Introduction
Colon cancer (CC) is the third most common cause of
cancer-related mortality, and is characterized by several stages of
inflammation, including tumor initiation, progression and malignant
invasion (1,2). The majority of clinical diagnoses of
CC are made during the late stages of progression, and thus, it is
associated with poor prognosis (3–5). As
the current treatment for CC, gene targeted therapy is an effective
strategy. Although a number of factors have been considered as
potential diagnostic and prognostic biomarkers in the process of
tumor proliferation and metastasis, the prevention of cancer
progression remains a challenge. Kallikrein-related peptidase 6
(KLK6), the kallikrein gene located on chromosome 19q13, is a
latent biomarker for colon and gastric cancer. Previous studies
have reported that KLK6, as an active trypsin-like serine protease,
is highly expressed in CC and is dysregulated in tumorigenesis
(6). Although KLK6, as a
tumor-associated protein, has been widely reported, the molecular
mechanisms underlying the regulation of the expression of KLK6 are
currently not well understood.
MicroRNAs (miRNAs/miRs), a class of small endogenous
non-coding RNAs, can silence target genes by specifically binding
to the 3′-untranslated regions (UTRs) and inhibiting
post-transcriptional gene expression. miRNAs, as tumor suppressors,
play a key role in cell processes, including cell development,
proliferation and apoptosis (7,8). Among
the different types of miRNAs, the let-7 family has been associated
with tumorigenesis in several types of cancer including, liver,
breast and CC. At present, the let-7 family, which consists of a
series of miRNAs, has been widely used as a tumor inhibitor and its
expression has been observed to be attenuated in several types of
human cancers (9). However, there
are few studies concerning the biological function and mechanisms
of let-7i-5p in CC, and thus, further investigation is
required.
In the present study, let-7i-5p was revealed to play
a tumor-suppressor role in CC cells. The results confirmed that the
KLK6 gene may be a direct target of let-7i-5p, and that KLK6 may
promote the proliferation and migration of CC cells. In addition,
the overexpression of let-7i-5p suppressed the growth of CC
xenografts in nude mice. These data indicated that let-7i-5p may be
a target for CC diagnostics and treatment.
Materials and methods
Clinical sample collection
The clinical samples were obtained with patient
written informed consent, and the study protocol was approved by
the Ethics Committee of the Affiliated Hospital of Medical College
of Qingdao University (Shandong, China). A total of 31 colon tissue
samples, including 27 tumor tissues and 4 normal colon tissues,
were obtained via surgical resection at the Affiliated Hospital of
Qingdao University between March 2015 to September 2016. The
average age of the patients was 50.3 (range, 42–75 years) and
66.67% were male patients. All patients did not receive any
antitumor treatment, such as chemotherapy or radiotherapy, before
surgery. Samples were immediately frozen in liquid nitrogen
following surgical resection and stored at −80°C for subsequent
study.
Cell line culture
The human CC cell lines SW480, HT-29, LoVo and
HCT-116, and the normal colon cell line FHC were obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA). All
cells were grown in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and
1% penicillin/streptavidin (Gibco; Thermo Fisher Scientific, Inc.),
and maintained at 37°C in a humidified atmosphere of 5%
CO2.
Small interfering (si)RNA
transfection
For siRNA transfection, 2×105 cells per
well were plated in a 6-well plate. After adhering for 24 h,
control miRNA (miR-con), let-7i-5p mimic and its corresponding
let-7i-5p inhibitor (Guangzhou RiboBio Co., Ltd., Guangzhou, China)
were added to the transfection medium for 6 h at 37°C in a
CO2 incubator. Loss of KLK6 expression was achieved
using KLK6 siRNA (sense, 5′-GUGCUGGGGAUGAGAAGUAdTdT-3′ and
antisense, 3′-dTdTCACGACCCCUACUCUUCAU-5′; Guangzhou RiboBio Co.,
Ltd.) (10), and cell transfection
was performed with Lipofectamine 2000™ reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Cells were supplemented with normal culture medium
and cultured at 37°C with 5% CO2 for up to 48 h before
harvesting.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
RNA was isolated from tissues or cells using a
mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific,
Inc.) following the manufacturer's instructions. First-strand cDNA
was synthesized using a PrimeScript First Strand cDNA Synthesis kit
(Takara Biotechnology Co., Ltd., Dalian, China). The extraction
concentration was determined with a NanoDrop spectrophotometer. The
products were kept at −80°C until further experimentation. The cDNA
was then amplified using the Power SYBR-Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with the
appropriate primers and an ABI 7500-fast thermocycler (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primers used were
as follows: KLK6 forward, 5′-GAAGCATAACCTTCGGCAAA-3′ and reverse,
5′-GGGAAATCACCATCTGCTGT-3′; GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′
(6). In addition, primers for
let-7i-5p and U6 were purchased from Tiangen Biotech Co., Ltd.
(Beijing, China). U6 and GAPDH served as the internal controls.
Relative expression was determined using the 2−∆∆Cq
method (11).
Western blot analysis
Following transfection experiments, the SW480,
HT-29, LoVo, HCT-116 and FHC cells were harvested and the protein
supernatants were isolated using cell lysis buffer (cat. no. 9803;
Cell Signaling Technology, Inc., Danvers, MA, USA) with added
phenylmethylsulfonyl fluoride. The extracted proteins were
determined using the BCA method. The total protein content (30 mg)
from cell lysates were resolved by 10% SDS-PAGE, and transferred to
a 0.45-mm nitrocellulose membrane (EMD Millipore, Billerica, MA,
USA) for 1 h. The membranes were washed with TBS-T containing 5%
(w/v) bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The membranes were incubated overnight with
specific primary antibodies [use 5% skim milk in TBST (10 mM
Tris-HCl, pH 8.0, 150 mM NaCl, 0.15% Tween-20) for 1:1,000
dilution] for KLK6 (cat. no. sc-374564; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), GAPDH (cat. no. 2118; Cell Signaling
Technology, Inc.), cleaved-caspase-3 (cat. no. 9664; Cell Signaling
Technology, Inc.) and cleaved peroxisome proliferator-activated
receptor (PARP; cat. no. 5625; Cell Signaling Technology, Inc.).
Membranes were then exposed to secondary antibodies conjugated to
horseradish peroxidase [anti-mouse IgG (H+L), cat. no. 14709; Cell
Signaling Technology, Inc.; anti-rabbit IgG (H+L), cat. no. 14708;
Cell Signaling Technology, Inc.; and use 5% skim milk in TBST (10
mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.15% Tween-20) for 1:2,000
dilution] for 2 h at room temperature. The membranes were washed
three times with TBS-T at room temperature. Chemiluminescent
signals were generated using the SuperSignal West Pico Trial kit
(Thermo Fisher Scientific, Inc.) and detected with the Vilber
Lourmat imaging system (Vilber Lourmat, Marne-la-Vallée,
France).
Cell viability assay
Cell proliferation was determined using an MTT
assay. Cells were seeded in a 96-well plate with density of the
optimized cell number (5,000 cells/well). Following 24 h of
seeding, cells were treated with siRNA or diluted chemicals at
indicated working concentration. Cells were incubated for indicated
time-points and then 20 µl MTT (5 mg/ml) was added into the wells
and 4 h later the mixed medium was replaced by 150 µl dimethyl
sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA). Subsequently, the
96-well plate was agitated for 15 min at room temperature. Then the
OD value of each well was determined using a fluorescence
microplate reader (Sunrise Remote; Tecan Austria GmbH, Grödig,
Austria) at a wavelength of 490 nm.
Luciferase reporter assay
Using the online platform TargetScan (http://www.targetscan.org), let-7i-5p was used to
search for candidate miRNAs that can bind to KLK6. The human colon
tumor cells were grown to 70–80% confluence in 24-well plates, and
then co-transfected with the recombinant plasmid containing the
wild-type/mutant KLK6 3′-UTR and miRNA mimics (50 nM) using
Lipofectamine 2000™ reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Luciferase
activity was analyzed 48 h after co-transfection using
Dual-Luciferase assays (Promega Corp.) and values were normalized
against Renilla luciferase activity.
Wound healing assay
Migration was analyzed by a wound healing assay.
Cells were seeded into 6-well plates (5×105/well) and
allowed to grow to 90–95% confluence. A linear wound was made by
scraping the cells using a 10-µl micropipette tip and debris was
washed away twice with PBS. At different time-points (0 and 24 h)
images of the cells were captured with an inverted microscope
(Olympus Corp., Tokyo, Japan), and the migration distances were
determined by ImageJ software (ImageJ 1.47v; http://imagej.nih.gov/ij/).
Apoptosis detection
Apoptosis was determined by Annexin V-fluorescein
isothiocyanate (FITC) and propidium iodide (PI) double staining.
Cells were cultured in 6-well plates at 37°C overnight in a
humidified atmosphere with 5% CO2, and then underwent
transfection for 48 h, as aforementioned. Cells were then
trypsinized and stained with Annexin V-FITC/PI. The levels of
apoptosis were measured using a FACSCalibur flow cytometer (BD
Biosciences, San Jose, CA, USA). Annexin
V+/PI− cells were considered to be apoptotic
cells.
Xenograft experiments
All mice were obtained from the Institute of
Laboratory Animal Science, Chinese Academy of Medical Sciences and
maintained under specific pathogen-free conditions in accordance
with the guidelines approved by the China Animal Welfare and
approval was granted by the Ethics Committee of the Medical College
of Qingdao University. The mice were divided into the following two
groups: The miR-con group and the let-7i-5p group. A total of
3×106 SW480 cells transfected either with let-7i-5p or
miR-con were subcutaneously injected into the right armpits of
4-week-old BALB/c mice. During these experiments, the mice were
weighed by table balance and the tumors were assessed with calipers
each week. After 5 weeks, the mice were euthanized using a
subcutaneous injection with sodium pentobarbital (150 mg/kg), and
the volume of the tumor was assessed. The tumor volume (V) was
calculated using the following formula: V (mm3) = 0.5 ×
length × (width)2.
Statistical analysis
All images were formatted for optimal presentation
using Adobe Illustrator CS4 (Adobe Systems, Inc., San Jose, CA,
USA). To determine statistical significance between two groups, a
Student's t-test was performed to calculate the associated
P-values. Statistical significance between multiple groups was
evaluated by one way analysis of variance (ANOVA) followed by a
Newman-Keuls post hoc test using GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA).
Results
KLK6 is significantly expressed in
colon tumors
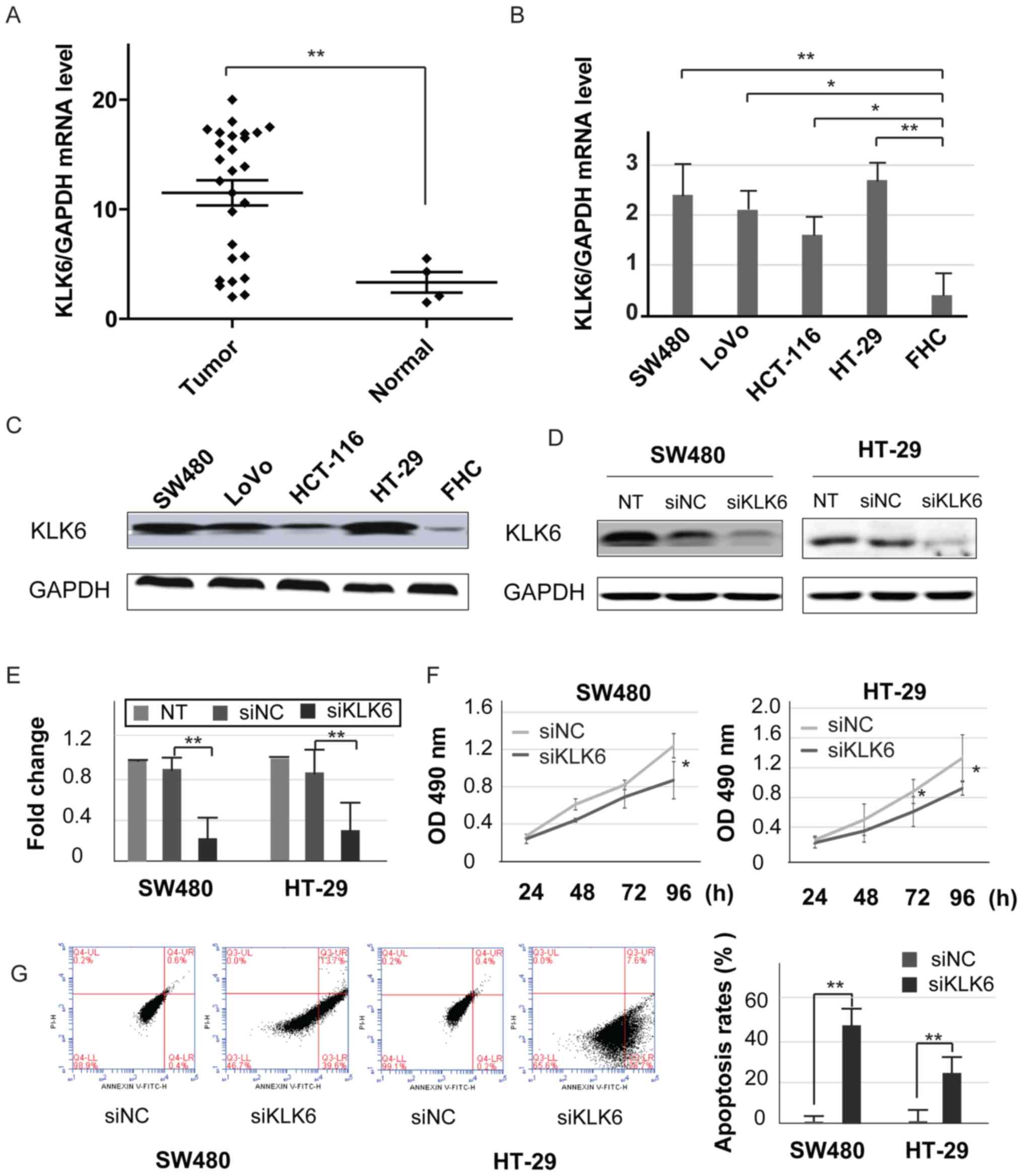
To examine the level of KLK6 in tissues and cell
lines, the present study quantified KLK6 expression in 27 colon
tumor tissues, 4 normal colon tissues, 4 CC cell lines and 1 normal
colon cell line. The results of qPCR revealed that the mRNA
expression level of KLK6 was significantly expressed in the colon
tumor tissues compared with that noted in the normal colon tissues
(Fig. 1A). Similarly, high mRNA and
protein KLK6 expression levels were demonstrated in the colon tumor
cell lines, including SW480, LoVo, HT-29 and HCT-116 cells
(Fig. 1B and C). These results were
consistent with those of a previous study (6). The expression levels of KLK6 were more
significantly increased in SW480 and HT-29 cells than in the other
two cell lines. To assess the biological role of KLK6,
KLK6-specific siRNAs or its corresponding control siRNA were
introduced into CC cells and the efficiency of KLK6 siRNAs was
evaluated (Fig. 1D and E).
Knockdown of KLK6 significantly decreased cell growth and enhanced
apoptosis in the SW480 and HT-29 cells. (Fig. 1F and G). These results indicated
that KLK6 may play a vital role in anti-apoptosis activity and cell
proliferation in CC cells.
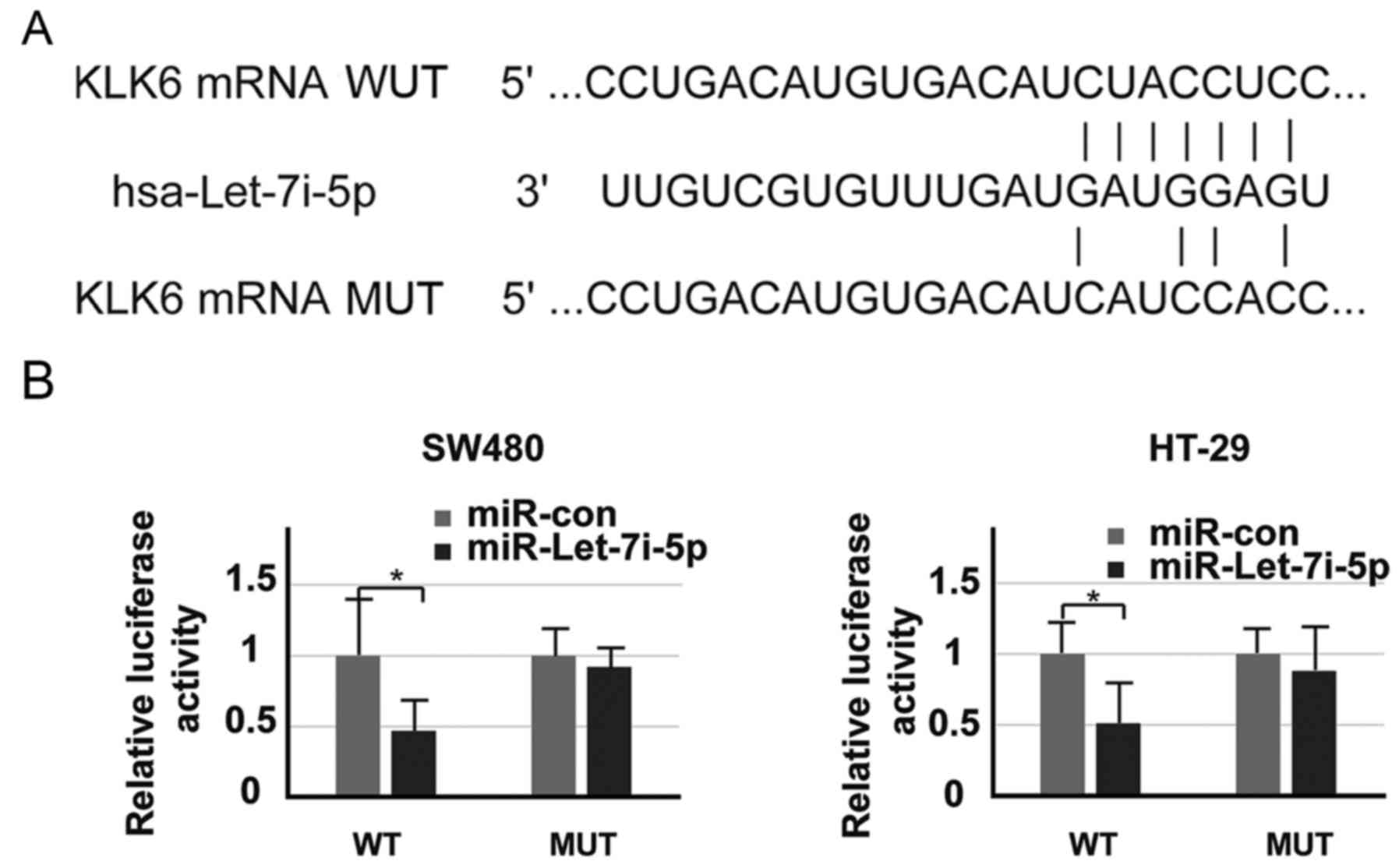
Let-7i-5p directly targets KLK6 3′-UTR
in colon cancer cells
Using the online platform TargetScan (http://www.targetscan.org), let-7i-5p was revealed to
be a potential KLK6 target gene. Let-7i-5p acts as an upstream
factor that may be able to directly bind to the 3′-UTR of KLK6 mRNA
(Fig. 2A). The present study cloned
the 3′-UTR wild-type or 3′-UTR mutant-type of KLK6 into a
pMIR-REPORT vector. As expected, the luciferase activity of the
3′-UTR wild-type in cells transfected with let-7i-5p was
significantly lower than that of those transfected with
miR-control, while the change in the luciferase activity of 3′-UTR
mutant-type cells was minimal (Fig.
2B). Thus, these results indicated that KLK6 directly targets
Let-7i-5p.
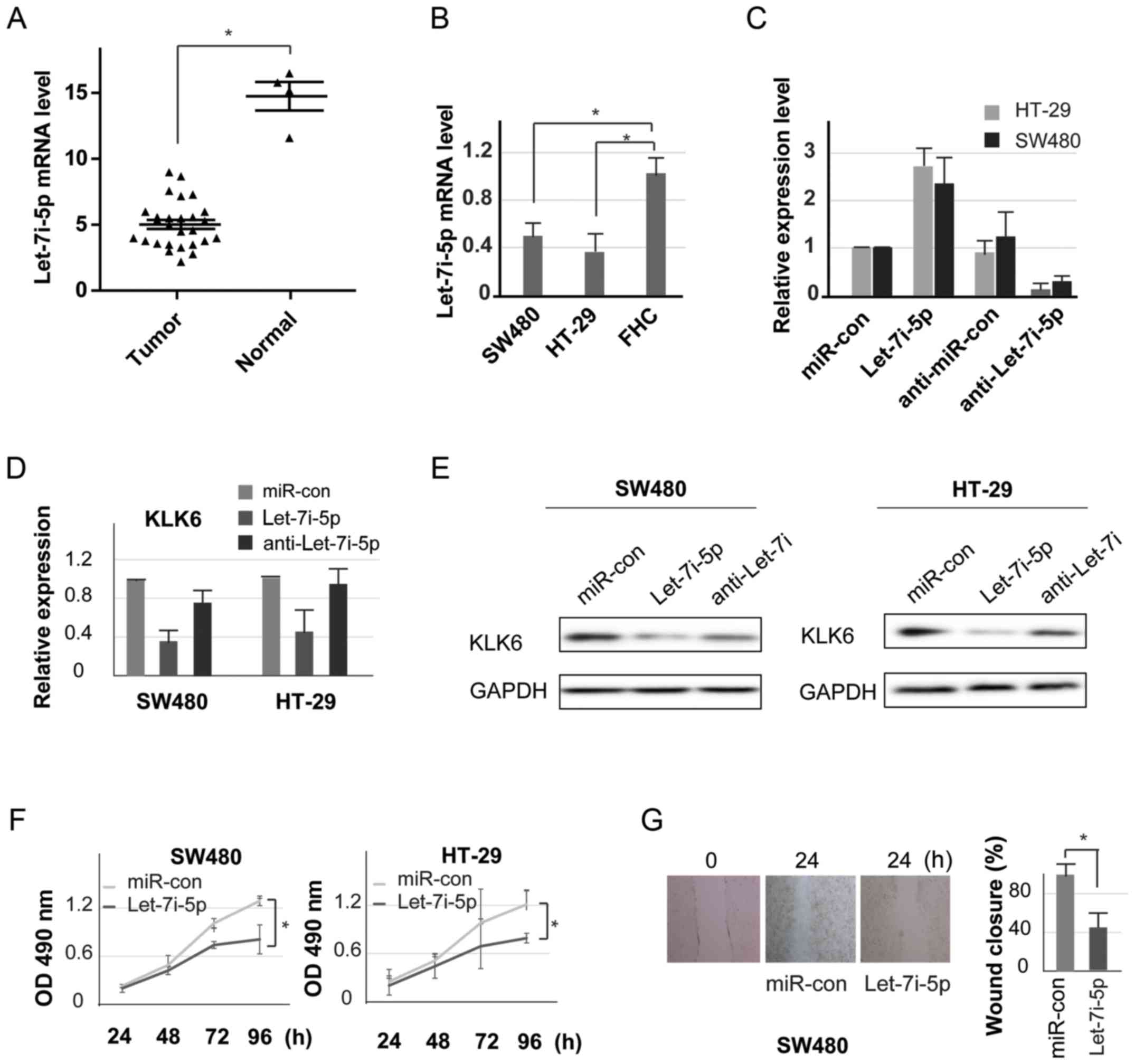
Let-7i-5p inhibits cell proliferation
and invasion
To observe the effect of let-7i-5p on CC, the
present study detected the expression of let-7i-5p in CC tissues
and cell lines by qPCR. The expression of let-7i-5p in tumor
samples was significantly lower when compared with that of the
normal samples (Fig. 3A).
Consistent with the results observed in cancer tissues, the
let-7i-5p mRNA level was significantly lower in CC cells compared
with the level noted in the normal colon cells (Fig. 3B). These data indicated that
let-7i-5p may serve as an important regulator in CC.
Furthermore, colon tumor cell lines were transfected
with let-7i-5p mimics, miR-con, anti-let-7i and anti-miR-con. qPCR
analysis was performed to determine the transfection efficiency in
the SW480 and HT-29 cells (Fig.
3C). As displayed in Fig. 3D and
E, there were markedly lower expression levels of KLK6 in cells
transfected with the let-7i-5p mimics than in the other groups. MTT
assay was used to evaluate cell proliferation. The results
demonstrated that overexpression of let-7i-5p inhibited cell growth
when compared with the negative control group in both SW480 and
HT-29 cells (Fig. 3F). Cell
migration is a crucial process in cancer metastasis (12), thus, a wound-healing assay was
employed to assess the migration and invasion of colon tumor cells.
High levels of let-7i-5p resulted in a significant decrease in cell
migration compared with the miR-con group (Fig. 3G). Collectively these results
indicated that let-7i-5p may directly target the KLK6 3′-UTR,
inhibiting cell proliferation and invasion in CC cells.
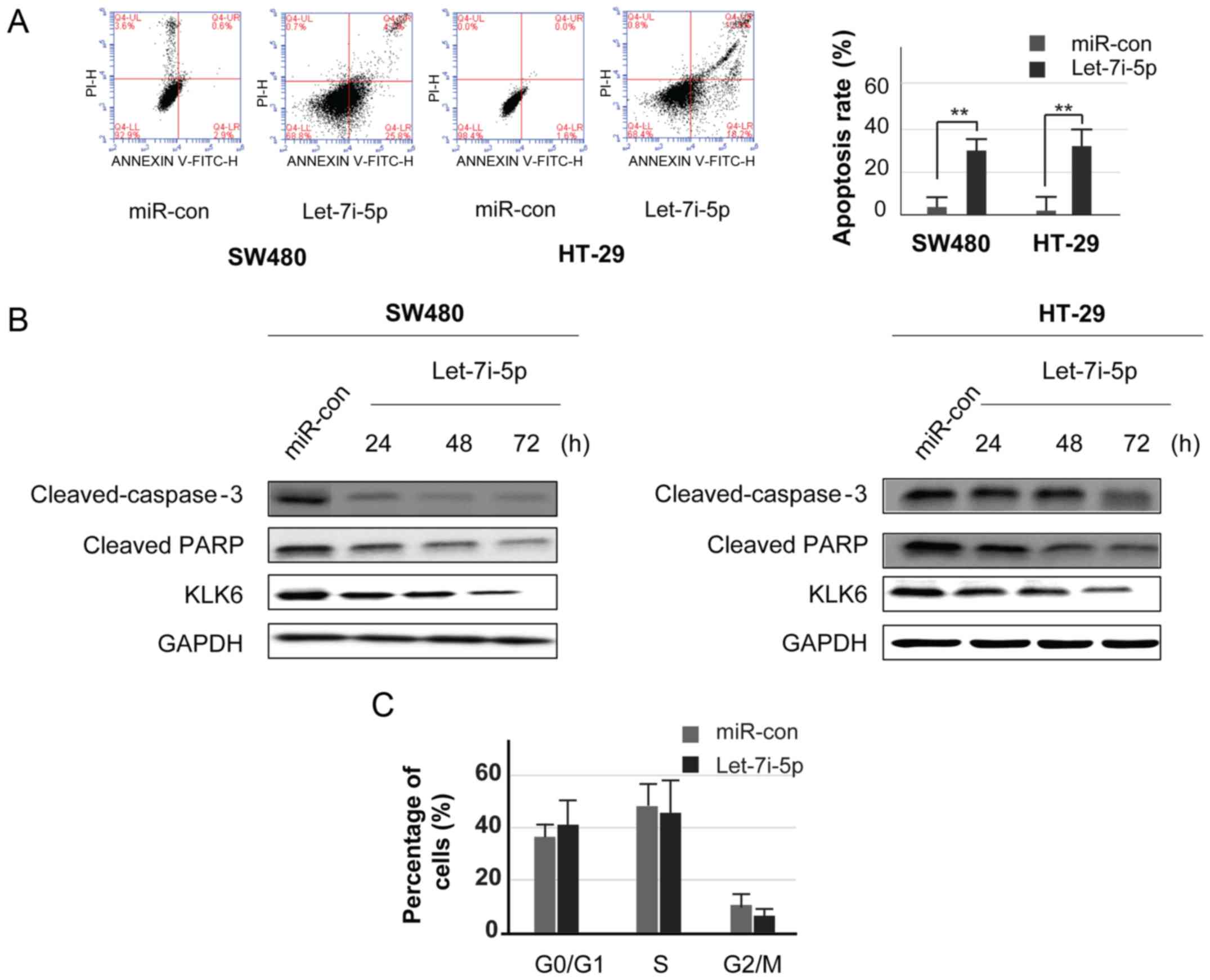
Let-7i-5p promotes apoptosis and
attenuates cell cycle progression in CC cell lines
Flow cytometry using Annexin V-FITC and PI double
staining demonstrated that endogenous let-7i-5p was increased, and
the rate of apoptosis was significantly increased in the SW480 and
HT29 cells (Fig. 4A). It was
revealed that the accumulation of let-7i-5p could increase the
number of apoptotic cells. Several studies have indicated that KLK6
is associated with a caspase-dependent pathway in many types of
tumor cells, including lung cancer, glioma and breast cancer.
However, the underlying mechanism is not clear in CC (13). To determine whether let-7i-5p
enhanced apoptosis via the caspase-dependent pathway, colon tumor
cell lines were transfected with let-7i-5p mimics or miR-con for 24
h, and then further cultured for 48 and 72 h. Western blotting was
carried out to analyze the levels of cleaved caspase-3 and PARP at
24, 48 and 72 h. The results revealed that let-7i-5p reduced the
levels of cleaved-caspase-3 (Fig.
4B). These results demonstrated that caspase-dependent
signaling may mediate the increased levels of apoptosis in colon
tumor cells.
Furthermore, flow cytometry was performed to
evaluate cell cycle distribution, and thereby, verify whether the
pro-apoptotic effect of let-7i-5p is associated with cell cycle
arrest. Cells were transfected with let-7i-5p mimics and miR-con
for 48 h. In the let-7i-5p group, there was a marked decrease in
the proportion of cells in the S and G2 phase, as well as cell
cycle arrest in the G1 phase. When compared with miR-con group, the
number of cells was decreased in S and G2 phases (Fig. 4C).
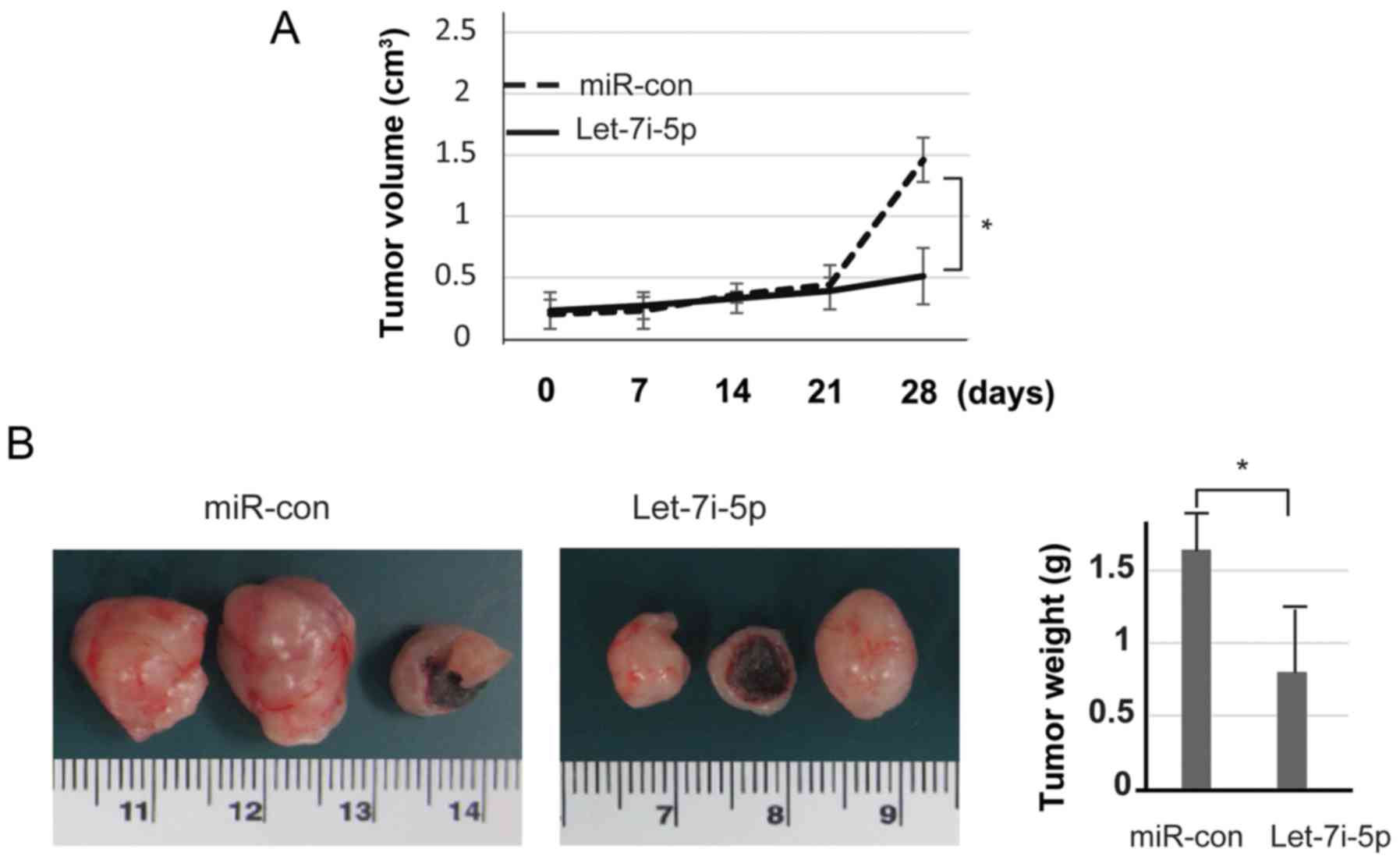
Let-7i-5p suppresses the tumor growth
of SW480 cells in vivo
The present study further evaluated the effects of
let-7i-5p in vivo by measuring colon xenograft growth. SW480
cells were transfected either with let-7i-5p or miR-con. To
establish the mouse xenograft models, stably transfected cells
(1×107 cells per condition) were injected into nude mice
subcutaneously; the tumors were then monitored every week. During
the 4-week period, the tumor volumes were assessed. The results
revealed that upregulation of let-7i-5p significantly inhibited
tumor growth (Fig. 5A). Let-7i-5p
markedly attenuated tumor growth compared with the miR-con group.
After 4 weeks, the mice were sacrificed and the tumors were
excised. As displayed in Fig. 5B,
tumor weight was assessed using calipers. These data indicated that
overexpression of let-7i-5p in SW480 cells markedly reduced their
ability to form tumors.
Discussion
Colorectal cancer is a common malignant tumor of the
digestive tract and is often clinically diagnosed at the late
stages of the disease. It has been reported that cancer is a
genetic disease induced by many complex factors (14,15).
The development of colon cancer (CC) is associated with common
genetic changes in Kirsten rat sarcoma viral proto-oncogene, B-Raf
proto-oncogene serine/threonine kinase, tumor protein p53 and some
genes related to the Wnt signaling pathway. Previous studies have
demonstrated that the serine protease KLK6 was found to be highly
expressed in several types of cancers, including pancreatic,
colorectal, gastric and breast cancers, and it may serve as a
biomarker for these cancers (16–19).
It has also been reported that KLK6 may play a vital role in both
cancer-inhibiting and cancer-promoting processes (20). The tissue type and the tumor
microenvironment have different effects on the process of
carcinogenesis due to the different roles of KLK6 (21). In the colon samples collected in the
present study, the majority of the tumor tissues expressed high
levels of KLK6 mRNA compared with the control tissue. In addition,
both high KLK6 mRNA and protein expression was observed in the CC
cell lines. Thus, the results indicated that high KLK6 expression
in the colon may be associated with cell proliferation and
malignant transformation. However, the underlying mechanism and
therapeutic targets of CC have yet to be fully elucidated.
miRNAs regulate a variety of biological processes
and they exert their functions under both physiological and
pathological conditions, including cell proliferation, apoptosis,
development and metabolism (22).
To date, many studies have reported that miRNAs are associated with
colon tumors. For example, miR-582-5p was observed to promote CC
cell proliferation by inhibiting the expression of Adenomatosis
polyposis coli (23). miR-552
is involved in the metastasis of CC cells by targeting a
disintegrin and metallopeptidase domain 28 (24). Currently, there are few studies that
have reported an association between let-7 and colon tumors, and
few results have elucidated the mechanistic relation between them.
In the present study, low expression of let-7i-5p was detected in
colon tumor tissue samples compared with that noted in normal colon
tissues. Similar results were observed in colon tumor cell lines.
In addition, upregulation of let-7i-5p expression markedly
inhibited colon tumor cell viability and invasion. Collectively,
these results indicated that let-7i-5p expression may gradually
decrease with increased colon tumor malignancy.
Previous research has revealed that miRNAs, as tumor
inhibitors, play a vital role in the development and progression of
cancer via complementary binding to the 3′-UTRs of target genes,
causing the degradation or translational suppression of mRNA
(25). The miRNA let-7 family,
which is widely considered to have tumor-suppressor activity, was
found to bind to the 3′-UTRs of target genes and serve an important
role in the regulation of the cell cycle, cell differentiation and
apoptosis in neuroblastoma, as well as liver and lung cancer
(9,26). The present study identified KLK6 as
a direct target gene of let-7i-5p. Enhanced expression of let-7i-5p
was observed to lower the expression level of KLK6 in colon tumor
cells and promoted malignant tumor progression. It was suggested
that let-7i-5p attenuates the viability and invasion of colon tumor
cells by targeting KLK6. The results of the present study also
demonstrated that the accumulation of let-7i-5p affected the cell
cycle, decreasing the proportion of cells in the S and G2 phases,
and inducing cell cycle arrest in the G1 phase. Furthermore,
upregulation of let-7i-5p was detected to alter several factors in
the caspase signaling pathways. Additionally, the present study
verified that mice injected with stably transduced let-7i-5p colon
tumor cells had fewer metastases in vivo. This was in
agreement with the data implicating let-7i-5p in the induction of
cell apoptosis and cell cycle arrest in colon tumor cells by
targeting KLK6, thereby inhibiting colon tumor cell growth.
Therefore, these results support the notion that the let-7i-5p/KLK6
axis may be an ideal therapeutic candidate for improving the
clinical outcomes of colon patients. However, further experiments
are required to determine the use of let-7i-5p/KLK6 as an effective
biomarker in the clinic.
In conclusion, the results of the present study
revealed that let-7i-5p may be a potential target of KLK6, with
regulatory roles in colon tumor cells. In addition, the results
demonstrated that let-7i-5p may serve a vital role in the growth
and metastasis of CC cells, affecting cell apoptosis and the cell
cycle. Therefore, these findings may provide the basis for
let-7i-5p as a diagnostic and prognostic marker for the treatment
of colon tumor. In addition, the results of the present study may
provide new methods for the diagnosis and treatment of CC patients.
As KLK6 has been reported to be a valuable biomarker in other types
of cancers, such as ovarian cancer (16–19),
additional studies are required to assess the role of the
let-7i-5p/KLK6 axis in other types of cancer. Furthermore, the
present study highlights that integrated bioinformatic approaches
on publicly available web tools, such as TargetScan, play critical
roles in identifying candidate targets of miRNAs and potential
mechanisms for cancers.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Nature
Science Foundation of China (grant no. 81602621) and the
Postdoctoral Applied Research Programs of Qingdao (grant no.
2016061).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
NH and BW conceived and designed the study; JS, LW,
NH and BW provided the study materials or patients; JS, QM, YY, ZY
and NH collected and assembled the data; JS, BW and NH analyzed and
interpreted the data. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The clinical samples were obtained with patient
written informed consent, and the study protocol was approved by
the Ethics Committee of the Affiliated Hospital of Medical College
of Qingdao University (Shandong, China). Animal experiments were
carried out in accordance with the guidelines approved by the China
Animal Welfare and approval.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fan NJ, Kang R, Ge XY, Li M, Liu Y, Chen
HM and Gao CF: Identification alpha-2-HS-glycoprotein precursor and
tubulin beta chain as serology diagnosis biomarker of colorectal
cancer. Diagn Pathol. 9:532014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Pisani P and Ferlay J: Global
cancer statistics. CA Cancer J Clin. 49:33–64, 31. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kinzler KW and Vogelstein B: Lessons from
hereditary colorectal cancer. Cell. 87:159–170. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsuchiya K: Remodeling of cancer stem
cells in gastrointestinal cancer. Nihon Rinsho. 73:855–859.
2015.PubMed/NCBI
|
|
6
|
Kim JT, Song EY, Chung KS, Kang MA, Kim
JW, Kim SJ, Yeom YI, Kim JH, Kim KH and Lee HG: Up-regulation and
clinical significance of serine protease kallikrein 6 in colon
cancer. Cancer. 117:2608–2619. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie Y, Yu F, Tang W, Alade BO, Peng ZH,
Wang Y, Li J and Oupický D: Synthesis and evaluation of
chloroquine-containing DMAEMA copolymers as efficient anti-miRNA
delivery vectors with improved endosomal escape and antimigratory
activity in cancer cells. Macromol Biosci. 18:2018.doi:
10.1002/mabi.201700194. View Article : Google Scholar
|
|
8
|
Zeng Y, Wang KX, Xu H and Hong Y:
Integrative miRNA analysis identifies hsa-miR-3154, hsa-miR-7-3,
and hsa-miR-600 as potential prognostic biomarker for cervical
cancer. J Cell Biochem. 119:1558–1566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barh D, Malhotra R, Ravi B and Sindhurani
P: MicroRNA let-7: An emerging next-generation cancer therapeutic.
Curr Oncol. 17:70–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cohen RJ, Dennis LH and Conrad ME:
Anticoagulant therapy in chronic idiopathic thrombocytopenic
purpura. Blood. 31:647–652. 1968.PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sells E, Pandey R, Chen H, Skovan BA, Cui
H and Ignatenko NA: Specific microRNA-mRNA regulatory network of
colon cancer invasion mediated by tissue kallikrein-related
peptidase 6. Neoplasia. 19:396–411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim TW, Lee SJ, Kim JT, Kim SJ, Min JK,
Bae KH, Jung H, Kim BY, Lim JS, Yang Y, et al: Kallikrein-related
peptidase 6 induces chemotherapeutic resistance by attenuating
auranofin-induced cell death through activation of autophagy in
gastric cancer. Oncotarget. 7:85332–85348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaback HR: The role of the
phosphoenolpyruvate-phosphotransferase system in the transport of
sugars by isolated membrane preparations of Escherichia coli. J
Biol Chem. 243:3711–3724. 1968.PubMed/NCBI
|
|
15
|
Quarles RH, Shin ML and Waksman BH: A
workshop on mechanisms of myelin breakdown. Ann Neurol. 19:971986.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Elbrønd H, Huniche B and Ostergaard L:
Rabbit sphincter of Oddi and duodenum are regulated by slow waves
with a common basic-mode activity. Scand J Gastroenterol.
25:534–540. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nagahara H, Mimori K, Utsunomiya T,
Barnard GF, Ohira M, Hirakawa K and Mori M: Clinicopathologic and
biological significance of kallikrein 6 overexpression in human
gastric cancer. Clin Cancer Res. 11:6800–6806. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ni X, Zhang W, Huang KC, Wang Y, Ng SK,
Mok SC, Berkowitz RS and Ng SW: Characterisation of human
kallikrein 6/protease M expression in ovarian cancer. Br J Cancer.
91:725–731. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tamir A, Jag U, Sarojini S, Schindewolf C,
Tanaka T, Gharbaran R, Patel H, Sood A, Hu W, Patwa R, et al:
Kallikrein family proteases KLK6 and KLK7 are potential early
detection and diagnostic biomarkers for serous and papillary serous
ovarian cancer subtypes. J Ovarian Res. 7:1092014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pampalakis G, Prosnikli E, Agalioti T,
Vlahou A, Zoumpourlis V and Sotiropoulou G: A tumor-protective role
for human kallikrein-related peptidase 6 in breast cancer mediated
by inhibition of epithelial-to-mesenchymal transition. Cancer Res.
69:3779–3787. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Diamandis EP, Yousef GM, Soosaipillai AR
and Bunting P: Human kallikrein 6 (zyme/protease M/neurosin): A new
serum biomarker of ovarian carcinoma. Clin Biochem. 33:579–583.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shu Z, Chen L and Ding D: miR-582-5P
induces colorectal cancer cell proliferation by targeting
adenomatous polyposis coli. World J Surg Oncol. 14:2392016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Li H, Wang Y, Wang L, Yan X, Zhang
D, Ma X, Du Y, Liu X and Yang Y: MicroRNA-552 enhances metastatic
capacity of colorectal cancer cells by targeting a disintegrin and
metalloprotease 28. Oncotarget. 7:70194–70210. 2016.PubMed/NCBI
|
|
24
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Vasconcellos JF, Byrnes C, Lee YT,
Allwardt JM, Kaushal M, Rabel A and Miller JL: Tough decoy
targeting of predominant let-7 miRNA species in adult human
hematopoietic cells. J Transl Med. 15:1692017. View Article : Google Scholar : PubMed/NCBI
|