Introduction
Prostate cancer (PC) is one of the most frequently
diagnosed cancers in men. One cause for neoplastic transformation
is abnormal gene expression, which is not determined by nucleotide
sequence changes in DNA, but by disturbances in epigenetic
mechanisms. The epigenetic changes in DNA structure are the result
of post-replication modification in DNA and/or post-translational
modification of proteins associated with DNA. In contrast to
mutations, epigenetic modifications are reversible and dynamically
occur. These epigenetic mechanisms include aberrant DNA methylation
(hypermethylation and hypomethylation) and modifications of
histones, chromatin remodeling and changes in gene expression
caused by non-coding RNAs (ncRNAs). Epigenetic mechanisms lead to
genomic instability and inappropriate gene expression and are the
best-characterized alteration in PC. Epigenetic mechanisms play an
important role in the initiation and development of PC. Global and
gene-specific hypermethylation and hypomethylation as well as
histone modifications have been found to be associated with PC
(1,2).
DNA methylation and prostate cancer
DNA methylation plays an important role in DNA
repair, recombination and replication, as well as regulation of
gene activity. DNA methylation involves the addition of a methyl
group to the 5′-carbon of cytosine in CpG dinucleotide sequences.
This process is catalyzed by a family of DNA methyltransferases
(DNMTs). CpG islands are CpG-rich areas of 200 bp to several
kilobases in length, usually located near the promoters of highly
expressed genes, and are the sites of common methylation in human
tumors, including the prostate. CpG islands, in more than 55% of
cases, form clusters and their methylation/demethylation results in
the inhibition/activation of transcription. Thus, methylation of
CpG islands in promoter regions of genes may prevent or deregulate
the synthesis of gene products (3–5).
Three active DNMTs have been identified (DNMT1,
DNMT3A and DNMT3B). A fourth enzyme previously known as DNMT2 is
not a DNA methyltransferase. DNMT2 or TRDMT1 has strong sequence
similarities with 5-methylcytosine methyltransferases, but the
enzyme was shown to methylate position 38 in aspartic acid transfer
RNA and does not methylate DNA. The smallest mammalian DNMT2 is
believed to participate in the recognition of DNA damage, DNA
recombination and mutation repair. DNMT1, the major enzyme
responsible for maintenance of the DNA methylation pattern is
located at the replication fork and methylates newly biosynthesized
DNA. DNMT3A and DNMT3B cannot differentiate between unmethylated
and hemi-methylated CpG sites, and they cannot copy a specific
pattern of methylation or contribute to the maintenance of the
methylation pattern (6).
Proteins that bind to methylated CpG islands (MBP,
methyl-CpG binding proteins) which include: MBP1, MBP2, MBP3 and
methyl CpG-binding protein-2 (MeCP2) recognize methylated CpG
islands in the promoter regions of the genes. These proteins at the
N-terminal contain methyl-CpG-binding domain (MBD) and in the
central portion transcriptional repression domain (TRD). MBP
proteins can activate histone deacetylase (HDAC), and/or
co-repressors of transcription, after recognizing methylated DNA.
Then, chromatin undergoes strong condensation, preventing access of
transcription factors to the promoter's genes (7).
Disturbances of DNA methylation can lead to
malignant transformation. In PC, hypermethylation and
hypomethylation of DNA can be observed. In normal cells, CpG
islands are protective against excessive methylation by active
transcription, DNA demethylation, regulation of replication and the
establishment of appropriate local chromatin structure, impeding
access to methyl-DNA transferase. In tumor cells, the mechanisms as
described above are altered, resulting in abnormal DNA methylation
regulation (2,8).
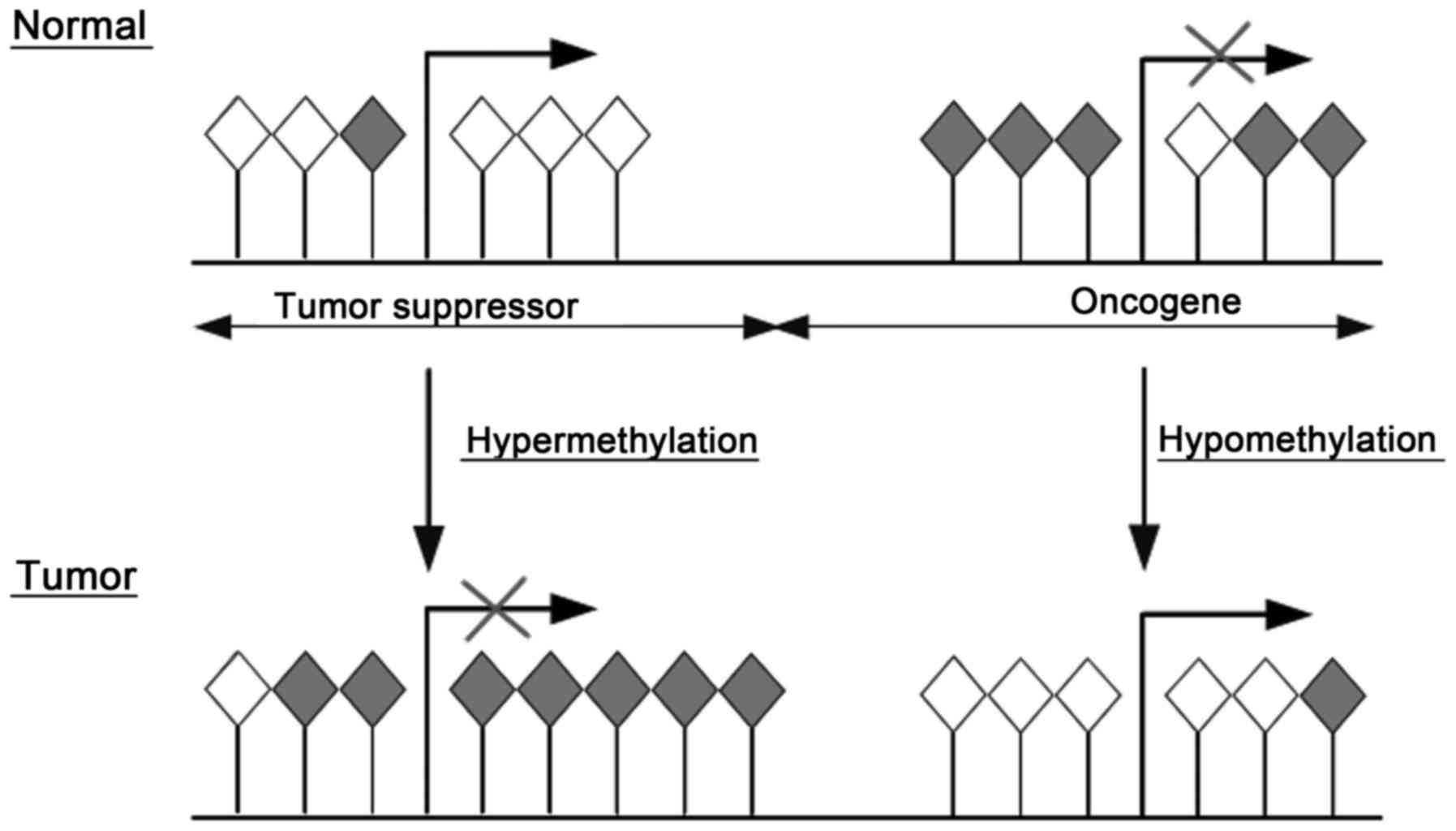
A common molecular feature associated with
tumorigenesis including PC is hypermethylation of cytosines 5′ in
CpG islands within the regulatory (promoter) region of suppressor
genes resulting in gene silencing. In contrast, hypomethylation in
the promoter region of oncogenes in tumors reactivates
transcription (Fig. 1). Moreover,
5-methyl cytosine is unstable and mutates to thymine and methylated
CpG sites degrade to TpG/CpA (9).
DNA hypermethylation
In PC, many CpG islands exhibit aberrant
hypermethylation. Genes undergoing methylation in PC include genes
involved in key cellular functions such as DNA repair (GSTP1
and MGMT), cell adhesion (CDH1 and CD44), cell
cycle control (CDKN2A), apoptosis (PYCARD) and also
are related to suppressor genes (APC, RARβ,
RARRES1 and RASSF1) (5,8,10).
Specific genes implicated within each category are summarized in
Table I.
 | Table I.Genes hypermethylated in prostate
cancer. |
Table I.
Genes hypermethylated in prostate
cancer.
| Gene | Chromosomal
location | Function | Hypermethylation in
prostate cancer (%) | Refs. |
|---|
| GSTP1 | 11q13 | Detoxification, DNA
repair | 13–100 | (13,16,72–82) |
| MGMT | 10q26 | Detoxification, DNA
repair | 0–76 | (17–19,35) |
| CDH1 | 16q22.1 | Cell adhesion | 0–72 | (22,29,83) |
| CD44 | 11p13 | Cell-cell
interactions, cell adhesion and migration | 20–78 | (21,23,84) |
| CCND2 | 12p13 | Regulation of
cyclin-dependent protein serine/threonine kinase activity | 8.4–99 | (19,73,75) |
| APC | 5q21-q22 | Tumor suppressor;
antagonist of the Wnt signaling pathway; regulator of cell
migration, adhesion, transcriptional activation and apoptosis | 14.5–100 | (19,27,75,76,80,82,85–87) |
| RARβ | 17q21 | Tumor suppressor;
regulation of development, differentiation, apoptosis,
granulopoeisis, and transcription of clock genes | 32.6–100 | (19,73,75–77) |
| RASSF1A | 3p21.3 | Tumor suppressor;
Ras protein signal transduction | 19.2–100 | (19,72,73,86) |
The glutathione S-transferase π 1 gene
(GSTP1) encodes glutathione S-transferase belonging to the π
class. It is responsible for protecting cells from oxidative stress
and chemical attacks. GSTP1 is involved in the metabolism,
detoxification and elimination of compounds potentially genotoxic,
and prevents DNA damage and initiation of neoplastic
transformation. GSTP1 is one of the earliest identified
genes which is hypermethylated in PC. GSTP1 hypermethylation
has been found in 13.3–100% of PC cases, whereas methylation of
GSTP1 within CpG dinucleotides has not been noted in normal
cells. GSTP1 hypermethylation is present at all stages of PC
development, and the assessment of the methylation profile of this
gene allows the differentiation of types and subtypes of PC which
may facilitate early diagnosis. GSTP1 gene methylation
profile can be assessed not only in serum but also in other body
fluids. In 39–83.2% of PC patients, GSTP1 DNA methylation
was detected in urine (11,12). Hypermethylation of GSTP1 DNA
was detected in plasma samples from 27 of 31 (92.86%) patients with
PC (13). Methylated GSTP1
DNA in serum is present in 28–32% of men with metastatic PC
(14,15). Mahon et al (16) identified plasma methylated
GSTP1 DNA as a potential prognostic marker in men with
castration-resistant PC as well as a potential surrogate
therapeutic efficacy marker for chemotherapy.
The O-6-methylguanine-DNA methyltransferase gene
(MGMT) encoding enzyme removes alkyl adducts from the O6
position of guanine. Methylation of the MGMT gene promoter
region has been detected in PC patients and cell lines (17–19).
DNA hypermethylation of CpG dinucleotides of the MGMT gene
has been associated with decrease and/or total loss of expression
of this gene, and results in the development of carcinoma. A study
conducted by Sidhu et al (18) indicated that development of prostate
carcinoma is correlated with modification of the MGMT methylation
pattern.
PC tumor cell invasion and progression have been
found to be associated with E-cadherin-1 (CDH1) gene
promoter hypermethylation. The effect of increased methylation of
CDH1 is a decrease in E-cadherin expression, which is one of
the main factors associated with dysfunction of intercellular
adhesion, inhibition of cell adhesion and promotion of neoplastic
transformation and metastasis (5,20).
The CD44 gene which encodes cluster of
differentiation CD44 may be another important mediator of prostate
carcinogenesis, since it is hypermethylated in 78% of patients with
PC in compared to only 10% of patients without cancer. Based on the
morphology of fibroblasts, increased CD44 gene promoter
hypermethylation was found to be a characteristic feature of
epithelial-to-mesenchymal transition (EMT) in non-prostate
malignant tumors (8,21–23).
The cyclin D2 (CCND2) gene encodes a protein
belonging to the conserved family of cyclin D, which are
characterized by cyclic occurrence in the cell. Cyclins D form
complexes with cyclin-dependent kinases CDK4 and CDK6. These
complexes regulate cell cycle transition from G1 to S phase.
CCND2 gene hypermethylation was found in 32% of PC cases
which was significantly higher as compared to the hypermethylation
noted in 6% of nonmalignant prostate tissues. In addition, high
levels of CCND2 methylation were found to correlate with
tumor aggressiveness (8,24). In a study by Henrique et al
(25), cyclin D2 promoter
methylation was found in 80% of benign prostatic hyperplasia (BPH),
in 99.2% of PC and in 100% of high-grade prostatic intraepithelial
neoplasia.
PYCARD also known as the target of
methylation induced silencing 1 (TMS1) gene, encodes a
proapoptotic protein, which contains a pyrin domain (PYD) in the
N-terminus and caspase recruitment domain (CARD) in the C-terminus.
Both domains are members of the death domain-fold superfamily.
PYCARD plays an important role in the development of many diseases,
including cancer. It is considered that PYCARD induces apoptosis
via caspase 9. Hypermethylation of PYCARD is a common event
in PC, and decreased PYCARD expression has been associated with
complete methylation of the promoter region in human prostate
carcinoma cell line LNCaP (26).
The adenomatosis polyposis coli (APC) gene
encodes a protein that is involved in the regulation of many
cellular processes including division, migration, adhesion and
differentiation of cells. Promoter methylation in the APC
gene has been associated with high grade and advanced stages of PC
(27,28).
The retinoic acid receptor β (RARβ) gene
encodes β retinoid acid (RA) receptor, which forms complexes with
two nuclear receptor families i.e. retinoid acid receptors (RARα, β
and γ) and X retinoid receptors (RXRα, β and γ). Retinoid acid
belongs to the retinoids which induce numerous cellular pathways
involving kinases responsible for activation or inhibition of
transcription of many genes. A study conducted by Jerónimo et
al (29) showed that
RARβ2 is hypermethylated in 97.5% of PC and in 94.7% of
prostate intraepithelial neoplasia (PIN), while only in 23.3% of
BPH. Thus, hypermethylation of genes encoding retinoid receptors
may cause changes in their expression, which can affect PC
progression.
The retinoic acid receptor responder gene 1
(RARRES1) also known as tazarotene-induced gene 1
(TIG1) was first identified as a gene responsive to retinoic
acid. Decreased levels of RARRES1 expression have been
demonstrated in PC. Furthermore, it has been suggested that
RARRES1 gene silencing may be a consequence of receptor
binding retinoid RARβ methylation and is correlated with PC
progression. Decreased RARRES1 expression affects cell-cell
interactions and results in increased proliferation and
invasiveness of tumor cells. Loss of RARRES1 expression may
explain the low sensitivity to retinoid-induced growth regulation
(30,31).
Ras-associated domain family 1 (RASSF1) is a
putative tumor-suppressor gene (TSG) located in the chromosomal
region 3p21.3. RASSF1 is functionally associated with cell cycle
control, microtubule stabilization, cellular adhesion, motility and
apoptosis. Hypermethylation of RASSF1A is not specific to PC
and has been identified in 35.5–96% of various cancers (32–34).
Hypermethylation of RASSF1A in 53% of PC cases was found to
be associated with a higher level of prostate-specific antigen
(PSA) and aggressive PC. The effects of changes in the expression
of RASSF1A include disturbances of the cell cycle and cell
proliferation (35). A
meta-analysis suggested that RASSF1A promoter methylation is
significantly associated with PC risk and its clinicopathological
variables (Gleason score, serum PSA level and tumor stage)
(36).
However, Pellacani et al (37) suggested that for a set of genes
(including GSTP1) that are hypermethylated in PC, gene
downregulation appears to be the result of cell differentiation and
is not cancer-specific. Hypermethylation is observed in more
differentiated cancer cells and is promoted by hyperproliferation.
These genes are maintained as actively expressed and
methylation-free in undifferentiated PC cells, and their
hypermethylation is not essential for either tumor development or
expansion.
DNA hypomethylation
The mechanism leading to hypomethylation of DNA in
cancer is not fully understood. Many studies have suggested diverse
causes for the hypomethylation of DNA, including shortages of
S-adenosylmethionine precursors or folic acid in the diet or
genetic abnormalities in the metabolic pathway of the donor of
methyl groups. DNA hypomethylation can also be attributed to a
deficiency in methyltransferases. Hypomethylation in promoter
regions of genes can lead to a reduction in genome stability
through an increase in the expression of transposons, which under
physiological conditions are silenced by methylation. Reduction in
methylation may also cause a reduction in chromosomal stability and
activation of proto-oncogenes (3,5).
Hypomethylation of DNA is more often observed at
metastasis than in the early stages of neoplastic transformation in
PC. Hypomethylation in PC is observed among repetitive sequences of
LINE-1 (long interspersed nuclear element-1), which is found in 64%
of cases of advanced PC with metastases to lymph nodes (38,39).
Reduction in the level of methylation of the genome
reduces the stability of chromosomes leading to activation of the
proto-oncogene MYC (homolog of viral myelocytomatosis oncogene) and
RAS (homolog of viral rat sarcoma oncogene). A strong correlation
between overexpression of proto-oncogene MYC in PC, and increased
degree of invasiveness of the tumor has been shown. Hypomethylation
of promotor regions of proto-oncogenes contributes to the
activation and overexpression of their products, and thus excessive
cell proliferation (8,40).
In PC, hypomethylation and higher expression of
several genes, among them PLAU, HPSE or CYP1B1
have been identified (Table
II).
 | Table II.Genes hypomethylated in prostate
cancer. |
Table II.
Genes hypomethylated in prostate
cancer.
| Gene | Chromosomal
location | Function | Hypermethylation in
prostate cancer (%) | Refs. |
|---|
| uPA,
PLAU | 10q22.2 | Blood coagulation,
fibrinolysis, hemostasis, plasminogen activation |
23.2–96.6 | (88,89) |
| HPSE | 4q21.3 | Cell adhesion |
8.5–30 | (43) |
| CYP1B1 | 2p22.2 | Oxidation |
5.7–17.1 | (44) |
| WNT5A | 3p21-p14 | Cell signaling | 65 | (45) |
| S100P | 4p16 | Cellular calcium
signaling | 50 | (45) |
| CRIP1 | 14q32.33 | Cellular repair and
intracellular zinc transport | 65 | (45) |
Urokinase plasminogen activator (uPA; PLAU) plays a
key role in tissue degradation, cell migration, angiogenesis,
cancer invasion and metastasis. uPA is a member of the serine
protease family and is strongly implicated as a promoter of tumor
progression in various human malignancies. It is synthesized and
secreted as a pro-enzyme, whose activation is markedly accelerated
upon binding to specific membrane-bounded or soluble cell surface
uPA receptors (uPARs). Binding to uPAR, uPA efficiently converts
the inactive zymogen, plasminogen, into the active serine protease,
plasmin, which then directly or indirectly cleaves extracellular
matrix components including laminin, fibronectin, fibrin,
vitronectin and collagen (41,42).
The link between the hypomethylation of the PLAU gene and
alterations in chromosome 10 and the invasiveness and metastasis of
different cancers has been confirmed, including PC (5,8).
Heparanase (HPSE), an endo-β-D-glucuronidase, can
cleave the heparan sulfate chain of heparan sulfate proteoglycans,
and is actively involved in the process of extracellular matrix
degradation. Heparanase activity is detectable in platelets,
neutrophils, activated T lymphocytes, and various malignancies
including PC. Hypomethylation of the HPSE gene has been
identified in 8.5–30% of PC cases (43).
Cytochrome P450s are a multigene family of
constitutively expressed and inducible enzymes involved in the
oxidative metabolic activation and deactivation of carcinogens and
cancer therapeutics. Cytochrome P450 1B1 (CYP1B1) is encoded by a
member of the CYP1 gene family CYP1B1 and is one of the major
enzymes involved in the hydroxylation of estrogens and activation
of potential carcinogens. CYP1B1 has been found to be
hypomethylated in 5.7–17.1% of PC cases (44).
Wingless-related MMTV integration site 5A (WNT5A),
S100 calcium-binding protein P (S100P) and cysteine-rich protein 1
(CRIP1) genes are also found to be hypomethylated in primary PC
tissues. The encoded proteins similarly as other hypomethylated
genes are associated with tumorigenesis and metastasis (3,45).
WNT5A activates the WNT/β-catenin-independent
pathway and its overexpression is associated with tumor
aggressiveness enhancing invasive activity. For this action,
WNT5A-induced receptor endocytosis with clathrin is required. WNT5A
promotes the aggressiveness of PC and its expression is involved in
relapse after prostatectomy (46,47).
S100P protein regulates calcium signal transduction
and mediates cytoskeletal interaction, protein phosphorylation and
transcriptional control. It is suggested that S100P could be
considered as a potential drug target or a chemosensitization
target, and could also serve as a biomarker for aggressive,
hormone-refractory and metastatic PC (48).
Cysteine-rich intestinal protein 1 (CRIP1) belongs
to the LIM/double-zinc finger protein family and has been shown to
be overexpressed in several tumor types, including breast,
endometrial and prostate (45,49,50).
Histone modifications and prostate
cancer
Histones are highly conserved alkaline proteins that
can become post-translationally modified at amino acid residues
located on their N- and C-terminal tails. There are four core
histones: histone 2A (H2A), histone 2B (H2B), histone 3 (H3) and
histone 4 (H4), and one linker histone, histone 1 (H1).
Approximately 146/7 base pairs of DNA are wound around each histone
octamer, which consists of two copies of each of the core histones,
in left-handed superhelical turns (51).
Modifications involving covalent
attachment/detachment of the molecules or functional groups to the
N-terminal tails of histone contribute to changes of chromatin
structure. The main histone modifications include: methylation,
acetylation and phosphorylation. Changes in chromatin structure are
the result of the remodeling caused by looseness, shifting
nucleosomes along the DNA and/or replacement of histones comprising
the nucleosome core. Post-translational modifications of histones
play an important role in the organization of chromatin structure,
particularly in the homeostasis between euchromatin,
transcriptionally active and heterochromatin, transcriptionally
inactive. Histone modifications can lead to activation or
repression of gene expression, depending on the type and position
of attached functional groups and amino acid residues which are
modified (2,51).
In tumor cells, changes in the pattern of the
modification and genomic location of modified histone occur. These
alterations overlap in the early stages of tumorigenesis and
accumulate as the disease progresses. Changes in the pattern of
histone modification in the promoter sequences may be the cause of
altered gene expression by activation of oncogenes and TSG
repression. These mechanisms may contribute to oncogenesis through
upregulation of transcription, replication, DNA repair or cell
cycle progression (51,52).
Histone acetylation
Acetylation is a key histone modification and
introduction of an acetyl group to the lysine residues of the
histone tail. Histone acetyltransferases (HATs) and histone
deacetylases (HDACs) are a family of enzymes that
acetylate/deacetylate the histone tails of the nucleosome. Linking
these reversible modifications with changes in gene expression
contribute to the recognition of HATs as co-activators and HDACs as
co-repressors of transcription. Mutations in genes encoding the
above-mentioned enzymes may result in neoplastic transformation
(2).
In prostate tumor cells, increased activity of
HDACs, and particularly isoforms HDAC1, HDAC2, and HDAC3, which is
correlated with elevated serum PSA levels and with increased
invasiveness of tumor cells has been observed. In cancer therapy,
HDAC inhibitors of the activity of HDACs are used to inhibit
proliferation and induce differentiation or apoptosis of neoplastic
cells. Hyperacetylation resulting from treatment with HDAC
inhibitors has been observed in both normal and neoplastic cells;
however, proapoptotic and antiproliferative activity is increased
in tumor cells. In addition, studies have demonstrated a lower
level of serum PSA in patients who were treated with HDAC
inhibitors. HDAC inhibitors activate transcription of the gene
encoding the protein p21, which is an inhibitor of cyclin-dependent
kinases, and simultaneously inhibits expression of cyclins D1, A
and E. The presence of p21 protein affects cell cycle arrest at the
G1 phase or G2/M in tumor cells (1,2,8).
Histone methylation
Histone methylation is a modification catalyzed by
histone methyltransferase (HMT) carrying a methyl group
(-CH3) derived from S-adenosylmethionine (SAM) on a
lysine or arginine residue. Methyl groups from lysine or arginine
residues are removed by histone demethylase (HDM). For the lysine
residue in histone mono-, di- and trimethyl groups may be attached.
The methylation of histones is associated with activation, as well
as repression of gene expression, depending on the position of
amino acid residues in a protein and the level of methylation
(53,54).
In PC cells, reduced levels of histone 3 lysine 4
trimethylation (H3K4me3), and histone 3 lysine 18 monoacetylation
(H3K18Ac) are observed. Low levels of these two modifications in
patients with PC are negative prognostic indicators as well as
indicators of an increased risk of relapse in comparison with
patients with high levels of these modifications. Elevated levels
of histone 3 lysine 9 and 18 monoacetylation (H3K9Ac, H3K18Ac) and
histone 3 lysine 4 dimethylation (H3K4me2) allow discrimination
between normal and PC cells (51,53).
Phosphorylation of histones
Phosphorylation of histones is catalyzed by a number
of kinases such as the cyclin-dependent kinases, called mitogen-
and stress-activated protein kinase (MSK) and the kinase Aurora B.
Phosphorylation is closely associated with the cell cycle. Most
phosphorylation take place on histone 3 serine residues at position
10 and 28, as well as threonine residue at positions 3, 6 and 11
(55,56).
DNA double-strand breaks which may lead to
chromosomal aberrations and/or apoptosis contribute to the
phosphorylation of serine residue at position 139 of histone
subtype 2A, called H2AX. The phosphorylated form of histone H2AX is
termed γH2AX. Histone H2AX phosphorylation is catalyzed by protein
kinase ataxia teleangiectasia mutated (ATM) and DNA-dependent
protein kinase (DNA-PK) which belong to the family of
phosphatidylinositol 3-kinase-related protein kinases (PI3KK/PIKK
kinases). Histone γH2AX causes relaxation of chromatin and thereby
facilitates the accumulation of repair factors at the site of
damage. Nanni et al (57)
demonstrated the presence of histone γH2AX at different stages of
PC. Detailed knowledge of the function of histone H2AX may be
important for the diagnosis of cancers, including PC and tumor
therapy (2,55,58,59).
In PC, overexpression of histone H2AZ, which is a variant of
histone 2A was observed (60).
H2A.Z is also associated with androgen receptor (AR) gene
transactivation and progression of PC. In PC histone 2AZ is
ubiquitinated in K120, K121 and K125 and is associated with
transcriptional silencing (61).
Recent studies have attempted to determine the
relationship between the activity of the proliferation marker Ki-67
(Kiel) in PC, and the level of phosphorylation of histone H3. The
expression of Ki-67 protein was found in all cell cycle phases
except G0 phase, whereas the phosphorylation of histone H3 was
identified only in G2 phase and mitosis. One study showed a higher
frequency of phosphorylation of histone H3 in PC, compared to BPH.
Furthermore, histone H3 phosphorylation in PC was found to
correlate with the proliferation index and expression of the Ki-67
protein and the level of serum PSA (62).
Role of histone modification in MDR1
gene silencing in prostate cancer
Multidrug resistance receptor 1 (MDR1) gene
encodes a transmembrane P-glycoprotein (P-gp), which is an
ATP-dependent transporter. P-glycoprotein plays an important role
in the removal of xenobiotics captured in the cell membrane and
their removal outside of the cell, which in the case of tumor cells
determines their resistance to the chemotherapeutic agents used.
MDR1 promoter methylation is common in prostate carcinoma.
However, studies indicate no correlation between MDR1 gene
promoter methylation and reduced transcription of the gene in the
early stages of carcinogenesis while post-translational
modifications of histones appear to be the primary mechanism for
suppression of the MDR1 gene. MDR1 promoter
methylation and P-gp expression in 121 PC, 39 high-grade prostatic
intraepithelial neoplasia (HGPIN), 28 BPH and 10 morphologically
normal prostate tissue (NPT) samples were studied, using
quantitative methylation-specific PCR and immunohistochemistry,
respectively. PC cell lines were exposed to a DNMT inhibitor,
5-aza-2′deoxycytidine (DAC), and histone deacetylase inhibitor
trichostatin A (TSA). Methylation and histone post-transcriptional
modification statuses were characterized and correlated with mRNA
and protein expression. MDR1 promoter methylation levels and
frequency were significantly increased from NPTs, to HGPIN and to
PC. Conversely, decreased or absent P-gp immunoexpression was
observed in HGPIN and PC, inversely correlating with methylation
levels. Exposure to DAC alone did not significantly alter
methylation levels, although increased expression was apparent.
However, P-gp mRNA and protein re-expression were higher in cell
lines exposed to TSA alone or combined with DAC. Accordingly,
histone active markers H3Ac, H3K4me2, H3K4me3, H3K9Ac and H4Ac were
increased at the MDR1 promoter after exposure to TSA alone
or combined with DAC (63).
Modifications of histones and proteins
polycomb and Trithorax in the development of prostate cancer
Polycomb group (PcG) proteins as well as trithorax
group (TrxG) proteins have been discovered to cooperatively
maintain the desirable histone patterns by methylating the histone
tails for precise gene expression in various cellular processes.
Playing opposing roles although both act to modify lysine residues
within histone tails, activities of the PcG proteins are associated
with repression of transcription while those of Trx proteins are
associated with enhancement (64).
PcG proteins form repressive complexes (PRCs). One
of them is the mammalian PRC1 complex consisting of the following
proteins: three Ph homologues [polyhomeotic-like protein 1 (PHC1),
PHC2 and PHC3], five Pc homologues [chromobox protein homologue 2
(CBX2), CBX4, CBX6, CBX7 and CBX8], two Psc homologues (BMI1 and
MEL18) and four other polycomb group RING finger proteins (PCGFs)
(65). Mammalian PRC2 complexes
contain the direct homologues enhancer of zeste called homologue 2
(EZH2) (or, in some cases, EZH1), embryonic ectoderm development
(EED), suppressor of zeste called 12 proteins (SUZ12) and RBBP4 and
RBBP7 (histone-binding protein, also known as retinoblastoma
associated protein 46 and 48 RBAP48 and RBAP46). PcG repressor
complexes inactivate specific regions of chromatin, inhibiting
transcription by specific modification of histone tails, then
recognized by specific effector proteins (65–67).
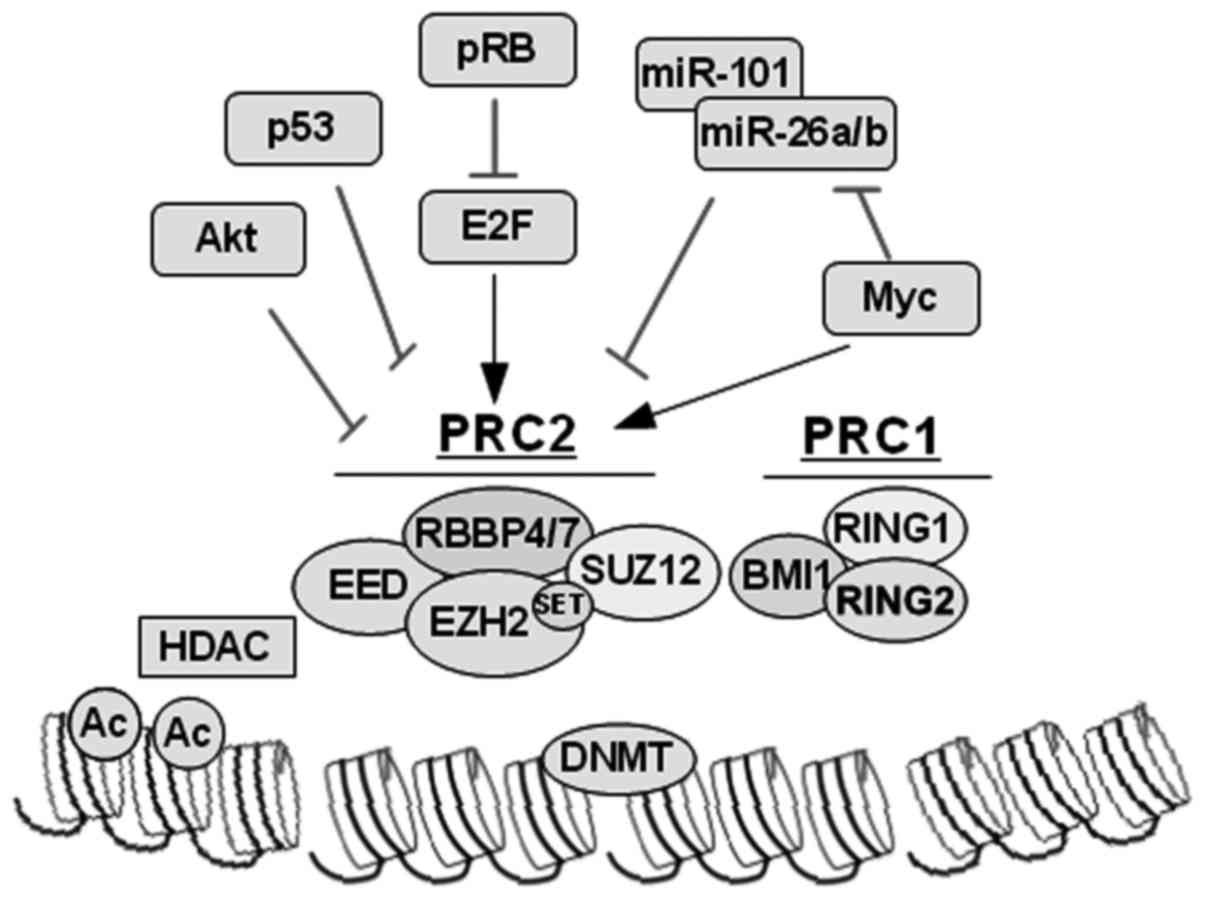
EZH2 catalyzes trimethylation of the histone H3 at
lysine 27 and is the enzymatic member of the polycomb repressor
complex PRC2. In addition, EZH2 induces chromatin compaction and
epigenetic silencing of key TSGs in cooperation with PRC1, HDAC and
DNMT, which subsequently result in tumorigenesis and metastasis.
EZH2 itself can be regulated post-translationally (by Akt-mediated
phosphorylation), post-transcriptionally (by microRNAs), and
through multiple pathways transcriptionally (by E2F, p53 and Myc).
Molecular targeting of EZH2, as well as HDAC and DNMT, provides
important lines for the development of therapeutic strategies with
which to target EZH2-high tumors such as late-stage metastatic PC
(Fig. 2) (64).
Moison et al (68) found that RARβ, a TSG frequently
silenced in PC, was hypermethylated in all studied prostate tumors
and methylation levels were positively correlated with H3K27me3
enrichments. Thus, using bisulfite conversion and pyrosequencing of
immunoprecipitated H3K27me3 chromatin, they demonstrated that DNA
methylation and polycomb repression co-exist in vivo at this
locus. They found this repressive association in 6/6 patient tumor
samples of different Gleason score, suggesting a strong interplay
of DNA methylation and EZH2 to silence RARβ during prostate
tumorigenesis.
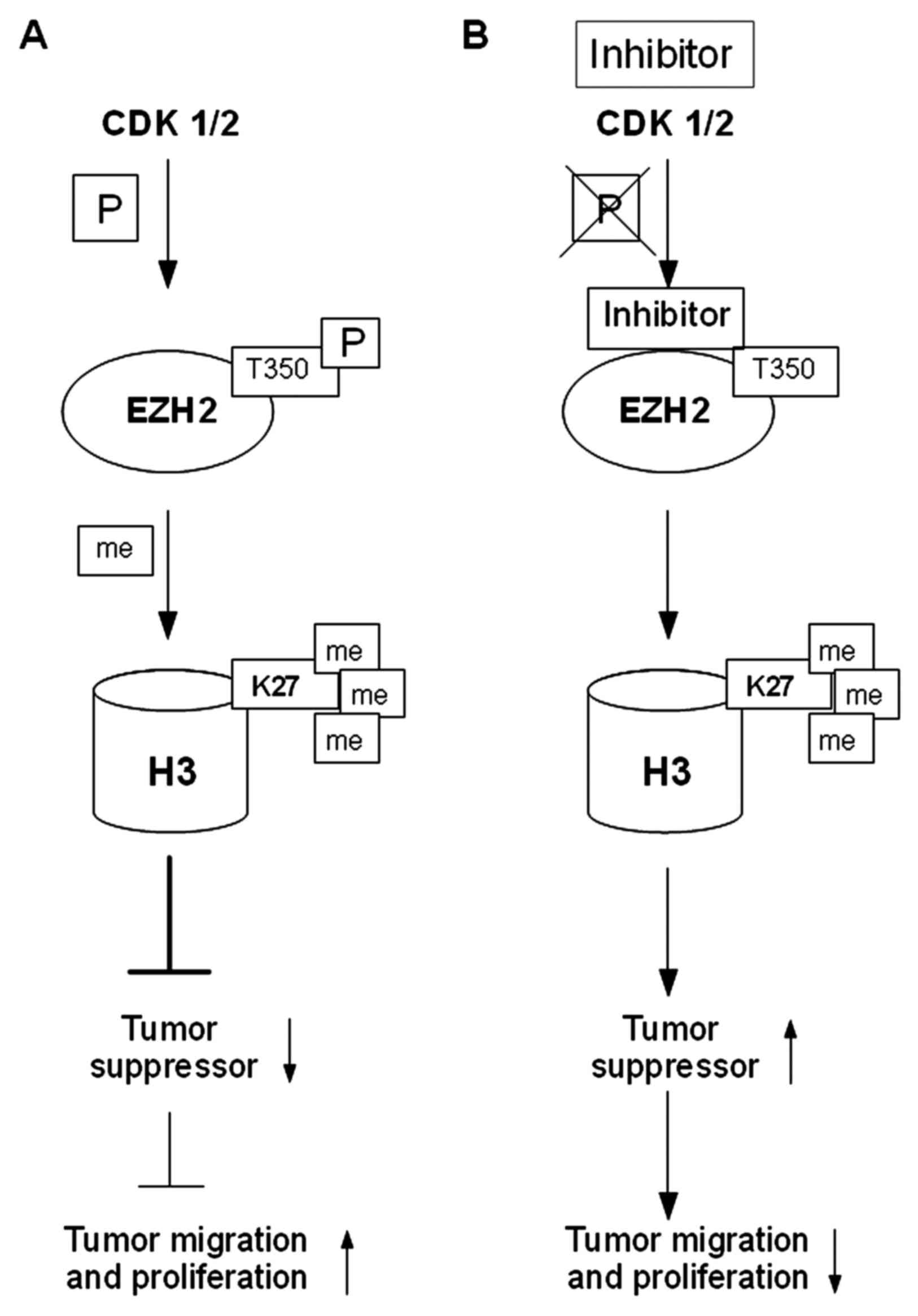
Cyclin-dependent kinases 1 and 2 (CDK1 and CDK2)
phosphorylate threonine at position 350 (T350) of the EZH2 histone
methyltransferase, which via SET domain (histone methyltransferase
domain), catalyzes trimethylation of lysine at position 27 of
histone H3 (H3K27me3). EZH2 phosphorylation at T350 neither affects
the assembly of the PRC2 complex nor the intrinsic HMTase activity
of PRC2. Phosphorylation at T350 is important for the recruitment
of EZH2 to the promoters of its target genes. Trimethylated lysine
residue at position 27 of histone H3 (H3K27me3) carried by the EZH2
protein, correlates with silencing of expression of the
SLIT2 gene (Slit homolog 2; SLIT2) in most cases of
metastatic PC (69,70).
In PC, enhanced transcription of the EZH2
gene was observed, resulting in an increased level of methylation
of the lysine residue at position 27 of histone H3 (H3K27me), which
inhibits the expression of TSGs, such as DAB2 protein interaction
(DAB2IP), adrenoceptor β2 (ADRβ2) and CDH1.
Epigenetic silencing of uppressor genes was found to lead to the
activation of the RAS proto-oncogene and the gene encoding the
nuclear transcription factor-κB (NF-κB), which contributes to
increased proliferation and migration of tumor cells. An increase
in the EZH2 expression level was found to correlate with the
development of PC and may indicate an aggressive nature of cancer.
In the presence of an inhibitor of CDK1/2 and EZH2/T350,
3-dezaneplanocin A (DZNeP), a decrease in H4K20 followed by an
increase in expression of TSGs, and subsequent blocking of the
growth and invasion of PC cells were observed (Fig. 3) (8,69).
Antagonists of polycomb proteins are trithorax group
(TrxG) proteins, which prevent the inhibition of transcription,
resulting in the relaxation of chromatin and a consequent increase
in access of transcriptional activators. TrxGs include both
ATP-dependent chromatin remodeling factors and histone modifiers.
Histone modifier TrxGs in mammals are grouped into 3 major
complexes: complex protein associated with SET domain (COMPASS),
COMPASS-like and ASH (absent small and homeotic discs). COMPASS
contains a histone methyltransferase domain, which is shared with
PRC2. COMPASS mediates H3K4me, unlike PRC2. COMPASS-like complexes
also display the SET domain, which is used to silence a more
restricted group of genes. Through H4K16 acetylation, COMPASS-like
can also activate gene expression. This complex is also able to
demethylate H3K27me3, depending on subunit composition thereby
directly counteracting PRC2. ASH1 protein can catalyze H3K36me3
giving a signal for activation (66,71).
Conclusion
Epigenetics is the study of the modification of DNA
and associated proteins without changing the nucleotide sequence.
DNA methylation, histone modification, nucleosome remodeling, and
RNA-mediated targeting regulate many biological processes that play
an important role in normal cell function, and trends occurring in
the process of carcinogenesis. These steps result in changes in
gene expression and disruption of cellular processes, initiating
the promotion of the development of cancer, including PC. The most
common epigenetic changes used as biomarkers of cancer are DNA
methylation and histone modifications.
References
|
1
|
Damaschke NA, Yang B, Bhusari S, Svaren JP
and Jarrard DF: Epigenetic susceptibility factors for prostate
cancer with aging. Prostate. 73:1721–1730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Willard SS and Koochekpour S: Regulators
of gene expression as biomarkers for prostate cancer. Am J Cancer
Res. 2:620–657. 2012.PubMed/NCBI
|
|
3
|
Chin SP, Dickinson JL and Holloway AF:
Epigenetic regulation of prostate cancer. Clin Epigenetics.
2:151–169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Day TK and Bianco-Miotto T: Common gene
pathways and families altered by DNA methylation in breast and
prostate cancers. Endocr Relat Cancer. 20:R215–R232. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Majumdar S, Buckles E, Estrada J and
Koochekpour S: Aberrant DNA methylation and prostate cancer. Curr
Genomics. 12:486–505. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Subramaniam D, Thombre R, Dhar A and Anant
S: DNA methyltransferases: A novel target for prevention and
therapy. Front Oncol. 4:802014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buck-Koehntop BA and Defossez PA: On how
mammalian transcription factors recognize methylated DNA.
Epigenetics. 8:131–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Albany C, Alva AS, Aparicio AM, Singal R,
Yellapragada S, Sonpavde G and Hahn NM: Epigenetics in prostate
cancer. Prostate Cancer. 2011:5803182011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang M and Park JY: DNA methylation in
promoter region as biomarkers in prostate cancer. Methods Mol Biol.
863:67–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashour N, Angulo JC, Andrés G, Alelú R,
González-Corpas A, Toledo MV, Rodríguez-Barbero JM, López JI,
Sánchez-Chapado M and Ropero S: A DNA hypermethylation profile
reveals new potential biomarkers for prostate cancer diagnosis and
prognosis. Prostate. 74:1171–1182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gonzalgo ML, Pavlovich CP, Lee SM and
Nelson WG: Prostate cancer detection by GSTP1 methylation analysis
of postbiopsy urine specimens. Clin Cancer Res. 9:2673–2677.
2003.PubMed/NCBI
|
|
12
|
Rouprêt M, Hupertan V, Yates DR, Catto JW,
Rehman I, Meuth M, Ricci S, Lacave R, Cancel-Tassin G, de la Taille
A, et al: Molecular detection of localized prostate cancer using
quantitative methylation-specific PCR on urinary cells obtained
following prostate massage. Clin Cancer Res. 13:1720–1725. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dumache R, Puiu M, Motoc M, Vernic C and
Dumitrascu V: Prostate cancer molecular detection in plasma samples
by glutathione S-transferase P1 (GSTP1) methylation analysis. Clin
Lab. 60:847–852. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bastian PJ, Palapattu GS, Lin X,
Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW
and Nelson WG: Preoperative serum DNA GSTP1 CpG island
hypermethylation and the risk of early prostate-specific antigen
recurrence following radical prostatectomy. Clin Cancer Res.
11:4037–4043. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reibenwein J, Pils D, Horak P, Tomicek B,
Goldner G, Worel N, Elandt K and Krainer M: Promoter
hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate
cancer patients and its clinical relevance. Prostate. 67:427–432.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mahon KL, Qu W, Devaney J, Paul C,
Castillo L, Wykes RJ, Chatfield MD, Boyer MJ, Stockler MR, Marx G,
et al: PRIMe consortium: Methylated Glutathione S-transferase 1
(mGSTP1) is a potential plasma free DNA epigenetic marker of
prognosis and response to chemotherapy in castrate-resistant
prostate cancer. Br J Cancer. 111:1802–1809. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang GH, Lee S, Lee HJ and Hwang KS:
Aberrant CpG island hypermethylation of multiple genes in prostate
cancer and prostatic intraepithelial neoplasia. J Pathol.
202:233–240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sidhu S, Deep JS, Sobti RC, Sharma VL and
Thakur H: Methylation pattern of MGMT gene in relation to age,
smoking, drinking and dietary habits as epigenetic biomarker in
prostate cancer patients. GEBJ. 8:1–11. 2010.
|
|
19
|
Tang D, Kryvenko ON, Mitrache N, Do KC,
Jankowski M, Chitale DA, Trudeau S, Rundle A, Belinsky SA and
Rybicki BA: Methylation of the RARB gene increases prostate cancer
risk in black Americans. J Urol. 190:317–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keil KP, Abler LL, Mehta V, Altmann HM,
Laporta J, Plisch EH, Suresh M, Hernandez LL and Vezina CM: DNA
methylation of E-cadherin is a priming mechanism for prostate
development. Dev Biol. 387:142–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kito H, Suzuki H, Ichikawa T, Sekita N,
Kamiya N, Akakura K, Igarashi T, Nakayama T, Watanabe M, Harigaya
K, et al: Hypermethylation of the CD44 gene is associated with
progression and metastasis of human prostate cancer. Prostate.
49:110–115. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Singal R, Ferdinand L, Reis IM and
Schlesselman JJ: Methylation of multiple genes in prostate cancer
and the relationship with clinicopathological features of disease.
Oncol Rep. 12:631–637. 2004.PubMed/NCBI
|
|
23
|
Woodson K, Hayes R, Wideroff L, Villaruz L
and Tangrea J: Hypermethylation of GSTP1, CD44, and E-cadherin
genes in prostate cancer among US Blacks and Whites. Prostate.
55:199–205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Padar A, Sathyanarayana UG, Suzuki M,
Maruyama R, Hsieh JT, Frenkel EP, Minna JD and Gazdar AF:
Inactivation of cyclin D2 gene in prostate cancers by aberrant
promoter methylation. Clin Cancer Res. 9:4730–4734. 2003.PubMed/NCBI
|
|
25
|
Henrique R, Costa VL, Cerveira N, Carvalho
AL, Hoque MO, Ribeiro FR, Oliveira J, Teixeira MR, Sidransky D and
Jerónimo C: Hypermethylation of Cyclin D2 is associated with loss
of mRNA expression and tumor development in prostate cancer. J Mol
Med. 84:911–918. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Das PM, Ramachandran K, Vanwert J,
Ferdinand L, Gopisetty G, Reis IM and Singal R: Methylation
mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer.
5:282006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Delgado-Cruzata L, Hruby GW, Gonzalez K,
McKiernan J, Benson MC, Santella RM and Shen J: DNA methylation
changes correlate with Gleason score and tumor stage in prostate
cancer. DNA Cell Biol. 31:187–192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Kron KJ, Pethe VV, Demetrashvili N,
Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L,
van der Kwast TH, et al: Association of tissue promoter methylation
levels of APC, TGFβ2, HOXD3 and RASSF1A with prostate cancer
progression. Int J Cancer. 129:2454–2462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jerónimo C, Henrique R, Hoque MO, Ribeiro
FR, Oliveira J, Fonseca D, Teixeira MR, Lopes C and Sidransky D:
Quantitative RARbeta2 hypermethylation: A promising prostate cancer
marker. Clin Cancer Res. 10:4010–4014. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Youssef EM, Chen XQ, Higuchi E, Kondo Y,
Garcia-Manero G, Lotan R and Issa JP: Hypermethylation and
silencing of the putative tumor suppressor Tazarotene-induced gene
1 in human cancers. Cancer Res. 64:2411–2417. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang J, Liu L and Pfeifer GP: Methylation
of the retinoid response gene TIG1 in prostate cancer correlates
with methylation of the retinoic acid receptor beta gene. Oncogene.
23:2241–2249. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choudhury JH and Ghosh SK: Promoter
hypermethylation profiling identifies subtypes of head and neck
cancer with distinct viral, environmental, genetic and survival
characteristics. PLoS One. 10:e01298082015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin JC, Wu YC, Wu CC, Shih PY, Wang WY and
Chien YC: DNA methylation markers and serum α-fetoprotein level are
prognostic factors in hepatocellular carcinoma. Ann Hepatol.
14:494–504. 2015.PubMed/NCBI
|
|
34
|
Zhang CY, Zhao YX, Xia RH, Han J, Wang BS,
Tian Z, Wang LZ, Hu YH and Li J: RASSF1A promoter hypermethylation
is a strong biomarker of poor survival in patients with salivary
adenoid cystic carcinoma in a Chinese population. PLoS One.
9:e1101592014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maruyama R, Toyooka S, Toyooka KO, Virmani
AK, Zöchbauer-Müller S, Farinas AJ, Minna JD, McConnell J, Frenkel
EP and Gazdar AF: Aberrant promoter methylation profile of prostate
cancers and its relationship to clinicopathological features. Clin
Cancer Res. 8:514–519. 2002.PubMed/NCBI
|
|
36
|
Ge YZ, Xu LW, Jia RP, Xu Z, Feng YM, Wu R,
Yu P, Zhao Y, Gui ZL, Tan SJ, et al: The association between
RASSF1A promoter methylation and prostate cancer: Evidence from 19
published studies. Tumour Biol. 35:3881–3890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pellacani D, Kestoras D, Droop AP, Frame
FM, Berry PA, Lawrence MG, Stower MJ, Simms MS, Mann VM, Collins
AT, et al: DNA hypermethylation in prostate cancer is a consequence
of aberrant epithelial differentiation and hyperproliferation. Cell
Death Differ. 21:761–773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Florl AR, Steinhoff C, Müller M, Seifert
HH, Hader C, Engers R, Ackermann R and Schulz WA: Coordinate
hypermethylation at specific genes in prostate carcinoma precedes
LINE-1 hypomethylation. Br J Cancer. 91:985–994. 2004.PubMed/NCBI
|
|
39
|
Schulz WA, Elo JP, Florl AR, Pennanen S,
Santourlidis S, Engers R, Buchardt M, Seifert HH and Visakorpi T:
Genomewide DNA hypomethylation is associated with alterations on
chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer.
35:58–65. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gurel B, Iwata T, Koh CM, Jenkins RB, Lan
F, Van Dang C, Hicks JL, Morgan J, Cornish TC, Sutcliffe S, et al:
Nuclear MYC protein overexpression is an early alteration in human
prostate carcinogenesis. Mod Pathol. 21:1156–1167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
LeBeau AM, Sevillano N, Markham K, Winter
MB, Murphy ST, Hostetter DR, West J, Lowman H, Craik CS and
VanBrocklin HF: Imaging active urokinase plasminogen activator in
prostate cancer. Cancer Res. 75:1225–1235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y and Cozzi PJ: Targeting uPA/uPAR in
prostate cancer. Cancer Treat Rev. 33:521–527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ogishima T, Shiina H, Breault JE,
Tabatabai L, Bassett WW, Enokida H, Li LC, Kawakami T, Urakami S,
Ribeiro-Filho LA, et al: Increased heparanase expression is caused
by promoter hypomethylation and up-regulation of transcriptional
factor early growth response-1 in human prostate cancer. Clin
Cancer Res. 11:1028–1036. 2005.PubMed/NCBI
|
|
44
|
Tokizane T, Shiina H, Igawa M, Enokida H,
Urakami S, Kawakami T, Ogishima T, Okino ST, Li LC, Tanaka Y, et
al: Cytochrome P450 1B1 is overexpressed and regulated by
hypomethylation in prostate cancer. Clin Cancer Res. 11:5793–5801.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Q, Williamson M, Bott S,
Brookman-Amissah N, Freeman A, Nariculam J, Hubank MJ, Ahmed A and
Masters JR: Hypomethylation of WNT5A, CRIP1 and S100P in prostate
cancer. Oncogene. 26:6560–6565. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shojima K, Sato A, Hanaki H, Tsujimoto I,
Nakamura M, Hattori K, Sato Y, Dohi K, Hirata M, Yamamoto H, et al:
Wnt5a promotes cancer cell invasion and proliferation by
receptor-mediated endocytosis-dependent and -independent
mechanisms, respectively. Sci Rep. 5:80422015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yamamoto H, Oue N, Sato A, Hasegawa Y,
Yamamoto H, Matsubara A, Yasui W and Kikuchi A: Wnt5a signaling is
involved in the aggressiveness of prostate cancer and expression of
metalloproteinase. Oncogene. 29:2036–2046. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Basu GD, Azorsa DO, Kiefer JA, Rojas AM,
Tuzmen S, Barrett MT, Trent JM, Kallioniemi O and Mousses S:
Functional evidence implicating S100P in prostate cancer
progression. Int J Cancer. 123:330–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lambropoulou M, Deftereou TE, Kynigopoulos
S, Patsias A, Anagnostopoulos C, Alexiadis G, Kotini A, Tsaroucha
A, Nikolaidou C, Kiziridou A, et al: Co-expression of galectin-3
and CRIP-1 in endometrial cancer: Prognostic value and patient
survival. Med Oncol. 33:82016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ludyga N, Englert S, Pflieger K, Rauser S,
Braselmann H, Walch A, Auer G, Höfler H and Aubele M: The impact of
cysteine-rich intestinal protein 1 (CRIP1) in human breast cancer.
Mol Cancer. 12:282013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chervona Y and Costa M: Histone
modifications and cancer: Biomarkers of prognosis? Am J Cancer Res.
2:589–597. 2012.PubMed/NCBI
|
|
52
|
Kurdistani SK: Histone modifications in
cancer biology and prognosis. Prog Drug Res. 67:91–106.
2011.PubMed/NCBI
|
|
53
|
Chen S and Sang N: Histone deacetylase
inhibitors: The epigenetic therapeutics that repress
hypoxia-inducible factors. J Biomed Biotechnol. 2011:1979462011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Crea F, Clermont PL, Mai A and Helgason
CD: Histone modifications, stem cells and prostate cancer. Curr
Pharm Des. 20:1687–1697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cohen I, Poręba E, Kamieniarz K and
Schneider R: Histone modifiers in cancer: Friends or foes? Genes
Cancer. 2:631–647. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sawicka A and Seiser C: Histone H3
phosphorylation - a versatile chromatin modification for different
occasions. Biochimie. 94:2193–2201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nanni S, Priolo C, Grasselli A, D'Eletto
M, Merola R, Moretti F, Gallucci M, De Carli P, Sentinelli S,
Cianciulli AM, et al: Epithelial-restricted gene profile of primary
cultures from human prostate tumors: A molecular approach to
predict clinical behavior of prostate cancer. Mol Cancer Res.
4:79–92. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sedelnikova OA and Bonner WM: GammaH2AX in
cancer cells: A potential biomarker for cancer diagnostics,
prediction and recurrence. Cell Cycle. 5:2909–2913. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shaheen FS, Znojek P, Fisher A, Webster M,
Plummer R, Gaughan L, Smith GC, Leung HY, Curtin NJ and Robson CN:
Targeting the DNA double strand break repair machinery in prostate
cancer. PLoS One. 6:e203112011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Baptista T, Graça I, Sousa EJ, Oliveira
AI, Costa NR, Costa-Pinheiro P, Amado F, Henrique R and Jerónimo C:
Regulation of histone H2A.Z expression is mediated by sirtuin 1 in
prostate cancer. Oncotarget. 4:1673–1685. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Monteiro FL, Baptista T, Amado F, Vitorino
R, Jerónimo C and Helguero LA: Expression and functionality of
histone H2A variants in cancer. Oncotarget. 5:3428–3443. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nowak M, Svensson MA, Carlsson J, Vogel W,
Kebschull M, Wernert N, Kristiansen G, Andrén O, Braun M and Perner
S: Prognostic significance of phospho-histone H3 in prostate
carcinoma. World J Urol. 32:703–707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Henrique R, Oliveira AI, Costa VL,
Baptista T, Martins AT, Morais A, Oliveira J and Jerónimo C:
Epigenetic regulation of MDR1 gene through post-translational
histone modifications in prostate cancer. BMC Genomics. 14:8982013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang YA and Yu J: EZH2, an epigenetic
driver of prostate cancer. Protein Cell. 4:331–341. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Schwartz YB and Pirrotta V: A new world of
Polycombs: Unexpected partnerships and emerging functions. Nat Rev
Genet. 14:853–864. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Piunti A and Shilatifard A: Epigenetic
balance of gene expression by Polycomb and COMPASS families.
Science. 352:aad97802016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang QT: Epigenetic regulation of cardiac
development and function by polycomb group and trithorax group
proteins. Dev Dyn. 241:1021–1033. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Moison C, Senamaud-Beaufort C, Fourrière
L, Champion C, Ceccaldi A, Lacomme S, Daunay A, Tost J, Arimondo PB
and Guieysse-Peugeot AL: DNA methylation associated with polycomb
repression in retinoic acid receptor β silencing. FASEB J.
27:1468–1478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Simon JA and Lange CA: Roles of the EZH2
histone methyltransferase in cancer epigenetics. Mutat Res.
647:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zeng X, Chen S and Huang H:
Phosphorylation of EZH2 by CDK1 and CDK2: A possible regulatory
mechanism of transmission of the H3K27me3 epigenetic mark through
cell divisions. Cell Cycle. 10:579–583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Clermont PL, Crea F and Helgason CD:
Trithorax Genes in Prostate CancerAdvances in Prostate Cancer.
Hamilton G: InTech; Croatia: pp. 541–564. 2013
|
|
72
|
Daniunaite K, Jarmalaite S, Kalinauskaite
N, Petroska D, Laurinavicius A, Lazutka JR and Jankevicius F:
Prognostic value of RASSF1 promoter methylation in prostate cancer.
J Urol. 192:1849–1855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gurioli G, Salvi S, Martignano F, Foca F,
Gunelli R, Costantini M, Cicchetti G, De Giorgi U, Sbarba PD,
Calistri D, et al: Methylation pattern analysis in prostate cancer
tissue: Identification of biomarkers using an MS-MLPA approach. J
Transl Med. 14:2492016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ikromov O, Alkamal I, Magheli A, Ratert N,
Sendeski M, Miller K, Krause H and Kempkensteffen C: Functional
epigenetic analysis of prostate carcinoma: A role for seryl-tRNA
synthetase? J Biomark 2014. 3621642014.
|
|
75
|
Litovkin K, Van Eynde A, Joniau S, Lerut
E, Laenen A, Gevaert T, Gevaert O, Spahn M, Kneitz B, Gramme P, et
al: DNA methylation-guided prediction of clinical failure in
high-risk prostate cancer. PLoS One. 10:e01306512015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Moritz R, Ellinger J, Nuhn P, Haese A,
Müller SC, Graefen M, Schlomm T and Bastian PJ: DNA
hypermethylation as a predictor of PSA recurrence in patients with
low- and intermediate-grade prostate cancer. Anticancer Res.
33:5249–5254. 2013.PubMed/NCBI
|
|
77
|
Serenaite I, Daniunaite K, Jankevicius F,
Laurinavicius A, Petroska D, Lazutka JR and Jarmalaite S:
Heterogeneity of DNA methylation in multifocal prostate cancer.
Virchows Arch. 466:53–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tan HL, Haffner MC, Esopi DM, Vaghasia AM,
Giannico GA, Ross HM, Ghosh S, Hicks JL, Zheng Q, Sangoi AR, et al:
Prostate adenocarcinomas aberrantly expressing p63 are molecularly
distinct from usual-type prostatic adenocarcinomas. Mod Pathol.
28:446–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tsvetkova A, Todorova A, Todorov T,
Georgiev G, Drandarska I and Mitev V: Molecular and
clinicopathological aspects of prostate cancer in Bulgarian
probands. Pathol Oncol Res. 21:969–976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yoon HY, Kim SK, Kim YW, Kang HW, Lee SC,
Ryu KH, Shon HS, Kim WJ and Kim YJ: Combined hypermethylation of
APC and GSTP1 as a molecular marker for prostate cancer:
Quantitative pyrosequencing analysis. J Biomol Screen. 17:987–992.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yoon HY, Kim YW, Kang HW, Kim WT, Yun SJ,
Lee SC, Kim WJ and Kim YJ: DNA methylation of GSTP1 in human
prostate tissues: Pyrosequencing analysis. Korean J Urol.
53:200–205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang W, Jiao H, Zhang X, Zhao R, Wang F,
He W, Zong H, Fan Q and Wang L: Correlation between the expression
of DNMT1, and GSTP1 and APC, and the methylation status of GSTP1
and APC in association with their clinical significance in prostate
cancer. Mol Med Rep. 12:141–146. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Bastian PJ, Ellinger J, Heukamp LC, Kahl
P, Müller SC and von Rücker A: Prognostic value of CpG island
hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in
patients undergoing radical prostatectomy. Eur Urol. 51:665–674;
discussion 674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Müller A and Florek M:
5-Azacytidine/Azacitidine. Recent Results Cancer Res. 184:159–170.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yoon HY, Kim YW, Kang HW, Kim WT, Yun SJ,
Lee SC, Kim WJ and Kim YJ: Pyrosequencing analysis of APC
methylation level in human prostate tissues: A molecular marker for
prostate cancer. Korean J Urol. 54:194–198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yaqinuddin A, Qureshi SA, Pervez S, Bashir
MU, Nazir R and Abbas F: Frequent DNA hypermethylation at the
RASSF1A and APC gene loci in prostate cancer patients of Pakistani
Origin. ISRN Urol. 2013:6272492013.PubMed/NCBI
|
|
87
|
Olkhov-Mitsel E, Zdravic D, Kron K, van
der Kwast T, Fleshner N and Bapat B: Novel multiplex MethyLight
protocol for detection of DNA methylation in patient tissues and
bodily fluids. Sci Rep. 4:44322014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Pakneshan P, Szyf M and Rabbani SA:
Hypomethylation of urokinase (uPA) promoter in breast and prostate
cancer: Prognostic and therapeutic implications. Curr Cancer Drug
Targets. 5:471–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hagelgans A, Menschikowski M, Fuessel S,
Nacke B, Arneth BM, Wirth MP and Siegert G: Deregulated expression
of urokinase and its inhibitor type 1 in prostate cancer cells:
Role of epigenetic mechanisms. Exp Mol Pathol. 94:458–465. 2013.
View Article : Google Scholar : PubMed/NCBI
|