Introduction
Glioblastoma, one of the most common primary brain
tumors is the most lethal intracranial malignant tumor accounting
for the majority of gliomas occurring in the human brain. Recent
statistics report that approximately 20.59 per 100,000 patients
were diagnosed each year in the United States between 2005–2009
(1). Glioblastoma is characterized
by poor prognosis due to its biological characteristics of rapid
proliferation, uncontrolled migration, infiltration, resistance to
chemotherapy, as well as high recurrence even after surgical
resection. Accumulating evidence shows that the basis of glioma
migration and infiltration is often closely related to the
excessive proliferation of cells. In recent years, a deeper
understanding of the molecular mechanism underlying glioma
development has led to the discovery of many molecular markers as
indicators of clinical diagnosis and treatment. Among them are
adhesion molecules involved in the migration and metastasis of
gliomas (2), which are mostly
clinically applied (3). However,
little is known about the effect of adhesion molecules on the
proliferation of tumor cells.
Cadherins are calcium-dependent cell adhesive
glycoproteins which play important roles in regulating cell
recognition, migration and tissue differentiation during embryonic
development (4,5). N-cadherin as a classic member of
cadherins is a homophilic transmembrane adhesion glycoprotein that
is widely distributed in the central nervous system, especially in
neurons and glial cells (2). In a
variety of tumors, the abnormal expression of N-cadherin enhances
cell activity and invasive ability (6) such as in breast (7), prostate (8) and bladder cancer (9). Similarly, the expression of N-cadherin
in glioma tissues is significantly higher compared with normal
brain tissues (10). Maret et
al (11) reported that the
precursors of N-cadherin (proN-cadherin) and N-cadherin were
present on the cell membrane and the proportion of precursors on
the tumor cells was higher (11).
Our previous study doubted recent studies on N-cadherin,
criticizing that these studies were actually about precursors of
N-cadherin, and we demonstrated that GDNF can promote the adhesion
of glioma cells to the matrix by promoting the expression of
proN-cadherin in glioma cells, which successively amplified the
process of migration and invasion (2).
Glial cell-line derived neurotrophic factor (GDNF),
a member of the transforming growth factor-β (TGF-β) superfamily,
is a soluble extracellular factor initially found to be a
protective factor for the survival and differentiation of
dopaminergic neurons (12). GDNF
plays important roles in neuronal survival, growth, differentiation
and migration (13,14). However, it has been reported to be
strongly expressed in human gliomas (15). During neurogenesis, GDNF regulates
cell differentiation and organ formation by promoting self-renewal
and proliferation of stem cells (12,16,17).
In our previous studies, we reported that GDNF was abnormally
highly expressed in glioma tissues and we suggested that high
concentration of GDNF promoted glioma development (2,18).
Since the specific mechanisms underlying glioma development are
constantly updated, GDNF has been identified as a major force of
attraction and has being studied extensively regarding its roles in
gliomas. One of these studies reported that GDNF could directly
stimulate the membrane receptor-Neuropilin-1 (19), and activate proliferation-related
signaling pathways, however, it is unknown whether GDNF can
indirectly activate other growth factor-related receptors.
Furthermore, it has been reported that GDNF can bind with the
adhesion molecule NCAM and RET on the cell membrane as a
co-receptor transduction signal to regulate the growth and
migration of Schwann neurons (20).
In addition, this study revealed that GDNF
indirectly stimulated other family receptors on the cell membrane
and strengthened their signal transduction processes, hence
promoting the growth and proliferation of glioblastoma cells. This
is a relatively new and original viewpoint, which indicates a new
direction for research on the relationship between adhesion
molecules and membrane receptors.
Materials and methods
Cell culture and transfection
The human malignant glioma cell line U251MG was
obtained from the Shanghai Institute of Biological Sciences
(Shanghai, China). It was verified that the cells we used matched
the profile of U251MG cells. The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; HyClone Laboratories, Logan, UT,
USA), supplemented with 10% fetal bovine serum (FBS; HyClone
Laboratories) and 0.1% penicillin-streptomycin at 37°C in a
humidified 5% CO2 atmosphere. According to the
experimental protocol, the cells were treated with human GDNF (50
ng/ml; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for
30 min, while untreated cells were left in medium (control).
The EF1A-proN-cadherin-IRES-EGFP and vector plasmids
were constructed based on the proN-cadherin sequence [(National
Center for Biotechnology Information (NCBI) reference sequence:
BC036470.1]. In addition, we also designed a highly effective small
interfering RNA (siRNA) plasmid and one negative control RNAi
vector plasmid. The target sequence of proN-cadherin siRNA and the
control were as follows: siRNA-sense, (5′-3′) GUGCAGUCUUAUCGAAG
GATT and antisense (5′-3′) UCCUUCGAUAAGACUGCA CTT; control sense,
(5′-3′) UUCUC CGAACGUGUCACGUTT and antisense, (5′-3′)
ACGUGACACGUUCGGAGAATT.
The proN-cadherin overexpression plasmid and siRNA
with their respective control plasmids were transfected into
serum-starved U251MG cells for 24 h by Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), continuously cultured
in 6-well plates.
Cell viability assays
The U251MG cells from different groups were seeded
into 96-well plates at a density of 1×104 cells/well.
The first MTT assay was performed after 12 h when the cells had
grown to 50% confluency. The above-mentioned results were regarded
as starting value (0 h). Concurrently, we treated cells with GDNF
50 ng/ml for 30 min according to the experimental grouping
protocol.
At different indicated time-points, MTT solution was
added to the wells and incubated for 4 h at 37°C. Subsequently, the
supernatant was discarded and 150 µl dimethylsulfoxide (DMSO) per
well was added. The optical density was assessed with a microplate
reader at a wavelength of 570 nm.
Co-immunoprecipitation and western
blot analysis
U251MG cell membrane protein was extracted using an
eukaryotic membrane protein extraction kit (ProteoExtract™, M-PEK;
Merck KGaA, Darmstadt, Germany). The primary anti-proN-cadherin
antibody (GTX101141; GeneTex, Irvine, CA, USA) and protein A/G
agarose (sc-2003; Santa Cruz Biotechnology, Santa Cruz, CA, USA)
for immunoprecipitation were added into the lysates with the
membrane protein at 4°C overnight on a low-speed rotating shaker.
The beads were washed three times with lysis buffer and boiled in
1X SDS loading buffer to elute the antibody bound protein. SDS-PAGE
gels (10%) were used to separate samples and polyvinylidene
fluoride (PVDF) membranes were used to transfer the protein blots.
Membranes were blocked with 5% skimmed milk, washed and incubated
with primary antibody [rabbit anti-FGFR1, 1:1,000; mouse anti-FGFR1
(pY653+pY654), 1:1,000] at 4°C overnight. The membranes were
incubated with IRdye secondary antibodies (goat anti-rabbit;
1:10,000; cat. no. 92632211; goat anti-mouse; 1:10,000; cat. no.
92632210; LI-COR Biosciences, Lincoln, NE, USA) and scanned by an
Odyssey imaging system (LI-COR Biosciences). In addition, total
protein from U251MG cells was extracted by RIPA lysis buffer
(Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) containing a
mixture of protease inhibitors.
U251MG cells were divided into 8 groups as follows:
normal, normal with 50 ng/ml GDNF, overexpression proN-cadherin
with or without 50 ng/ml GDNF treated for 30 min, the control
plasmid group, proN-cadherin siRNA with or without 50 ng/ml GDNF
treated for 30 min. Prior to this study, we had applied three types
of siRNA to verify the downregulation of proN-cadherin. The type
used in the present study could realize the downregulation of
proN-cadherin and ensure cell survival without considerable
cytotoxicity. The siRNA sequences were based on the 477 bp sequence
(pro-domain) on the N-terminal of proN-cadherin mRNA (ref. seq.,
BC036470.1). After blocking by 5% skimmed milk, the samples were
incubated with primary antibody (rabbit anti-proN-cadherin
antibody; 1:1,000; cat. no. GTX101141; GeneTex; mouse anti-GAPDH
antibody; 1:1,000; cat. no. sc-365062; Santa Cruz Biotechnology;
rabbit anti-caveolin1, 1:1,000; cat. no. ab17052; Abcam) at 4°C
overnight. Then, the samples were incubated with IRdye secondary
antibodies (goat anti-rabbit, 1:1,000; goat anti-mouse, 1:1,000;
LI-COR Biosciences) at room temperature for 2h. Finally, the
protein bands were scanned by Odyssey imaging system (LI-COR
Biosciences) and quantifed with ImageJ software (National
Institutes of Health, Bethesda, MD, USA).
Immunofluorescence assay
Coverslips were put into 24-well plates as U251MG
cells were seeded into different wells and cultured for 24 h. Cells
were then monitored until 50% confluent and treated with GDNF 50
ng/ml for 30 min. Subsequently, cells were washed with PBS three
times, followed by fixation for 30 min at room temperature in 4%
paraformaldehyde. Fixed cells were permeabilized for 5 min at room
temperature using 0.3% Triton X-100 and blocked for 30 min using 5%
goat serum diluted with PBS. Furthermore, the samples were
incubated with GDNF antibody (rabbit anti-GDNF, 1:250; cat. no.
ab18956; Abcam) overnight at 4°C, followed by a series of washing
with PBS and finally incubation with secondary antibody: goat
anti-rabbit IgG (H+L)-DyLight 594 (1:1,000; EarthOx Life Sciences,
Millbrae, CA, USA) for 2 h in the dark and
4′6-diamidino-2-phenylindole (DAPI). Successively, cells were
stained by proN-cadherin antibody (rabbit anti-proN-cadherin
antibody, 1:250; cat. no. GTX101141; GeneTex) and incubated with
goat anti-rabbit IgG (H+L)-DyLight 488 (at 1:1,000; EarthOx Life
Sciences). Fluorescence images were captured with a fluorescent
inverted microscope (Leica Microsystems, Wetzlar, Germany).
Real-time quantitative PCR
Total RNA was extracted by TRIzol reagent
(15596-026; Invitrogen) and the first-strand cDNA was synthesized
by RevertAidTM H Minus First Strand cDNA Synthesis kit (K1631;
Fermentas; Thermo Fisher Scientific, Inc.), followed by qPCR using
the SYBR-Green PCR Master Mix (ABI 4309155; Applied Biosystems;
Thermo Fisher Scientific). qPCR conditions were as follows: 5 min
at 95°C; 20 sec at 94°C, 20 sec at 61°C and 20 sec at 72°C for 40
cycles followed by 72°C for 5 min. The above procedure was
implemented on the Real-Time-PCR system (ABI 7900). β-actin was
used as a reference gene and qRT-PCR was performed in triplicates
for each sample. The relative expression level of target genes was
calculated by the 2−∆∆Ct method. Upstream and downstream
primer sequences for the amplification of the target gene and
internal reference are listed in Table
I.
 | Table I.Primer information. |
Table I.
Primer information.
| Gene name | Forward primer | Reverse primer | Sequence length
(bp) |
|---|
|
Homo-proN-cadherin |
5′-agcagtgagcctgcagattt-3′ |
5′-gtggccactgtgcttactga-3′ | 243 |
| Homo-β-actin |
5′-cattaaggagaagctgtgct-3′ |
5′-gttgaaggtagtttcgtgga-3′ | 208 |
Statistical analysis
The quantitative data were presented as the mean ±
standard deviation (SD) of two independent experiments and analyzed
by the Student's t-test. Multiple comparisons between groups were
performed using one-way analysis of variance followed by the
Student-Newman-Keuls test for statistical analysis. Statistical
analyses were performed using SPSS version 19.0 (IBM Corp., Armonk,
NY, USA). For all statistical analyses, P<0.05 was considered to
indicate a statistically significant difference.
Results
GDNF amplifies the expression of
proN-cadherin and promotes U251MG cell viability
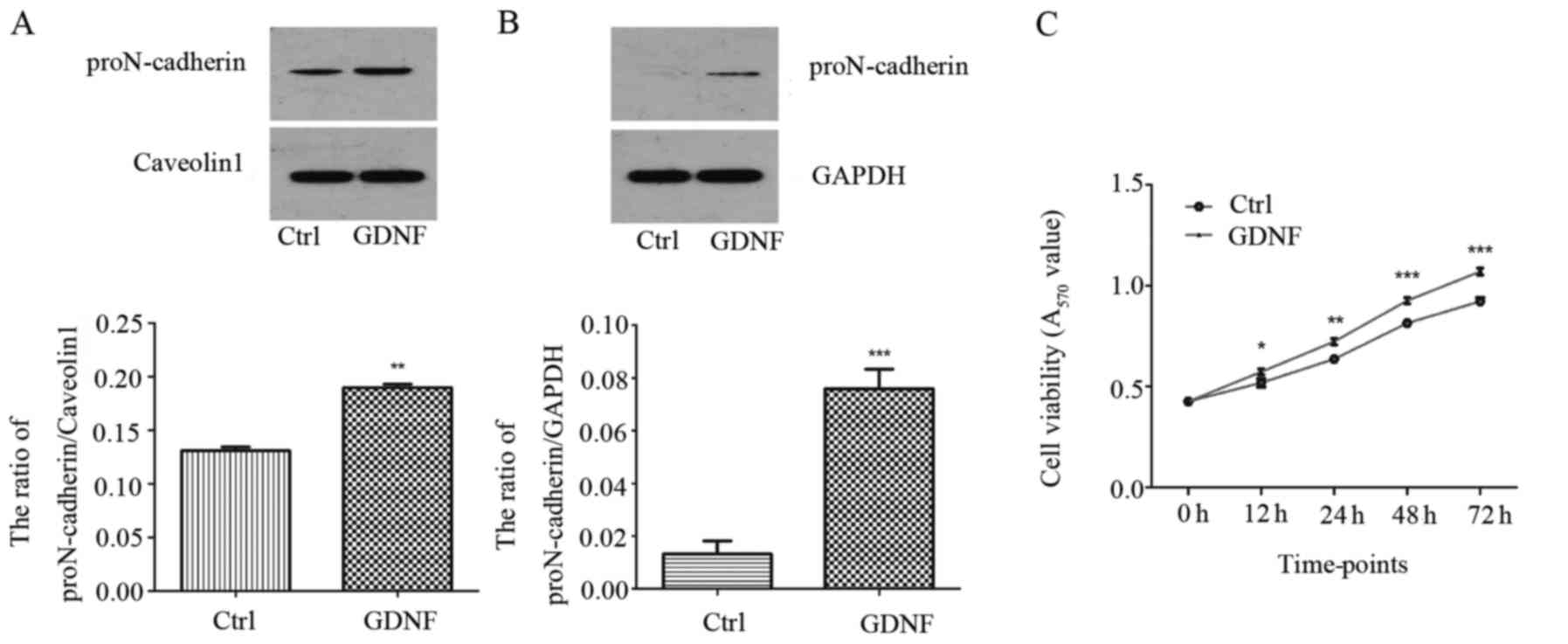
The proN-cadherin and N-cadherin expression level
were evaluated in U251MG cells treated with or without 50 ng/ml
GDNF for 30 min, comparing both the membrane and the cytoplasm
proteins. As displayed in Fig. 1A and
B proN-cadherin was mainly expressed on the cell membrane and
GDNF enhanced the expression level of proN-cadherin in both the
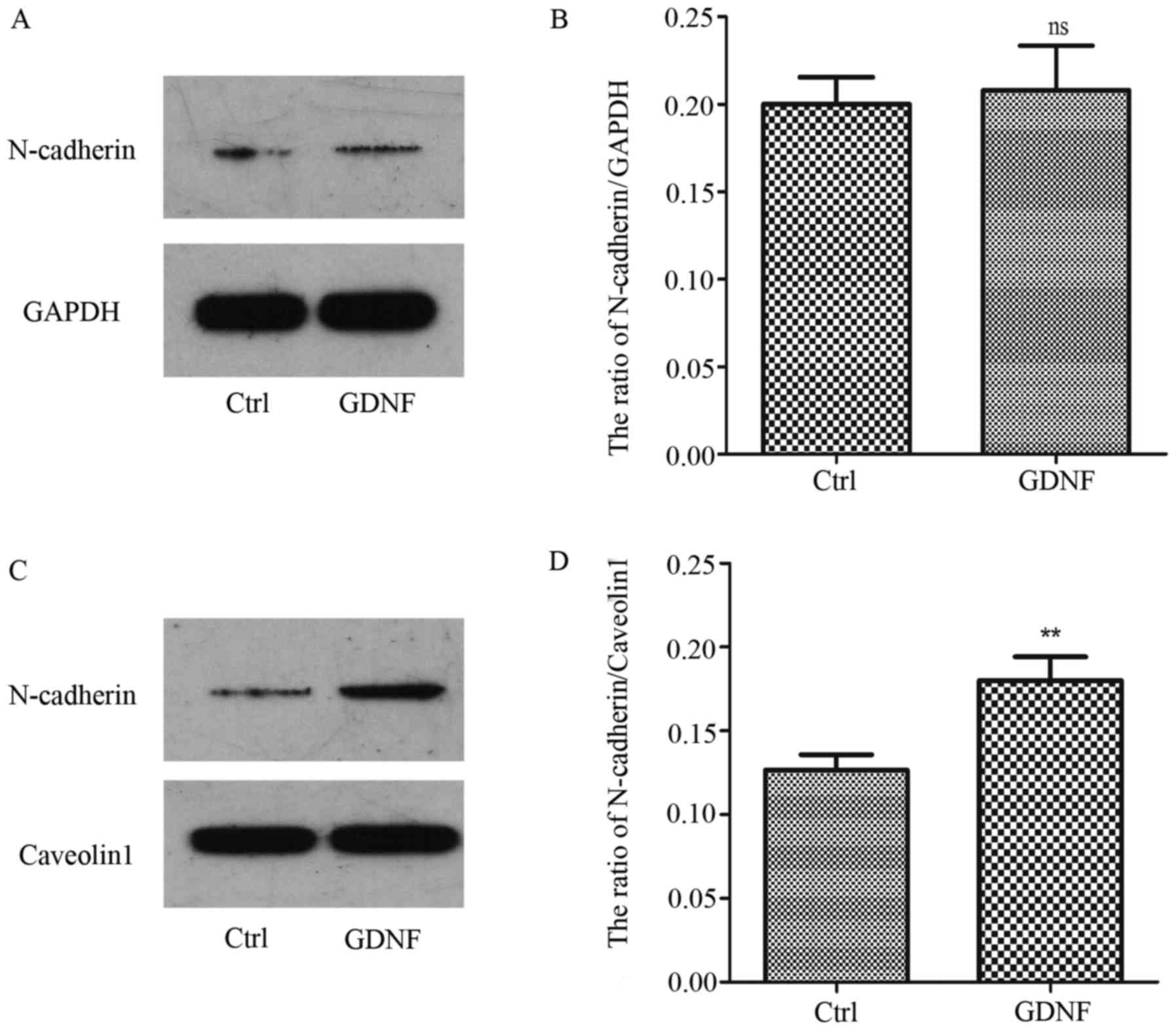
cytoplasm and the cell membrane. However, the level of N-cadherin
in the membrane was obviously increased with GDNF treatment.
Concurrently, we observed that the N-cadherin level in the
cytoplasm was almost not changed (Fig.
2).
Concurrently, we analyzed the proliferative effects
of GDNF on the U251MG cells using an MTT assay. The results
revealed that GDNF promoted the viability of U251MG cells in a
significant manner (Fig. 1C).
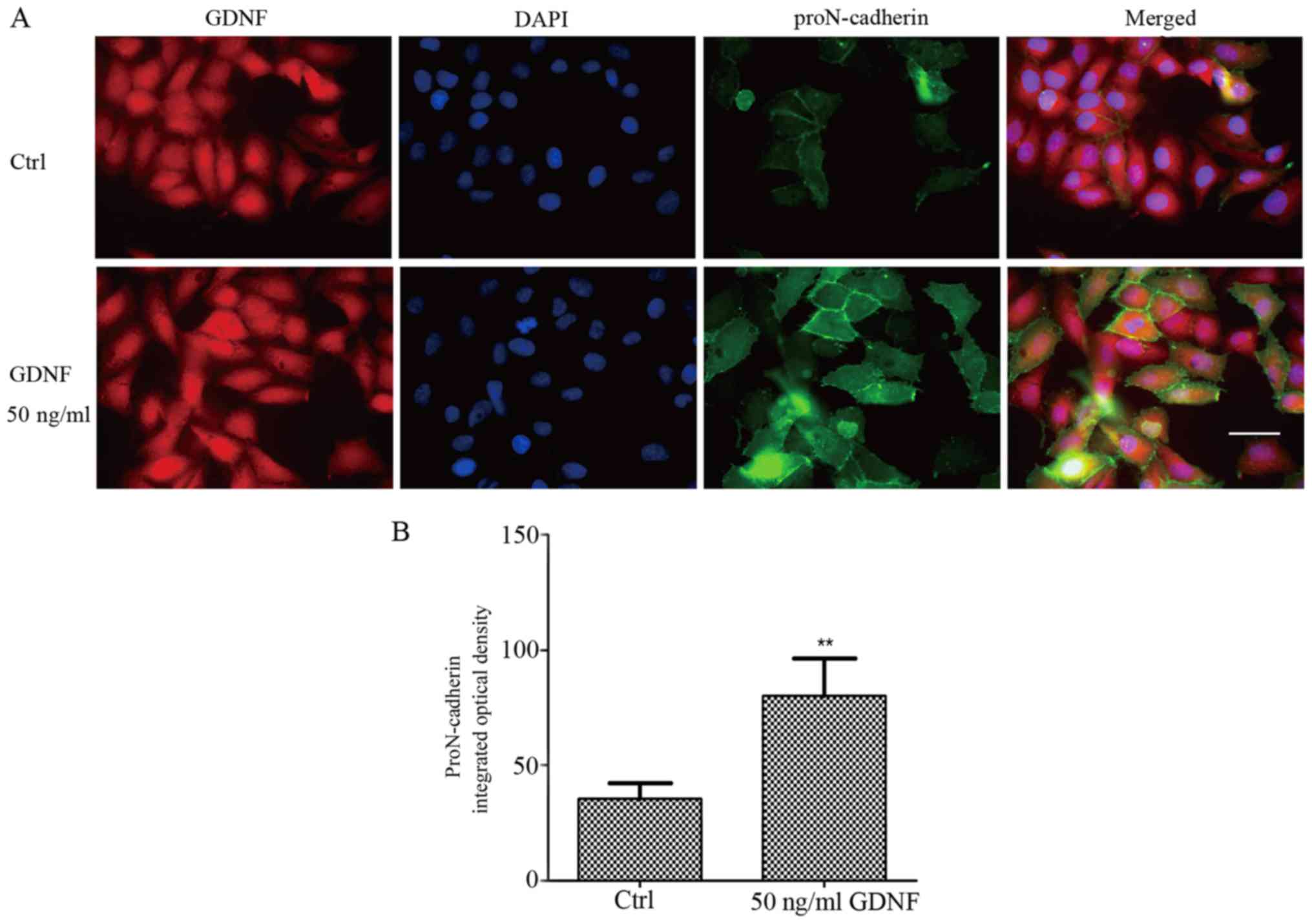
In our previous study, proN-cadherin was reported to
be abundantly present in the cytomembrane and could interact with
GDNF (2). As displayed in Fig. 1, we confirmed that proN-cadherin was
mainly expressed in the cytomembrane, however, the percentage of
proN-cadherin in the cytoplasm was extremely low. To further
validate these results, we performed immunofluorescence (IF)
experiments to localize the protein expression within the cells.
The fluorescence intensity of proN-cadherin was significantly
higher in U251MG cells with 50 ng/ml GDNF than in cells without
GDNF (Fig. 3). This finding
indicated that proN-cadherin was more abundant in the membrane of
U251MG cells due to the high expression of GDNF in the cells.
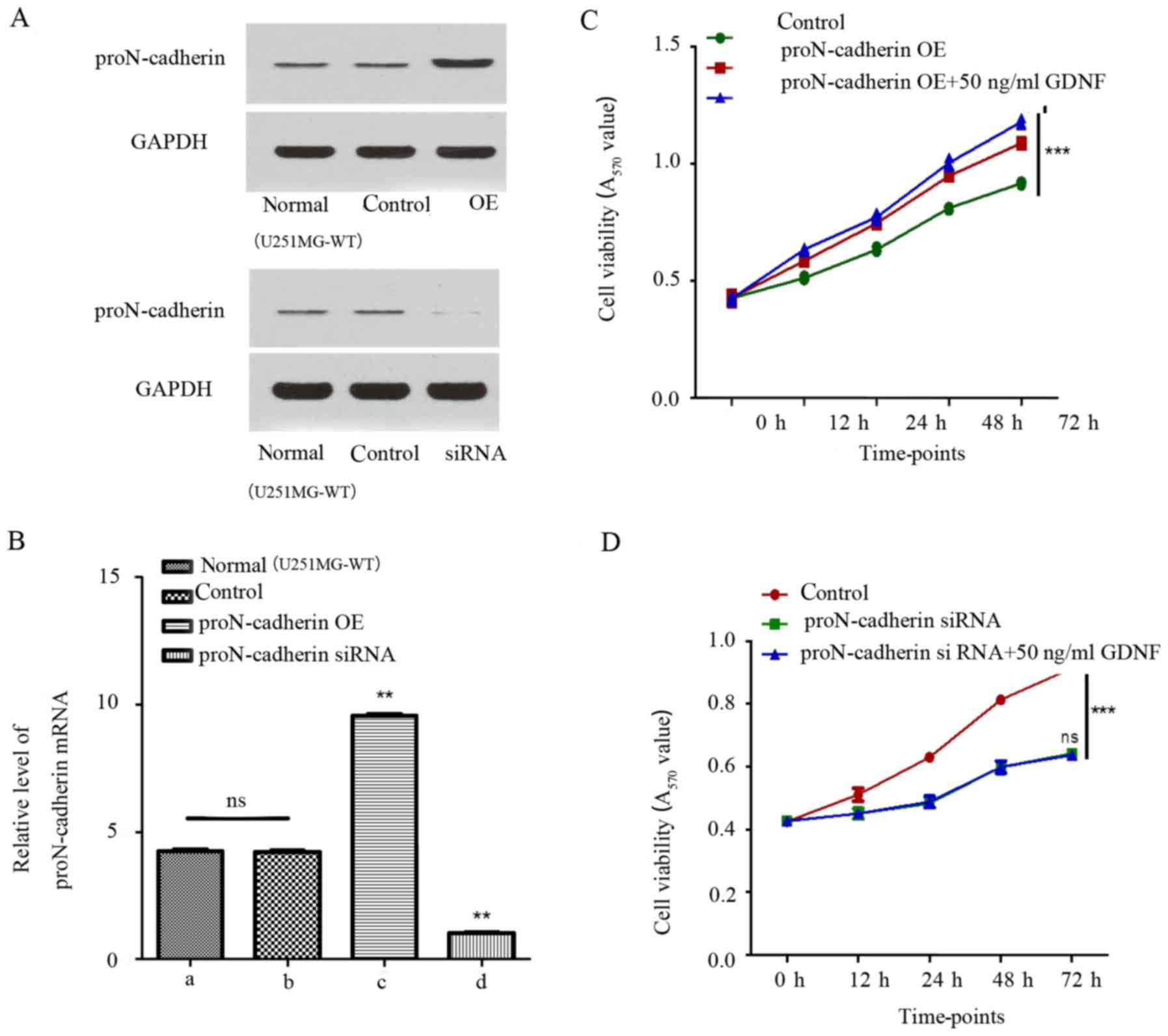
GDNF promotes proN-cadherin-induced
viability of U251MG cells
Since the relationship between proN-cadherin and
cell viability was unclear, the expression of proN-cadherin was
altered by constructing plasmids to overexpress proN-cadherin and
proN-cadherin siRNA to implement the variation of protein and mRNA
expression level. ProN-cadherin protein and mRNA expression was
verified by western blot analysis and qPCR respectively in samples
from transfected and untransfected cells (Fig. 4A and B). Consequently, while
comparing the control group with the proN-cadherin overexpressed
group using the MTT assay, we observed an obviously increased rate
of cell viability in the proN-cadherin OE group (Fig. 4C). Notably, proN-cadherin OE group
with exogenous GDNF facilitated cell viability more obviously.
Furthermore, siRNA of proN-cadherin reduced the rate of cell
viability. The viability could not be improved despite treatment
with 50 ng/ml GDNF, which indicated that it was more difficult for
GDNF to play a role in promoting the cell viability under the low
proN-cadherin expression state (Fig.
4D). Detailed measurement data are listed in Tables II and III.
 | Table II.The OD570-difference
comparison between proN-cadherin OE and control groups at different
time-points (mean ± SD, n=3). |
Table II.
The OD570-difference
comparison between proN-cadherin OE and control groups at different
time-points (mean ± SD, n=3).
| Time-point (h) | Control | proN-cadherin
OE | proN-cadherin
OE+GDNF |
|---|
| 0 | 0.426±0.017 | 0.427±0.020 | 0.425±0.012 |
| 12 | 0.512±0.010 | 0.585±0.011 | 0.633±0.008 |
| 24 | 0.632±0.011 | 0.745±0.006 | 0.771±0.016 |
| 48 | 0.810±0.007 | 0.947±0.008 | 1.003±0.020 |
| 72 | 0.916±0.008 | 1.087±0.009 | 1.178±0.015 |
 | Table III.The OD570-difference
comparison between proN-cadherin siRNA and control groups at
different time-points (mean ± SD, n=3). |
Table III.
The OD570-difference
comparison between proN-cadherin siRNA and control groups at
different time-points (mean ± SD, n=3).
| Time-point (h) | Control | proN-cadherin
siRNA | proN-cadherin
siRNA+GDNF |
|---|
| 0 | 0.425±0.010 | 0.426±0.013 | 0.427±0.009 |
| 12 | 0.511±0.021 | 0.450±0.017 | 0.451±0.014 |
| 24 | 0.630±0.008 | 0.483±0.014 | 0.488±0.018 |
| 48 | 0.814±0.014 | 0.599±0.014 | 0.600±0.019 |
| 72 | 0.914±0.011 | 0.642±0.011 | 0.643±0.010 |
Based on these results we concluded that the
increasing level of proN-cadherin on the membrane of U251MG cells
improved the ability of cell viability to a considerable extent. In
addition, the synergistic effect of GDNF and proN-cadherin would
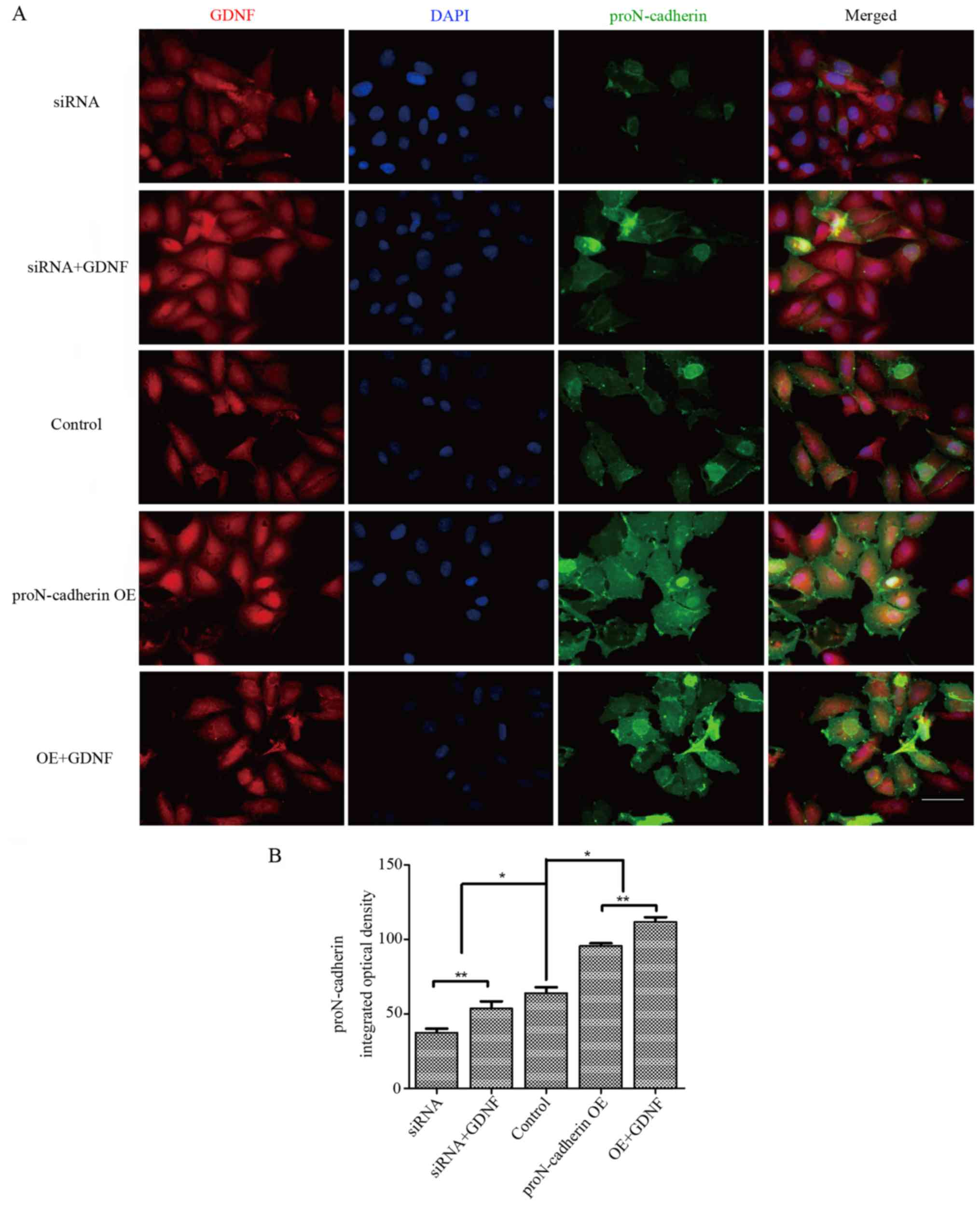
reinforce its effect on cell viability. To identify changes in the
orientation and expression level of proN-cadherin, we provided
morphological evidence by performing immunofluorescence assay. In
the overexpression group with 50 ng/ml GDNF, the integrated optical
density of proN-cadherin was higher than in other groups
(P<0.05) (Fig. 5) and it was
clearly observed that proN-cadherin on the cell membrane was
significantly increased under the effect of GDNF whether in the
siRNA or in the proN-cadherin OE group.
GDNF increases the phosphorylation
level of FGFR1 and strengthens proN-cadherin and FGFR1 (pY653+Y654)
interaction on the cell membrane
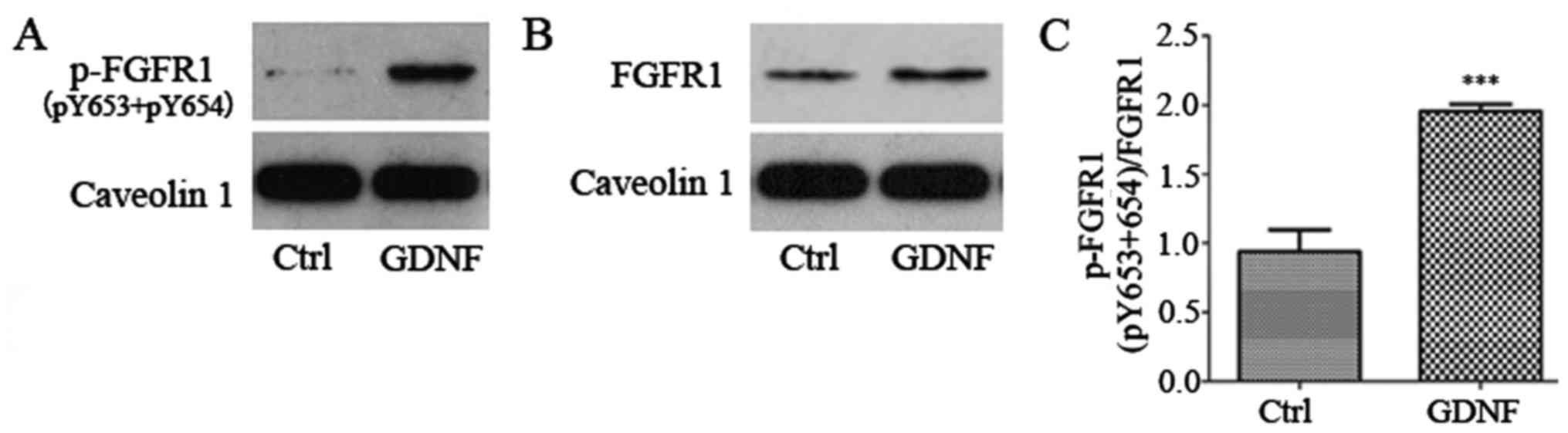
Subsequently, we explored how GDNF-induced
proN-cadherin activation in the cell membrane exerts a role in
regulating cell viability. Caveolin 1 was used as a suitable
reference of the membrane protein, and we observed that the
phosphorylation level of FGFR1(pY653+pY654) increased significantly
(Fig. 6). Although the expression
of FGFR1 was slightly enhanced by GDNF, the increasing ratio of
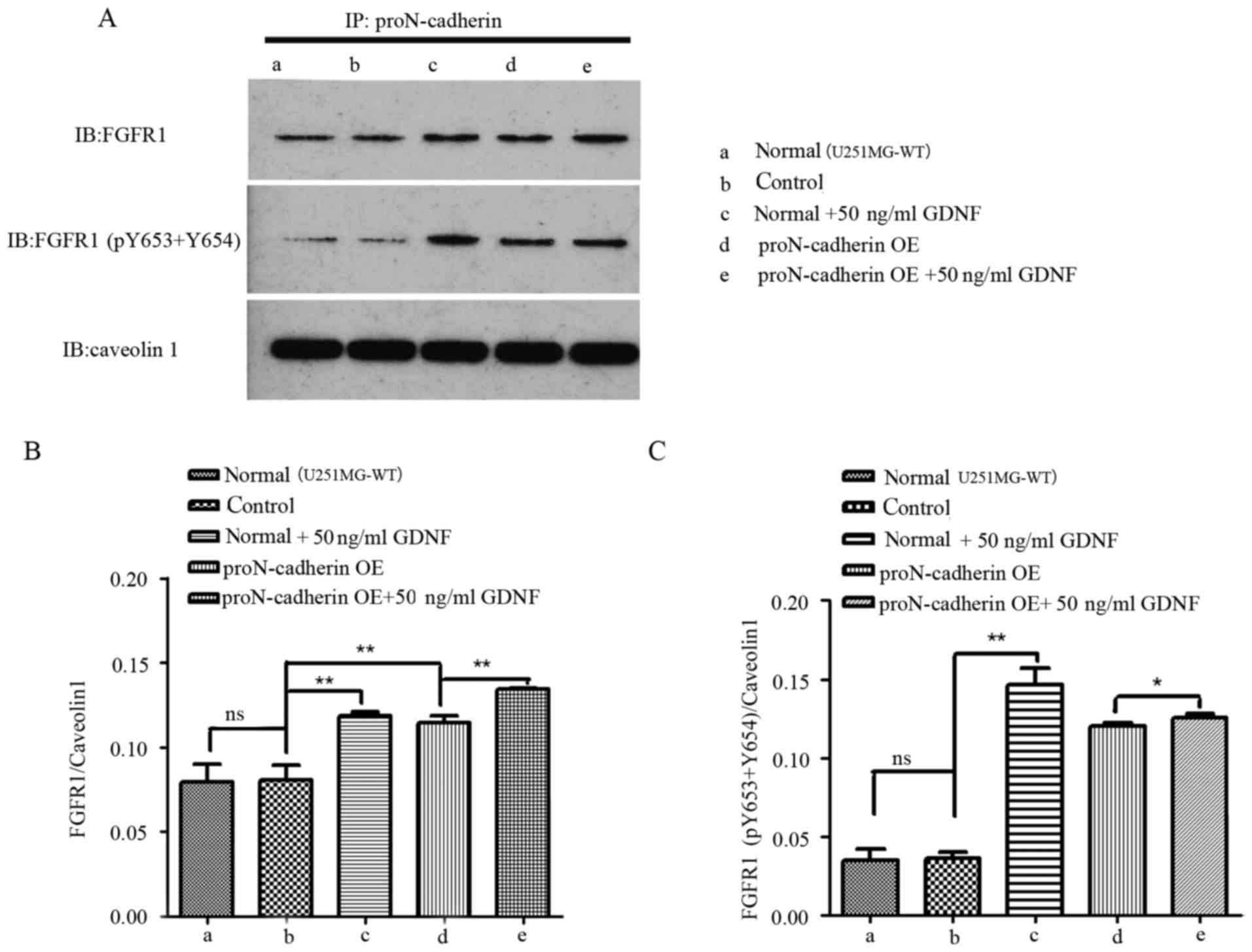
phosphorylated FGFR1 was still greater. After immunoprecipitation
protein spectrum analysis, we observed that FGFR1 interacted with
proN-cadherin. Furthermore, we examined two phosphorylation sites
on FGFR1 (Y653 and Y654). The conclusion of GDNF interacting with
proN-cadherin has been demonstrated in our previous study (2), but the interaction between
proN-cadherin and FGFR1 was reported in the present study for the
first time. We speculated that the potential mechanism employed by
GDNF-induced proN-cadherin interaction with FGFR1 was an effort to
improve signal transmission and cell viability. Under the influence
of exogenous GDNF, the combining amount of these two membrane
proteins would be enhanced, especially the interaction between the
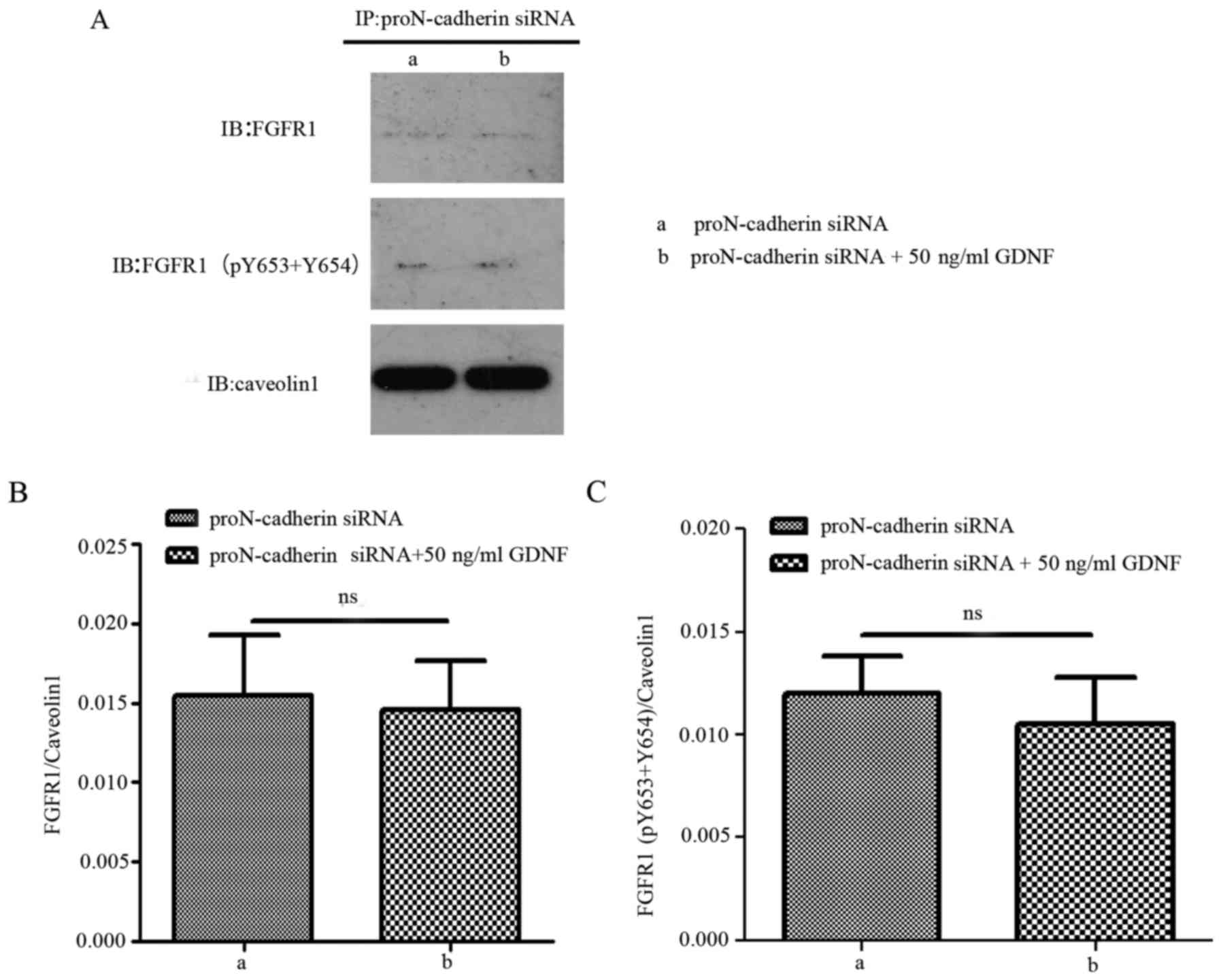
phosphorylated FGFR1 and proN-cadherin (Fig. 7). When proN-cadherin was
downregulated, there was no significant change observed in the
interaction between proN-cadherin and FGFR1/FGFR1 (pY653+pY654)
regardless of the presence or absence of GDNF (Fig. 8). These results indicated that the
presence of proN-cadherin was vital for this process.
The above-mentioned results indicated that
proN-cadherin interacted with FGFR1/FGFR1 (pY653+pY654) and this
combined capacity could be enhanced by exogenous GDNF treatment and
the high expression of membrane proN-cadherin protein. However,
once proN-cadherin protein was downregulated on the cell membrane,
GDNF would not play a role in promoting interactions between these
two proteins. For further speculation, the proliferative effects
may not be realized without GDNF mediating the connection between
proN-cadherin and FGFR1.
Discussion
We have previously reported that GDNF exhibited
protective effects on dopaminergic neurons by interacting with
transmembrane proteins such as integrin β1 (21), NCAM (22) and N-cadherin (23). Furthermore, according to a previous
study GDNF was approximately five times more highly expressed in
human malignant gliomas compared to normal human brain tissues
(15). Based on these data, we
recently reported the interactions between GDNF and precursors
N-cadherin by molecular docking analysis, co-immunoprecipitation
and immunofluorescence analysis, and provided evidence that GDNF
interacted with five AA residues in the EC3 region of
proN-cadherins (2).
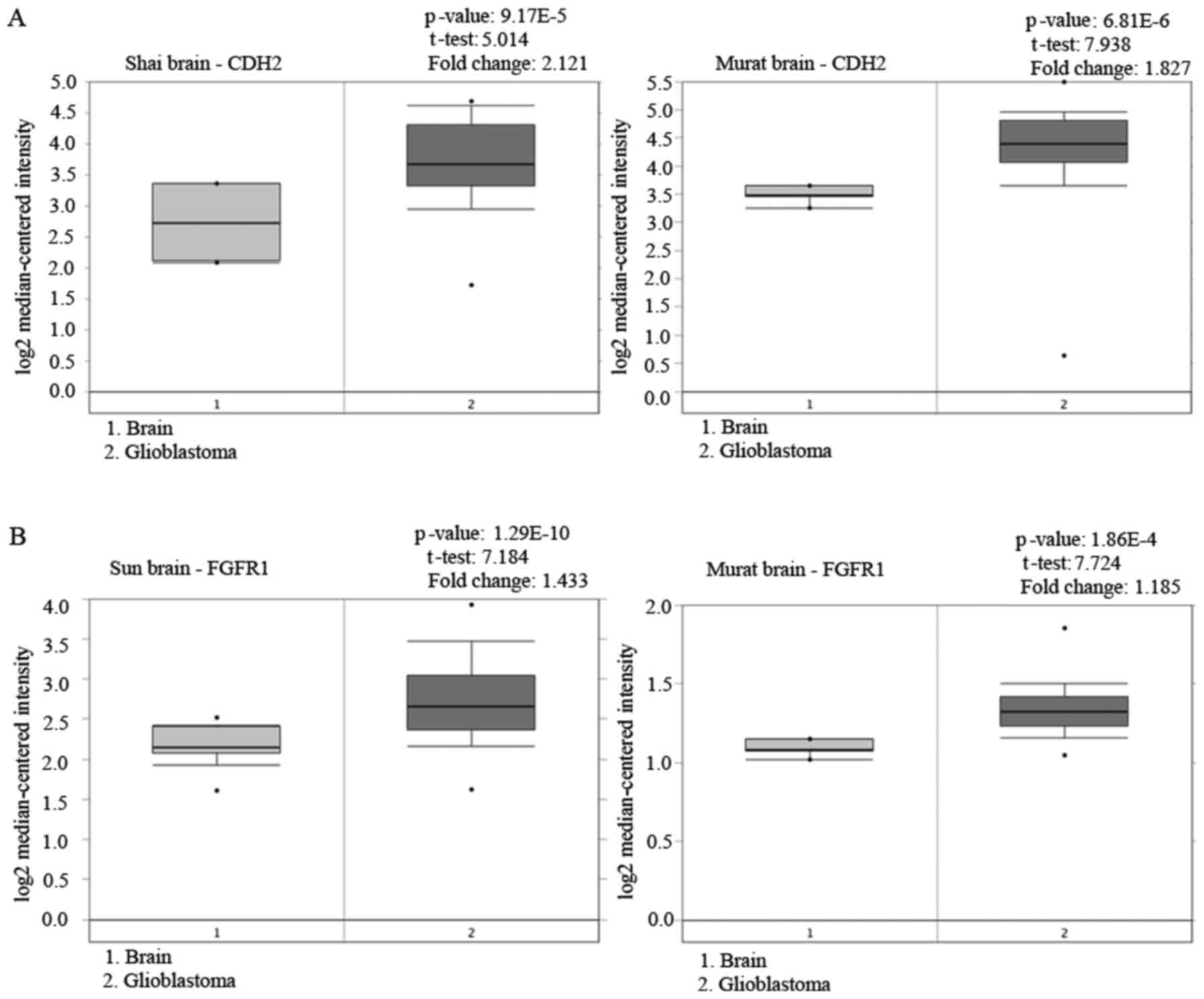
In the present study, we presented stronger evidence
to support our recent study on the proN-cadherin expression
(2) with data from the Oncomine
database (http://www.oncomine.org) acknowledging
the expression of N-cadherin in glioblastoma tissue samples
(Fig. 9A). Concurrently, we
observed that GDNF could promote the expression of proN-cadherin on
the cell membrane as well as glioma cell viability. Accumulating
evidence indicated that GDNF could directly mediate signal
transduction via membrane receptors to regulate gliomas cell
viability. However, the present study focused on the interaction
among proN-cadherin and other receptors mediated by GDNF, which
would enhance cell viability indirectly. We observed the changes in
cell viability on the basis of knockdown and overexpression of
proN-cadherin complemented with the exogenous GDNF. Furthermore,
the relationship between proN-cadherin and FGFR1 receptor on the
membrane was demonstrated, where proN-cadherin was more likely to
bind to phosphorylated FGFR1. Based on the analysis of the results,
we concluded that both overexpression of proN-cadherin and
exogenous GDNF promoted U251MG cell viability and if these two
parameters were achieved combined, cell viability would be more
obvious.
Fibroblast growth factors (FGF) are a family of
ligands that bind to four different types of cell surface receptors
(FGFR1, FGFR2, FGFR3 and FGFR4) (24). FGF ligand binding to the FGFR caused
receptor dimerization, transphosphorylation and activation of an
intracellular tyrosine kinase domain (25). FGFR1 binds to the ligand FGF to
activate the PI3K-AKT, IP3-PLC/DAG, JAK-STAT (26) and other signaling pathways which
regulate cell self-renewal, metabolism, proliferation, EMT and
angiogenesis (27,28). Recent studies revealed that the FGFR
family, especially FGFR1, was abnormally highly expressed in a
variety of tumor tissues like prostate, pancreatic and cervical
cancer, as well as gliomas (29)
(Fig. 9B). Xian et al
(30) reported that abnormal
expression of FGFR1 activated the downstream ERK pathway and
significantly promoted the proliferation of epithelial cells of
breast cancer (30). Furthermore,
FGFR1-mediated signaling pathways are known to modulate key
cellular activities like proliferation, differentiation and
survival (25,31). The phosphorylation of tyrosine 653
and tyrosine 654 in the FGFR1 leads to a large conformational
change in the activated portion of the FGFR1. In addition, pY653
and pY654 interacted with surrounding residues favorably. Further
studies revealed that the phosphorylation of Y653 and Y654 in FGFR1
would facilitate the binding of the receptor to the phospholipase
Cγ through the SH2 domain, which is more favorable for downstream
signaling activation (32). In this
study, GDNF promoted the expression of proN-cadherin on the glioma
cell membrane. GDNF was linked to proN-cadherin, which enhanced
cell to cell interaction, however, overexpression of proN-cadherin
promoted the phosphorylation of FGFR1 and the interaction between
these two proteins was enhanced under the influence of exogenous
GDNF. Therefore, we proposed that GDNF indirectly activated the
FGFR1 receptor and modulated the relationship between proN-cadherin
and FGFR1 synergistically to stimulate the signal transduction
pathway involved in glioma cell viability.
In conclusion, we elucidated a potential GDNF
mechanism of action in promoting glioma cell viability. The
development of gliomas may be through the cross-linking effect of
membrane adhesion molecules and growth factor receptor family
prompted by GDNF, thereby increasing the degree of activation of
the growth factor receptors, which helped signal transduction and
prolonged the response time of FGF-FGFR. The interaction of FGFR1
and proN-cadherin was enhanced by GDNF stimulation and
phosphorylation of FGFR1 was increased. Subsequently, sustained
activation of FGFR1 would undoubtedly activate the downstream
signal pathway, which would explain the cell viability. This
mechanism may offer a new perspective. Concurrently, we proposed
that this new viewpoint concerning the correlation of adhesion
molecules and membrane signaling receptors, would provide new
insights in the field of signal transduction research. In addition,
whether proN-cadherin of adjacent cells activated FGFR1 in other
cells remained undetermined. The present study set a new precedence
for studies on cell-cell communication since it revealed that
resistance to proN-cadherin cross-linking with FGFR1 may provide a
new perspective for cancer therapeutic treatment.
Acknowledgements
We wish to thank the Neurobiology Research Center of
Xuzhou Medical University for providing the research platform. We
would like to express our deep gratitude to the research team. CXT
also expresses his thanks to Miss Chun Yan Mu for her encouragement
and spiritual support.
Funding
The present study was funded by the National Natural
Science Research Fundation of China (grant nos. 81372698 and
81402918) and the Natural Science Foundation of Jiangsu Province,
China (BK20140228).
Availability of data and material
All data generated or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
CXT conceived and designed the study, conducted the
project administration, the drafting and submission of the
manuscript. YXG performed the majority of the experiments and data
collection. XFL performed most experiments, data analysis and
literature search. SYT assisted in the experiments. AAA assisted in
the experiments and offered critical review of the manuscript. YG
performed the statistical analysis and data processing. GQJ
organized the images and tables. YX conducted the experiment
guidance. LYH received funding and conducted experiment guidance.
DSG received funding and contributed in the project supervision,
study conception and design. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dolecek TA, Propp JM, Stroup NE and
Kruchko C: CBTRUS statistical report: Primary brain and central
nervous system tumors diagnosed in the United States in 2005–2009.
Neuro Oncol. 14 Suppl 5:v1–v49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xiong Y, Liu L, Zhu S, Zhang B, Qin Y, Yao
R, Zhou H and Gao DS: Precursor N-cadherin mediates glial cell
line-derived neurotrophic factor-promoted human malignant glioma.
Oncotarget. 8:24902–24914. 2017.PubMed/NCBI
|
|
3
|
Kourtidis A, Lu R, Pence LJ and
Anastasiadis PZ: A central role for cadherin signaling in cancer.
Exp Cell Res. 358:78–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Halbleib JM and Nelson WJ: Cadherins in
development: Cell adhesion, sorting, and tissue morphogenesis.
Genes Dev. 20:3199–3214. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goodwin M and Yap AS: Classical cadherin
adhesion molecules: Coordinating cell adhesion, signaling and the
cytoskeleton. J Mol Histol. 35:839–844. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosivatz E, Becker I, Bamba M, Schott C,
Diebold J, Mayr D, Höfler H and Becker KF: Neoexpression of
N-cadherin in E-cadherin positive colon cancers. Int J Cancer.
111:711–719. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han AC, Soler AP, Knudsen KA and Salazar
H: Distinct cadherin profiles in special variant carcinomas and
other tumors of the breast. Hum Pathol. 30:1035–1039. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomita K, van Bokhoven A, van Leenders GJ,
Ruijter ET, Jansen CF, Bussemakers MJ and Schalken JA: Cadherin
switching in human prostate cancer progression. Cancer Res.
60:3650–3654. 2000.PubMed/NCBI
|
|
9
|
Giroldi LA, Bringuier PP, Shimazui T,
Jansen K and Schalken JA: Changes in cadherin-catenin complexes in
the progression of human bladder carcinoma. Int J Cancer. 82:70–76.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Asano K, Duntsch CD, Zhou Q, Weimar JD,
Bordelon D, Robertson JH and Pourmotabbed T: Correlation of
N-cadherin expression in high grade gliomas with tissue invasion. J
Neurooncol. 70:3–15. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maret D, Gruzglin E, Sadr MS, Siu V, Shan
W, Koch AW, Seidah NG, Del Maestro RF and Colman DR: Surface
expression of precursor N-cadherin promotes tumor cell invasion.
Neoplasia. 12:1066–1080. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin LF, Doherty DH, Lile JD, Bektesh S and
Collins F: GDNF: A glial cell line-derived neurotrophic factor for
midbrain dopaminergic neurons. Science. 260:1130–1132. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sariola H and Saarma M: Novel functions
and signalling pathways for GDNF. J Cell Sci. 116:3855–3862. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Airaksinen MS and Saarma M: The GDNF
family: Signalling, biological functions and therapeutic value. Nat
Rev Neurosci. 3:383–394. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wiesenhofer B, Stockhammer G, Kostron H,
Maier H, Hinterhuber H and Humpel C: Glial cell line-derived
neurotrophic factor (GDNF) and its receptor (GFR-alpha 1) are
strongly expressed in human gliomas. Acta Neuropathol. 99:131–137.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sainio K, Suvanto P, Davies J, Wartiovaara
J, Wartiovaara K, Saarma M, Arumäe U, Meng X, Lindahl M, Pachnis V,
et al: Glial-cell-line-derived neurotrophic factor is required for
bud initiation from ureteric epithelium. Development.
124:4077–4087. 1997.PubMed/NCBI
|
|
17
|
Meng X, Lindahl M, Hyvönen ME, Parvinen M,
de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H,
Lakso M, et al: Regulation of cell fate decision of
undifferentiated spermatogonia by GDNF. Science. 287:1489–1493.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang BL, Guo TW, Gao LL, Ji GQ, Gu XH,
Shao YQ, Yao RQ and Gao DS: Egr-1 and RNA POL II facilitate glioma
cell GDNF transcription induced by histone hyperacetylation in
promoter II. Oncotarget. 8:45105–45116. 2017.PubMed/NCBI
|
|
19
|
Sun S, Lei Y, Li Q, Wu Y, Zhang L, Mu PP,
Ji GQ, Tang CX, Wang YQ, Gao J, et al: Neuropilin-1 is a glial cell
line-derived neurotrophic factor receptor in glioblastoma.
Oncotarget. 8:74019–74035. 2017.PubMed/NCBI
|
|
20
|
Paratcha G, Ledda F and Ibáñez CF: The
neural cell adhesion molecule NCAM is an alternative signaling
receptor for GDNF family ligands. Cell. 113:867–879. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao JP, Wang HJ, Yu JK, Yang H, Xiao CH
and Gao DS: Involvement of NCAM in the effects of GDNF on the
neurite outgrowth in the dopamine neurons. Neurosci Res.
61:390–397. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao JP, Yu JK, Li C, Sun Y, Yuan HH, Wang
HJ and Gao DS: Integrin beta1 is involved in the signaling of glial
cell line-derived neurotrophic factor. J Comp Neurol. 509:203–210.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zuo T, Qin JY, Chen J, Shi Z, Liu M, Gao X
and Gao D: Involvement of N-cadherin in the protective effect of
glial cell line-derived neurotrophic factor on dopaminergic neuron
damage. Int J Mol Med. 31:561–568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jaye M, Schlessinger J and Dionne CA:
Fibroblast growth factor receptor tyrosine kinases: Molecular
analysis and signal transduction. Biochim Biophys Acta.
1135:185–199. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Turner N and Grose R: Fibroblast growth
factor signalling: From development to cancer. Nat Rev Cancer.
10:116–129. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang S and Ding Z: Fibroblast growth
factor receptors in breast cancer. Tumour Biol.
39:10104283176983702017.PubMed/NCBI
|
|
27
|
Carter EP, Fearon AE and Grose RP:
Careless talk costs lives: Fibroblast growth factor receptor
signalling and the consequences of pathway malfunction. Trends Cell
Biol. 25:221–233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ornitz DM and Itoh N: The fibroblast
growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol.
4:215–266. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kelleher FC, O'Sullivan H, Smyth E,
McDermott R and Viterbo A: Fibroblast growth factor receptors,
developmental corruption and malignant disease. Carcinogenesis.
34:2198–2205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xian W, Schwertfeger KL, Vargo-Gogola T
and Rosen JM: Pleiotropic effects of FGFR1 on cell proliferation,
survival, and migration in a 3D mammary epithelial cell model. J
Cell Biol. 171:663–673. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dienstmann R, Rodon J, Prat A,
Perez-Garcia J, Adamo B, Felip E, Cortes J, Iafrate AJ, Nuciforo P
and Tabernero J: Genomic aberrations in the FGFR pathway:
Opportunities for targeted therapies in solid tumors. Ann Oncol.
25:552–563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bae JH, Lew ED, Yuzawa S, Tomé F, Lax I
and Schlessinger J: The selectivity of receptor tyrosine kinase
signaling is controlled by a secondary SH2 domain binding site.
Cell. 138:514–524. 2009. View Article : Google Scholar : PubMed/NCBI
|