Introduction
Colorectal cancer (CRC) is one of the most common
malignant cancers worldwide. In the United States, it has been
reported to be the third most common type of cancer with
cancer-associated mortality ranking second in men and third in
women (1). Various advances have
been made in the diagnosis and treatment of CRC, however, the
prognosis of patients with CRC remains very poor, with metastasis
as the leading cause of cancer-associated mortality among such
patients (2,3). Unfortunately, no effective therapeutic
strategies for patients with metastasis are currently available,
and the underlying molecular mechanism of CRC-associated metastasis
remains unclear (4). An improved
understanding of the molecular mechanisms that mediate
CRC-associated metastasis may contribute to increasing the
effectiveness of current therapies for the treatment of CRC.
Metastasis is a multiple-step process. The
activation of numerous signaling pathways can lead to metastasis
(5–7). The canonical Wnt/β-catenin signaling
pathway has been demonstrated to play a key role in the promotion
of cancer metastasis (8–10). Binding of the Wnt ligand to its
receptor leads to inhibition of the cytoplasmic degradation complex
and stabilization of β-catenin. β-catenin then accumulates in the
cytoplasm and translocates into the nucleus, resulting in the
induction of downstream Wnt genes, including LCF-1, MMP7 and Slug,
which are factors for cancer metastasis (10–12).
Hairy and enhancer of split family basic
helix-loop-helix transcription factor 6 (HES6; 24 kDa), a member of
the HES family of proteins, is located on chromosome 2q37 and the
amplification of this region has been reported in prostate, breast
and lung cancers (13–15). In addition, the overexpression of
HES6 has been detected in various human cancers, including prostate
and ovarian cancers, hepatic carcinoma and glioma (16–18).
HES6 functions as a basic helix-loop-helix transcription repressor
and has been demonstrated to play a vital role in the progression
of various tumors (19–21). However, the clinical significance
and biological role of HES6 in human CRC, and its association with
the canonical Wnt/β-catenin signaling pathway requires further
investigation.
In the present study, the expression of HES6 was
evaluated within CRC cell lines and tissues. In addition, the
present study investigated the association of the expression of
HES6 with the clinicopathological characteristics and prognosis of
patients with CRC, and the effect of HES6 on the metastasis of CRC,
as well as the underlying molecular mechanism.
Materials and methods
Cell linesand treatment
A total of 9 CRC cell lines (HT-29, COLO 205, LoVo,
SW480, SW620, HCT116, HCT-15, Caco-2 and LS174T) and a normal colon
epithelial cell line (FHC) were obtained from the American Type
Culture Collection (ATCC; Rockville, MD, USA). All CRC cell lines
as well as the FHC cell line were cultured in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; HyClone
Laboratories; GE Healthcare Life Sciences, Logan, UT, USA), 100
µg/ml streptomycin and 100 µg/ml penicillin (Invitrogen; Thermo
Fisher Scientific, Inc.) in a humidified incubator containing 5%
CO2 at 37°C.
Patients and tissue specimens
A total of 213 paraffin-embedded CRC samples were
collected at the Sun Yat-sen University Cancer Center (Guangzhou,
China), from patients with a median age of 56 years (ranging from
20 to 85 years) who were histopathologically and clinically
diagnosed from January 2005 to December 2007. The present study was
approved by the Sun Yat-sen University Cancer Center Institutional
Board. Prior patient consent was also obtained. The
clinicopathological characteristics of the 213 patients are
summarized in Table I. In addition,
15 pairs of primary CRC tissues and the adjacent normal tissues (5
cm away from the cancer lesions) were collected from patients with
CRC who were enrolled between July 2015 and December 2016 and
underwent colorectal resection at the Third Affiliated Hospital of
Guangzhou Medical University. The clinicopathological features of
15 patients are summarized in Table
II. Tissue samples were snap-frozen in liquid nitrogen and then
stored at −80°C until further use. The pathological diagnosis and
confirmation of tissue specimens were performed by at least two
pathologists. The tumor, node and metastasis (TNM) staging system
was used according to the National Comprehensive Cancer Network
(NCCN) guidelines (22). None of
the patients received chemotherapy or radiotherapy prior to sample
collection and patients with other malignancies were excluded. The
research protocols were approved by the Clinical Research Ethics
Committee of the Third Affiliated Hospital of Guangzhou Medical
University (reference no. 2017, no. 117). All patients provided
written informed consent for the analysis of their tissue for
research purposes.
 | Table I.Clinicopathological features and
tumor expression of HES6 in 213 patients with colorectal
cancer. |
Table I.
Clinicopathological features and
tumor expression of HES6 in 213 patients with colorectal
cancer.
| Features | No. of cases
(%) |
|---|
| Total no. of
patients | 213 |
| Age (years) |
|
<56 | 93
(43.7) |
|
≥56 | 120 (56.3) |
| Sex |
|
Male | 123 (57.7) |
|
Female | 90
(42.3) |
|
Differentiation |
| Well
and moderate | 174 (81.7) |
| Poor
and undifferentiated | 39
(18.3) |
| Chemotherapy |
|
Yes | 91
(42.7) |
| No | 122 (57.3) |
| T stage |
| 1 | 7
(3.3) |
| 2 | 15
(7.0) |
| 3 | 158 (74.2) |
| 4 | 33
(15.5) |
| N stage |
| 0 | 122 (57.3) |
| 1 | 66
(31.0) |
| 2 | 25
(11.7) |
| Metastasis |
| M0 | 149 (70.0) |
| M1 | 64
(30.0) |
| Prognosis |
|
Survival | 122 (57.3) |
|
Death | 91
(42.7) |
| Expression of
HES6 |
|
High | 138 (64.8) |
|
Low | 75
(35.2) |
 | Table II.Clinicopathological features of 15
patients with colorectal cancer. |
Table II.
Clinicopathological features of 15
patients with colorectal cancer.
| Features | No. of cases
(%) |
|---|
| Total no. of
patients | 15 |
| Age (years) |
|
<56 | 6
(40.0) |
|
≥56 | 9
(60.0) |
| Sex |
|
Male | 8
(53.3) |
|
Female | 7
(46.7) |
|
Differentiation |
| Well
and moderate | 7
(46.7) |
| Poor
and undifferentiated | 8
(53.3) |
| Chemotherapy |
|
Yes | 5
(33.3) |
| No | 10 (66.7) |
| T Stage |
| 1 | 2
(13.3) |
| 2 | 3
(20.0) |
| 3 | 7
(46.7) |
| 4 | 3
(20.0) |
| N Stage |
| 0 | 5
(33.3) |
| 1 | 7
(46.7) |
| 2 | 3
(20.0) |
| Metastasis |
| M0 | 11 (73.3) |
| M1 | 4
(26.7) |
RNA extraction, reverse transcription
and quantitative real-time PCR (RT-qPCR)
Total RNA from cultured cells and CRC tissue samples
were isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
present study examined the mRNA expression levels of HES6 in CRC
cell lines relative to normal colon epithelial cells, as well as in
each of the primary CRC tissues relative to the adjacent normal
tissues obtained from the same patient, by PCR with published
primers (16). Complementary DNA
(cDNA) was synthesized from 2 µg total RNA using M-MLV Reverse
Transcriptase (Promega Corp., Madison, WI, USA). Quantitation and
amplification were performed using SYBR Green I (Roche Diagnostics
GmbH, Manheim, Germany) and an ABI 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction
conditions were as follows: initial denaturation at 95°C for 5 min,
followed by 40 cycles of denaturation at 95°C for 10 sec, primer
annealing at 60°C for 30 sec and extension at 72°C for 1 min. The
expression levels of HES6 were normalized to GAPDH. The procedures
were performed as previously described and expression levels were
analyzed by the 2−∆∆Cq method used for relative
quantification (23). Each
experiment was performed in triplicate and repeated at least three
times. The primer sequences of HES6 and GAPDH are provided in
Table III.
 | Table III.Primer and shRNA sequences used in
present study. |
Table III.
Primer and shRNA sequences used in
present study.
| Name | Sequence (5′ to
3′) |
|---|
| HES6-F |
TGCCGAGCTCCTGAACCATC |
| HES6-R |
TGGTTCAGGAGCTCGGCAGCGACG |
| GAPDH-F |
CGCTGAGTACGTCGTGGAGTC |
| GAPDH-R |
GCTGATGATCTTGAGGCTGTTGTC |
| β-catenin-F |
CATCCTAGCTCGGGATGTTCAC |
| β-catenin-R |
TCCTTGTCCTGAGCAAGTTCAC |
| HES6-shRNA1-F |
GATCCCCCAGCCTGACCACAGCCCAAATTTCAAGAGAATTTGGGCTGTGGTCAGGCTGTTTTTA |
| HES6-shRNA1-R |
AGCTTAAAAACAGCCTGACCACAGCCCAAATTCTCTTGAAATTTGGGCTGTGGTCAGGCTGGGG |
| HES6-shRNA2-F |
GATCCCCCGAGCTCCTGAACCATCTGCTTTCAAGAGAAGCAGATGGTTCAGGAGCTCGTTTTTA |
| HES6-shRNA2-R |
AGCTTAAAAACGAGCTCCTGAACCATCTGCTTCTCTTGAAAGCAGATGGTTCAGGAGCTCGGGG |
Western blot analysis
The cells were harvested in sampling buffer (62.5
mmol/l Tris-HCl pH 6.8, 10% glycerol, 2% SDS) and then heated at
100°C for 5 min. Protein concentration was determined via a
Bradford assay using Bio-Rad protein assay (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Equal quantities of protein (30 µg) were
electrophoretically separated via 9% SDS-PAGE and transferred to
polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica,
MA, USA). Following blocking for 1 h in Tris-buffered saline
containing 0.1% Tween-20 at room temperature with 5% fat-free milk,
the membranes were incubated with anti-HES6 antibody (dilution
1:3,000; cat. no. ab66461; Abcam, Cambridge, UK) overnight at 4°C,
followed by incubation with horseradish peroxidase-conjugated
anti-rabbit IgG antibody (dilution 1:4,000; cat. no. SC-2004; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA). Finally, the enhanced
chemiluminescence (ECL) Prime Western Blotting Detection reagent
(GE Healthcare, Chicago, IL, USA) was used to detect the expression
of HES6 according to the manufacturer's instructions. An
anti-α-tubulin antibody (dilution 1:4,000; cat. no. T9026;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used as a
loading control.
Immunohistochemistry (IHC)
IHC was used to assess the protein expression
profile of HES6 in 213 formalin-fixed, paraffin-embedded CRC
tissues. The procedures followed standard protocols as previously
described (23). The degree of
immunostaining for each sample was reviewed and scored
independently by two pathologists. The scores were based on both
the proportion of positively-stained tumor cells and the intensity
of staining. The proportions of tumor cells were scored as follows:
0, <5% positive tumor cells; 1, 6–10% positive tumor cells; 2,
11–50% positive tumor cells; 3, 51–75% positive tumor cells and 4,
>75% positive tumor cells. The grade of staining intensity was
as follows: 0, no staining; 1, weak staining (light yellow); 2,
moderate staining (yellow brown) and 3, strong staining (brown).
The staining index (SI) was calculated by multiplying the score of
the staining intensity by the proportion of positive tumor cells,
which ranged from 0–12. Samples with an SI ≥6 were denoted as the
high expression group and those with an SI<6 were classified as
the low expression group.
Gene set enrichment analysis
(GSEA)
In the present study, a CRC cohort was downloaded
from The Cancer Genome Atlas (https://cancergenome.nih.gov/) and GSEA 2.0.9
(http://www.broadinstitute.org/gsea/)
was used. Gene set permutations were performed 1,000 times for each
analysis. The pathways enriched in each phenotype were sorted by
the nominal P-value and enrichment score (ES).
Overexpression and knockdown
experiments
The coding sequence of the human HES6 gene was
amplified by PCR and cloned into the pSin-EF2 lentiviral vector.
Two short hairpin RNA (shRNA) oligonucleotides targeting HES6 were
cloned into the pSuper-retro-puro vector, and the target sequences
were as follows: (NM_018645.4)
5′-CCGGGCTGAACTGAGTCAGGCTCCTCTCGAGAGGAGCCTGACTCAGTTCAGCTTTTT-3′.
Transfection of plasmids or shRNAs was performed using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The procedures
of retroviral production and infection were conducted as described
in a previous study (24). All
stable cell lines expressing HES6 or HES6 shRNA were selected with
0.5 µg/ml puromycin for 48 h or 10 days following infection. The
TCF4-dn (pLX303) plasmid (cat. no. 42592) was purchased from
Addgene (Cambridge, MA, USA). The sequences of the shRNAs are
listed in Table III.
Wound healing assay
Cell migration ability was determined using the
scratch-wound assay. In brief, cells were cultured in 6-well plates
with RPMI-1640 medium and 10% FBS until a monolayer of cells was
formed. Then, a straight linear wound was created in the middle of
the cell monolayer with a sterile pipette tip. Images of the cells
along the wound line were captured at 0 and 24 h following wounding
under an inverted Olympus IX50 microscope (Olympus Corp., Tokyo,
Japan) with a ×10 objective lens.
Transwell matrix invasion assay
Cells (2×105) were seeded into the upper
chamber of polycarbonate Transwell filters coated with Matrigel (BD
Biosciences, San Jose, CA, USA), and 20% FBS was added into the
lower chamber to induce invasion. After incubation at 37°C for 24
h, the cells that had invaded to the bottom surface of the membrane
were fixed in 1% paraformaldehyde, stained with hematoxylin, and
counted in 10 random fields of view/well. All experiments were
performed in triplicate.
Three-dimensional (3D) spheroid
invasion assay
Cells (1×104) were seeded into 24-well
plates coated with 2% Matrigel (BD Biosciences), and the medium was
replaced every other day. Images of the cells were captured at
2-day intervals for 2 weeks under a light microscope with a
magnification of ×200.
Dual-Luciferase assay
Cells (2×104) were seeded in triplicate
in 24-well plates and allowed to settle for 24 h. Subsequently, 150
ng luciferase reporter plasmids or the control luciferase plasmid
plus 5 ng pRL-TK Renilla plasmid (Promega Corp.) were
transfected into CRC cells using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h following
transfection, a Dual-Luciferase reporter assay (Promega Corp.) was
performed according to the manufacturer's instructions. Three
independent experiments were performed and the data are presented
as the mean ± standard deviation.
Immunofluorescence analysis
Cells (2×105) were seeded on coverslips
for 48 h. The cells were incubated with a primary antibody against
β-catenin (dilution 1:1,000; cat. no. 8480; Cell Signaling
Technology, Inc., Danvers, MA, USA), and then incubated with
rhodamine-conjugated or FITC-conjugated goat antibodies against
rabbit IgG (dilution 1:5,00; cat. no. 4412; Cell Signaling
Technology). Coverslips were counter stained with DAPI and
visualized under a confocal laser-scanning microscope (Olympus
FV1000; Olympus Corp.). Data were processed with FV10-ASW 1.7
Viewer. The sequences of β-catenin are listed in Table III.
Statistical analysis
SPSS 19.0 statistical software (IBM Corp., Armonk,
NY, USA) and GraphPad Prism 5.0 software (GraphPad Software, Inc.,
La Jolla, CA, USA) were used for all statistical analyses. The
χ2 test was used for comparisons between groups, while
Cox regression analysis was performed for univariate and
multivariate survival analyses. The Kaplan-Meier method was used to
plot the survival curves followed by a log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
HES6 is upregulated in CRC cell
lines
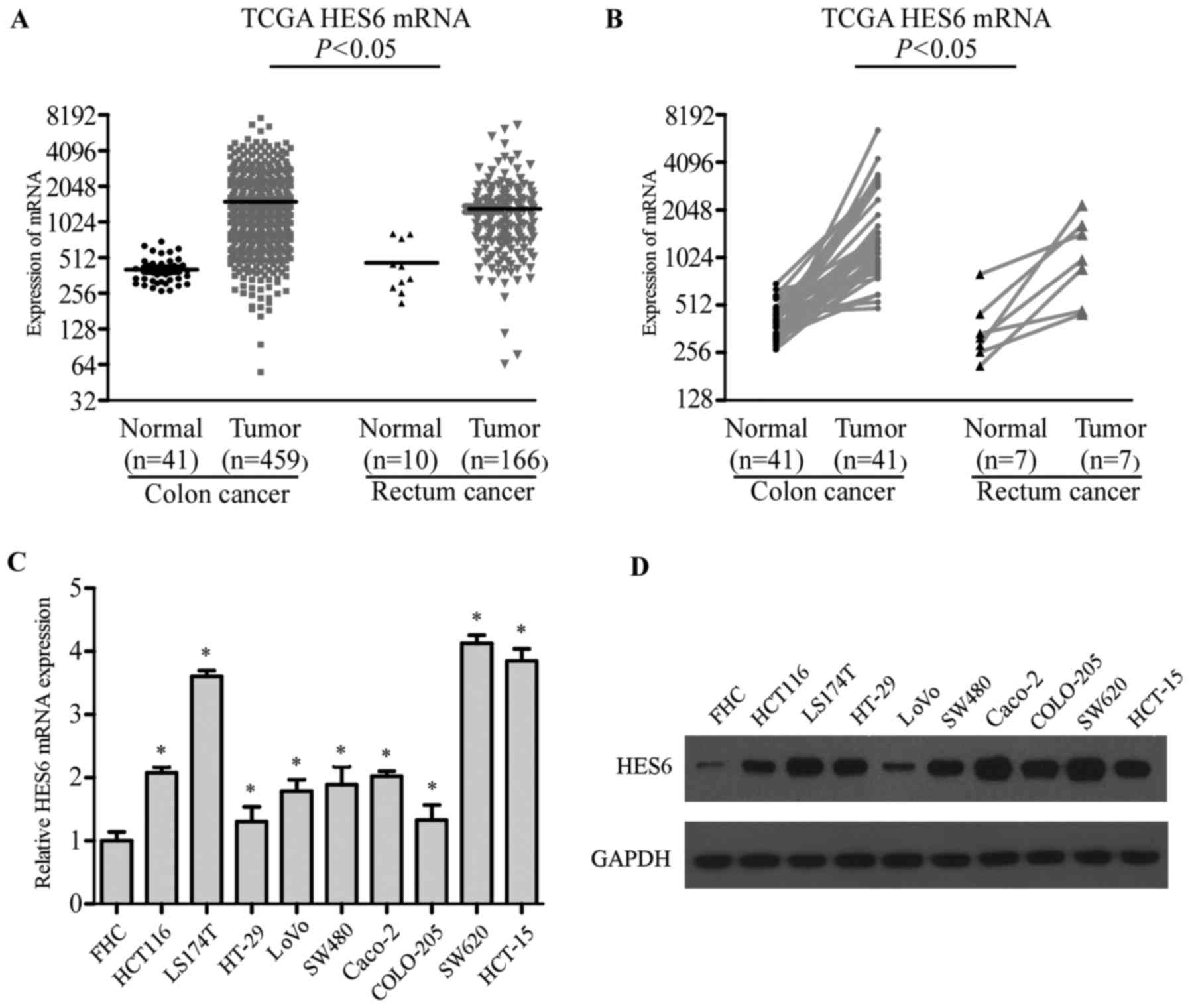
The data of the Cancer Genome Atlas (TCGA) revealed
that HES6 mRNA expression levels were elevated in CRC tissues
compared with normal and matched adjacent non-cancerous tissues,
respectively (Fig. 1A and B).
The present study examined the expression of HES6 in
9 CRC cell lines (HT-29, COLO-205, LoVo, SW480, SW620, HCT 116,
HCT-15, Caco-2 and LS174T) and a normal colon epithelial cell line
(FHC). HES6 mRNA expression levels were upregulated at least 2-fold
in CRC cell lines compared with FHC (Fig. 1C). Western blotting revealed the
levels of HES6 protein expression were significantly higher in CRC
cell lines compared with FHC (Fig.
1D). Collectively, these results demonstrated that the
expression of HES6 was elevated in CRC cell lines.
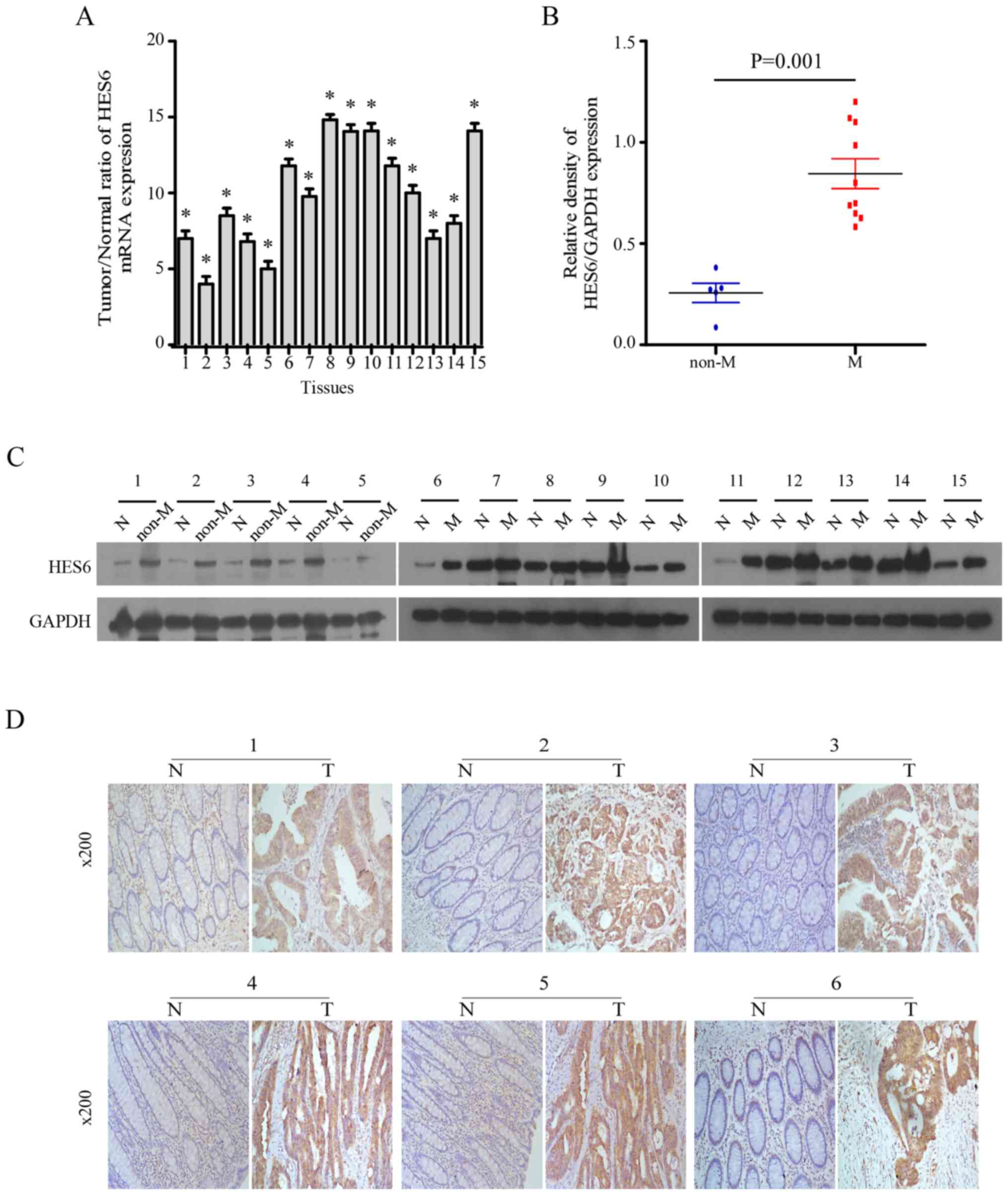
HES6 expression is elevated in primary
human CRC lesions
Subsequently, the present study investigated the
expression of HES6 in 15 primary CRC tissues (T) and matched
adjacent non-cancerous tissues (N) from the same patients. HES6
mRNA expression levels in the CRC tissues were at least 3-fold
higher than in the paired normal tissues (Fig. 2A). Western blotting revealed that
HES6 protein expression levels were significantly higher in the 15
CRC tissues compared with adjacent non-cancerous tissues. Of note,
HES6 expression levels were higher in the CRC tissues from patients
with distant metastasis compared with primary lesions (Fig. 2B and C). Consistent with these
results, IHC analysis demonstrated that the extent of HES6 staining
was negative to low in the adjacent normal tissues however,
positive staining was observed in the tumor tissues of patients
with or without metastasis (Fig.
2D). These results indicated that HES6 expression was elevated
in CRC lesions at both the mRNA and protein levels, and higher HES6
expression may be considered as a predictor for the risk of
metastasis.
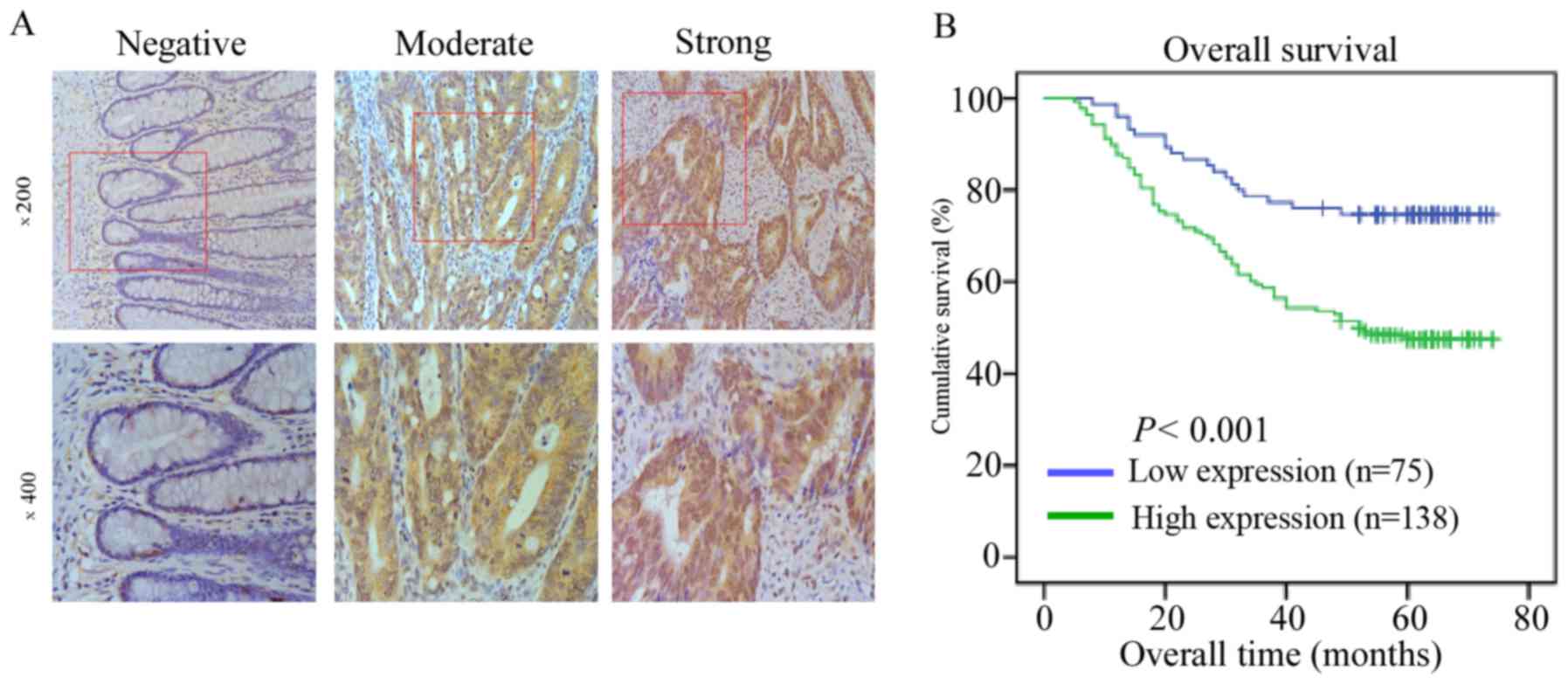
Association between the increased
expression of HES6 and the clinical features of CRC
In the present study, the expression level of HES6
in 213 paraffin-embedded, archived CRC tissues was analyzed via
IHC. A total of 138 cases (64.8%) exhibited positive staining for
HES6 protein in the 213 registered patient samples. The remaining
75 cases (35.2%) demonstrated weak staining for HES6 protein
expression. Examples of negative, moderate and strong HES6 staining
by IHC in human CRC tissues are presented in Fig. 3A. The samples were divided into low
and high HES6 expression groups as summarized in Table I. High HES6 protein expression
levels were strongly associated with T and N stages, and distant
metastasis (P<0.05). However, no significant association was
reported between HES6 protein expression and patient age, sex,
histological differentiation grade or chemotherapy in patients with
CRC (Table IV).
 | Table IV.Relation between the expression of
HES6 and the clinicopathological features of colorectal
carcinoma. |
Table IV.
Relation between the expression of
HES6 and the clinicopathological features of colorectal
carcinoma.
| Features | Total | HES6 weak
expression (%) | HES6 strong
expression (%) | P-value
(Chi-squared test) | P-value (Fisher's
exact test) |
|---|
| Age (years) |
|
|
| 0.279 | 0.313 |
|
<56 | 93 | 29 (13.6) | 64 (30.0) |
|
|
|
≥56 | 120 | 46 (21.6) | 74 (34.7) |
|
|
| Sex |
|
|
| 0.841 | 0.885 |
|
Male | 123 | 44 (20.7) | 79 (37.1) |
|
|
|
Female | 90 | 31 (14.6) | 59 (27.7) |
|
|
| T Stage |
|
|
| <0.001 | <0.001 |
|
T1-T2 | 27 | 19 (8.9) | 8 (3.8) |
|
|
|
T3-T4 | 186 | 56 (26.3) | 130 (61.0) |
|
|
| N Stage |
|
|
| 0.020 | 0.021 |
| N0 | 122 | 51 (23.9) | 71 (33.3) |
|
|
|
N1-N2 | 91 | 24 (11.3) | 67 (31.5) |
|
|
| Metastasis |
|
|
| <0.001 | <0.001 |
| M0 | 149 | 65 (30.5) | 84 (39.4) |
|
|
| M1 | 64 | 10 (4.7) | 54 (25.4) |
|
|
|
Differentiation |
|
|
| 0.638 | 0.711 |
|
Well/moderate | 174 | 60 (28.2) | 114 (53.5) |
|
|
|
Poor/undifferentiated | 39 | 15 (7.0) | 24 (11.3) |
|
|
| Chemotherapy |
|
|
| 0.781 | 0.885 |
|
Yes | 91 | 33 (15.5) | 58 (27.2) |
|
|
| No | 122 | 42 (19.7) | 80 (37.6) |
|
|
| Prognosis |
|
|
| <0.001 | <0.001 |
|
Survival | 122 | 56 (26.3) | 66 (31.0) |
|
|
|
Death | 91 | 19 (8.9) | 72 (33.8) |
|
|
HES6 is associated with poor prognosis
in patients with CRC
Kaplan-Meier survival analysis and a log-rank test
were used to evaluate the prognostic value of HES6 protein
expression in CRC. A significant difference in survival time was
demonstrated between the high and low HES6 protein expression
groups via the log-rank test (P<0.001; Fig. 3B). Compared with the low HES6
expression group, patients with higher HES6 expression levels
exhibited significantly shorter overall survival. In addition, the
cumulative overall survival was only 47.8% in the high HES6
expression group compared to 74.7% in the low HES6 expression
group.
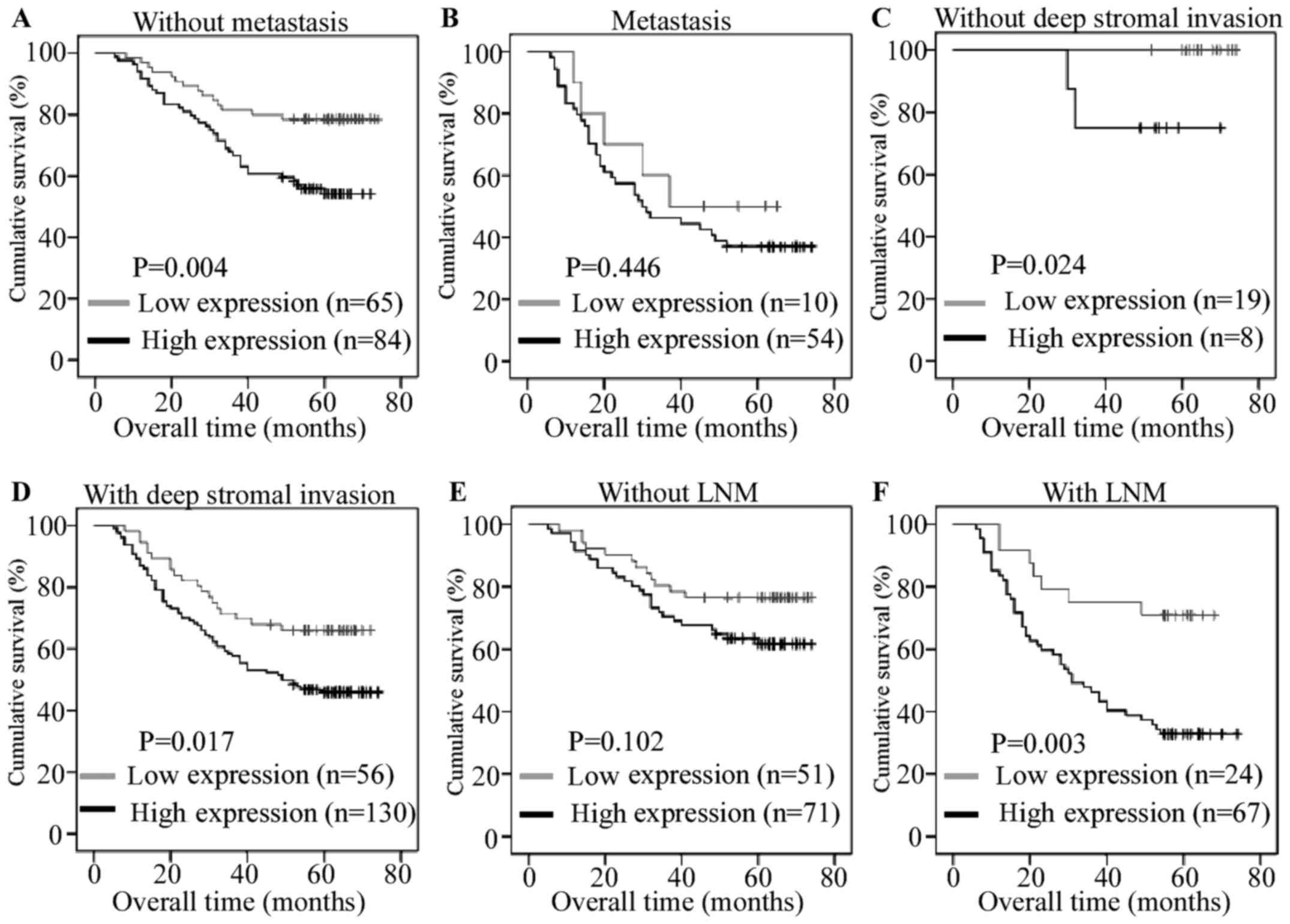
The prognostic value of HES6 in the T and N stages
and distant metastasis subgroups were also investigated in the
present study. As presented in Fig.
4, the expression of HES6 was significantly associated with the
overall survival of patients without metastasis (log-rank test,
P=0.004), without deep stromal invasion (log-rank test, P=0.024),
with deep stromal invasion (log-rank test, P=0.017) and with lymph
node metastasis (log-rank test, P=0.003). Univariate Cox regression
and multivariate analysis revealed that higher HES6 expression
levels (P=0.027), N stage (P=0.015) and metastasis (P=0.025) may be
predictors of poor clinical outcomes in patients with CRC (Table V). These data indicated that high
HES6 protein expression levels may be a novel prognostic indicator
for CRC.
 | Table V.Univariate and multivariate analyses
of prognostic factors in colorectal cancer using a Cox-regression
model. |
Table V.
Univariate and multivariate analyses
of prognostic factors in colorectal cancer using a Cox-regression
model.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Factors | Number of
patients | P-value | Regression
coefficient (SE) | P-value | Relative risk | 95% confidence
interval |
|---|
| HES6
expression |
| <0.001 | 2.478 (0.258) | 0.027 | 1.803 | 1.069–3.039 |
|
Low | 75 |
|
|
|
|
|
|
High | 138 |
|
|
|
|
|
| T Stage |
| 0.003 | 8.446 (0.715) | 0.052 | 4.184 | 0.989–17.696 |
|
T1-T2 | 27 |
|
|
|
|
|
|
T3-T4 | 186 |
|
|
|
|
|
| N Stage |
| <0.001 | 2.228 (0.212) | 0.015 | 1.703 | 1.108–2.617 |
| N0 | 122 |
|
|
|
|
|
|
N1-N2 | 91 |
|
|
|
|
|
| Metastasis |
| <0.001 | 2.316 (0.213) | 0.025 | 1.641 | 1.063–2.535 |
| M0 | 149 |
|
|
|
|
|
| M1 | 64 |
|
|
|
|
|
|
Differentiation |
| 0.045 | 1.648 (0.249) | 0.146 | 1.446 | 0.879–2.377 |
|
Well/moderate | 174 |
|
|
|
|
|
|
Poor/undifferentiated | 39 |
|
|
|
|
|
| Chemotherapy |
| 0.010 | 1.716 (0.210) | 0.056 | 1.514 | 0.989–2.317 |
|
Yes | 91 |
|
|
|
|
|
| No | 122 |
|
|
|
|
|
HES6 promotes the migration and
invasion of CRC cells
Since the phenotypes of migration and invasion are
key characteristics of cancer metastasis, the present study
investigated whether HES6 mediates the migration and invasion of
CRC cells. Firstly, TCGA profiles were analyzed by gene set
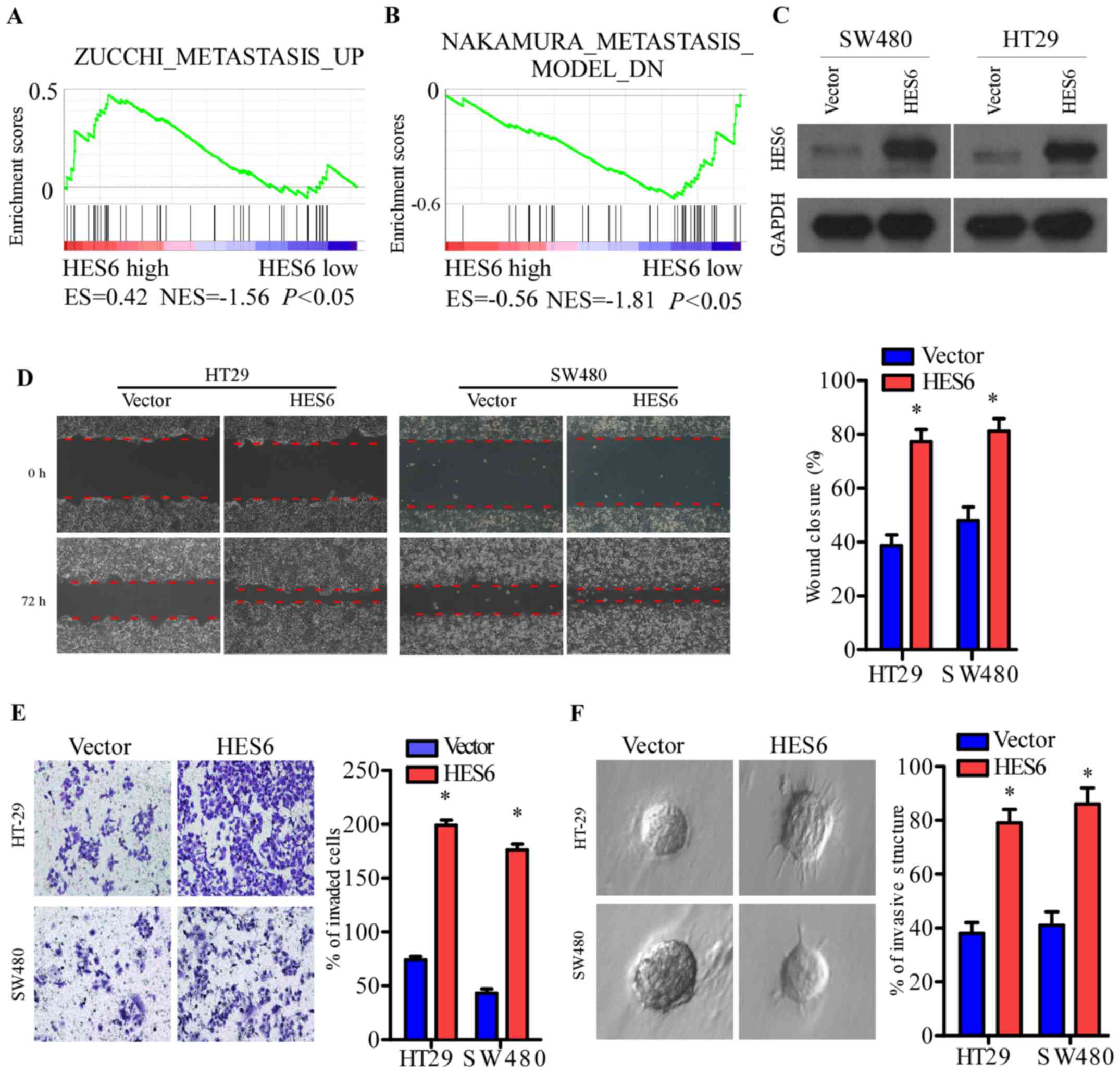
enrichment analysis (GSEA) (25,26),
which revealed that HES6 expression levels were related with cancer
metastasis (Fig. 5A and B). In the
present study, stably-expressing HES6 cell lines were established
using the human CRC cell lines, HT-29 and SW480 (Fig. 5C). The wound healing and Transwell
assays revealed that HES6 overexpression promoted the migration and
invasion of CRC cells compared with control cells (Fig. 5D and E). However, no significant
increase in cell numbers was observed following HES6
overexpression. In the 3D spheroid invasion assay, a better model
of tumor invasion in vivo, HES6 expression promoted the
invasive phenotype, as indicated by an increased number of outward
projections from a single cell (Fig.
5F). These results demonstrated that elevated HES6 expression
levels may promote the migration and invasion of CRC cells, which
supports the association between HES6 expression and T, N, and M
classification observed in a large cohort of clinical
specimens.
Knockdown of HES6 inhibits the
migration and invasion of CRC cells
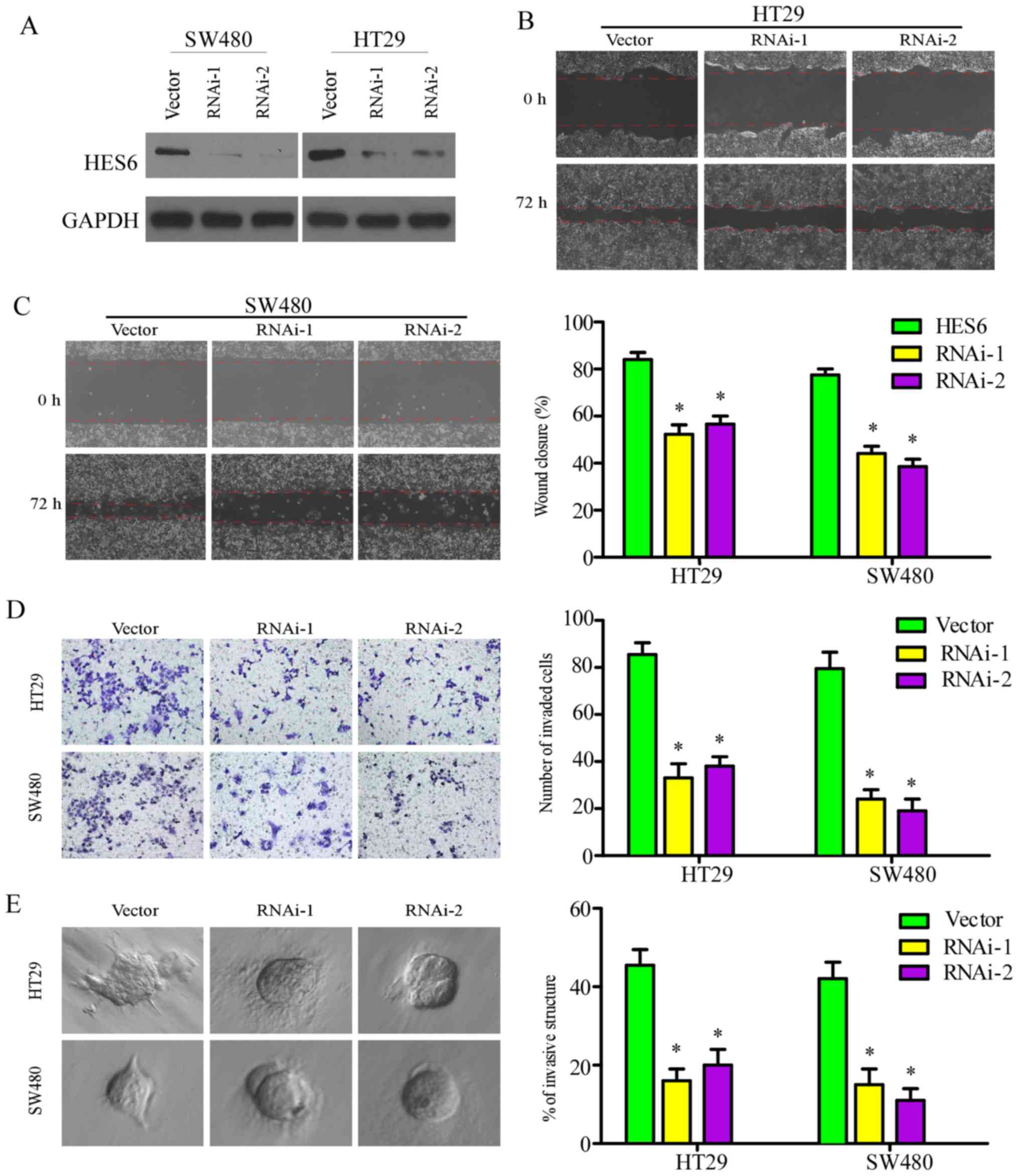
The role of HES6 in CRC metastasis was investigated
in the present study by silencing endogenous HES6 expression with
specific shRNAs (Fig. 6A). In the
wound-healing, Transwell and 3D spheroid invasion assays, cell
migration and invasion were significantly reduced in the SW480 and
HT29 cell lines following knockdown of HES6 (Fig. 6B-E). These results revealed that
downregulation of HES6 inhibited the migration and invasion of CRC
cells in vitro.
HES6 activates the Wnt/β-catenin
signaling pathway in CRC
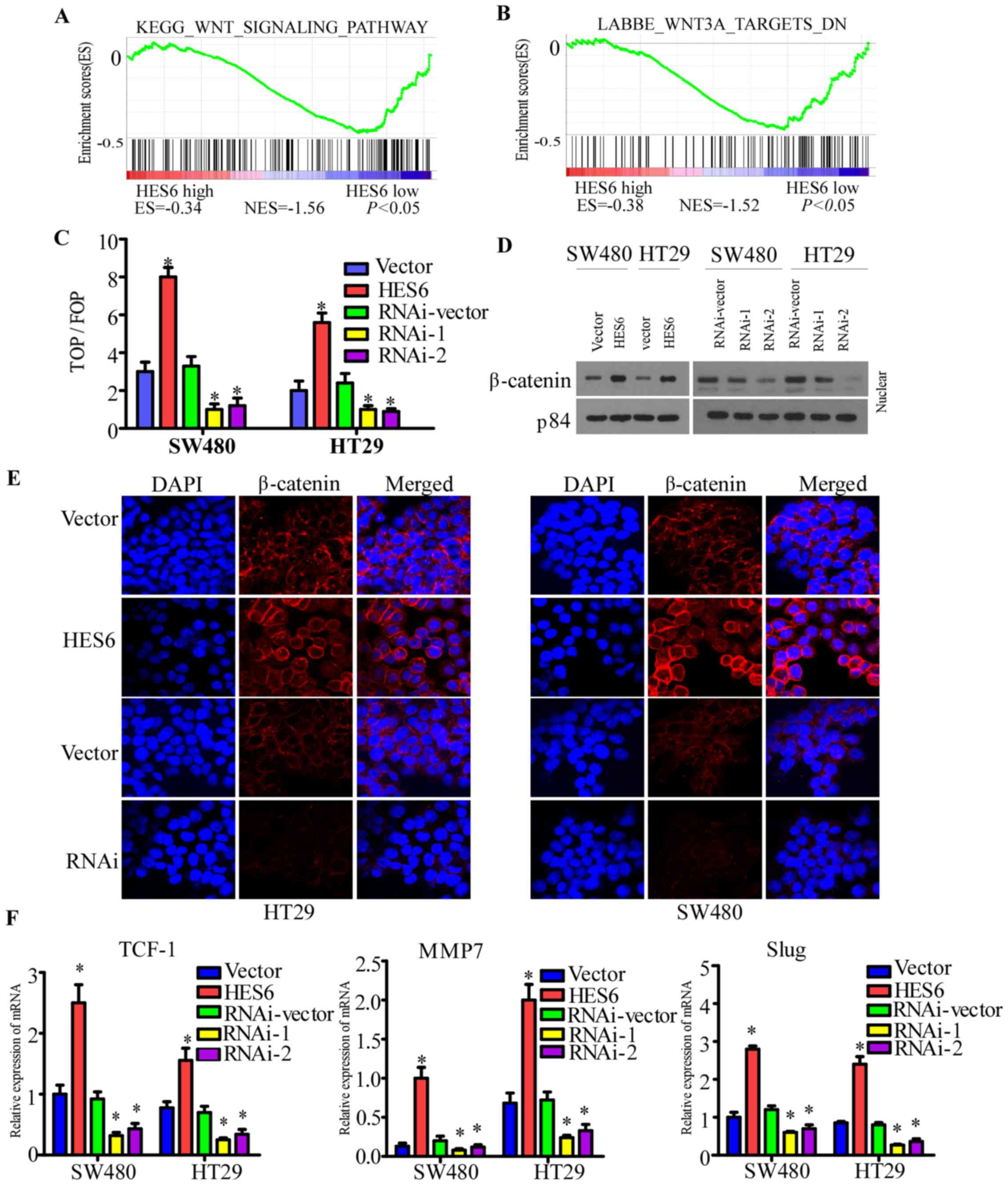
The GSEA revealed that the HES6 expression profiles
in TCGA were associated with the Wnt/β-catenin signaling pathway
(Fig. 7A and B). In addition,
whether HES6 enhances cell invasion and migration in CRC via the
activation of the Wnt/β-catenin signaling pathway was investigated
in the present study. The luciferase reporter assay demonstrated
that overexpression of HES6 enhanced, while HES6 silencing reduced,
the ratio of TOP flash to FOP flash activity in the indicated cells
(Fig. 7C). Western blotting
revealed that overexpression of HES6 upregulated the expression of
β-catenin levels in the nuclei (Fig.
7D). The immunofluorescence staining assays demonstrated that
overexpression of HES6 led to the nuclear accumulation of
β-catenin, while the opposite results were observed following the
silencing of HES6 (Fig. 7E).
To confirm the effects of HES6 on the Wnt/β-catenin
signaling pathway in CRC, RT-qPCR was used to examine the
expression of several Wnt target genes. Overexpression of HES6
upregulated, while HES6 silencing downregulated, the expression
levels of TCF-1, MMP7 and Slug in the indicated cells (Fig. 7F). The Wnt/β-catenin signaling
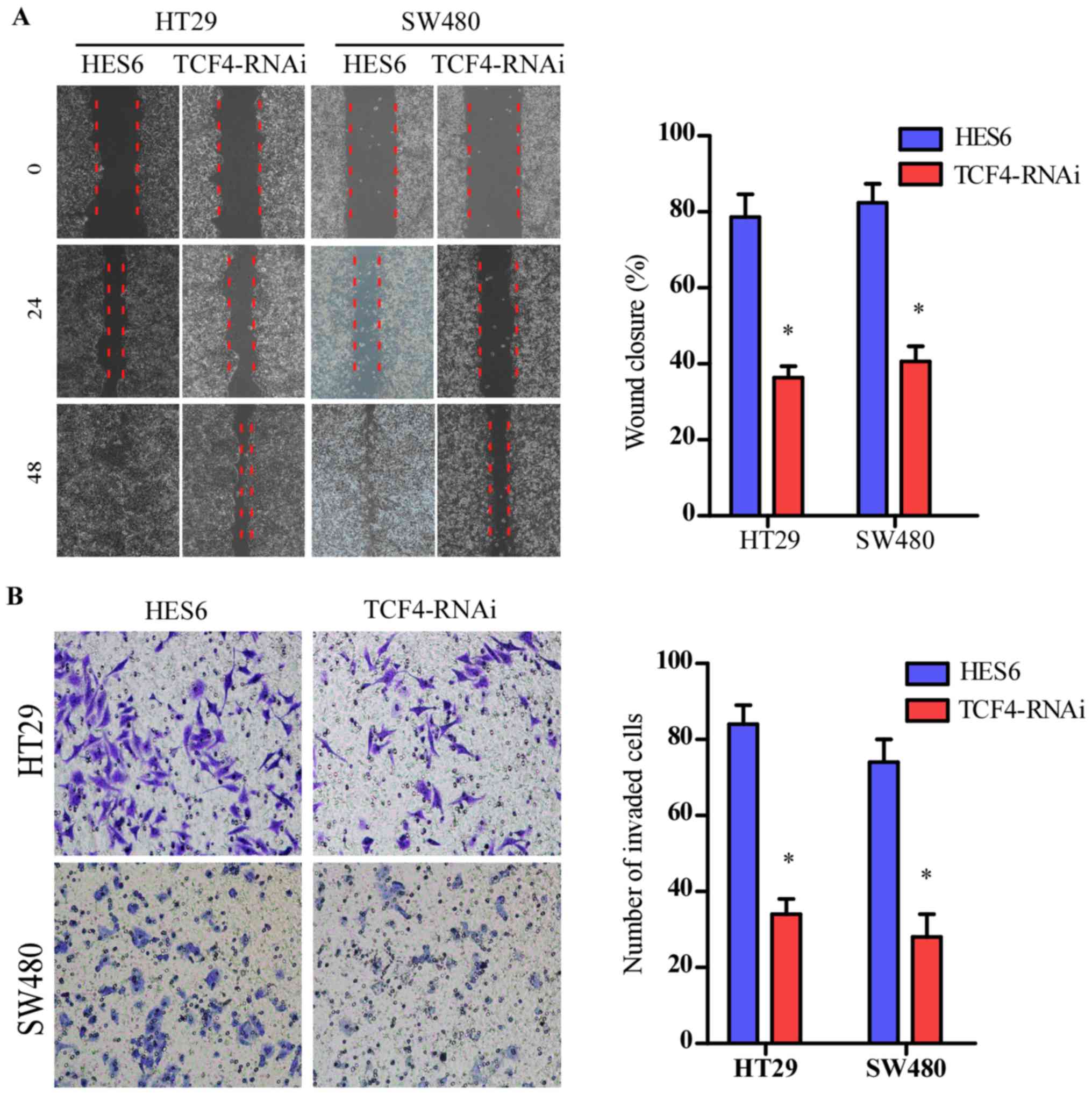
pathway was inhibited by silencing of TCF-4 expression, as
previously described (10). The
migration and invasive abilities of the indicated cells were
attenuated (Fig. 8A and B).
Collectively, these results revealed that HES6 promoted migration
and invasion by activating the Wnt/β-catenin pathway in CRC.
Discussion
In the present study, it was reported that HES6
expression was elevated in primary human CRC lesions at the protein
and mRNA levels. HES6 protein expression was associated with T
stage, lymph node metastasis and distant metastasis in patients
with CRC. CRC patients with elevated levels of HES6 expression
exhibited a poor prognosis. In addition, the multivariate analysis
conducted in the present study demonstrated that HES6 may be a
potential novel prognostic indicator for survival in patients with
CRC. Finally, ectopic HES6 expression was proposed to enhance the
migration and invasion of CRC cells by activating the Wnt/β-catenin
signaling pathway in the present study.
The oncogenic role of HES6, a helix-loop-helix
transcriptional suppressor, has been previously reported in several
studies: for example, HES6 was demonstrated to regulate the
differentiation of numerous cell types during myogenesis (27). Nam et al (28) proposed that HES6 binds to HES1,
suppressing the function of HES1 and promoting the differentiation
of neural stem cells. Additionally, elevated HES6 expression was
reported to lead to a significant increase in the invasive
phenotype of prostate cancer and glioma (14,29).
The depletion of HES6 decreased cell migration in alveolar
rhabdomyosarcoma, as determined by scratch-wound assays (30). In addition, high HES6 expression
levels have been correlated with poor survival in patients with
prostate, breast and ovarian cancers (16,17,31).
However, to the best of our knowledge, this is the first study that
has demonstrated the association between HES6 and the pathogenesis
of CRC.
Swearingen et al (15) previously reported that HES6 was
upregulated only at the transcriptional level in a xenograft model
of metastatic CRC. The results of the present study, however,
demonstrated that HES6 was upregulated at both the protein and mRNA
levels in CRC cell lines and primary CRC lesions. Furthermore,
analysis of TCGA data revealed that HES6 was upregulated in primary
lesions of patients with CRC. These differing results may be
reflective of the small sample size employed in the study of
Swearingen et al (15), as
only three isogenic lung tumor metastases were analyzed.
Based on the IHC analyses of the present study, high
HES6 expression levels were significantly correlated with T stage,
lymph node metastasis, distant metastasis and survival status in
patients with CRC. These results indicated that patients with CRC
possessing high HES6 expression levels tended to have a poorer
prognosis. In addition, univariate and multivariate analyses in the
present study indicated that HES6 expression was a significant
independent predictor of poor prognosis in CRC patients, as
previously described for other types of cancer (16,17,31).
Furthermore, an association was observed between poor overall
survival and high HES6 protein expression in patients with CRC
without metastasis, with and without deep stromal invasion, or
lymph node metastasis, which indicated that HES6 may be a novel
biomarker for the prediction of overall survival in these
subgroups. However, no correlation between HES6 expression and
survival was observed in patients with metastasis. This may be due
to the small number of patients in this subgroup (n=64). Therefore,
the results of the present study should be further verified with a
larger cohort of patients in the future.
At present, the main therapy applied to patients
with CRC is surgical resection. However, patients may experience
various effects following resection even in the same TNM stage
(32). It has been reported that
~50% of patients will experience a recurrence within the first 3
years following surgery (33).
Therefore, the results of the present study indicated that
additional radiotherapy and chemotherapy to reduce malignancy or
metastasis, may be beneficial for patients with high HES6
expression levels.
The Wnt/β-catenin signaling pathway is crucial for
the progression of CRC (34,35).
In the present study, HES6 was reported to enhance cell migration
and invasion in CRC by activating the Wnt/β-catenin signaling
pathway, and upregulating the expression of downstream target
genes, including Slug and MMP7, which have been associated with
cancer metastasis (36,37). The in vitro investigations
conducted in the present study, in which HES6 was overexpressed in
CRC cells, demonstrated that HES6 upregulation increased cell
migration and invasive abilities. This suggested that HES6 may be
involved in epithelial-mesenchymal transition, whereby cells lose
their cell-cell adhesion, and gain migratory and invasive
properties. Thus, the results of the present study revealed a novel
pathological mechanism underlying CRC, in which the Wnt/β-catenin
signaling pathway is activated via the overexpression of HES6.
Therefore, targeting the activation of the HES6-mediated
Wnt/β-catenin signaling pathway may represent a novel
anti-metastasis therapy for CRC.
In the present study, the clinical significance of
HES6 expression in patients with CRC was investigated and revealed
that HES6 may enhance the migration and invasive abilities of CRC
cell lines by activating the Wnt/β-catenin signaling pathway.
However, these findings require further validation in a larger
cohort of samples. In addition, the precise mechanism underlying
the HES6-associated activation of the Wnt/β-catenin signaling
pathway requires further study. In conclusion, HES6 protein levels
may be considered as a novel predictor of clinical outcome and
metastasis in CRC patients.
Acknowledgements
We express our thanks to the use of the facility and
the academic advice from the staff of the State Key Laboratory of
Oncology in South China, as well as the Key Laboratory of Major
Obstetric Diseases in Guangdong Province and the Key Laboratory for
Reproduction and Genetics of Guangdong Higher Education
Institutes.
Funding
The present study was supported by the Science and
Technology Planning Projects of Guangdong Province, China (grant
no. 2016B090918130).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. Still, more details about the
datasets used during the present study are available from the
corresponding author upon reasonable request.
Authors' contributions
YX and XX conceived and designed the study. YX, XL,
HZ and ZZ performed the experiments. YX and XL wrote the
manuscript. YX, LS and XX reviewed and edited the manuscript.
XianW, XiaoW, SL and HZ performed data acquisition and curation.
Supervision throughout this manuscript was done by LS and XX. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Ethical approval of the study was obtained by the
Clinical Research Ethics Committee of the Third Affiliated Hospital
of Guangzhou Medical University and the Sun Yat-sen University
Cancer Center Institutional Board. Written informed consent was
obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen HN, Yuan K, Xie N, Wang K, Huang Z,
Chen Y, Dou Q, Wu M, Nice EC, Zhou ZG, et al: PDLIM1 stabilizes the
E-cadherin/β-catenin complex to prevent epithelial-mesenchymal
transition and metastatic potential of colorectal cancer cells.
Cancer Res. 76:1122–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun J, Ding C, Yang Z, Liu T, Zhang X,
Zhao C and Wang J: The long non-coding RNA TUG1 indicates a poor
prognosis for colorectal cancer and promotes metastasis by
affecting epithelial-mesenchymal transition. J Transl Med.
14:422016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Agarwal E, Robb CM, Smith LM, Brattain MG,
Wang J, Black JD and Chowdhury S: Role of Akt2 in regulation of
metastasis suppressor 1 expression and colorectal cancer
metastasis. Oncogene. 36:3104–3118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nürnberg A, Kitzing T and Grosse R:
Nucleating actin for invasion. Nat Rev Cancer. 11:177–187. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie R, Wang J, Tang W, Li Y, Peng Y, Zhang
H, Liu G, Huang X, Zhao J, Li A, et al: Rufy3 promotes metastasis
through epithelial-mesenchymal transition in colorectal cancer.
Cancer Lett. 390:30–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu Y, Zheng S, An N, Athanasopoulos T,
Popplewell L, Liang A, Li K, Hu C and Zhu Y: β-catenin as a
potential key target for tumor suppression. Int J Cancer.
129:1541–1551. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan
J, Wu J and Li M: MicroRNA-374a activates Wnt/β-catenin signaling
to promote breast cancer metastasis. J Clin Invest. 123:566–579.
2013.PubMed/NCBI
|
|
11
|
Liu C, Li Y, Semenov M, Han C, Baeg GH,
Tan Y, Zhang Z, Lin X and He X: Control of beta-catenin
phosphorylation/degradation by a dual-kinase mechanism. Cell.
108:837–847. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brabletz T, Jung A, Reu S, Porzner M,
Hlubek F, Kunz-Schughart LA, Knuechel R and Kirchner T: Variable
beta-catenin expression in colorectal cancers indicates tumor
progression driven by the tumor environment. Proc Natl Acad Sci
USA. 98:10356–10361. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Höglund M, Frigyesi A, Säll T, Gisselsson
D and Mitelman F: Statistical behavior of complex cancer
karyotypes. Genes Chromosomes Cancer. 42:327–341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carvalho FL, Marchionni L, Gupta A,
Kummangal BA, Schaeffer EM, Ross AE and Berman DM: HES6 promotes
prostate cancer aggressiveness independently of Notch signalling. J
Cell Mol Med. 19:1624–1636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Swearingen ML, Sun D, Bourner M and
Weinstein EJ: Detection of differentially expressed HES-6 gene in
metastatic colon carcinoma by combination of suppression
subtractive hybridization and cDNA library array. Cancer Lett.
198:229–239. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ramos-Montoya A, Lamb AD, Russell R,
Carroll T, Jurmeister S, Galeano-Dalmau N, Massie CE, Boren J, Bon
H, Theodorou V, et al: HES6 drives a critical AR transcriptional
programme to induce castration-resistant prostate cancer through
activation of an E2F1-mediated cell cycle network. EMBO Mol Med.
6:651–61. 2014.PubMed/NCBI
|
|
17
|
Chiaramonte R, Colombo M, Bulfamante G,
Falleni M, Tosi D, Garavelli S, De Simone D, Vigolo E, Todoerti K,
Neri A, et al: Notch pathway promotes ovarian cancer growth and
migration via CXCR4/SDF1α chemokine system. Int J Biochem Cell
Biol. 66:134–140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gramantieri L, Giovannini C, Lanzi A,
Chieco P, Ravaioli M, Venturi A, Grazi GL and Bolondi L: Aberrant
Notch3 and Notch4 expression in human hepatocellular carcinoma.
Liver Int. 27:997–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koyano-Nakagawa N, Kim J, Anderson D and
Kintner C: Hes6 acts in a positive feedback loop with the
neurogenins to promote neuronal differentiation. Development.
127:4203–4216. 2000.PubMed/NCBI
|
|
20
|
Bae S, Bessho Y, Hojo M and Kageyama R:
The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal
differentiation. Development. 127:2933–2943. 2000.PubMed/NCBI
|
|
21
|
Drenzek JG, Seiler NL, Jaskula-Sztul R,
Rausch MM and Rose SL: Xanthohumol decreases Notch1 expression and
cell growth by cell cycle arrest and induction of apoptosis in
epithelial ovarian cancer cell lines. Gynecol Oncol. 122:396–401.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benson AB III, Arnoletti JP, Bekaii-Saab
T, Chan E, Chen YJ, Choti MA, Cooper HS, Dilawari RA, Engstrom PF,
Enzinger PC, et al: National Comprehensive Cancer Network: Anal
Carcinoma, Version 2.2012: Featured updates to the NCCN guidelines.
J Natl Compr Canc Netw. 10:449–454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−∆∆C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Ouyang F, Liu X, Wu S, Wu HM, Xu
Y, Wang B, Zhu J, Xu X and Zhang L: Overexpressed CISD2 has
prognostic value in human gastric cancer and promotes gastric
cancer cell proliferation and tumorigenesis via AKT signaling
pathway. Oncotarget. 7:3791–3805. 2016.PubMed/NCBI
|
|
25
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Malone CMP, Domaschenz R, Amagase Y,
Dunham I, Murai K and Jones PH: Hes6 is required for actin
cytoskeletal organization in differentiating C2C12 myoblasts. Exp
Cell Res. 317:1590–1602. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nam SM, Kim YN, Kim JW, Kyeong DS, Lee SH,
Son Y, Shin JH, Kim J, Yi SS, Yoon YS, et al: Hairy and enhancer of
split 6 (Hes6) deficiency in mouse impairs neuroblast
differentiation in dentate gyrus without affecting cell
proliferation and integration into mature neurons. Cell Mol
Neurobiol. 36:57–67. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Haapa-Paananen S, Kiviluoto S, Waltari M,
Puputti M, Mpindi JP, Kohonen P, Tynninen O, Haapasalo H, Joensuu
H, Perälä M, et al: HES6 gene is selectively overexpressed in
glioma and represents an important transcriptional regulator of
glioma proliferation. Oncogene. 31:1299–1310. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wickramasinghe CM, Domaschenz R, Amagase
Y, Williamson D, Missiaglia E, Shipley J, Murai K and Jones PH:
HES6 enhances the motility of alveolar rhabdomyosarcoma cells. Exp
Cell Res. 319:103–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hartman J, Lam EW, Gustafsson JA and Ström
A: Hes-6, an inhibitor of Hes-1, is regulated by 17 beta-estradiol
and promotes breast cancer cell proliferation. Breast Cancer Res.
11:R792009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuipers EJ, Grady WM, Lieberman D,
Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ and Watanabe T:
Colorectal cancer. Nat Rev Dis Primers. 1:150652015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okada-Iwasaki R, Takahashi Y, Watanabe Y,
Ishida H, Saito J, Nakai R and Asai A: The discovery and
characterization of K-756, a novel Wnt/β-catenin pathway inhibitor
targeting tankyrase. Mol Cancer Ther. 15:1525–1534. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Banday MZ, Sameer AS, Mir AH, Mokhdomi TA,
Chowdri NA and Haq E: Matrix metalloproteinase (MMP) −2, −7 and −9
promoter polymorphisms in colorectal cancer in ethnic Kashmiri
population - A case-control study and a mini review. Gene.
589:81–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Komiya Y, Onodera Y, Kuroiwa M, Nomimura
S, Kubo Y, Nam JM, Kajiwara K, Nada S, Oneyama C, Sabe H, et al:
The Rho guanine nucleotide exchange factor ARHGEF5 promotes tumor
malignancy via epithelial-mesenchymal transition. Oncogenesis.
5:e2582016. View Article : Google Scholar : PubMed/NCBI
|