Long non-coding RNA (lncRNA) is a type of non-coding
RNA (ncRNA) that is comprised of over 200 nucleotides (nt) and
lacks an open reading region and the capacity for protein coding
(1). More than 70% of the human
genome has been transcribed into ncRNAs (2). In addition to lncRNAs, other ncRNAs
have been identified, such as microRNAs (miRNA), which are small
interfering RNAs (siRNA) made up of 20–24 nucleotides. miRNA was
the first ncRNA found, though recently lncRNA has garnered more
attention. It has been demonstrated that lncRNAs play crucial roles
by influencing and changing cell growth, survival, cell cycle,
differentiation and apoptosis; they also play a vital part in many
diseases, including cancer (3).
However, despite the above findings, the biological functions and
molecular mechanisms of lncRNAs remain largely unknown (4). Utilizing next generation sequencing
(NGS), three generations of sequencing, RNA-Seq, RIP-Seq and RNA
array, the biological functions of lncRNAs could be gradually
discovered (5), which would be
indispensable for optimizing diagnostic and treatment methods for
cervical cancer patients.
Cervical cancer is one of the most serious cancers.
Each year, there are approximately 500,000 newly diagnosed cases,
and 200,000 deaths due to cervical cancer occur world-wide
(6). Without appropriate early
diagnostic methods, particularly in developing areas, cervical
cancer has developed into invasive cancers in a large number of
patients, which has led to lower survival rates (7). Traditional radiotherapy and
chemotherapy are still the most common therapies for the treatment
of advanced cervical cancer; however, these methods are not always
effective and can cause severe side-effects (8,9).
Therefore, it is urgent that biomarkers and novel treatment targets
are found for effective diagnosis and treatment of cervical cancer.
Many lncRNAs have been shown to be molecular regulatory factors in
cancer and may provide therapeutic targets for improving survival
in cases of cervical cancer. In this review, these aspects are
explored: the interaction between lncRNAs and miRNA, and the
molecular mechanisms involved; the interactions among the proteins
(or the mRNAs encoding these proteins at the protein or
transcriptome levels). In addition, an overview of research done on
the function of lncRNAs in cervical cancer is provided along with
prospective clinical applications of lncRNAs in cervical
cancer.
Studies of lncRNA molecular mechanisms have shown
that lncRNAs are related to tumorigenesis (10). lncRNAs interact with proteins,
miRNAs and mRNAs, developing complex mechanistic networks during
tumor growth (11). Therefore,
clarifying how lncRNAs regulate the process of gene transcription
and post-transcription can lead to a better understanding of the
pathogenesis of cervical cancer.
Cervical cancer-related lncRNAs have been
demonstrated to directly bind to target proteins or mRNAs to
conduct post-transcriptional modification. Reportedly, lncRNA
HOXA11-AS is involved in carcinogenesis through regulating the
expression of HOXA11 (12).
Besides, lncRNA-TI17313 was named lncRNA-EBIC, due to the fact that
it is an EZH2-binding lncRNA in cervical cancer (13). EZH2 is an important member of PRC2,
which is involved in several important regulatory mechanisms in
cancer. Besides lncRNA-EBIC other lncRNAs, such as HOTAIR,
lncRNA-HEIH PVT1 and H19 have been shown to bind to EZH2 and take
part in cancer epigenome modulation (14–17).
Tseng et al (18) shows gain
of PVT1 long non-coding RNA expression was required for high MYC
protein levels in 8q24-amplified human cancer cells. PVT1 RNA and
MYC protein expression correlated in primary human tumours, and
copy number of PVT1 was co-increased in >98% of
MYC-copy-increase cancers. C-Myc is an oncoprotein, which is
upregulated in cervical cancer (19,20).
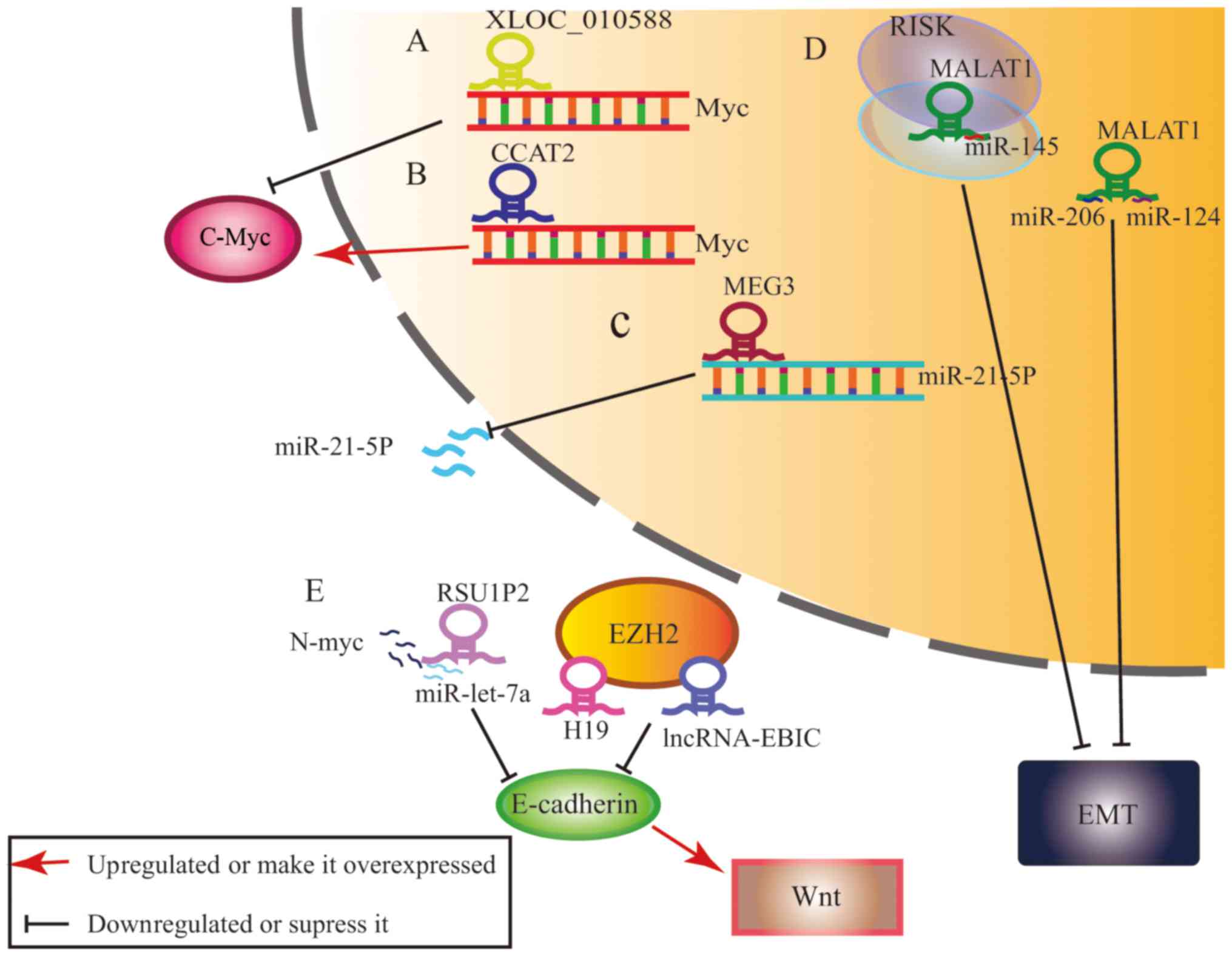
C-Myc acts as a downstream effector of XLOC_010588, CCAT2 and
RSU1P2 (21–23) and can also bind and stabilize the
Myc via inhibiting its phosphorylation at threonine 58 (24). E-cadherin expression is repressed in
cancer and has been found to be upregulated by MALAT1, but
repressed by lncRNA-EBIC (25). In
addition, lncRNA-CCHE1 increases the expression of PCNA in cervical
cancer by associating with PCNA mRNA (26). These studies illustrate that lncRNAs
play a role in cervical cancer by interacting with mRNAs and/or
proteins.
Since the function of competing endogenous RNA
(ceRNA) was discovered, lncRNAs have been regarded as one of the
most striking ceRNA and ‘talk’ to mRNAs or transcribed pseudogenes
using microRNA response elements (MREs) as letters (27). lncRNAs work as ‘miRNA sponges’,
which inhibit normal miRNAs targeting vitality on mRNAs (28–30).
It has been reported that MEG3 acts as a cancer suppressor via
lessening the expression of miR-21-5p in cervical cancer, in
vitro (31). MALAT1 sponges
miR-124, miR-145 and miR-375 promote the malignant behavior of
cervical cancer (32–34). In other cancers, H19 has been shown
to react with miR-675, miR-140 and miR-200, sponges circRNA MYLK
and binds competitively with miRNA-29a-3p to suppress cervical
cancer (35–38). These findings demonstrate the
interactions between lncRNAs, miRNAs and circRNAs in cervical
cancer.
Human papillomavirus (HPV) infection is a critical
factor in the development of cervical cancer (39,40)
and highest risk of developing cervical cancer come from HPV type
16 and 18 (41,42). Moreover, some studies have suggested
that lncRNAs and miRNAs play significant roles in the progression
of cervical cancer by sponging the miRNAs combined with HPV
proteins. Other studies have shown evidence of crosstalk between
the HPV16 E7 oncoprotein and lncRNA, such as HOTAIR (43). Increasing evidence suggests that
miRNAs-HPV protein like miR-135a-E6/E7 has an important function in
cervical cancer (44). Because of
the significance of HPV proteins in cervical cancer oncogenicity,
finding effective lncRNAs that inhibit HPV protein transcription
and translation is necessary (Fig.
1).
HOTAIR is one of most well-studied lncRNAs in
cervical cancer. It is 2.2 kb expressed from the HOXC cluster
located in chromosome 12q13.3 (45). In patients with invasive FIGO stage
IA-IVB cervical cancer, it was found to be an independent
prognostic factor for reduced survival. Furthermore, higher HOTAIR
levels have been observed in malignant tissue compared with normal
cervix tissue (46,47). A case-control study, including 510
cervical cancer patients (cases) and 713 none-cancer individuals
(controls), further indicated that the rs920778T allele conferred
elevated HOTAIR transcriptional activity, thus, increasing the risk
of developing cervical cancer (48). In addition, rs2366152C was
significantly over-represented and affected HOTAIR expression in
HPV16-positive cervical cancer cases (49). Another study demonstrated that
HOTAIR and HPV16 E7 (oncoprotein HPV E7 is the major transforming
agent, which leads to carcinogenesis) strongly interact with each
other, but not through the PRC2-complex (a complex that is related
to a large number of lncRNAs) (43). The mammalian target of rapamycin
(mTOR) has emerged as an important effector in cell-signaling
pathways (50). Zhang et al
(51), found that HOTAIR
overexpression upregulates the mTOR pathway in cervical carcinoma
cells. A higher level of HOTAIR was detected in the serum from
cervical patients compared to normal women, indicating that HOTAIR
may be a useful new circulating biomarker in serum. The study also
showed that HOTAIR overexpression promoted cervical cancer cell
growth, invasion and migration and that HOTAIR knockdown increased
cell apoptosis, via the epithelial-mesenchymal transition (EMT) and
Notch signaling pathways (52). In
a study by Kim et al (46),
HOTAIR overexpression in SiHa cells and cervical cancer tissue
promoted VEGF and mmP-9 protein expression. VEGF and mmP-9 not only
play critical roles in the malignant behavior of cervical cancer,
but knockdown of HOTAIR upregulates p21 and increases the
radio-sensitivity of HeLa cells (53). Many other studies have found that
HOTAIR was able to silence some tumor suppressors, such as HOXD10,
PTEN and RBM38 and activate some significant signaling pathways,
like STAT3, wnt/β-catenin and PI3K/AKT (54–58).
H19 is involved in many kinds of cancer, including
ovarian, lung cancer and hepatocellular carcinoma. In spite of
this, fewer studies have been done examining the relationship
between H19 and cervical cancer than in investigating the
relationship between HOTAIR and cervical cancer. Under normal
conditions, only fetal tissue and adult muscle express H19, thus it
often functions as ncRNA (59). H19
is upregulated in various human cancers, including bladder and
breast cancer, and in lung carcinoma cells suggesting an oncogenic
function (60). Conversely, in
hepatocellular carcinoma, H19-mediated metastasis is suppressed by
epigenetic activation of miR-200 suggesting that H19 may function
to suppress cancer (35,61,62).
In ovarian cancer, H19 upregulates SLUG expression via miR-675,
suppressing E-cadherin and activating EMT (36). Kim et al (63), explained that in cervical cancer,
H19 and IGF2 are expressed abnormally and that this abnormal
expression might be associated with the progression of cervical
cancer.
XLOC_010588 is a 1950nt lncRNA, which has thus far
been described in only one publication. It is located in chr13 on
the downstream side of TGFβ-stimulated clone-22 (TSC-22). Studies
have found that TSC-22 has DNA binding sites and acts as a tumor
suppressor. It inferred that XLOC_010588 may suppress the invasive
behavior of cancer as well. They found that XLOC_010588 expression
suppressed the progression of hepatocellular carcinoma, gastric,
colon, breast, cervical and ovarian cancer. In addition, they found
that XLOC_010588 inhibited the proliferation of cervical cancer
cells via downregulating the expression of oncoprotein c-Myc
(21).
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) was first mentioned in association with lung cancer, but
has now been found to be largely expressed in most cancers, working
as a decoy for splicing factors leading to splicing malfunctioning
(64). It has been demonstrated
that there is a reciprocal regulation between miR-375 and MALAT1
through suppression of the EMT pathway. MALAT1 also acts as an
miR-206 sponge (34). It has also
been shown that downregulating MALAT1 in CaSki, HeLa and SiHa
cells, as well as in cervical cancer tissues, weakens cancer cell
invasion and metastasis by inhibiting EMT and regulation of the
MALAT1-miR-124-RBG2 axis (25,33).
MALAT1 expression is an independent prognostic factor in addition
to tumor size, FIGO stage and lymph node metastasis (65). In radiotherapy to treat cervical
cancer, MALAT1 may result in radioresistance by working as a
miR-145 sponge (32).
CCAT2 has been shown to be overexpressed in breast
cancer and high levels of CCAT2 indicate poor prognosis and CMF
adjuvant chemoresistance (66).
CCAT2 has also been found to be upregulated in cervical cancer
cells and tissues. In HeLa, CaSki and SiHa cervical cancer cells,
CCAT2 knockdown inhibits cervical cancer cell proliferation at the
G1 phase and triggers cell apoptosis (22,67).
However, high CCAT2 expression is indicative of a more advanced
FIGO stage, lymph node metastasis and deep cervical invasion and
lower survival (68).
SPRY4-IT1 is an unspliced, polyadenylated 687nt4
transcript derived from the second intron of the SPRY4 gene. Its
potential for carcinogenesis was first (69,70)
found in melanoma and high levels of SPRY4-IT1 have since been
confirmed in gastric, non-small cell lung cancer (NSCLC),
esophageal squamous cell carcinoma (ESCC) and other cancers
(71–76). Cao et al (77), showed that SPRY4-IT1 expression is a
good candidate marker for discriminating between tumor tissue and
normal tissue and for predicting poor prognoses in patients with
cervical cancer.
Ras suppressor protein 1 pseudogene 2 (RSU1P2)
upregulation promotes the malignant phenotype of cervical cancer.
It was revealed that RSU1P2 acts as a ceRNA of miRNA let-7a and
regulates IGF1R and N-myc expression. The transcription factor,
N-myc forms a positive feedback loop with RSU1P2, in turn
activating its expression (23). To
the best of our knowledge, the N-myc positive feedback loop has not
been reported in studies of other types of cancer and requires
further research.
CCHE1 is located in an intergenic region on
chromatin 10. It physically binds to proliferating cell nuclear
antigen (PCNA) mRNA and upregulates PCNA expression, which promotes
the proliferation of cervical cancer cells. In contrast, decreasing
the CCHE1 level via RNA pull-down assays inhibits the proliferation
of cervical cancer cells. The present review also report that
higher CCHE1 expression was significantly associated with large
tumor size, advanced FIGO stage, invasion and low survival
(26). There is a similar
phenomenon, which occurs in hepatocellular carcinoma. Peng et
al (82), further confirmed
that CCHE1 knockdown inactivates the ERK/MAPK pathway, arresting
growth and promoting cell apoptosis.
PAX8-AS1 PAX8 antisense RNA 1 (PAX8-AS1), an
important regulator in the upstream region of PAX8 (on chromosome
2q13) (83). PAX8-AS1 contains
specific single nucleotide polymorphisms (SNPs), which can
represent expression quantitative trait loci (eQTLs) for PAX8. Two
eQTLs SNPs (rs4848320 and rs1110839) in PAX8-AS1 decrease the risk
of cervical cancer. In addition, PAX8 expression has been
recognized as a novel biomarker for fallopian tubes and uterus
cancer diagnoses (84,85).
The potential function of MEG3 has been studied in a
number of cancer types. Downregulation and overexpression of MEG3
alter pituitary tumor cell proliferation, suggesting that MEG3 may
be a potential biomarker (86).
Likewise, re-expression of MEG3 prevents the proliferation of
glioma tumor cells, in vitro (87) and in meningioma (88). Expression of MEG3 is lower in
cervical cancer tissue compared with non-neoplastic tissue. In
human cervical carcinoma cell lines, high levels of MEG3 inhibit
cell proliferation, induce G2/M cell cycle arrest and promote
apoptosis (89) via regulation of
miR-21-5p. Knockdown of MEG3 results in significant upregulation of
miR-21-5p expression in HeLa and CaSki cells (31). These results indicate that MEG3
functions as a tumor suppressor, resulting in the inhibition of
tumor growth in cervical cancer.
To the best of our knowledge, there has been only
one study examining lncRNA-LET. In that study, low levels of
lncRNA-LET expression led to significantly poorer overall survival
compared to higher lncRNA-LET expression in patients with cervical
cancer. Downregulation of lncRNA-LET was associated with a poor
prognosis in patients with cervical cancer (90). However, more research is necessary
in order to elucidate the lncRNA-LET mechanisms associated with
cancer.
A recent study reported that one-fifth of all human
lncRNAs is physically related to polycomb repressive complex 2
(PRC2, comprised of histone H3 lysine 27 methylase EZH2, SUZ12 and
EED), suggesting that lncRNAs may play a general role in leading to
transcriptional repression by recruiting polycomb-group proteins to
their target genes (91). Several
lncRNAs have been shown to physically bind to EZH2, such as
lncRNA-HEIH, H19 and HOTAIR, and play important roles in regulating
cancer epigenetics (14–16). High levels lncRNA-EBIC and EZH2
promote migration and invasion of cervical cancer cells in
vitro, resulting in a decrease in E-cadherin expression
(13). lncRNA-EBIC has been
suggested a key molecule in the migration and invasion of cells and
cervical cancer metastasis.
Plasmacytoma variant translocation 1 (PVT1) is a
highly conserved lncRNA, which is located downstream of MYC (an
oncoprotein). PVT1 has attracted great attention due to its
frequent co-amplification with MYC in several types of cancer
(92–94). In breast and hepatocellular
carcinoma, PVT1 function has been attributed to the binding and
stabilization of the Myc (18) and
Nop2 (95) proteins, respectively.
In gastric cancer cells, PVT1 represses the expression of p15 and
p16 via co-reaction with EZH2 (14m). Iden et al (24), utilized siRNA and LNA-mediated
knockdown for detecting the suppressant effects of low PVT1 levels
on cervical cancer cell proliferation, migration and invasion,
apoptosis and cisplatin resistance. In contrast, high PVT1 levels
were correlated with poorer survival. PVT1 also binds directly to
EZH2, recruiting EZH2 to the miR-200b promoter and inhibiting
miR-200b expression, which also plays an effective role in
regulating the behavior of cervical cancer (17) (Table
I).
It has been demonstrated that many circulating RNAs
have diagnostic potential for cancer and are surprisingly stable in
blood (96–99). In a study of colorectal cancer,
lncRNAs, such as GAS5, HOTAIR, H19 and MEG3 were found in
extracellular vesicles (small, phospholipid-enclosed vesicles
released by cells into their environment), BCAR4, MEG3 and other 19
lncRNAs were found to be significantly different between the
exosomes of samples from healthy people compared to those with
colorectal cancer. These have the potential for use as biomarkers
for the diagnosis of colorectal cancer (100). In other types of cancer, lncRNAs
can be detected in the plasma. PCA3, MALAT-1 and lncRNA-PCAT-18
have been identified as potential biomarkers for patients with
metastatic prostate cancer (101–103). It has also been reported that the
H19 plasma level was significantly higher, with a sensitivity of
74% and a specificity of 58%, in patients diagnosed with gastric
cancer (GC) compared with healthy controls (104,105). In hepatocellular carcinoma
patients the levels of the lncRNAs, HULC, LINC00152, RP11-160H22.5,
XLOC_014172, LOC149086 and lncRNA-AF085935 are upregulated in the
plasma, particularly lncRNA-AF085935, which was found not only in
the plasma from cancer patients, but also in patients infected with
hepatitis B (106–109). Only one circulating lncRNA,
HOTAIR, has been demonstrated in patients with cervical cancer. Li
et al (110), identified
plasma HOTAIR overexpression in the serum of cervical cancer
patients. However, the study of circulating lncRNAs is new and
there are still several challenges, such as finding better ways to
collect good quality plasma/serum from whole-blood, handling lncRNA
quantification and cancer-related genes may be expressed highly in
cancer tissues, but lower expressed in serum, which must be
overcome in order to identify lncRNAs, which can be relied upon as
novel, specific and sensitive cancer biomarkers.
Exosomes are identified as important members of
circulating tumor biomarkers and can be found in blood, urine and
other extracellular fluids (111,112) that contain a large variety of
biological components such as proteins, mRNAs, miRNAs and lncRNAs
(113). The expression levels of
HOTAIR, MALAT1 and MEG3 were significantly different in exosomes
isolated from cervical cancer patients compared to those isolated
from normal controls (114).
Exosomes can also transmit lncARSR, acting as a ceRNA, in renal
cancer. The above results indicate that exosome could play a vital
role in finding more sensitive and specific circulating tumor
biomarkers (Fig. 2).
Although only a few lncRNAs have been functionally
characterized, they play a novel role in the regulation of gene
expression. In cervical cancer, lncRNAs are important as potential
biomarkers for cervical cancer prognosis, invasion, metastasis,
chemo-resistance and radio-resistance. At the same time, the
lncRNAs interact with circRNA, miRNA, proteins/mRNAs and can be
detected in plasma, serum, exosomes and other vesicles in
extracellular fluid, which has opened up new avenues for their use
as easily accessible biomarkers. However, the potentially
significant regulatory mechanisms of lncRNAs in cervical cancer
need further exploration and characterization.
Inhibiting oncogenic lncRNA might be the most direct
approach for the treatment of cervical cancer. As yet, no RNA
interference-based drug has been approved; achieving viable and
efficient inhibition of lncRNA via siRNAs, miRNAs or by another
method, while avoiding side-effects, presents a great challenge and
necessitates further study.
The present review was supported by grants from the
National Natural Science Foundation of China (81302242), the Jilin
Province Science and Technology Funds (20150204007YY and
20130102094JC and 20140204022YY), the Jilin Province Development
and Reform Commission Funds (2014G073 and 2016C046-2).
|
1
|
Ernst C and Morton CC: Identification and
function of long non-coding RNA. Front Cell Neurosci. 7:1682013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ling H, Vincent K, Pichler M, Fodde R,
Berindan-Neagoe I, Slack FJ and Calin GA: Junk DNA and the long
non-coding RNA twist in cancer genetics. Oncogene. 34:5003–5011.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang CL: Cidofovir inhibits cervical
cancer cell Siha proliferation. Chin Gen M. 12:2012–1024. 2009.
|
|
7
|
Peterson EB, Ostroff JS, DuHamel KN,
DAgostino TA, Hernandez M, Canzona MR and Bylund CL: Impact of
provider-patient communication on cancer screening adherence: A
systematic review. Prev Med. 93:96–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
An JS, Huang MN, Song YM, Li N, Wu LY and
Zhan QM: A preliminary study of genes related to concomitant
chemoradiotherapy resistance in advanced uterine cervical squamous
cell carcinoma. Chin Med J (Engl). 126:4109–4115. 2013.PubMed/NCBI
|
|
9
|
Gadducci A, Tana R, Cosio S and Cionini L:
Treatment options in recurrent cervical cancer (Review). Oncol
Lett. 1:3–11. 2010.PubMed/NCBI
|
|
10
|
Wang GY, Zhu YY and Zhang YQ: The
functional role of long non-coding RNA in digestive system
carcinomas. Bull Cancer. 101:E27–E31. 2014.PubMed/NCBI
|
|
11
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen J, Fu Z, Ji C, Gu P, Xu P, Yu N, Kan
Y, Wu X, Shen R and Shen Y: Systematic gene microarray analysis of
the lncRNA expression profiles in human uterine cervix carcinoma.
Biomed Pharmacother. 72:83–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long noncoding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9:e1003402014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang S, Zhang G and Liu J: Long noncoding
RNA PVT1 promotes cervical cancer progression through
epigenetically silencing miR-200b. APMIS. 124:649–658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014.PubMed/NCBI
|
|
19
|
Yuan Y, Zhang J, Cai L, Ding C, Wang X,
Chen H, Wang X, Yan J and Lu J: Leptin induces cell proliferation
and reduces cell apoptosis by activating c-myc in cervical cancer.
Oncol Rep. 29:2291–2296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rughooputh S, Manraj S, Eddoo R and
Greenwell P: Expression of the c-myc oncogene and the presence of
HPV: 18: Possible surrogate markers for cervical cancer? Br J
Biomed Sci. 66:74–78. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao LM, Sun XY, Liu AW, Wu JB, Cheng XL,
Lin JX, Zheng M and Huang L: Low expression of long noncoding
XLOC_010588 indicates a poor prognosis and promotes proliferation
through upregulation of c-Myc in cervical cancer. Gynecol Oncol.
133:616–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Q, Guo X, Que S, Yang X, Fan H, Liu M,
Li X and Tang H: LncRNA RSU1P2 contributes to tumorigenesis by
acting as a ceRNA against let-7a in cervical cancer cells.
Oncotarget. Jul 26–2016.(Epub ahead of print). doi:
10.18632/oncotarget.10844.
|
|
24
|
Iden M, Fye S, Li K, Chowdhury T,
Ramchandran R and Rader JS: The lncRNA PVT1 contributes to the
cervical cancer phenotype and associates with poor patient
prognosis. PLoS One. 11:e01562742016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun R, Qin C, Jiang B, Fang S, Pan X, Peng
L, Liu Z, Li W, Li Y and Li G: Down-regulation of MALAT1 inhibits
cervical cancer cell invasion and metastasis by inhibition of
epithelial-mesenchymal transition. Mol Biosyst. 12:952–962. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang M, Zhai X, Xia B, Wang Y and Lou G:
Long noncoding RNA CCHE1 promotes cervical cancer cell
proliferation via upregulating PCNA. Tumour Biol. 36:7615–7622.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang J, Yao T, Wang Y, Yu J, Liu Y and
Lin Z: Long noncoding RNA MEG3 is downregulated in cervical cancer
and affects cell proliferation and apoptosis by regulating miR-21.
Cancer Biol Ther. 17:104–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu H, He Y, Lin L, Qi Z, Ma L, Li L and Su
Y: Long non-coding RNA MALAT1 modulates radiosensitivity of
HR-HPV+ cervical cancer via sponging miR-145. Tumour
Biol. 37:1683–1691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu S, Song L, Zeng S and Zhang L:
MALAT1-miR-124-RBG2 axis is involved in growth and invasion of
HR-HPV-positive cervical cancer cells. Tumour Biol. 37:633–640.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu S, Song L, Yao H, Zhang L, Xu D, Gao F
and Li Q: MiR-375 is epigenetically downregulated by HPV-16 E6
mediated DNMT1 upregulation and modulates EMT of cervical cancer
cells by suppressing lncRNA MALAT1. PLoS One. 11:e01634602016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the MiR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matouk IJ, Raveh E, Abu-lail R, Mezan S,
Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M,
et al: Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys
Acta. 1843:1414–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan
J, Peng G and Liao Y: The lncRNA H19 interacts with miR-140 to
modulate glioma growth by targeting iASPP. Arch Biochem Biophys.
610:1–7. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bosch FX, Manos MM, Muñoz N, Sherman M,
Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R and Shah KV:
Prevalence of human papillomavirus in cervical cancer: A worldwide
perspective. International biological study on cervical cancer
(IBSCC) Study Group. J Natl Cancer Inst. 87:796–802. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Greco D, Kivi N, Qian K, Leivonen SK,
Auvinen P and Auvinen E: Human papillomavirus 16 E5 modulates the
expression of host microRNAs. PLoS One. 6:e216462011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sasagawa T, Takagi H and Makinoda S:
Immune responses against human papillomavirus (HPV) infection and
evasion of host defense in cervical cancer. J Infect Chemother.
18:807–815. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Khorasanizadeh F, Hassanloo J, Khaksar N,
Taheri Mohammad S, Marzaban M, Rashidi HB, Sari Akbari A and
Zendehdel K: Epidemiology of cervical cancer and human papilloma
virus infection among Iranian women - analyses of national data and
systematic review of the literature. Gynecol Oncol. 128:277–281.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sharma S, Mandal P, Sadhukhan T, Chowdhury
Roy R, Ranjan Mondal N, Chakravarty B, Chatterjee T, Roy S and
Sengupta S: Bridging links between long noncoding RNA HOTAIR and
HPV oncoprotein e7 in cervical cancer pathogenesis. Sci Rep.
5:117242015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Leung CO, Deng W, Ye TM, Ngan HY, Tsao SW,
Cheung AN, Pang RT and Yeung WS: miR-135a leads to cervical cancer
cell transformation through regulation of β-catenin via a
SIAH1-dependent ubiquitin proteosomal pathway. Carcinogenesis.
35:1931–1940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim
SW and Kim YT: Long non-coding RNA HOTAIR is associated with human
cervical cancer progression. Int J Oncol. 46:521–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo L, Lu X, Zheng L, Liu X and Hu M:
Association of long non-coding RNA HOTAIR polymorphisms with
cervical cancer risk in a chinese population. PLoS One.
11:e01600392016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sharma Sweta S, Roy Rahul C and Ranjan
Nidhu M: Identification of genetic variation in the lncRNA HOTAIR
associated with HPV16-related cervical cancer pathogenesis. Cell
Oncol. 9:282016.
|
|
50
|
Guertin DA and Sabatini DM: Defining the
role of mTOR in cancer. Cancer Cell. 12:9–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang D, Zhou XH, Zhang J, Zhou YX, Ying
J, Wu GQ and Qian JH: Propofol promotes cell apoptosis via
inhibiting HOTAIR mediated mTOR pathway in cervical cancer. Biochem
Biophys Res Commun. 468:561–567. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee M, Kim HJ, Kim SW, Park SA, Chun KH,
Cho NH, Song YS and Kim YT: The long non-coding RNA HOTAIR
increases tumour growth and invasion in cervical cancer by
targeting the Notch pathway. Oncotarget. 7:44558–44571. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jing L, Yuan W, Ruofan D, Jinjin Y and
Haifeng Q: HOTAIR enhanced aggressive biological behaviors and
induced radio-resistance via inhibiting p21 in cervical cancer.
Tumour Biol. 36:3611–3619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li L, Liu B, Wapinski OL, Tsai MC, Qu K,
Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al: Targeted
disruption of Hotair leads to homeotic transformation and gene
derepression. Cell Rep. 5:3–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du
C, Peng C, Xie H, Zhou L, Wu J, et al: Long non-coding RNA HOTAIR
promotes cell migration and invasion via down-regulation of RNA
binding motif protein 38 in hepatocellular carcinoma cells. Int J
Mol Sci. 15:4060–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J
and Liu M: Long intergenic noncoding RNA HOTAIR is overexpressed
and regulates PTEN methylation in laryngeal squamous cell
carcinoma. Am J Pathol. 182:64–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Carrion K, Dyo J, Patel V, Sasik R,
Mohamed SA, Hardiman G and Nigam V: The long non-coding
HOTAIR is modulated by cyclic stretch and WNT/β-CATENIN in
human aortic valve cells and is a novel repressor of calcification
genes. PLoS One. 9:e965772014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lee DF, Su J, Kim HS, Chang B, Papatsenko
D, Zhao R, Yuan Y, Gingold J, Xia W, Darr H, et al: Modeling
familial cancer with induced pluripotent stem cells. Cell.
161:240–254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yoshimizu T, Miroglio A, Ripoche MA,
Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B,
Jammes H, et al: The H19 locus acts in vivo as a tumor suppressor.
Proc Natl Acad Sci USA. 105:12417–12422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kim SJ, Park SE, Lee C, Lee SY, Jo JH, Kim
JM and Oh YK: Alterations in promoter usage and expression levels
of insulin-like growth factor-II and H19 genes in cervical
carcinoma exhibiting biallelic expression of IGF-II. Biochim
Biophys Acta. 1586:307–315. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yang L, Bai HS, Deng Y and Fan L: High
MALAT1 expression predicts a poor prognosis of cervical cancer and
promotes cancer cell growth and invasion. Eur Rev Med Pharmacol
Sci. 19:3187–3193. 2015.PubMed/NCBI
|
|
66
|
Redis RS, Sieuwerts AM, Look MP, Tudoran
O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al:
CCAT2, a novel long non-coding RNA in breast cancer: Expression
study and clinical correlations. Oncotarget. 4:1748–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wu L, Jin L, Zhang W and Zhang L: Roles of
long non-coding RNA CCAT2 in cervical cancer cell growth and
apoptosis. Med Sci Monit. 22:875–879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen X, Liu L and Zhu W: Up-regulation of
long non-coding RNA CCAT2 correlates with tumor metastasis and poor
prognosis in cervical squamous cell cancer patients. Int J Clin Exp
Pathol. 8:13261–13266. 2015.PubMed/NCBI
|
|
69
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu T, Shen SK, Xiong JG, Xu Y, Zhang HQ,
Liu HJ and Lu ZG: Clinical significance of long noncoding RNA
SPRY4-IT1 in melanoma patients. FEBS Open Bio. 6:147–154. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou
P, De W and Liu XH: Decreased long noncoding RNA SPRY4-IT1
contributing to gastric cancer cell metastasis partly via affecting
epithelial-mesenchymal transition. J Transl Med. 13:2502015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong
R, Yang JS, Xu TP, Liu YW, Zou YF, et al: EZH2-mediated epigenetic
suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell
proliferation and metastasis by affecting the
epithelial-mesenchymal transition. Cell Death Dis. 5:e12982014.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li
SQ, Wang CM, Tong YS, Tuo L, Wu M, et al: Long noncoding RNA
SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and
associated with poor prognosis. Tumour Biol. 35:7743–7754. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Peng W, Wu G, Fan H, Wu J and Feng J: Long
noncoding RNA SPRY4-IT1 predicts poor patient prognosis and
promotes tumorigenesis in gastric cancer. Tumour Biol.
36:6751–6758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhao XL, Zhao ZH, Xu WC, Hou JQ and Du XY:
Increased expression of SPRY4-IT1 predicts poor prognosis and
promotes tumor growth and metastasis in bladder cancer. Int J Clin
Exp Pathol. 8:1954–1960. 2015.PubMed/NCBI
|
|
76
|
Zhang HM, Yang FQ, Yan Y, Che JP and Zheng
JH: High expression of long non-coding RNA SPRY4-IT1 predicts poor
prognosis of clear cell renal cell carcinoma. Int J Clin Exp
Pathol. 7:5801–5809. 2014.PubMed/NCBI
|
|
77
|
Cao Y, Liu Y, Lu X, Wang Y, Qiao H and Liu
M: Upregulation of long noncoding RNA SPRY4-IT1 correlates with
tumor progression and poor prognosis in cervical cancer. FEBS Open
Bio. 6:954–960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wu Y, Lyu H, Liu H, Shi X, Song Y and Liu
B: Downregulation of the long noncoding RNA GAS5-AS1 contributes to
tumor metastasis in non-small cell lung cancer. Sci Rep.
6:310932016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Liang W, Lv T, Shi X, Liu H, Zhu Q, Zeng
J, Yang W, Yin J and Song Y: Circulating long noncoding RNA GAS5 is
a novel biomarker for the diagnosis of nonsmall cell lung cancer.
Medicine (Baltimore). 95:e46082016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhang N, Wang AY, Wang XK, Sun XM and Xue
HZ: GAS5 is downregulated in gastric cancer cells by promoter
hypermethylation and regulates adriamycin sensitivity. Eur Rev Med
Pharmacol Sci. 20:3199–3205. 2016.PubMed/NCBI
|
|
81
|
Cao S, Liu W, Li F, Zhao W and Qin C:
Decreased expression of lncRNA GAS5 predicts a poor prognosis in
cervical cancer. Int J Clin Exp Pathol. 7:6776–6783.
2014.PubMed/NCBI
|
|
82
|
Peng W and Fan H: Long noncoding RNA CCHE1
indicates a poor prognosis of hepatocellular carcinoma and promotes
carcinogenesis via activation of the ERK/MAPK pathway. Biomed
Pharmacother. 83:450–455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Han J, Zhou W, Jia M, Wen J, Jiang J, Shi
J, Zhang K, Ma H, Liu J, Ren J, et al: Expression quantitative
trait loci in long non-coding RNA PAX8-AS1 are associated with
decreased risk of cervical cancer. Mol Genet Genomics.
291:1743–1748. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Tacha D, Zhou D and Cheng L: Expression of
PAX8 in normal and neoplastic tissues: A comprehensive
immunohistochemical study. Appl Immunohistochem Mol Morphol.
19:293–299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Waters L, Crumley S, Truong L, Mody D and
Coffey D: PAX2 and PAX8: Useful markers for metastatic effusions.
Acta Cytol. 58:60–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang P, Ren Z and Sun P: Overexpression of
the long non-coding RNA MEG3 impairs in vitro glioma cell
proliferation. J Cell Biochem. 113:1868–1874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Qin R, Chen Z, Ding Y, Hao J, Hu J and Guo
F: Long non-coding RNA MEG3 inhibits the proliferation of cervical
carcinoma cells through the induction of cell cycle arrest and
apoptosis. Neoplasma. 60:486–492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Jiang S, Wang HL and Yang J: Low
expression of long non-coding RNA LET inhibits carcinogenesis of
cervical cancer. Int J Clin Exp Pathol. 8:806–811. 2015.PubMed/NCBI
|
|
91
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales Rivea D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Barsotti AM1, Beckerman R, Laptenko O,
Huppi K, Caplen NJ and Prives C: P53-dependent induction of PVT1
and miR-1204. J Biol Chem. 287:2509–2519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu
QH, Weng WW, Tan C, Sheng WQ, Zhou XY, et al: Circulating CUDR,
LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish
patients with gastric cancer from healthy controls. Int J Cancer.
137:1128–1135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Ronnau CG, Verhaegh GW, Luna-Velez MV and
Schalken JA: Noncoding RNAs as novel biomarkers in prostate cancer.
BioMed Res Int. 2014:5917032014.https://doi.org/10.1155/2014/591703 View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Isin M, Ozgur E, Cetin G, Erten N, Aktan
M, Gezer U and Dalay N: Investigation of circulating lncRNAs in
B-cell neoplasms. Clin Chim Acta. 431:255–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lorenzen JM, Schauerte C, Kielstein JT,
Hübner A, Martino F, Fiedler J, Gupta SK, Faulhaber-Walter R,
Kumarswamy R, Hafer C, et al: Circulating long noncoding RNATapSaki
is a predictor of mortality in critically ill patients with acute
kidney injury. Clin Chem. 61:191–201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S,
Huang D, Weng WW, Tan C, Sheng W, et al: Circulating long RNAs in
serum extracellular vesicles: Their characterization and potential
application as biomarkers for diagnosis of colorectal cancer.
Cancer Epidemiol Biomarkers Prev. 25:1158–1166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y,
Zhang W, Jing T, Zhang C, Shen J, et al: Development and
prospective multicenter evaluation of the long noncoding RNA
MALAT-1 as a diagnostic urinary biomarker for prostate cancer.
Oncotarget. 5:11091–11102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Crea F, Watahiki A, Quagliata L, Xue H,
Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al:
Identification of a long non-coding RNA as a novel biomarker and
potential therapeutic target for metastatic prostate cancer.
Oncotarget. 5:764–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Merola R, Tomao L, Antenucci A, Sperduti
I, Sentinelli S, Masi S, Mandoj C, Orlandi G, Papalia R,
Guaglianone S, et al: PCA3 in prostate cancer and tumor
aggressiveness detection on 407 high-risk patients: A National
Cancer Institute experience. J Exp Clin Cancer Res. 34:152015.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
105
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. BioMed Res Int. 2013:1361062013.https://doi.org/10.1155/2013/136106 View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Lu J, Xie F, Geng L, Shen W, Sui C and
Yang J: Investigation of serum lncRNA-uc003wbd and lncRNA-AF085935
expression profile in patients with hepatocellular carcinoma and
HBV. Tumour Biol. 36:3231–3236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Tang J, Jiang R, Deng L, Zhang X, Wang K
and Sun B: Circulation long non-coding RNAs act as biomarkers for
predicting tumorigenesis and metastasis in hepatocellular
carcinoma. Oncotarget. 6:4505–4515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 act as novel biomarkers in
predicting diagnosis of hepatocellular carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li J, Wang Y, Yu J, Dong R and Qiu H: A
high level of circulating HOTAIR is associated with progression and
poor prognosis of cervical cancer. Tumour Biol. 36:1661–1665. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ahadi A, Brennan S, Kennedy PJ, Hutvagner
G and Tran N: Long non-coding RNAs harboring miRNA seed regions are
enriched in prostate cancer exosomes. Sci Rep. 6:249222016.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Berrondo C, Flax J, Kucherov V, Siebert A,
Osinski T, Rosenberg A, Fucile C, Richheimer S and Beckham CJ:
Expression of the long non-coding RNA HOTAIR correlates with
disease progression in bladder cancer and is contained in bladder
cancer patient urinary exosomes. PLoS One. 11:e01472362016.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M
and Tian L: Combined detection of serum exosomal miR-21 and HOTAIR
as diagnostic and prognostic biomarkers for laryngeal squamous cell
carcinoma. Med Oncol. 31:1482014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhang J, Liu SC, Luo XH, Tao GX, Guan M,
Yuan H and Hu DK: Exosomal long noncoding RNAs are differentially
expressed in the cervicovaginal lavage samples of cervical cancer
patients. J Clin Lab Anal. 30:1116–1121. 2016. View Article : Google Scholar : PubMed/NCBI
|