels
were examined in Rapa with/without SAMC-treated tumors. Rapa and
SAMC increased the Bax expression, while downregulated the Bcl-2
expression. The Bax/Bcl-2 ratio increased ~40-fold in the Rapa and
SAMC combination group compared to the control (Fig. 3). Tumor suppressor p53 plays a
critical role in the induction of apoptosis, which was also
examined by western blotting in this study. Rapa increased p53
expression, and further enhanced in the combination group (Fig. 3), indicating that the apoptosis may
be p53-dependent.
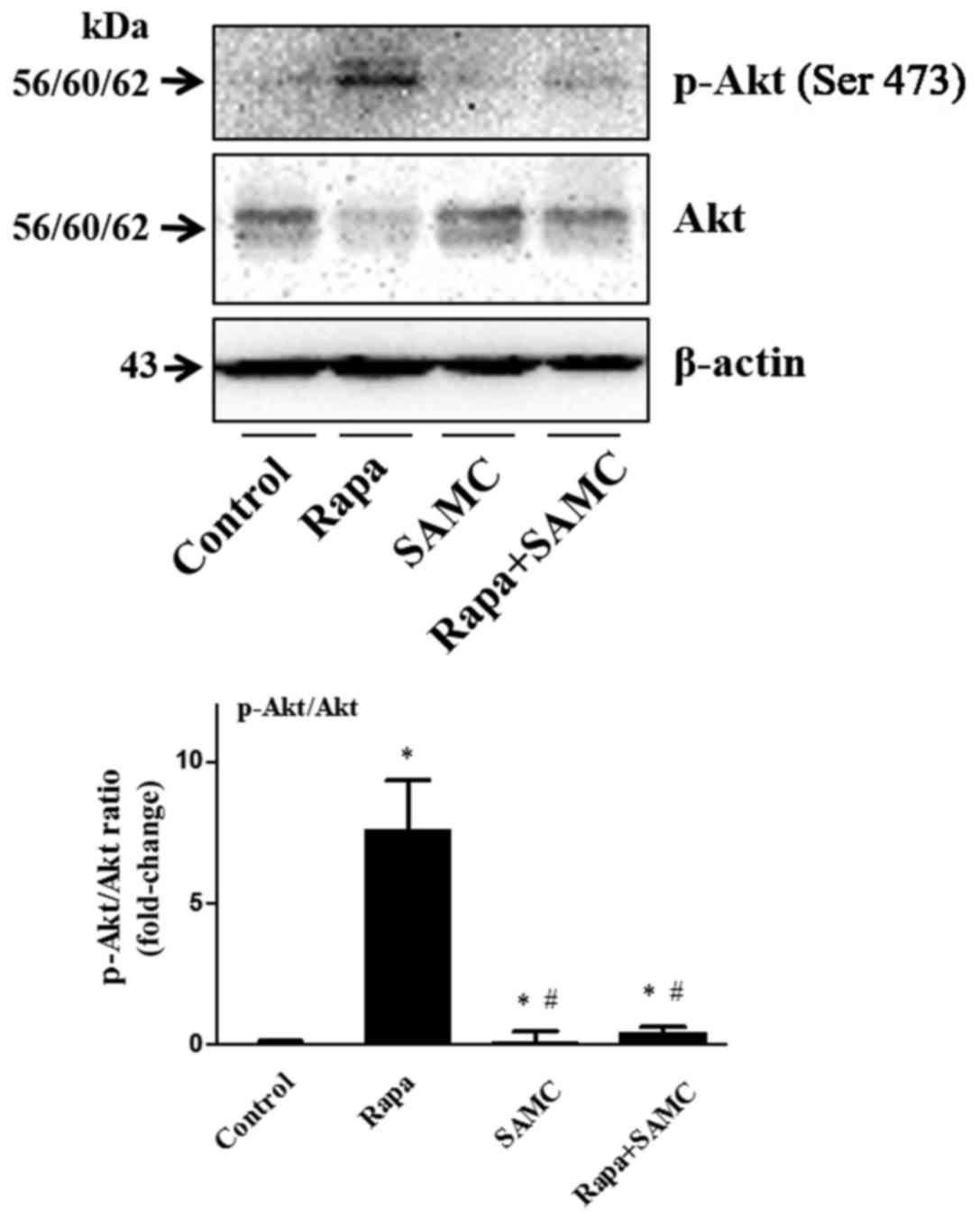
Rapamycin and SAMC combination
reverses rapamycin-induced upregulation of Akt signaling
The serine/threonine kinase mTOR is a downstream
effector of the PI3K/AKT pathway, and mTOR inhibition consequently
reactivates Akt signaling (24).
The Akt signaling activation plays a role of drug-resistance in
rapamycin-related cancer therapy, so we examined the Akt
phosphorylation in tumor tissues. Rapa treatment alone increased
Akt phosphorylation, but SAMC alone did not change the Akt status.
However, the Akt activation was inhibited in the combination group
(Fig. 4).
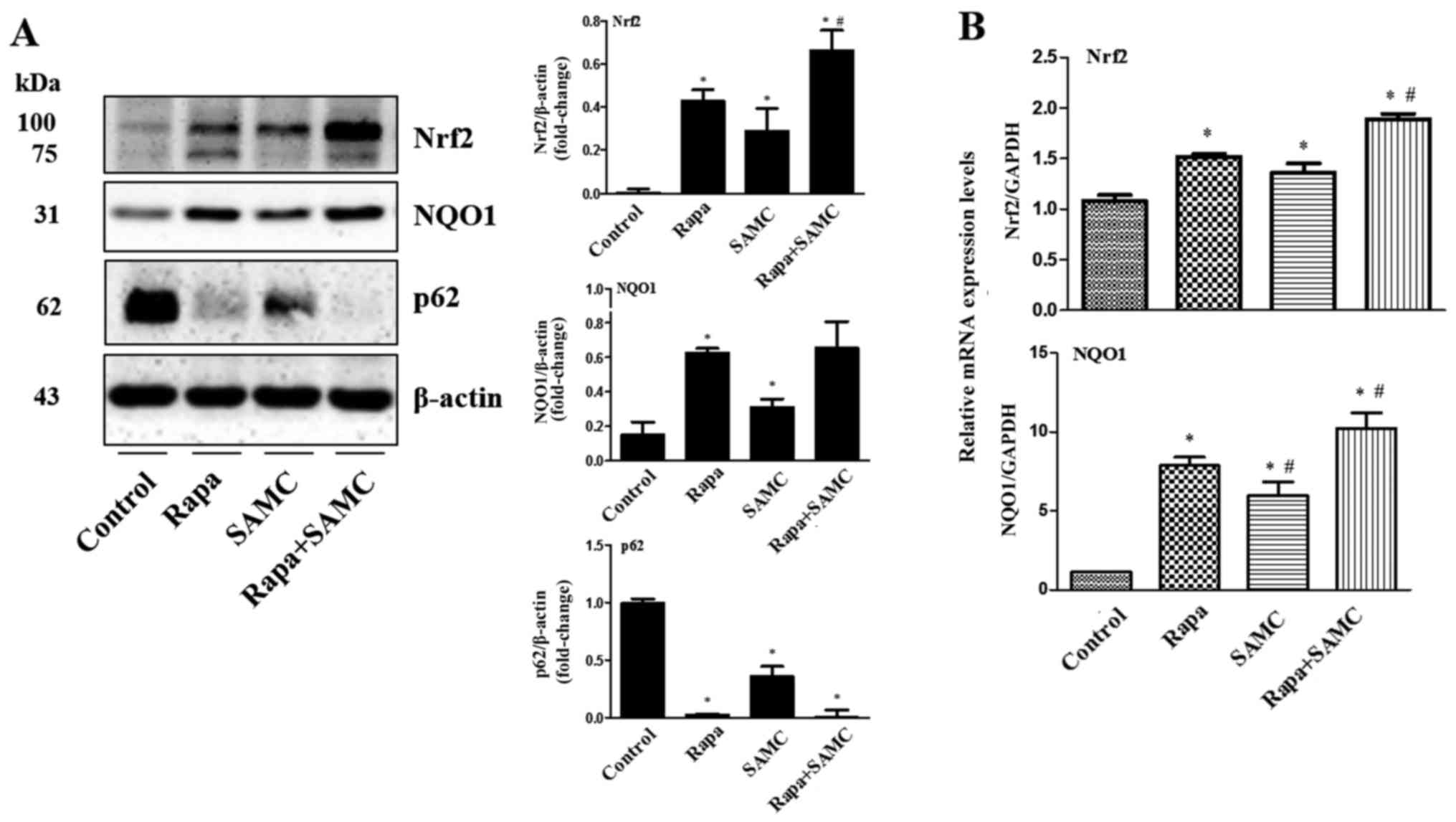
Co-treatment of rapamycin and SAMC
suppresses tumor growth by activating antioxidant system via
regulating p62
To further explore the mechanism of
tumor-suppressing effects by the combination of rapamycin and SAMC,
we examined the antioxidant transcription factor Nrf2 and its
downstream gene expression such as NQO1. In Fig. 5, western blotting and q-PCR results
show that the Nrf2 expression was elevated in the Rapa-treated
group compared to the control, and further enhanced when combined
with SAMC. While the downstream gene NQO1 expression was consistent
with Nrf2 expression.
Autophagy has been shown to suppress tumorigenesis
and the key associate point relied on p62/SQSTM, promoting
oxidative stress status (25). Our
results indicate that the Rapa, SAMC and combination groups
downregulated p62 level compared to the control (Fig. 5A).
Effects of rapamycin and SAMC on SOD,
GSH-Px, CAT and MDA
To further explore the mechanisms of rapamycin, SAMC
and combination for tumor growth inhibition, we determined the
activities of antioxidant enzyme SOD, GSH-Px and CAT as well as the
lipid peroxidation marker MDA content in mouse liver tissues. The
Rapa alone and combination treatment decreased oxidative stress as
indicated by enhanced SOD, GSH-Px and CAT contents and diminishment
of MDA level in liver tissues (Table
II). The levels of SOD, GSH-Px and CAT were increased by 43.97,
58.81 and 11.08%, respectively, and the MDA was decreased by 47.05%
in the Rapa-treated mice compared to the control group. The
antioxidant effect of the combination treatment was further
strengthened, the SOD, GSH-Px and CAT values were 287.19±4.53,
183.74±22.05 and 18.30±2.67 (U/mg protein), respectively, and MDA
content was diminished to 2.81±0.04 (nmol/mg protein), which were
significantly different from the control group.
 | Table II.SOD, GSH-Px, CAT activity and MDA
level in HCT-116 xenograft mouse liver tissue. |
Table II.
SOD, GSH-Px, CAT activity and MDA
level in HCT-116 xenograft mouse liver tissue.
| Group | SOD (U/mg
protein) | GSH-Px (U/mg
protein) | CAT (U/mg
protein) | MDA (nmol/mg
protein) |
|---|
| Control |
167.42±11.26 |
103.40±12.33 |
11.08±0.33 |
5.76±0.18 |
| Rapa |
241.04±12.27a |
164.21±10.38a |
16.20±0.38a |
3.05±0.06a |
| SAMC |
216.44±9.43a |
136.74±20.05 |
13.74±0.05a |
3.28±0.13a |
| Rapa+SAMC |
287.19±4.53a,b |
183.74±22.05a |
18.30±2.67a |
2.81±0.04a,b |
Discussion
In cancer therapy, the combination therapeutic
regimen regulating autophagic pathway has important therapeutic
effects (26). Mechanistic target
of rapamycin (mTOR) is an essential mediator in tumorigenesis, and
rapamycin has been shown to induce cancer cell death by stimulating
autophagy or apoptosis (9).
Autophagy inhibition has been examined by many research
laboratories, indicating that it may be a novel way of increasing
the efficacy of anticancer agents (27).
Garlic has been used in traditional Chinese medicine
(TCM) for centuries for its anti-inflammatory, cardiovascular
protection, and anticancer effects (28). SAMC, a water-soluble garlic
derivative, has been shown to have antiproliferation ability in
many cancer cell lines, and tumor suppressing effect under in
vivo conditions (29). Thus, we
explored the anticancer effects of SAMC and combination use with
rapamycin as well as their molecular mechanisms.
In this study, we showed that rapamycin suppressed
tumor growth in the HCT-116 xenograft mouse model, and this effect
was enhanced when combined with SAMC. In brief, the SAMC treatment
upregulated the expression level of Bax, whereas rapamycin
downregulated Bcl-2. As a consequence, the combination of rapamycin
and SAMC markedly raised the Bax/Bcl-2 ratio and significantly
increased the induction of apoptosis in colorectal cancer. The
p53-dependent apoptosis observed was in agreement with published
results that the activation of p53 mediates the upregulation of Bax
and downregulation of Bcl-2 (30).
In cancer cells and its microenvironment, hypoxia
and nutrient-deprivation condition could induce the autophagy
process. Autophagy breaks down cellular damaged organelles and
accumulates proteins for recycling, and the catabolites are
recycled and used for biosynthesis and energy-metabolism as a
cytoprotective response, which is essential for cancer cell
survival (31). SAMC has been
demonstrated to enhance autophagy in a liver disease model
(32). We tested the autophagy
activity in colorectal cancer tumor tissues. Western blotting
results show that rapamycin and SAMC treatments increased the
autophagic marker LC3-II protein expression. These results
demonstrate that co-treatment of rapamycin and SAMC in colorectal
cancer tumors can activate the autophagy pathway.
Because mTOR signaling contributes to drug
resistance in patients (33), and
one suggestion is that there is a mTOR-mediated negative feedback
loop to Akt (34), we tested the
Akt phosphorylation in colorectal cancer. The phosphorylation of
Akt (p-Akt) expression was elevated in the rapamycin treatment
group, which was reversed with the combination treatment. These
results demonstrate that SAMC could be a good adjuvant with
rapamycin in cancer treatment.
The hypoxia condition in tumors increases oxidative
stress, which will activate the master regulator of antioxidant
defense regulator Nrf2 which participates in tumor growth (35,36).
Garlic and DATS has been shown to participate in Nrf2-antioxidant
responses (37). In our study,
western blotting and q-PCR analyses both confirmed that rapamycin
increased Nrf2 expression and the downstream gene NQO-1, the
antioxidant protein expression was further enhanced with the
combination treatment, indicating that rapamycin and SAMC could
suppress tumor growth by the Nrf2 signaling pathway. Since the
interaction between Nrf2 and autophagy plays a key role in
tumorigenesis (38), we explored
the autophagosome cargo protein p62/SQSTM1 expression. Previous
study has demonstrated that autophagy can suppress tumorigenesis
through elimination of p62. Our results indicated that p62 in tumor
was downregulated by the rapamycin treatment with/without SAMC,
which was consistent with the recent report that Nrf2-Keap1 binding
competed p62 (39) for autophagy
degradation (40). So, the
Nrf2-Keap1 system activated by rapamycin and SAMC co-treatment in
HCT-116 xenograft mice could be related to autophagy through p62,
which is a key pathway in tumorigenesis and cancer therapy.
It is well-known that levels of SOD, GSH-Px, CAT and
MDA are considered as common indexes of tissue antioxidant status
(41). The MDA level is widely used
as a marker of lipid peroxidation damage. Furthermore, inhibition
of antioxidant SOD, MDA and GSH enzymes is implicated in the
pathogenesis conditions. SAMC has been reported to reduce oxidative
stress and inflammation in liver-injury mice (42). Our study results show that rapamycin
with/without SAMC treatment effectively increased the liver
antioxidative capacity and alleviated the oxidative stress
conditions.
In conclusion, these results showed that the
rapamycin and SAMC combination induced apoptosis, activation of
autophagy, and downregulation of p62. Additionally, this
combination reversed the oxidative stress condition by activating
antioxidative transcriptor Nrf2 and downstream gene NQO1 as well as
increased SOD, GSH-Px, and CAT activities and decreased the MDA
level in liver tissues. Therefore, combining rapamycin and SAMC for
the treatment of colorectal cancer might be feasible in clinical
use. The underlying mechanism of autophagy/p62/Nrf2 pathway
revealed in this study may provide a new direction for drug
development.
Acknowledgements
This study was supported by the funds from National
‘Major Science and Technology Project-Prevention and Treatment of
AIDS, Viral Hepatitis, and Other Major Infectious Diseases’ (grant
no. 2013ZX10005004), Major Project of Science and Technology of
Shandong Province (grant no. 2015ZDJS04001), Science and Technology
Enterprise Technology Innovation Fund of Jiangsu Province (grant
no. BC2014172), Small and Medium Enterprise Technology Innovation
Project of Lianyungang City (grant no. CK1333).
Glossary
Abbreviations
Abbreviations:
|
SAMC
|
S-allylmercaptocysteine
|
|
mTOR
|
mammalian target of rapamycin
|
|
OSCs
|
organosulfur compounds
|
|
Nrf2
|
nuclear factor (erythroid-derived
2)-like 2
|
|
HO-1
|
heme oxyegenase-1
|
|
NQO1
|
NAD(P)H: quinone oxidoreductase 1
|
|
TCM
|
traditional Chinese medicine
|
References
|
1
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ouakrim D Ait, Pizot C, Boniol M, Malvezzi
M, Boniol M, Negri E, Bota M, Jenkins MA, Bleiberg H and Autier P:
Trends in colorectal cancer mortality in Europe: Retrospective
analysis of the WHO mortality database. BMJ. 351:h49702015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
White E: The role for autophagy in cancer.
J Clin Invest. 125:42–46. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
White E: Deconvoluting the
context-dependent role for autophagy in cancer. Nat Rev Cancer.
12:401–410. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu YL, Jahangiri A, Delay M and Aghi MK:
Tumor cell autophagy as an adaptive response mediating resistance
to treatments such as antiangiogenic therapy. Cancer Res.
72:4294–4299. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim YC and Guan KL: mTOR: A pharmacologic
target for autophagy regulation. J Clin Invest. 125:25–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu K, Liu P and Wei W: mTOR signaling in
tumorigenesis. Biochim Biophys Acta. 1846:638–654. 2014.PubMed/NCBI
|
|
8
|
Mita MM, Mita A and Rowinsky EK: The
molecular target of rapamycin (mTOR) as a therapeutic target
against cancer. Cancer Biol Ther. 2:(Suppl 1). S169–S177. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abraham RT and Gibbons JJ: The mammalian
target of rapamycin signaling pathway: Twists and turns in the road
to cancer therapy. Clin Cancer Res. 13:3109–3114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun SY, Rosenberg LM, Wang X, Zhou Z, Yue
P, Fu H and Khuri FR: Activation of Akt and eIF4E survival pathways
by rapamycin-mediated mammalian target of rapamycin inhibition.
Cancer Res. 65:7052–7058. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rahman K and Lowe GM: Garlic and
cardiovascular disease: A critical review. J Nutr. 136:(Suppl).
736S–740S. 2006.PubMed/NCBI
|
|
12
|
Lee Y: Induction of apoptosis by
S-allylmercapto-L-cysteine, a biotransformed garlic derivative, on
a human gastric cancer cell line. Int J Mol Med. 21:765–770.
2008.PubMed/NCBI
|
|
13
|
Hu H, Zhang XP, Wang YL, Chua CW, Luk SU,
Wong YC, Ling MT, Wang XF and Xu KX: Identification of a novel
function of Id-1 in mediating the anticancer responses of SAMC, a
water-soluble garlic derivative, in human bladder cancer cells. Mol
Med Rep. 4:9–16. 2011.PubMed/NCBI
|
|
14
|
Howard EW, Ling MT, Chua CW, Cheung HW,
Wang X and Wong YC: Garlic-derived S-allylmercaptocysteine is a
novel in vivo antimetastatic agent for androgen-independent
prostate cancer. Clin Cancer Res. 13:1847–1856. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee Y, Kim H, Lee J and Kim K: Anticancer
activity of S-allylmercapto-L-cysteine on implanted tumor of human
gastric cancer cell. Biol Pharm Bull. 34:677–681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shirin H, Pinto JT, Kawabata Y, Soh JW,
Delohery T, Moss SF, Murty V, Rivlin RS, Holt PR and Weinstein IB:
Antiproliferative effects of S-allylmercaptocysteine on colon
cancer cells when tested alone or in combination with sulindac
sulfide. Cancer Res. 61:725–731. 2001.PubMed/NCBI
|
|
17
|
Zhang L, Li J, Ma J, Chen X, Chen K, Jiang
Z, Zong L, Yu S, Li X, Xu Q, et al: The relevance of Nrf2 pathway
and autophagy in pancreatic cancer cells upon stimulation of
reactive oxygen species. Oxid Med Cell Longev. 2016:38972502016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hiramatsu K, Tsuneyoshi T, Ogawa T and
Morihara N: Aged garlic extract enhances heme oxygenase-1 and
glutamate-cysteine ligase modifier subunit expression via the
nuclear factor erythroid 2-related factor 2-antioxidant response
element signaling pathway in human endothelial cells. Nutr Res.
36:143–149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu XH, Li GL, Wang BA, Qin Y, Bai SR, Rong
J, Deng T and Li Q: Diallyl trisufide protects against oxygen
glucose deprivation -induced apoptosis by scavenging free radicals
via the PI3K/Akt -mediated Nrf2/HO-1 signaling pathway in B35
neural cells. Brain Res. 1614:38–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Menegon S, Columbano A and Giordano S: The
dual roles of NRF2 in cancer. Trends Mol Med. 22:578–593. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Starkenmann C, Niclass Y and Troccaz M:
Nonvolatile S-alk(en)ylthio-L-cysteine derivatives in fresh onion
(Allium cepa L. cultivar). J Agric Food Chem. 59:9457–9465.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Li HY, Zhang ZH, Bian HL and Lin
G: Garlic-derived compound S-allylmercaptocysteine inhibits cell
growth and induces apoptosis via the JNK and p38 pathways in human
colorectal carcinoma cells. Oncol Lett. 8:2591–2596.
2014.PubMed/NCBI
|
|
23
|
Tanida I, Minematsu-Ikeguchi N, Ueno T and
Kominami E: Lysosomal turnover, but not a cellular level, of
endogenous LC3 is a marker for autophagy. Autophagy. 1:84–91. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Willems L, Tamburini J, Chapuis N, Lacombe
C, Mayeux P and Bouscary D: PI3K and mTOR signaling pathways in
cancer: New data on targeted therapies. Curr Oncol Rep. 14:129–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mathew R, Karp CM, Beaudoin B, Vuong N,
Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al:
Autophagy suppresses tumorigenesis through elimination of p62.
Cell. 137:1062–1075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alayev A, Berger SM, Kramer MY, Schwartz
NS and Holz MK: The combination of rapamycin and resveratrol blocks
autophagy and induces apoptosis in breast cancer cells. J Cell
Biochem. 116:450–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amaravadi RK, Lippincott-Schwartz J, Yin
XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT and White E:
Principles and current strategies for targeting autophagy for
cancer treatment. Clin Cancer Res. 17:654–666. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thomson M and Ali M: Garlic [Allium
sativum]: A review of its potential use as an anticancer agent.
Curr Cancer Drug Targets. 3:67–81. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang D, Qin Y, Zhao W, Zhai X, Guo Z,
Wang R, Tong L, Lin L, Chen H, Wong YC, et al:
S-allylmercaptocysteine effectively inhibits the proliferation of
colorectal cancer cells under in vitro and in vivo conditions.
Cancer Lett. 310:69–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang XW: Role of p53 and apoptosis in
carcinogenesis. Anticancer Res. 19A:4759–4771. 1999.
|
|
31
|
Viry E, Paggetti J, Baginska J,
Mgrditchian T, Berchem G, Moussay E and Janji B: Autophagy: An
adaptive metabolic response to stress shaping the antitumor
immunity. Biochem Pharmacol. 92:31–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao J, Guo R, Fung ML, Liong EC, Chang
RC, Ching YP and Tipoe GL: Garlic-derived S-Allylmercaptocysteine
ameliorates nonalcoholic fatty liver dsease in a rat model through
inhibition of apoptosis and enhancing autophagy. Evid Based
Complement Alternat Med. 2013:6429202013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang BH and Liu LZ: Role of mTOR in
anticancer drug resistance: Perspectives for improved drug
treatment. Drug Resist Updat. 11:63–76. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feldman ME and Shokat KM: New inhibitors
of the PI3K-Akt-mTOR pathway: Insights into mTOR signaling from a
new generation of Tor Kinase Domain Inhibitors (TORKinibs). Curr
Top Microbiol Immunol. 347:241–262. 2010.PubMed/NCBI
|
|
35
|
Leinonen HM, Kansanen E, Pölönen P,
Heinäniemi M and Levonen AL: Role of the Keap1-Nrf2 pathway in
cancer. Adv Cancer Res. 122:281–320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moon EJ and Giaccia A: Dual roles of NRF2
in tumor prevention and progression: Possible implications in
cancer treatment. Free Radic Biol Med. 79:292–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Padiya R, Chowdhury D, Borkar R, Srinivas
R, Bhadra M Pal and Banerjee SK: Garlic attenuates cardiac
oxidative stress via activation of PI3K/AKT/Nrf2-Keap1 pathway in
fructose-fed diabetic rat. PLoS One. 9:e942282014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen N, Wu L, Yuan H and Wang J:
ROS/Autophagy/Nrf2 pathway mediated low-dose radiation induced
radio-resistance in human lung adenocarcinoma A549 Cell. Int J Biol
Sci. 11:833–844. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Komatsu M, Kurokawa H, Waguri S, Taguchi
K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et
al: The selective autophagy substrate p62 activates the stress
responsive transcription factor Nrf2 through inactivation of Keap1.
Nat Cell Biol. 12:213–223. 2010.PubMed/NCBI
|
|
40
|
Jiang T, Harder B, de la Rojo Vega M, Wong
PK, Chapman E and Zhang DD: p62 links autophagy and Nrf2 signaling.
Free Radic Biol Med. 88B:199–204. 2015. View Article : Google Scholar
|
|
41
|
Cigremis Y, Turel H, Adiguzel K, Akgoz M,
Kart A, Karaman M and Ozen H: The effects of acute acetaminophen
toxicity on hepatic mRNA expression of SOD, CAT, GSH-Px, and levels
of peroxynitrite, nitric oxide, reduced glutathione, and
malondialdehyde in rabbit. Mol Cell Biochem. 323:31–38. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiao J, Liong EC, Ling MT, Ching YP, Fung
ML and Tipoe GL: S-allylmercaptocysteine reduces carbon
tetrachloride-induced hepatic oxidative stress and
necroinflammation via nuclear factor kappa B-dependent pathways in
mice. Eur J Nutr. 51:323–333. 2012. View Article : Google Scholar : PubMed/NCBI
|