Introduction
Renal cell carcinoma (RCC) is the third most common
urological malignancy in the USA and represents 2–3% of all adult
malignancies. Furthermore, the incidence rate has progressively
increased, particularly in young patients and or those with
high-grade disease (1–3). In 2017, ~63,990 cases were diagnosed
with kidney and renal pelvis cancer and 14,400 cases were expected
to succumb to disease in the USA alone (4). Various risk factors have been
associated with RCC, including active and passive cigarette
smoking, obesity, hypertension and absent fruit and vegetable
consumption (5). Although the
morbidity of treatment has decreased with the use of
nephron-sparing surgery, laparoscopic and robotic surgery and
minimally invasive procedures, and biologic response modifiers are
applied to patients with metastatic RCC, the prognosis of terminal
cancer remains poor, with a 5-year survival rate of 5–10% (6,7).
Therefore, it's imperative to identify the clinical and molecular
phenotype of occurrence and development of RCC in order to uncover
novel targets for the diagnosis, treatment and prognosis of
RCC.
MicroRNAs (miRs), which are small noncoding RNA
molecules (21–25 bases in length), regulate the gene expression in
a sequence-specific manner and promote the suppression of protein
synthesis or mRNA degradation (8).
The uniqueness of miRs is that one miR can regulate numerous
protein-coding RNAs (9). In the
human genome, miRs have been correlated with the regulation of
30–60% of protein-coding genes (10,11).
Accumulating evidence has suggested that miRs, prominently those
upregulated or downregulated in other types of malignancies, act as
either oncogenes or anti-oncogenes, respectively, in RCC. For
example, miR-367 exerts oncogenic effects in osteosarcoma and
promotes the proliferation, migration and invasion of tumor cells
with the regulation of DOC-2/DAB2 interactive protein, which was
consistent with the oncogenic role of miR-367 in RCC (12,13).
miR-572 is a type of miR that was recently confirmed
to be involved in various types of cancer, including chronic
lymphocytic leukemia (14),
nasopharyngeal carcinoma (15),
ovarian cancer (16) and basal cell
carcinoma (17). Results from a
study by Wang et al (18)
revealed that, compared with corresponding non-tumor controls,
miR-572 was markedly elevated in the serum of patients with RCC.
However, the expression and the function of miR-572 in RCC remains
elusive. Thus, the present study was performed with the purpose of
ascertaining the expression of miR-572 in RCC and corresponding
normal tissues. Furthermore, the role of miR-572 in the
proliferation, apoptosis and prognosis of RCC cell was further
explored.
Materials and methods
Sample collection
RCC tissues (n=52) and corresponding non-cancerous
renal tissues (n=52) from patients that were pathologically
diagnosed with RCC and had received no therapy were collected from
the Peking University Shenzhen Hospital between January 2016 and
June 2017. (Guangdong, China). The corresponding non-cancerous
renal tissues were collected >2 cm away from the visible tumor
range. Written informed consent was obtained from every patient.
All the patients were pathologically reviewed and diagnosed with
RCC and had not received any chemoradiotherapy. The present study
was approved by the Ethics Committees of Peking University Shenzhen
Hospital. The collected tissues were checked and classified by
hematoxylin and eosin staining. Subsequently, the samples were
immersed in RNAlater reagent (Qiagen GmbH, Inc., Hilden, Germany)
for ~0.5 h and stored at −80°C. The clinical and pathological
information of the patients is presented in Table I.
 | Table I.Clinicopathological features in
patients with renal cell carcinoma. |
Table I.
Clinicopathological features in
patients with renal cell carcinoma.
|
Characteristics | Number of
cases |
|---|
| Mean age range
(years) | 52 (27–72) |
| Sexual
distinction |
|
|
Male/female | 30/18 |
| Histological
type |
|
| Clear
cell/papillary | 39/9 |
| pT-stage |
|
|
T1/T2/T3 + T4 | 27/19/2 |
| Fuhrman grade |
|
|
I/II/III/IV | 15/22/8/3 |
| AJCC clinical
stages |
|
|
I/II/III+IV | 27/18/3 |
Formalin-fixed paraffin-embedded
(FFPE) tissue specimens
The FFPE RCC samples obtained from the Department of
Pathology of Peking University Shenzhen Hospital originated from
patients with RCC who had undergone surgery at Peking University
Shenzhen Hospital between January 2011 and January 2014. For
fixation, all the tissue sample was fixated with 10% formalin
concentration at 4°C for 48 h. For RNA extraction, ~10-µm-thick
FFPE sections were used. The clinical and pathological
characteristics of the patients, which are presented in Table II, were analyzed on the basis of
the 2010 American Joint Committee on Cancer staging system
(19). Total RNA of FFPE samples
was extracted using an miRNeasy FFPE Kit (Qiagen GmbH, Inc.).
 | Table II.Association between microRNA-572
expression levela and
clinical information in FFPE renal cancer samples. |
Table II.
Association between microRNA-572
expression levela and
clinical information in FFPE renal cancer samples.
|
|
| No. of patients
(%) |
|
|---|
|
|
|
|
|
|---|
| Variable | Total | High miR-572
expression | Low miR-572
expression |
P-valueb |
|---|
| Sex |
|
Male | 26 | 14 | 12 | 0.751 |
|
Female | 16 | 7 | 9 |
|
| Age (years) |
|
≤60 | 33 | 17 | 16 | 1.000 |
|
>60 | 9 | 4 | 5 |
|
| Tumor size
(cm) |
|
≤4.0 | 17 | 6 | 11 | 0.208 |
|
>4.0 | 25 | 15 | 10 |
|
| Tumor stage |
| I+
II | 27 | 12 | 15 | 0.520 |
|
III+IV | 15 | 9 | 6 |
|
Cell culture and transfection
The cells used in the present study included human
embryo kidney cells (293T; Cell Bank of Type Culture Collection of
the Chinese Academy of Medical Sciences, Shanghai, China) and RCC
cell lines (786-O, ACHN and Caki-1; American Type Culture
Collection, Manassas, VA, USA). Cells were maintained at 37°C in an
atmosphere containing 5% CO2. The composition of the
medium was as follows: 89% Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc.), 0.5% antibiotics (100 µl/ml penicillin and 100 mg/ml
streptomycin sulfates) and 0.5% glutamine. A total of 100 pmol
miR-572 mimic, inhibitor, negative control (NC) and inhibitor
negative control (all Shanghai GenePharma Co., Ltd., Shanghai,
China) were transfected into RCC cells for 5 h using Lipofectamine
2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. Subsequent experiments were
performed 24–48 h following transfection. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
performed to evaluate the transfected effect. The sequences were
presented in Table III.
 | Table III.Sequences of primers and miRs. |
Table III.
Sequences of primers and miRs.
| Primer/miR | Sequence |
|---|
| miR-572 | F: |
5′-GTCCGCTCGGCGGTGGCCCA-3′ |
|
| R: | Universal primers
(miScript SYBR-Green PCR kit) |
| U6 | F: |
5′-CTCGCTTCGGCAGCACA-3′ |
|
| R: |
5′-ACGCTTCACGAATTTGCGT-3′ |
| miR-572 | F: |
5′-GUCCGCUCGGCGGUGGCCCA-3′ |
| mimic | R: |
5′-GGCCACCGCCGAGCGGACUU-3′ |
| miR-572
inhibitor |
|
5′-UGGGCCACCGCCGAGCGGAC-3′ |
| NC | F: |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
|
| R: |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| NC inhibitor |
|
5′-CAGUACUUUUGUGUAGUACAA-3′ |
RNA extraction, cDNA synthesis and
RT-qPCR
Total RNA of tissues and cell lines (786-O, ACHN,
Caki-1 and 293T) were extracted and purified with the using of
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the
RNeasy Maxi Kit (Qiagen GmbH, Inc.), respectively, according to the
manufacturer's protocol. For improved experimental results, the
extractive RNA samples were measured using a NanoDrop 2000c (Thermo
Fisher Scientific, Inc.) to ensure the concentration and the
optical density ratio (260/280) of 1.8–2.0 were taken into account.
Following this, the eligible RNAs were synthesized to cDNA with the
miScript II RT Kit (Qiagen GmbH). qPCR reactions were performed to
ascertain the expression levels of miR-572 using the Roche
LightCycler 480 Real-Time PCR System with a miScript SYBR-Green PCR
Kit (Qiagen GmbH) under the following conditions: 95°C for 1 min,
followed by 40 cycles at 95°C for 15 sec, 55°C for 30 sec and 72°C
for 30 sec. RNU6B (U6) was used as internal reference for
normalization. The primers used in the qPCR are indicated in
Table III. Relative expression
levels were calculated using the 2−ΔΔCq method (20).
Wound healing assay
Assessment of the cell migratory aptitude was
performed using the wound healing assay. In the assay, a 12-well
plate was inoculated with 786-O and ACHN cells (~3×105
cells/each well) transfected with miR-572 mimic, inhibitor, NC or
inhibitor NC (Shanghai GenePharma, Co., Ltd.). At 6 h
post-transfection, a scratch was introduced using a sterile 200-µl
pipette tip. Subsequently, the floating cells were washed away
using phosphate-buffered saline (PBS; Gibco; Thermo Fisher
Scientific, Inc.). At 0, 12 and 24 h, A Leica DMIRB inverted
fluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany)
with a digital camera system (Olympus Corporation, Tokyo, Japan)
was used to capture images of the scratches.
Cell Counting Kit-8 (CCK-8) assay
Assessment of the proliferation capacity of 786-O
and ACHN cells was performed using the CCK-8 assay. A total of 5
pmol of miR-572 mimics, inhibitors, NC or inhibitor NC was
transfected into the cells that had seeded for 24 h in 96-well
plats with 4×103 cells/well. Following gene
transfection, 10 µl CCK-8 (Everbright lnc., Sayreville, NJ, USA)
was added to each well and incubated for 0, 24, 48 and 72 h. The
optical density value of each well was calculated 1 h later with an
ELISA microplate reader (model 680; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) at a wavelength of 490 nm.
Transwell assay
The Transwell assay was used to assess the migratory
and invasive aptitude of ACNH and 786-O cells. According to the
manufacturer's recommendation, Transwell chamber inserts (BD
Biosciences, New York, NJ, USA) with Matrigel were used to
determine cell migration, whereas the inserts without Matrigel were
used to assay cell invasion. A total of 200 µl serum-free DMEM
containing 2×104 transfected cells was added into the
upper chamber of the inserts, whereas 500 µl DMEM with 10% FBS was
added into the lower chamber of the inserts. Following 48 h of
incubation, the cells migrated or invaded to the lower chamber of
the inserts were stained with crystal violet at room temperature
for 25 min, washed with PBS three times and counted using a
microscope under the white light at a magnification of ×100 (Leica
Microsystems GmbH), successively.
Flow cytometric assay
Assessment of cell apoptosis rates depended on flow
cytometry assay. The cells plated in 6-well plates with
~3×105 cells/plate were transfected with 200 pmol of
miR-572 mimics, miR-572 inhibitors, NC inhibitor or NC according to
the manufacturer's instructions. Following incubated for 48 h, all
cells were harvested, washed by cold PBS twice and re-suspended in
100 µl 1X binding buffer. Subsequently, the re-suspended cells were
mixed with 5 µl propidium iodide (Invitrogen; Thermo Fisher
Scientific, Inc.) and 5 µl Annexin V-fluorescein isothiocyanate
(Invitrogen; Thermo Fisher Scientific, Inc.), and cultured in
darkness at room temperature for 15 min. Each tube was treated with
400 µl binding buffer. The apoptotic rates were measured using a
flow cytometer (EPICS, Xl-4; Beckman Coulter, Inc., Brea, CA, USA).
FlowJo software (version, X; FlowJo LLC, Ashland, OR, USA) was used
for data analysis.
Statistical analysis
All assays were repeated three times. All
statistical analysis was executed with the use of SPSS 19.0 (IBM
Corp., Armonk, NY, USA). Paired Student's t-test was applied to
assess the significance of the differences between two groups of
data in matched tumor and non-cancerous tissues. The significance
between cell lines was analyzing using one-way analysis of variance
and Dunnett's post hoc test. Student's t-test was used the analysis
of assays for characterizing phenotypes of cells. Analysis of the
correlation between miR-572 expression and clinicopathological
variables was performed using Fisher's exact test or Pearson's
χ2 test. The univariate and multivariate levels between
miR-572 expression and clinicopathological variables or survival
were determined using Cox proportional hazard regression analysis.
The survival curves were plotted with Kaplan-Meier curves. The
log-rank test was applied to evaluate the differences between these
curves. P<0.05 was considered to indicate a statistically
significant difference.
Results
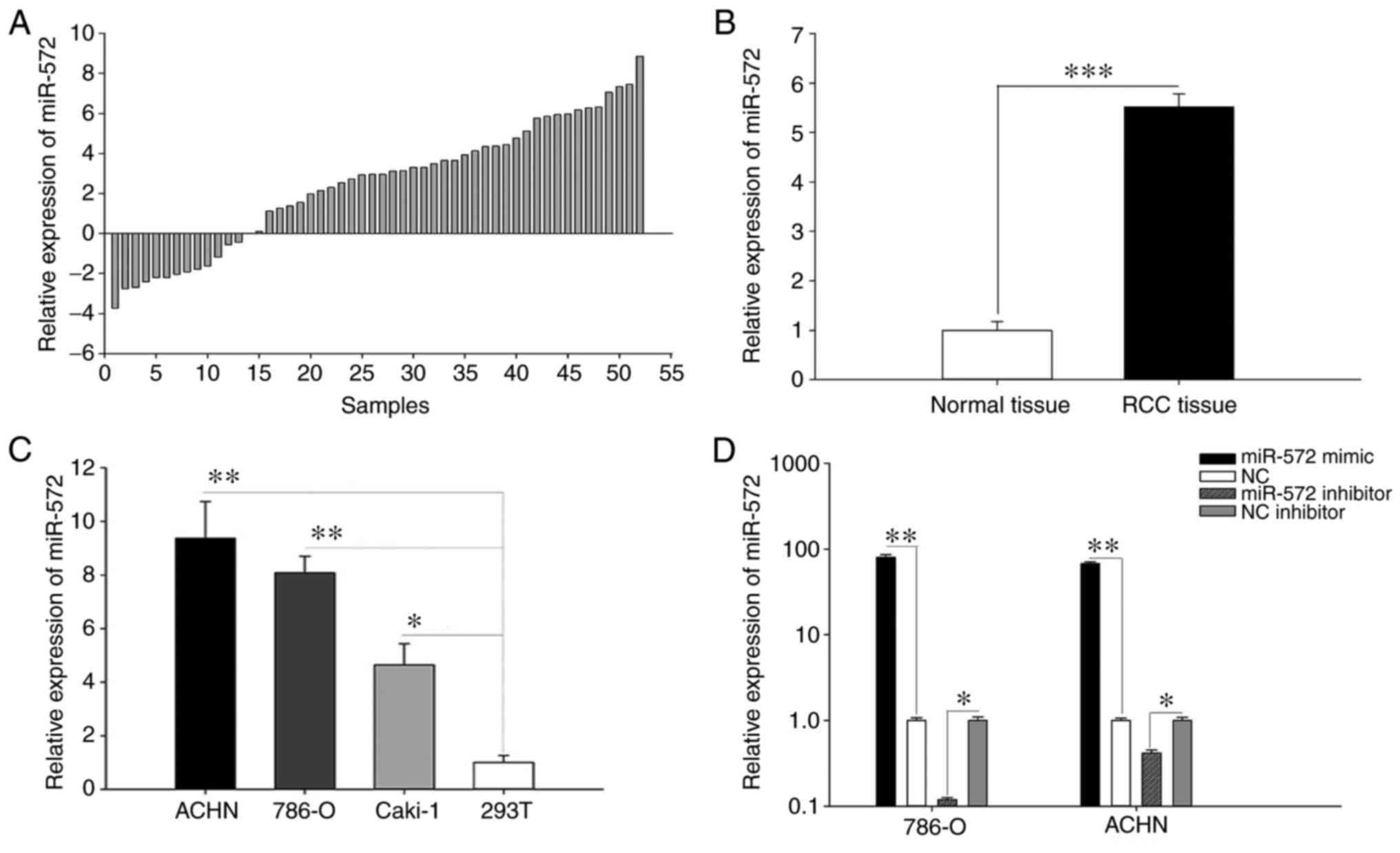
Upregulation of miR-572 was observed
in RCC tissues and cell lines
RT-qPCR was performed with 52 pairs of RCC tissues
and corresponding non-cancerous renal tissues with the purpose of
exploring the clinical association of miR-572 in RCC. As indicated
in Fig. 1A, the relative expression
levels of miR-572 in 52 paired tissues [log2(T/N)]) were
demonstrated and a total of 39 RCC tissues exhibited upregulation
of miR-572. Fig. 1B indicated the
mean expression of miR-572 and the results suggested that miR-572
was significantly upregulated in RCC tissues (5.515±0.263) compared
with corresponding non-cancerous renal tissues (1.000±0.179;
P<0.001). The relative expression levels of miR-572 in RCC cell
lines (786-O, Caki-1 and ACHN) and 293T cell lines were also
assessed (Fig. 1C). The results
indicated that the expression levels of miR-572 were significantly
increased in RCC cell lines (786-O, 8.084±0.618, P<0.01; ACHN,
9.372±1.371, P<0.01; and Caki-1, 4.643±0.793, P<0.05)
compared with 293T cells (1.000±0.265). These results indicated
that miR-572 may be up regulated in RCC tissues and possibly
functions as an oncogene in RCC.
Validation of cell transfection
efficiency
As indicated in Fig.
1C, the expression of miR-572 in ACHN and 780-O was most
different from 293T. Subsequently, ACHN and 780O cells were
selected for analysis in further functional assays. Validation of
the transfection efficiency of miR-572 mimic or inhibitor was
assessed using RT-qPCR. The outcomes revealed that the expression
levels of miR-572 in cells transfected with miR-572 mimic were
79.893 times higher (786-O cells, P<0.01) and 67.649 times
higher (ACHN cells, P<0.01) compared with NC. The expression
levels of miR-572 in cells transfected with miR-572 inhibitor
exhibited the opposite effect and were 0.119 times higher (786-O
cells, P<0.05) and 0.418 times higher (ACHN cells, P<0.05)
compared with NC inhibitor (Fig.
1D).
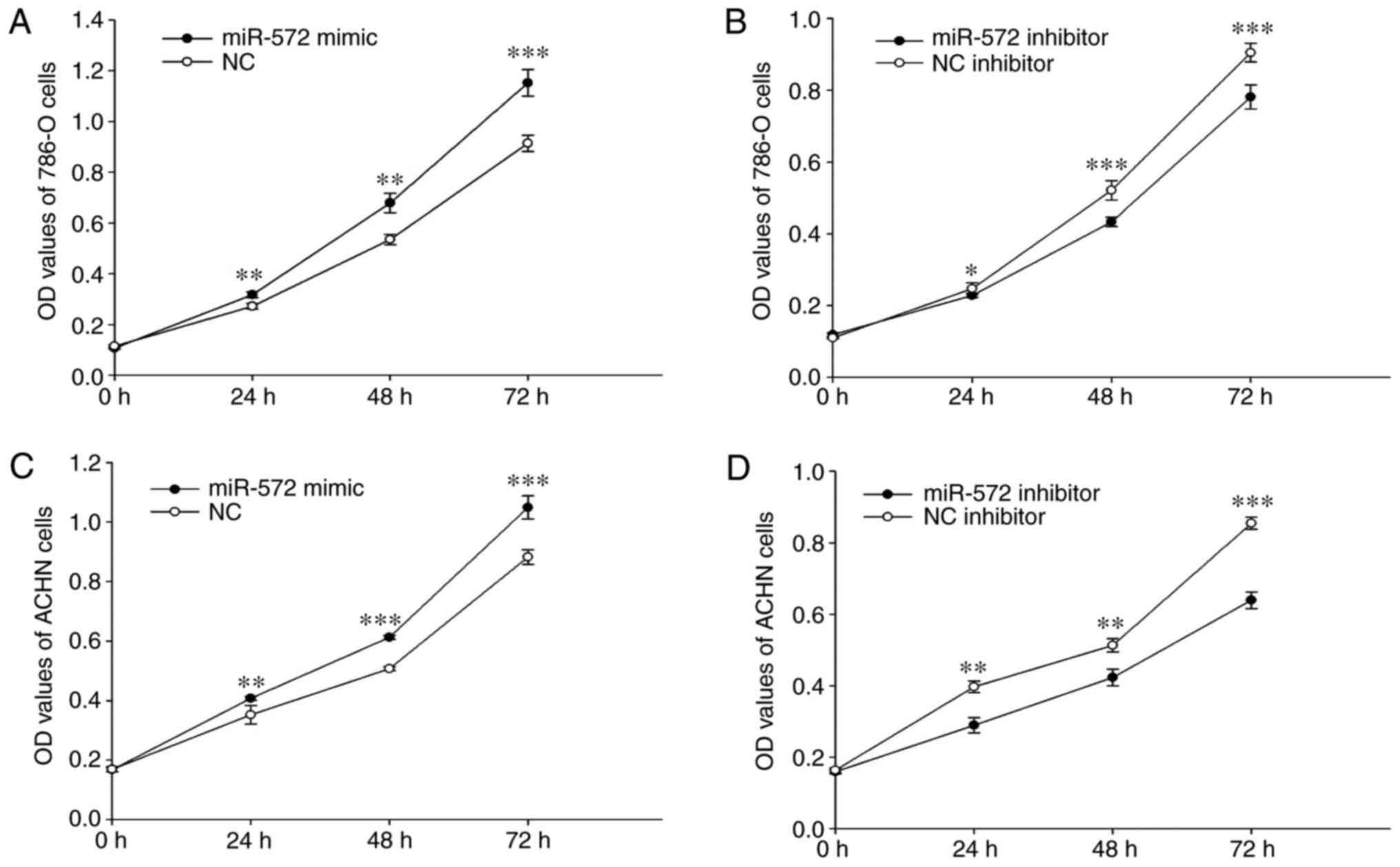
miR-572 promotes cell
proliferation
The role of miR-572 in cell proliferation was
investigated using the CCK-8 assay. The outcomes indicated that the
proliferation of 786-O significantly increased by 33.74%
(P<0.01), 37.58% (P<0.01) and 24.72% (P<0.001) following
transfection with miR-572 mimic compared with NC. However,
proliferation was significantly decreased by 25.18% (P<0.05),
33.90% (P<0.001) and 10.01% (P<0.001) following transfection
with miR-572 inhibitor compared with NC inhibitor at 24, 48 and 72
h (Fig. 2A and B). Similar outcomes
were obtained for ACHN cells. Notably, the proliferation was
significantly increased by 30.99% (P<0.01), 32.39% (P<0.001),
16.27% (P<0.001) but significantly decreased by 79.54%
(P<0.01), 3.37% (P<0.01), 57.70% (P<0.001) at 24, 48 and
72 h following transfection with miR-572 mimic or inhibitor
compared with the respective NC (Fig.
2C and D). These outcomes demonstrated that miR-572 may promote
cell proliferation.
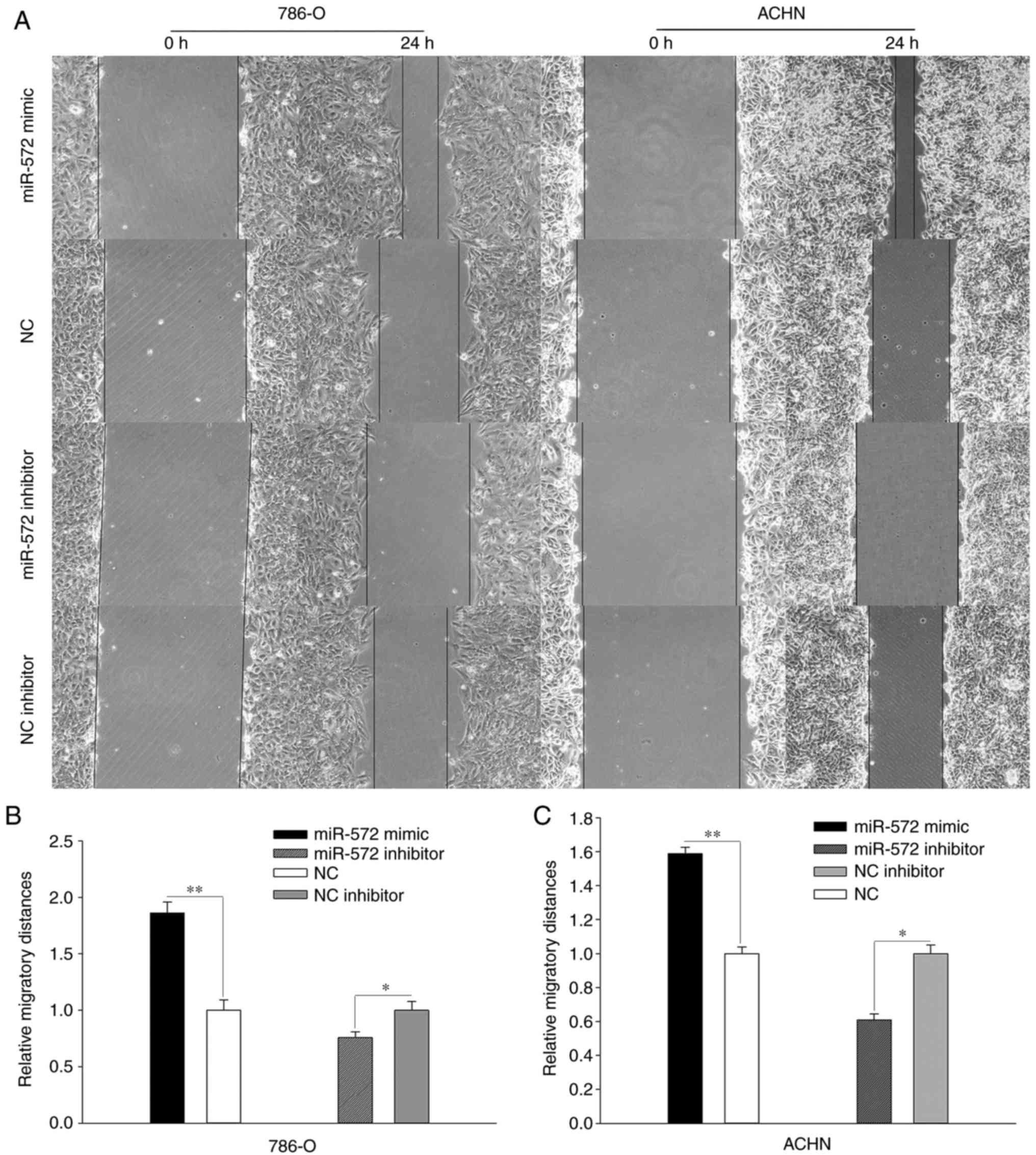
miR-572 promotes cell mobility
Assessment of the role of miR-572 of cell mobility
was performed using wound healing assays and Transwell assays
(Figs. 3 and 4). In the wound healing assay (Fig. 3A), 786-O and ACHN cells transfected
with miR-572 mimic exhibited significantly increased migration
compared with NC (Fig. 3B and C;
786-O, 1.862±0.096 vs. 1.000±0.092, P<0.01; ACHN, 1.589±0.038
vs. 1.000±0.038, P<0.01), whereas transfection with miR-572
inhibitor had an opposite result when compared with NC (Fig. 3B and C; 786-O, 0.756±0.052 vs.
1.000±0.079, P<0.05; ACHN, 0.609±0.034 vs. 1.000±0.051,
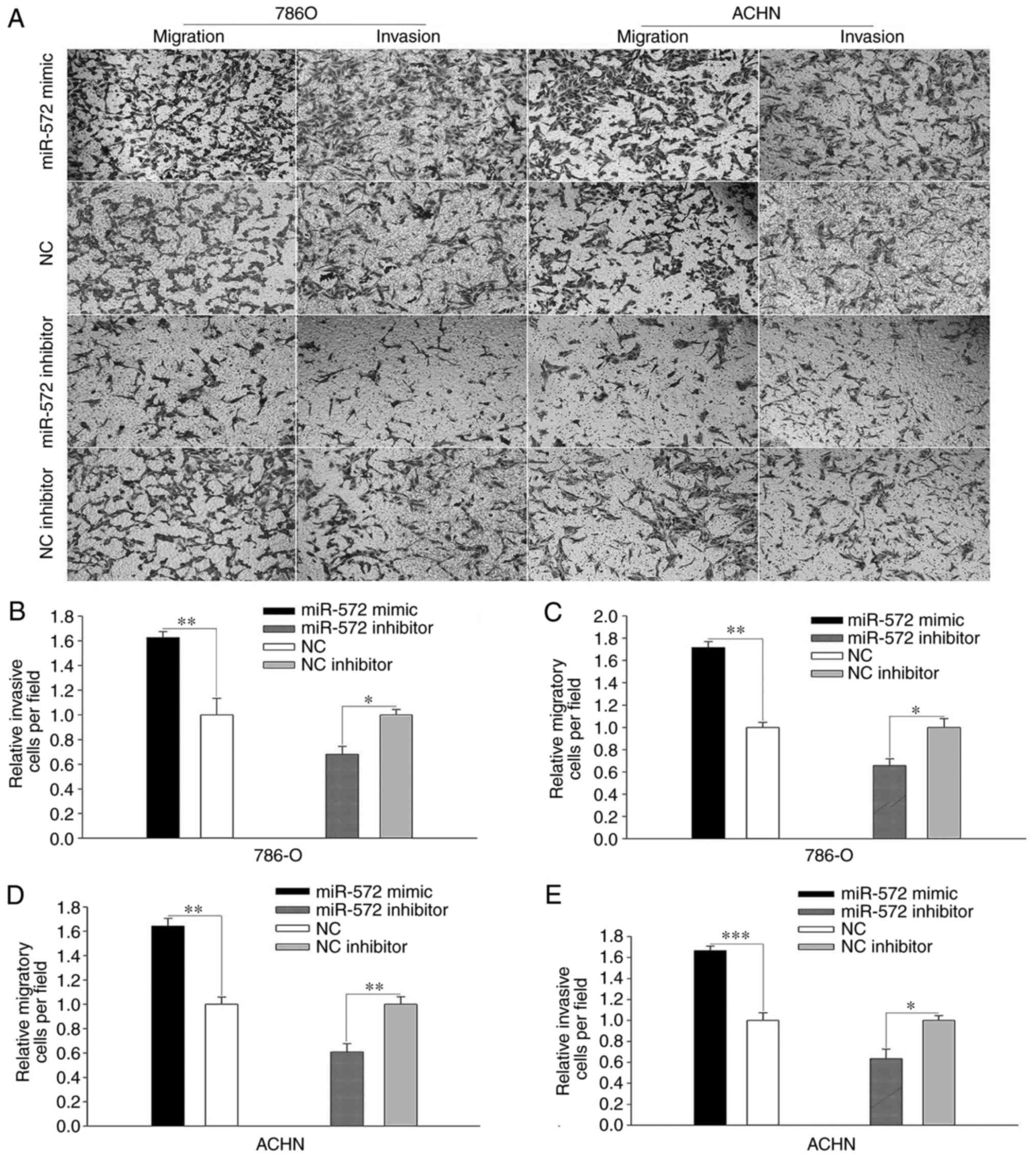
P<0.05). In the Transwell assay (Fig. 4A), the number of relative migratory
786-O and ACHN cells per field was significantly increased
following treatment with miR-572 mimic (Fig. 4C; 786-O, 1.717±0.053 vs.
1.000±0.046, P<0.01; Fig. 4E;
ACHN, 1.642±0.064 vs. 1.000±0.060, P<0.001). However, the number
of migratory cells was significantly decreased following treatment
with miR-572 inhibitor (Fig. 4C and
E, respectively; 786-O, 0.656±0.060 vs. 1.000±0.079, P<0.05;
and ACHN, 0.609±0.070 vs. 1.000±0.062, P<0.05). Notably,
elevated levels of miR-572 significantly promoted cell invasion
compared with the NC (Fig. 4B;
786-O, 1.626±0.048 vs. 1.000±0.133, P<0.05; Fig. 4D; ACHN, 1.667±0.043 vs. 1.000±0.074,
P<0.01), whereas these effects were significantly reversed by
miR-572 inhibitor when compared with NC inhibitor (Fig. 4B, 786-O, 0.680±0.065 vs.
1.000±0.043, P<0.05; Fig. 4D;
ACHN, 0.635±0.091 vs. 1.000±0.047, P<0.01). These outcomes
demonstrated that miR-572 may promote cell mobility.
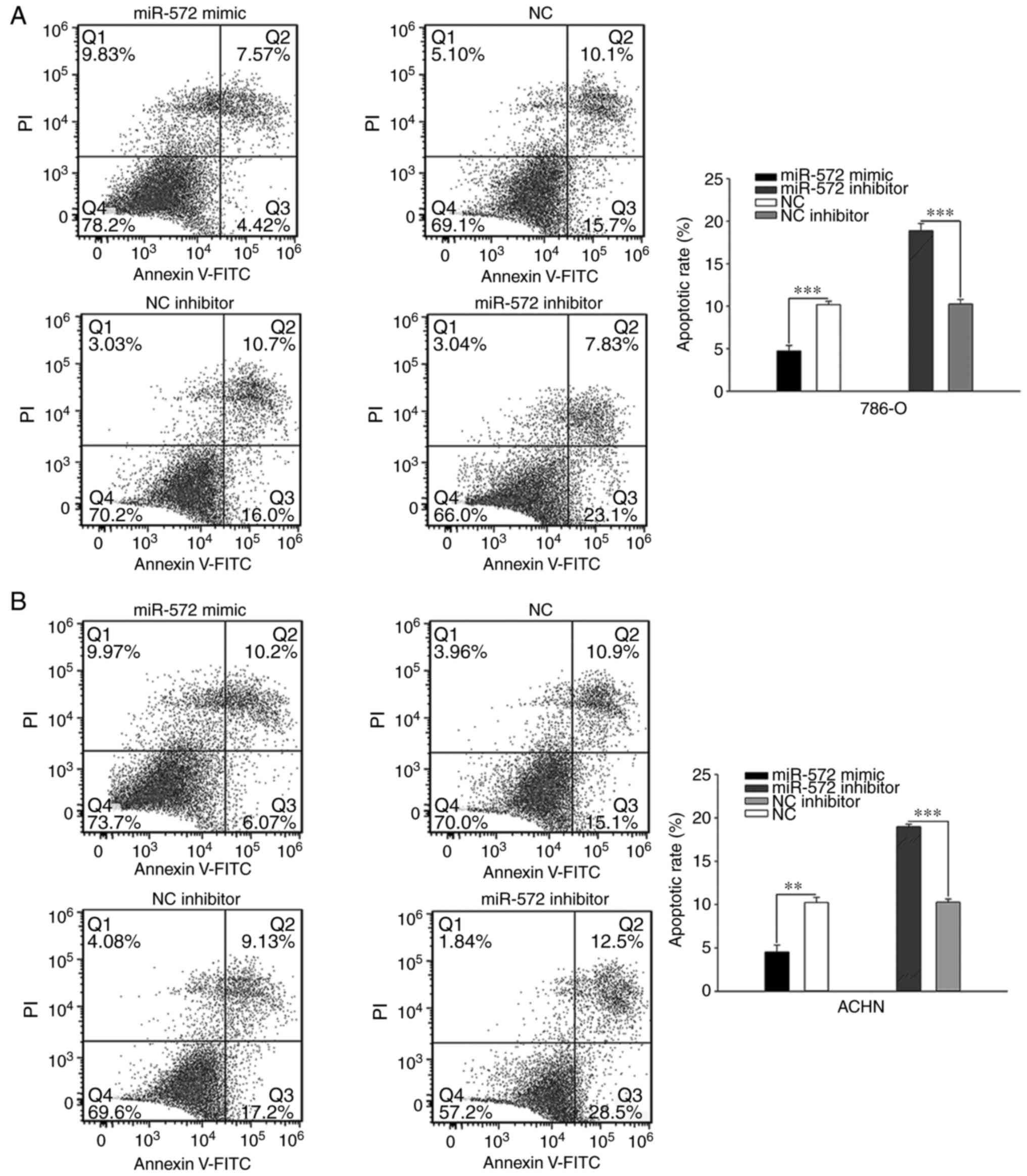
miR-572 inhibits early apoptosis in
cells
Assessment of cell apoptosis was performed using
flow cytometry. Elevated levels of miR-572 significantly inhibited
cell early apoptosis compared with NC (786-O, 4.73±0.635% vs.
10.17±0.404%, P<0.001; ACHN, 4.52±0.800% vs. 10.22±0.595%,
P<0.01), whereas these effects could be significantly reversed
by miR-572 inhibitor (786-O, 18.87±0.862% vs. 10.26±0.530%,
P<0.001; ACHN, 10.30±0.367% vs. 18.96±0.306%, P<0.001;
Fig. 5). These outcomes
demonstrated that miR-572 may inhibit early apoptosis in cells.
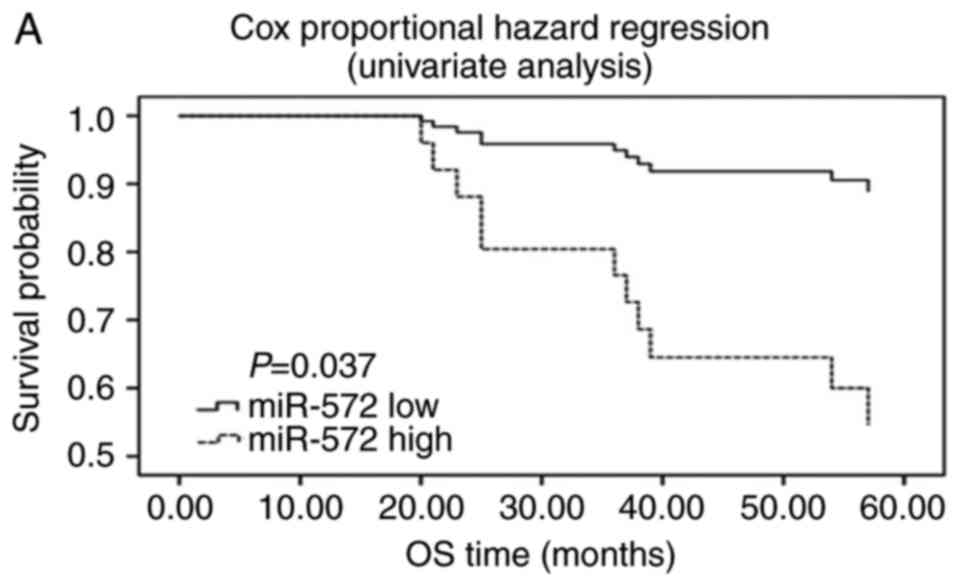
miR-572 serves as a potential
independent prognostic marker for RCC
The association between the expression levels of
miR-572 and clinicopathological variables or overall survival was
analyzed with the FFPE renal cancer samples. As presented in
Table II, no significant
difference was observed between miR-572 expression level and sex,
age, tumor size and tumor stage. However, univariate analysis
(Fig. 6A) suggested that the
expression level of miR-572 was inversely correlated with overall
survival (HR=0.195, 95% CI=0.042–0.903, P=0.037; Table IV). Similar outcomes were indicated
in the survival curves of Cox proportional hazard regression
analysis (Fig. 6B), which suggested
that the patients with low miR-572 expression levels had a
significantly increased overall survival after adjusting for sex,
age, tumor size and tumor stage in the multivariate analysis
(HR=0.174, 95% CI=0.034–0.878, P=0.034; Table IV). Furthermore, data of the
Kaplan-Meier survival curves demonstrated that the patients with
low expression of miR-572 had a significantly longer overall
survival (P=0.019, Fig. 6C). These
outcomes demonstrated that miR-572 may serve as a potential
independent prognostic marker for RCC.
 | Table IV.miR-572 expression and patients'
survival. |
Table IV.
miR-572 expression and patients'
survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex (female vs.
male) | 2.037
(0.537–7.720) | 0.295 | 4.672
(0.830–26.287) | 0.080 |
| Age (≤60 vs. >60
years) | 0.760
(0.164–3.519) | 0.726 | 2.054
(0.301–14.012) | 0.462 |
| Tumor size (≤4.0
vs. >4.0 cm) | 8.458
(1.081–66.188) | 0.042 | 10.184
(1.213–85.507) | 0.033 |
| Tumor stage (I+II
vs. III+IV) | 1.014
(0.296–3.473) | 0.982 | 0.415
(0.112–1.533) | 0.187 |
| miR-572 (low vs.
high expression) | 0.195
(0.042–0.903) | 0.037 | 0.174
(0.034–0.878) | 0.034 |
Discussion
RCC, which accounts for the 7th and the 9th most
common malignancy in men and women worldwide, respectively, gives
rise to ~20,9000 new cases and 102,000 fatalities every year
(5,21). There is a general consent that
modulation of miR expression serves a momentous role in oncogene
and anti-oncogene proteins by mediating their effects (22). miRs have been indicated to be
implicated in numerous biological processes, including
proliferation, apoptosis, metabolic reprogramming and pluripotency
(23). In recent years, an
increasing number of tumor signaling pathways that interact with
miRs were revealed to be associated with RCC, including the HIF-1
signaling pathway, mTOR signaling pathway and c-MET signaling
pathway (24). Results from Dey
et al (25) demonstrated
that miR-21 functions as an oncogene in RCC and overexpression of
miR-21 increases tumor cell proliferation and invasion via
Akt/TORC1 signaling. Another study revealed that the oncomir
miR-23b promotes kidney tumor growth by inhibiting proline oxidase,
which functions as a mitochondrial tumor suppressor by decreasing
hypoxia-inducible factor signaling (26).
Accumulating evidence has suggested that miRs serve
an irreplaceable role in the genesis and development of RCC. For
example, tumor suppressor miR-338-3p suppresses the expression of
ALK5 in RCC, which results in the inhibition of cell invasion
(27). Furthermore, miR-766-3p,
mediated by DNA-methylation, attenuates the proliferation ability
of RCC and overexpression of miR-766-3p is associated with clinical
stage and poor survival (28).
Circulating miR-572 has been demonstrated to be significantly
increased in RCC by TaqMan Low Density Array and RT-qPCR analysis
in a previous study (18). However,
the expression and roles of miR-572 in RCC tissues and cell lines
remain unclear.
In the present study, results of RT-qPCR suggested
that the expression of miR-572 in RCC tissues and 786-O and ACHN
cells was significantly increased compared with that in
corresponding non-cancerous renal tissues and 293T cells,
respectively. These result are in accordance with the expression
levels of miR-572 determined in nasopharyngeal carcinoma (15), ovarian cancer (16) and human neuroblastoma (29). Further experiments with wound
healing assays, CCK-8 assays, Transwell assays and flow cytometric
assays suggested that overexpression of miR-572 results in a
positive effect on RCC cell proliferation, migration and invasion
and inhibition of early apoptosis, respectively. Notably, opposite
effects were observed in the case of suppressing the expression of
miR-572, suggesting that miR-572 functions as an oncogene in RCC.
Further prognostic analysis demonstrated that patients with
downregulated miR-572 had a significantly longer overall survival,
suggesting miR-572 may be a promising independent prognostic marker
for RCC.
The molecular mechanisms concerning miR-572 in
mediating the genesis and development of tumors are well documented
in numerous types of cancer and diseases. Results from Yu et
al (30) provided evidence that
miRNA-572 accelerates the recovery of early post-operative
cognitive dysfunction by means of inhibiting neural cell adhesion
molecule1. In human ovarian cancer, miR-572 was revealed to be
upregulated in cancer tissues as well as cell lines, which was
associated with poor prognosis (31). Furthermore, miRNA-572 was indicated
to directly inhibit SOCS1 and p21, which promoted the proliferation
of ovarian cancer cells (31). In
another study concerning ovarian cancer, suppression of PPP2R2C
expression made miR-572 conducive to tumor cell proliferation,
which suggests that miR-572 may be a target for treating ovarian
cancer (16). Similarly, Yan et
al (15) demonstrated that
overexpression of miR-572 and downregulation of PPP2R2C had the
same effect on nasopharyngeal carcinoma (accelerated nasopharyngeal
carcinoma cell proliferation and invasion). Furthermore, this study
indicated that miR-572 binds to the 3′-untranslated region of
PPP2R2C, leading to the inhibition of its expression, which
suggests that miR-572 exerts its functions by downregulating
PPP2R2C.
The molecular mechanism mediated by miR-572 in RCC
will be explored in our future study. To understand the molecular
mechanism of miR-572 in RCC, two databases (targetScan and miRWalk)
will be used and three genes (PPP2R2C, CIB2 and BAG1) will be
selected as potential targets that are regulated by miR-572.
In conclusion, upregulation of miR-572 was indicated
in RCC tissues and cell lines. Further experiments revealed that
overexpression of miR-572 results in a positive effect on the
proliferation, migration and invasion of cells and inhibition of
early apoptosis, which suggests that miR-572 acts as oncogene in
RCC. Further prognostic analysis demonstrated that patients with
downregulated miR-572 expression have a significantly reduced
overall survival, which suggests that miR-572 may be a promising
independent prognostic marker for RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Development Fund Project of Shenzhen (grant no.
JCYJ20170307111334308), Clinical Research Project of Shenzhen
Health Commission (grant no. SZLY2018023) and the Fund of
‘San-ming’ Project of Medicine in Shenzhen (grant no.
SZSM201612066).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XP and ZL performed experimental work (RT-qPCR,
wound scratch assay, cell proliferatin assay, Transwell assay and
flow cytometry assay) and aided in writing the manuscript. LiwZ
collected tissue samples. JQ, LiaZ and JX designed the study,
coordinated the molecular biology experiment and assisted in
writing the manuscript. WX, XG and HL designed the study and
observed the patients. SY designed the study, recruited patients
and examined controls. YG extracted the total RNA from the FFPE
samples. YL collected tissue samples and edited the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Peking University Shenzhen Hospital (Shenzhen, China).
Patient consent for publication
Written informed consent was obtained from all
patients for publication of data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Petrozza V, Pastore AL, Palleschi G, Tito
C, Porta N, Ricci S, Marigliano C, Costantini M, Simone G, Di Carlo
A, et al: Secreted miR-210-3p as non-invasive biomarker in clear
cell renal cell carcinoma. Oncotarget. 8:69551–69558. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
King SC, Pollack LA, Li J, King JB and
Master VA: Continued increase in incidence of renal cell carcinoma,
especially in young patients and high grade disease: United States
2001 to 2010. J Urol. 191:1665–1670. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patel C, Ahmed A and Ellsworth P: Renal
cell carcinoma: A reappraisal. Urol Nurs. 32:182–190; quiz 191.
2012.PubMed/NCBI
|
|
7
|
Hadoux J, Vignot S and De La Motte Rouge
T: Renal cell carcinoma: Focus on safety and efficacy of
temsirolimus. Clin Med Insights Oncol. 4:143–154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Homami A and Ghazi F: MicroRNAs as
biomarkers associated with bladder cancer. Med J Islamic Repub
Iran. 30:4752016.
|
|
9
|
Enokida H, Yoshino H, Matsushita R and
Nakagawa M: The role of microRNAs in bladder cancer. Investig Clin
Urol. 1 Suppl 57:S60–S76. 2016. View Article : Google Scholar
|
|
10
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding D, Zhang Y, Wen L, Fu J, Bai X, Fan
Y, Lin Y, Dai H, Li Q, Zhang Y, et al: MiR-367 regulates cell
proliferation and metastasis by targeting metastasis-associated
protein 3 (MTA3) in clear-cell renal cell carcinoma. Oncotarget.
8:63084–63095. 2017.PubMed/NCBI
|
|
13
|
Cai W, Jiang H, Yu Y, Xu Y, Zuo W, Wang S
and Su Z: miR−367 regulation of DOC-2/DAB2 interactive
protein promotes proliferation, migration and invasion of
osteosarcoma cells. Biomed Pharmacother. 95:120–128. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang
YH, Liu P, Hong M, Miao KR, Liu P, et al: miR-181a/b significantly
enhances drug sensitivity in chronic lymphocytic leukemia cells via
targeting multiple anti-apoptosis genes. Carcinogenesis.
33:1294–1301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yan L, Cai K, Liang J, Liu H, Liu Y and
Gui J: Interaction between miR-572 and PPP2R2C, and their effects
on the proliferation, migration, and invasion of nasopharyngeal
carcinoma (NPC) cells. Biochem Cell Biol. 95:578–584. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu AH, Huang YL, Zhang LZ, Tian G, Liao QZ
and Chen SL: MiR-572 prompted cell proliferation of human ovarian
cancer cells by suppressing PPP2R2C expression. Biomed
Pharmacother. 77:92–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Tian H, Yuan J, Wu H, Wu J and Zhu
X: CONSORT: Sam68 is directly regulated by MiR-204 and promotes the
self-renewal potential of breast cancer cells by activating the
wnt/beta-catenin signaling pathway. Medicine. 94:e22282015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hatziapostolou M, Polytarchou C and
Iliopoulos D: miRNAs link metabolic reprogramming to oncogenesis.
Trends Endocrinol Metab. 24:361–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sellitti DF and Doi SQ: MicroRNAs in renal
cell carcinoma. Microrna. 4:26–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dey N, Das F, Ghosh-Choudhury N, Mandal
CC, Parekh DJ, Block K, Kasinath BS, Abboud HE and Choudhury GG:
microRNA-21 governs TORC1 activation in renal cancer cell
proliferation and invasion. PLoS One. 7:e373662012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu W, Zabirnyk O, Wang H, Shiao YH,
Nickerson ML, Khalil S, Anderson LM, Perantoni AO and Phang JM:
miR-23b* targets proline oxidase, a novel tumor suppressor protein
in renal cancer. Oncogene. 29:4914–4924. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Wang C, Li H, Niu X, Liu X, Pei
D, Guo X, Xu X and Li Y: miR-338-3p inhibits the invasion of renal
cell carcinoma by downregulation of ALK5. Oncotarget.
8:64106–64113. 2017.PubMed/NCBI
|
|
28
|
Chen C, Xue S, Zhang J, Chen W, Gong D,
Zheng J, Ma J, Xue W, Chen Y, Zhai W, et al:
DNA-methylation-mediated repression of miR-766-3p promotes cell
proliferation via targeting SF2 expression in renal cell carcinoma.
Int J Cancer. 141:1867–1878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mundalil Vasu M, Anitha A, Takahashi T,
Thanseem I, Iwata K, Asakawa T and Suzuki K: Fluoxetine increases
the expression of miR-572 and miR-663a in human neuroblastoma cell
lines. PLoS One. 11:e01644252016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu X, Liu S, Li J, Fan X, Chen Y, Bi X,
Liu S and Deng X: MicroRNA-572 improves early post-operative
cognitive dysfunction by down-regulating neural cell adhesion
molecule 1. PLoS One. 10:e01185112015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu
J, Wu G, Li J and Jiang L: Upregulation of miR-572
transcriptionally suppresses SOCS1 and p21 and contributes to human
ovarian cancer progression. Oncotarget. 6:15180–15193.
2015.PubMed/NCBI
|