Introduction
Pancreatic cancer is malignancy of the digestive
system. The overall survival outlook for pancreatic cancer
worldwide is still grim. Although the survival rate of pancreatic
cancer patients has steadily increased, progress in research is
very slow and there is still no treatment that is effective and no
early diagnosis that is reliable (1–3).
Pancreatic cancer remains the most severe cancer worldwide, with a
total survival rate in 5 years below 5% (4). Thus, searching for more effective
treatments and strategies to improve the therapeutic effect, and
discovering the generation and molecular mechanisms of metastasis
in pancreatic cancer is urgently needed. The family of Krupple-like
transcription factor (KLF) is comprised of 17 distinctive members,
which have a variety of cellular functions (5). KLF9 is one of the most important
genes. KLF9 is an important nuclear transcription factor expressed
widely and conserved highly in higher mammals and human tissues
such as the brain, liver and uterus (6,7). KLF9
is a member of the KLF transcription factor family (8). Studies in tumors have revealed that
the role of KLFs is conditionally dependent (9), and possess tumor suppressive and
proto-oncogenic effects on tumors (10,11). A
recent study (12) revealed that
KLF9 exhibited low expression in the esophagus and could bind to
TCF4 to inhibit the β-catenin/TCF signaling pathway, thus
suppressing tumor growth. In colorectal cancer, compared with
normal tissue, KLF9 transcription and protein levels were revealed
to be significantly downregulated (13). In endometrial cancer, KLF9
expression was also significantly downregulated. In these tumors,
inhibition of downstream molecules of actin skeletal protein
regulatory factors was detected after overexpression of KLF9
(14). However, the expression of
KLF9 in pancreatic cancer has not been investigated. In addition,
the effects of KLF9 on apoptosis, the cell cycle and
epithelial-mesenchymal transition (EMT) in pancreatic cancer remain
to be fully elucidated.
More and more evidence has revealed that KLF9 is not
only involved in many important biological events, including cell
proliferation growth (15) and
immune response (16), but is also
closely related to the occurrence and development of multiple
tumors (17–20). We observed that upregulation of KLF9
expression had an obvious effect on proliferation, apoptosis,
invasion and migration of PANC-1 and BxPC-3 cells in vitro
and this study may provide data on KLF9 as a prognostic marker and
therapeutic target drug on pancreatic cancer.
Materials and methods
Patients and tissue samples
The Second Affiliated Hospital of Nanchang
University provided 60 pairs of specimens of pancreatic cancer and
matched paracancerous tissue. Using histopathology according to the
World Health Organization (WHO) standards, these patients (32 male
and 28 female patients; 26 patients under the age of 60 and 34
patients over 60 years) were diagnosed with pancreatic cancer
(21) and underwent surgery from
January 2011 to March 2015. Fresh tissue samples were cut into 4-mm
cubes, and snap-frozen in liquid nitrogen and stored at −80°C.
According to the American Cancer Joint Committee (AJCC), clinical
data, including distant metastasis, lymph node metastasis, depth of
invasion, tumor differentiation, tumor location, tumor size, age
and sex, were obtained from medical records (22). The Ethics Committee of the Second
Affiliated Hospital of Nanchang University approved our study, and
all participants signed and provided written informed consent. This
study was conducted by the ethical standards of the Helsinki
Declaration.
Immunohistochemistry
Paraffin-embedded tissue were cut into 4-µm thick
sections and stained with hematoxylin and eosin. Using a two-step
immunohistochemistry method, we first deparaffinized the tissue
sections two times in xylene, 10 min each time, rehydrated with a
fractional alcohol series (100–50%) for 2 min, and then the
sections underwent antigen repair at 130°C for 10 min in a pressure
cooker. To block the potential endogenous peroxidase activity, we
cultured the sections with hydrogen peroxide (0.3%), washed them 3
times with phosphate-buffered saline (PBS) for 3 min, and then
placed them at room temperature with 20% normal goat serum for 30
min. Then, we cultured the sections with mouse monoclonal anti-KLF9
antibody (cat. no. sc-517075; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) at a dilution of 1:200 at 4°C overnight. The
following day it was washed three times with PBS. Next, the
sections were cultured with biotin-labeled goat anti-mouse IgG
(dilution 1:200; cat. no. PV-6000ZSGB-Bio; OriGene Technologies,
Inc., Beijing, China) at 37°C for 30 min. The slices were reacted
with 3,3′-diaminobenzidine solution, stained with hematoxylin and
covered with coverslips after being washed with PBS three times.
Immunohistochemistry sections were then examined and scored
independently using blind pathology on a light microscope (Nikon
Corp., Tokyo, Japan). The scoring system was calculated according
to the dyeing intensity (grade 0 indicated no staining; grade 1
indicated mild staining; grade 2 indicated moderate staining; grade
3 indicated strong staining) and the percentage of dyeing (grade 0
indicated no staining; grade 1 indicated staining percentage
<10%; grade 2 indicated dyeing percentage 10–40%; grade 3
indicated dyeing percentage >40%). Then, according to previous
studies, the total staining index was acquired by multiplying each
immune staining score by 0 to 9, as follows: 0–1 indicated KLF9
negative expression, or 2–9 indicated KLF9 positive expression
(23).
Culture of cell lines
The Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China) provided Sw1990, BxPC-3, Capan-1 and
PANC-1 pancreatic cancer cell lines. Rui-Lu Biotech (Shanghai,
China) provided the non-transformed pancreatic epithelial cell line
HPDE6c7 (non-cancerous line). We cultured cells in high glucose
medium DMEM (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 9 and 10% fetal bovine serum (FBS; Hyclone;
GE Healthcare Life Sciences, Logan, UT, USA) and 1% sodium pyruvate
(Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified
incubator with 95% air and 5% CO2 maintained at 37°C. We
grew cells in the exponential phase and sub-cultured them at nearly
80% confluence.
Protein extraction and
immunoblotting
Using RIPA buffer (Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) with 1% phenylmethanesulfonyl fluoride, we
extracted total cell proteins. We quantified them with the Bradford
method. Next, we separated the protein samples (20 µg per lane) in
SDS-PAGE capsules with 10% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis (SDS-PAGE). We then transferred them to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). In the immunoblotting process, we first cultured the
membranes with 5% bovine serum albumin (BSA) in Tris-buffered
saline-Tween-20 (TBS-T). Next, we cultured them overnight at 4°C
with the corresponding primary antibody. The following day, we
washed the membranes three times with TBS-T, each time for 10 min.
Then, the membranes were cultured with horseradish peroxidase
conjugated secondary antibody for 1 h at ambient temperature
followed by enhanced chemiluminescent reagent (ECL; EMD Millipore).
The proteins CDK4 (cat. no. PKA-325), cyclin B (cat. no. 7518) and
cyclin D1 (cat. no. P4190-10) were obtained from bioWORLD (Dublin,
OH, USA); p53 (cat. no. 2527), MMP-9 (cat. no. 13667), MMP-2 (cat.
no. 13132), N-cadherin (cat. no. 13116), E-cadherin (cat. no.
3195), Bcl-2 (cat. no. 4223) and Bax (cat. no. 2772) were purchased
from Cell Signaling Technology, Inc. (Danvers, MA, USA). We applied
all the antibodies following the instructions of the manufacturers.
Anti-GAPDH (ZSGB-Bio; OriGene Technologies, Inc.) served as an
endogenous reference. All the primary antibodies were diluted at a
ratio of 1:1,000 and the secondary antibody was diluted at a ratio
of 1:5,000.
saRNA and gene transfection
We obtained Human KLF9 saRNA reagent from Shanghai
GenePharma Co., Ltd. (Shanghai, China), with the sequences:
KLF9_s1, 5′-UGUGCAGUAAUCCUUCCAGTT-3′, KLF9_s2,
5′-UUCGAUCGCUUGAUCAUGCTT-3′ and KLF9_s3,
5′-UUAACGUGAUUCAAGAGAGTT-3′ respectively. GAPDH saRNA served as the
positive control sequence (5′-UGACCUCAACUACAUGGUUTT-3′) and the
negative control sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The
pancreatic cancer BxPC-3 and PANC-1 cell lines grew in a six-well
plate with 95% air and 5% CO2 using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) reagent following the
manufacturer's instructions on transient transfection of saRNAs.
After 48 h, the transfected cells could be used for further assay
or protein extraction.
Flow cytometric cell cycle and
apoptosis assays
An saRNA fragment was transfected transiently by
using Lipofectamine 2000 and the cell cycle distribution was
analyzed as follows 48 h later. Cells (1×106) were
harvested with trypsin digestion, washed using PBS, and fixed using
70% ethanol overnight at 4°C. The following day, we re-suspended
the cells in 500 µl of propidium iodide (PI)/RNase staining
solution (Sungene Biotech Co., Ltd., Tianjin, China) and cultured
for 30 min at 37°C. Next, we studied the samples using a FACScan
flow cytometer (BD Immunocytometry Systems, San Jose, CA, USA). For
the evaluation of cell apoptosis, using the Annexin V-FITC/PI
Apoptosis Detection kit (Nanjing KeyGen Biotech Co., Ltd.), we
assessed the cells duplicated from the cell cycle analysis,
following the manufacturer's instructions. In brief, we washed
these cells in ice-cold PBS and cultured them with Annexin V-FITC
and PI solutions for 15 min in the dark. Next, using FACScan flow
cytometry, we studied them for apoptosis. We studied no less than
105 cells for each sample, which were analyzed by flow cytometry
(BD Biosciences, San Jose, CA, USA) within 1 h. Finally, we
summarized the data as the mean ± standard error (SD).
Cell proliferation assay
Using CCK-8 assay kit (KeyGen Biotech Co., Ltd.)
after cell KLF9 overexpression, we assessed cell proliferation. We
seeded the cells in 96-well plates at a density of 5,000
cells/well, cultured them for 12 h, and then promptly transfected
them using the saRNAs aforementioned. The cells were cultured for
24, 48, 72 and 96 h, respectively. At the end of each experiment,
10 µl of the CCK-8 reagent cell culture medium was added to each
well; then the mixture was cultured for another hour and assessed
under a microplate reader at an optical density of 450 nm
(PerkinElmer, Inc., Waltham, MA, USA). The data was expressed as
the mean ± standard error. Each group was set up with three wells
and repeated at least 3 times.
Transwell migration and invasion of
tumors
Migration and invasion of tumor cells were assessed
using a modified 24-well filter Boyden chamber with the filter
either precoated or uncoated with Matrigel (BD Biosciences,
Shanghai, China). At a concentration of 1×106 cells/ml,
saRNA cells grew and were transfected transiently with pancreatic
cancer cells in serum-free DMEM. Next, 200 µl of cell solution was
added into the upper chamber of a filter with 8-µm pores and DMEM
with 20% FBS was added in the bottom chamber. After being cultured
for 48 h, the non-invaded cells and Matrigel were wiped away
carefully. Cells that had migrated or invaded the lower chamber of
the filter were fixed in 4% paraformaldehyde and stained with 0.5%
crystal violet. At an ×10 magnification under inverted microscope,
we counted the number of invaded or migrated cells in 5 randomly
selected fields (Nikon Corp.).
Statistical analysis
Using the Kaplan-Meier curve, we assessed total
survival and KLF9 expression was assessed using the log rank test.
The in vitro data are expressed as the means ± standard
error and performed using one-way analysis of variance. Multiple
comparison between the groups was performed using a
Student-Newman-Keuls (S-N-K) test. All assays were performed
independently three times. Using SPSS v17.0 software (SPSS, Inc.,
Chicago, IL, USA), we conducted all statistical analyses. A value
of P<0.05 was considered to indicate a statistically significant
difference.
Results
The relationship between the clinicopathological
features of pancreatic cancer and the expression of KLF9 was
revealed in Table I. Our data
indicated that the expression of KLF9 in tumor tissue was related
to the depth of vascular invasion (P=0.016) and differentiation
(P<0.001). However, KLF9 expression was not related to tumor
location, age, sex, metastasis of lymph node, nerve invasion and
TNM stage. The Kaplan-Meier curve was used to analyze the
relationship between the expression of KLF9 in pancreatic cancer
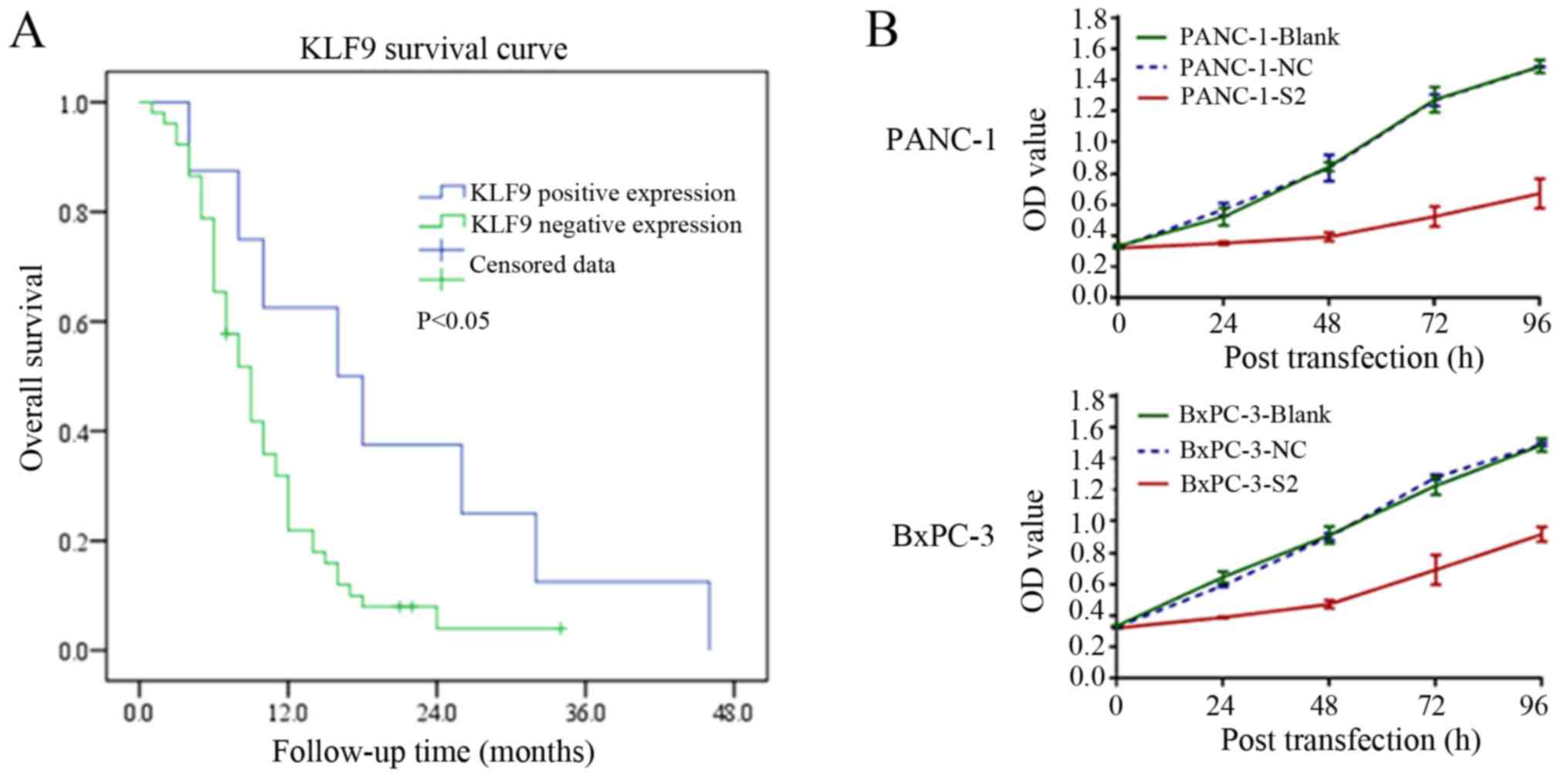
and the total survival of patients. Our data revealed that
overexpression of KLF9 was related to the total survival of these
patients (P<0.05; Fig. 1A).
 | Table I.Relationship between the expression
level of KLF9 and the clinicopathological parameters of pancreatic
cancer patients. |
Table I.
Relationship between the expression
level of KLF9 and the clinicopathological parameters of pancreatic
cancer patients.
|
|
| KLF9
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameters | n | High [n (%)] | Low [n (%)] | P-value |
|---|
| Sex |
|
|
| 0.712 |
|
Male | 32 | 5 (15.6) | 27 (84.4) |
|
|
Female | 28 | 3 (10.7) | 25 (89.3) |
|
| Age (years) |
|
|
| 0.717 |
|
<60 | 26 | 4 (15.4) | 22 (84.6) |
|
|
≥60 | 34 | 4 (11.8) | 30 (88.2) |
|
| Tumor location |
|
|
| 0.067 |
| Head
and neck | 34 | 2 (5.9) | 32 (94.1) |
|
| Body
and tail | 26 | 6 (23.1) | 20 (76.9) |
|
| Tumor
differentiation |
|
|
| <0.001 |
|
High | 5 | 5 (100) | 0 (0) |
|
|
Medium | 18 | 3 (16.7) | 15 (83.3) |
|
|
Low | 37 | 0 (15) | 37 (100) |
|
| AJCC stage |
|
|
| 0.698 |
|
I–II | 37 | 6 (16.2) | 31 (83.8) |
|
|
III–IV | 23 | 2 (8.7) | 21 (91.3) |
|
| Nerve invasion |
|
|
| 0.429 |
| No | 16 | 3 (18.8) | 13 (81.2) |
|
|
Yes | 44 | 5 (11.4) | 39 (88.6) |
|
| Vascular
invasion |
|
|
| 0.016 |
| No | 35 | 8 (22.9) | 27 (77.1) |
|
|
Yes | 25 | 0 (0) | 25 (100) |
|
| Lymph node
metastasis |
|
|
| 0.130 |
| Absent
(N0) | 32 | 2 (6.3) | 30 (93.7) |
|
| Present
(N1-3) | 28 | 6 (21.4) | 22 (78.6) |
|
We evaluated the effect of KLF9 overexpression on
the proliferation, cell cycle distribution, and apoptosis of
pancreatic cancer cells. In particular, KLF9 overexpression
significantly reduced the proliferation abilities of PANC-1 and
BxPC-3 cells in comparison with positive saRNA control cells
(P<0.01; Fig. 1B). The
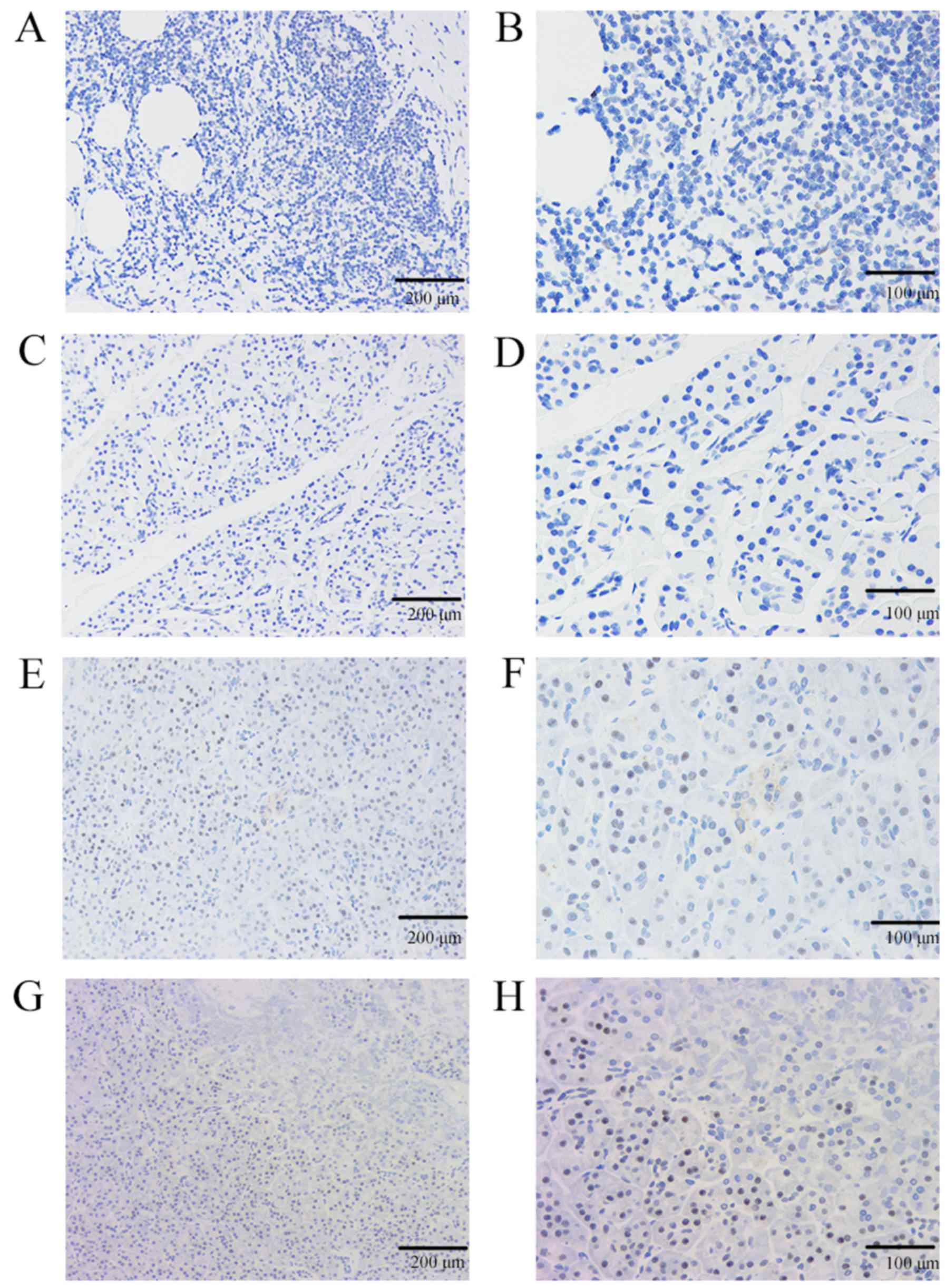
expression of KLF9 in pancreatic cancer and matched paracancerous
tissue samples was first evaluated here, and KLF9 revealed low
expression or no expression (52/60, 86.67%) in pancreatic cancer
tissues, whereas it exhibited a high expression in adjacent tissues
of pancreatic cancer (56/60, 93.33%) (Fig. 2). Thus, compared with the expression
in matched paracancerous tissues the expression of KLF9 in 52
pancreatic cancer tissues was lower (P<0.001).
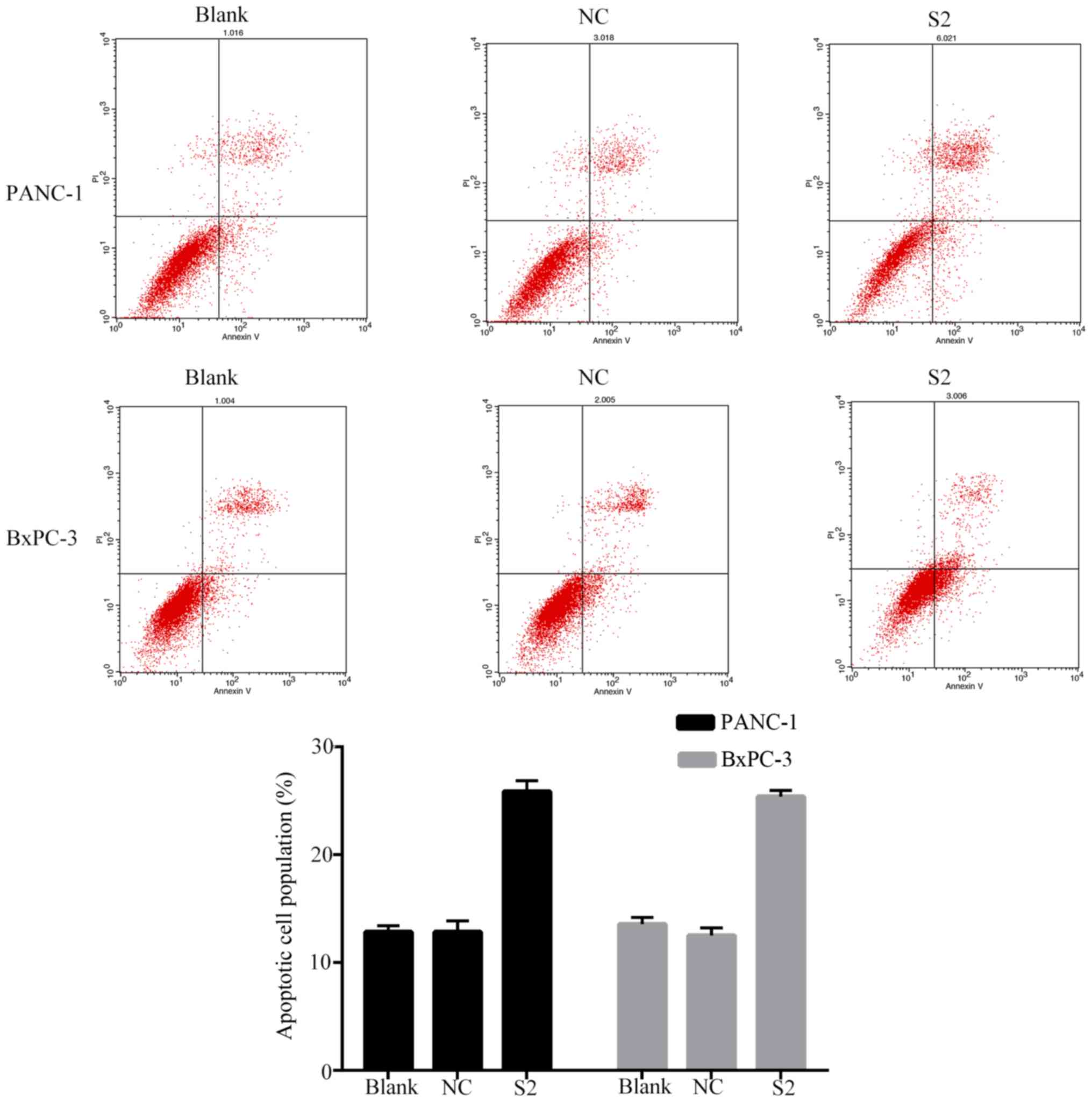
Furthermore, we found that transfection of KLF9
saRNA into BxPC-3 and PANC-1 cells significantly increased the
number of apoptotic (P<0.01; Fig.
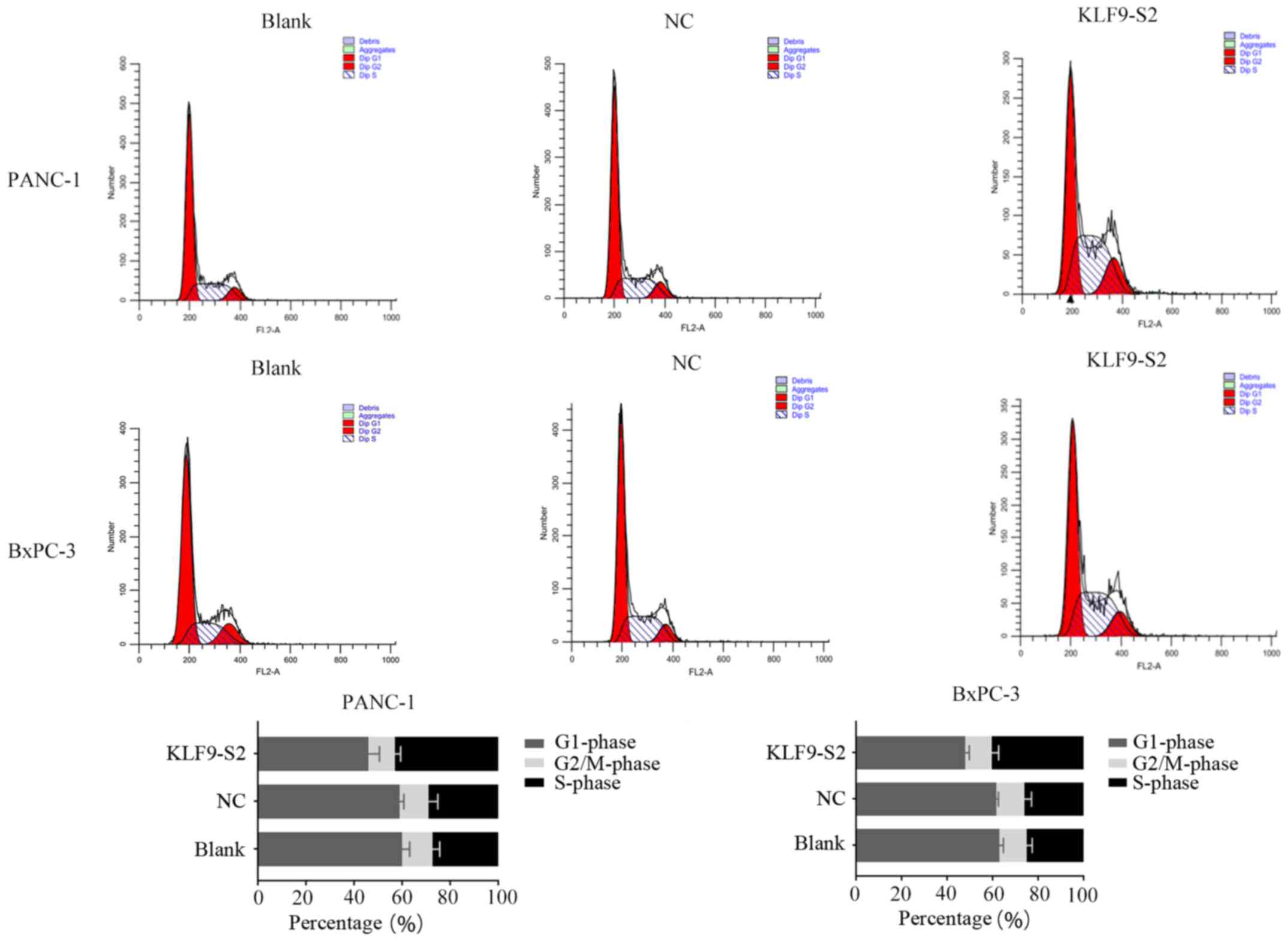
3) cells. Cell cycle profile data revealed that overexpression
of KLF9 significantly increased the number of cells in the S phase
from <30% to >40% (P<0.01; Fig. 4).
Since our in vitro data revealed that low
expression of KLF9 was related to metastasis of pancreatic cancer,
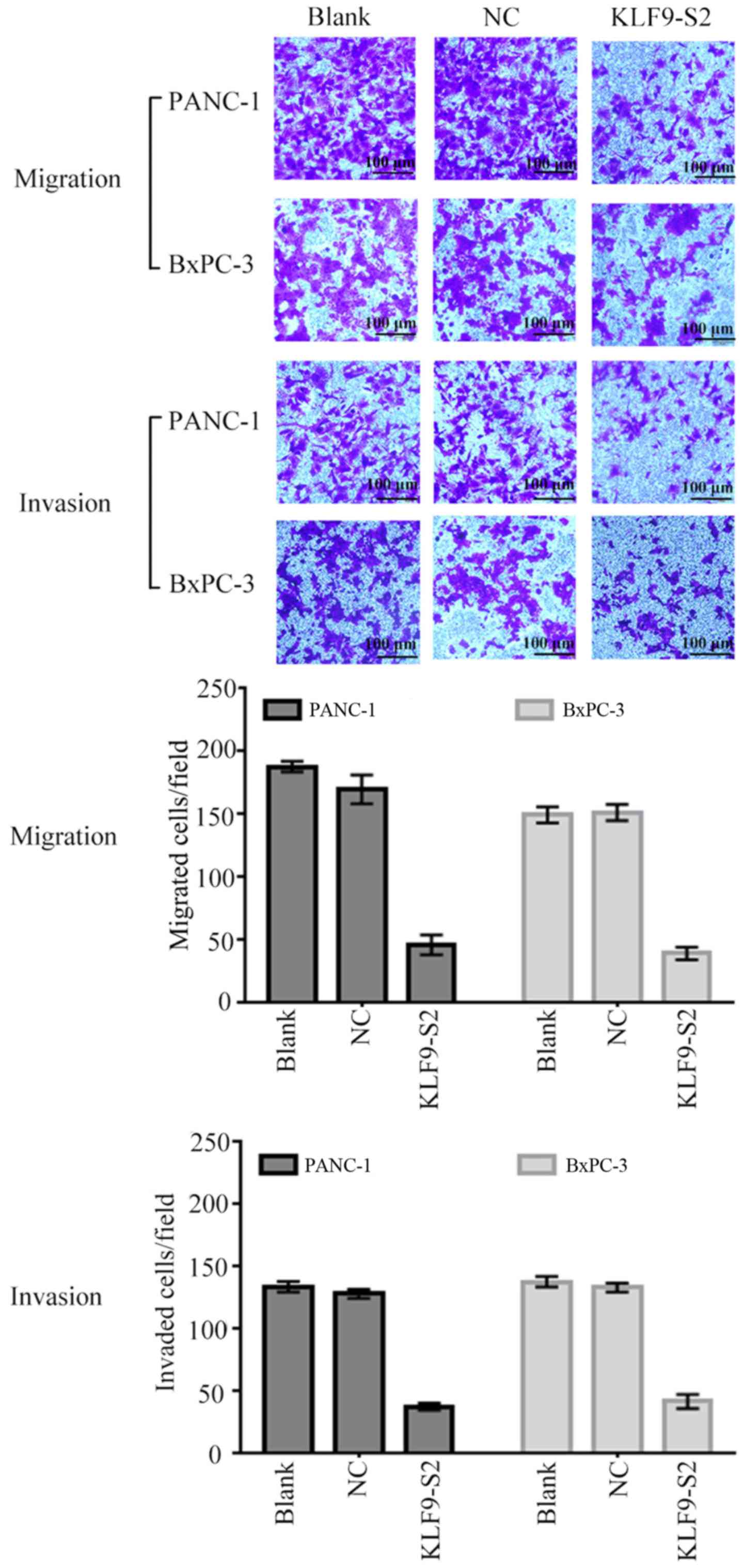
we assessed whether KLF9 overexpression inhibited the invasion and
migration of pancreatic cancer cells. It was revealed by the data
that KLF9 saRNA significantly decreased the migration and invasive
abilities of BxPC-3 and PANC-1 cells (P<0.05; Fig. 5).
The potential molecular events in KLF9
overexpression in pancreatic cancer cells was further explored. The
data indicated that KLF9 regulated the cell cycle progression of
pancreatic cancer cells. In addition, we found that overexpression
of KLF9 increased the expression of p53 and Bax and downregulated
Bcl-2 in PANC-1 and BxPC-3 cells (Fig.
6A). The data further indicated that KLF9 overexpression
significantly upregulated the levels of proteins cyclin D1 and
CDK4, and downregulated cyclin B (Fig.
6B). Next, tumor cell EMT-related gene expression was assessed.
In the course of cell EMT, epithelial marker expression of
E-cadherin was increased, yet cell mobility markers, MMP-9 and
MMP-2, and mesenchymal marker expression N-cadherin were reduced.
The data revealed that KLF9 overexpression could significantly
modulate protein expression (Fig.
6C).
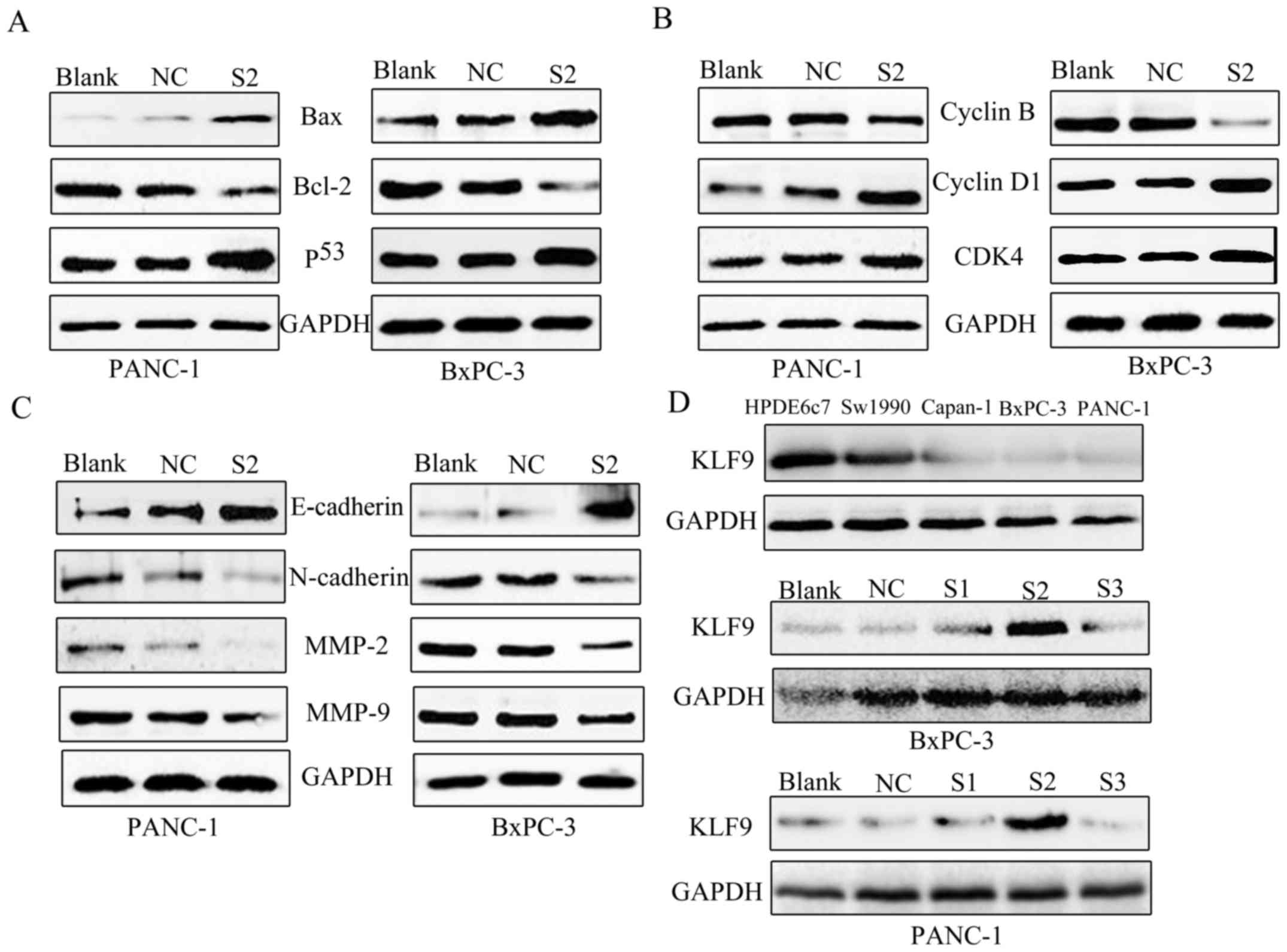 | Figure 6.Western blotting. (A) The expression
of apoptosis-related proteins. Western blotting revealed
downregulated Bcl-2, and upregulated Bax and p53 following
overexpression of KLF9 in PANC-1 and BxPC-3 cells. (B) The
expression of cell cycle distribution proteins. Western blotting
revealed downregulated cyclin B, and upregulated cyclin D1 and CDK4
following overexpression of KLF9 in PANC-1 and BxPC-3 cells. (C)
The effect of KLF9 on EMT. Overexpression of KLF9 markedly
suppressed N-cadherin, MMP-2 and MMP-9, while it increased
E-cadherin expression in PANC-1 and BxPC-3 cells. (D) Western blot
analysis of KLF9 protein in pancreatic cancer cell lines and
screening of optimal saRNA from KLF9-S1, KLF9-S2 and KLF9-S3. KLF9
was expressed at high levels in HPDE6c7 cells, however in Sw1990
cells it was expressed at a lower level when compared to HPDE6c7,
and was almost absent in Capan-1, PANC-1 and BxPC-3 cells. KLF9-S2
was expressed at the highest level among three interference
fragments (KLF9-S1, KLF9-S2 and KLF9-S3). KLF9, Kruppel-like factor
9; saRNA, small activating RNA; S1, saRNA 1; S2, saRNA 2; S3, saRNA
3; NC, negative control. |
KLF9 protein expression in PC cell lines (Sw1990,
BxPC-3, Capan-1, and PANC-1) was significantly lower than that in
the non-transformed pancreatic epithelial cell line HPDE6c7
(Fig. 6D). This indicated an
association between low expression of KLF9 and pancreatic cancer.
To demonstrate the role of KLF9 in the development and progression
of pancreatic cancer, we used a KLF9 saRNA construct to promote the
expression of KLF9 in PANC-1 and BxPC-3 cell lines. We found that
KLF9 was significantly overexpressed after 48 h of saRNA
transfection and KLF9-S2 was screened from the three interference
fragments (KLF9-S1, KLF9-S2, and KLF9-S3) (Fig. 6D).
Discussion
Pancreatic cancer refers to a type of malignancy of
the digestive system, whose prognosis is very poor (24). Despite progress in immunotherapy,
radiotherapy, chemotherapy and surgery in recent decades, effective
treatment of pancreatic cancer still poses a main clinical
challenge. Thus, a new treatment program for pancreatic cancer is
required. However, the precise development and occurrence
mechanisms of pancreatic cancer are far from fully elucidated. It
is still the most aggressive cancer in the world, and the total
5-year survival rate is <5%. Accordingly, it is urgent to
further explore the molecular mechanisms of pancreatic cancer and
develop more effective therapeutic strategies to improve the
treatment effects. The molecular markers in this study may be
helpful for the early diagnosis of pancreatic cancer, which can
significantly improve the survival of patients.
In the present study, we found that the expression
of KLF9 in pancreatic tumors was related to pancreatic cancer
behaviors and the total survival of patients. We then evaluated the
effect of the low expression of KLF9 on the malignant behavior of
pancreatic cancer cells in vitro. We also determined that
KLF9 exhibited low expression in tumor tissue in comparison with
adjacent paracancerous tissue, which is relevant to tumor
differentiation and vascular invasion. Furthermore, our data
revealed that low expression of KLF9 was associated with a poor
total survival rate, which is an independent prognostic indicator
for pancreatic cancer.
Our data in vitro demonstrated that
overexpression of KLF9 could reduce the viability of pancreatic
cancer cells and induce arrest of tumor cell apoptosis at the S
phase of the cell cycle. Overexpression of KLF9 could also inhibit
migration and invasion capacities of tumor cells in vitro.
At the gene level, overexpression of KLF9 upregulated the
expression of cyclin D1, CDK4, p53, Bax and E-cadherin, while it
downregulated the levels of cyclin B, N-cadherin, MMP-2 and MMP-9.
Our study revealed that KLF9 may be used as a prognostic indicator
and therapeutic target for pancreatic cancer. In fact, cancer
growth is characterized by uncontrolled cell proliferation and
transformation. In addition, at the molecular level, numerous genes
including proteins that regulate cell proliferation and death as
well as genomic stability, are altered (25,26).
In the present study, the human KLF9 gene is located
on chromosome 9q13 and its coding sequence. The coding region is as
long as 735 bp and contains two exons that encode a polypeptide
chain containing 144 amino acid residues. KLF9 peptide chain amino
terminal ~84-116 is rich in Asp/Glu acidic amino acid residues, the
carboxyl terminal (143 to 167, 173 to 197 and amino acids 203 to
225) is a conserved DNA binding domain containing three
tandem/adjacent Cys2/His2 zinc fingers (27). Studies have shown that the DNA copy
number of chromosome 9q is reduced in various cancers, which is
associated with low expression of KLF9 (13,28).
Though our study did not assess changes in DNA copy number, our
data revealed the expression of KLF9 in pancreatic cancer tissues,
for the first time.
The low expression of KLF9 was associated with poor
progression and total survival in patients with pancreatic cancer,
which was consistent with previous studies in cancers of other
organs (29,30). Pancreatic cancer is a heterogeneous
and complex disease since the development of pancreatic cancer
involves a variety of genetic changes (31). Our research focused on the
overexpression of KLF9 which inhibited the malignant behavior of
pancreatic cancer in vitro, consistent with previous
published studies (17,32). For example, when KLF9 was
upregulated, the proliferation of pancreatic cells was
significantly reduced. As a mitotic modulator, KLF9 can regulate
the formation of mitotic spindles and cell mitosis. Tumor cells
cannot form a normal level of mitotic spindles and stagnate at the
S phase of the cell cycle. At the molecular level, overexpression
of KLF9 can regulate cell proliferation, apoptosis, and expression
of multiple genes of EMT, which play an important role in invasion
and metastasis of tumor cells. During EMT, tumor cells lose more
polarity and obtain the ability to migrate and invade by
degradation of the extracellular matrix (ECM) (33). MMP is a family of zinc-dependent
endopeptidases whose function is the reduction of the ECM
composition in tissues (34,35).
Our data revealed that overexpression of KLF9 inhibited migration
and invasion of pancreatic cancer cells by downregulating the
expression of MMP-2 and MMP-9. We also found that KLF9
overexpression upregulated the level of E-cadherin and
downregulated that of N-cadherin.
A recent study revealed that KLF9 was downregulated
in esophageal cancer and could be combined with TCF4 to inhibit
beta-catenin/TCF signaling pathways, thus stimulating tumor
suppression (12). In addition,
KLF9 is considered to be a tumor suppressor gene in many tumors and
targeting KLF9 expression may be used as a therapeutic strategy for
pancreatic cancer in future (36).
Acknowledgements
We are grateful to the Molecular Center Laboratory
staff of the Second Affiliated Hospital of Nanchang University.
Funding
The present study was supported from grants from the
National Natural Science Foundation of China (no. 81060187) and was
partially supported by grants from the Nanchang University Graduate
Innovation Foundation (no. cx2016337) of Jiangxi Province,
China.
Availability of data and materials
All data yielded or studied in the present study are
included in this published article.
Authors' contributions
ZZ, FZ, DW, WZ, MW, YZ, JL, LW and XY conceived and
designed the experiments. ZZ, FZ, DW, XY and YZ performed the
experiments. YZ, JL, WZ, MW and LW analyzed the data. ZZ, FZ, DW,
WZ and XY drafted the manuscript. All the authors modified and
agreed with the results and conclusions of the study. XY obtained
the funding. XY supervised the study. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The Ethics Committee of the Second Affiliated
Hospital of Nanchang University approved our study, and all
participants signed written informed consent. The present study was
conducted in accordance with the ethical standards of the Helsinki
Declaration.
Patient consent for publication
Not applicable.
Competing interests
There authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
KLF9
|
Kruppel-like transcription factor
9
|
|
EMT
|
epithelial-mesenchymal transition
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
Yang Y, Yan S, Tian H and Bao Y:
Macrophage inhibitory cytokine-1 versus carbohydrate antigen 19-9
as a biomarker for diagnosis of pancreatic cancer: A
PRISMA-compliant meta-analysis of diagnostic accuracy studies.
Medicine (Baltimore). 97:e99942018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
German RR, Fink AK, Heron M, Stewart SL,
Johnson CJ, Finch JL and Yin D: Accuracy of Cancer Mortality Study
Group: The accuracy of cancer mortality statistics based on death
certificates in the United States. Cancer Epidemiol. 35:126–131.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Limame R, de Beeck Op K, Lardon F, De
Wever O and Pauwels P: Krüppel-like factors in cancer progression:
Three fingers on the steering wheel. Oncotarget. 5:29–48. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heard ME, Simmons CD, Simmen FA and Simmen
RC: Krüppel-like factor 9 deficiency in uterine endometrial cells
promotes ectopic lesion establishment associated with activated
notch and hedgehog signaling in a mouse model of endometriosis.
Endocrinology. 155:1532–1546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohguchi H, Tanaka T, Uchida A, Magoori K,
Kudo H, Kim I, Daigo K, Sakakibara I, Okamura M, Harigae H, et al:
Hepatocyte nuclear factor 4alpha contributes to thyroid hormone
homeostasis by cooperatively regulating the type 1 iodothyronine
deiodinase gene with GATA4 and Kruppel-like transcription factor 9.
Mol Cell Biol. 28:3917–3931. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bieker JJ: Krüppel-like factors: Three
fingers in many pies. J Biol Chem. 276:34355–34358. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tetreault MP, Yang Y and Katz JP:
Krüppel-like factors in cancer. Nat Rev Cancer. 13:701–713. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yori JL, Johnson E, Zhou G, Jain MK and
Keri RA: Kruppel-like factor 4 inhibits epithelial-to-mesenchymal
transition through regulation of E-cadherin gene expression. J Biol
Chem. 285:16854–16863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yori JL, Seachrist DD, Johnson E, Lozada
KL, Abdul-Karim FW, Chodosh LA, Schiemann WP and Keri RA:
Krüppel-like factor 4 inhibits tumorigenic progression and
metastasis in a mouse model of breast cancer. Neoplasia.
13:601–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiao F, Yao F, Chen L, Lu C, Ni Y, Fang W
and Jin H: Krüppel-like factor 9 was down-regulated in esophageal
squamous cell carcinoma and negatively regulated beta-catenin/TCF
signaling. Mol Carcinog. 55:280–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kang L, Lü B, Xu J, Hu H and Lai M:
Downregulation of Krüppel-like factor 9 in human colorectal cancer.
Pathol Int. 58:334–338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Simmen FA, Su Y, Xiao R, Zeng Z and Simmen
RC: The Krüppel-like factor 9 (KLF9) network in HEC-1-A endometrial
carcinoma cells suggests the carcinogenic potential of
dys-regulated KLF9 expression. Reprod Biol Endocrinol. 6:412008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pabona JM, Velarde MC, Zeng Z, Simmen FA
and Simmen RC: Nuclear receptor co-regulator Krüppel-like factor 9
and prohibitin 2 expression in estrogen-induced epithelial cell
proliferation in the mouse uterus. J Endocrinol. 200:63–73. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pabona JM, Simmen FA, Nikiforov MA, Zhuang
D, Shankar K, Velarde MC, Zelenko Z, Giudice LC and Simmen RC:
Krüppel-like factor 9 and progesterone receptor coregulation of
decidualizing endometrial stromal cells: Implications for the
pathogenesis of endometriosis. J Clin Endocrinol Metab.
97:E376–E392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fu DZ, Cheng Y, He H, Liu HY and Liu YF:
The fate of Krüppel-like factor 9-positive hepatic carcinoma cells
may be determined by the programmed cell death protein 5. Int J
Oncol. 44:153–160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen P, Sun J, Xu G, Zhang L, Yang Z, Xia
S, Wang Y, Liu Y and Shi G: KLF9, a transcription factor induced in
flutamide-caused cell apoptosis, inhibits AKT activation and
suppresses tumor growth of prostate cancer cells. Prostate.
74:946–958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Y, Ju B, Tian J, Liu F, Yu H, Xiao
H, Liu X, Liu W, Yao Z and Hao Q: Ovarian cancer stem cell-specific
gene expression profiling and targeted drug prescreening. Oncol
Rep. 31:1235–1248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ying M, Tilghman J, Wei Y,
Guerrero-Cazares H, Quinones-Hinojosa A, Ji H and Laterra J:
Kruppel-like factor-9 (KLF9) inhibits glioblastoma stemness through
global transcription repression and integrin α6 inhibition. J Biol
Chem. 289:32742–32756. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang M, Ke NW, Zeng L, Zhang Y, Tan CL,
Zhang H, Mai G, Tian BL and Liu XB: Survival analyses for patients
with surgically resected pancreatic neuroendocrine tumors by World
Health Organization 2010 Grading Classifications and American Joint
Committee on Cancer 2010 Staging Systems. Medicine (Baltimore).
94:e21562015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kamarajah SK, Burns WR, Frankel TL, Cho CS
and Nathan H: Validation of the American Joint Commission on Cancer
(AJCC) 8th Edition Staging System for Patients with Pancreatic
Adenocarcinoma: A Surveillance, Epidemiology and End Results (SEER)
analysis. Ann Surg Oncol. 24:2023–2030. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Warner SL, Stephens BJ, Nwokenkwo S,
Hostetter G, Sugeng A, Hidalgo M, Trent JM, Han H and Von Hoff DD:
Validation of TPX2 as a potential therapeutic target in pancreatic
cancer cells. Clin Cancer Res. 15:6519–6528. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frič P, Šedo A, Škrha J, Bušek P, Laclav
M, Škrha P and Zavoral M: Early detection of sporadic pancreatic
cancer: Time for change. Eur J Gastroenterol Hepatol. 29:885–891.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sieber O, Heinimann K and Tomlinson I:
Genomic stability and tumorigenesis. Semin Cancer Biol. 15:61–66.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kang L and Lai MD: BTEB/KLF9 and its
transcriptional regulation. Yi Chuan. 29:515–522. 2007.(In
Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu XJ, Shi Y, Chen JL and Ma S:
Krüppel-like factors in hepatocellular carcinoma. Tumour Biol.
36:533–541. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun J, Wang B and Liu Y, Zhang L, Ma A,
Yang Z, Ji Y and Liu Y: Transcription factor KLF9 suppresses the
growth of hepatocellular carcinoma cells in vivo and positively
regulates p53 expression. Cancer Lett. 355:25–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Limame R, de Beeck KO, Van Laere S, Croes
L, De Wilde A, Dirix L, Van Camp G, Peeters M, De Wever O, Lardon
F, et al: Expression profiling of migrated and invaded breast
cancer cells predicts early metastatic relapse and reveals
Krüppel-like factor 9 as a potential suppressor of invasive growth
in breast cancer. Oncoscience. 1:69–81. 2013.PubMed/NCBI
|
|
31
|
Tang H, Wei P, Chang P, Li Y, Yan D, Liu
C, Hassan M and Li D: Genetic polymorphisms associated with
pancreatic cancer survival: A genome-wide association study. Int J
Cancer. 141:678–686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brown AR, Simmen RC, Raj VR, Van TT,
MacLeod SL and Simmen FA: Krüppel-like factor 9 (KLF9) prevents
colorectal cancer through inhibition of interferon-related
signaling. Carcinogenesis. 36:946–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bogachek MV, De Andrade JP and Weigel RJ:
Regulation of epithelial-mesenchymal transition through SUMOylation
of transcription factors. Cancer Res. 75:11–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ying M, Sang Y, Li Y, Guerrero-Cazares H,
Quinones-Hinojosa A, Vescovi AL, Eberhart CG, Xia S and Laterra J:
Krüppel-like family of transcription factor 9, a
differentiation-associated transcription factor, suppresses Notch1
signaling and inhibits glioblastoma-initiating stem cells. Stem
Cells. 29:20–31. 2011. View
Article : Google Scholar : PubMed/NCBI
|