Introduction
Prostate cancer (PCa) is a disease with genomic,
pathological and clinical heterogeneity (1). Routine prostate-specific antigen (PSA)
screening for PCa aids in the detection of localized early stage
tumors in most cases (2); however,
it can also lead to unnecessary diagnoses or overtreatment of
indolent PCa (3). In addition,
traditional clinicopathological parameters fail to precisely
distinguish cases, again leading to inappropriate treatment
(4). Thus, molecular biomarkers for
PCa diagnosis, prognosis and treatment response have been
investigated over previous decades, in order to offer more
personalized medicine.
MicroRNAs (miRNAs/miRs) are evolutionarily
conserved, short (~18–22 nucleotides), non-coding, single-stranded
RNA molecules that act as posttranscriptional gene regulators by
targeting the 3′untranslated region (3′ UTR) of target mRNAs
(5). Many studies have shown that
miRNAs serve a critical role in various tumors (6–8). In
particular, studies have investigated the potential use of miRNAs
as biomarkers in the diagnosis, prognosis and treatment of cancers
(7) including PCa (8). In PCa, a number of miRNAs, such as
miR-205, miR-221, miR-222 and miR-145, have been demonstrated to be
consistently dysregulated (9–12).
miRNAs or miRNA signatures have potential clinical use in almost
all aspects of PCa management (13). Notably, miR-195, located on
chromosome 17p13.1 (positioned from 6,881,953 to 6,862,065 bp) and
belonging to the miR-15 family (14), has been demonstrated to be a
critical regulator in the development and progression of tumors
(15–20). In our previous study, we identified
hsa-miR-195-5p (miR-195) as a novel prognostic indicator for PCa.
We also found that miR-195 exerted its molecular effects by
targeting RPS6KB1 (21). Therefore,
in this study, we aimed to continue our investigations into the
molecular function of miR-195 and to identify its targets in
PCa.
Materials and methods
Patients and tissue samples
The tissue microarrays (TMAs), including 126 primary
PCa tissues and 22 adjacent non-cancerous prostate tissues along
with detailed clinical information of the samples, were purchased
from Xi'an Alenabio Biotech Co., Ltd. (cat no.: PR752 & PR753).
All human tissues were collected under Institutional Review Board
(IRB) and Health Insurance Portability and Accountability (HIPPA)
approved protocols. Patients known to have undergone chemotherapy
and/or radiotherapy prior to tissue isolation were excluded from
the study.
Additionally, a cohort from the Memorial
Sloan-Kettering Cancer Center (the Taylor cohort) with publicly
available data (GEO accession no. GSE21032), including 150 primary
PCa tissues and 29 adjacent non-cancerous prostate tissues, was
assessed in regards to clinicopathological parameters. Of these
cases, 111 of the primary PCa tissues and all of the non-cancerous
tissues had available miRNA microarray expression data (22). Biochemical recurrence (BCR) was
defined as PSA ≥0.2 ng/ml on two consecutive measurements after
radical prostatectomy. BCR-free survival was defined as the time
interval between initial surgery and the date of BCR. Overall
survival was determined as the time interval between the initial
surgery and the date of the last follow-up or patient death.
Animals
Animal housing and the experiments in this study
were performed in compliance with the guidelines of the Institute
for Laboratory Animal Research at Guangzhou Medical University
(Guangzhou, China). A total of 32 BALB/c nude mice (4- to
5-week-old males) were purchased from Guangdong Medical Laboratory
Animal Center and were housed in wire-top cages (5 mice per cage)
with sawdust bedding in an isolated, clean, air-conditioned room at
a temperature of 25–26°C and relative humidity of ~50% under a 12-h
light/dark cycle.
Cell culture
The human PCa cell lines, LNCaP (cat. no. 63462566)
and DU145 (cat. no. 61761869) were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA) in 2015 and were
cultured in RPMI-1640 medium (Hyclone, USA) supplemented with 10%
fetal bovine serum (Gibco, USA), 2 mM L-glutamine, and antibiotics.
Cell line characterization and passaging for ~3 months were
performed prior to purchase. Therefore, we did not carry out
reauthentication of the cell lines. ATCC used Short Tandem Repeat
(STR) profiling to detect the misidentified, cross-contaminated, or
genetically drifted cells. A Promega PowerPlex® 18D
System was used to amplify 17 STR loci plus amelogenin. Human
umbilical vein endothelial cells (HUVECs) were obtained from the
Cell Bank, Chinese Academy of Sciences (Shanghai, China). All cell
lines were maintained at 37°C in a humidified chamber supplemented
with 5% CO2.
Oligonucleotide and plasmid
transfection
The miRNA mimics (miR-195, cat. no. miR10000461-1-5)
and negative control miRNA mimics (miR-NC, cat. no. miR01201-1-5)
used for transient transfection were designed and synthesized by
RiboBio (Guangzhou, China). The PRR11 coding sequence (without the
3′ UTR) was cloned into a pCDNA3.1 (+)-Vector (cat. no. V790-20;
Invitrogen, USA), while a blank vector was used as a negative
control. Short interfering RNA (siRNA) against PRR11 (si-PRR11) and
negative control siRNA with non-specific sequences (si-NC) were
synthesized by Sigma-Aldrich (USA). The targeting sequences of the
si-PRR11 were as follows: 5′-CUGCAUAACCCAGAGUUUAdTdT-3′ (sense) and
5′-UAAACUCUGGGUUAUGCAGdTdT-3′ (antisense). The sequences of the
si-NC were as follows: 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and
5′-ACGUGACACGUUCGGAGAATT-3′ (antisense). Cells were transfected
with the miRNA mimics, siRNA and pCDNA3.1(+)-PRR11 using
Lipofectamine 2000 Reagent (cat. no. 11668019; Invitrogen,
USA)according to the manufacturer's protocol. At 48 h after
transfection, the cells were used in the cell cycle, CCK-8, RNA
extraction and western blotting assays.
Knockdown of PRR11 by lentiviral shRNA
in DU145 and LNCaP cells
Packaging of lentiviral particles was performed as
follows: Briefly, three lentivirus expression plasmids containing
siRNA against PRR11 were constructed by GeneChem Corporation
(Shanghai, China) and were used to infect DU145 and LNCaP cells in
the presence of 6 µg/ml Polybrene: PRR11-shRNA1:
5′-TCAGATGGATCTGCGGAAACTTCCTGTCAGATTTCCGCAGATCCATCTGA-3′;
PRR11-shRNA2: GGATCTGCGGAAACTGCTTCTTCCTGTCAGAAAGCAGTTTCCGCAGATCC;
and PRR11-shRNA3:
CCTAGAAGCCCAACTCCAACTTCCTGTCAGATTGGAGTTGGGCTTCTAGG. Infected cells
were selected for using puromycin, and the knockdown of PRR11 was
confirmed via western blotting using anti-PRR11 antibody (cat. no.
HPA023923; Sigma-Aldrich, USA). One of the PRR11-shRNAs was chosen
for further experiments.
Microarray analysis
Total RNA was extracted using TRIzol reagent (cat.
no. 15596-018; Life Technologies, Carlsbad, CA, USA) following the
manufacturer's instructions, and RNA integrity was checked against
an RIN threshold in an Agilent Bioanalyzer 2100 (Agilent
Technologies, Santa Clara, CA, USA). Qualified total RNA was
further purified with an RNeasy Micro kit (cat. no. 74004; Qiagen,
GmBH, Germany) and RNase-Free DNase Set (cat. no. 79254; Qiagen).
Total RNA was then amplified, labeled and purified using a GeneChip
3′IVT Express Kit (cat. no. 901229; Affymetrix, Santa Clara, CA,
USA) following the manufacturer's instructions to obtain
biotin-labeled cRNA. For array hybridization, the hybridization
procedure was performed using a GeneChip® Hybridization,
Wash and Stain Kit (cat. no. 900720; Affymetrix) in a Hybridization
Oven 645 (cat. no. 00-0331-220V) and Fluidics Station 450 (cat. no.
00-0079; Affymetrix) following the manufacturer's instructions. For
data acquisition, slides were scanned with a GeneChip®
Scanner 3000 (cat. no. 00-00212; Affymetrix) using Command Console
Software 3.1 (Affymetrix) at default settings. Raw data were
normalized by the MAS 5.0 algorithm using Gene Spring Software 11.0
(Agilent Technologies).
Bioinformatic miRNA target
prediction
The online program TargetScan (release 6.2)
(23) was used to predict potential
target genes for miR-195.
RT-qPCR
The expression levels of miR-195 and PRR11 mRNA in
the PCa cell lines were detected by RT-qPCR analysis according to
the protocol in our previous study (24). The oligonucleotide sequences (5′-3′)
of the primers used in the present study were as follows: PRR11 (F,
CCTGCTAGCTACATTTACA, R, GAATGGTCAAGTCATTTAGC); GAP DH (F,
CATGGGTGTGAACCATGAGAAGTA, R, CAGTAGAGGCAGGGATGATGTTCT);
hsa-miR-195-5p (cat. no. HmiRQP0283; GeneCopoeia, USA) and U6 (cat.
no. HmiRQP9001; GeneCopoeia, USA).
Western blot analysis
The expression levels of PRR11 protein in the PCa
cell lines were detected by western blot analysis according to the
protocol in our previous study (24). The antibodies used in the present
study were as follows: Anti-PRR11 (polyclonal rabbit, cat. no.
HPA023923; Sigma-Aldrich, USA), anti-GAPDH (HRP-conjugated
monoclonal mouse, cat. no. KC-5G5; KangChen Bio-Tech, Shanghai,
China).
Immunohistochemistry
The expression pattern and subcellular localization
of PRR11 protein in clinical PCa tissues, and of CD31 and Ki-67 in
the subcutaneous tumor xenografts of nude mice, were detected by
immunohistochemistry. The specimens were fixed in 10% neutral
buffered formalin and subsequently embedded in paraffin. The
paraffin-embedded tissues were cut to a thickness of 4 µm and
deparaffinized with xylene, and then rehydrated for further
peroxidase (DAB) immunohistochemistry staining employing a Dako
EnVision System (Dako Diagnostics, Switzerland). For this staining
assay, following a brief proteolytic digestion and peroxidase
blocking of the tissue slides, the slides were incubated overnight
at 4°C with the abovementioned anti-PRR11 antibody at a dilution of
1:600, along with anti-CD31 (rabbit monoclonal antibody; cat. no.
ZA-0568; ZSGB-BIO, China) at a dilution of 1:200, and anti-Ki-67
(rabbit monoclonal antibody; cat. no. ZA-0502; ZSGB-BIO, China) at
a dilution of 1:200. After washing, peroxidase-labeled polymer and
chromogen substrate were used to visualize the staining of the
proteins of interest. In each immunohistochemistry run, negative
controls were included by omitting the primary antibody.
Following hematoxylin counterstaining,
immunostaining was scored by two independent experienced
pathologists who were blinded to the clinicopathological data and
clinical outcomes. The scores of the two pathologists were compared
and any discrepant scores were reviewed through re-examination of
the staining by both pathologists to achieve a consensus score.
Scores were assigned by evaluating the immunolabeling of tumor
cells. The number of positively stained cells in 10 representative
microscopic fields was counted, along with the percentage of
positive cells. Given the heterogeneity of the target protein
staining, tumor specimens were scored in a semi-quantitative
manner. The scoring system based on the percentage of
immunoreactive tumor cells was as follows: 0 (0–5%), 1 (6–25%), 2
(26–50%), 3 (51–75%) and 4 (>75%). Additionally, staining
intensity was visually scored and stratified as follows: 0
(negative), 1 (weak), 2 (moderate) and 3 (strong). Final
immunoreactivity scores (IRS) for PRR11 and Ki-67 were then
obtained for each sample by multiplying the percentage of positive
cells by the intensity score. Vasculature density in the tumor
xenografts was also determined from the number of CD31-positive
vessels.
Generation of the in vivo xenograft
model
For the in vivo tumor formation assays, DU145
or LNCaP cells transfected with lentivirus expression plasmid
containing PRR11-shRNA or negative control (scramble) were
trypsinized and suspended in PBS. Subsequently, the cells were
subcutaneously injected into the right flank of each nude mouse (8
mice per group); DU145 cells were injected with 0.2 ml PBS at a
concentration of 2.5×107 cells/ml, while LNCaP cells
were injected as a mixture of 0.1 ml PBS at a concentration of
5×108 cells/ml and an equal volume of Matrigel (cat. no.
356234; BD Biosciences). The tumor sizes were measured at 2-day
intervals as soon as the tumors were measurable, and the tumor
volumes were calculated as follows: V (mm3) = width
(mm2) × length (mm)/2. On day 42, all mice in the LNCaP
and DU145 groups were sacrificed.
Luciferase reporter assay
The expression of the target gene of miR-195 was
evaluated in LNCaP cells by a luciferase reporter assay. The
putative miR-195 complementary site in the 3′ UTR of PRR11 mRNA
(NCBI reference sequence: NM_018304.3; 3′ UTR-1: 3490–3496 and 3′
UTR-2: 4827–4833) or a mutant sequence was cloned into a psiCHECK-2
luciferase reporter vector (Promega, Madison, WI, USA). LNCaP cells
were co-transfected with 50 nM miR-195 mimic or miR-NC and 0.5 µg
of psi-PRR11-3′ UTR-1-WT, psi-PRR11-3′ UTR-2-WT, psi-PRR11-3′
UTR-1-MUT or psi-PRR11-3′ UTR-2-MUT. Cells were collected 48 h
after transfection and analyzed with a Dual-Luciferase Reporter
Assay System (Promega). The firefly and Renilla luciferase
signals were detected with a GloMax fluorescence reader (Promega),
and the Renilla luciferase signal was normalized to the
firefly luciferase signal.
Cell viability assay
For cell viability assays, 2×103 cells
were seeded in 96-well plates and cultured for 24, 48 and 72 h.
Cells were then incubated with 20 µl of CCK-8 solution (cat. no.
C0038; Beyotime, China) for 4 h at 37°C. The absorbance was
measured at a wavelength of 495 nm with a spectrophotometer, and
data were expressed as means ± SD of three independent
experiments.
HUVEC tube formation assay
A total of 200 µl human umbilical vein endothelial
cells (HUVECs; 2×104 cells) were seeded in 48-well
plates containing 200 µl BD Matrigel Basement Membrane Matrix (cat.
no. 356234; BD Biosciences) for 8–12 h at 37°C. LNCaP and DU145
cells transfected with oligonucleotide and/or plasmid were seeded
in the upper Transwell chambers (cat. no. 3495; Corning
Incorporated, Corning, NY, USA), in which the conditioned medium
permeated through the 0.4-µm micropores to the Matrigel, which
established a non-contact co-culture system. Images were acquired
with a phase-contrast microscope. The numbers of tubes were counted
in three individual wells and presented as the mean ± SD.
Cell cycle analysis
A flow cytometry assay (kit cat. no. KGA511) was
performed to assess the cell cycle distributions of the DU145 and
LNCaP cells. At 48 h after cell transfection, attached and
suspended cells were harvested with a pipette, washed once with 1
ml PBS, and resuspended in 500 µl PBS containing 50 µg/ml propidium
iodide (PI). RNase A (100 µg/ml) and 0.2% Triton X-100 were then
added to the cells, which was followed by incubation at 4°C for 30
min in the dark prior to flow cytometric analysis (BD FACSCaliber).
Data analysis was performed using ModFit software (Verity Software
House, Inc., Topsham, ME, USA.).
Statistical analysis
Continuous variables are expressed as means ± SD.
SPSS version 20.0 for Windows (SPSS, Inc., IL, USA) and SAS 9.1
(SAS Institute, Cary, NC, USA) were used for all statistical
analyses, which were performed by two independent biostatisticians.
The RT-qPCR and western blot data were analyzed by Wilcoxon
signed-rank tests. The Fisher's exact test was used for any 2×2
tables and the Pearson χ2 test was used for non-2×2
tables. Mann-Whitney U and Kruskal-Wallis H tests were performed to
examine the associations between PRR11 expression and the
clinicopathological characteristics of PCa patients in the Taylor
cohort. The Kaplan-Meier method was used for survival analysis, and
Cox regression analysis was used for univariate and multivariate
analyses. Finally, Spearman correlations were calculated for the
expression levels of miR-195 and PRR11 in the Taylor cohort. A
P-value of <0.05 was considered to indicate a statistically
significant difference.
Results
Upregulation of miR-195 suppresses
angiogenesis and proliferation in vitro
In our previous study, we found that miR-195
suppressed invasion and migration while promoting apoptosis in PCa
cells (21). To verify the
biological role of miR-195 in angiogenesis and proliferation in
vitro, we used LNCaP (P<0.001) and DU145 (P<0.001) PCa
cell lines overexpressing miR-195 mimics (miR-195 group) or
negative control miR (miR-NC group) via transient transfection.
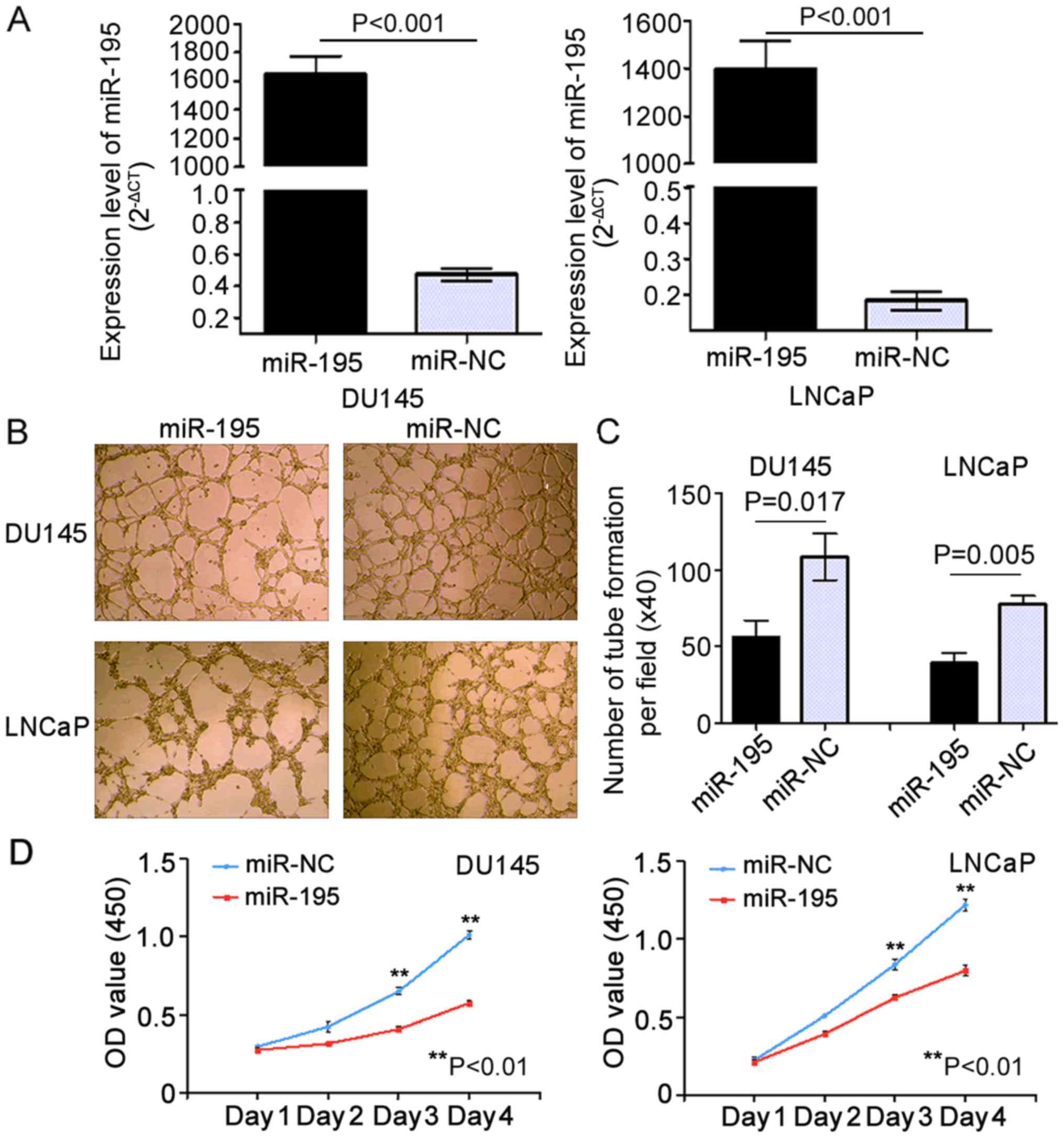
RT-qPCR analysis confirmed that transient transfection of the cell
lines was successfully established (Fig. 1A). To investigate whether miR-195
has biological effects on tumor angiogenesis, HUVEC tube formation
assays were carried out. The results indicated that the tube
formation of HUVECs was strongly inhibited by the conditioned media
from DU145 and LNCaP cells overexpressing miR-195 when compared
with their negative control (miR-NC) media (Fig. 1B). Notably, the relative
tube-forming abilities of miR-195-overexpressing DU145 and LNCaP
cells, determined by comparing the number of complete tubes per
field, were significantly increased when compared with their miR-NC
counterparts (P=0.017 and P=0.005, respectively; Fig. 1C). Moreover, in both the DU145 and
LNCaP cells, overexpression of miR-195 strongly inhibited cell
proliferation (Day 4: P=0.008 and P=0.006, respectively; Fig. 1D) compared with the controls.
Microarray analysis of differentially
expressed mRNAs induced by miR-195
Since miRNAs in most cases only cause modest
decreases in protein translation, the miRNA-mediated regulation of
proteins with long half-lives may not be detected by measuring
steady-state protein levels using standard proteomic quantification
(25), which was used in our
previous study (21). Therefore, to
solve the shortage of the standard proteomic quantification and
identify more novel targets of miR-195 in PCa, we performed
microarray analysis on the same cells (LNCaP cells overexpressing
miR-195 or miR-NC) to detect miR-195-induced changes in mRNA
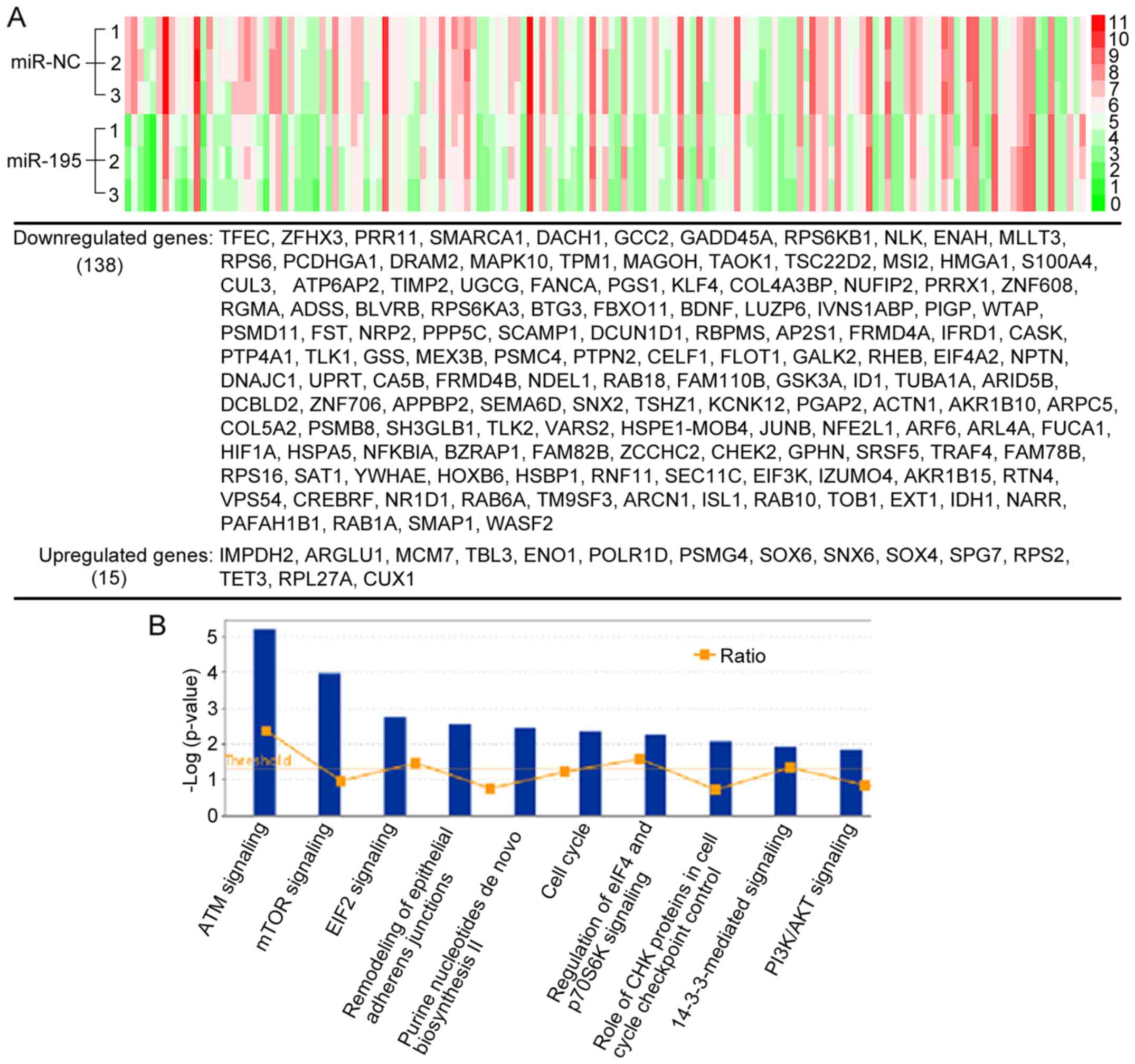
levels. As shown in Fig. 2A, a
total of 153 genes differentially regulated with fold changes ≤-1.5
or ≥1.5 (miR-195 vs. miR-NC) were identified in at least two
independent experiments. miRNAs typically destabilize
post-transcriptional mRNAs. Thus, as expected, among the 153 genes,
there were 138 (90.2%) downregulated genes and 15 (9.8%)
upregulated genes. We then investigated the molecular roles of the
differentially expressed genes regulated by miR-195 by using
Ingenuity Pathway Analysis (IPA). Among the pathways enriched for
differentially expressed genes, there were a number of
tumor-related pathways, including mTOR signaling, EIF-2 signaling,
cell cycle signaling, and remodeling of epithelial adherens
junctions signaling (Fig. 2B),
which all have broad effects on cell behavior. These results
suggested that the biological pathways and functions regulated by
miR-195 were closely associated with PCa progression.
PRR11 is a direct target of
miR-195
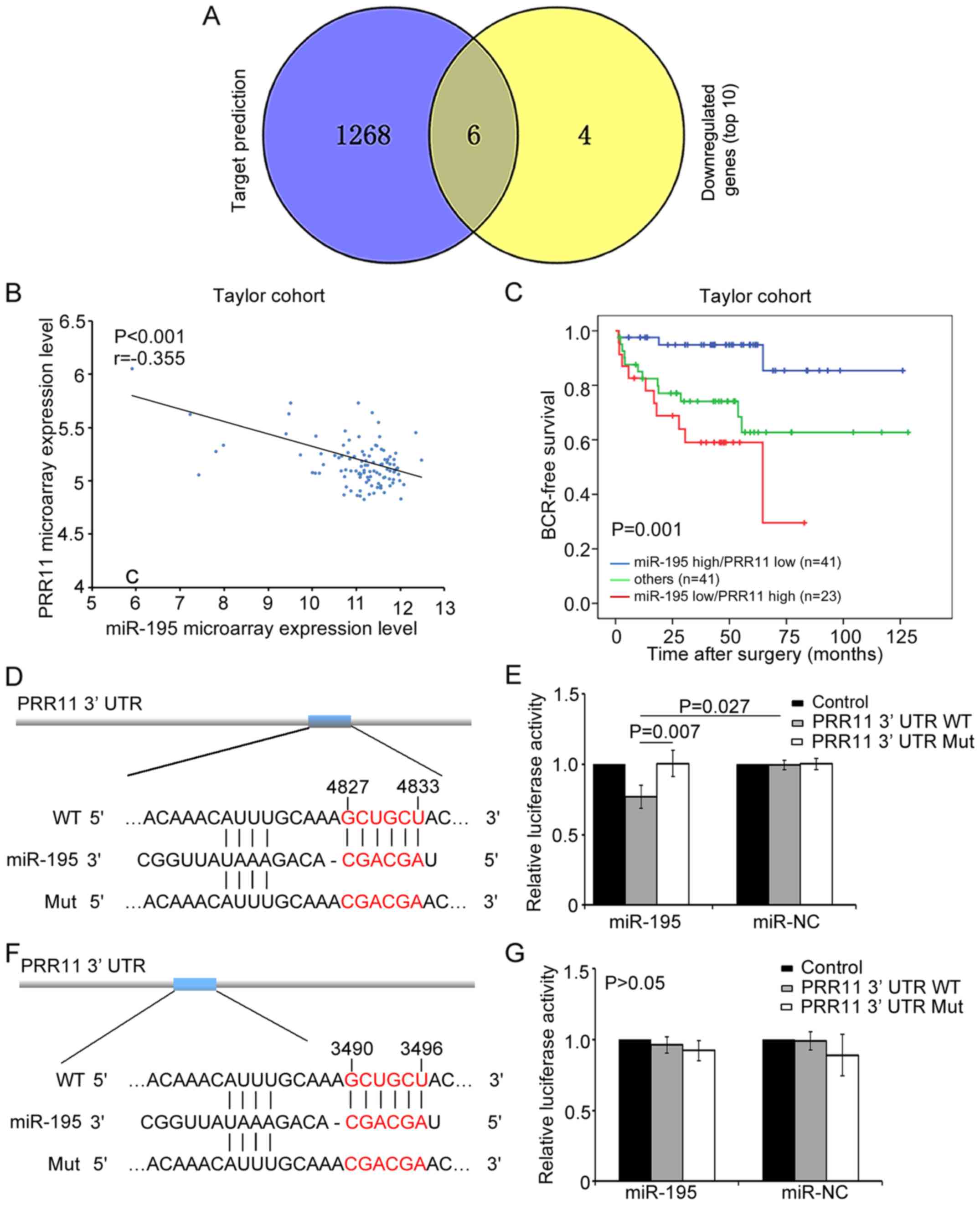
We additionally used an miRNA target prediction
program (TargetScan) to predict the top 10 candidate targets of the
downregulated genes identified by microarray. TFEC, ZFHX3, PRR11,
GCC2, RPS6KB1 and ENAH were predicted, which demonstrated that
miR-195 may bind to the 3′ UTR sequences of these genes (Fig. 3A). We further found that PRR11 and
RPS6KB1 were involved in the top 10 enriched pathways (data not
shown). RPS6KB1 is a critical component of mTOR signaling and was
identified as a target gene of miR-195 in our previous study
(21). Meanwhile, the present
results identified PRR11 as a key component in cell cycle
signaling, implying that PRR11 might also be an important mediator
of the biological role of miR-195 in PCa.
We continued to investigate the relationship between
miR-195 expression and PRR11 mRNA expression in the Taylor cohort,
comprised of mRNA and miRNA expression profiles for 113 primary PCa
tissues (22). The results
demonstrated that PRR11 expression was negatively correlated with
miR-195 expression (r=−0.355, P<0.001; Fig. 3B), which corresponded with the
targeting data for miR-195.
More notably, we found that miR-195 downregulation
combined with PRR11 upregulation was associated with aggressive
clinicopathological features and poor prognosis of PCa patients in
the Taylor cohort. As shown in Table
I, miR-195 downregulation and PRR11 upregulation were
frequently observed in cases presenting with the most aggressive
features, including those with higher Gleason scores (P=0.005),
higher risk of metastasis (P<0.001) and BCR (P=0.003). By
contrast, miR-195 upregulation and PRR11 downregulation often
occurred in the most indolent tumors, while other regulation
states, including simultaneous upregulation or downregulation of
miR-195 and PRR11, could be seen in the patients with intermediate
risk of progression. Kaplan-Meier analysis also indicated that
miR-195 expression combined with PRR11 expression could
significantly stratify patients into three groups based on BCR-free
survival time (P=0.001; Fig. 3C).
Notably, patients with low miR-195 and high PRR11 expression were
likely to have the shortest BCR-free survival time. Additionally,
univariate and multivariate Cox regression analysis revealed that
miR-195 expression combined with PRR11 expression served as an
independent predictor for BCR-free survival (P=0.001 and P=0.034,
respectively; Table II). These
results indicated that PRR11, in an opposing manner to miR-195,
might act as a promotive factor in PCa progression. Thus, PRR11 may
be a potential target of miR-195 in PCa.
 | Table I.Association of miR-195/PRR11
expression and the clinicopathological features of the prostate
cancer (PCa) patients. |
Table I.
Association of miR-195/PRR11
expression and the clinicopathological features of the prostate
cancer (PCa) patients.
|
|
|
hsa-miR-195/PRR11 |
|
|---|
|
|
|
|
|
|---|
|
| N | miR-195 high/PRR11
low (n=42) | Others (n=42) | miR-195 low/PRR11
high (n=27) | P-value |
|---|
| Mean age
(years) | 111 | 58.57±6.17 | 57.34±8.07 | 59.48±8.45 | 0.495 |
| Preoperative PSA
(ng/ml) |
|
|
|
|
|
|
<4 | 26 | 9 (34.6) | 14 (53.8) | 3 (11.5) | 0.126 |
| ≥4 | 83 | 33 (39.8) | 28 (33.7) | 22 (26.5) |
|
| Gleason score |
|
|
|
|
|
|
<8 | 87 | 38 (43.7) | 34 (39.1) | 15 (17.2) | 0.005 |
| ≥8 | 17 | 3 (17.6) | 5 (29.4) | 9 (52.9) |
|
| Clinical stage |
|
|
|
|
|
|
<T2A | 64 | 23 (35.9) | 24 (37.5) | 17 (26.6) | 0.456 |
|
≥T2A | 43 | 18 (41.9) | 18 (41.9) | 7 (16.3) |
|
| Pathological
stage |
|
|
|
|
|
|
T2A-T2C | 69 | 31 (44.9) | 24 (34.8) | 14 (20.3) | 0.197 |
|
T3A-T4 | 37 | 10 (27) | 17 (45.9) | 10 (27) |
|
| Metastasis |
|
|
|
|
|
|
Negative | 93 | 40 (43) | 37 (39.8) | 16 (17.2) | <0.001 |
|
Positive | 18 | 2 (11.1) | 5 (27.8) | 11 (61.1) |
|
| Biochemical
recurrence |
|
|
|
|
|
|
Negative | 80 | 38 (47.5) | 29 (36.3) | 13 (16.3) | 0.003 |
|
Positive | 25 | 3 (12) | 12 (48) | 10 (40) |
|
| Overall
survival |
|
|
|
|
|
|
Alive | 99 | 38 (38.4) | 40 (40.4) | 21 (21.2) | 0.07 |
|
Died | 12 | 4 (33.3) | 2 (16.7) | 6 (50.0) |
|
 | Table II.Prognostic value of miR-195/PRR11
expression for the biochemical recurrence-free survival in
univariate and multivariate analyses by Cox regression. |
Table II.
Prognostic value of miR-195/PRR11
expression for the biochemical recurrence-free survival in
univariate and multivariate analyses by Cox regression.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) | 1.04
(0.98–1.09) | 0.186 | 1.02
(0.96–1.08) | 0.581 |
| Clinical tumor
stage | 0.85
(0.37–1.94) | 0.700 | 0.80
(0.32–2.02) | 0.641 |
| Pathological tumor
stage | 5.83
(2.50–13.56) | <0.001 | 2.70
(0.92–7.93) | 0.070 |
| Preoperative
PSA | 2.45
(0.73–8.22) | 0.366 | 1.30
(0.35–4.80) | 0.700 |
| Gleason score | 11.97
(5.31–27.01) | <0.001 | 6.06
(2.07–17.71) | 0.001 |
| miR-195/PRR11 | 2.59
(1.51–4.42) | 0.001 | 1.98
(1.05–3.71) | 0.034 |
According to the TargetScan prediction, two putative
binding sites for miR-195 were found in the 3′ UTR of PRR11 at
4827–4833 and 3490–3496 bp (Fig. 3D and
F). To confirm these predictions, a luciferase reporter assay
was carried out in LNCaP cells. In this assay, relative luciferase
activity was markedly reduced in cells co-transfected with
psi-PRR11-3′ UTR-2-WT luciferase reporter and miR-195 mimic
compared with negative control cells (P=0.027; Fig. 3E). In contrast, the expression of
the luciferase reporter containing a mutated sequence of the PRR11
fragment (psi-PRR11-3′ UTR-2-MUT) was not affected by
co-transfection with hsa-miR-195 mimics. Meanwhile, relative
luciferase activity was not significantly changed in cells
co-transfected with psi-PRR11-3′ UTR-1-WT luciferase reporter and
miR-195 mimic relative to the negative control cells (P>0.05;
Fig. 3G). These results indicated
that the sequence at 4827–4833 bp in the 3′ UTR of PRR11 was the
complementary site for the miR-195 seed region, and further
demonstrated that PRR11 is a direct target of miR-195.
miR-195 exerts its tumor suppressive
role partially by downregulating PRR11 expression
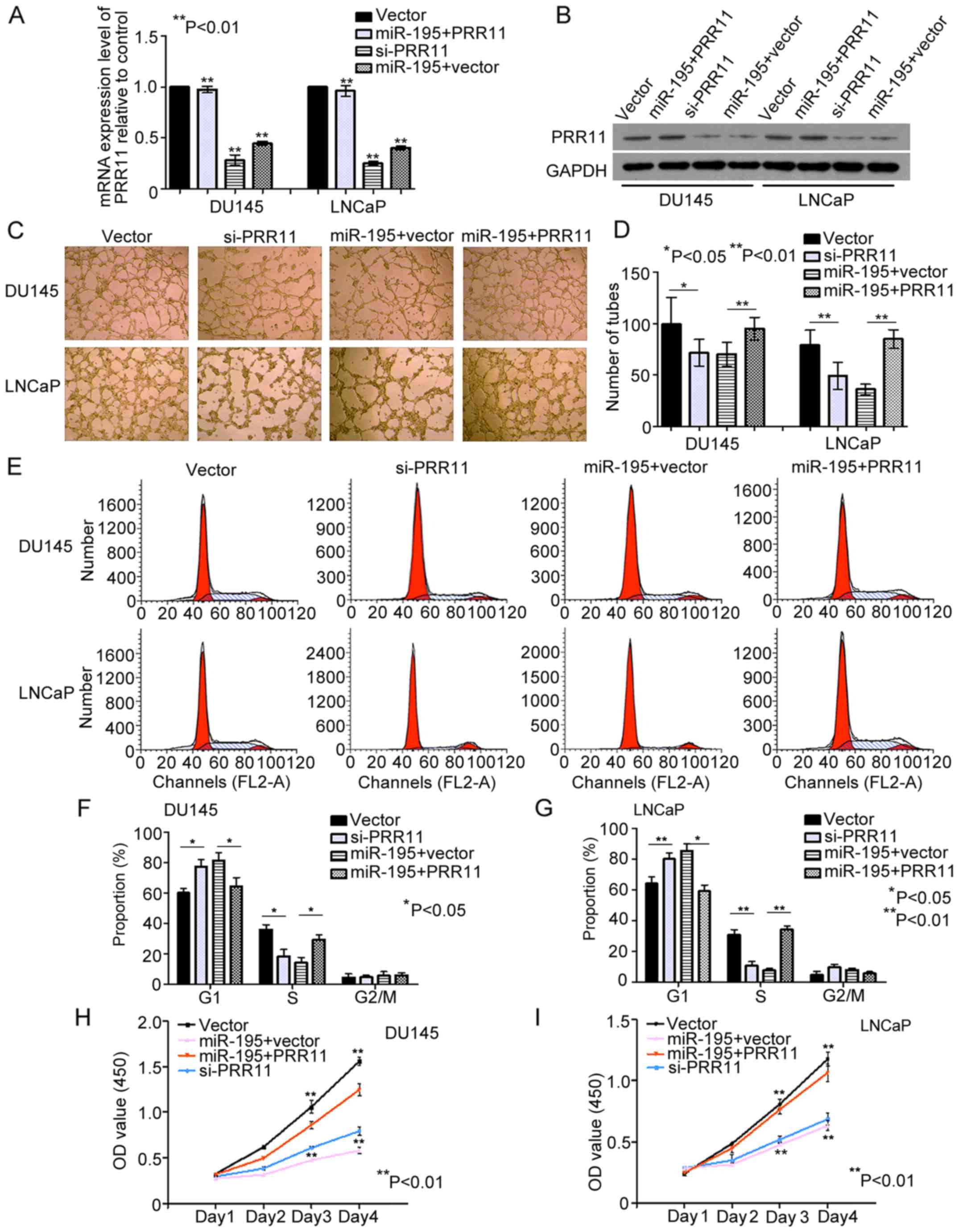
The direct targeting of PRR11 by miR-195 was
evaluated in the PCa cell lines DU145 and LNCaP. We found that the
mRNA and protein levels of PRR11 were significantly decreased by
miR-195 overexpression in DU145 and LNCaP cells (both P<0.001;
Fig. 4A and B). Subsequently, we
knocked down the endogenous expression of PRR11 in DU145 and LNCaP
cells with PRR11 siRNA (both P<0.001; Fig. 4A and B). Compared with the negative
controls, knockdown of PRR11 in the DU145 and LNCaP cells inhibited
the tube formation abilities of HUVECs (P=0.046 and P=0.006,
respectively; Fig. 4C and D),
suppressed cell cycling (G1 phase: both P=0.045; S phase: both
P=0.001; Fig. 4E-G) and suppressed
cell proliferation (Day 4: P=0.003 and P=0.005, respectively;
Fig. 4H and I). Furthermore, we
transfected cells with pCDNA3.1 (+)-vectors expressing PRR11
without its 3′ UTR. As shown in Fig. 4A
and B, the endogenous PRR11 expression levels were detected by
qRT-PCR and western blot analysis in the DU145 and LNCaP cells
transfected with the miR-195 mimics in the presence of PRR11 or
vector control for 48 h (P<0.001 and P<0.002, respectively).
In turn, restoration of PRR11 expression markedly attenuated the
effects of miR-195 on HUVEC tube formation (P=0.008 and P<0.001,
respectively; Fig. 4C and D), cell
cycling (G1 phase: P=0.033 and P=0.027, respectively; S phase:
P=0.041 and P=0.003, respectively; Fig.
4E-G) and cell proliferation (Day 4: P=0.007 and P=0.009,
respectively; Fig. 4H and I).
Collectively, these findings indicated that miR-195 serves a tumor
suppressive role by downregulating PRR11 expression.
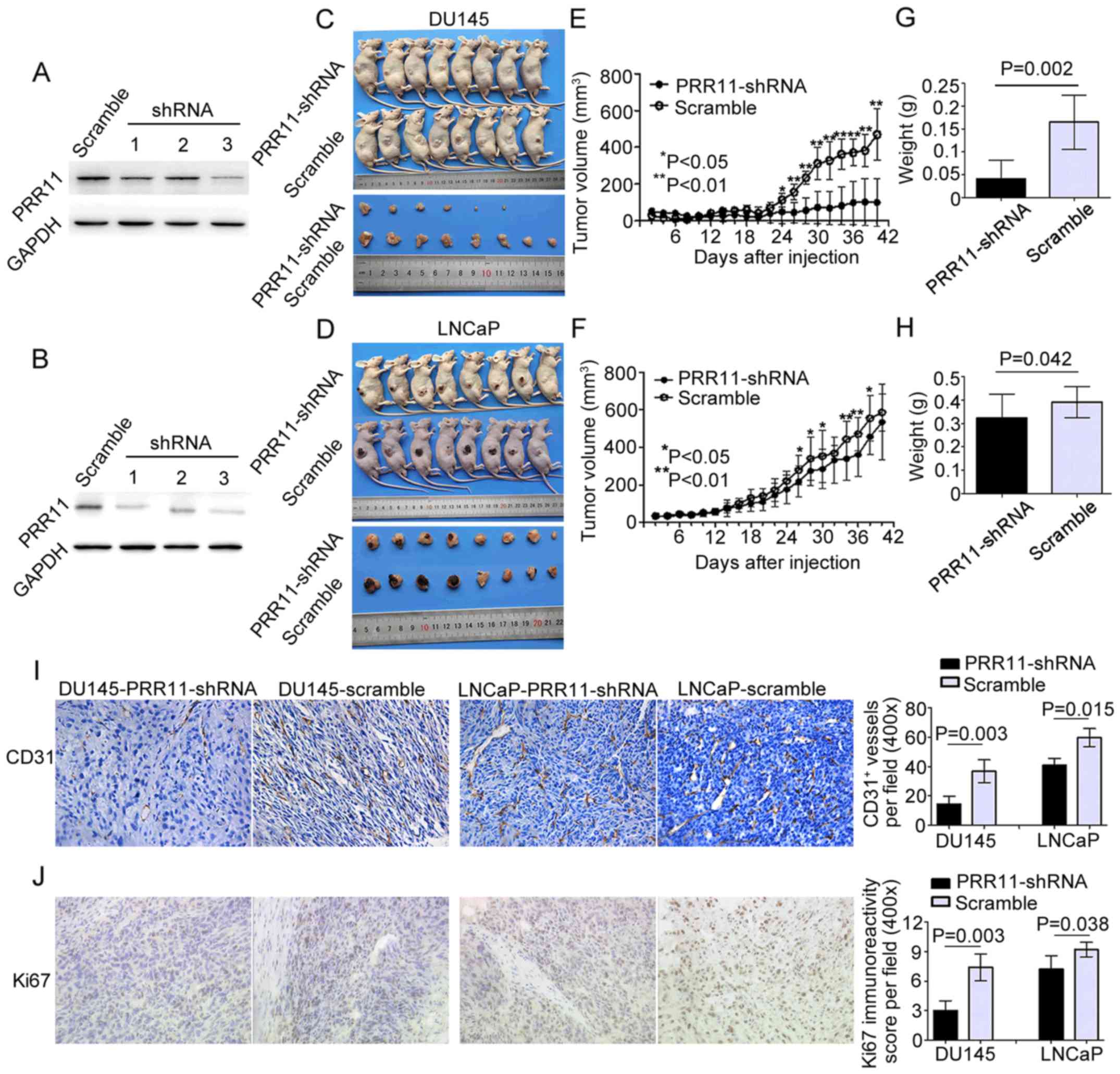
Knockdown of PRR11 suppresses tumor
growth and angiogenesis in vivo
The in vitro assays indicated that PRR11
downregulation inhibited the proliferative and angiogenic
activities of tumor cells. To evaluate the biological functions of
PRR11 in vivo, we stably suppressed PRR11 expression in
DU145 and LNCaP cells through transfection with a lentivirus
expression plasmid containing siRNA against PRR11. As shown in
Fig. 5A and B, we chose the DU145
and LNCaP cells transfected with the third shRNA for subsequent
experiments. The PRR11-suppressed cell lines (PRR11-shRNA) and
control PCa cell lines (scramble) were subcutaneously injected into
the right side of male nude mice (8 mice per group). Compared with
the controls, the PCa cells with suppressed PRR11 expression formed
significantly smaller tumor nodules (Fig. 5C and D). Additionally, the knockdown
of PRR11 markedly reduced the growth of tumor nodules and the
weight of tumors in the PRR11-shRNA groups on day 42 when compared
with the scramble control groups (Fig.
5E-H). The proliferative and angiogenic abilities of the tumor
xenografts were subsequently evaluated via histopathological
staining for the proliferative marker Ki67 and pan-endothelial
marker CD31. The results showed that in the tumor xenografts of
each cell line, downregulation of PRR11 reduced the number of
vessels (P=0.003 and P=0.015, respectively; Fig. 5I) and suppressed cell proliferation
(P=0.003 and P=0.038, respectively; Fig. 5J).
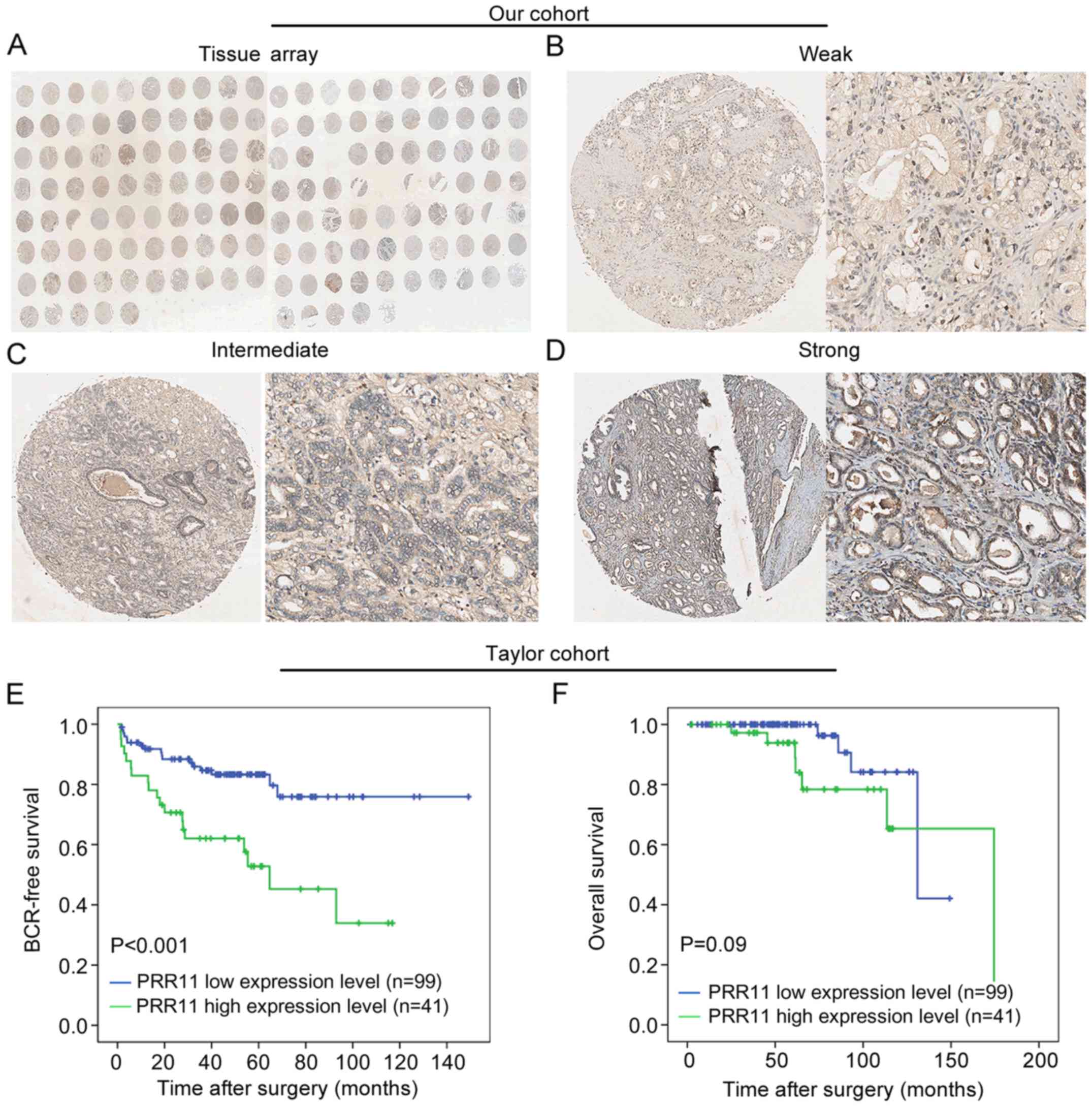
PRR11 upregulation occurs in
aggressive tumors and is associated with poor clinical outcome
To investigate the expression pattern of PRR11 in
PCa, immunohistochemical staining for PRR11 was employed to detect
the expression pattern and subcellular localization of PRR11
protein in 126 primary PCa tissues and 22 adjacent non-cancerous
prostate tissues (Fig. 6A). As
shown in Fig. 6B-D, we identified
PRR11-positive staining in the cytoplasm and cellular membrane of
PCa and benign glandular epithelium cells, with evenly distributed
staining patterns observed at weak, intermediate and strong
intensities. PRR11 was recorded as low if the final IRS was no
>4. As shown in Table III, we
found that high PRR11 expression frequently occurred in the PCa
tissues (P<0.001) and in patients with advanced pathological
stage PCa (P=0.008).
 | Table III.Association of PRR11 expression and
the clinicopathological characteristics of the PCa in two
cohorts. |
Table III.
Association of PRR11 expression and
the clinicopathological characteristics of the PCa in two
cohorts.
|
| Taylor cohort | Our cohort |
|---|
|
|
|
|
|---|
|
| N | Mean ± SD | P-value | Low | High | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
<70 | 144 | 5.25±0.36 | 0.330 |
|
|
|
|
≥70 | 6 | 5.62±0.83 |
|
|
|
|
| Preoperative PSA
(ng/ml) |
|
|
|
|
|
|
|
<4 | 25 | 5.27±0.44 | 0.674 |
|
|
|
| ≥4 | 122 | 5.24±0.32 |
|
|
|
|
| Gleason score |
|
|
|
|
|
|
|
<8 | 117 | 5.18±0.27 | 0.013 |
|
|
|
| ≥8 | 22 | 5.50±0.54 |
|
|
|
|
| Clinical stage |
|
|
|
|
|
|
|
<T2A | 80 | 5.21±0.31 | 0.182 |
|
|
|
|
≥T2A | 65 | 5.28±0.39 |
|
|
|
|
| Pathological
stage |
|
|
|
|
|
|
|
T2A-T2C | 86 | 5.17±0.25 | 0.012 | 26 | 34 | 0.008 |
|
T3A-T4 | 55 | 5.33±0.42 |
| 14 | 52 |
|
| Metastasis |
|
|
|
|
|
|
|
Negative | 122 | 5.17±0.23 | <0.001 |
|
|
|
|
Positive | 28 | 5.70±0.60 |
|
|
|
|
| Biochemical
recurrence |
|
|
|
|
|
|
|
Negative | 104 | 5.16±0.23 | 0.003 |
|
|
|
|
Positive | 36 | 5.42±0.48 |
|
|
|
|
| Overall
survival |
|
|
|
|
|
|
|
Alive | 131 | 5.21±0.31 | 0.007 |
|
|
|
|
Died | 19 | 5.66±0.63 |
|
|
|
|
| Tissue type |
|
|
|
|
|
|
|
Cancer | 150 | 5.27±0.39 | <0.001 | 41 | 85 | <0.001 |
|
Benign | 29 | 5.09±0.14 |
| 18 | 4 |
|
Finally, we validated the clinical value of PRR11 in
the Taylor cohort. As shown in Table
III, PRR11 was significantly upregulated in PCa tissues
relative to non-cancerous tissues (P<0.001). More notably, PRR11
overexpression was significantly associated with higher Gleason
score (P=0.013), more advanced pathological stage (P=0.012),
positive metastasis (P<0.001), positive BCR (P=0.003) and
shorter overall survival time (P=0.007) in the PCa patients
(Table III). Kaplan-Meier
analysis was also conducted to assess the prognostic value of PRR11
expression in human PCa. The data demonstrated that PRR11
expression level could markedly stratify the patients into high
risk and low risk groups regarding BCR (P<0.001; Fig. 6E). However, there was no difference
between the overall survival times of patients with high and low
PRR11 expression (P=0.09; Fig. 6F).
Meanwhile, univariate analysis revealed that PRR11 expression
(P=0.001), pathological tumor stage (P<0.001) and Gleason score
(P<0.001) were significant prognostic factors for BCR-free
survival time in patients with PCa (Table IV). Additionally, Cox proportional
hazards multivariate analysis also indicated that PRR11 expression
level (P=0.018), pathological tumor stage (P=0.003) and Gleason
score (P<0.001) were independent predictors of BCR-free survival
time in PCa patients (Table IV).
Taken together, these findings demonstrated that PRR11
overexpression is associated with aggressive tumor behavior and
poor prognosis.
 | Table IV.Prognostic value of PRR11 expression
for the biochemical recurrence-free survival in univariate and
multivariate analyses by cox regression. |
Table IV.
Prognostic value of PRR11 expression
for the biochemical recurrence-free survival in univariate and
multivariate analyses by cox regression.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) | 1.02
(0.97–1.07) | 0.434 | 0.99
(0.94–1.06) | 0.882 |
| Clinical tumor
stage | 1.03
(0.53–2.01) | 0.926 | 0.73
(0.35–1.52) | 0.405 |
| Pathological tumor
stage | 5.23
(2.56–10.68) | <0.001 | 3.34
(1.52–7.33) | 0.003 |
| Preoperative
PSA | 1.34
(0.52–3.46) | 0.545 | 1.29
(0.47–3.56) | 0.625 |
| Gleason score | 11.59
(5.84–23.01) | <0.001 | 5.31
(2.70–10.48) | <0.001 |
| PRR11 | 3.09
(1.60–5.95) | 0.001 | 2.39
(1.16–4.92) | 0.018 |
Discussion
Traditional predicted methods are unable to
accurately stratify PCa patients according to actual clinical
outcome, which can lead to overtreatment. There is an urgent need
to investigate the molecular mechanism and genetic abnormity
underlying PCa progression. miRNAs have been extensively studied
over the last two decades, and it has been well established that
miRNAs serve essential regulatory roles in virtually all cellular
processes, and that altered miRNA expression is involved in many
human cancers, including PCa (26).
In PCa, miRNAs tend to be preferentially downregulated during PCa
progression and metastasis (27).
In our previous study, we firstly reported that miR-195 plays an
important role in PCa progression and is involved in tumor
invasion, migration and apoptosis. In the present study, we
continued to investigate the molecular function and identify novel
targets of miR-195 in PCa. The results showed that miR-195
upregulation significantly inhibited angiogenesis and
proliferation. Moreover, PRR11 was identified as a novel target of
miR-195, and miR-195 expression combined with PRR11 expression was
associated with aggressive tumor behavior and poor clinical
outcome.
miR-195 has been proven to be involved in many tumor
cell processes, including angiogenesis, invasion, migration,
proliferation, apoptosis and epithelial mesenchymal transition
(EMT) (15–20). Additionally, each individual miRNA
can modulate the expression of multiple mRNAs (6), and miRNAs exert their biological
functions by regulating target genes. In PCa, miR-195 has been
reported to play a regulatory role in the migration, invasion,
proliferation, EMT, angiogenesis and metastasis of tumor cells by
targeting the 3′ UTR sequence of RPS6KB1, FGF2, Fra-1 and BCOX1
(21,28–30).
In the present study, we reported that miR-195 could suppress the
proliferation and cell cycling of PCa cells, and reduce HUVEC tube
formation, by downregulating its novel target PRR11.
PRR11, located on chromosome 17q22, has been
reported to be closely associated with cell cycle progression
(31,32). Multiple highly conserved sequence
motifs in the C terminus of PRR11 protein can be targeted by the
anaphase-promoting complex (APC/C) and FBW7-SCF, which may cause
PRR11 protein degradation and subsequently cell cycle arrest. This
regulatory mechanism is considered to control cell cycle
progression (33). Ji et al
further found that PRR11 might be involved in cell cycle
regulation, especially S phase progression, by altering the
expression of cyclin A1, RRM1, MAP4K4, and DHRS2 (32). PRR11 was also demonstrated to
participate in various biological processes in tumor cells,
including cell invasion, migration and proliferation, by acting as
an oncogene (32,34). Moreover, recent studies have shown
that PRR11 overexpression is significantly associated with
aggressive clinicopathological features and poor clinical outcome
in lung cancer, hilar cholangiocarcinoma, gastric cancer and breast
cancer (32,34–36).
However, the molecular role and clinical relevance of PRR11 in PCa
remains unclear. Here, we firstly reported that PRR11 was
upregulated in PCa tissues. More importantly, PRR11 overexpression
was frequently observed in patients with higher Gleason scores,
tumors of more advanced pathological stage and positive metastasis.
Additionally, survival analysis revealed that PRR11 could be an
independent predictor of the risk of BCR. However, PRR11 expression
level was unable to stratify patients according to overall survival
time, although this may have been due to the lack of data
concerning PCa-specific survival in the Taylor cohort. In
vitro assays indicated that the knockdown of PRR11 expression
could markedly suppress cell proliferation, the cell cycle and
angiogenesis in PCa cells. Meanwhile, the subcutaneous xenograft
model further showed that the knockdown of PRR11 significantly
suppressed tumor growth and angiogenesis in vivo. These
findings demonstrated that PRR11 functions as a promotive factor in
PCa progression. Furthermore, critical pathways and differentially
expressed genes affected by PRR11 knockdown in PCa were analyzed by
microarray analysis employing bioinformatics software. Multiple
dysregulated genes were enriched in tumor invasion and
metastasis-related pathways, such as CCNA1, RRM1, MAP4K4 and CCL2,
which indicated that PRR11 might exert its oncogene role via these
important downstream genes (32).
However, the definitive mechanism underlying the effects of PRR11
on PCa progression requires further investigation.
In conclusion, our research indicated the role of
PRR11 in PCa and identified PRR11 as a new target of miR-195.
Notably, PRR11 is an important downstream mediator of the
suppressive effects of miR-195 on PCa progression. In addition,
miR-195 expression combined with PRR11 expression could accurately
stratify PCa patients into different groups according to clinical
outcome, and thus the use of these markers may aid to achieve more
effective management of PCa in future clinical practice.
Acknowledgements
This study was supported by grants from China
Postdoctoral Science Foundation (2015M580710), Natural Science
Foundation of Guangdong Province (2016A030310276), National Key
Basic Research Program of China (2015CB553706), National Natural
Science Foundation of China (81602541, 81370804, 81272813,
81571427, 81402430, 81602541, 81641102), Hospital Young Program
Foundation (201523-gyfyy), Special Research Fund for the Doctoral
Program of Higher Education (20134423110004), Science and
Technology Project in Guangdong (2014A020209085), Colleges and
Universities in Guangzhou Yangcheng Scholars Research Project
(12A017S), Distinguished Young Talents in Higher Education
Foundation of Guangdong Province (2014KQNCX121).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fraser M, Berlin A, Bristow RG and van der
Kwast T: Genomic, pathological, and clinical heterogeneity as
drivers of personalized medicine in prostate cancer. Urol Oncol.
33:85–94. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cooperberg MR, Broering JM, Litwin MS,
Lubeck DP, Mehta SS, Henning JM and Carroll PR;
CaPSUREInvestigators, : The contemporary management of prostate
cancer in the United States: Lessons from the cancer of the
prostate strategic urologic research endeavor (CapSURE), a national
disease registry. J Urol. 171:1393–1401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dall'Era MA, Cooperberg MR, Chan JM,
Davies BJ, Albertsen PC, Klotz LH, Warlick CA, Holmberg L, Bailey
DE Jr, Wallace ME, et al: Active surveillance for early-stage
prostate cancer: Review of the current literature. Cancer.
112:1650–1659. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Capitanio U, Briganti A, Gallina A, Suardi
N, Karakiewicz PI, Montorsi F and Scattoni V: Predictive models
before and after radical prostatectomy. Prostate. 70:1371–1378.
2010.PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim WT and Kim WJ: MicroRNAs in prostate
cancer. Prostate Int. 1:3–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fabris L, Ceder Y, Chinnaiyan AM, Jenster
GW, Sorensen KD, Tomlins S, Visakorpi T and Calin GA: The potential
of MicroRNAs as prostate cancer biomarkers. Eur Urol. 70:312–322.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bhattacharya A, Schmitz U, Wolkenhauer O,
Schönherr M, Raatz Y and Kunz M: Regulation of cell cycle
checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene.
32:3175–3183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang
Y, Zou C, Zhang X, Liu S, Wang X, et al: Analysis of MiR-195 and
MiR-497 expression, regulation and role in breast cancer. Clin
Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ding J, Huang S, Wang Y, Tian Q, Zha R,
Shi H, Wang Q, Ge C, Chen T, Zhao Y, et al: Genome-wide screening
reveals that miR-195 targets the TNF-α/NF-κB pathway by
down-regulating IκB kinase alpha and TAB3 in hepatocellular
carcinoma. Hepatology. 58:654–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Soon PS, Tacon LJ, Gill AJ, Bambach CP,
Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson
BG and Sidhu SB: miR-195 and miR-483-5p identified as predictors of
poor prognosis in adrenocortical cancer. Clin Cancer Res.
15:7684–7692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jia LF, Wei SB, Gong K, Gan YH and Yu GY:
Prognostic implications of micoRNA miR-195 expression in human
tongue squamous cell carcinoma. PLoS One. 8:e566342013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu MG, Li S, Yu TT, Qian LJ, Cao RS, Zhu
H, Xiao B, Jiao CH, Tang NN, Ma JJ, et al: Differential expression
of miR-195 in esophageal squamous cell carcinoma and miR-195
expression inhibits tumor cell proliferation and invasion by
targeting of Cdc42. FEBS Lett. 587:3471–3479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai C, Chen QB, Han ZD, Zhang YQ, He HC,
Chen JH, Chen YR, Yang SB, Wu YD, Zeng YR, et al: miR-195 inhibits
tumor progression by targeting RPS6KB1 in human prostate cancer.
Clin Cancer Res. 21:4922–4934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maragkakis M, Alexiou P, Papadopoulos GL,
Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis
K, Simossis VA, et al: Accurate microRNA target prediction
correlates with protein repression levels. BMC Bioinformatics.
10:2952009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC,
Ling XH, Fu X, Dai QS, Cai C, Chen JH, et al: MicroRNA-224 inhibits
progression of human prostate cancer by downregulating TRIB1. Int J
Cancer. 135:541–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ong SE, Blagoev B, Kratchmarova I,
Kristensen DB, Steen H, Pandey A and Mann M: Stable isotope
labeling by amino acids in cell culture, SILAC, as a simple and
accurate approach to expression proteomics. Mol Cell Proteomics.
1:376–386. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bolton EM, Tuzova AV, Walsh AL, Lynch T
and Perry AS: Noncoding RNAs in prostate cancer: The long and the
short of it. Clin Cancer Res. 20:35–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu C, Guan H, Wang Y, Chen M, Xu B, Zhang
L, Lu K, Tao T, Zhang X and Huang Y: miR-195 inhibits EMT by
targeting FGF2 in prostate cancer cells. PLoS One. 10:e01440732015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu J, Ji A, Wang X, Zhu Y, Yu Y, Lin Y,
Liu Y, Li S, Liang Z, Xu X, et al: MicroRNA-195-5p, a new regulator
of Fra-1, suppresses the migration and invasion of prostate cancer
cells. J Transl Med. 13:2892015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo J, Wang M and Liu X: MicroRNA-195
suppresses tumor cell proliferation and metastasis by directly
targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res.
34:912015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang C, Zhang Y, Li Y, Zhu H, Wang Y, Cai
W, Zhu J, Ozaki T and Bu Y: PRR11 regulates late-S to G2/M phase
progression and induces premature chromatin condensation (PCC).
Biochem Biophys Res Commun. 458:501–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ji Y, Xie M, Lan H, Zhang Y, Long Y, Weng
H, Li D, Cai W, Zhu H, Niu Y, et al: PRR11 is a novel gene
implicated in cell cycle progression and lung cancer. Int J Biochem
Cell Biol. 45:645–656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Larance M, Ahmad Y, Kirkwood KJ, Ly T and
Lamond AI: Global subcellular characterization of protein
degradation using quantitative proteomics. Mol Cell Proteomics.
12:638–650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y, Cha Z, Fang W, Qian B, Yu W, Li W,
Yu G and Gao Y: The prognostic potential and oncogenic effects of
PRR11 expression in hilar cholangiocarcinoma. Oncotarget.
6:20419–20433. 2015.PubMed/NCBI
|
|
35
|
Song Z, Liu W, Xiao Y, Zhang M, Luo Y,
Yuan W, Xu Y, Yu G and Hu Y: PRR11 is a prognostic marker and
potential oncogene in patients with gastric cancer. PLoS One.
10:e01289432015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou F, Liu H, Zhang X, Shen Y, Zheng D,
Zhang A, Lai Y and Li H: Proline-rich protein 11 regulates
epithelial-to-mesenchymal transition to promote breast cancer cell
invasion. Int J Clin Exp Pathol. 7:8692–8699. 2014.PubMed/NCBI
|