Introduction
The Wnt signaling pathway controls various cellular
events such as migration, proliferation and differentiation
throughout development (1).
Deregulation of Wnt signaling has been found in colon cancer where,
most frequently, mutations in APC lead to cytoplasmic accumulation
of β-catenin and its translocation to the nucleus (2,3).
Following translocation to the nucleus, β-catenin binds to T-cell
factor/lymphoid enhancer factor (TCF/LEF), which activates
transcription of target genes and ultimately results in the
uncontrolled cell proliferation of tumor cells (4). Although nuclear β-catenin expression
level is a biomarker associated with invasion, metastasis and poor
prognosis of colon cancer, it cannot explain the phenotypic
heterogeneity of colon cancer.
Na+/H+ exchanger regulatory
factor 1 (NHERF1/EBP50), an adaptor molecule that interacts with
β-catenin, has been recently implicated in the progression of colon
cancer (5,6). This complex regulates Wnt signaling
and may cooperate in the development of colon cancer. Other
influences that enhance the activation of the Wnt/β-catenin pathway
have been documented, either from components of the Wnt pathway
itself or from parallel pathways that connect to the Wnt pathway,
such as the PI3K/PTEN/Akt pathway (7) or the Hippo-YAP/TAZ pathway (8). Notably, NHERF1 also interacts with
Frizzled, PTEN and YAP (6) and
suppresses the Wnt signaling pathway, the PI3K/PTEN/Akt pathway and
the Hippo-YAP/TAZ pathway, respectively. Moreover, there is some
evidence for a dual role of NHERF1 in cancer, depending on its
intracellular localization. However, NHERF1−/−
immortalized mouse embryonic fibroblasts (MEFs), in contrast to
their wild-type counterparts, present anchorage-independent growth
and increased migration (5).
Decreased survival and increased intestinal tumor burden were
observed in ApcMin/+NHERF1−/− double-mutant
mice (6). In addition, during the
steps involved in early colon carcinogenesis, NHERF1 is gradually
lost at the expense of nuclear and cytoplasmic expression in
adenoma and is a new marker of colorectal cancer (9). As a whole, NHERF1 appears to behave as
a tumor suppressor in colon cancer. However, the molecular
mechanism of NHERF1 downregulation in colon cancer is not well
understood.
In the present study, we revealed that NHERF1 was
significantly downregulated in colon adenocarcinoma (CA) tissues
and was correlated with patient outcomes. Meanwhile, NHERF1
expression was associated with epithelial-to-mesenchymal transition
(EMT) phenotype in CA cells. A predicted CpG island was located
within the NHERF1 promoter and NHERF1 was found to be silenced by
promoter methylation in CA. DNA methyltransferase 1 (DNMT1) is one
of the key transmethylases leading to the hypermethylation status
of the NHERF1 promoter and silencing of NHERF1 in CA. Inhibition of
the Wnt signaling pathway upregulated NHERF1 and significantly
downregulated DNMT1 expression. In addition, regulation of NHERF1
expression was found to be dependent on DNMT1 expression in CA.
These results have elucidated the molecular mechanisms underlying
the downregulation of NHERF1 and occurrence/progression of CA.
Materials and methods
The Cancer Genome Atlas (TCGA)
data
The mRNA data (RNA Seq v2) and clinical information
for patients in the TCGA_COAD dataset were downloaded from
https://www.synapse.org and cBioPortal database
(www.cbioportal.org), respectively and
used for differential mRNA expression, correlation, prognosis and
gene set enrichment analysis.
GEO dataset collection and
differential expression analysis
Microarray data were obtained from 4 datasets. The 4
series were accessed at the National Centers for Biotechnology
Information (NCBI) Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/) which
served as a public repository for gene expression datasets, and the
accession numbers were GSE21510, GSE32323, GSE91370, GSE79740,
GSE77955, GSE60620, GSE18560 and GSE97023. Differentially expressed
genes were obtained using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/). GEO2R is an
interactive web tool that compares two groups of samples under the
same experimental conditions and can analyze almost any GEO series
(10).
Gene set enrichment analysis
The association between phenotypes, pathways and
NHERF1 expression was analyzed using Gene Set Enrichment Analysis
(GSEA v2.2, http://www.broad.mit.edu/gsea/). GSEA calculates a
gene set Enrichment Score (ES) that estimates whether genes from
pre-defined gene set [obtained from the Molecular Signatures
Database, MSigDB, http://software.broadinstitute.org,
POSITIVE_REGULATION_ OF _EPITHELIAL_TO_MESENCHYMAL_TRANSITION, H
ALLMARK_WNT_BETA_CATENIN_SIGNALING] are enriched among the highest-
(or lowest-) ranked genes or distributed randomly. Default settings
were used. Thresholds for significance were determined by
permutation analysis (1,000 permutations). False discovery rate
(FDR) was calculated. A gene set was considered significantly
enriched when the FDR score was <0.05.
The IHC data in colon cancer
The IHC-based NHERF1 protein expression data
including high-resolution images were viewed and downloaded from
The Human Protein Atlas web portal (www.proteinatlas.org). ImageJ (National Institutes of
Health, Bethesda, MD, USA) was used to evaluate the NHERF1
expression and localization.
Cell culture
Human colon cancer cell lines were purchased from
the Cell Resource Center of Beijing Xiehe (Beijing, China) and
cultivated in an incubator at 37°C with 5% CO2. HCT116,
SW480, LoVo and HT29 cells were maintained in high-glucose
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with HyClone™ 10%
fetal bovine serum (FBS; GE Healthcare Life Sciences, Logan, UT,
USA) as well as penicillin (100 U/ml; Thermo Fisher
Scientific).
Methylation-specific PCR
The CpG island of the NHERF1 gene was predicted by
Database of CpG Islands (http://dbcat.cgm.ntu.edu.tw/). Genomic DNA was
extracted for methylation analysis from cells in culture by using
the Genomic DNA Miniprep Kit (Sigma-Aldrich; Merck KGaA) One
microgram of genomic DNA was modified with sodium bisulfite using
the DNA Bisulfite Conversion kit (Tiangen Biotech, Co., Ltd.,
Beijing, China) according to the specifications of the
manufacturer. Methylation-specific PCR (MSP) was run in a total
volume of 20 µl. MSP reactions were subjected to initial incubation
at 95°C for 5 min, followed by 35 cycles of 94°C for 20 sec, and
annealing at 60°C for 30 sec and 72°C for 20 sec. Final extension
was conducted by incubation at 72°C for 5 min. MSP products were
separated on 2% agarose gels and visualized after ethidium bromide
staining. The following primers were used: Unmethylated forward,
5′-TAGTTTAGTTTGGGTGATAGAGTGA-3′ and unmethylated reverse,
5′-CAACCTAATTTCCCCTACCTATAACA-3′; methylated forward,
5′-GTAGTTTAGTTTGGGCGATAGAGC-3′ and methylated reverse,
5′-GACCTAATTTCCCCTACCTATAACG-3′.
RNA isolation and real-time
quantitative PCR analyses
Total RNA was isolated using TRIzol (Sigma-Aldrich;
Merck KGaA) and cDNAs were synthesized using a Reverse
Transcription kit (cat. no. 1708842; Bio-Rad Laboratories,
Hercules, CA, USA). Real-time quantitative RT-PCR was performed
using Applied Biosystems® SYBR-Green Master Mix on the
7900HT Fast Real-Time PCR System (Thermo Fisher Scientific).
Demethylation treatment
Cells were treated with 2 µM 5-aza-2′-deoxycytidine
or 3 µM 6-thioguanine (Sigma-Aldrich; Merck KGaA) for 3–4 days,
with medium and 5-aza-2′-deoxycytidine replenished every day. After
treatment, the cells were collected for RNA isolation.
Western blotting
Samples were run on 10% sodium dodecyl sulfate
(SDS)-polyacrylamide gels (PAGE) and transferred to NC membranes.
The blots were blocked in blocking buffer (5% non-fat dry milk in
TBST buffer) for 1 h at room temperature and then incubated with
primary antibodies (1:1,000) in blocking buffer overnight at 4°C.
The blots were washed 3 times with TBST buffer and incubated for 1
h at room temperature with a horseradish peroxidase
(HRP)-conjugated anti-mouse IgG (cat. no. A4416) or anti-rabbit IgG
(cat. no. A4914) secondary antibody (1:3,000; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) in blocking buffer. Finally, the blots
were washed 3 times with TBST buffer and visualized via
enzyme-linked chemiluminescence using the electrochemiluminescence
(ECL) kit (Applygen Technologies Inc., Beijing, China).
The primary antibody specific for NHERF1 (cat. no.
611160) was purchased from BD Biosciences (San Jose, CA, USA), the
primary antibodies specific for GAPDH (cat. no. 5174) was purchased
from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Cancer cell line Encyclopedia
database
The data concerning APC and β-catenin mutation in
colorectal cancer cell lines were obtained from the Cancer Cell
Line Encyclopedia (https://portals.broadinstitute.org/ccle).
Statistical analysis
Statistical analyses were performed using SPSS 19.0
(IBM Corp., Armonk, NY, USA) and GraphPad Prism 5 (GraphPad
Software, Inc., San Diego, CA, USA). Group distributions were
compared using the Student's t-test. A value of P<0.05 was
considered to indicate a statistically significant result. The
correlation of NHERF1 and EMT marker expression levels were
analyzed by Pearson's correlation analysis.
Results
NHERF1 mRNA expression is
downregulated in CA tissues and is correlated with patient
outcomes
To investigate the role of NHERF1 in CA patients, we
compared the NHERF1 expression level in adjacent normal tissues vs.
tumour tissues using the colon adenocarcinoma (COAD) mRNASeq v2
data set in The Cancer Genome Atlas (TCGA) and revealed that NHERF1
was significantly downregulated in CA tissues (Fig. 1A). Consistent with the above
results, downregulation of NHERF1 in CA was validated from 2
independent GEO datasets (Fig. 1B;
GSE21510 and GSE32323). In addition, NHERF1 expression was also
able to significantly distinguish CA tissues and adjacent normal
tissues, revealing that NHERF1 has the advantage for acting as a
prognostic marker in CA (Fig. 1C).
To further analyze the prognostic value of NHERF1 in CA tissues,
the Kaplan-Meier analyses was conducted on the TCGA_COAD dataset.
As shown in Fig. 1D, low NHERF1
expression was found to be significantly associated with worse
disease-free survival (DFS) (log-rank P=0.0284) of CA patients.
These results demonstrated that CA tissues had low NHERF1
expression and that patients with low NHERF1 expression had poor
prognoses. Our findings suggest that NHERF1 may play an important
role in CA occurrence and progression in patients in the clinical
setting.
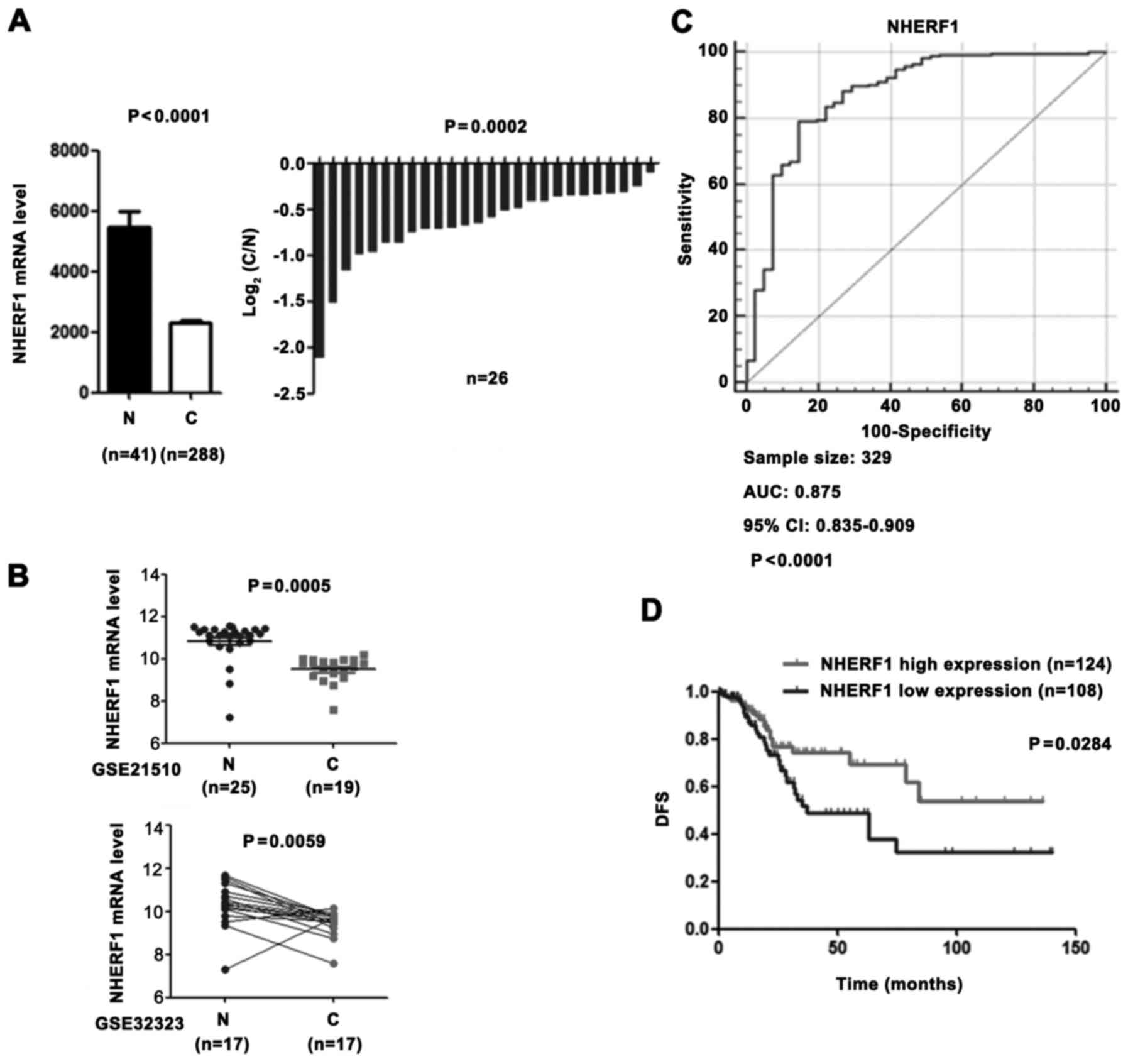 | Figure 1.The low expression level of NHERF1 in
CA is associated with a poor prognosis. (A) NHERF1 mRNA level in CA
(C) and adjacent normal tissues (N) from the TCGA COAD dataset.
Left panel, bar graph shows NHERF1 expression in CA (n=288) and
adjacent normal tissues (n=41, independent sample t-test,
P<0.0001). Error bar represent standard error of the mean (SEM).
Right panel, relative NHERF1 expression in CA patients (n=26) and
their matched normal tissues (n=26, paired sample t-test,
P=0.0002). (B) NHERF1 expression in CA tissues from independent
validation sets. GSE21510: 19 CA tissues and 25 normal controls
(independent sample t-test, P=0.0005). GSE32323: 17 CA tissues and
17 paired adjacent normal tissues (paired sample t-test, P=0.0059).
(C) NHERF1 showed ROC curves and an AUC with diagnostic power to
distinguish CA patients from healthy controls. The area under the
ROC curve was 0.875 (95% CI, 0.835–0.909). (D) KM curve for DFS of
patients with relative high (n=124) and low (n=108) NHERF1 mRNA
level. The curve indicated a shorter DFS time (P<0.05) with low
NHERF1 mRNA level. CA, colon adenocarcinoma; ROC, receiver
operating characteristic curve; AUC, area under the ROC curve; KM,
Kaplan-Meier; DFS, disease-free survival; CI, confidence
interval. |
Low NHERF1 expression is associated
with EMT phenotype in CA
In a previous study, NHERF1 depletion induced
EMT-like changes in a colorectal cancer cell line (9). In order to further determine the role
of NHERF1 in clinical patients, we performed additional analyses on
TCGA_COAD dataset. GSEA result revealed that the NHERF1 expression
level was negatively correlated with the EMT phenotype (Fig. 2A) and levels of mesenchymal markers
(N-cadherin, Snail and fibronectin), but was positively correlated
with the level of epithelial marker E-cadherin in CA tissues
(Fig. 2B-E), suggesting a close
correlation between the NHERF1 level and EMT phenotype. These
results suggest that NHERF1 possibly acts as a potential tumor
suppressor in CA by inhibiting the EMT process. In addition, we
also found that the apical membrane NHERF1 was significantly
reduced in tumor tissues, implying that apical membrane NHERF1
expression was negatively correlated with EMT and the malignant
phenotype in colon cancer patients (Fig. 2F). These findings revealed the
clinical role of NHERF1 in colon cancer.
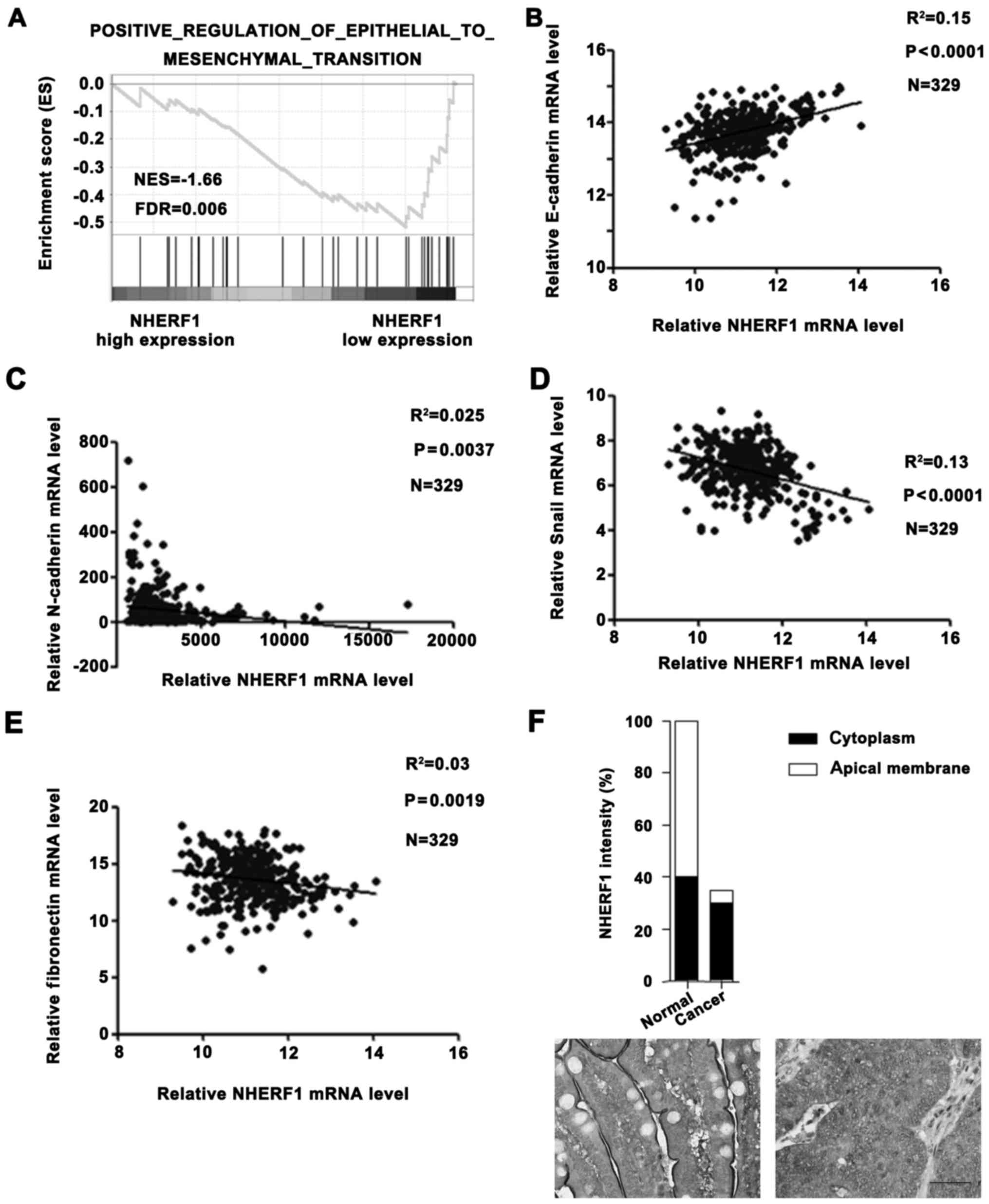 | Figure 2.NHERF1 expression is correlated with
EMT status in CA tissues. (A) Enrichment plot of gene expression
signatures for EMT according to NHERF1 mRNA levels by gene set
enrichment analysis (GSEA) of TCGA dataset. Samples were divided
into high and low NHERF1 expression groups. False discovery rate
(FDR) provides the estimated probability that a gene set with a
given normalized ES (NES) represents a false-positive finding; FDR
<0.05 is a widely accepted cut-off for the identification of
biologically significant gene sets. (B-E) Correlation between
NHERF1 and expression of EMT markers in CA. NHERF1 expression level
was negatively correlated with the expression level of epithelial
marker E-cadherin (B; R2=0.15, P<0.0001) and
positively correlated with expression levels of mesenchymal
markers: N-cadherin (C; R2=0.025, P=0.0037), Snail (D;
R2=0.13, P<0.0001) and fibronectin (E;
R2=0.03, P=0.0019) in CA tissues (n=329). (F) The Human
Protein Atlas database results revealed that the expression level
of NHERF1 was downregulated in colon cancer tissues compared with
normal tissues. In normal tissue, NHERF1 was mainly located in the
apical membrane. In contrast, NHERF1 was mainly located in the
cytoplasm in colon cancer tissue. Scale bar, 50 µm. CA, colon
adenocarcinoma; EMT, epithelial-to-mesenchymal transition. |
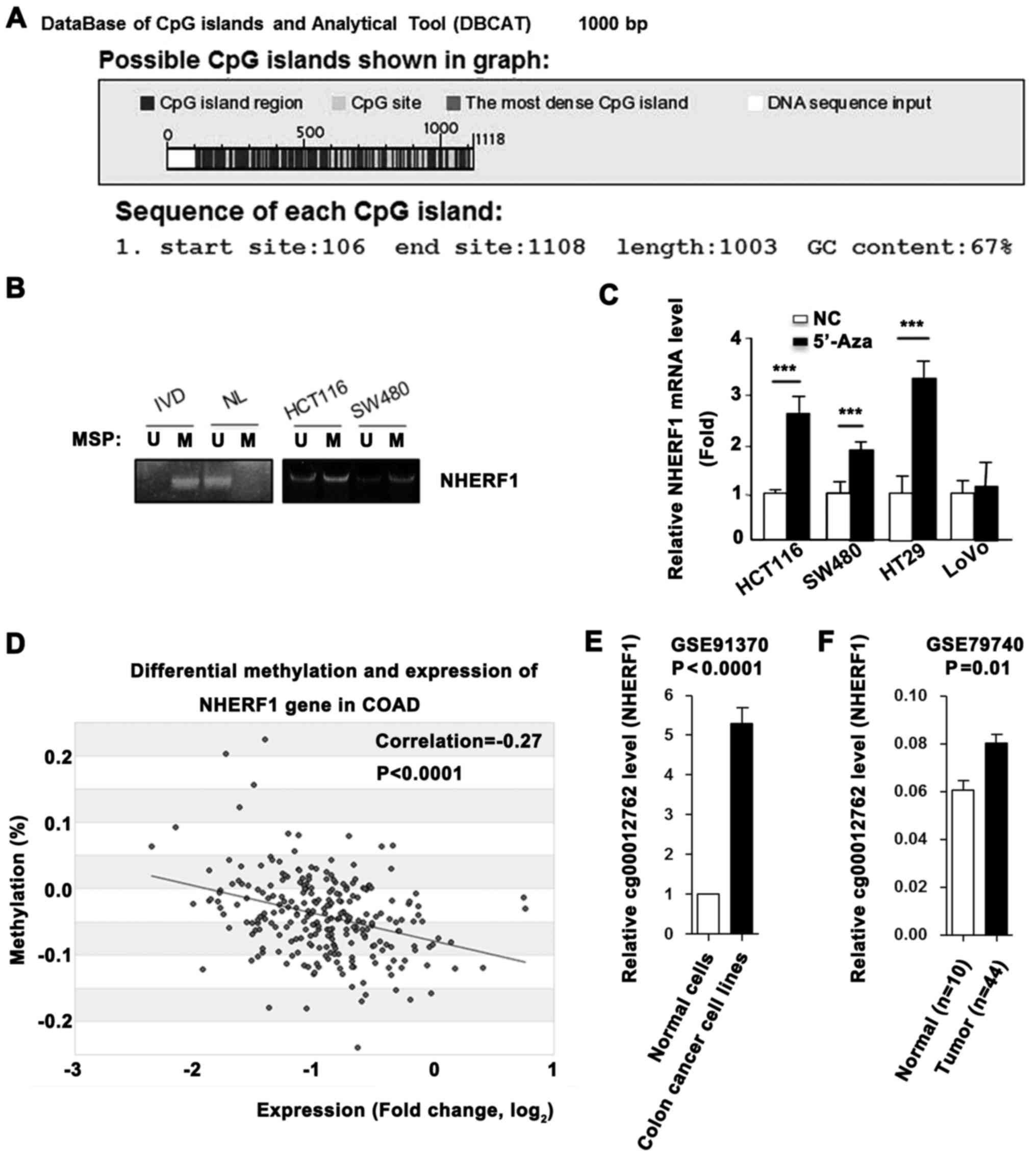
NHERF1 is silenced by promoter
methylation in CA
High promoter methylation level always leads to
transcriptional gene silencing. Sequence analysis using Database of
CpG islands (http://dbcat.cgm.ntu.edu.tw/) revealed that a CpG
island (or CpG-rich sequence) is located within the NHERF1 promoter
(Fig. 3A). Next, we performed MSP
assays to detect the DNA methylation status of the NHERF1 promoter
in CA cell lines. As shown in Fig.
3B, DNA was hypermethylated in the HCT116 and SW480 cells.
Furthermore, demethylation treatment by 5-aza-2-deoxycytidine
(5′-Aza) successfully restored the mRNA level of NHERF1 in the
colon cancer cell lines (Fig. 3C).
To further evaluate the correlation between methylation and
transcription of NHERF1 in CA, we retrieved methylation and mRNA
expression data on NHERF1 from the MethHC database (http://methhc.mbc.nctu.edu.tw/php/index.php). Linear
regression analysis demonstrated a significant negative correlation
between mRNA and promoter methylation levels of NHERF1 (Fig. 3D). In addition, high levels of
promoter methylation of NHERF1 in colon cancer cell lines, and low
methylation in normal epithelial cells were noted (Fig. 3E). The promoter methylation level of
NHERF1 was also higher in colon cancer tissues than the level noted
in the normal tissues (Fig. 3F).
These findings demonstrated that NHERF1 is downregulated by
promoter hypermethylation in CA.
DNMT1 expression correlates with low
NHERF1 mRNA level in CA
With the highest reduction of DNMT1 in colon cancer
cells, extensive loss of DNA methylation occurred in the promoter
and hypermethylated CpG islands, indicating a major role for DNMT1
in maintaining genomic DNA methylation (11). A high DNMT1 expression level in
tumors suggests a causal link to DNA hypermethylation and silenced
NHERF1 expression. To test this hypothesis, we looked at the
association between DNMT1 expression and the NHERF1 mRNA level from
GEO and TCGA databases. As shown in Fig. 4A, the NHERF1 mRNA level was
upregulated in DNMT1-knockout (KO) mice. To further evaluate the
correlation between DNMT1 and NHERF1, TCGA_COAD dataset was used.
We revealed that the expression of DNMT1 was significantly
upregulated and was negatively correlated with the expression of
NHERF1 in CA tissues (Fig. 4B and
C). To further investigate whether NHERF1 expression is reduced
by DNMT1, the DNMT1 inhibitor 6-thioguanine (6-TG), causing
downregulation of DNMT1 through ubiquitin-targeted degradation
(12), was used. As shown in
Fig. 4D, the mRNA levels of NHERF1
were significantly upregulated in HCT116, HT29 and SW480 cells
treated with 6-TG. However, NHERF1 mRNA did not change in the LoVo
cells treated with 6-TG. In addition, the protein levels of NHERF1
were consistent with the mRNA levels in the HCT116 and HT29 cells
(Fig. 4E). It is known that APC and
β-catenin are usually mutated in colon cancer cell lines and are
involved in colon cancer progression. To detect the regulatory
effect of DNMT1 on NHERF1 in different mutated cell lines, GEO
database was used. As shown in Fig.
4F, the expression of NHERF1 and DNMT1 were not significantly
different in the non-mutated group, the APC-mutated group and the
APC/β-catenin-mutated group. In addition, NHERF1 was negatively
correlated with DNMT1 expression in 34 colon cancer cell lines
(Fig. 4G). These results suggest
that DNMT1 may inhibit NHERF1 expression in most colon cancer cell
lines and this effect is not related with APC or β-catenin
mutations.
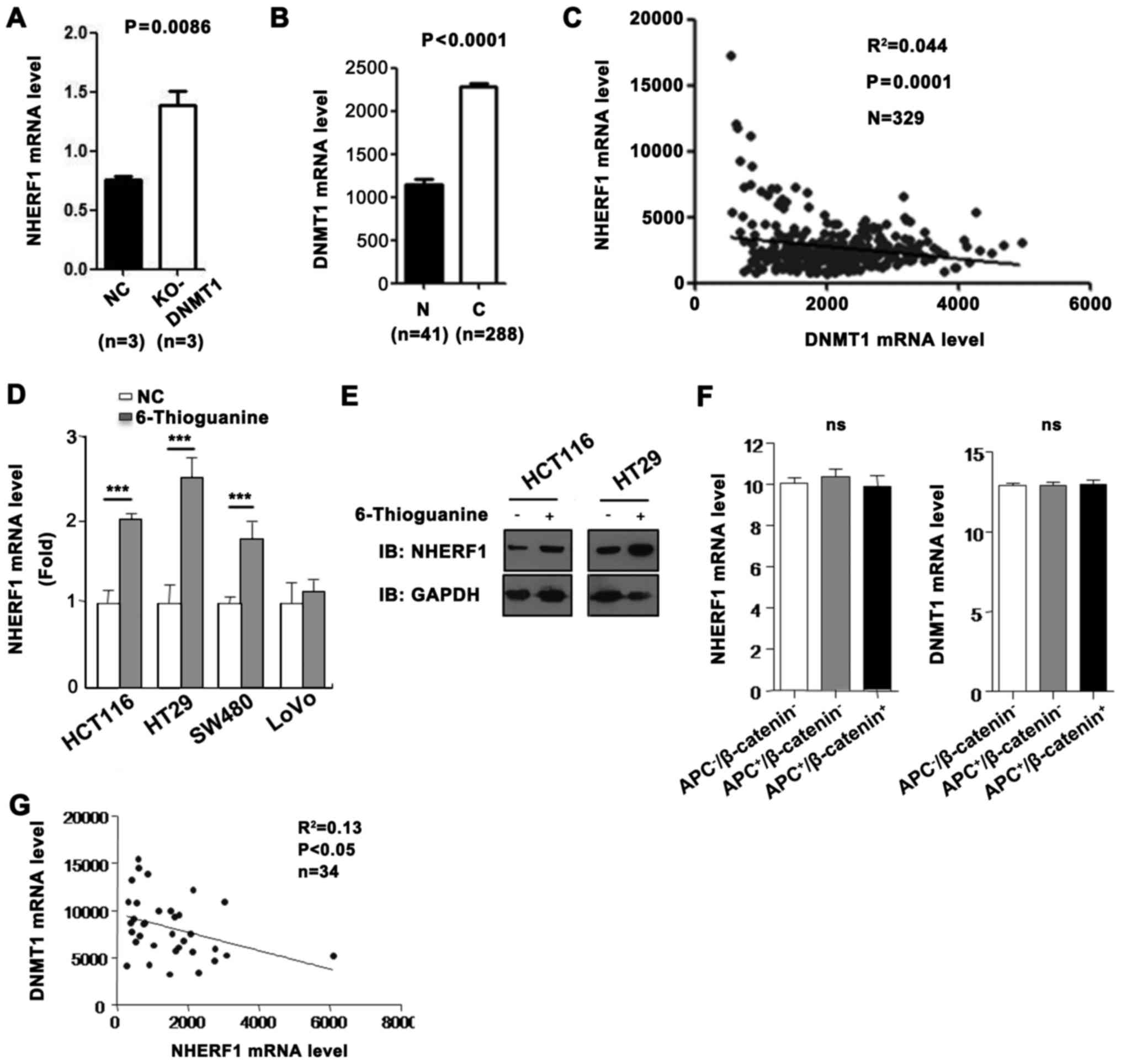 | Figure 4.NHERF1 was reduced by transmethylase
DNMT1 in CA. (A) NHERF1 expression was upregulated in
DNMT1-knockout (KO) mice comparing with negative controls
(GSE60620, n=3, paired sample t-test, P=0.0086). (B) DNMT1 mRNA
level in CA and adjacent normal tissues (N) from the TCGA_COAD
dataset. Bar graph shows DNMT1 expression in CA (C) (n=288) and
adjacent normal tissues (N) (n=41, independent sample t-test,
P<0.0001). (C) Correlation between NHERF1 and DNMT1 expression
in CA (n=329; R2=0.044, P=0.0001) in CA tissues from the TCGA_COAD
dataset. (D) 6-Thioguanine (6-TG) treatment resulted in increased
NHERF1 mRNA level in colon cancer cell lines. The NHERF1 expression
was analyzed by RT-PCR before and after 6-TG treatment
(***P<0.0001). (E) 6-TG treatment resulted in increased NHERF1
protein level in colon cancer cell lines. Cell lysates were
detected by western blotting with anti-NHERF1. GAPDH served as a
loading control. (F) NHERF1 (left) and DNMT1 (right) expression in
the non-mutated group (APC−/β-catenin−;
n=20), APC-mutated group (APC+/β-catenin−;
n=10) and the APC/β-catenin-mutated group
(APC+/β-catenin+; n=3). ns, not significant.
(G) Correlation between NHERF1 and DNMT1 expression in colon cancer
cell lines (n=34; R2=0.13, P<0.05). CA, colon
adenocarcinoma. |
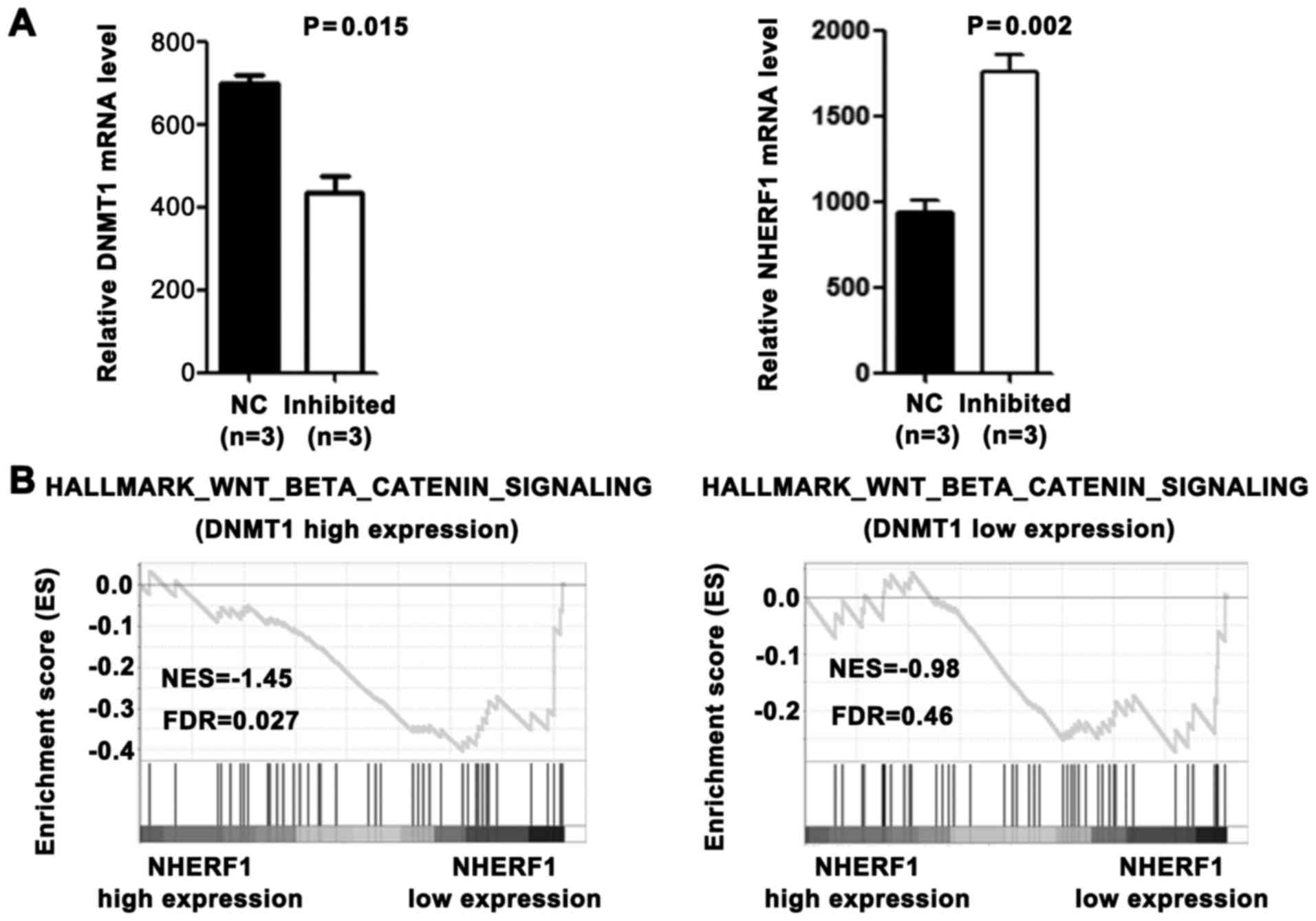
Wnt signaling pathway promotes DNMT1
expression and inhibits NHERF1 expression in CA
Colorectal cancer arises from activating mutations
in the Wnt signaling pathway that converge with additional
molecular changes to shape tumor development and patient prognosis
(9). We demonstrated that DNMT1 was
upregulated in CA tissues and caused the downregulation of NHERF1.
To investigate whether it is associated with overactivated Wnt
signaling pathway in CA, the GEO database was used. As shown in
Fig. 5A, the DNMT1 mRNA level was
significantly downregulated and the NHERF1 mRNA level was
significantly upregulated in colon cancer cell line Ls174T after
Wnt signaling inhibition (GSE18560). To further evaluate the
relationship between the Wnt signaling pathway and these 2 genes,
we performed GSEA. As shown in Fig.
5B, among DNMT1 high expression CA patients, the Wnt signaling
pathway activation gene set was highly enriched in the NHERF1 low
expression group. On the contrary, among DNMT1 low expression CA
patients, Wnt signaling pathway activation gene set was not
correlated with NHERF1 expression. These results suggest that the
overactivated Wnt signaling pathway promotes DNMT1 expression and
may inhibit NHERF1 expression in a DNMT1-dependent manner in
CA.
Discussion
The scaffold protein NHERF1/EBP50 contains an
ERM-binding region and two PDZ domains. NHERF1 always exerts its
function via association with multiple partners and by regulating
multiple signaling pathways, such as the Wnt signaling pathway
(1,5,13), the
PI3K/PTEN/Akt pathway (7,14,15),
the Hippo-YAP/TAZ pathway (8) and
parathyroid hormone 1 receptor signalling (16). In NHERF1−/− mice,
Ezrin-radixin-moesin (ERM) proteins are significantly reduced in
brush border membranes from kidney and small intestine (17). In addition, NHERF1−/−
females were generally smaller and showed impaired mobility
consistent with muscle weakness that was apparent in several
performance coordination tests. Most of the small
NHERF1−/− female animals died 30–35 days after birth.
Autopsies of dead NHERF1−/− females suggested that some
developed hydrocephaly (18).
Recently, NHERF1 has been implicated in the
progression of various human malignancies. NHERF1 gene mutations
found with low frequency (3%) in breast cancer cell lines and
primary tumors were associated with loss of heterozygosity and
increased aggressiveness (19),
suggesting a tumor-suppressor role for NHERF1. NHERF1−/−
immortalized mouse embryonic fibroblasts (MEFs), in contrast to
their wild-type counterparts, presented anchorage-independent
growth and increased migration (5),
supportive of a tumor-suppressor function for NHERF1. NHERF1 acts
as a tumor suppressor in vivo for intestinal adenoma
development via suppressing the Wnt signalling pathway. Decreased
survival and increased intestinal tumor burden were observed in
ApcMin/+NHERF1−/− double-mutant mice
(6). In contrast, Shibata et
al (20) reported that the
NHERF1/β-catenin complex promotes Wnt signaling and may cooperate
in the development of liver cancer. There has been increasing
evidence for a dual role of NHERF1 in cancer, depending on its
intracellular localization. In a previous study, apical membrane
NHERF1 depletion induced EMT-like changes and cytoplasmic NHERF1
promoted cell proliferation in colon cancer cells (9). In addition, nuclear NHERF1 is involved
during colon cancer carcinogenesis (21–23).
In fact, phosphorylation has been shown to regulate assembly and
disassembly of cellular target complexes, NHERF1 subcellular
localization and oligomerization. In addition, it has been shown
that NHERF1 phosphorylation status depends on the cell cycle and
also plays a role in its progression (24–26).
Although NHERF1 is tightly controlled in cells, NHERF1 appears to
behave as a tumor suppressor in various human malignancies.
It was reported that NHERF1 is significantly
downregulated in multiple cancers, including colorectal cancer
(9), particularly in early
carcinogenesis (21), cervical
(27), pancreatic cancer (28) and high-grade glioblastoma (14). Recently, NHERF1 is regarded as a new
marker in colorectal cancer (9). In
the present study, our results indicated that NHERF1 is
downregulated in colon adenocarcinoma which is consistent with
previous reports. Although the clinical role of NHERF1 in colon
cancer is somewhat clear, the mechanism underlying the
downregulation of NHERF1 is not well understood. In previous
studies, the NHERF1 promoter contains 13 half-estrogen-responsive
elements and NHERF1 regulation by estrogens has been evidenced in
several carcinoma cell types including endometrial, mammary
(29) and biliary epithelial cells
(30). In vitro, positive
regulation of NHERF1 by estrogens was illustrated in ER-positive
breast carcinoma cell lines; whereas no NHERF1 stimulation was
noted in ER-negative ones (31–33).
In addition, there is no direct evidence, but several studies have
suggested that NHERF1 may potentially be subjected to epigenetic
modifications (24).
The present study is the first direct demonstration
that NHERF1 expression is regulated by epigenetic modifications in
cancer. It is known that global DNA hypomethylation is a general
characteristic of cancer (34) and
the progression of DNA hypomethylation may be driven by ‘oncogenes’
(35). However, genome-wide studies
of cancer epigenomes reveal that 1–10% of CpG islands are
aberrantly methylated, which suggests that thousands of gene
promoters may be hypermethylated in the average cancer (36). The aberrant hypermethylation of
genes appears to be a common molecular mechanism for silencing
tumor-suppressor genes and can contribute to cancer formation
through the transcriptional repression of these genes (37,38).
We found that NHERF1 was downregulated by promoter hypermethylation
in colon cancer. Activated Wnt signaling-mediated upregulation of
DNMT1 led to a hypermethylation status of the NHERF1 promoter and
silenced NHERF1 expression in colon adenocarcinoma. In addition,
activated Wnt signaling appears to inhibit NHERF1 expression in a
DNMT1- dependent manner. These results suggest that activated Wnt
signaling by mutations in APC inhibits NHERF1 expression by
promoting DNMT1 expression and loss of NHERF1 overactivated Wnt
signaling pathway in colon adenocarcinoma, identifying a feedback
loop in the Wnt/DNMT1/NHERF1 pathway. In addition, we also observed
that NHERF1 is correlated with the EMT phenotype which is
associated with poor prognosis in colon adenocarcinoma patients.
Collectively, the data reported in this study demonstrated, for the
first time, a novel relationship between the Wnt signaling pathway
and NHERF1. NHERF1 did not only inhibit the Wnt signaling pathway
but was also downregulated by the Wnt signaling pathway in colon
adenocarcinoma.
In summary, our data may provide a new insight in
mechanism of NHERF1 downregualtion in early occurrence steps of
carcinogenesis, suggesting that Wnt/DNMT1/NHERF1 axis acts as a
crucial role in occurrence and progression of colon cancer.
Acknowledgements
The authors thank Dr Z. Peng (State Key Laboratory
of Proteomics, Beijing Proteome Research Center, Beijing Institute
of Radiation Medicine, Collaborative Innovation Center for Cancer
Medicine, Beijing, China) for kindly providing materials and
technical assistance.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LY conceived and designed the experiments; YG
performed the experiments, MW analyzed the data and XJ wrote the
paper; HZ and YZ contributed by procuring the reagents, materials
and analytical tools. In addition, HZ performed TCGA data and IHC
analysis and YZ performed GSEA and methylation data analysis. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NHERF1
|
Na+/H+ exchanger
regulatory factor 1
|
|
CA
|
adenocarcinoma of the colon
|
|
DNMT1
|
DNA methyltransferase 1
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
6-TG
|
6-thioguanine
|
References
|
1
|
Lin YY, Hsu YH, Huang HY, Shann YJ, Huang
CY, Wei SC, Chen CL and Jou TS: Aberrant nuclear localization of
EBP50 promotes colorectal carcinogenesis in xenotransplanted mice
by modulating TCF-1 and β-catenin interactions. J Clin Invest.
122:1881–1894. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giles RH, van Es JH and Clevers H: Caught
up in a wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
3
|
Wangefjord S, Brandstedt J, Lindquist KE,
Nodin B, Jirstrom K and Eberhard J: Associations of beta-catenin
alterations and MSI screening status with expression of key cell
cycle regulating proteins and survival from colorectal cancer.
Diagn Pathol. 8:102013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stanczak A, Stec R, Bodnar L, Olszewski W,
Cichowicz M, Kozlowski W, Szczylik C, Pietrucha T, Wieczorek M and
Lamparska-Przybysz M: Prognostic significance of Wnt-1, β-catenin
and E-cadherin expression in advanced colorectal carcinoma. Pathol
Oncol Res. 17:955–963. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kreimann EL, Morales FC, de Orbeta-Cruz J,
Takahashi Y, Adams H, Liu TJ, McCrea PD and Georgescu MM: Cortical
stabilization of beta-catenin contributes to NHERF1/EBP50 tumor
suppressor function. Oncogene. 26:5290–5299. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Georgescu MM, Gagea M and Cote G:
NHERF1/EBP50 suppresses wnt-β-catenin pathway-driven intestinal
neoplasia. Neoplasia. 18:512–523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parsons DW, Wang TL, Samuels Y, Bardelli
A, Cummins JM, DeLong L, Silliman N, Ptak J, Szabo S, Willson JK,
et al: Colorectal cancer: Mutations in a signalling pathway.
Nature. 436:7922005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Azzolin L, Panciera T, Soligo S, Enzo E,
Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V,
et al: YAP/TAZ incorporation in the β-catenin destruction complex
orchestrates the Wnt response. Cell. 158:157–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hayashi Y, Molina JR, Hamilton SR and
Georgescu MM: NHERF1/EBP50 is a new marker in colorectal cancer.
Neoplasia. 12:1013–1022. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai Y, Tsai HC, Yen RC, Zhang YW, Kong X,
Wang W, Xia L and Baylin SB: Critical threshold levels of DNA
methyltransferase 1 are required to maintain DNA methylation across
the genome in human cancer cells. Genome Res. 27:533–544. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flesner BK, Kumar SR and Bryan JN:
6-Thioguanine and zebularine down-regulate DNMT1 and globally
demethylate canine malignant lymphoid cells. BMC Vet Res.
10:2902014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wheeler DS, Barrick SR, Grubisha MJ,
Brufsky AM, Friedman PA and Romero G: Direct interaction between
NHERF1 and Frizzled regulates β-catenin signaling. Oncogene.
30:32–42. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Molina JR, Agarwal NK, Morales FC, Hayashi
Y, Aldape KD, Cote G and Georgescu MM: PTEN, NHERF1 and PHLPP form
a tumor suppressor network that is disabled in glioblastoma.
Oncogene. 31:1264–1274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Molina JR, Morales FC, Hayashi Y, Aldape
KD and Georgescu MM: Loss of PTEN binding adapter protein NHERF1
from plasma membrane in glioblastoma contributes to PTEN
inactivation. Cancer Res. 70:6697–6703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mahon MJ, Donowitz M, Yun CC and Segre GV:
Na+/H+ exchanger regulatory factor 2 directs
parathyroid hormone 1 receptor signalling. Nature. 417:858–861.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morales FC, Takahashi Y, Kreimann EL and
Georgescu MM: Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50
organizes ERM proteins at the apical membrane of polarized
epithelia. Proc Natl Acad Sci USA. 101:17705–17710. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shenolikar S, Voltz JW, Minkoff CM, Wade
JB and Weinman EJ: Targeted disruption of the mouse NHERF-1 gene
promotes internalization of proximal tubule sodium-phosphate
cotransporter type IIa and renal phosphate wasting. Proc Natl Acad
Sci USA. 99:11470–11475. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dai JL, Wang L, Sahin AA, Broemeling LD,
Schutte M and Pan Y: NHERF (Na+/H+ exchanger
regulatory factor) gene mutations in human breast cancer. Oncogene.
23:8681–8687. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shibata T, Chuma M, Kokubu A, Sakamoto M
and Hirohashi S: EBP50, a beta-catenin-associating protein,
enhances Wnt signaling and is over-expressed in hepatocellular
carcinoma. Hepatology. 38:178–186. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mangia A, Saponaro C, Malfettone A,
Bisceglie D, Bellizzi A, Asselti M, Popescu O, Reshkin SJ, Paradiso
A and Simone G: Involvement of nuclear NHERF1 in colorectal cancer
progression. Oncol Rep. 28:889–894. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Malfettone A, Silvestris N, Paradiso A,
Mattioli E, Simone G and Mangia A: Overexpression of nuclear NHERF1
in advanced colorectal cancer: Association with hypoxic
microenvironment and tumor invasive phenotype. Exp Mol Pathol.
92:296–303. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gu Y, Yu H, Hao C, Martin TA, Hargest R,
He J, Cheng S and Jiang WG: NHERF1 regulates the progression of
colorectal cancer through the interplay with VEGFR2 pathway.
Oncotarget. 8:7753–7765. 2017.PubMed/NCBI
|
|
24
|
Vaquero J, Nguyen Ho-Bouldoires TH,
Claperon A and Fouassier L: Role of the PDZ-scaffold protein
NHERF1/EBP50 in cancer biology: From signaling regulation to
clinical relevance. Oncogene. 36:3067–3079. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song GJ, Leslie KL, Barrick S, Mamonova T,
Fitzpatrick JM, Drombosky KW, Peyser N, Wang B, Pellegrini M, Bauer
PM, et al: Phosphorylation of ezrin-radixin-moesin-binding
phosphoprotein 50 (EBP50) by Akt promotes stability and mitogenic
function of S-phase kinase-associated protein-2 (Skp2). J Biol
Chem. 290:2879–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He J, Lau AG, Yaffe MB and Hall RA:
Phosphorylation and cell cycle-dependent regulation of
Na+/H+ exchanger regulatory factor-1 by Cdc2
kinase. J Biol Chem. 276:41559–41565. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peng Z, Wang Q, Zhang Y, He J and Zheng J:
EBP50 interacts with EGFR and regulates EGFR signaling to affect
the prognosis of cervical cancer patients. Int J Oncol.
49:1737–1745. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ji MY, Fan DK, Lv XG, Peng XL, Lei XF and
Dong WG: The detection of EBP50 expression using quantum dot
immunohistochemistry in pancreatic cancer tissue and down-regulated
EBP50 effect on PC-2 cells. J Mol Histol. 43:517–526. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smith PM, Cowan A, Milgram SL and White
BA: Tissue-specific regulation by estrogen of ezrin and
ezrin/radixin/moesin-binding protein 50. Endocrine. 22:119–126.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fouassier L, Rosenberg P, Mergey M,
Saubaméa B, Clapéron A, Kinnman N, Chignard N, Jacobsson-Ekman G,
Strandvik B, Rey C, et al: Ezrin-radixin-moesin-binding
phosphoprotein (EBP50), an estrogen-inducible scaffold protein,
contributes to biliary epithelial cell proliferation. Am J Pathol.
174:869–880. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ediger TR, Kraus WL, Weinman EJ and
Katzenellenbogen BS: Estrogen receptor regulation of the
Na+/H+ exchange regulatory factor.
Endocrinology. 140:2976–2982. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harrington WR, Sheng S, Barnett DH, Petz
LN, Katzenellenbogen JA and Katzenellenbogen BS: Activities of
estrogen receptor alpha- and beta-selective ligands at diverse
estrogen responsive gene sites mediating transactivation or
transrepression. Mol Cell Endocrinol. 206:13–22. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stemmer-Rachamimov AO, Wiederhold T,
Nielsen GP, James M, Pinney-Michalowski D, Roy JE, Cohen WA, Ramesh
V and Louis DN: NHE-RF, a merlin-interacting protein, is primarily
expressed in luminal epithelia, proliferative endometrium, and
estrogen receptor-positive breast carcinomas. Am J Pathol.
158:57–62. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Diala ES and Hoffman RM: Hypomethylation
of HeLa cell DNA and the absence of 5-methylcytosine in SV40 and
adenovirus (type 2) DNA: Analysis by HPLC. Biochem Biophys Res
Commun. 107:19–26. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mudbhary R, Hoshida Y, Chernyavskaya Y,
Jacob V, Villanueva A, Fiel MI, Chen X, Kojima K, Thung S, Bronson
RT, et al: UHRF1 overexpression drives DNA hypomethylation and
hepatocellular carcinoma. Cancer Cell. 25:196–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Costello JF, Fruhwald MC, Smiraglia DJ,
Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P,
Lang JC, et al: Aberrant CpG-island methylation has non-random and
tumour-type-specific patterns. Nat Genet. 24:132–138. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hoffman RM: Altered methionine metabolism,
DNA methylation and oncogene expression in carcinogenesis. A review
and synthesis. Biochim Biophys Acta. 738:49–87. 1984.
|
|
38
|
Lao VV and Grady WM: Epigenetics and
colorectal cancer. Nat Rev Gastroenterol Hepatol. 8:686–700. 2011.
View Article : Google Scholar : PubMed/NCBI
|