Introduction
Oral squamous cell carcinoma (OSCC) accounts for
>90% of oral malignancies, with ~250,000 new cases each year
worldwide (1,2). Approximately two-thirds of these cases
occur in developing countries, especially in the South and
Southeast Asia. Despite consistent efforts of therapy development
including surgery, chemotherapy and radiotherapy, the prognosis of
OSCC is still poor because of tumor recurrence and metastasis
(3). Hence, it is important to seek
potential therapeutic biomarkers of OSCC, which are critical to
early prediction and treatment evaluation (4).
Runt-related transcription factor 3 (RUNX3), one
member of RUNX family of DNA-binding transcription factors, has
been shown closely related to tumorigenesis (5). The inactivation of RUNX3 was observed
in various human cancer types, such as esophageal (6), gastric (7), colorectal (8), lung (9) and breast cancer (10), caused by promoter hypermethylation
and protein mislocalization. The restoration of RUNX3 could inhibit
the metastasis and angiogenesis of many cancer cells in
vitro (11–14). These studies supported that RUNX3
acted as a tumor suppressor. However, other studies provided
evidence that RUNX3 may have an oncogenic role in head and neck
squamous cell carcinoma (HNSCC) (15,16)
and skin cancers (15).
In OSCC, the RUNX3 function as either an oncogene or
a tumor suppressor gene is controversial. Tanji et al found
higher labeling indexes (LIs) of RUNX3 expression in OSCC tissues
compared with that in the normal epithelia, but they also pointed
out that RUNX3 LIs correlated with the histological grades of OSCC,
being the highest in the well differentiated OSCC (17). On the other hand, Gao et al
discovered that the expression of RUNX3 protein was markedly
reduced in OSCC specimens compared with the matched adjacent normal
tissues. Furthermore, they found that only 14.7% of OSCC samples
(22 of 150) showed a normal nuclear localization of RUNX3 protein
(18). All these findings suggested
the function of RUNX3 and the underlying mechanism in oral
carcinogenesis are still unclear.
The objectives of the present study were to examine
the RUNX3 staining in 232 OSCC samples using tissue microarray
(TMA) technology and analyze its correlation with
clinicopathological parameters. Also, we tried to clarify the
potential molecular mechanisms.
Materials and methods
Ethics statement
This study was performed under a protocol approved
by the Institutional Review Boards of Affiliated Stomatological
Hospital of Nanjing Medical College and all examinations were
performed after obtaining written informed consents.
Patients and samples
A total of 232 OSCC patients hospitalized in
Department of Oral and Maxillofacial Surgery, Stomatological
Hospital of Nanjing Medical College from January 2008 to January
2014 were included in this study. None of the patients received
preoperative radiotherapy or chemotherapy. Each patient was
pathologically diagnosed as squamous cell carcinoma and graded
according to WHO criteria by two pathologists. All patients were
followed up at least 24 months and the complete clinicopathological
data including recurrence and metastasis were collected.
These OSCC samples were sent to Jiangsu Key
Laboratory of Biological Cancer Therapy, Xuzhou Medical College
(Xuzhou, China) to manufacture the OSCC TMA. The array dot diameter
was 1.5 mm, and each dot represented a tissue spot from one
individual specimen that was selected and pathologically
confirmed.
Immunohistochemistry of OSCC TMA
Immunohistochemistry of OSCC TMA was performed with
the streptavidin-peroxidase (Sp) method using a standard Sp kit
(Zhongshan Biotech, Beijing, China). The TMA slide was incubated
with monoclonal mouse anti-RUNX3 antibody (1:250, Abcam, USA)
overnight at 4°C, and diaminobenzidine (DAB; Zhongshan Biotech,
China) was used to produce a brown precipitate. Negative controls
were obtained by substituting primary antibodies with non-immune
serum. The immunoreactivity was assessed blindly by two independent
observers using Zeiss Imager Z1 microscope, and the image was
collected by AxioCam MRc5 camera. The expression of RUNX3 was
graded as positive when 5% of tumor cells showed immunopositivity,
while the biopsies with <5% were considered negative (18).
Cell lines and transfection
Human OSCC cell lines HN6 and Cal27 were purchased
from the Shanghai Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences (Shanghai, China). Human umbilical
vascular endothelial cells (HUVECs) were obtained from KeyGen
Biotech (Nanjing, China). HN6 and Cal27 cells were cultured in
RPMI-1640 medium supplemented with 10% fetal calf serum
(Invitrogen, Shanghai, China), and HUVECs were cultured in DMEM
medium supplemented with 10% fetal calf serum. Cells were placed at
37°C in humidified incubator with 95% air, 5% CO2.
The pFlag-control and pFlag-RUNX3 expression
plasmids were obtained from Jiangsu Key Laboratory of Biological
Cancer Therapy, Xuzhou Medical College (Xuzhou, China).
Transfection of the pFlag-control and the pFlag-RUNX3 plasmids into
HN6 and Cal27 was carried out using Lipofectamine 2000 transfection
reagent (Invitrogen) following the manufacturer's protocol.
Scratch wound healing assay
Cells were seeded in 6-well plates at a density of
5×104 cells/well and cultured to confluence. The cell
monolayer was serum-free starved overnight. Confluent cell
monolayer was then scraped with a yellow pipette tip for scratch
wounds and washed twice with PBS. After 24-h incubation, the cell
images were captured in the same position. The wound areas were
evaluated by AxioVision 4.8 software.
Migration assay
Cell migration was determined using Transwell
migration assay (8-µm pore size; Cell Biolabs). Cells were seeded
into the upper chamber in serum-free medium at a density of
5×104. After a 12-h incubation at 37°C, cells in the
upper chamber were carefully removed with a cotton swab. Cells
traversed the membrane were fixed in methanol and stained with
leucocrystal violet. Images were taken with an inverted microscope.
Five random selected fields of view were captured and the invasive
cells were counted.
Invasion assay
The Transwell filter inserts were coated with
Matrigel (BD Biosciences, NJ, USA). Transfected 0.5×105
HN6 cells and 1×105 Cal27 cells were seeded in
serum-free medium in the upper chamber. After a 24-h incubation at
37°C, cells at the top of the Matrigel were gently removed with a
cotton swab. Invasive cells at the bottom of Matrigel were fixed in
methanol, stained with leucocrystal violet and counted.
Cell proliferation assay
Transfected HN6 and Cal27 cells (1×106)
were cultured in 6-well plate with serum-free medium for 24 h. The
medium was collected as a conditioned medium. Cellular
proliferation was performed using Cell counting kit-8 (CCK-8)
purchased from Beyotime Institute of Biotechnology (Nanjing,
China). Briefly, 2×104 HUVECs were suspended in
conditioned medium or vehicle control and seeded at a density of
2×104 per well in a 96-well plate. After incubation at
37°C for 24 h, cell proliferation was detected according to the
manufacturer's instructions.
Endothelial cell tube formation
assay
HUVECs were starved overnight and then seeded at a
density of 2×104 per well in a 48-well plate pre-coated
with Matrigel. After incubation at 37°C for 30 min, the medium was
replaced with conditioned medium or vehicle control. After a 24-h
incubation, three random selected fields of view were captured.
Quantified evaluation of tube formation was obtained from measuring
the length of tube-like structures. The experiments were repeated
three times.
Western blot analysis
Cells were washed with PBS and lysed in
radioimmunoprecipitation lysis buffer (Beyotime, China) containing
1 mM phenylmethylsulfonyl fluoride based on the manufacturer's
instructions. The cell protein (10 µg) was separated on a 12%
SDS-polyacrylamide gel. The protein was then transferred to
nitrocellulose membrane and incubated overnight at 4°C with the
following antibodies: mouse anti-RUNX3 (1:2,000, Abcam), mouse
anti-VEGF (1:1,000, Abcam), rabbit anti-MMP9 and mouse anti-β-actin
(1:1,000, Boster Biotechnology, China). Membranes were then washed
in PBS and incubated with secondary antibody (goat anti-rabbit and
goat anti-mouse IgG) for 2 h and then washed in PBST (PBS
containing Tween-20) at room temperature. After that, the membrane
was stained by coloration fluid which contains 10 ml alkaline
phosphatase buffer, 33 µl BCIP, and 66 µl NBT. Finally, the protein
bands were detected using SuperSignal West Pico Chemiluminescent
Substrate (Thermo Fisher Scientific, Waltham, MA, USA) and exposed
to Kodak X-ray film. Three independent trials of each experiment
were carried out.
ELISA for MMP-9 and VEGF
HN6 and Cal27 cells were seeded in 6-well plates at
a density of 1×106 cells per well. Then, cells were
transfected with pFlag-control and pFlag-RUNX3 with serum-free
medium. The supernatants were collected 24 h after transfection.
MMP-9 and VEGF concentration were determined using Quantikine ELISA
kits according to the manufacturer's instructions (R&D Systems,
MN, USA).
Statistical analysis
Statistical analysis was performed with SPSS 20
software (SPSS Inc., IL, USA) and data were expressed as the mean ±
SD. The associations between RUNX3 staining and the
clinicopathologic parameters were evaluated by two-sided Fisher's
exact and χ2 tests. The correlation between statue of
RUNX3 expression and patient's survival was analyzed by
Kaplan-Meier survival analysis. For migration and invasion assays,
endothelial cell tube formation and CCK-8 cell proliferation
assays, and data of western blotting and ELISA, Student's t-test
was used. Differences were considered significant when
P<0.05.
Results
Aberrant expression of RUNX3 protein
in OSCC
We first determined whether RUNX3 expression was
changed in human OSCC. Immunohistochemistry of the OSCC TMA was
performed to detect the expression of RUNX3 protein and the
distribution of RUNX3-positive cells. We observed RUNX3 staining
displayed different patterns: 93 (40.1%) cases presented with
positive nuclear staining and 85 (36.6%) cases with positive
cytoplasmic staining, and only 54 (23.2%) cases showed
underexpression of RUNX3 protein with undetectable or low levels of
staining (Fig. 1A). The cytoplasmic
staining, termed as ‘RUNX3 mislocalization’ in the cytoplasm,
suggested RUNX3 protein was inactive in a non-functional form as a
tumor suppressor (7,10). Both underexpression and
mislocalization of RUNX3 protein were aberrant and represented
RUNX3 gene impairment in cancer cells; hence we analyzed these two
patterns together as negative expression.
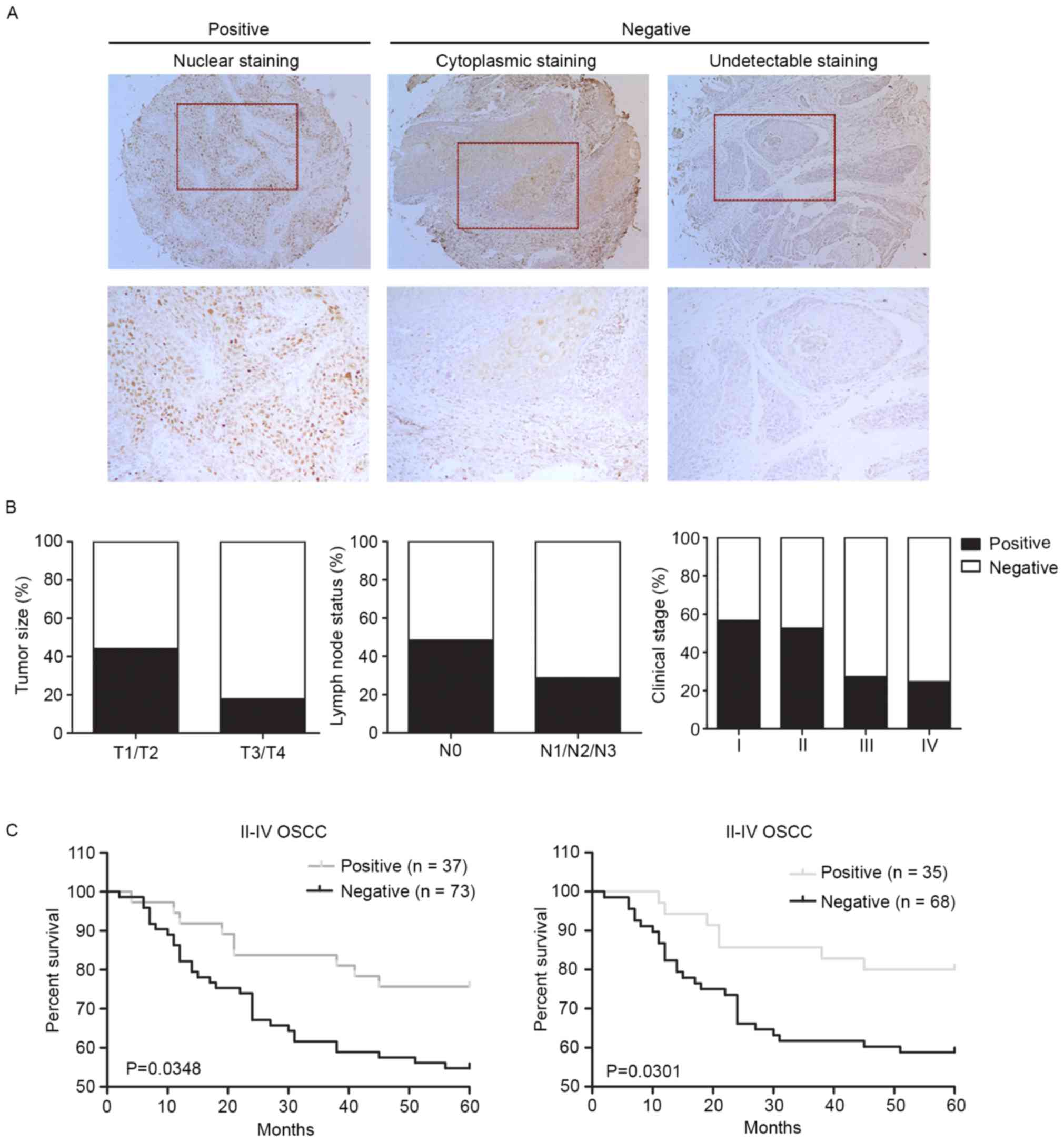 | Figure 1.Negative expression of RUNX3 is
associated with tumor size, lymph node metastasis and clinical
stage, and worsens patients' survival. (A) Representative images of
RUNX3 immunohistochemical staining in OSCC tissues. Left panel,
positive expression of nuclear staining in OSCC tissue. Middle
panel, negative expression of cytoplasmic staining in OSCC tissue.
Right panel, negative expression of undectectable staining in OSCC
tissue. Top panel, magnification, ×100. Bottom panel,
magnification, ×200. (B) Reduced RUNX3 expression correlates with
high tumor size (P=0.0021, χ2 test), lymph node
metastasis (P<0.0001, χ2 test), (P=0.000, by
two-sided Fisher's exact test). (C) Negative RUNX3 expression
correlates with a poorer 5-year overall survival (P=0.0349,
log-rank test) and 5-year disease-specific survival (P=0.0301,
log-rank test) for 110 II–IV OSCC patients. |
Correlation of RUNX3 expression with
clinicopathological parameters
Based on TMA results, we further investigated the
relationship between RUNX3 expression and the clinicopathological
parameters of OSCC. We found that positive expression of RUNX3 was
more frequent in T1/T2 compared with T3/T4 cases (P=0.0021). Also,
we found that negative expression of RUNX3 was significantly
correlated with lymph node statue and clinical stage (P<0.0001
and P=0.000, respectively). The correlations between expression of
RUNX3 with other clinicopathological variables, including patient
age, sex, invasion stage and histological grade, were not
significant (Fig. 1B and Table I).
 | Table I.RUNX3 expression and
clinicopathological characteristics of 232 OSCCs. |
Table I.
RUNX3 expression and
clinicopathological characteristics of 232 OSCCs.
|
| RUNX3 staining |
|
|
|---|
|
|
|
|
|
|---|
| Variables | Negative (%) | Positive (%) | Total |
P-valuea |
|---|
| Age |
| ≥60
years | 75
(60.0) | 50 (40.0) | 125 |
1.0 |
| <60
years | 64
(59.8) | 43 (40.2) | 107 |
|
| Sex |
|
Male | 82
(58.2) | 59 (41.8) | 141 |
0.5833 |
|
Female | 57
(62.6) | 34 (37.4) | 91 |
|
| Tumor size |
| T1 and
T2 | 107 (55.4) | 86 (44.6) | 193 |
0.0021 |
| T3 and
T4 | 32
(82.2) | 7
(17.8) | 39 |
|
| Lymph node
status |
| N0 | 62
(48.4) | 66 (51.6) | 128 | <0.0001 |
| N1, 2
and 3 | 77
(74.0) | 27 (26.0) | 104 |
|
| Invasion
status |
|
Yes | 128 (61.8) | 79 (38.2) | 207 |
0.1290 |
| No | 11
(44.0) | 14 (56.0) | 25 |
|
| Histological
grade |
| I | 70
(53.8) | 60 (46.2) | 130 |
0.1030 |
| II | 53
(67.9) | 25 (32.1) | 78 |
|
|
III | 16
(66.7) | 8
(33.3) | 24 |
|
| Clinical stage |
| I | 33
(43.4) | 43 (56.6) | 76 |
0.0001 |
| II | 19
(47.5) | 21 (52.5) | 40 |
|
|
III | 40
(72.7) | 15 (27.3) | 55 |
|
| IV | 46
(77.0) | 14 (23.0) | 61 |
|
Negative expression of RUNX3
correlates with poor patient survival
We then analyzed whether the RUNX3 expression was
associated with the survival of patients by Kaplan-Meier survival
analysis. The cases of clinical stage I were excluded because of
favorable prognosis and then 110 cases (among 156 cases of clinical
stage II–IV) with at least 60 months follow-up were chosen for
prognosis analysis. Our data revealed that negative expression of
RUNX3 in OSCC correlated with both 5-year overall and
disease-specific patient survival (P=0.0348 and P=0.0301,
respectively, log-rank test; Fig.
1C).
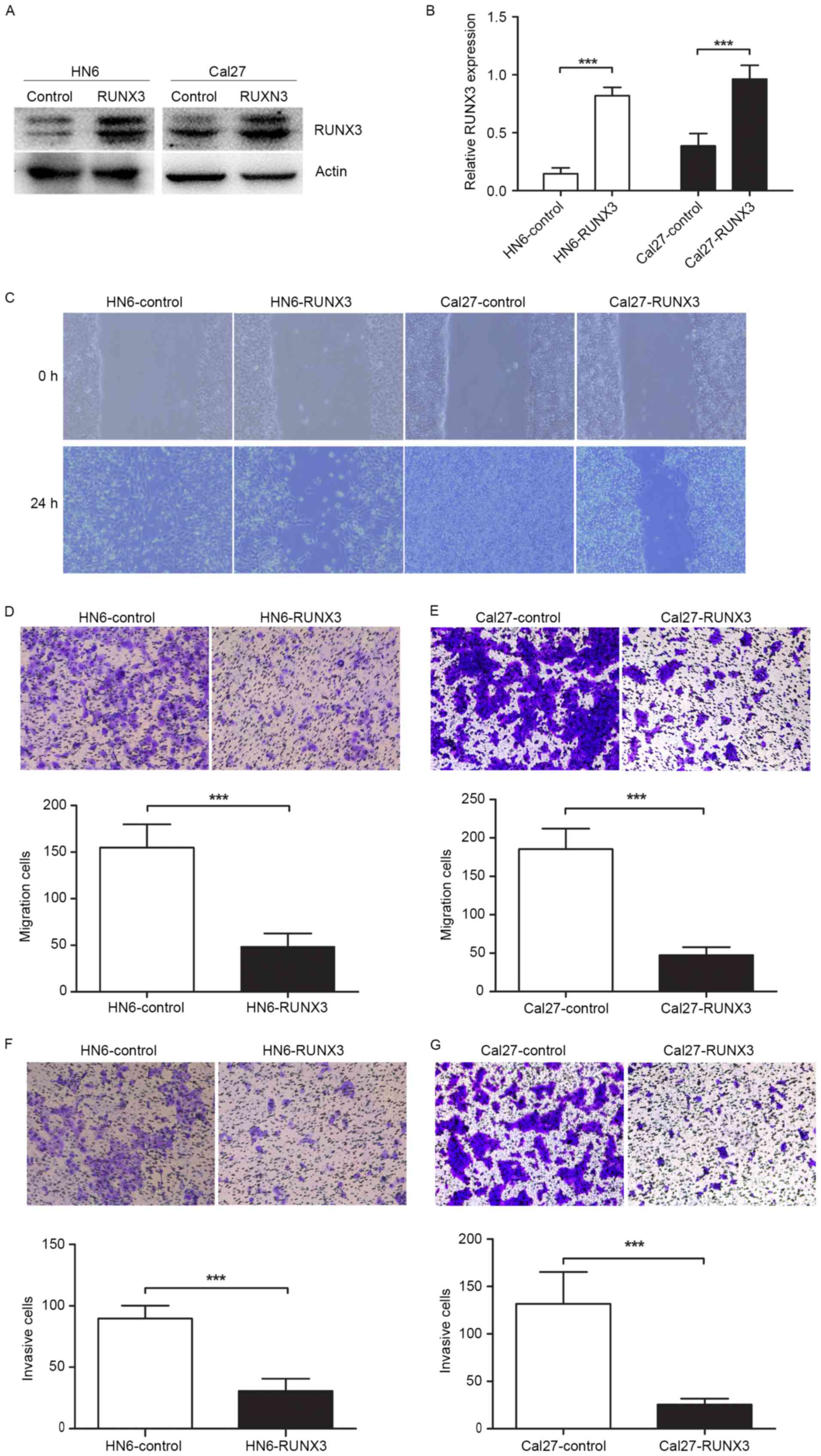
Inhibition of cells migration and
invasion in RUNX3 transfected OSCC cells in vitro
To determine the effects of RUNX3 restoration on
OSCC cell migration and invasion, we transiently transfected HN6
and Cal27 cells with pFlag-control and pFlag-RUNX3 plasmids.
Twenty-four hours after transfection, RUNX3 protein was
significantly overexpressed in cancer cells (Fig. 2A and B). The protein detection of
RUNX3 by western blotting showed two bands, and this could be due
to protein mislocalization, which was reported in a previous study
(7). The results of wound healing
and Transwell assays showed that RUNX3 tansfected cells showed less
migration ability compared with vehicle control (Fig. 2C-E). Moreover, the invasion assay
proved that RUNX3 restoration inhibited cell invasive ability of
HN6 and Cal27 cells in Matrigel-coated Transwell (Fig. 2F and G).
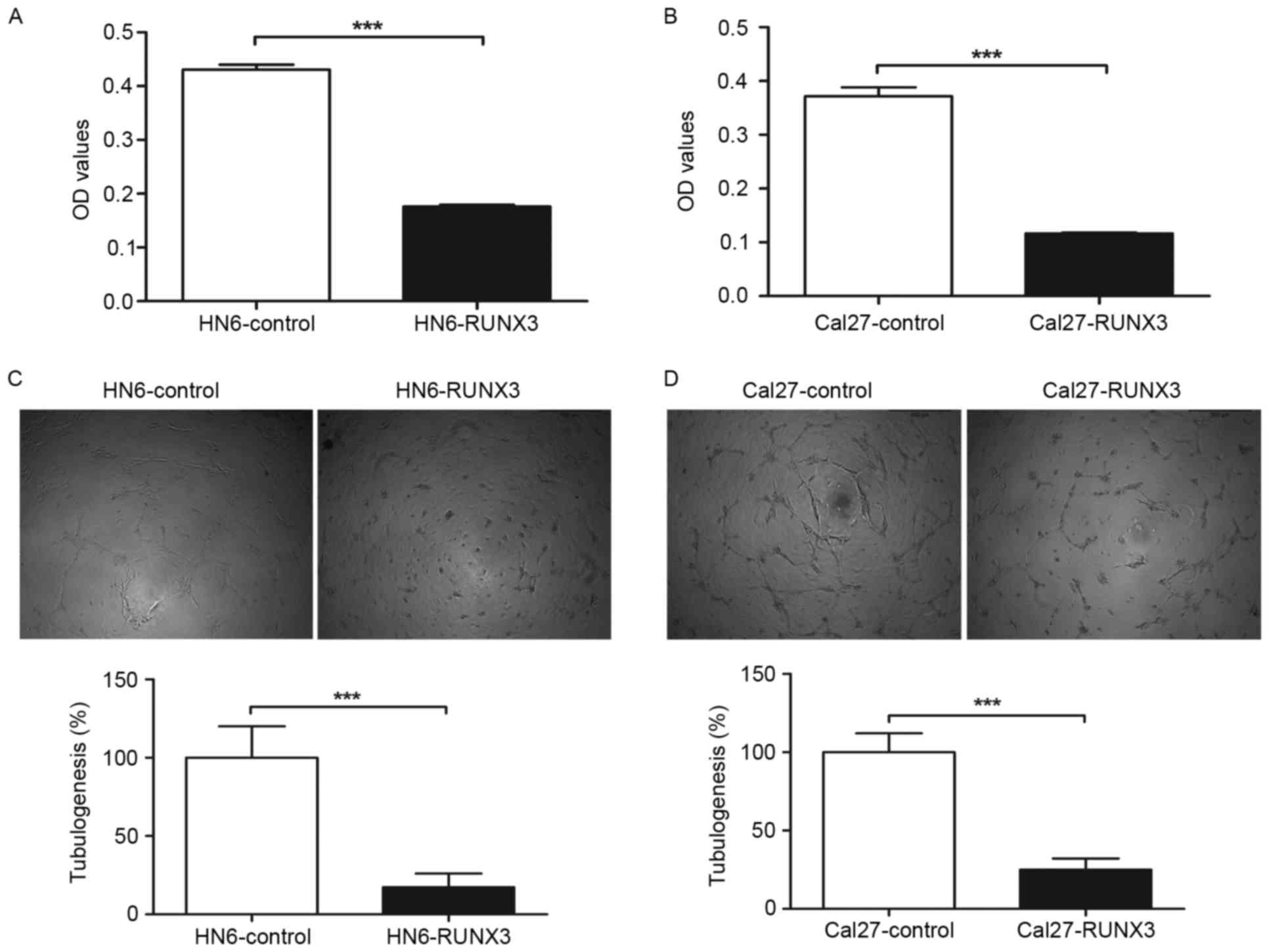
Reduction of HUVEC proliferation and
tube formation with conditioned medium from RUNX3 transfected OSCC
cells in vitro
To further determine the effect of restored RUNX3
expression on angiogenic potential of human OSCC cells, we used the
supernatant of HN6 and Cal27 cells as conditioned medium to culture
the endothelial cell line HUVECs. We observed that conditioned
medium from HN6 and Cal27 cells transfected with pFlag-RUNX3
inhibited proliferation of HUVECs compared with those of
pFlag-control transfected cells (Fig.
3A and B). In tube formation assay, the average tube length of
HUVECs cultured with supernatant from pFlag-RUNX3 transfected HN6
and Cal27 cells was significantly decreased in contrast with that
of vehicle control, as shown in Fig. 3C
and D.
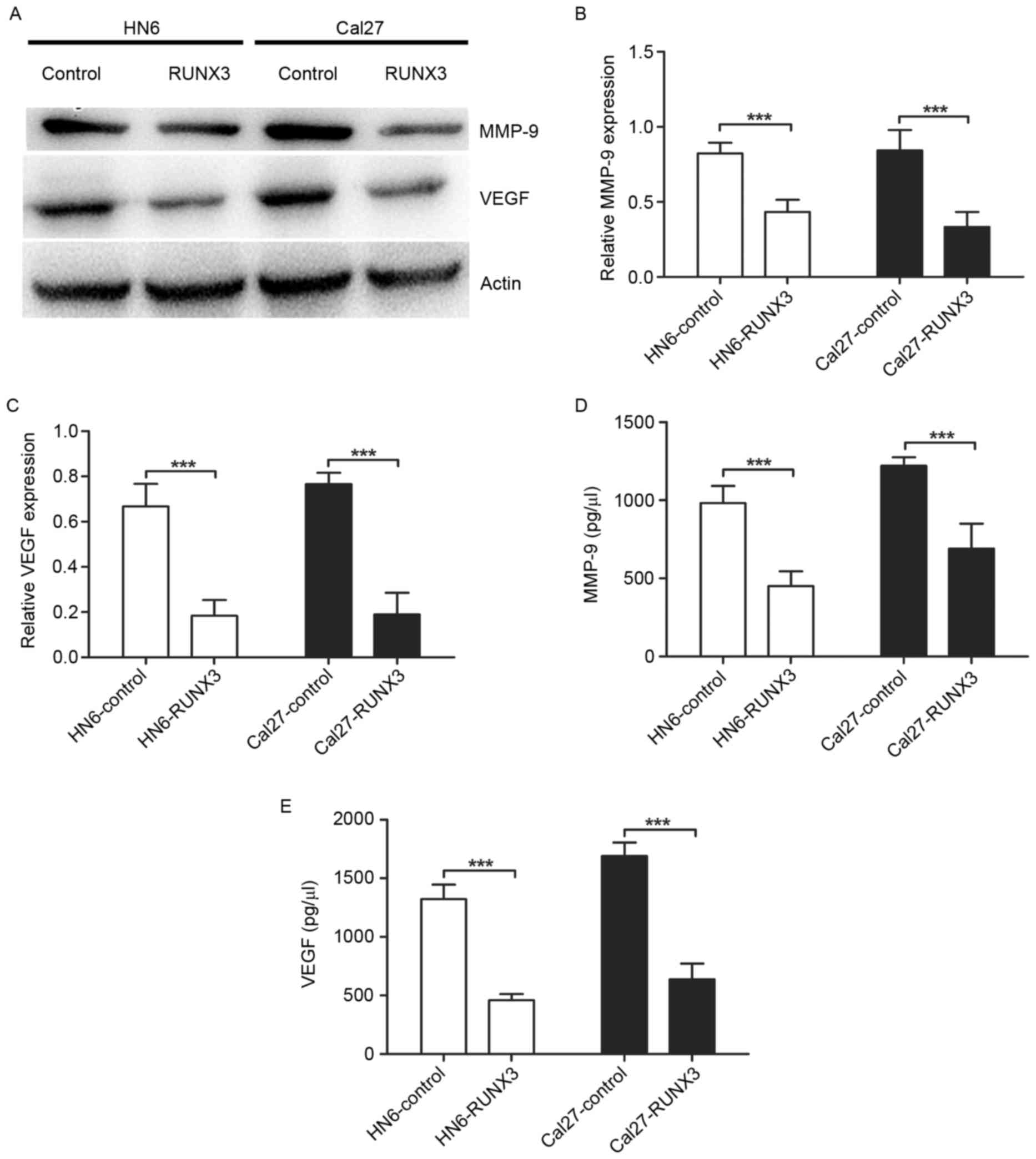
Suppression of MMP-9 and VEGF
expression and secretion in RUNX3 transfected OSCC cells
To investigate the mechanisms of RUNX3 regulating
invasion and angiogenesis, we performed western blotting and ELISA
to detect the MMPs and VEGF levels in OSCC. We found expression of
MMP-9 and VEGF protein was downregulated significantly in HN6 and
Cal27 cells transfected with pFlag-RUNX3 (Fig. 4A-C). Also, the restoration of RUNX3
in HN6 and Cal27 cells led to a significant reduction of MMP-9 and
VEGF secretion in conditioned medium (Fig. 4D and E).
Discussion
This study examined the status of RUNX3 expression
in OSCC by TMA technology and found RUNX3 was inactivated in 139 of
232 OSCC specimens. Among these 139 cases, RUNX3 protein could be
underexpressed (54 cases) or mislocalized in the cytoplasm (85
cases). RUNX3 was frequently inactivated by two mechanisms of
protein mislocalization and promoter hypermethylation, which had
been proved in many cancers including OSCC (7,10,18–20).
In OSCC, Gao et al (18)
reported that 30% OSCC specimens presented with exclusive RUNX3
cytoplasmic retention, and 55.3% showed underexpression of RUNX3.
Also, both of these aberrant RUNX3 statues were correlated with
tumor differentiation, but not with local lymph node metastasis
(18). Supic et al found
that RUNX3 gene promoter hypermethylation was significantly
associated with lymph node involvement and tumor stage, but not
with the overall survival of tongue carcinoma (20). In the present study, we analyzed
these two types together as negative expression of RUNX3, which was
significantly correlated with tumor size, lymph node metastasis and
clinical stage, but not with tumor differentiation. All these
findings strongly supported the notion that RUNX3 acted as a tumor
suppressor in OSCC.
Loss of RUNX3 expression was proved to worsen poor
survival in breast, gastric cancer, and esophageal cancer (11,21,22).
These studies indicated that the loss of RUNX3 expression may
contribute to tumor metastasis. In the present study, negative
expression of RUNX3 was associated with some clinical aspects which
were crucial to OSCC prognosis. Based on the follow-up analysis,
the data of 110 cases with at least 5-year follow-up demonstrated
that negative expression of RUNX3 worsened the patient survival.
Therefore, RUNX3 could be a potential prognostic factor of
OSCC.
Cumulated studies proved that the restoration of
RUNX3 could inhibit malignant behavior of various cancer cells
(12,13). RUNX3 is a downstream target of
transforming growth factor-β (TGF-β) mediated tumor suppressor
pathway, the key regulator of cellular proliferation, invasion and
migration in a variety of different cancer types (23). Moreover, RUNX3 is a negative
modulator of Wnt (wingless type)/β-catenin signaling pathway, which
is involved in the cancer initiation and malignant transformation
in a wide range of human cancers, including OSCC (24). In our previous studies, we reported
that attenuation of RUNX3 expression could suppress the cell
migration and invasion ability in breast, renal tumor and glioma
(11,14,25).
In the present study, we confirmed that these effects of RUNX3
restoration were equally important on OSCC cell lines. The
abilities of migration and invasion were significantly inhibited by
reintroduction of RUNX3 in HN6 and Cal27 cells.
There are reports that TGF-β signaling pathway could
influence cancer cells migration and invasion by regulating matrix
metalloproteinases (MMPs) such as MMP-2 and MMP-9 (26). Being a key downstream molecule of
TGF-β signaling pathway, RUNX3 may have the potential role in
controlling MMPs which has been reported to participate closely in
tumor progression (27). RUNX3
overexpression inhibited cell migration and invasion by
downregulation of MMP-9 expression in human esophageal squamous
cell carcinoma (13). We previously
determined the relationship between RUNX3 and MMP-2 in glioma and
breast cancer (11,14), as well as MMP-9 in renal cancer
cells (25). Here, we found
downregulation of MMP-9 protein and decreased secretion of MMP-9 in
HN6 and Cal27 cells transfected with RUNX3. Our data indicated that
RUNX3 may suppress invasion and migration through downregulating
MMP-9 expression and secretion in OSCC cell lines in
vitro.
Besides the inhibition of invasion and migration,
RUNX3 played the role of tumor suppressor through antiangiogenesis
(12,25,28).
In the present study, we found that supernatant from HN6 and Cal27
cells transfected with RUNX3 had less ability to induce HUVEC
proliferation and tube formation compared with that from vehicle
control. These results implied that the angiogenic potential of
OSCC cells was reduced by RUNX3 restoration. Furthermore, we
detected the expression and secretion of VEGF, which had been
proved to be a vital angiogenic factor of tumor blood vessel
formation in solid tumors (29).
The relationship between RUNX3 and VEGF was determined in previous
studies (12,25). In western blotting and ELISA
analysis, our results also demonstrated that VEGF expression and
secretion was decreased by restoration of RUNX3 in OSCC cell lines
in vitro.
Recently, the role of RUNX3 as an oncogene was
promoted in skin cancer and HNSCC (15,16,30).
Tsunematsu et al presumed this distinct oncogenic role of
RUNX3 attributed to the pathogenesis of skin cancer and HNSCC, both
of which arose from squamous epithelium. Also, the functional
mutation or protein mislocalization of RUNX3 was not found in HNSCC
cells (15). However,
hypermethylation of RUNX3 gene promoter was observed in 35% of
tongue cancer, one type of OSCC origin from squamous epithelium,
and was significantly associated with lymph node involvement and
tumor stage (20). In addition, a
reduced expression and protein mislocalization of RUNX3 were
observed in esophageal squamous cell carcinoma (13,31).
Our TMA results demonstrated that protein mislocalization of RUNX3
was frequent in OSCC tissues. These studies indicated that the
function of RUNX3 may be different even in the same pathogenesis of
cancer cells. Further experiments are required to clarify this
phenomenon.
In conclusion, our results verified that reduced
RUNX3 expression significantly correlated with tumor size, local
lymph node metastasis and clinical stage, and predicted poor
prognosis in OSCC. RUNX3 regulated OSCC cancer cell migration,
invasion and angiogenesis through suppressing MMP-9 and VEGF
expression and activity. Our clinical and mechanistic data
indicated that RUNX3 played a tumor suppressor role in OSCC, and
targeting of RUNX3 pathway could be a potential therapy for
OSCC.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81672678), The Project Funded by the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (PAPD, 2014-37), The Project Funded by Jiangsu
Provincial Commission of Health and Family Planning (Z201511), and
The Open Project of Jiangsu Key Laboratory of Oral Diseases
(JSKLOD-KF-1501).
References
|
1
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lambert R, Sauvaget C, de Camargo Cancela
M and Sankaranarayanan R: Epidemiology of cancer from the oral
cavity and oropharynx. Eur J Gastroenterol Hepatol. 23:633–641.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weatherspoon DJ, Chattopadhyay A,
Boroumand S and Garcia I: Oral cavity and oropharyngeal cancer
incidence trends and disparities in the United States: 2000–2010.
Cancer Epidemiol. 39:497–504. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ghantous Y, Yaffi V and Abu-Elnaaj I: Oral
cavity cancer: Epidemiology and early diagnosis. Refuat Hapeh
Vehashinayim. 32(55–63): 712015.(In Hebrew).
|
|
5
|
Ito Y: Oncogenic potential of the RUNX
gene family: ‘Overview’. Oncogene. 23:4198–4208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tonomoto Y, Tachibana M, Dhar DK, Onoda T,
Hata K, Ohnuma H, Tanaka T and Nagasue N: Differential expression
of RUNX genes in human esophageal squamous cell carcinoma:
Downregulation of RUNX3 worsens patient prognosis. Oncology.
73:346–356. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada
K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al: RUNX3, a
novel tumor suppressor, is frequently inactivated in gastric cancer
by protein mislocalization. Cancer Res. 65:7743–7750. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soong R, Shah N, Peh BK, Chong PY, Ng SS,
Zeps N, Joseph D, Salto-Tellez M, Iacopetta B and Ito Y: The
expression of RUNX3 in colorectal cancer is associated with disease
stage and patient outcome. Br J Cancer. 100:676–679. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu GP, Ji Y, Chen GQ, Huang B, Shen K, Wu
S and Shen ZY: Application of RUNX3 gene promoter methylation in
the diagnosis of non-small cell lung cancer. Oncol Lett. 3:159–162.
2012.PubMed/NCBI
|
|
10
|
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito
K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al: RUNX3 is
frequently inactivated by dual mechanisms of protein
mislocalization and promoter hypermethylation in breast cancer.
Cancer Res. 66:6512–6520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bai J, Yong HM, Chen FF, Song WB, Li C,
Liu H and Zheng JN: RUNX3 is a prognostic marker and potential
therapeutic target in human breast cancer. J Cancer Res Clin Oncol.
139:1813–1823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim BR, Kang MH, Kim JL, Na YJ, Park SH,
Lee SI, Kang S, Joung SY, Lee SY, Lee DH, et al: RUNX3 inhibits the
metastasis and angiogenesis of colorectal cancer. Oncol Rep.
36:2601–2608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen H, Wang Z, Wang S, Zhang Z and Shi S:
Effect and mechanism of RUNX3 gene on biological characteristics of
human esophageal squamous cell carcinoma (ESCC). Med Oncol.
32:3572015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mei PJ, Bai J, Liu H, Li C, Wu YP, Yu ZQ
and Zheng JN: RUNX3 expression is lost in glioma and its
restoration causes drastic suppression of tumor invasion and
migration. J Cancer Res Clin Oncol. 137:1823–1830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsunematsu T, Kudo Y, Iizuka S, Ogawa I,
Fujita T, Kurihara H, Abiko Y and Takata T: RUNX3 has an oncogenic
role in head and neck cancer. PLoS One. 4:e58922009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kudo Y, Tsunematsu T and Takata T:
Oncogenic role of RUNX3 in head and neck cancer. J Cell Biochem.
112:387–393. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanji Y, Osaki M, Nagahama Y, Kodani I,
Ryoke K and Ito H: Runt-related transcription factor 3 expression
in human oral squamous cell carcinomas; implication for tumor
progression and prognosis. Oral Oncol. 43:88–94. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao F, Huang C, Lin M, Wang Z, Shen J,
Zhang H, Jiang L and Chen Q: Frequent inactivation of RUNX3 by
promoter hypermethylation and protein mislocalization in oral
squamous cell carcinomas. J Cancer Res Clin Oncol. 135:739–747.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subramaniam MM, Chan JY, Soong R, Ito K,
Ito Y, Yeoh KG, Salto-Tellez M and Putti TC: RUNX3 inactivation by
frequent promoter hypermethylation and protein mislocalization
constitute an early event in breast cancer progression. Breast
Cancer Res Treat. 113:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Supic G, Kozomara R, Jovic N, Zeljic K and
Magic Z: Hypermethylation of RUNX3 but not WIF1 gene and its
association with stage and nodal status of tongue cancers. Oral
Dis. 17:794–800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi M, Wang Z, Liu XY and Chen D:
Inactivation of RUNX3 predicts poor prognosis in esophageal
squamous cell carcinoma after Ivor-Lewis esophagectomy. Med Oncol.
31:3092014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng HC, Liu YP, Shan YS, Huang CY, Lin
FC, Lin LC, Lee L, Tsai CH, Hsiao M and Lu PJ: Loss of RUNX3
increases osteopontin expression and promotes cell migration in
gastric cancer. Carcinogenesis. 34:2452–2459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramaniam MM, Chan JY, Yeoh KG, Quek T,
Ito K and Salto-Tellez M: Molecular pathology of RUNX3 in human
carcinogenesis. Biochim Biophys Acta. 1796:315–331. 2009.PubMed/NCBI
|
|
24
|
Romana PG: WITHDRAWN: Cell alterations and
molecular mechanisms in oral carcinogenesis. Int J Oral Maxillofac
Surg. Aug 4–2010.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen F, Bai J, Li W, Mei P, Liu H, Li L,
Pan Z, Wu Y and Zheng J: RUNX3 suppresses migration, invasion and
angiogenesis of human renal cell carcinoma. PLoS One. 8:e562412013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krstic J and Santibanez JF: Transforming
growth factor-beta and matrix metalloproteinases: Functional
interactions in tumor stroma-infiltrating myeloid cells. Sci World
J. 2014:5217542014. View Article : Google Scholar
|
|
27
|
Martin MD and Matrisian LM: The other side
of MMPs: Protective roles in tumor progression. Cancer Metastasis
Rev. 26:717–724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng Z, Wei D, Wang L, Tang H, Zhang J, Le
X, Jia Z, Li Q and Xie K: RUNX3 inhibits the expression of vascular
endothelial growth factor and reduces the angiogenesis, growth, and
metastasis of human gastric cancer. Clin Cancer Res. 12:6386–6394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cao EG, Wang E, Pal K, Dutta SK, Bar-Sagi
D and Mukhopadhyay D: VEGF exerts an angiogenesis-independent
function in cancer cells to promote their malignant progression.
Cancer Res. 72:3912–3918. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS,
Kang SG, Kim CH, Lee YJ, Chun JS and Cho MK: Expression of RUNX3 in
skin cancers. Clin Exp Dermatol. 36:769–774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hiramatsu T, Osaki M, Ito Y, Tanji Y,
Tokuyasu N and Ito H: Expression of RUNX3 protein in human
esophageal mucosa and squamous cell carcinoma. Pathobiology.
72:316–324. 2005. View Article : Google Scholar : PubMed/NCBI
|