Introduction
Colorectal cancer (CRC) is one of the most common
types of cancer worldwide, which affects the gastrointestinal tract
and is associated with a high mortality rate due to its rapid
progression from the initial time period of the diagnosis (1–3). CRC
is the fifth leading cause of cancer-associated mortality for men
and women in China (4). Although
the disease can be successfully treated following an early
diagnosis, the majority of patients with CRC present at an advanced
pathological stage. Consequently, the survival rate of these
patients is considerably low. The identification of novel molecular
markers can assist in the early detection of CRC and the prevention
of the disease.
MicroRNAs (miRNAs) comprise a class of small,
non-coding RNA molecules, 20–22 nucleotides in length, which
regulate the post-transcriptional processes of several genes. To
date, ~1,000 miRNAs have been identified that regulate the function
of >50% of human genes (5).
Several miRNAs have elevated expression in human tumors and act in
an oncogenic manner, whereas others have reduced expression and act
as tumor suppressors (6,7). miRNAs have been identified to possess
altered expression in CRC (8).
miRNA-31 (miR-31) was shown to be upregulated in BRAF-mutated CRC
compared with wild-type CRC, and an association was identified
between the expression of miR-31 and proximal tumor location
(9). Despite investigations that
have been performed in the field of miRNAs and CRC, investigations
of changes in the expression levels of miR-31 in different regions
of colorectal tissue, and putative associations with other factors
involved in the regulation of gene transcription have been limited.
The LOC554202 gene, which is located on human chromosome 9, has
been shown to possess an intron region responsible for the
transcription of miR-31 (10).
LOC554202 is transcribed into a long non-coding RNA (lncRNA).
lncRNAs are a class of RNA sequences abundantly expressed in the
nucleus and cytoplasm, which possess >200 nucleotides and are
involved in the regulation of translation, although they do not
possess protein-coding potential (11). lncRNAs can be targeted for tumor
diagnosis and/or treatment, suggesting a broad prospect for
diagnosis, prognosis, and targeted therapy (12–14).
lncRNAs have been found to exist in tumors and are important in
their development and progression (15–17).
It is important to note that LOC554202 and miR-31 are derived from
the same major gene locus, 9p21.3, which indicates that the
transcriptional fragment of miR-31 is in the first intron of
LOC554202. Both of these sequences share a common promoter. As a
consequence, LOC554202 is the host gene of miR-31, and miR-31 is
the classic-intron miRNA of LOC554202. Intron miRNAs and their
corresponding host genes typically exhibit the same expression
profile, indicating that they exert a similar function (18). The expression levels of LOC554202
were found to be upregulated in breast cancer, and the knockdown of
this RNA was associated with decreased cellular proliferation and
induction of apoptosis, which suggested an oncogenic function
(19). The contradictory results of
the majority of studies performed on miR-31 and cancer are
summarized in Table I. LOC554202
and miR-31 are abnormally expressed in multiple types of tumor,
including CRC (20,21). LOC554202 and miR-31 have been shown
to be involved in the development and progression of CRC (20,22,23).
However, the expression levels of LOC554202 and miR-31 in CRC and
their effect on the progression of CRC following oxaliplatin-based
chemotherapy remain to be elucidated.
 | Table I.Summary of studies that have assessed
the associations between the expression of human miRs in human
cancer and the incidence of survival, clinicopathological
characteristics and cell function. |
Table I.
Summary of studies that have assessed
the associations between the expression of human miRs in human
cancer and the incidence of survival, clinicopathological
characteristics and cell function.
| Author, year | miR | Cases | Results | Variable | Study type | (Refs.) |
|---|
| Augoff et
al | miR-31 | 7 TNBC cell lines
(1 normal, 6 cancer) | miR-31 and host
gene LOC554202 downregulated in TNBC lines of basal subtype and
overexpressed in luminal counterparts, deacetylation and/or
demethylation resulting in increased expression of miR-31 and
LOC554202 | Sequencing, miR
expression by PCR, methylation-specific PCR | ES | (5) |
| Slaby et
al | miR-21, miR-31,
miR-143 and miR-145 | 29 CRC, 6 non-tumor
specimens | miR-21 correlated
with CRC clinical stage | miR expression by
PCR | CC | (8) |
| Nosho et
al | miR-31 | 721 CRCs, 381
serrated lesions and 251 non-serrated adenomas | High expression of
miR-31 associated with BRAF and KRAS mutations and proximal
location; cancer-specific miR-31 inhibition decreased cell invasion
and proliferation | miR expression by
PCR | CC, ES | (9) |
| Tazawa et
al | miR-34a | HCT116 and RKO cell
lines, 25 colon cancer and adjacent normal samples | 9/25 human colon
cancer (36%) showed decreased expression of miR-34a compared with
counterpart normal tissues, miR-34a suppressed in vitro and
in vivo growth of HCT116 and RKO cells | miR expression by
PCR, cell proliferation assays, in vivo tumor growth
xenografts | ES, CC | (15) |
| Wang et
al | miR-31 | 40 cases of gastric
cancer tissues and adjacent normal tissues | miR-31
downregulated in 40 cases of gastric cancer tissues compared to
adjacent normal tissues | miR expression by
PCR, IHC | CC | (34) |
| Wang et
al | miR-31, miR-143 and
miR-145 | 98 primary CRC
specimens, with corresponding normal mucosa specimens | miR-31 upregulated
in CRC, vs. normal, miR-31 expression positively correlated with
advanced TNM stage and deeper tumor invasion | miR expression by
PCR | CC | (36) |
Therefore, the aim of the present study was to
examine the expression levels of miR-31 and its host gene
LOC554202, and their impact on the overall survival rate of
patients with oxaliplatin-treated CRC.
Materials and methods
Study population
Patients with CRC who received oxaliplatin-based
chemotherapy were recruited between June 2005 and March 2010 at the
First Affiliated Hospital of China Medical University (Shenyang,
China). The inclusion criteria were as follows: i) Initially
diagnosed with CRC; ii) tissue specimens were approved for
experimental analysis; iii) patients who received oxaliplatin-based
FOLFOX and/or XELOX regimens. The principle of chemotherapy is
based on the body surface area of the patient and was calculated
according to the National Comprehensive Cancer Network (NCCN)
guidelines (www.nccn.org). The FOLFOX regimen
included 200 mg of oxaliplatin (intravenous infusion) with 600 mg
of calcium folinate (intravenous infusion) and 6 g of fluoride
(intravenous infusion 48 h). The XELOX regimen included 200 mg of
oxaliplatin (intravenous infusion) with capecitabine (oral, 2,000
mg/m2 body surface area per day, twice orally, 14 days
per course). The interval time of the chemotherapy was 21 days. The
medication regimen did not significantly change with the NCCN
update, thus the medication for all sample patients was considered
as an application of the same chemotherapeutic regimen.
The exclusion criteria were as follows: i)
Incomplete patient information and/or lack of follow-up; ii) lack
of LOC554202 and/or miR-31 detection; iii) loss and/or damage of
clinical samples; iv) patients with history of myocardial
infarction, cerebral infarction, atrial myxoma, aortic valve
regurgitation, pulmonary infarction, renal failure dialysis
treatment history, high risk of hypertension, respiratory failure,
chronic obstructive pulmonary disease, and/or human
immunodeficiency virus; v) serious postoperative complications,
including intestinal fistula, anastomotic obstruction and pulmonary
infarction; and vi) preoperative adjuvant chemotherapy and/or
radiotherapy.
Cancer tissues and the intestinal mucosa were
obtained postoperatively. The clinical staging of the patients was
performed in accordance with the WHO (World Health Organization)
and tumor-node-metastasis staging guidelines (24). The patients included in the present
study were phase III, phase IV and/or phase II at high risk.
According to the differentiation of the tumor, the patients were
classified into Grade 1 (poorly differentiated, medium-poorly
differentiated), Grade 2 (medium differentiation), and Grade 3
(medium-well differentiated, well differentiated). All patients
underwent enhanced CT examination of pulmonary and abdominal tissue
regions prior to surgery. The patients received routine CT scanning
and colonoscopy during follow-up in order to detect the metastasis
and/or recurrence of the tumor. The tumor adhesion and vascular
infiltration were investigated by intraoperative and postoperative
pathological examination. The postoperative follow-up duration was
5 years. The average maximum diameter for tumor foci was ~6 cm,
which was used as the dividing line of the tumor size.
The present study was approved by the Medical Ethics
Committee of the China Medical University at the First Hospital of
China Medical University. Written informed consent was required by
the patients for their participation in the study, as requested by
the ethical guidelines of the First Hospital of China Medical
University and the Medical Ethics Committee of the Chinese Medical
Universities. All patients were anonymized.
Tissue microarray (TMA)
Paraformaldehyde-fixed fresh tissues from the
central zone of the medial lobe were embedded in paraffin and
sliced into 4-µm sections. The sections were stained with
hematoxylin and eosin (H&E) and placed on coverslips with
specific mounting medium for observation using an Nikon ECLIPSE 80i
microscope (Nikon, Tokyo, Japan). Subsequently, the central zone of
tissues was stained, and the paraformaldehyde-fixed tissues were
re-organized to a new TMA. A total of six TMAs, comprising three
for the cancer tissue and three for the adjacent mucosa, were
obtained and sliced into 4-µm sections for in situ
hybridization (ISH) analysis.
ISH analysis
The ISH was performed as previously described
(25). Briefly, the tissues were
treated with hydrogen peroxide at room temperature for 10 min.
Subsequently, the sections were boiled (95–100°C) in citric buffer
for 15 min and incubated with protease at 40°C for 30 min. The
slides were hybridized with the SVV probe in hybridization buffer
[6 SSC (1 SSC is 0.15 mol/l NaCl, 0.015 mol/l Na-citrate), 25%
formamide, 0.2% lithium dodecyl sulfate, and blocking reagents] at
40°C for 2 h. The sequence biotin-digoxin and streptavidin-biotin
complex amplifiers were added for 60 min at 37°C, and chromogen
3,3′-diaminobenzidine tetrahydrochloride (DAB) was applied as a
0.02% solution containing 0.005% H2O2 in 50
mM ammonium acetate-citrate acid buffer (pH 6.0). The sections were
lightly counterstained with Mayer's hematoxylin and mounted on an
Nikon ECLIPSE 80i microscope (Nikon). The probes used for LOC554202
and miR-31 detection were as follows:
5′-Dig-CTGTGGCTCTTCTGGATCTCTAGG-Dig-3′ and
5′-Dig-AGCTATGCCAGCATCTTGCCT-Dig-3′, respectively.
Microarray analysis
The TMA was scanned using a Pannoramic MIDI scanner
(3D HISTECH, Budapest, Hungary) and analyzed by Quant Center
software (3D HISTECH, Budapest, Hungary). An H-score evaluation was
performed. The histochemistry score (H-score) was evaluated
according to previous studies (26,27).
The process was performed as follows: The number of cells in each
slice with positive staining and its staining intensity were
converted into the corresponding values in order to semi-quantify
the staining of the tissues. The H-score score was obtained
following scanning of the tissue with Pannoramic viewer 1.15.4
software (3D HISTECH, Budapest, Hungary). The image was enlarged,
and the image of the cancer tissue was captured. The associated
Quant Center Center V2.0 analysis software DensitoQuant program (3D
HISTECH) was used to automatically identify and set the dark brown
color on the section to as strongly positive and the brownish
yellow staining as medium. Positive, light yellow staining was set
as weakly positive and blue-violet staining was negative. The
percentage of positive cells was identified and analyzed for each
interception area. H-score = Σ (PI × I) = (negative percentage × 0)
+ (weak positive percentage × 1) + (medium positive percentage × 2)
+ (strong positive percentage × 3), where PI represents the
percentage of positive cell number, I represents the intensity of
staining, and the H-score score ranges between 0 and 300. When all
cells are strongly positive, a maximum value of 300 can be reached,
whereas when all cells are negative, a minimum value of 0 is
reached. In this method, the number of positive cells in each
section and its staining intensity are converted into corresponding
values to enable the quantification of tissue staining, which is
more accurate than the traditional human microscopic assessment of
the degree of positivity.
Statistical analysis
Statistical analysis was performed using SPSS 20.0
(IBM Corp., Armonk, NY, USA) and GraphPad Prism 7.0 (GraphPad
Software, Inc., La Jolla, CA, USA). The classification data were
analyzed using Pearson's χ2 and/or Fisher's tests. The
survival rate was calculated using the Kaplan-Meier method and the
log-rank test. The multivariate COX proportional hazard regression
model was used to evaluate the association of multiple variables.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Baseline characteristics
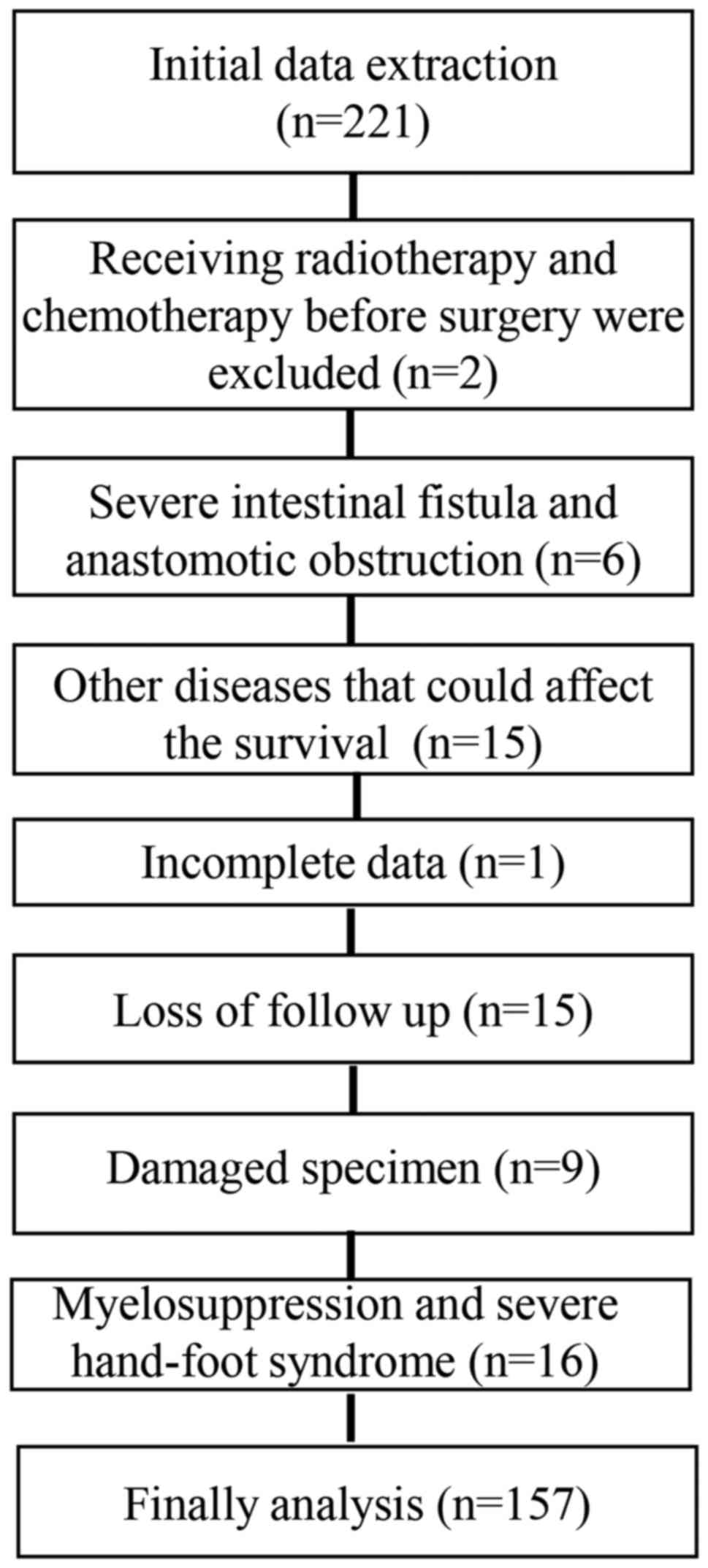
A total of 221 patients with CRC treated with
oxaliplatin-based chemotherapy were enrolled between June 2005 and
March 2010 at the First Affiliated Hospital of China Medical
University. A total of 64 patients were excluded for the following
reasons: Two patients were excluded due to radiotherapy and
chemotherapy treatment prior to surgery, six were excluded due to
severe intestinal fistula and anastomotic obstruction, 15 were
excluded for having diseases that may affect the survival rate. A
total of 15 patients lost contact information during follow-up, 9
had damaged specimens, and 1 was omitted due to incomplete clinical
information. In addition, 16 failed to complete chemotherapy, being
excluded due to myelosuppression and severe hand-foot syndrome
(Fig. 1). In total, 157 patients
were included for analysis, with an average age of 64 years old
(17–88 years), comprising 87 (55.4.0%) men and 70 (44.6%) women.
The demographic data and clinical parameters, including age, sex,
family history, tumor differentiation, pathological type, primary
location, bowel mass, degree of invasion, tumor size, adhesions,
organ metastasis and vascular infiltration, are listed in Table II. The numbers of patients
presenting with Grade 1, 2 and 3 tumors were 85 (54.1%), 60 (38.2%)
and 12 (7.6%), respectively. The majority of the pathological types
were adenocarcinoma (142 cases), whereas the remaining 15 cases
were mucinous adenocarcinoma. The majority of the tumor samples
were larger in the intestinal perimeter and penetrated the serosal
layer; the primary location of cancer was in the colon for 75 cases
(47.8%) and the rectum for 82 cases (52.2%). In 70 (44.6%) cases,
tumor diameter was ≥6 cm, whereas 87 cases (55.4%) exhibited a
tumor diameter of <6 cm. The majority of the tumors indicated no
adhesion, organ metastasis and/or vessel infiltration. A total of
49 cases were classified as disease-free survival (DFS) subjects
accounting for 31.2% of the total sample size, whereas 108 cases
(68.8%) exhibited CRC progression. The average DFS time was 41.90
months (1–110 months); a total of 59 (37.6%) patients survived,
whereas 98 (62.4%) patients succumbed to the disease. The mean OS
time was 53.75 months (6–110 months).
 | Table II.Clinicopathological characteristics
of patients with colorectal cancer treated with platinum-based
chemotherapy. |
Table II.
Clinicopathological characteristics
of patients with colorectal cancer treated with platinum-based
chemotherapy.
| Characteristic | Number of
cases | % |
|---|
| Total | 157 | 100 |
| Average age, years
(range) | 64 (17–88) |
|
| Age (years) |
|
≤64 | 77 | 49.0 |
|
>64 | 80 | 51.0 |
| Sex |
|
Male | 87 | 55.4 |
|
Female | 70 | 44.6 |
| Family history |
|
Positive | 14 |
8.9 |
|
Negative | 143 | 91.1 |
|
Differentiation |
| Grade
1 | 85 | 54.1 |
| Grade
2 | 60 | 38.2 |
| Grade
3 | 12 |
7.6 |
| Pathological
pattern |
|
Adenocarcinoma | 142 | 90.4 |
|
Mucinous adenocarcinoma | 15 |
9.6 |
| Primary organ |
|
Colon | 75 | 47.8 |
|
Rectum | 82 | 52.2 |
| Intestinal tract
occupation |
|
≤1/2 | 45 | 28.7 |
|
>1/2 | 112 | 71.3 |
| Infiltration |
| Invaded
serosa | 128 | 81.5 |
|
Non-invaded serosa | 29 | 18.5 |
| Size (cm) |
| ≤6 | 87 | 55.4 |
|
>6 | 70 | 44.6 |
| Adhesion |
|
Negative | 149 | 94.9 |
|
Positive | 8 |
5.1 |
| Metastasis |
|
Negative | 128 | 81.5 |
|
Positive | 29 | 18.5 |
| Vascular
invasion |
|
Negative | 141 | 89.8 |
|
Positive | 16 | 10.2 |
| DFS |
|
Negative | 49 | 31.2 |
|
Positive | 108 | 68.8 |
| OS |
|
Survived | 59 | 37.6 |
|
Deceased | 98 | 62.4 |
| Follow-up, months
(range) |
|
DFS | 41.97 (1–110) |
|
| OS | 53.75 (6–110) |
|
Expression and correlation of
LOC554202 and miR-31
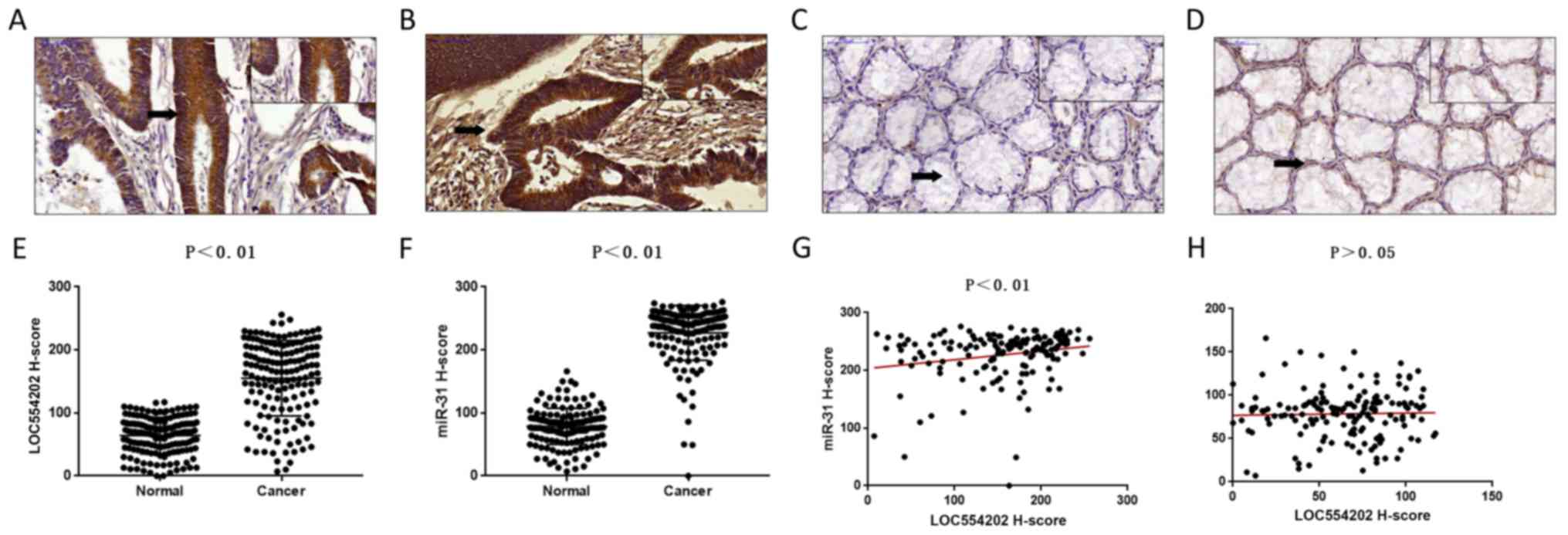
LOC554202 and miR-31 were positively expressed in
the nucleus and cytoplasm in cancer tissues (Fig. 2A and B), whereas their expression
was relatively weak in the adjacent tissues (Fig. 2C and D). The expression score of
LOC554202 in CRC was 155.24±59.94, which was significantly higher
than that noted in the adjacent tissues [64.16±28.95 (0–117)]
(Fig. 2E). The expression of miR-31
in CRC cancer was 226.80±43.47 (0–276), which was significantly
higher compared with that observed in the adjacent tissues
[78.59±28.70 (7–166)] (Fig. 2F).
Linear regression analysis indicated a significant positive
correlation between the expression of LOC554202 and miR-31 in CRC
(P<0.01, Fig. 2G). No
correlation was noted between the expression of LOC554202 and
miR-31 in normal mucosal tissues (Fig.
2H). The results indicated that the expression levels of
LOC554202 and miR-31 were correlated in CRC but were not correlated
in adjacent normal tissues.
Analysis of LOC554202 and miR-31 in
CRC by the receiver operating characteristic (ROC) curve
The H-score scores of LOC554202 and miR-31 in the
CRC tissues were quantified and classified. In the present study,
the H-score of LOC554202 was determined as follows: Scores of 0–70
were classified as 1, scores of 71–140 as 2, scores of 141–210 as
3, and scores of >211 as 4; the miR-31 H-scores were determined
as follows: 0–85 was classified as 1, 86–170 was classified as 2,
171–230 was classified as 3, and scores >230 were classified as
4.
An ROC curve was used to evaluate the optimal
cut-off value for LOC554202 and miR-31. The results indicated that
it was possible to distinguish the expression levels of LOC554202
by the primary position of the CRC (P=0.008), the pathological type
(P=0.010), the DFS (P=0.025) and the OS (P=0.016) (Fig. 3A-D). In addition, it was possible to
distinguish the expression levels of miR-31 by the pathological
type (P=0.040), the DFS (P=0.038) and the OS (P=0.036) (Fig. 3F-H), although the primary position
was not effectively distinguished (P=0.421). The largest area under
the curve (AUC) of the ROC curve corresponded to the pathological
type (Fig. 3B and F). Therefore,
the ROC curve of the pathological type was selected for reference.
Based on these data, a 65% cut-off (H-score of 165) was defined for
the expression level of LOC554202, and an 83% cut-off value
(H-score of 230) was defined for the expression level of
miR-31.
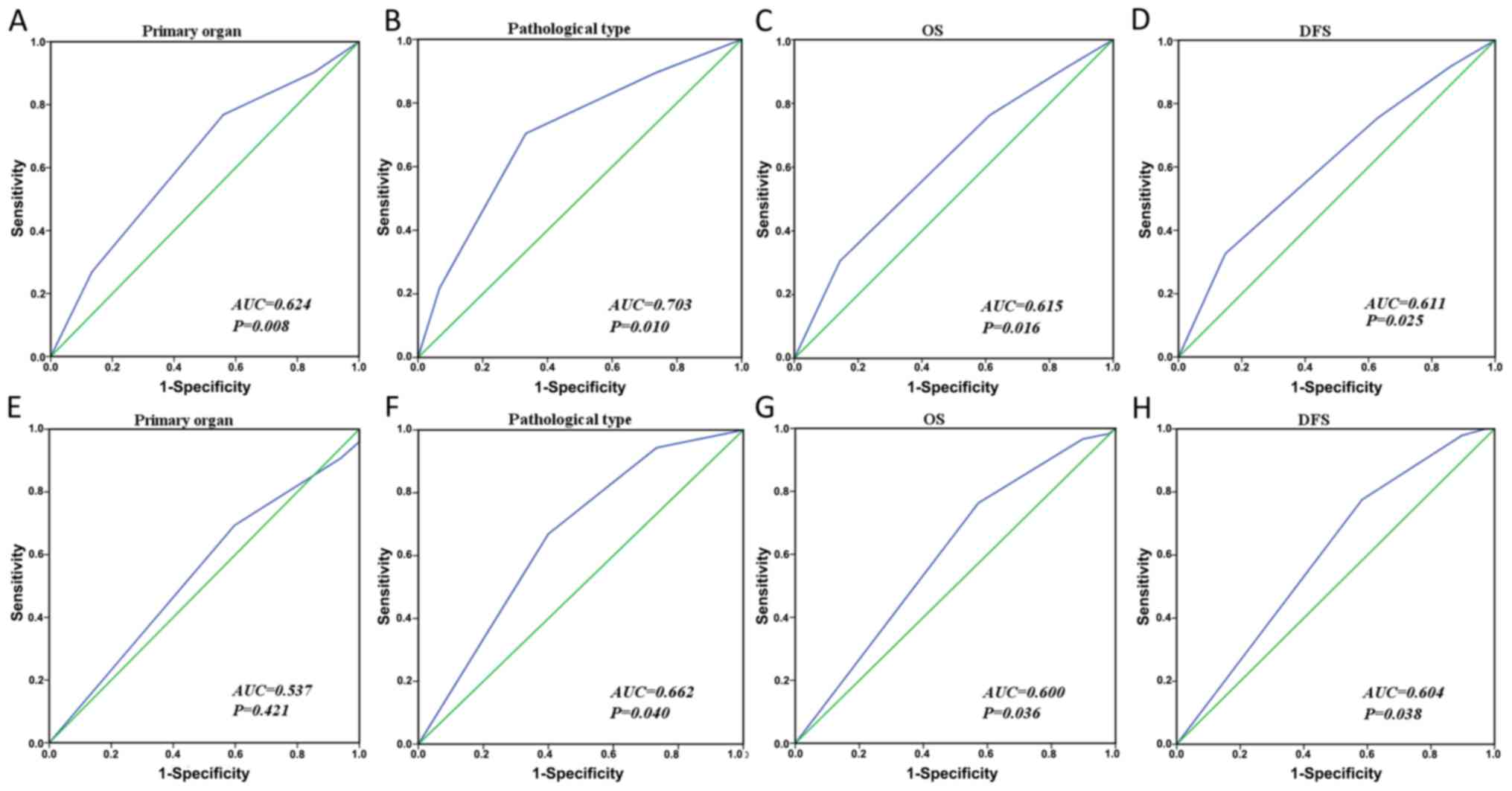 | Figure 3.ROC curves were used to determine the
cut-off scores for LOC554202 and miR-31 in colorectal cancer. The
sensitivity and specificity for each outcome were plotted and the
AUC and P-values were calculated. ROC curves for LOC554202 and (A)
primary organ, (B) pathological type, (C) OS, and (D) DFS. ROC
curves for miR-31 and (E) primary organ, (F) pathological type, (G)
OS and (H) DFS. miR, microRNA; ROC, receiver operating character;
AUC, area under the curve; DFS, disease-free survival; OS, overall
survival. |
Association between the expression
levels of LOC554202 and miR-31 and clinicopathological parameters
of patients with CRC treated with platinum-based chemotherapy
In order to investigate whether the expression
levels of LOC554202 and miR-31 in CRC tissues correlated with the
clinicopathological parameters of the patients with CRC, Pearson's
χ2 test was used and logistic regression was adjusted to
analyze the correlation between the expression of LOC554202 and/or
miR-31, the co-expression of LOC554202 and miR-31, and the
clinicopathological parameters.
Following stratification of the analysis of the
pathological types, it was concluded that 54.2% of the patients
with adenocarcinoma exhibited a high expression of LOC554202,
whereas only 13.3% of the patients with mucinous adenocarcinoma
expressed LOC554202 [P=0.003, adjusted logistic regression analysis
P=0.007, adjusted OR (95% CI)=0.116 (0.025–0.548)]. The expression
of miR-31 was high in 66.9% of the CRC tissues from the patients
with adenocarcinoma, and patients with high expression of miR-31
accounted for 40% of patients with mucinous adenocarcinoma
(P=0.039). Following adjustment by logistic regression analysis,
the parameters were as follows: P=0.038, adjusted OR (95% CI)=0.308
(0.101–0.937). These data suggested that the elevated expression
levels of miR-31 and LOC554202 were likely to occur in patients
with adenocarcinoma (Table
III).
 | Table III.Correlation of the expression
LOC554202 and miR-31 with clinicopathological parameters in
patients with colorectal cancer treated with platinum-based
chemotherapy. |
Table III.
Correlation of the expression
LOC554202 and miR-31 with clinicopathological parameters in
patients with colorectal cancer treated with platinum-based
chemotherapy.
|
| LOC554202
expression, n (%) |
|
| miR-31 expression,
n (%) |
|
|
|---|
|
|
|
|
|
|
|
|
|---|
|
Characteristics | Low | High |
P-valuea,b | Adjusted OR
(95%CI) | Low | High |
P-valuea,b | Adjusted OR
(95%CI)c |
|---|
| Age (years) |
|
|
|
|
|
|
|
|
|
≤64 | 35 (45.5) | 42 (54.5) | 0.299a | 1.000 | 25 (32.5) | 52 (67.5) | 0.411a | 1.000 |
|
>64 | 43 (53.8) | 37 (46.2) | 0.299b | 0.717
(0.383–1.341) | 31 (38.8) | 49 (61.2) | 0.412b | 0.760
(0.394–1.464) |
| Sex |
|
|
|
|
|
|
|
|
|
Male | 41 (46.5) | 46 (53.5) | 0.475a | 1.000 | 28 (32.2) | 59 (67.8) | 0.310a | 1.000 |
|
Female | 37 (53.5) | 33 (46.5) | 0.496b | 0.803
(0.426–1.511) | 28 (40.0) | 42 (60.0) | 0.322b | 0.717
(0.371–1.385) |
| Family history |
|
|
|
|
|
|
|
|
|
Negative | 71 (49.7) | 72 (50.3) | 0.980a | 1.000 | 51 (35.7) | 92 (64.3) | 0.997a | 1.000 |
|
Positive | 7 (50.0) | 7 (50.0) | 0.975b | 0.983
(0.327–2.956) | 5 (35.7) | 9 (64.3) | 0.993b | 0.995
(0.316–3.136) |
|
Differentiation |
|
|
|
|
|
|
|
|
|
Low | 42 (49.4) | 43 (50.6) | 0.997a | 1.000 | 30 (35.3) | 55 (64.7) | 0.971a | 1.000 |
|
Medium | 30 (50.0) | 30 (50.0) | 0.914b | 1.052
(0.310–3.565) | 22 (36.7) | 38 (63.3) | 0.822b | 1.160
(0.319–4.218) |
|
High | 6 (50.0) | 6 (50.0) | 0.950b | 1.034
(0.298–3.592) | 4 (33.3) | 8 (66.7) | 0.794b | 1.205
(0.320–4.435) |
| Pathological
pattern |
|
|
|
|
|
|
|
|
|
Adenocarcinoma | 65 (45.8) | 77 (54.2) |
0.003a | 1.000 | 47 (33.1) | 95 (66.9) |
0.039a | 1.000 |
|
Mucinous adenocarcinoma | 13 (86.7) | 2 (13.3) |
0.007b | 0.116
(0.025–0.548) | 9 (60.0) | 6 (40.0) |
0.038b | 0.308
(0.101–0.937) |
| Primary organ |
|
|
|
|
|
|
|
|
|
Colon | 46 (61.3) | 29 (38.7) | 0.0051 | 1.000 | 23 (30.7) | 52 (69.3) |
0.2111 | 1.000 |
|
Rectum | 32 (39.0) | 50 (61.0) | 0.0072 | 2.502
(1.290–4.850) | 33 (40.2) | 49 (59.8) |
0.1652 | 0.616
(0.311–1.221) |
| Intestinal tract
occupation |
|
|
|
|
|
|
|
|
|
≤1/2 | 18 (40.0) | 27 (60.0) | 0.124a | 1.000 | 15 (33.3) | 30 (66.7) | 0.699a | 1.000 |
|
>1/2 | 60 (53.6) | 52 (46.4) | 0.142b | 0.586
(0.287–1.197) | 41 (36.6) | 71 (63.4) | 0.749b | 0.885
(0.422–1.857) |
| Infiltration |
|
|
|
|
|
|
|
|
| Invaded
serosa | 66 (51.6) | 62 (48.4) | 0.322a | 1.000 | 41 (36.7) | 81 (63.3) | 0.564a | 1.000 |
|
Non-invaded serosa | 12 (41.4) | 17 (58.6) | 0.318b | 1.518
(0.669–3.445) | 9 (31.0) | 20 (69.0) | 0.559b | 1.295
(0.544–3.081) |
| Size (cm) |
|
|
|
|
|
|
|
|
| ≤6 | 39 (44.8) | 48 (55.2) | 0.175a | 1.000 | 34 (39.1) | 53 (60.9) | 0.320a | 1.000 |
|
>6 | 39 (55.7) | 31 (44.3) | 0.177b | 0.646
(0.342–1.219) | 22 (31.4) | 48 (68.6) | 0.316b | 1.406
(0.723–2.734) |
| Adhesion |
|
|
|
|
|
|
|
|
|
Negative | 74 (49.7) | 75 (50.3) | 0.985a | 1.000 | 53 (35.6) | 96 (64.4) | 0.912a | 1.000 |
|
Positive | 4 (50.0) | 4 (50.0) | 0.931b | 0.939
(0.224–3.939) | 3 (37.5) | 5 (62.5) | 0.869b | 0.883
(0.201–3.881) |
| Metastasis |
|
|
|
|
|
|
|
|
|
Negative | 62 (48.4) | 66 (51.6) | 0.512a | 1.000 | 43 (33.6) | 85 (66.4) | 0.254a | 1.000 |
|
Positive | 16 (55.2) | 13 (44.8) | 0.451b | 0.730
(0.321–1.656) | 13 (44.8) | 16 (55.2) | 0.222b | 0.596
(0.260–1.366) |
| Vascular
invasion |
|
|
|
|
|
|
|
|
|
Negative | 69 (48.9) | 72 (51.1) | 0.579a | 1.000 | 52 (36.9) | 89 (63.1) | 0.347a | 1.000 |
|
Positive | 9 (56.3) | 7 (43.8) | 0.573b | 0.741
(0.260–2.107) | 4 (25.0) | 12 (75.0) | 0.354b | 1.752
(0.536–5.728) |
| DFS |
|
|
|
|
|
|
|
|
|
Negative | 18 (36.7) | 31 (63.3) |
0.029a | 1.000 | 11 (22.4) | 38 (77.6) |
0.020a | 1.000 |
|
Positive | 60 (55.6) | 48 (44.4) |
0.033b | 0.467
(0.232–0.940) | 45 (41.7) | 63 (58.3) |
0.023b | 0.406
(0.186–0.883) |
| OS |
|
|
|
|
|
|
|
|
|
Survives | 22 (37.9) | 36 (62.1) |
0.024a | 1.000 | 14 (23.7) | 45 (76.3) |
0.015a | 1.000 |
|
Deceased | 56 (56.6) | 43 (43.4) |
0.029b | 0.474
(0.243–0.925) | 42 (42.9) | 56 (57.1) |
0.018b | 0.418
(0.203–0.862) |
In the primary location stratification analysis, 61%
of patients with rectal cancer exhibited high expression of
LOC554202, whereas only 38.7% of patients with colon cancer
exhibited high expression of LOC554202 (P=0.005). The adjusted
logistic regression analysis demonstrated the following parameters:
P=0.007, corresponding adjusted OR (95% CI)=2.502 (1.290–4.850).
Based on these results, it was deduced that higher expression
levels of LOC554202 were more likely to occur in rectal cancer than
in colon cancer (Table III).
Analysis of DFS indicated that 44.4% of the patients
with tumor progression exhibited high expression of LOC554202,
whereas the percentage of patients without disease progression was
63.3%. The data suggested that DFS exhibited a significant
correlation with the expression of LOC554202 (P=0.029). The
parameters estimated by the adjusted logistic regression analysis
were as follows: P=0.033, corresponding adjusted OR (95% CI)=0.467
(0.232–0.940). In addition, miR-31 exhibited high expression in
58.6% of patients with tumor progression and 77.6% of patients
without tumor progression. The proportion with elevated expression
of miR-31 was higher in the population without tumor progression
(P=0.020). The P-value and OR derived by logistic regression
analysis were P=0.023 and adjusted OR (95% CI)=0.406 (0.186–0.883),
respectively. The aforementioned results indicated that patients
with low expression levels of LOC554202 and miR-31 were more prone
to tumor progression (Table
III).
Following assessment of the variable OS, the results
indicated that 62.5% of patients exhibited high expression levels
of LOC554202 in surviving patients, whereas only 43.4% of the
patients who succumbed to mortality exhibited high expression of
LOC554202 (P=0.024). The data were analyzed by logistic regression
analysis and the results were as follows: P=0.029 and adjusted OR
(95% CI)=0.474 (0.243–0.925). The expression of miR-31 was high in
57.1% of patients who succumbed to mortality and 76.3% of patients
who survived. The elevated expression of miR-31 was significantly
different between these two groups of patients [P=0.015, adjusted
by logistic regression analysis: P=0.018, adjusted OR (95%
CI)=0.418 (0.203–0.862). These data indicated that patients with
high expression levels of miR-31 and LOC554202 were more likely to
survive (Table III).
No significant differences were found between the
expression of miR-31 and/or LOC554202 and age, sex, family history,
tumor differentiation, tumor size, infiltration, adhesions and/or
organ metastasis were noted (Table
III).
The correlation between the elevated expression
levels of LOC554202 and miR-31 and the clinicopathological
parameters of patients treated with platinum-based chemotherapy for
CRC were analyzed. The patients that exhibited high expression of
miR-31 had high expression of LOC554202 in the colon and in the
rectum at percentages of 44.2 and 67.3%, respectively. This
association was significant (P=0.019) and the P-value was decreased
following adjusted logistic regression analysis [P=0.015, adjusted
OR (95% CI)=2.857 (1.127–6.651)]. LOC554202 and miR-31 were
expressed at high levels concomitantly in the rectum (Table IV). In the tumor progression group
of subjects, 71.1% of patients with high expression of LOC554202
presented without tumor progression, whereas 46.0% of patients with
high expression of LOC554202 were noted in the tumor progression
group (P=0.014). The corresponding statistical parameters following
adjustment by logistic regression analysis were as follows:
P=0.008, adjusted OR (95% CI)=0.301 (0.124–0.726) (Table IV). Therefore, LOC554202 and miR-31
exhibited synergistic high expression, which was correlated with
the inhibition of tumor progression. Among patients who survived
and those who succumbed to mortality, expression of LOC554202 was
high in 71.1 and 42.5%, respectively (P=0.005). The corresponding
statistical parameters following adjustment by logistic regression
analysis were as follows: P=0.004, adjusted OR (95% CI)=0.283
(0.121–0.662). The co-expression of LOC554202 and miR-31 were
associated with higher survival rates (Table IV).
 | Table IV.Correlation between the expression of
LOC554202 and miR-31 status in patients with colorectal cancer
treated with platinum-based chemotherapy |
Table IV.
Correlation between the expression of
LOC554202 and miR-31 status in patients with colorectal cancer
treated with platinum-based chemotherapy
|
| LOC554202
expression |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Low | High |
P-valuea,b | Adjusted OR (95%
CI)c |
|---|
| All patients |
| miR-31 |
| Low
expression | 33 (58.9) | 23 (41.1) | 0.084a | 1 (reference) |
| High
expression | 45 (44.6) | 56 (55.4) | 0.097b | 1.754
(0.903–3.408) |
| miR-31 low
expression |
| Primary organ |
|
Colon | 17 (73.9) | 6
(26.1) | 0.057a | 1 (reference) |
|
Rectum | 16 (48.5) | 17 (51.5) | 0.216b | 2.141
(0.642–7.143) |
| DFS |
|
Negative | 7
(63.6) | 4
(36.4) | 0.723a | 1 (reference) |
|
Positive | 26 (57.8) | 19 (42.2) | 0.683b | 1.345
(0.325–5.572) |
| OS |
|
Negative | 9
(64.3) | 5
(35.7) | 0.638a | 1 (reference) |
|
Positive | 24 (57.1) | 18 (42.9) | 0.511b | 1.551
(0.419–5.743) |
| miR-31 high
expression |
| Primary organ |
|
Colon | 29 (55.8) | 23 (44.2) |
0.019a | 1 (reference) |
|
Rectum | 16 (32.7) | 33 (67.3) |
0.015b | 2.857
(1.127–6.651) |
| DFS |
|
Negative | 11 (28.9) | 27 (71.1) |
0.014a | 1 (reference) |
|
Positive | 34 (54.0) | 29 (46.0) |
0.008b | 0.301
(0.124–0.726) |
| OS |
|
Negative | 13 (28.9) | 32 (71.1) |
0.005a | 1 (reference) |
|
Positive | 32 (57.1) | 24 (42.9) |
0.004b | 0.283
(0.121–0.662) |
| Primary organ:
Colon |
| miR-31 |
| Low
expression | 17 (73.9) | 6
(26.1) | 0.137a | 1 (reference) |
| High
expression | 29 (55.8) | 23 (44.2) | 0.143b | 2.257
(0.759–6.710) |
| DFS |
|
Negative | 11 (44.0) | 14 (56.0) |
0.029a | 1 (reference) |
|
Positive | 35 (70.0) | 15 (30.0) |
0.034b | 0.334
(0.121–0.921) |
| OS |
|
Negative | 15 (48.4) | 16 (51.6) | 0.053a | 1 (reference) |
|
Positive | 31 (70.5) | 13 (29.5) | 0.060b | 0.394
(0.150–1.039) |
| Primary organ:
Rectum |
| miR-31 |
| Low
expression | 16 (48.5) | 17 (51.5) | 0.150a | 1 (reference) |
| High
expression | 16 (32.7) | 33 (67.3) | 0.256b | 1.708
(0.678–4.301) |
| DFS |
|
Negative | 7
(29.2) | 17 (70.8) | 0.239a | 1 (reference) |
|
Positive | 25 (43.1) | 33 (56.9) | 0.177b | 0.492
(0.175–1.378) |
| OS |
|
Negative | 7
(25.0) | 21 (75.0) | 0.061a | 1 (reference) |
|
Positive | 25 (46.3) | 29 (53.7) |
0.040b | 0.343
(0.124–0.951) |
In patients with colon cancer, 56.0% of the subjects
had high expression of LOC554202 in the absence of tumor
progression. However, only 30.0% of patients with tumor progression
exhibited high expression of LOC554202 [P=0.029, adjusted by
logistic regression P=0.034, corresponding adjusted OR (95%
CI)=0.334 (0.121–0.921)] (Table
IV). Therefore, it was hypothesized that the elevated
expression of LOC554202 is associated with the inhibition of the
progression of colon cancer, whereas the low expression of this
gene in colon cancer is more likely to facilitate the tumor
progression process.
Based on the aforementioned analysis, the expression
levels of miR-31, which were associated with high and/or low
expression of LOC554202 in patients with CRC exhibited a
significant correlation with the parameters of CRC primary
location, DFS and OS (Table V).
 | Table V.miR-31 correlation with DFS and OS in
relation to high and/or low expression of LOC554202 in patients
with colorectal cancer. |
Table V.
miR-31 correlation with DFS and OS in
relation to high and/or low expression of LOC554202 in patients
with colorectal cancer.
|
| miR-31
expression |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Low | High |
P-valuea,b | Adjusted OR (95%
CI)c |
|---|
| Loc554202 low
expression |
| Primary organ |
| Colon | 17 (37.0) | 29 (63.0) | 0.251a | 1 (reference) |
| Rectum | 16 (50.0) | 16 (50.0) | 0.078b | 0.409
(0.151–1.104) |
| DFS |
| Negative | 7 (38.9) | 11 (61.1) | 0.738a | 1 (reference) |
| Positive | 26 (43.3) | 34 (56.7) | 0.867b | 0.910
(0.301–2.749) |
| OS |
| Negative | 9 (40.9) | 13 (59.1) | 0.875a | 1 (reference) |
| Positive | 24 (42.9) | 32 (57.1) | 0.921b | 0.949
(0.339–2.655) |
| Loc554202 high
expression |
| Primary organ |
| Colon | 6 (20.7) | 23 (79.3) | 0.209a | 1 (reference) |
| Rectum | 17 (33.3) | 33 (64.7) | 0.284b | 0.548
(0.182–1.645) |
| DFS |
| Negative | 4 (12.9) | 27 (87.1) |
0.011a | 1 (reference) |
| Positive | 19 | 29 |
0.007b | 0.185
(0.055–0.626) |
| OS |
| Negative | 5 | 32 |
0.004a | 1 (reference) |
| Positive | 18 | 24 |
0.003b | 0.176
(0.056–0.553) |
| Primary organ:
Colon |
| Loc554202 |
| Low expression | 17 | 29 | 0.137a | 1 (reference) |
| High
expression | 6 | 23 | 0.143b | 2.254
(0.759–6.698) |
| DFS |
| Negative | 3 | 22 |
0.013a | 1 (reference) |
| Positive | 20 | 30 |
0.023b | 0.208
(0.054–0.804) |
| OS |
| Negative | 4 | 27 |
0.005a | 1 (reference) |
| Positive | 19 | 25 |
0.008b | 0.185
(0.054–0.638) |
| Primary organ:
Rectum |
| Loc554202 |
| Low expression | 16 | 16 | 0.150a | 1 (reference) |
| High
expression | 17 | 33 | 0.256b | 1.708
(0.678–4.301) |
| DFS |
| Negative | 8 | 16 | 0.412a | 1 (reference) |
| Positive | 25 | 33 | 0.309b | 0.591
(0.215–1.629) |
| OS |
| Negative | 10 | 18 | 0.547a | 1 (reference) |
| Positive | 23 | 31 | 0.404b | 0.664
(0.254–1.738) |
Association between the expression of
LOC554202 and/or miR-31 and the survival rates of patients with CRC
treated with platinum-based chemotherapy
The single factor log-rank test and the multivariate
Cox survival risk regression model were used to analyze the
correlation between LOC554202 and miR-31 and the prognosis of
patients with CRC treated with oxaliplatin.
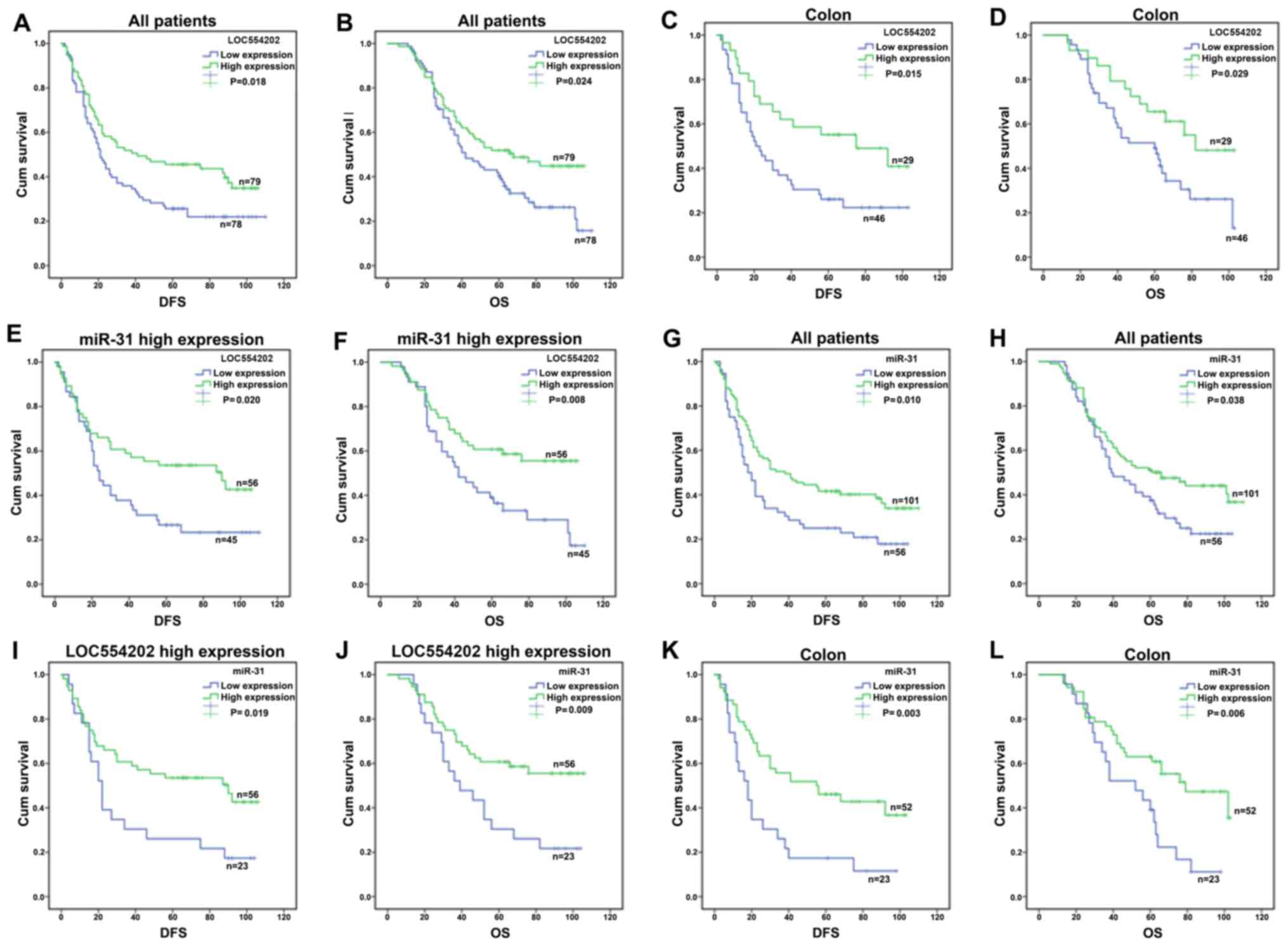
The mean DFS of patients with high expression of
LOC554202 was 56.246 months, and the 95% CI was 46.733–65.759
months. The mean DFS of patients with low expression of LOC554202
was 40.68 months and the 95% CI was 31.765–49.492 months (log-rank:
P=0.018, Fig. 4A). The results
indicated that a high expression of LOC554202 significantly
prolonged the patient DFS (95% CI)=0.647 (0.441–0.949), P=0.026
(Table VI); the expression of
LOC554202 was significantly higher than that in the control group
(P<0.05). Further analysis of the correlation between LOC554202
and OS indicated that the mean survival time (MST) of patients with
high expression of LOC554202 who received oxaliplatin was 67.12
months. The 95% CI was 58.72–75.53 months. Patients with a low
expression of LOC554202 exhibited an MST of 56.19 months (95% CI
48.37–64.01 months). There was a statistically significant
difference according to the log-rank test results (P=0.024,
Fig. 4B). Cox analysis further
indicated that a high expression of LOC554202 was a protective
factor for OS prolongation, corresponding to HR (95% CI)=0.64
(0.43–0.96), P=0.031 (Table
VI).
 | Table VI.Multivariate COX regression analysis
of the association of LOC554202 and miR-31 expression and clinical
pathological features with DFS and OS in patients with colorectal
cancer treated with platinum-based chemotherapy. |
Table VI.
Multivariate COX regression analysis
of the association of LOC554202 and miR-31 expression and clinical
pathological features with DFS and OS in patients with colorectal
cancer treated with platinum-based chemotherapy.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Variable | Total (n) | Events, n (%) | Adjusted HR (95%
CI) | P-value | Total (n) | Events, n (%) | Adjusted HR (95%
CI) | P-value |
|---|
| All patients |
|
|
|
|
|
|
|
|
|
LOC554202 |
|
|
|
|
|
|
|
|
|
Low | 78 | 60 (76.9) | 1 (reference) | − | 78 | 56 (63.6) | 1 (reference) | − |
|
High | 79 | 48 (60.8) | 0.647
(0.441–0.949) | 0.026 | 79 | 42 (53.2) | 0.643
(0.430–0.961) | 0.031 |
| miR-31 |
|
|
|
|
|
|
|
|
|
Low | 56 | 45 (80.4) | 1 (reference) | − | 56 | 42 (75.0) | 1 (reference) | − |
|
High | 101 | 63 (62.4) | 0.620
(0.421–0.911) | 0.015 | 101 | 56 (55.4) | 0.666
(0.446–0.995) | 0.047 |
| LOC554202
expression |
|
|
|
|
|
|
|
|
| miR-31 low
expression |
|
|
|
|
|
|
|
|
|
Low | 33 | 26 (78.8) | 1 (reference) | − | 33 | 24 (72.7) | 1 (reference) | − |
|
High | 23 | 19 (82.6) | 0.905
(0.485–1.689) | 0.754 | 23 | 18 (78.3) | 1.230
(0.646–2.343) | 0.529 |
| miR-31 high
expression |
|
|
|
|
|
|
|
|
|
Low | 45 | 34 (75.5) | 1 (reference) | − | 45 | 32 (71.1) | 1 (reference) | − |
|
High | 56 | 29 (51.8) | 0.520
(0.314–0.861) | 0.011 | 56 | 24 (42.9) | 0.467
(0.272–0.800) | 0.006 |
| miR-31
expression |
|
|
|
|
|
|
|
|
| LOC554202 low
expression |
|
|
|
|
|
|
|
|
|
Low | 33 | 26 | 1 (reference) | − | 33 | 24 | 1 (reference) | − |
|
High | 45 | 34 | 0.834
(0.492–1.411) | 0.498 | 45 | 32 | 1.024
(0.590–1.778) | 0.933 |
| LOC554202 high
expression |
|
|
|
|
|
|
|
|
|
Low | 23 | 19 | 1 (reference) | − | 23 | 18 | 1 (reference) | − |
|
High | 56 | 29 | 0.431
(0.233–0.795) | 0.007 | 56 | 24 | 0.390
(0.204–0.746) | 0.004 |
| Colon |
|
|
|
|
|
|
|
|
|
LOC554202 |
|
|
|
|
|
|
|
|
|
Low | 46 | 35 | 1 (reference) |
| 46 | 31 | 1 (reference) |
|
|
High | 29 | 15 | 0.495
(0.269–0.912) | 0.024 | 29 | 13 | 0.490
(0.255–0.940) | 0.032 |
| miR-31 |
|
|
|
|
|
|
|
|
|
Low | 23 | 20 | 1 (reference) |
| 23 | 19 | 1 (reference) |
|
|
High | 52 | 30 | 0.463
(0.258–0.834) | 0.010 | 52 | 25 | 0.446
(0.240–0.829) | 0.011 |
In addition, significant correlations between the
differential expression of LOC554202 in patients with colon cancer
(n=75) and the DFS and OS were noted (Fig. 4C and D). The mean DFS of the
patients with high expression of LOC554202 was 64.19 months and the
95% CI was 49.34–78.67 months. The mean DFS of patients with low
expression of LOC554202 was 39.74 months and the 95% CI was
28.89–50.59 months. The data indicated that the high expression of
LOC554202 significantly prolonged the DFS in patients with colon
cancer. Furthermore, the results indicated that the MST of patients
with high expression of LOC554202 was 74.78 months and the 95% CI
was 62.988–86.761 months, whereas the MST of patients with a low
expression of LOC554202 was 58.3 months with a 95% CI range of
48.48–67.58 months (P=0.029, Fig.
4D).
The patients were further stratified and analyzed
according to expression levels of miR-31. A significant correlation
between the differential expression of LOC554202 and the DFS and OS
in patients with high expression of miR-31 was noted (n=101)
(Fig. 4E and F). The mean DFS of
patients with high co-expression of miR-31 and LOC554202 (n=56) was
63.61 months (95% CI was 52.18–75.05 months), whereas the mean DFS
of patients with low expression of LOC554202 and high expression of
miR-31 (n=45) was 43.70 months with a 95% CI of 31.839–55.094
months (P=0.020, Fig. 4E).
Corrected multivariate Cox analysis further suggested that a high
expression of LOC554202 was a protective factor for DFS
prolongation in patients with high expression of miR-31
[corresponding HR (95% CI)=0.520 (0.314–0.861)] (P=0.011). The
analysis of OS demonstrated that the MST in patients with high
expression of miR-31 and LOC554202 was 73.74 months, and the 95% CI
was 63.70–83.78 months, whereas that of the patients with low
expression of LOC554202 and high expression of miR-31was 56.69
months with a 95% CI range of 46.04–67.34 months. These parameters
were statistically different according to the log-rank test
(P=0.008, Fig. 4F).
The analysis of the association between the miRNA
differential expression and prognosis indicated that patients with
high expression of miR-31 exhibited an average DFS of 55.582
months, with a 95% CI range of 46.93–64.24 months; the patients
with low expression of miR-31 had an average DFS of 36.63 months
and the 95% CI was 27.02–46.24 months (log-rank: P=0.010, Fig. 4G). Further analysis of the
correlation between miR-31 and OS demonstrated that the MST in
patients with high expression of miR-31 was 67.38 months with a 95%
CI range of 59.63–75.13 months, whereas the corresponding MST of
the patients with low expression of miR-31 was 53.63 months and the
95% CI was 45.18–62.08 months (P=0.038, Fig. 4H). In addition, the corrected
multivariate Cox analysis indicated that a high expression of
miR-31 was a protective factor for extended OS [HR (95% CI)=0.666
(0.446–0.995)] (P=0.047, Table
VI). The same as previous studies, the mean DFS of patients
with high co-expression of miR-31 and LOC554202 (n=56) was 63.61
months (95% CI was 52.18–75.05 months), whereas the mean DFS of
patients with low expression of miR-31 and high expression of
LOC554202 (n=23) was 38.565 months with a 95% CI of 23.838–53.293
months (P=0.019; Fig. 4I).
Corrected multivariate Cox analysis further suggested that a high
expression of miR-31 was a protective factor for DFS prolongation
in patients with high expression of LOC554202 [corresponding HR
(95% CI)=0.431 (0.233–0.795)] (P=0.007). The analysis of OS
indicated that the MST in patients with high expression of miR-31
and LOC554202 was 73.74 months, and the 95% CI was 63.70–83.78
months, whereas that of the patients with low expression of miR-31
and high expression of LOC554202 was 51.391 months with a 95% CI
range of 38.208–64.574 months. These parameters were statistically
different according to the log-rank test (P=0.009; Fig. 4J). Corrected multivariate Cox
analysis further suggested that a high expression of miR-31 was a
protective factor for MST in patients with high expression of
LOC554202 [corresponding HR (95% CI)=0.390 (0.204–0.746)]
(P=0.004).
The expression of miR-31 was further analyzed with
regard to the primary location of the tumor. In patients with CRC,
the mean DFS of the patients with a high expression of miR-31 was
57.62 months and the 95% CI was 46.46–68.77 months. The mean DFS of
patients with a low expression of miR-31 was 29.49 months and the
95% CI was 17.24–41.75 months (log-rank: P=0.003, Fig. 4K). The data indicated that miR-31
and LOC554202 not only significantly improved the DFS of patients
with platinum-based chemotherapy, but also improved the OS. The
association between the increase in DFS and OS and the disease
incidence was present in patients with colon cancer but not in
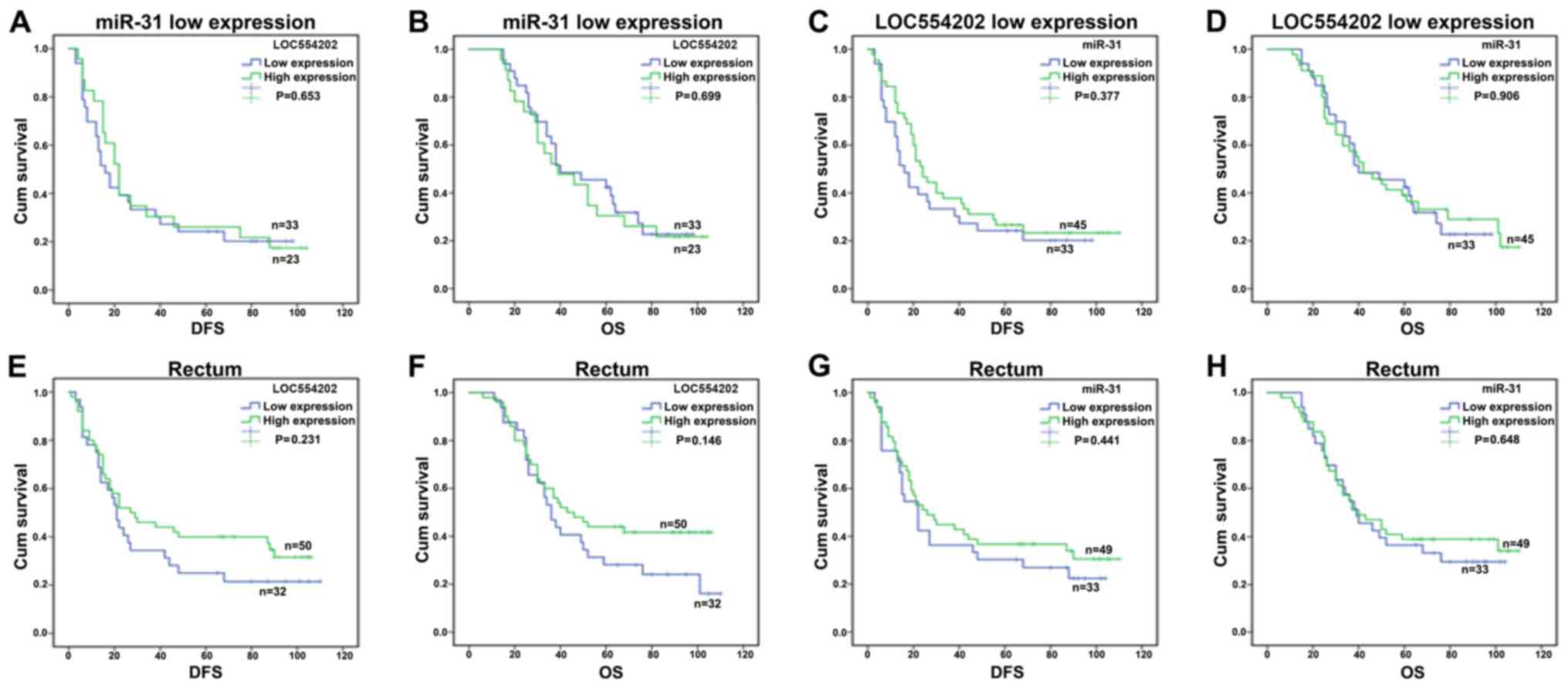
patients with rectal cancer (Fig.
5). In addition, the correlations between the remaining
clinicopathological parameters and DFS and OS were analyzed by
multiple factors Cox analysis. The data demonstrated that family
history (P=0.037) and tumor organ metastasis (P=0.033) were risk
factors for the prognosis of patients (Table VII).
 | Table VII.Correlation between other
clinicopathological parameters and DFS and OS. |
Table VII.
Correlation between other
clinicopathological parameters and DFS and OS.
|
| DFS | OS |
|---|
|
|
|
|
|---|
| Feature | Total | Events | Adjusted HR
(95%CI) | P-value | Total | Events | Adjusted HR
(95%CI) | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
|
|
|
≤64 | 77 | 51 | 1 (reference) | − | 77 | 46 | 1 (reference) | − |
|
>64 | 80 | 57 | 1.118
(0.766–1.632) | 0.562 | 80 | 52 | 1.202
(0.809–1.785) | 0.362 |
| Family history |
|
|
|
|
|
|
|
|
|
Negative | 143 | 96 | 1 (reference) | − | 143 | 88 | 1 (reference) | − |
|
Positive | 14 | 12 | 1.901
(1.041–3.474) | 0.037 | 14 | 10 | 1.654
(0.858–3.189) | 0.133 |
| Sex |
|
|
|
|
|
|
|
|
|
Male | 86 | 59 | 1 (reference) | − | 86 | 51 | 1 (reference) | − |
|
Female | 71 | 49 | 1.161
(0.794–1.697) | 0.442 | 71 | 47 | 1.148
(0.771–1.710) | 0.498 |
| Pathological
pattern |
|
|
|
|
|
|
|
|
|
Adenocarcinoma | 142 | 95 | 1 (reference) | − | 142 | 87 | 1 (reference) | − |
|
Mucinous adenocarcinoma | 15 | 13 | 1.360
(0.756–2.450) | 0.305 | 15 | 11 | 1.133
(0.597–2.151) | 0.702 |
| Tumor size
(cm) |
|
|
|
|
|
|
|
|
| ≤6 | 87 | 65 | 1 (reference) | − | 87 | 50 | 1 (reference) | − |
|
>6 | 70 | 43 | 0.702
(0.475–1.037) | 0.075 | 70 | 48 | 0.757
(0.505–1.134) | 0.177 |
| Intestinal tract
occupation |
|
|
|
|
|
|
|
|
|
≤1/2 | 45 | 29 | 1 (reference) | − | 45 | 26 | 1 (reference) | − |
|
>1/2 | 112 | 79 | 1.258
(0.816–1.939) | 0.299 | 112 | 72 | 1.229
(0.779–1.938) | 0.376 |
| Infiltration |
|
|
|
|
|
|
|
|
| Invaded
serosa | 128 | 87 | 1 (reference) | − | 128 | 79 | 1 (reference) | − |
|
Non-invaded serosa | 29 | 21 | 1.045
(0.648–1.684) | 0.857 | 29 | 19 | 1.027
(0.621–1.697) | 0.918 |
| Metastasis |
|
|
|
|
|
|
|
|
|
Negative | 128 | 86 | 1 (reference) | − | 128 | 77 | 1 (reference) | − |
|
Positive | 29 | 22 | 1.673
(1.042–2.688) | 0.033 | 29 | 21 | 1.874
(1.147–3.061) | 0.012 |
| Vascular
invasion |
|
|
|
|
|
|
|
|
|
Negative | 141 | 98 | 1 (reference) | − | 141 | 91 | 1 (reference) | − |
|
Positive | 16 | 10 | 0.736
(0.383–1.417) | 0.359 | 16 | 7 | 0.681
(0.314–1.475) | 0.329 |
| Adhesion |
|
|
|
|
|
|
|
|
|
Negative | 149 | 101 | 1 (reference) | − | 149 | 92 | 1 (reference) | − |
|
Positive | 8 | 7 | 1.715
(0.793–3.713) | 0.171 | 8 | 6 | 2.004
(0.872–4.603) | 0.102 |
| Primary organ |
|
|
|
|
|
|
|
|
|
Colon | 75 | 50 | 1 (reference) | − | 75 | 44 | 1 (reference) | − |
|
Rectum | 82 | 58 | 1.229
(0.828–1.825) | 0.307 | 82 | 54 | 1.414
(0.931–2.148) | 0.104 |
Discussion
To the best of our knowledge, the present study is
the first to report that the elevated expression of miR-31 and
LOC554202 are optimal predictors for the prognosis of patients with
oxaliplatin-treated CRC. These findings may provide novel insights
into CRC treatment.
It has been reported that at least 25% of miRNA
sequences are located in the host gene, a number of which are
highly conserved (28). Baskerville
and Bartel reported that the majority of intron miRNAs and their
host genes possess a related expression profile, where their
expression levels are consistent with each other, and both are
controlled by the promoter of the host gene, suggesting that they
are related at a functional and/or translational level (18). However, the expression levels of
certain miRNA-coding genes are inconsistent with the expression
levels of their corresponding host genes, and/or the opposite
pattern is observed (29). In the
present study, the expression of LOC554202 was shown to be present
in CRC tissues (H-score=155.24), compared with normal intestinal
mucosal tissues, and the expression of miR-31 was high in CRC
(H-score=226.80). In addition, the expression levels of LOC554202
and miR-31 were correlated in CRC tissues in a concomitant pattern,
which indicated increased and/or decreased co-expression of the two
sequences. These results suggested that miR-31 may exert a
regulatory role and/or a positive feedback towards the host gene
LOC554202. The association between intron miRNAs and host genes
remains to be fully elucidated, and different expression patterns
exist between the two. It has been reported that ~1/3 of intron
miRNAs have independent promoters and certain miRNAs have different
processing pathways following transcription (30), and that these factors can lead to
inconsistent expression levels of miRNAs compared with their host
genes. LOC554202 and miR-31 share a common promoter (31), although additional possible
mechanisms may affect the coherence of their expression.
In the present study, LOC554202 was expressed at
high levels in CRC tissues compared with normal intestinal mucosal
tissues, in contrast to a previous study by Ding et al
(32). The levels of miR-19a,
miR-21, miR-29a, miR-92, miR-148a and miR-200b were increased in
CRC tissues according to miRNA expression profiles, whereas the
expression of miR-30c, miR-133a and miR-145 were decreased. By
comparison, it was found that the miRNA expression profiles from
cell lines and cancer tissues were different. Therefore, the miRNA
expression profile obtained from cell lines may not be suitable for
deducing the corresponding expression profile of clinical samples.
This may account for the contradictory findings reported by Augoff
et al, where it was shown that both miR-31 and LOC554202
were downregulated in triple negative breast cancer cell lines
(5), whereas the upregulation of
these two markers was observed in the present study. It is
important to note that Augoff et al investigated breast and
cancer, not CRC, cell lines, thus the findings may be attributed to
the different tissue origin of the tumor to that in the present
study. In the low invasive breast cancer, LOC554202 and miR-31 were
expressed at low levels (5).
Therefore, the roles of LOC554202 in the development of the tumor
are complex, depending on the different tissues of the tumor, and
the different environments and conditions, all of which may have a
regulatory effect on cell behavior. Bandre et al used
polymerase chain reaction analysis to detect 156 miRNAs from 15 CRC
cell lines. The expression levels were compared with the
corresponding expression levels in the CCD-18Co human normal
colonic cell line. The authors reported that the expression levels
of miR-31 were significantly different (17). In addition, the expression levels of
miR-31 were associated with the clinical stage of CRC, which was
significantly higher in stage IV CRC than stage II CRC. The probe
hybridization technique used in the present study may be
responsible for the conflicting results compared with the
experimental methods applied by Yang et al (21) and Ding et al (32). However, the study by Ding et
al confirmed that the expression level of LOC554202 was
associated with the occurrence and development of colon cancer and
its high expression was a predictor of colon cancer.
In the present study, the high expression of
LOC554202 and miR-31ncRNAs prolonged the survival rate of patients
with colon cancer who received chemotherapy following surgery,
although no correlation in patients with rectal cancer was noted.
Previous studies have shown that the role of LOC554202 in the
development of tumors is complex. Shi et al demonstrated
that LOC554202 was expressed at high levels in MDA-MB-231 and
MDA-MB-435S breast cancer cell lines, and LOC554202 was associated
with tumor staging (19). Following
silencing of the LOC554202 gene, cell proliferation was decreased,
apoptosis was increased, and migratory ability was decreased
(19). The results of the study by
Yang et al support the hypothesis that the expression of
LOC554202 correlates with prognosis (21). In addition, Ding et al
indicated that the expression of LOC554202 was low in CRC, and
inhibited cell proliferation and induced apoptosis by activating
the caspase signaling pathway (32). The results of the present study
suggested that LOC554202 can be used as an independent biomarker
for the prediction of the postoperative prognosis of CRC.
The high expression of miR-31 in breast cancer
inhibits the activity and aggressiveness of breast cancer cells and
improves the OS rate of patients (33). Studies performed in patients with
gastric cancer have shown that miR-31 was expressed at low levels
in cancer tissues and can inhibit cell invasion and migration
(34); studies in hepatocellular
carcinoma have further shown that the overexpression of miR-31 can
induce the inhibition of cell proliferation and migration (35). In CRC, miR-31 is expressed at high
levels in cancer tissues and cells, and its expression levels may
be associated with the pathogenesis, invasion, staging, and
migration of CRC (8,9,23,36).
The results of the present study demonstrated that patients with
high expression levels of LOC554202 and miR-31, who were treated
with platinum-based chemotherapy postoperatively, exhibited longer
DFS and OS times and reduced tumor progression, compared with
patients without treatment. These findings suggested that
platinum-based chemotherapeutic drugs can regulate the two
lncRNA-related genes and/or signaling pathways, which may result in
enhanced growth inhibition and the inhibition of CRC cell
invasion.
The effects of miR-31 on the efficacy of platinum
compounds have been discussed extensively. It has been shown that
the expression levels of miR-31 are negatively correlated with E2F
transcription factor 2 (E2F2) and Enhancer of zeste homolog 2
(EZH2) in malignant tumors including gastric, ovarian and bladder
cancer, and CRC (37–41). E2F2 and EZH2 can enhance drug
resistance by inhibiting platinum-drug-mediated apoptosis, whereas
low expression levels of E2F2 and EZH2 enhance platinum
chemosensitivity (39,42–45).
Li et al reported that miR-31 in gallbladder cancer
sensitized tumor cells to platinum-based chemotherapy via control
of the Src proto-oncogene (40).
The results of the present study suggested that miR-31 was
expressed at a high level in CRC and was associated with longer DFS
and OS times following platinum-based chemotherapy. The data
suggested that this effect may be associated with miR-31 and/or
other associated factors that enhance drug sensitivity. A high
expression of miR-31 in CRC may improve the cell sensitivity to
platinum-based agents and enhance platinum-induced apoptosis via
E2F2, EZH2 and/or associated signaling transcription factors, which
may facilitate the efficacy of chemotherapeutic agents in order to
increase the survival rate of the patients following
chemotherapy.
The present study is in alignment with previous
studies that have reported LOC554202 as a putative biomarker for
CRC (21,32). Although LOC554202 was shown to be
downregulated in colon tumors, its expression pattern was
associated with the OS and DFS times of 178 patients with CRC,
which was in agreement with the other results of the present study.
In addition, miR-31 has been demonstrated to exert an oncogenic
role in colon cancer, which may partly explain the elevated
expression of LOC554202 in colon tumors noted in the present study
(46). It is important to note that
the present study is one of the few studies to have investigated
the concomitant expression of miR-31 and LOC554202 in patients with
CRC treated with oxaliplatin and their putative associations with
DFS and OS times.
Although colonic and rectal mucosal cells exert
various identical and/or similar cellular functions and signaling
pathways, differences have been reported. For example, Ayiomamitis
et al demonstrated that high expression levels of mutL
homolog 1 (MLH1) in colon cancer tissues and adjacent normal
tissues were higher than those in patients with rectal cancer and
adjacent tissues (41). The
silencing of MLH1 in lung cancer cells leads to increased
expression of miR-31, which may be responsible for the different
expression levels of miR-31 between rectal and colon cancer
(47). Concomitantly, the high
expression levels of miR-31 and LOC554202 in rectal cancer may be
due to the feedback regulation of miR-31 toward the host gene
LOC554202. However, as the length of the tumor was not counted from
the distance of the anus, the hypothesis that high expression
levels of LOC554202 and miR-31 are associated with specific tissue
regions of the colorectal area requires further investigation.
Although the majority of studies have concentrated
on the expression analysis of miRNA-31 in clinical samples, few
studies have demonstrated a plausible mechanism by which these
effects are mediated. Tazawa et al showed that no induction
of miR-34 was noted in p53−/− cells following treatment
with DNA-damaging agent Adriamycin (15). This tumor suppressive role was
promoted via the E2F1 and E2F3 proteins and was independent of
apoptosis and caspase 3 activation (15).
It is important to note that the control of the
expression of miR-31 and its host gene LOC554202 may be attributed
to epigenetic regulation via mechanisms involving methylation
and/or acetylation. It has been shown that the proximal CpG islands
located within the promoter or 5′ untranslated regions of miRNAs
can epigenetically silence gene expression through DNA
hypermethylation (48). Previous
studies have demonstrated that certain miRNA genes with their
corresponding host genes (hsa-mir-10a/HOXB4, hsa-mir-126/EGFL7,
hsa-mir-152/COPZ2, hsa-mir-191/DALRD3, and
hsa-mir-342/EVL) were found to be epigenetically
downregulated, and/or silenced either by histone modification
and/or CpG island hypermethylation in the promoter region in cancer
cells (48). Methylated mir-155,
mir-152, mir-137, and mir-31 host genes are considered as promising
diagnostic and/or prognostic biomarkers of prostate cancer, whereas
the methylation status of particular miRNA host genes as
independent variables and/or in combinations may assist in the
prediction of poor prognosis in patients with prostate cancer
preoperatively (49). With regard
to miR-31, it has been shown that promoter methylation was
predictive of biochemical prostate cancer recurrent survival rate
(49). This suggests that the
silencing of this gene can affect its expression and corresponding
function with regard to cancer development (49). In addition, Augoff et al
demonstrated that loss of miR-31 expression in triple negative
breast cancer cell lines was attributed to hypermethylation of its
promoter-associated CpG island (5).
Further investigations on the epigenetic regulation of miRNA/genes,
including miR-31, may reveal novel approaches for the prevention or
treatment of human cancer. Similarly, the contribution of miR-31
polymorphisms to the survival rates of patients with CRC cannot be
ruled out and requires investigation in future investigations. To
date, polymorphisms of miR-31 have been described in the
involvement of this RNA in the regulation of angiotensinogen
expression and body fat distribution (50). The miR-31 rs13283671 polymorphism
was previously described in lung cancer, although it was not
correlated with survival rate and/or certain clinicopathological
parameters (51).
The findings of the present study may initially
appear contradictory to the general hypothesis that a high
expression of miR-31 contributes to CCR development. LOC554202 is
an lncRNA, reported to be expressed at a high level in breast
cancer and associated with advanced pathologic stage and tumor size
(19). However, in CRC cells,
LOC554202 has a tumor suppressor role in the activation of specific
caspase cleavage cascades, and low expression of LOC554202 was
associated with advanced pathologic stage and increased tumor size
(32). miR-31 is a widely
investigated miRNA and studies have reported controversial
conclusions regarding its role in CRC cells (52–56).
These studies suggest that a high expression of this miRNA in
cancer does not always result in a decreased tumor growth,
including that noted in cases of the oncosuppressors p53 and
phosphatase and tensin homolog. In the present study, it was found
that LOC554202 and miR-31 were expressed at high levels in
oxaliplatin-treated CRC specimens, and their higher expression
predicted optimal outcomes. As double-edged genes, the high
expression levels of LOC554202 and miR-31 may be a responsive
adaptation to oxaliplatin treatment. Investigations of the
underlying mechanism of LOC554202 and miR-31 in oxaliplatin-treated
CRC are ongoing.
Previously published studies have produced
contradictory results with regard to the application of plasma
levels of miR-31. It has been shown that miR-31 levels in plasma
are upregulated in patients with CRC compared with healthy
subjects, and that clinical analysis of the expression of miR-31 in
patients with colon cancer may facilitate the diagnosis of colon
cancer (57). By contrast, Jo et
al investigated the miR profiles of plasma from patients with
rectal cancer during chemoradiotherapy and found that plasma miRNA
expression levels did not necessarily represent the miRNA
expression levels in tumor tissues (58). This indicates that plasma miRs
cannot be used with certainty for prognosis as their expression may
be different with that in tumor tissues. Therefore, further
investigations are required to validate their application as
potential biomarkers in human plasma. The mechanism underlying the
interaction between miR-31 and LOC554202 was not examined in the
present study and warrants further investigation in the future.
Evidence in the literature remains limited, and no information
exists, to the best of our knowledge, with regard to CRC. A
previous study revealed an association with the oncogene RNF144B in
chordoma. Specifically, EZH2, a binding protein of LOC554202, was
positively regulated by LOC554202, leading to the reduced
expression of miR-31 (59).
Furthermore, the impaired function of miR-31 restored expression of
the RNF144B oncogene and maintained the metastasis-promoting
activity in vitro (59).
These results were also shown in vivo (59). This mechanism requires investigation
in CRC in the future.
The present study had several limitations. The
sample size of the study was small, and the nature of the study was
retrospective. Further prospective studies are required to confirm
the prediction efficiency of miR-31 and LOC554202. In addition,
functional in vitro assays can provide insight into the
mechanism of action of LOC554202 in CRC. The implication of
LOC554202 in cancer metastasis can be examined, as it has been
shown that miR-31 contributes to the invasion and migration of CRC
cells by the miR-31/factor-inhibiting hypoxia-inducible factor 1
nexus (60). Loss of function and
gain of function experiments can determine whether the expression
of LOC554202 that is regulated by miR-31 is responsible for cancer
invasion. In addition, a meta-analysis is likely to be a
significant addition to this line of investigation in examining the
expression of miR-31 and LOC554202 in CRC. However, the studies
cited above contain significant heterogeneity for inclusion in a
preliminary meta-analysis. This may be examined in a separate
report to include a careful selection of studies with similar
parameters for investigation, so as to minimize bias.
In conclusion, miR-31 and its homologous host gene
LOC554202 were expressed at high levels and positively correlated
with CRC prognosis. The increased expression of the latter RNA
sequences prolonged the survival rate of patients with CRC, notably
those with colon cancer. miR-31 and LOC554202 may be potential
markers for the evaluation of the prognosis of postoperative
patients treated with oxaliplatin-based chemotherapy. In
vivo and in vitro assays, and a large randomized
controlled trial with a large sample size are warranted to further
elucidate the roles of LOC554202 and miR-31 in CRC.
Acknowledgements
We gratefully appreciate the efforts and
contributions of doctors, nurses and technical staff at the First
Hospital of China Medical University.
Funding
The present study was supported in part by a grant
from the Liaoning Province Education Department Foundation (nos.
LS201617 and LK201646).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YL participated in the literature search, designed
the study, collected the data, analysed and interpreted the data
and wrote the manuscript. SX participated in the design of the and
provided the critical revision. HW, CX, LD, WS, XH, RL and FZ
carried out the data collection and data analysis, and provided the
critical revision. All authors read and approved the final
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of the China Medical University at the First Hospital of
China Medical University. Written informed consent was required by
the patients for their participation in the study, as requested by
the ethical guidelines of the First Hospital of China Medical
University and the Medical Ethics Committee of the Chinese Medical
Universities. All patients were anonymized.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nosho K, Igarashi H, Nojima M, Ito M,
Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, et
al: Association of microRNA-31 with BRAF mutation, colorectal
cancer survival and serrated pathway. Carcinogenesis. 35:776–783.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Corcoran DL, Pandit KV, Gordon B,
Bhattacharjee A, Kaminski N and Benos PV: Features of mammalian
microRNA promoters emerge from polymerase II chromatin
immunoprecipitation data. PLoS One. 4:e52792009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yao Y, Li J and Wang L: Large intervening
non-coding RNA HOTAIR is an indicator of poor prognosis and a
therapeutic target in human cancers. Int J Mol Sci. 15:18985–18999.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao XL, Zhao ZH, Xu WC, Hou JQ and Du XY:
Increased expression of SPRY4-IT1 predicts poor prognosis and
promotes tumor growth and metastasis in bladder cancer. Int J Clin
Exp Pathol. 8:1954–1960. 2015.PubMed/NCBI
|
|
15
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–14477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xi Y, Shalgi R, Fodstad O, Pilpel Y and Ju
J: Differentially regulated micro-RNAs and actively translated
messenger RNA transcripts by tumor suppressor p53 in colon cancer.
Clin Cancer Res. 12:2014–2024. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bandres E, Cubedo E, Agirre X, Malumbres
R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, et
al: Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi Y, Lu J, Zhou J, Tan X, He Y, Ding J,
Tian Y, Wang L and Wang K: Long non-coding RNA Loc554202 regulates
proliferation and migration in breast cancer cells. Biochem Biophys
Res Commun. 446:448–453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schee K, Boye K, Abrahamsen TW, Fodstad O
and Flatmark K: Clinical relevance of microRNA miR-21, miR-31,
miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC
Cancer. 12:5052012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang L, Wei H and Xiao HJ: Long non-coding
RNA Loc554202 expression as a prognostic factor in patients with
colorectal cancer. Eur Rev Med Pharmacol Sci. 20:4243–4247.
2016.PubMed/NCBI
|
|
22
|
Xi S, Yang M, Tao Y, Xu H, Shan J,
Inchauste S, Zhang M, Mercedes L, Hong JA, Rao M, et al: Cigarette
smoke induces C/EBP-β-mediated activation of miR-31 in normal human
respiratory epithelia and lung cancer cells. PLoS One.
5:e137642010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang MH, Yu J, Chen N, Wang XY, Liu XY,
Wang S and Ding YQ: Elevated microRNA-31 expression regulates
colorectal cancer progression by repressing its target gene SATB2.
PLoS One. 8:e853532013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dindo D, Demartines N and Clavien PA:
Classification of surgical complications: A new proposal with
evaluation in a cohort of 6336 patients and results of a survey.
Ann Surg. 240:205–213. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang F, Flanagan J, Su N, Wang LC, Bui S,
Nielson A, Wu X, Vo HT, Ma XJ and Luo Y: RNAscope: A novel in situ
RNA analysis platform for formalin-fixed, paraffin-embedded
tissues. J Mol Diagn. 14:22–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yeo W, Chan SL, Mo FK, Chu CM, Hui JW,
Tong JH, Chan AW, Koh J, Hui EP, Loong H, et al: Phase I/II study
of temsirolimus for patients with unresectable Hepatocellular
Carcinoma (HCC)- a correlative study to explore potential
biomarkers for response. BMC Cancer. 15:3952015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Azim HA Jr, Peccatori FA, Brohee S,
Branstetter D, Loi S, Viale G, Piccart M, Dougall WC, Pruneri G and
Sotiriou C: RANK-ligand (RANKL) expression in young breast cancer
patients and during pregnancy. Breast Cancer Res. 17:242015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li SC, Tang P and Lin WC: Intronic
microRNA: discovery and biological implications. DNA Cell Biol.
26:195–207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Y and Gorski DH: Regulation of
angiogenesis through a microRNA (miR-130a) that down-regulates
antiangiogenic homeobox genes GAX and HOXA5. Blood. 111:1217–1226.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yi R, Pasolli HA, Landthaler M, Hafner M,
Ojo T, Sheridan R, Sander C, O'Carroll D, Stoffel M, Tuschl T, et
al: DGCR8-dependent microRNA biogenesis is essential for skin
development. Proc Natl Acad Sci USA. 106:498–502. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ozsolak F, Poling LL, Wang Z, Liu H, Liu
XS, Roeder RG, Zhang X, Song JS and Fisher DE: Chromatin structure
analyses identify miRNA promoters. Genes Dev. 22:3172–3183. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding J, Lu B, Wang J, Wang J, Shi Y, Lian
Y, Zhu Y, Wang J, Fan Y, Wang Z, et al: Long non-coding RNA
Loc554202 induces apoptosis in colorectal cancer cells via the
caspase cleavage cascades. J Exp Clin Cancer Res. 34:1002015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo LJ, Yang F, Ding JJ, Yan DL, Wang DD,
Yang SJ, Ding L, Li J, Chen D, Ma R, et al: MiR-31 inhibits
migration and invasion by targeting SATB2 in triple negative breast
cancer. Gene. 594:47–58. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang H, Zhang X, Liu Y, Ni Z, Lin Y, Duan
Z, Shi Y, Wang G and Li F: Downregulated miR-31 level associates
with poor prognosis of gastric cancer and its restoration
suppresses tumor cell malignant phenotypes by inhibiting E2F2.
Oncotarget. 7:36577–36589. 2016.PubMed/NCBI
|
|
35
|
Du Z, Niu S, Xu X and Xu Q:
MicroRNA31-NDRG3 regulation axes are essential for hepatocellular
carcinoma survival and drug resistance. Cancer Biomark. 19:221–230.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B,
Gu J, Chen HY and Sun XF: Clinicopathological significance of
microRNA-31, −143 and −145 expression in colorectal cancer. Dis
Markers. 26:27–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun Y, Jin L, Liu JH, Sui YX, Han LL and
Shen XL: Interfering EZH2 expression reverses the cisplatin
resistance in human ovarian cancer by inhibiting autophagy. Cancer
Biother Radiopharm. 31:246–252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou W, Wang J, Man WY, Zhang QW and Xu
WG: siRNA silencing EZH2 reverses cisplatin-resistance of human
non-small cell lung and gastric cancer cells. Asian Pac J Cancer
Prev. 16:2425–2430. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu S, Yu L, Li Z, Shen Y, Wang J and Cai
J: Overexpression of EZH2 contributes to acquired cisplatin
resistance in ovarian cancer cells in vitro and in vivo. Cancer
Biol Ther. 10:788–795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li M, Chen W, Zhang H, Zhang Y, Ke F, Wu
X, Zhang Y, Weng M, Liu Y and Gong W: MiR-31 regulates the
cisplatin resistance by targeting Src in gallbladder cancer.
Oncotarget. 7:83060–83070. 2016.PubMed/NCBI
|
|
41
|
Ayiomamitis GD, Notas G, Zaravinos A,
Zizi-Sermpetzoglou A, Georgiadou M, Sfakianaki O and Kouroumallis
E: Differences in telomerase activity between colon and rectal
cancer. Can J Surg. 57:199–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wen L, Cheng F, Zhou Y and Yin C: MiR-26a
enhances the sensitivity of gastric cancer cells to cisplatin by
targeting NRAS and E2F2. Saudi J Gastroenterol. 21:313–319. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu H, Li W, Yu X, Gao F, Duan Z, Ma X,
Tan S, Yuan Y, Liu L, Wang J, et al: : EZH2-mediated Puma gene
repression regulates non-small cell lung cancer cell proliferation
and cisplatin-induced apoptosis. Oncotarget. 7:56338–56354.
2016.PubMed/NCBI
|
|
44
|
Cai L, Wang Z and Liu D: Interference with
endogenous EZH2 reverses the chemotherapy drug resistance in
cervical cancer cells partly by up-regulating Dicer expression.
Tumour Biol. 37:6359–6369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Q, Padi SK, Tindall DJ and Guo B:
Polycomb protein EZH2 suppresses apoptosis by silencing the
proapoptotic miR-31. Cell Death Dis. 5:e14862014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ito M, Mitsuhashi K, Igarashi H, Nosho K,
Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, et
al: : MicroRNA-31 expression in relation to BRAF mutation, CpG
island methylation and colorectal continuum in serrated lesions.
Int J Cancer. 135:2507–2515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhong Z, Dong Z, Yang L, Chen X and Gong
Z: MicroRNA-31-5p modulates cell cycle by targeting human mutL
homolog 1 in human cancer cells. Tumour Biol. 34:1959–1965. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Godnic I, Zorc M, Jevsinek Skok D, Calin
GA, Horvat S, Dovc P, Kovac M and Kunej T: Genome-wide and
species-wide in silico screening for intragenic MicroRNAs in human,
mouse and chicken. PLoS One. 8:e651652013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Daniunaite K, Dubikaityte M, Gibas P,
Bakavicius A, Rimantas Lazutka J, Ulys A, Jankevicius F and
Jarmalaite S: Clinical significance of miRNA host gene promoter
methylation in prostate cancer. Hum Mol Genet. 26:2451–2461. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Machal J, Novak J, Hezova R, Zlamal F,
Vasku A, Slaby O and Bienertova-Vasku J: Polymorphism in miR-31 and
miR-584 binding site in the angiotensinogen gene differentially
influences body fat distribution in both sexes. Genes Nutr.
10:4882015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen Z, Xu L, Ye X, Shen S, Li Z, Niu X
and Lu S: Polymorphisms of microRNA sequences or binding sites and
lung cancer: A meta-analysis and systematic review. PLoS One.
8:e610082013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tateishi Y, Okudela K, Mitsui H, Umeda S,
Suzuki T, Kojima Y, Watanabe K, Kawano N, Endo I and Ohashi K: The
potential role of microRNA-31 expression in early colorectal
cancer. Pathol Int. 65:513–518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Carames C, Cristobal I, Moreno V, Marin
JP, Gonzalez-Alonso P, Torrejon B, Minguez P, Leon A, Martín JI,
Hernández R, et al: MicroRNA-31 Emerges as a predictive biomarker
of pathological response and outcome in locally advanced rectal
cancer. Int J Mol Sci. 17(pii): E8782016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang X, Xu X, Zhu J, Zhang S, Wu Y, Wu Y,
Zhao K, Xing C, Cao J, Zhu H, et al: miR-31 affects colorectal
cancer cells by inhibiting autophagy in cancer-associated
fibroblasts. Oncotarget. 7:79617–79628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu Z, Bai J, Zhang L, Lou F, Ke F, Cai W
and Wang H: Conditional knockout of microRNA-31 promotes the
development of colitis associated cancer. Biochem Biophys Res
Commun. 490:62–68. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tian Y, Ma X, Lv C, Sheng X, Li X, Zhao R,
Song Y, Andl T, Plikus MV, Sun J, et al: Stress responsive miR-31
is a major modulator of mouse intestinal stem cells during
regeneration and tumorigenesis. Elife. 6(pii):
e295382017.PubMed/NCBI
|
|
57
|
Wang YN, Chen ZH and Chen WC: Novel
circulating microRNAs expression profile in colon cancer: A pilot
study. Eur J Med Res. 22:512017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jo P, Azizian A, Salendo J, Kramer F,
Bernhardt M, Wolff HA, Gruber J, Grade M, Beißbarth T, Ghadimi BM
and Gaedcke J: Changes of microrna levels in plasma of patients
with rectal cancer during chemoradiotherapy. Int J Mol Sci.
18(pii): E11402017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ma X, Qi S, Duan Z, Liao H, Yang B, Wang
W, Tan J, Li Q and Xia X: Long non-coding RNA LOC554202 modulates
chordoma cell proliferation and invasion by recruiting EZH2 and
regulating miR-31 expression. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
60
|
Chen T, Yao LQ, Shi Q, Ren Z, Ye LC, Xu
JM, Zhou PH and Zhong YS: MicroRNA-31 contributes to colorectal
cancer development by targeting factor inhibiting HIF-1alpha
(FIH-1). Cancer Biol Ther. 15:516–523. 2014. View Article : Google Scholar : PubMed/NCBI
|