Introduction
Lung cancer is the most common malignant tumor (13%
of all cancers) worldwide, both in terms of morbidity and
mortality. It is more common among men (18% of all cancers), while
in women it occupies the fourth place (9.5%). The morbidity of this
type of tumor shows a growing trend (1). Furthermore, a greater percentage of
cases in both sexes is observed in developing countries (2). The primary reasons for this include,
among others, an aging population and a change in lifestyle
observed over the past few decades, in which an inadequate diet,
the lack of physical activity and smoking are often found (1,3,4).
Recent research on lung cancer in never-smokers has
led to the distinction of a separate disease classification
referred to as lung cancer in never smokers (LCINS) (5–7). The
etiology of this cancer is not fully understood. However, a number
of possible risk factors have been described, e.g. lung disease
family history, passive smoking, diet or work-related exposure to
harmful substances (8). When it
comes to histological classification, two main groups of lung
cancer can be observed: small cell lung cancers (SCLCs) and
non-small cell lung cancers (NSCLCs) (9).
Adenocarcinoma (AC) is the most common NSCLC subtype
(approximately 50% of all diagnosed cases). It develops in the
smaller bronchi and bronchioles and in the alveolar epithelium
(10), and it is most often located
in the peripheral segments of the lung. It is equally common among
smokers and non-smokers, which indicates that its etiopathogenesis
is to a greater extent linked to factors other than tobacco smoke.
Among the different NSCLCs, AC has the most unfavorable prognosis
(10). Due to the complexity of the
molecular mechanisms initiating the development of NSCLCs, it is
necessary to investigate new potential cancer markers. In this
sense, the SOX18 protein appears to be an auspicious element in
future anticancer therapies.
The SOX family proteins (SRY-related HMG-box)
are important transcription factors involved, for example, in the
development of the cardiovascular system and the lymphatic ducts
(11,12). The SOX family is composed of
approximately 20 proteins divided into 10 groups, from A to J
(13,14). There is a low sequence homology
between the groups. However, proteins within the same group show at
least 80% homology in the HMG domain, and they possess other
conservative domains inside the group (13,15).
Group F proteins (SOX7, SOX17, SOX18) are primarily involved in the
development of the cardiovascular system during embryogenesis
(16–18).
It has been observed that SOX18 takes part in wound
healing processes and the development of atherosclerosis (19). Likewise, an increased level of this
protein has been found in melanomas and malignant pancreatic,
stomach and breast tumors (21).
Furthermore, high expression of SOX18 in gastric cancer stromal
cells has been correlated with a poorer patient prognosis (22). In breast cancer, SOX18 is associated
with tumor malignancy grade (G) and Ki-67 proliferation index;
therefore, it can be of particular significance in future prognosis
(21). It has been shown in
additional in vitro studies that the SOX18 protein
stimulates the migration and proliferation of human umbilical vein
endothelial cells (HUVECs), the result of which is intensive
angiogenesis. Moreover, it was demonstrated that inhibition of
SOX18 expression in the MCF-7 human breast cancer cell line
resulted in the weakening of the ability of these cells to migrate
due to the destabilization of the actin cytoskeleton structure,
which further confirms the role of SOX18 in cell migration
(23).
Studies aiming to determine the prognostic
significance of the SOX18 expression have been conducted in
gastric, breast and lung cancer to date (24–27).
An increased expression of all of the genes from the SOX F group
has been shown in gastric tumor tissue compared to unaltered
stomach tissue. Furthermore, it was discovered that cases with high
SOX18 expression are characterized by a shorter recurrence-free
survival time. The results of these studies suggest that an
increase in the SOX18 protein expression may be an adverse
prognostic factor in gastric cancer (22).
The role of SOX18 expression in NSCLC is not yet
fully understood. However, taking into account previous reports,
this protein may be an important factor in the development and
progression of NSCLCs. Our preliminary studies have shown a
disparity in the amount of SOX18 mRNA relative to the
quantity of protein (25–27), which may be associated with the
regulation of the translation by microRNAs (miRNAs). This has
allowed us to hypothesize that the SOX18 transcript level is
subject to control by miRNA molecules, since similar mechanisms are
observed in many other types of tumors in relation to different
types of proteins, as for example miRNA-34b in prostate cancer
(28,29).
miRNAs are involved in many important biological
processes, such as proliferation, cell differentiation, apoptosis,
embryogenesis and organogenesis. An increasingly visible role of
miRNAs in the regulation of cell proliferation and cell
differentiation and apoptosis has drawn the attention of scientists
to the relationship of miRNAs and tumor processes (30–34).
As discovered, miRNAs do not only regulate the expression of
multiple oncogenes and tumor-suppressor genes, but may also act
themselves as oncogenes and suppressors. The correlation between
miRNA expression profiles and patient survival may reveal their
potential role as prognostic markers. A relationship between the
expression level of different miRNAs and the survival of patients
with lung AC has been shown (31,34–36).
As is the case with miR-9500 molecules in lung cancer, miRNAs
affecting the transcription of the SOX18 gene may also be of
great prognostic importance (37).
Discovering a method for a sensitive and efficient determination of
the expression level of miRNA molecules could be useful in
elucidating the pathogenesis of lung cancer, and thereby also in
the reduction of lung cancer mortality.
Materials and methods
Patients and clinical samples
The present experiments were carried out using
archival paraffin blocks of lung AC as well as pairs of AC and
non-malignant lung tissue (NMLT) resected adjacent to the primary
tumor. The samples were obtained during surgical resection in
2012–2016 at the Lower Silesian Lung Diseases Centre in Wroclaw.
The paraffin sections of the AC samples were stained with
haematoxylin and eosin (H&E) in order to verify the
appropriateness of the immunohistochemical (IHC) analyses. The
study group consisted of 50 pairs of AC and NMLTs collected into
RNAlater solution (Qiagen, Hilden, Germany) and stored at −20°C for
RT-qPCR and ddPCR experiments. Additionally, the same samples were
collected, frozen in liquid nitrogen and stored at −80°C for
western blot analysis. Clinical data were derived from hospital
archives and are summarized in Table
I.
 | Table I.AC patient and tumor characteristics
(N=50). |
Table I.
AC patient and tumor characteristics
(N=50).
| Parameters | Data |
|---|
| Age (years) |
|
|
Mean | 64.58±7.72 |
|
Range | 52–81 |
| Sex, n (%) |
|
|
Male | 24 (48.0) |
|
Female | 26 (52.0) |
| Tumor size, n
(%) |
|
| T1 | 21 (42.0) |
| T2 | 22 (44.0) |
| T3 | 3 (6.0) |
| T4 | 4 (8.0) |
| Lymph nodes, n
(%) |
|
| N0 | 28 (56.0) |
| N1, N2,
N3 | 22 (44.0) |
| pTNM, n (%) |
|
| 1A | 16 (32.0) |
| 1B | 7 (14.0) |
| 2A | 13 (26.0) |
| 2B | 0 (0.0) |
| 3A | 12 (24.0) |
| 3B | 0 (0.0) |
| 4 | 2 (4.0) |
| Grade, n (%) |
|
| G1 | 2 (4.0) |
| G2 | 21 (42.0) |
| G3 | 27 (54.0) |
Cell lines
For the present study we used three lung cancer cell
lines (NCI-H1703, NCI-H522, A549) obtained from American Type
Culture Collection (ATCC, Manassas, VA, USA). The NCI-H1703 cell
line was derived from a stage I LSCC, the NCI-H522 cell line from a
stage II AC, and the A549 cell line from a highly malignant AC. The
NCI-H1703 and NCI-H522 cell lines were cultured in RPMI-1640 medium
with the addition of 2 mM L-glutamine (Lonza, Basel, Switzerland).
The A549 cell line was grown in a high glucose DMEM medium with the
addition of 2 mM L-glutamine (Sigma, St. Louis, MO, USA). All media
were supplemented with FBS (Sigma) up to a final concentration of
10%. The cell lines were cultured at 37°C in 5% CO2.
Immunohistochemistry (IHC)
The AC and NMLT samples fixed in 10% buffered
formalin and embedded in paraffin were used for the IHC reactions.
In order to determine SOX18 expression, the murine monoclonal mouse
antibody directed against SOX18 (D-8, Sc-166025; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) was used at a dilution of 1:100
according to a previously established protocol (25). The IHC procedure was performed on
the Autostainer Link 48 (DakoCytomation, Glostrup, Denmark) so as
to provide reliable and repeatable conditions.
RNA extraction, cDNA synthesis and
real-time PCR reactions
Total RNA was isolated from the RNAlater-fixed
samples of AC and their corresponding NMLT samples with the use of
the RNeasy Mini kit (Qiagen). It was later transcribed to cDNA by
means of the High Capacity Reverse Transcriptase kit (Applied
Biosystems, Foster City, CA, USA) according to the manufacturer's
protocol. RT-qPCR was carried out in 20 µl volumes using the TaqMan
Universal PCR MasterMix (Applied Biosystems) on a 7900HT Fast
Real-time PCR System. The TaqMan specific probes used in the
experiment (Hs00746079_s1 for SOX18, 000268 for hsa-miR-7a,
000402 for hsa-miR-24-3p as well as Hs00188166_m1 for SDHA
and 000490 for hsa-miR-191 as reference genes) were also obtained
from Applied Biosystems. The reactions were all performed in
triplicates under the following conditions: activation of
polymerase at 50°C for 2 min, initial denaturation at 94°C for 10
min followed by 40 cycles of denaturation at 94°C for 15 sec, and
annealing and elongation at 60°C for 1 min. The relative mRNA
expression of the markers studied was calculated using the ∆∆Ct
method.
miRNA quantification using Droplet
Digital PCR™ (ddPCR)
Small fractions of RNA containing miRNAs from the
cell lines studied and from the RNAlater-fixed samples of AC and
NMLT were isolated with the use of the mirVana miRNA Isolation kit
(Ambion, Waltham, MA, USA) according to the manufacturer's
protocol. For reverse transcription (RT-PCR), the TaqMan MicroRNA
Reverse Transcription kit (Applied Biosystems) was used together
with the aforementioned miRNA-specific stem-loop primers (Applied
Biosystems). An input of 30 ng of RNA from each sample was
reverse-transcribed using a C1000 Touch Thermal Cycler (Bio-Rad,
Hercules, CA, USA). The thermocycler parameters were as follows:
hold for 30 min at 16°C, for 30 min at 42°C, and finally for 5 min
at 85°C.
The ddPCR reaction mixtures contained 1.33 µl of RT
product, 1 µl of TaqMan miRNA specific probe (Thermo Fisher
Scientific, Walthman, MA, USA), 7.67 µl of molecular biology-grade
water and 10 µl of 2X ddPCR™ MasterMix for Probes (Bio-Rad). The 20
µl of the reaction mixtures were loaded into a plastic cartridge
(Bio-Rad) with 70 µl of Droplet Generation Oil for Probes (Bio-Rad)
in the QX100 Droplet Generator (Bio-Rad). The droplets obtained
from each sample were then transferred to a 96-well PCR plate
(Eppendorf, Hamburg, Germany). PCR amplifications were carried out
in a C1000 Touch thermal cycler at 95°C for 10 min, followed by 40
cycles of 95°C for 3 sec and 60°C for 1 min as well as 1 cycle of
98°C for 10 min ending at room temperature (RT). Finally, the plate
was loaded onto a Droplet Reader (Bio-Rad) and read automatically.
The absolute quantification of each miRNA was calculated from the
number of positive counts per panel by using the Poisson
distribution. The quantification of the target miRNAs is presented
as the number of copies/µl in the PCR reaction mixture.
SDS-PAGE and western blotting
Whole cell lysates were obtained from the AC and
NMLT samples by using the T-PER Tissue Protein Extraction kit
(Thermo Fisher Scientific) with the addition of a cocktail of
inhibitors (Sigma), 250 units of Benzonase® (Merck
Millipore, Bedford, MA, USA) and 2 mM PMSF (phenylmethanesulfonyl
fluoride). The lysates were mixed with 4X SDS-PAGE gel loading
buffer (200 mM Tris-HCl - pH 6.8, 400 mM DTT, 8% SDS, 0.4%
bromophenol blue, 40% glycerol), loaded on 10% acrylamide gel and
separated by SDS-PAGE under reducing conditions, and then
transferred onto a PVDF membrane in the XCell SureLock™ Mini Gel
Electrophoresis System (Thermo Fisher Scientific). After the
protein transfer, the membrane was incubated in a blocker solution
(4% BSA in TBST buffer) for 1 h at RT followed by overnight
incubation at 4°C with anti-SOX18 monoclonal mouse antibody,
diluted 1:100 (D-8, Sc-166025; Santa Cruz Biotechnology).
Subsequently, the membrane was washed with TBST buffer and
incubated for 1 h at RT with secondary donkey anti-mouse antibody
conjugated with HRP, diluted 1:3000 (709-035-149; Jackson
ImmunoResearch, Mill Valley, CA, USA), then rinsed and treated with
the Immun-Star HRP Chemiluminescent kit (Bio-Rad). Rabbit
anti-human β-actin monoclonal antibody (#4970; Cell Signaling
Technology, Danvers, MA, USA), diluted 1:1000, was used as an
internal control. The western blotting results were analyzed using
the ChemiDoc MP System (Bio-Rad).
In vitro studies - MirTrap System
In order to identify that miR-7a and miR-24-3p
interact with the SOX18 gene transcript, an in vitro
test was conducted using the MirTrap System kit (Clontech
Laboratories, Mountain View, CA, USA). The MirTrap System is based
on the DYKDDDDK (FLAG epitope) tag on the dominant-negative subunit
of the RISC protein, which allows for the capture and isolation of
the entire Ago/RISC complex containing the miRNA/target mRNA pair
of interest. After performing the in vitro experiments
(according to the manufacturer's protocol), the miR-7a's and
miR-24-3p's target in the lung cancer cell lines was quantified by
using the RT-qPCR technique. In order to perform the fold
enrichment analysis of the SOX18/miR-7a and
SOX18/miR-24-3p pairs, the positive (AcGFP1/miR-132) and
negative (hPlod3/miR-132) controls were analyzed according to the
manufacturer's protocol.
The NCI-H1703, NCI-H522 and A549 cell lines
expressing the MirTrap protein as well as the mRNA fusion of AcGFP1
with a miR-132 target sequence were cotransfected with the microRNA
mimics for miR-7a and miR-24-3p. The RISC/miRNA/SOX18
complexes were immunoprecipitated via anti-DYKDDDDK (FLAG) beads.
RNA was isolated, and the fold-enrichment of the miRNA target in
the complex was determined with the use of the qRT-PCR technique. A
2.5-fold enrichment was considered to be a positive result. The
enrichment was calculated by using the GAPDH transcript for
normalization purposes. The levels of mRNA were determined with the
quantitative RT-qPCR method with the use of the SYBR-Green kit
(Bio-Rad) and the 7900HT Fast Real-time System (Applied
Biosystems).
Statistical analysis
The Shapiro-Wilk test was used so as to evaluate the
normality assumption of the groups examined. In order to compare
the differences between the LSCC and the NMLT groups, the Wilcoxon
signed-rank test was used. Additionally, the Spearman correlation
test was carried out for the analysis of the existing correlations.
All the statistical analyses were performed using Prism 5.0
(GraphPad, La Jolla, CA, USA). The results were considered
statistically significant at P<0.05.
Results
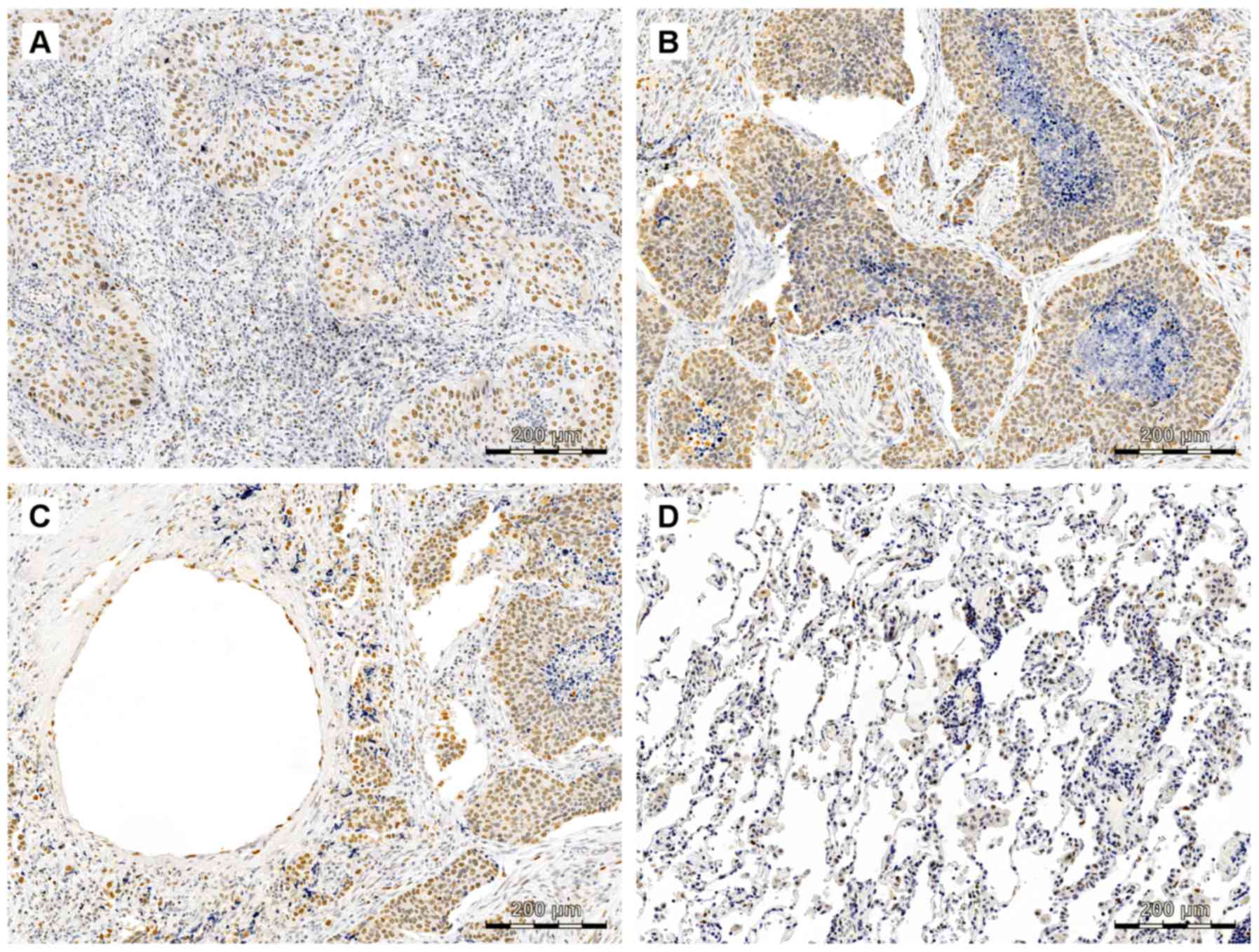
Immunohistochemistry
In the presented results, SOX18 expression was
observed mostly in the nuclei of both cancer and endothelial cells
of vessels (Fig. 1). A nuclear
localization of the SOX18 protein was observed only in AC cells and
in the epithelial cells of vessels, but no SOX18 protein expression
was noted in the tumor stromal and NMLT samples.
SOX18 mRNA expression levels in AC and
NMLT samples - RT-qPCR
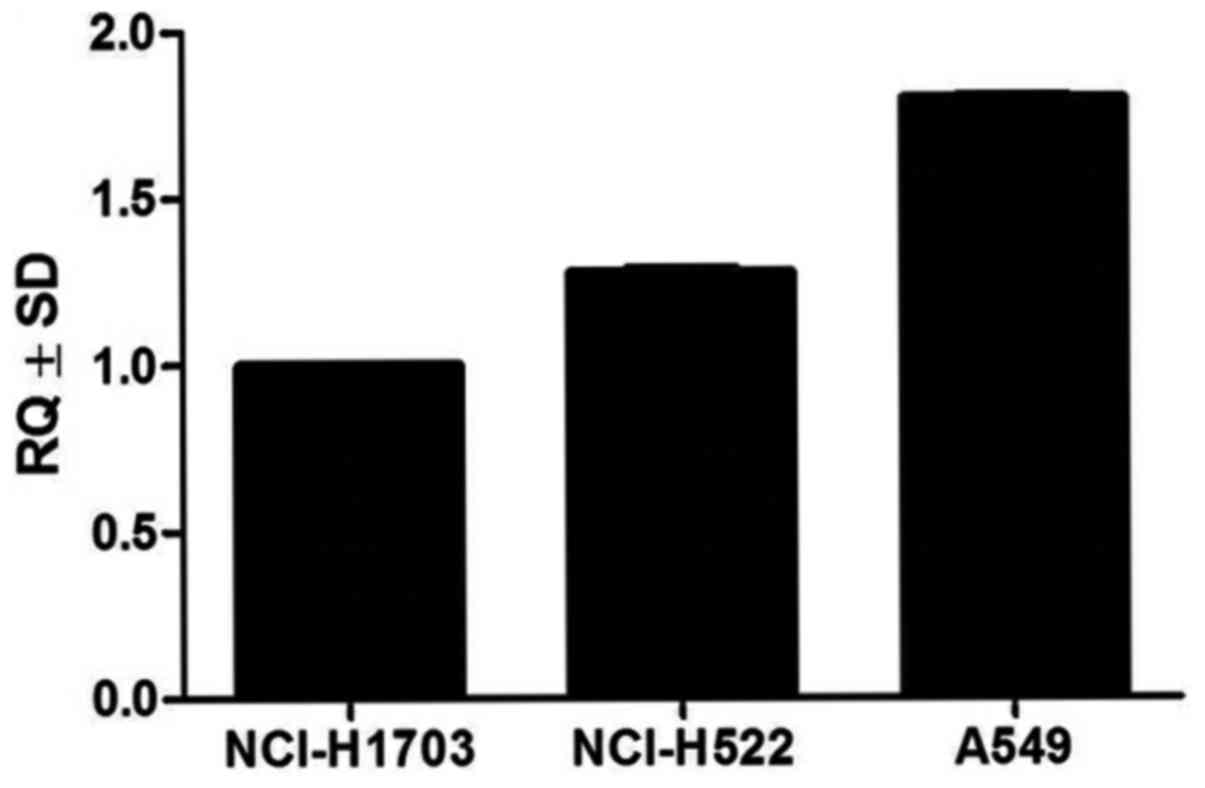
Analysis of SOX18 mRNA levels using the RT-qPCR
technique in the studied cell lines revealed increased expression
in the NCI-H1703, NCI-H522 and A549 cancer cell lines (Fig. 2). The SOX18 mRNA expression
level was determined in all 50 (100%) cases of AC and all 50 (100%)
cases of NMLT (data not shown).
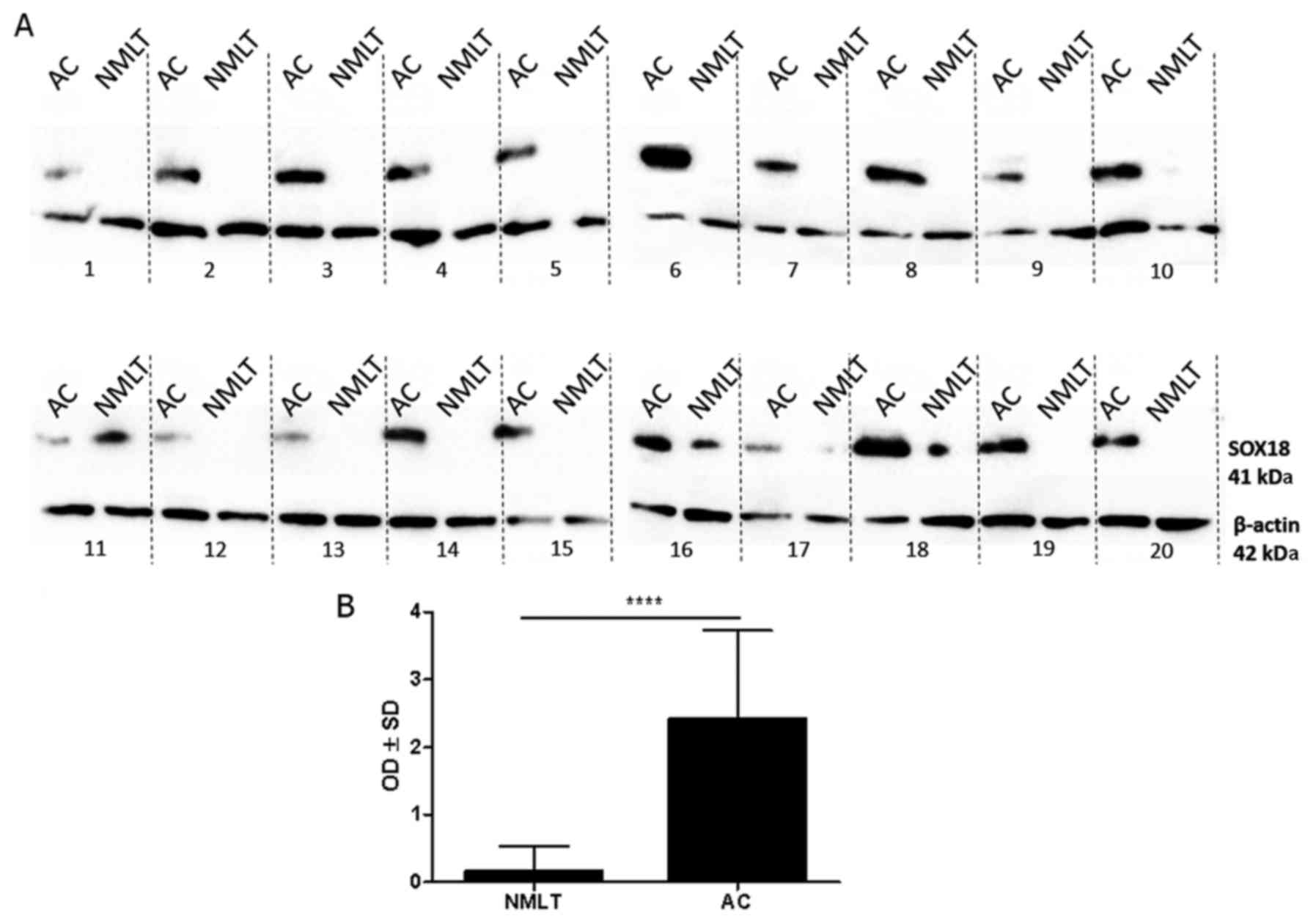
SOX18 protein level - western
blotting
Bands of SOX18 protein were observed at 41 kDa in
the whole-cell fractions of all 50 (100%) cases of AC, but only in
4 (8%) cases of NMLT (Fig. 3A). The
expression of the SOX18 protein was significantly higher in all of
the analyzed cases of AC compared to NMLT (mean OD±SD, 2.42±1.31
vs. 0.16±0.38, respectively; P<0.0001 Wilcoxon signed-rank test)
(Fig. 3B).
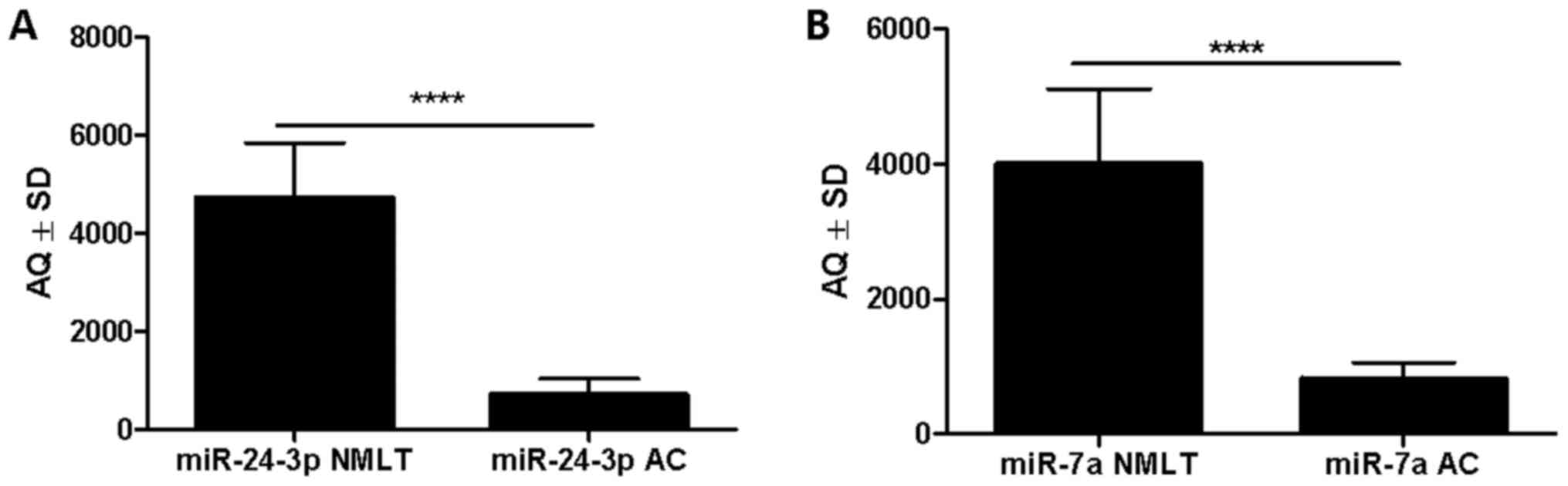
miRNA expression levels - ddPCR
According to the ddPCR absolute quantification
method, miR-7a was significantly more highly expressed in 48 NMLT
cases (96%) compared to that noted in AC (4%) (mean AQ±SD
4,008±1,104 vs. 827±234, respectively; P<0.0001, Wilcoxon
signed-rank test) (Fig. 4B). The
same observation was made for miR-24-3p: there was a higher
expression in 48 cases of NMLT (96%) compared to AC (4%) (mean
AQ±SD 4,726±1,114 vs. 708±325, respectively; P<0.0001, Wilcoxon
signed-rank test) (Fig. 4A).
Overall, both miR-7a and miR-24-3p had a
significantly higher copy number in NMLT samples compared to the AC
tissues. By using the Spearman correlation test, positive
correlations were observed between AQ values of miR-7a and
miR-24-3p in AC cases (r=0.4344, P=0.0016), AQ values of miR-7a and
SOX18 mRNA in AC samples (r=0.3813, P=0.0063), miR-7a and SOX18
mRNA in NMLT cases (r=0.4029, P=0.0023) and AQ values of miR-7a in
NMLT and AC samples (r= −0.3443, P=0.0144).
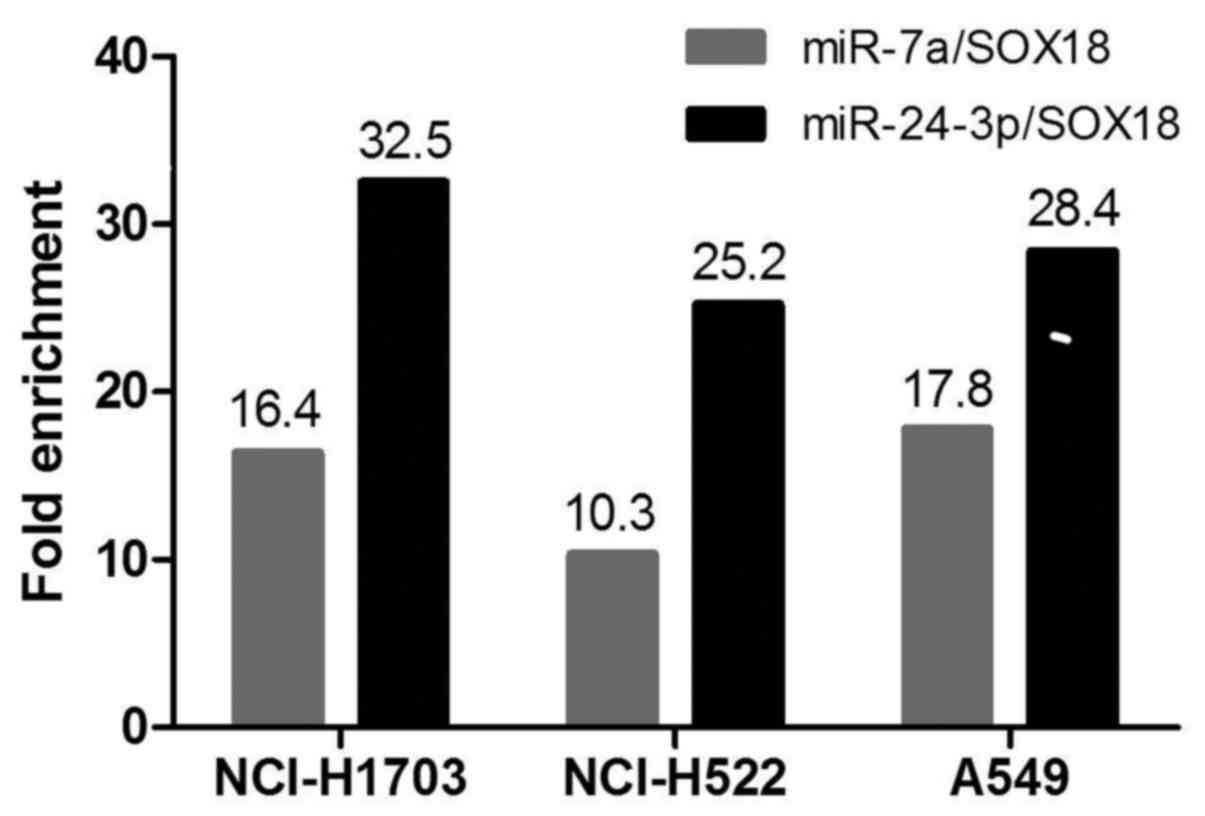
In vitro cell line studies - MirTrap
System
MirTrap System experiments followed by RT-qPCR
analyses validated SOX18 mRNA enriched against the miR-7a/MirTrap
and miR-24-3p/MirTrap complexes. The RT-qPCR results are shown as
relative fold-change between the studied miRNAs and miR-132
(positive control). The results presented in Fig. 5 show that miR-24-3p is classified as
highly enriched, and miR-7a as moderately enriched.
Discussion
SOX family genes (SRY-related HMG-box) were
isolated in mammals in 1990 on the basis of the presence of the
conservative HMG (high mobility group box) protein domain,
primarily occurring in the sex-determining region Y (SRY) (11). SOX proteins can be found in many
tissues at different stages of development, where they fulfill
important functions in a variety of processes occurring in the
body, such as embryonic development, disease processes e.g.
arteriosclerosis, carcinogenesis (14,17,38–40).
In recent years, their role in tumors has been intensively studied,
which has allowed for the demonstration of their participation in
the pathogenesis of many malignant tumors (41).
Based on the results of our previous research, we
observed a differential SOX18 expression both at the mRNA and
protein levels in NSCLC (25,42).
Of note, the level of mRNA did not reflect in any way the level of
protein, determined by the western blot method. This allowed us to
hypothesize that the SOX18 transcript level is subject to
control by miRNA molecules, because similar mechanisms are observed
in many other types of tumors (28,30,35,52).
There are some speculations on the role of the
methylation increment of CpG islands within the SOX18 gene
promoter in NSCLC progression (43). The presence of an altered
methylation profile of the SOX18 gene promoter in tumor and
tumor-surrounding tissue compared to unchanged tissue has been
shown by Azhikina et al in NSCLC cases (44). The authors suggest that the changes
in the methylation pattern of the SOX18 promoter in the
tissues surrounding the tumor (which do not exhibit malignancy
features in the morphological and histological image) may indicate
early genetic changes leading to carcinogenesis.
Varying expression levels of SOX proteins have been
attested depending on the type of cancer in which they occur. This
may indicate that the same protein can serve opposing functions in
different tumors (20). For
example, in a glioblastoma multiform cell line, a significant
overexpression of the SOX2 protein has been observed, while the
silencing of the SOX2 gene has been proved to result in reduced
invasiveness and mitigation of cell migration (45). In turn, a decreased SOX2 expression
has been observed in gastric cancer. All this resulted in the
inhibition of the cell growth by blocking the cell cycle and the
induction of apoptosis in the cells (46).
The molecular mechanisms responsible for the
disparity observed between SOX18 mRNA and the protein level
in NSCLCs are not well known. Even though the hypermethylation of
the SOX18 promoter has been described, the complexity of the
role of SOX18 in NSCLC progression has not yet been fully
explained. Another important feature of lung cancer development,
besides the epigenetic changes, are miRNAs, whose role in NSCLC has
been already well proven (28,31,34,47–52).
Based on our recent studies on lung squamous cell carcinoma (LSCC),
we have been able to determine two miRNA molecules from the
potential panel of miRNAs that most probably interact with the
SOX18 transcript (42). We
observed statistically higher expression levels of miR-7a and
miR-24-3p in the NMLT samples in comparison to the LSCC
samples.
In the present study, we aimed to ascertain whether
the SOX18 transcript modulation mechanism observed in LSCC
cells is also characteristic for adenocarcinoma (AC) cells and,
with the use of the RISC-Trap technique, we finally provide
evidence of the fact that miR-7a and miR-24-3p interact with the
SOX18 transcript in lung cancer cell lines.
The results obtained with the RT-qPCR technique show
a statistically higher expression level of SOX18 mRNA in
NMLT compared to that in the corresponding AC. Moreover, we
observed a statistically higher expression level of the SOX18
protein in AC samples compared to the NMLT ones. It appears that
the same mechanism exists both in LSCC and AC cells, which could be
characteristic of the progression mechanism in NSCLC cells. The
SOX18 transcription factor is expressed during embryonic
development, but its presence has been observed in different cells
of many organs such as the lungs, the skeletal muscles, the stomach
and the heart (21). Therefore, we
previously postulated that the SOX18 transcript may be
modulated by miRNA molecules after embryonic development. This
could explain the disparity observed between the mRNA and protein
levels of the SOX18 transcription factor.
During our several years of research, we have been
able to demonstrate the role of the SOX18 transcription factor in
several types of cancer: invasive ductal breast carcinoma, ovarian
cancer and non-small cell lung cancer (25–27).
The contribution of the SOX18 transcription factor in tumors is
currently being intensively studied on the grounds of the
demonstrated role of this protein in the processes of angiogenesis
and lymphangiogenesis as well as in cell proliferation (17,18,23,40,53–56).
Taking into account the fact that miR-7a and
miR-24-3p suppress the SOX18 transcript in NSCLC cells, the
variable levels of these miRNA molecules may be detected in the
blood of patients, which increases the availability and
universality of the use of these molecules in lung cancer
diagnosis, as they could provide information not only about the
type, but also about the stage of the tumor. A relationship between
the expression level of 8 miRNAs and the survival of patients with
lung AC has been shown (35).
Patients with an increased expression of miR-155, miR-17-3p,
miR-106a, miR-93 or miR-21 or with a reduced expression of
miR-7a-2, miR-7b or miR-145 exhibited a significantly lower
survival rate (52). The prospects
for the use of miRNAs in cancer therapy seem promising as well. It
has been shown that miRNA inhibition can lead in vitro to a
reduction in tumor cell proliferation (33,51,57).
Performing the experiments with the MirTrap System
gave us final proof of the miR-7a and miR-24-3p properties for
binding with the SOX18 transcript in lung cancer cells. The
cell lines studied in this study represent not only different types
of NSCLC (NCI-H1703 and A549, lung AC; NCI-H522, lung squamous cell
carcinoma), but also exhibit different potential for lung cancer
invasiveness. High values of fold enrichment of RISC/miRNA
complexes confirmed that the SOX18 transcript is supressed
by miR-7a and miR-24-3p in NSCLC cells.
The results presented in the present study are
similar to those describing the role and function of miR-7a and
miR-24-3p in NSCLC cells, where miR-7a was suppressed, BCL-2
(B-cell lymphoma 2 protein) was identified as a possible target and
miR-24-3p regulated the autophagy process (28). The fact that a single miRNA molecule
is able to control many different mRNAs is not new. Similar
observations have already been noted in many different types of
cancer (30,31,34–36).
Therefore, it is possible that miR-7a and miR-24-3p suppress the
SOX18 transcript together with some other transcripts in
NSCLCs.
As mentioned in our previous study, miR-7a and
miR-24-3p are successfully downregulated in NSCLC cells in order to
express the SOX18 transcription factor necessary for the tumor to
develop new blood vessels. This mechanism is still unclear, but
most probably miR-7a and miR-24-3p molecules are inhibited by some
endogenous siRNA molecules called antagomirs (anti-miRs). These
antisense transcripts (NATs) are transcribed from the opposite
strand of other protein-coding or non-protein-coding genes
(58–60). To date, it has not been confirmed
that the same suppression mechanism which has been identified in
Alzheimer's disease occurs in NSCLC pathology (61).
In the present study, we demonstrated that the
disparity between the mRNA and protein levels of the SOX18
transcription factor was caused by miR-7a and miR-24-3p, although
the mechanism through which lung cancer cells downregulate miRNA
molecule levels is still unclear. The molecular pathogenesis of the
development of non-small cell lung carcinoma is extremely complex
and complicated. Understanding the molecular basis of the
development of this malignant tumor may enable the use of targeted
therapy, which may result in a better patient outome. It is
therefore important to search for new and more effective
therapeutic strategoes as well as new proteins that could be
potential targets. In this sense, the SOX18 protein and the SOX
protein family appear to be auspicious elements for anticancer
therapy.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Saraiya M, Patel P, Cherala SS,
Barnholtz-Sloan J, Kim J, Wiggins CL and Wingo PA: Recent trends in
cutaneous melanoma incidence and death rates in the United States,
1992–2006. J Am Acad Dermatol. 65 Suppl 1:S17–S25.e1-3. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Thun M, Yu XQ, Hartman AM,
Cokkinides V, Center MM, Ross H and Ward EM: Changes in smoking
prevalence among U.S. adults by state and region: Estimates from
the Tobacco Use Supplement to the Current Population Survey,
1992–2007. BMC Public Health. 11:5122011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yano M, Sasaki H, Moriyama S, Hikosaka Y,
Yokota K, Kobayashi S, Hara M and Fujii Y: Post-operative acute
exacerbation of pulmonary fibrosis in lung cancer patients
undergoing lung resection. Interact Cardiovasc Thorac Surg.
14:146–150. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yano M, Sasaki H, Moriyama S, Kawano O,
Hikosaka Y and Fujii Y: Prognostic factors of pathologic stage IB
non-small cell lung cancer. Ann Thorac Cardiovasc Surg. 17:58–62.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yano T, Haro A, Shikada Y, Maruyama R and
Maehara Y: Non-small cell lung cancer in never smokers as a
representative ‘non-smoking-associated lung cancer’: Epidemiology
and clinical features. Int J Clin Oncol. 16:287–293. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brownson RC, Alavanja MC, Caporaso N,
Simoes EJ and Chang JC: Epidemiology and prevention of lung cancer
in nonsmokers. Epidemiol Rev. 20:218–236. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kołodziej Ł, Boczar T, Bohatyrewicz A and
Zietek P: Outcome of operative treatment for supination-external
rotation Lauge-Hansen stage IV ankle fractures. Chir Narzadow Ruchu
Ortop Pol. 75:231–235. 2010.(In Polish). PubMed/NCBI
|
|
10
|
Kadara H, Kabbout M and Wistuba II:
Pulmonary adenocarcinoma: A renewed entity in 2011. Respirology.
17:50–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gubbay J, Collignon J, Koopman P, Capel B,
Economou A, Münsterberg A, Vivian N, Goodfellow P and Lovell-Badge
R: A gene mapping to the sex-determining region of the mouse Y
chromosome is a member of a novel family of embryonically expressed
genes. Nature. 346:245–250. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakamoto Y, Hara K, Kanai-Azuma M, Matsui
T, Miura Y, Tsunekawa N, Kurohmaru M, Saijoh Y, Koopman P and Kanai
Y: Redundant roles of Sox17 and Sox18 in early cardiovascular
development of mouse embryos. Biochem Biophys Res Commun.
360:539–544. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wegner M: From head to toes: The multiple
facets of Sox proteins. Nucleic Acids Res. 27:1409–1420. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bowles J, Schepers G and Koopman P:
Phylogeny of the SOX family of developmental transcription factors
based on sequence and structural indicators. Dev Biol. 227:239–255.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chung MI, Ma AC, Fung TK and Leung AY:
Characterization of Sry-related HMG box group F genes in zebrafish
hematopoiesis. Exp Hematol. 39:986–998.e5. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
François M: Sox18 orchestrates the
commitment of the lymphatic vessels. Med Sci (Paris). 25:127–129.
2009.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
17
|
François M, Caprini A, Hosking B, Orsenigo
F, Wilhelm D, Browne C, Paavonen K, Karnezis T, Shayan R, Downes M,
et al: Sox18 induces development of the lymphatic vasculature in
mice. Nature. 456:643–647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Francois M, Koopman P and Beltrame M: SoxF
genes: Key players in the development of the cardio-vascular
system. Int J Biochem Cell Biol. 42:445–448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Milivojevic M, Petrovic I,
Kovacevic-Grujicic N, Popovic J, Mojsin M and Stevanovic M:
Construction and functional analysis of novel dominant-negative
mutant of human SOX18 protein. Biochemistry (Mosc). 78:1287–1292.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu Y, Li Y, Jun Wei JW and Liu X: The
role of Sox genes in lung morphogenesis and cancer. Int J Mol Sci.
13:15767–15783. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saitoh T and Katoh M: Expression of human
SOX18 in normal tissues and tumors. Int J Mol Med. 10:339–344.
2002.PubMed/NCBI
|
|
22
|
Eom BW, Jo MJ, Kook MC, Ryu KW, Choi IJ,
Nam BH, Kim YW and Lee JH: The lymphangiogenic factor SOX 18: a key
indicator to stage gastric tumor progression. Int J Cancer.
131:41–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Young N, Hahn CN, Poh A, Dong C, Wilhelm
D, Olsson J, Muscat GE, Parsons P, Gamble JR and Koopman P: Effect
of disrupted SOX18 transcription factor function on tumor growth,
vascularization, and endothelial development. J Natl Cancer Inst.
98:1060–1067. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
DeSantis C, Siegel R, Bandi P and Jemal A:
Breast cancer statistics, 2011. CA Cancer J Clin. 61:409–418. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jethon A, Pula B, Olbromski M, Werynska B,
Muszczynska-Bernhard B, Witkiewicz W, Dziegiel P and
Podhorska-Okolow M: Prognostic significance of SOX18 expression in
non-small cell lung cancer. Int J Oncol. 46:123–132. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pula B, Kobierzycki C, Solinski D,
Olbromski M, Nowak-Markwitz E, Spaczynski M, Kedzia W, Zabel M and
Dziegiel P: SOX18 expression predicts response to platinum-based
chemotherapy in ovarian cancer. Anticancer Res. 34:4029–4037.
2014.PubMed/NCBI
|
|
27
|
Pula B, Olbromski M, Wojnar A,
Gomulkiewicz A, Witkiewicz W, Ugorski M, Dziegiel P and
Podhorska-Okolow M: Impact of SOX18 expression in cancer cells and
vessels on the outcome of invasive ductal breast carcinoma. Cell
Oncol (Dordr). 36:469–483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kasinski AL and Slack FJ: miRNA-34
prevents cancer initiation and progression in a therapeutically
resistant K-ras and p53-induced mouse model of lung adenocarcinoma.
Cancer Res. 72:5576–5587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yin H, Sheng Z, Zhang X, Du Y, Qin C, Liu
H, Dun Y, Wang Q, Jin C, Zhao Y, et al: Overexpression of SOX18
promotes prostate cancer progression via the regulation of TCF1,
c-Myc, cyclin D1 and MMP-7. Oncol Rep. 37:1045–1051. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Devaraj S and Natarajan J: miRNA-mRNA
network detects hub mRNAs and cancer specific miRNAs in lung
cancer. In Silico Biol. 11:281–295. 2011-2012.
|
|
32
|
Ferracin M, Bassi C, Pedriali M, Pagotto
S, D'Abundo L, Zagatti B, Corrà F, Musa G, Callegari E, Lupini L,
et al: miR-125b targets erythropoietin and its receptor and their
expression correlates with metastatic potential and ERBB2/HER2
expression. Mol Cancer. 12:1302013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goswami S, Tarapore RS, Strong AM
Poenitzsch, TeSlaa JJ, Grinblat Y, Setaluri V and Spiegelman VS:
MicroRNA-340-mediated degradation of microphthalmia-associated
transcription factor (MITF) mRNA is inhibited by coding region
determinant-binding protein (CRD-BP). J Biol Chem. 290:384–395.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang C, Hu X, Alattar M and Zhao H: miRNA
expression profiles associated with diagnosis and prognosis in lung
cancer. Expert Rev Anticancer Ther. 14:453–461. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mairinger FD, Ting S, Werner R, Walter RF,
Hager T, Vollbrecht C, Christoph D, Worm K, Mairinger T,
Sheu-Grabellus SY, et al: Different micro-RNA expression profiles
distinguish subtypes of neuroendocrine tumors of the lung: results
of a profiling study. Mod Pathol. 27:1632–1640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yoo JK, Jung HY, Lee JM, Yi H, Oh SH, Ko
HY, Yoo H, Kim HR, Song H, Kim S, et al: The novel miR-9500
regulates the proliferation and migration of human lung cancer
cells by targeting Akt1. Cell Death Differ. 21:1150–1159. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lovell-Badge R: The early history of the
Sox genes. Int J Biochem Cell Biol. 42:378–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Downes M, François M, Ferguson C, Parton
RG and Koopman P: Vascular defects in a mouse model of
hypotrichosis-lymphedema-telangiectasia syndrome indicate a role
for SOX18 in blood vessel maturation. Hum Mol Genet. 18:2839–2850.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Luo M, Guo XT, Yang W, Liu LQ, Li LW and
Xin XY: Inhibition of tumor angiogenesis by cell-permeable dominant
negative SOX18 mutants. Med Hypotheses. 70:880–882. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Castillo SD and Sanchez-Cespedes M: The
SOX family of genes in cancer development: Biological relevance and
opportunities for therapy. Expert Opin Ther Targets. 16:903–919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Olbromski M, Grzegrzolka J,
Jankowska-Konsur A, Witkiewicz W, Podhorska-Okolow M and Dziegiel
P: MicroRNAs modulate the expression of the SOX18 transcript in
lung squamous cell carcinoma. Oncol Rep. 36:2884–2892. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dammann R, Strunnikova M, Schagdarsurengin
U, Rastetter M, Papritz M, Hattenhorst UE, Hofmann HS, Silber RE,
Burdach S and Hansen G: CpG island methylation and expression of
tumour-associated genes in lung carcinoma. Eur J Cancer.
41:1223–1236. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Azhikina T, Kozlova A, Skvortsov T and
Sverdlov E: Heterogeneity and degree of TIMP4, GATA4, SOX18, and
EGFL7 gene promoter methylation in non-small cell lung cancer and
surrounding tissues. Cancer Genet. 204:492–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Alonso MM, Diez-Valle R, Manterola L,
Rubio A, Liu D, Cortes-Santiago N, Urquiza L, Jauregi P, de Munain
A Lopez, Sampron N, et al: Genetic and epigenetic modifications of
Sox2 contribute to the invasive phenotype of malignant gliomas.
PLoS One. 6:e267402011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Otsubo T, Akiyama Y, Yanagihara K and
Yuasa Y: SOX2 is frequently downregulated in gastric cancers and
inhibits cell growth through cell-cycle arrest and apoptosis. Br J
Cancer. 98:824–831. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lang Y, Xu S, Ma J, Wu J, Jin S, Cao S and
Yu Y: MicroRNA-429 induces tumorigenesis of human non-small cell
lung cancer cells and targets multiple tumor suppressor genes.
Biochem Biophys Res Commun. 450:154–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F,
Bao G, Kong H, Ge C, Zhang F, et al: miRNA-200c inhibits invasion
and metastasis of human non-small cell lung cancer by directly
targeting ubiquitin specific peptidase 25. Mol Cancer. 13:1662014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Salim H, Akbar NS, Zong D, Vaculova AH,
Lewensohn R, Moshfegh A, Viktorsson K and Zhivotovsky B: miRNA-214
modulates radiotherapy response of non-small cell lung cancer cells
through regulation of p38MAPK, apoptosis and senescence. Br J
Cancer. 107:1361–1373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun Q, Hang M, Guo X, Shao W and Zeng G:
Expression and significance of miRNA-21 and BTG2 in lung cancer.
Tumour Biol. 34:4017–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xie C, Han Y, Liu Y, Han L and Liu J:
miRNA-124 down-regulates SOX8 expression and suppresses cell
proliferation in non-small cell lung cancer. Int J Clin Exp Pathol.
7:7518–7526. 2014.PubMed/NCBI
|
|
52
|
Zhang C, Ge S, Hu C, Yang N and Zhang J:
MiRNA-218, a new regulator of HMGB1, suppresses cell migration and
invasion in non-small cell lung cancer. Acta Biochim Biophys Sin
(Shanghai). 45:1055–1061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cermenati S, Moleri S, Cimbro S, Corti P,
Del Giacco L, Amodeo R, Dejana E, Koopman P, Cotelli F and Beltrame
M: Sox18 and Sox7 play redundant roles in vascular development.
Blood. 111:2657–2666. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Duong T, Koltowska K, Pichol-Thievend C,
Le Guen L, Fontaine F, Smith KA, Truong V, Skoczylas R, Stacker SA,
Achen MG, et al: VEGFD regulates blood vascular development by
modulating SOX18 activity. Blood. 123:1102–1112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Duong T, Proulx ST, Luciani P, Leroux JC,
Detmar M, Koopman P and Francois M: Genetic ablation of SOX18
function suppresses tumor lymphangiogenesis and metastasis of
melanoma in mice. Cancer Res. 72:3105–3114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fontaine F, Overman J, Moustaqil M,
Mamidyala S, Salim A, Narasimhan K, Prokoph N, Robertson AA, Lua L,
Alexandrov K, et al: Small-molecule inhibitors of the SOX18
transcription factor. Cell Chem Biol. 24:346–359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lv J and Xu L, Xu Y, Qiu M, Yang X, Wang
J, Yin R and Xu L: Expression of MiRNA-221 in non-small cell lung
cancer tissues and correlation with prognosis. Zhongguo Fei Ai Za
Zhi. 17:221–225. 2014.(Abstract in English). PubMed/NCBI
|
|
58
|
Brock M, Samillan VJ, Trenkmann M,
Schwarzwald C, Ulrich S, Gay RE, Gassmann M, Ostergaard L, Gay S,
Speich R, et al: AntagomiR directed against miR-20a restores
functional BMPR2 signalling and prevents vascular remodelling in
hypoxia-induced pulmonary hypertension. Eur Heart J. 35:3203–3211.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Selvamani A, Sathyan P, Miranda RC and
Sohrabji F: An antagomir to microRNA Let7f promotes neuroprotection
in an ischemic stroke model. PLoS One. 7:e326622012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Song MS and Rossi JJ: The anti-miR21
antagomir, a therapeutic tool for colorectal cancer, has a
potential synergistic effect by perturbing an
angiogenesis-associated miR30. Front Genet. 4:3012014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Faghihi MA, Zhang M, Huang J, Modarresi F,
Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G III and
Wahlestedt C: Evidence for natural antisense transcript-mediated
inhibition of microRNA function. Genome Biol. 11:R562010.
View Article : Google Scholar : PubMed/NCBI
|