Introduction
Head and neck squamous cell carcinoma (HNSCC) refers
to a group of biologically similar cancers arising from the mucous
squamous epithelia in the head and neck area. HNSCC is an
aggressive cancer with poor overall survival (1–4). In
spite of the recent advancements in treatment modalities for HNSCC,
the long-term survival rates have not significantly improved over
the past decade (5). Currently,
emerging evidence suggests that cancer stem cells (CSCs) are
responsible for local recurrence, metastatic spread, and treatment
resistance in HNSCC (6).
To date, research on CSCs has become profound, and
CSCs have been functionally defined as a subset of tumor cells that
exhibit the ability of self-renewal and multipotency in cancerous
malignancy (7). CSCs only account
for a minor proportion of the total cancerous burden but can play
paramount roles in determining the outcomes of cancers (8). Thus, identification of CSCs provides
novel therapeutic promise for improving cancer treatment (9,10).
Previous studies conducted in several types of cancer have reported
that these CSCs exhibit increased expression of certain biomarkers
resulting in the acquisition of stem-like properties (11,12).
Confirmation of these CSCs requires the identification of such
molecular biomarkers (9).
Discovering effective biomarkers is critical to a
better understanding of the biological features of CSCs. To date,
several putative protein molecules have been proposed to identify
the CSCs in HNSCC, including CD44, CD133, Nanog, Oct4, Sox2 and
ALDH1 (12–15). However, validity of these CSC
biomarkers has been questioned recently, and the clinical
significance of these molecules in HNSCC remains to be ascertained,
especially based on large cohort data. The Cancer Genome Atlas
(TCGA) project holds great promise for a comprehensive
understanding of human cancer with powerful and detailed data
(16,17). The UCSC Cancer Genomics Browser
presents the TCGA data in a coherent, integrated system with
genomic, clinical annotation data in multiple views (18). In this study, we managed to collect
the reported CSC biomarkers of HNSCC and analyze these biomarkers
via bioinformatics based on the TCGA primary HNSCC cohort. We
systematically demonstrated the expression patterns, clinical
significance, and potential targeted strategies for these molecules
in HNSCC.
Materials and methods
Searching for the reported CSC
biomarkers for HNSCC
Studies were scanned by searching the electronic
database PubMed with the terms ‘cancer stem-like cells’, ‘cancer
stem cells’, ‘tumor stem-like cells’, ‘tumor stem cells’, ‘CSCs’,
and ‘head and neck squamous cell carcinoma’, ‘HNSCC’. In order to
be included for further summary, the following criteria were met:
i) an original research paper in a peer-reviewed journal; ii)
studies in humans; iii) studies with validated evidence to
demonstrate the reported biomarkers tightly concerned with the CSC
characteristics of HNSCC. Conference abstracts, reviews, comments,
case reports, and letters to the editor were excluded.
Subsequently, all potentially eligible studies were retrieved and
the following information was extracted: i) name of the reported
CSC biomarker; ii) the reported clinical significance for each CSC
biomarker in HNSCC areas. In addition, the encoding genes for the
reported CSC biomarkers were annotated. We demonstrated the
cellular location and biological roles for each reported
CSC-related molecule based on The Human Protein Atlas. Search
results for the CSC biomarkers of HNSCC are listed in Table I.
 | Table I.Search results for the reported CSC
biomarkers of HNSCC. |
Table I.
Search results for the reported CSC
biomarkers of HNSCC.
| CSC biomarker | Encoded gene | Cellular
location | Biological
roles | Clinical
significance |
|---|
| CD44 | CD44 | Plasma
membrane | Signal
transducer | Lymph node
metastasis, recurrence |
| CD24 | CD24 |
Cellular vesicles | Signal
transducer | Tumorigenicity,
angiogenesis |
| CD98 | SLC7A5,
SLC3A2 | Nucleus, plasma
membrane, cytosol | Signal transducer;
Amino acid transport | Tumorigenicity,
recurrence |
| EpCAM | EPCAM | Plasma
membrane | Signal
transducer |
Chemoresistance |
| c-Met | MET | Plasma membrane,
cytosol | Signal
transducer | Chemoresistance,
metastasis |
| CD133 | PROM1 | Plasma membrane,
cytoplasm | Signal
transducer | Metastasis,
tumorigenicity, chemoresistance |
| CD166 | ALCAM | Plasma membrane,
cytoplasm | Signal
transducer | Recurrence |
| Notch1 | NOTCH1 | Nucleoplasm | Signal
transducer | Tumorigenicity,
chemoresistance |
| CD10 | MME | Plasma
membrane | Zinc-dependent
metalloendoprotease | Tumorigenicity,
chemoresistance |
| MT1-MMP | MMP14 | Cytoplasm | Zinc-dependent
metalloendoprotease | Recurrence,
chemoresistance, metastasis |
| ALDH1 | ALDH1A1 | Cytosol | Detoxifying
enzyme | Recurrence,
radiochemoresistance |
| SOX2 | SOX2 | Nucleoplasm | Transcription
factor | Lymph node
metastasis, recurrence, chemoresistance |
| Oct4 | POU5F1 | Nucleoplasm,
cytosol | Transcription
factor | Lymph node
metastasis, chemoresistance |
| Nanog | NANOG | Nucleoplasm | Transcription
factor | Chemoresistance,
recurrence, lymph node metastasis |
| KLF4 | KLF4 | Nucleoplasm | Transcription
factor | Lymph node
metastasis, distant metastasis |
| Brachyury | T | Nucleoplasm | Transcription
factor | Lymph node
metastasis, distant metastasis |
| Bmi-1 | BMI1 | Nucleus, nuclear
bodies, cytosol | Transcriptional
repressors | Chemoresistance,
metastasis |
| Topoisomerase I,
IIα, IIIα | TOP1, TOP2A,
TOP3A | Nucleus | Topoisomerase | Lymph node
metastasis |
| TAZ | TAZ | Plasma
membrane | Transcriptional
regulation | Tumor growth, lymph
node metastasis |
| EHMT2 | EHMT2 | Nucleoplasm | Euchromatic
methyltransferase | Lymph node
metastasis |
| JMJD6 | JMJD6 | Nucleoplasm | Arginine
demethylase, lysine hydroxylase | Recurrence,
chemoresistance |
| ABCG2 | ABCG2 | Plasma membrane,
nucleus | ABC transporter
protein | Lymph node
metastasis, recurrence, chemoresistance |
| ABCG5 | ABCG5 | Nucleus | ABC transporter
protein |
Chemoresistance |
| SLC2A13 | SLC2A13 | Nuclear
membrane |
H+-myo-inositol
transporter | Tumorigenicity |
| GRP78 | HSPA5 | Cytosol | Endoplasmic
reticulum chaperone | Recurrence,
radioresistance, tumorigenicity |
Bioinformatic analysis for the
reported CSC biomarkers of HNSCC based on the TCGA primary HNSCC
cohort
Bioinformatic analyses were performed based on the
TCGA primary HNSCC cohort using the UCSC Xena Browser. Totally, 604
cases were searched, and only cases of primary HNSCC were filtered
and included for further analysis for the gene expression patterns
of each reported CSC biomarker. Expression heat-maps and
Kaplan-Meier curves stratified by the defined gene were generated
and clustered online, and detailed data were downloaded for
subsequent statistical analysis. To illustrate the
clinicopathological features of the reported CSC biomarkers, we
downloaded and analyzed the detailed data for the expression level,
pathological nodal extracapsular spread, lymphovascular invasion,
neoplasm histologic grade, tumor size, nodal status, and pathologic
stage.
Searching for targeted treatment based
on the reported CSC biomarkers in cancer areas
Studies were scanned by searching electronic
database PubMed for the targeted treatment based on the reported
CSC biomarkers in the pan-cancer areas. Articles were reviewed to
figure out and summary the targeted strategies in cancer areas
based on the reported CSC biomarkers of HNSCC.
Statistical analysis
Statistical analyses were conducted with SPSS 20.0
software (IBM Corp., Armonk, NY, USA). Based on the detailed data
for each biomarker, all cases involved were divided equally into
two groups, a high-expression group and low-expression group. To
illustrate the underlying relationship among the reported CSC
biomarkers, crosstab analyses were performed and Chi-squared tests
were used to assess the statistical significance for correlations
between the gene expression level of CD44 and the gene expression
levels of other biomarkers. In addition, Chi-squared tests were
used to assess the statistical significance for correlations
between the gene expression level of each biomarker and each
clinicopathological variable. Xena Browser compares the different
Kaplan-Meier curves using the log-rank test. p<0.05 was
considered to indicate a statistically significant difference. Venn
diagrams were generated for clustering analyses.
Results
Detailed information for the reported
CSC biomarkers of HNSCC
A total of 27 molecules, encoded by 28 genes, were
demonstrated and reported to be tightly linked with the CSC
properties of HNSCC. Detailed information for each reported
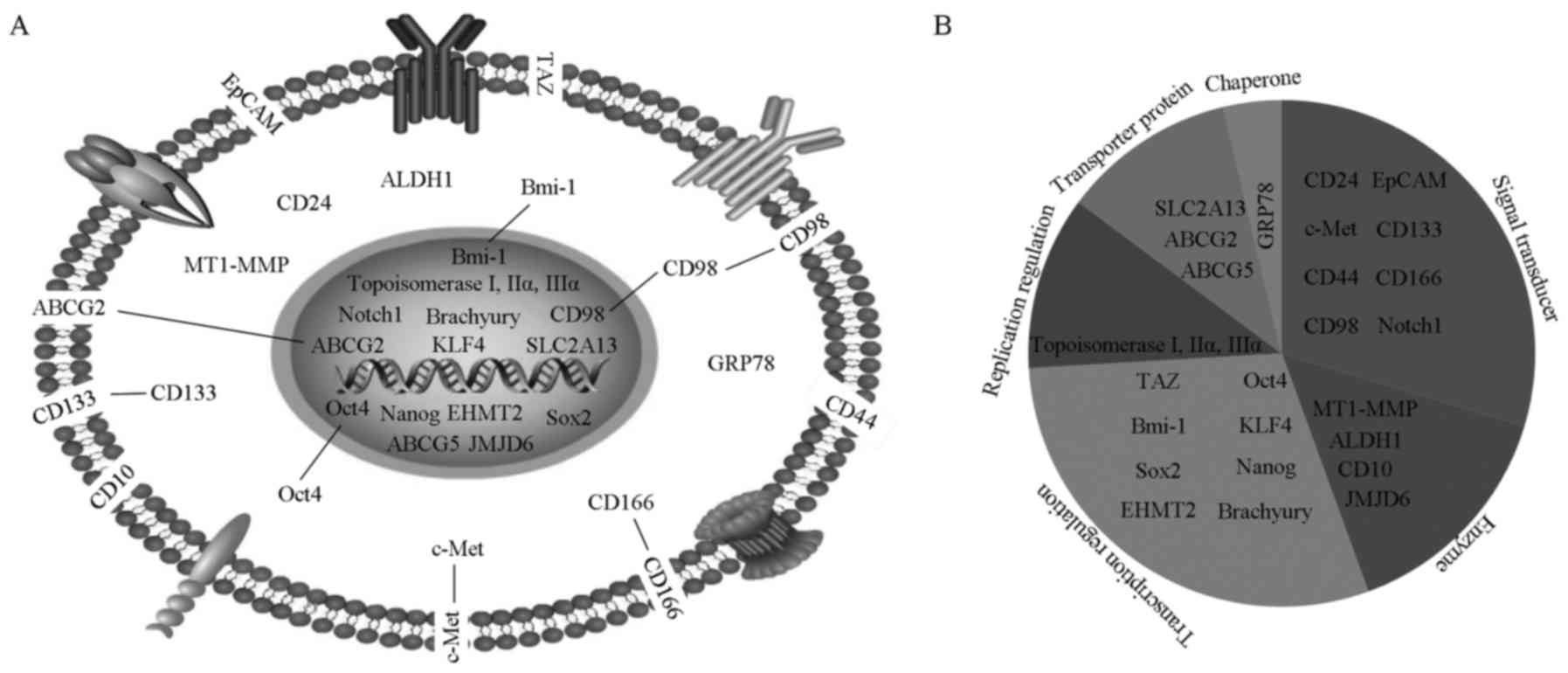
biomarker has been summarized. As shown in Fig. 1A, cellular locations for these
molecules are designated. Four molecules are located at the plasma
membrane (CD44, EpCAM, CD10 and TAZ), 4 molecules in the cytoplasm
(CD24, MT1-MMP, ALDH1 and GRP78), and 12 molecules in the nucleus
(topoisomerase I/IIα/IIIα, Notch1, Brachyury, ABCG5, Sox2, SLC2A13,
Nanog, KLF4, JMJD6 and EHMT2). In addition, there are 3 molecules
distributed at both the plasma membrane and cytoplasm (c-Met,
CD133, and CD166), 2 molecules at both the cytoplasm and nucleus
(Oct4 and Bmi-1), and 1 molecule at both the plasma membrane and
nucleus (ABCG2). Additionally, CD98 is widely scattered among the
plasma membrane, cytoplasma, and nucleus.
Furthermore, we also characterized the biological
roles for these molecules into 6 categories (Fig. 1B): replication regulation
(topoisomerase I, IIα and IIIα), transcription regulation (TAZ,
Oct4, Bmi-1, KLF4, Sox2, Nanog, EHMT2 and Brachyury), signal
transducer (CD24, EpCAM, c-Met, CD133, CD44, CD166, CD98 and
Notch1), transporter protein (SLC2A13, ABCG2 and ABCG5), enzyme
(MT1-MMP, ALDH1, CD10 and JMJD6) and chaperone protein (GRP78).
Accordingly, we observed some heterogeneity existing among these
biomarkers more or less, indicating that there might be variable
mechanisms in regulating the CSC behaviors of HNSCC for these
molecules.
Underlying relationship among the
reported CSC biomarkers based on the TCGA primary HNSCC cohort
Although these biomarkers have been reported to
regulate the stem-like ability of HNSCC cells, evidence for the
underlying relationship among these molecules has not been
demonstrated previously. Herein, we aimed to choose the TCGA
primary HNSCC cohort to comprehensively evaluate the underlying
relationship and validate the clinical significance for each
biomarker. To date, CD44 has reported to be the most frequently
used biomarker for identifying the CSCs in HNSCC (19,20).
In this study, we chose CD44 as a reference biomarker for the
reported CSC biomarkers in HNSCC to analyze the clinical
significance and underlying relationship among them. In total,
there are 604 cases of HNSCC in the TCGA cohort, and only 528 cases
of primary HNSCC were filtered and included for further analysis.
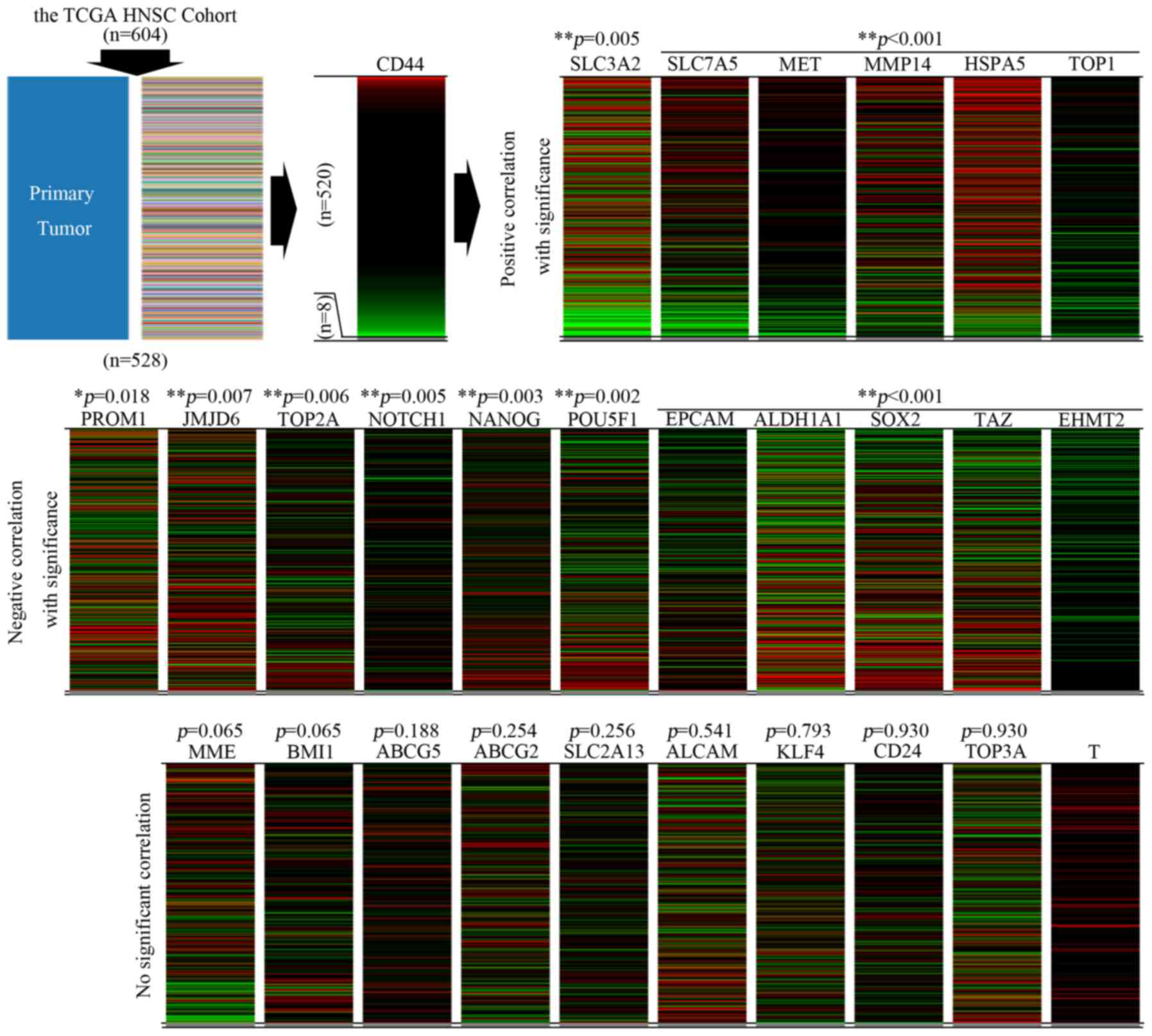
By using the UCSC Xena browser, we generated a series of heatmaps
referencing to the expression pattern of CD44 among the 520 cases
(Fig. 2).
By data mining, we explored the expression patterns
among these reported CSC biomarkers of HNSCC. We found that by
referring to the expression pattern of CD44, the expression
patterns of the remaining 26 biomarkers were clustered into three
subgroups (Fig. 2). In the subgroup
with a significantly positive correlation to the expression pattern
of CD44 (Group A), the following biomarkers were included: CD98,
c-Met, MT1-MMP, GRP78 and topoisomerase I. In the subgroup with a
significantly negative correlation to the expression pattern of
CD44 (Group B), the following molecules were included: CD133,
JMJD6, topoisomerase IIα, Notch1, Nanog, Oct4, EpCAM, ALDH1, Sox2,
TAZ and EHMT2. Moreover, the expression pattern of the following
molecules were observed without significant correlation to that of
CD44 (Group C): CD10, Bmi-1, ABCG5, ABCG2, SLC2A13, CD166, KLF4,
CD24 and topoisomerase IIIα. Among these, the detailed data
downloaded for Brachyury (encoded by T) was not enough for further
studies. Thus, the incomplete heat-map for T was added at the end
of Fig. 2 and no further analysis
was performed for this biomarker. The above data indicated that
great heterogeneity existed among the expression pattern of the
reported CSC biomarkers in HNSCC, suggesting that subgrouping
clusters might exist for all the CSCs in HNSCC.
Clinicopathological features and
overall survival evaluation for the reported CSC biomarkers based
on the TCGA primary HNSCC cohort
Based on the reported studies, the included 27
molecules have been identified to regulate cancer-stem like
behaviors of HNSCC. The above data showed powerful evidence to
indicate that these molecules might exert their roles with variable
mechanisms. However, the validity and clinical significance for
these biomarkers need to be further ascertained. Herein, we managed
to validate the clinical significance for each biomarker based on
the TCGA primary HNSCC cohort (Table
II, Fig. 3).
 | Table II.Clinicopathological evaluation for
the reported CSC biomarkers based on the TCGA primary HNSCC
cohort. |
Table II.
Clinicopathological evaluation for
the reported CSC biomarkers based on the TCGA primary HNSCC
cohort.
|
| Pathological nodal
extracapsular spread | Lymphovascular
invasion | Neoplasm histologic
grade | Tumor size | Nodal status | Pathologic
stage |
|---|
|
| Yes/No | Yes/No | G1+G2/G3+G4 | T1+T2/T3+T4 |
N0/N1+ | I+II/III+IV |
|---|
| CD44 | 0.932 | 0.275 | 0.074 | 0.848 | 0.403 | 0.011b |
| Subgroup with a
significantly positive correlation to the expression pattern of
CD44 (Group A) |
|
SLC3A2 | 0.004a | 0.778 | 0.001b | 0.004a | 0.451 | 0.154 |
|
SLC7A5 | 0.674 | 0.613 | 0.001b | 0.733 | 0.045b | 0.857 |
|
MET | 0.243 | 0.908 | 0.972 | 0.525 | 0.833 | 0.180 |
|
MMP14 | 0.714 | 0.392 | 0.993 | 0.455 | 0.298 | 0.986 |
|
HSPA5 | 0.022a | 0.754 | 0.177 | 0.324 | 0.267 | 0.785 |
|
TOP1 | 0.994 | 0.007b | 0.014b | 0.164 | 0.235 | 0.244 |
| Subgroup with a
significantly negative correlation to the expression pattern of
CD44 (Group B) |
|
TOP2A | 0.097 | 0.317 | 0.000a | 0.839 | 0.068 | 0.915 |
|
NOTCH1 | 0.732 | 0.087 | 0.068 | 0.216 | 0.264 | 0.160 |
|
NANOG | 0.327 | 0.032a | 0.043a | 0.775 | 0.934 | 0.812 |
|
PROM1 | 0.901 | 0.121 | 0.393 | 0.807 | 0.818 | 0.523 |
|
JMJD6 | 0.016a | 0.053 | 0.549 | 0.005* | 0.017a | 0.000a |
|
EPCAM | 0.375 | 0.000a | 0.077 | 0.338 | 0.118 | 0.025a |
|
ALDH1A1 | 0.369 | 0.179 | 0.085 | 1.000 | 0.198 | 0.429 |
|
SOX2 | 1.000 | 0.180 | 0.157 | 0.925 | 0.692 | 0.214 |
|
POU5F1 | 0.608 | 0.179 | 0.224 | 0.257 | 0.693 | 0.653 |
|
TAZ | 0.304 | 0.093 | 0.362 | 0.572 | 0.693 | 0.142 |
|
EHMT2 | 0.126 | 0.092 | 0.011a | 0.132 | 0.003a | 1.000 |
| Subgroup without a
significant correlation to the expression patter of CD44 (Group
C) |
|
MME | 0.522 | 0.145 | 0.015b | 0.220 | 0.489 | 0.736 |
|
BMI1 | 0.123 | 0.117 | 0.012a | 0.451 | 0.693 | 0.572 |
|
ABCG2 | 0.523 | 0.180 | 0.129 | 0.925 | 0.374 | 1.000 |
|
ABCG5 | 0.007a | 0.007a | 0.420 | 0.300 | 0.093 | 0.072 |
|
SLC2A13 | 0.608 | 0.823 | 1.000 | 0.637 | 0.767 | 0.822 |
|
CD24 | 0.248 | 0.014b | 0.001b | 0.132 | 0.094 | 0.115 |
|
KLF4 | 0.523 | 0.313 | 0.000b | 0.637 | 0.489 | 0.574 |
|
ALCAM | 0.441 | 0.315 | 0.020a | 0.707 | 0.553 | 0.142 |
|
TOP3A | 0.029* | 0.092 | 0.362 | 0.851 | 0.093 | 0.258 |
As summarized in Table
II, the molecules represented significantly positive
correlation to the clinicopathological features were filtered and
analyzed based on the TCGA primary HNSCC cohort. For the evaluation
of nodal extracapsular spread, the molecules CD98 and GRP78 (Group
A), JMJD6 (Group B), ABCG5 and topoisomerase IIIα (Group C) were
identified as significant. For the evaluation of lymphovascular
invasion, the molecules Nanog and EpCAM (Group B), and ABCG5 (Group
C) were identified as significant. For the evaluation of histologic
grade, the molecules topoisomerase IIα, Nanog and EHMT2 (Group B),
and BMI-1 and CD166 (Group C) were identified as significant. For
the evaluation of tumor size, the molecules CD98 (Group A) and
JMJD6 (Group B) were identified as significant. For the evaluation
of nodal status, the molecules JMJD6 and EHMT2 (Group B) were
identified as significant. For the evaluation of pathologic stage,
the molecules JMJD6 and EpCAM (Group B) were identified as
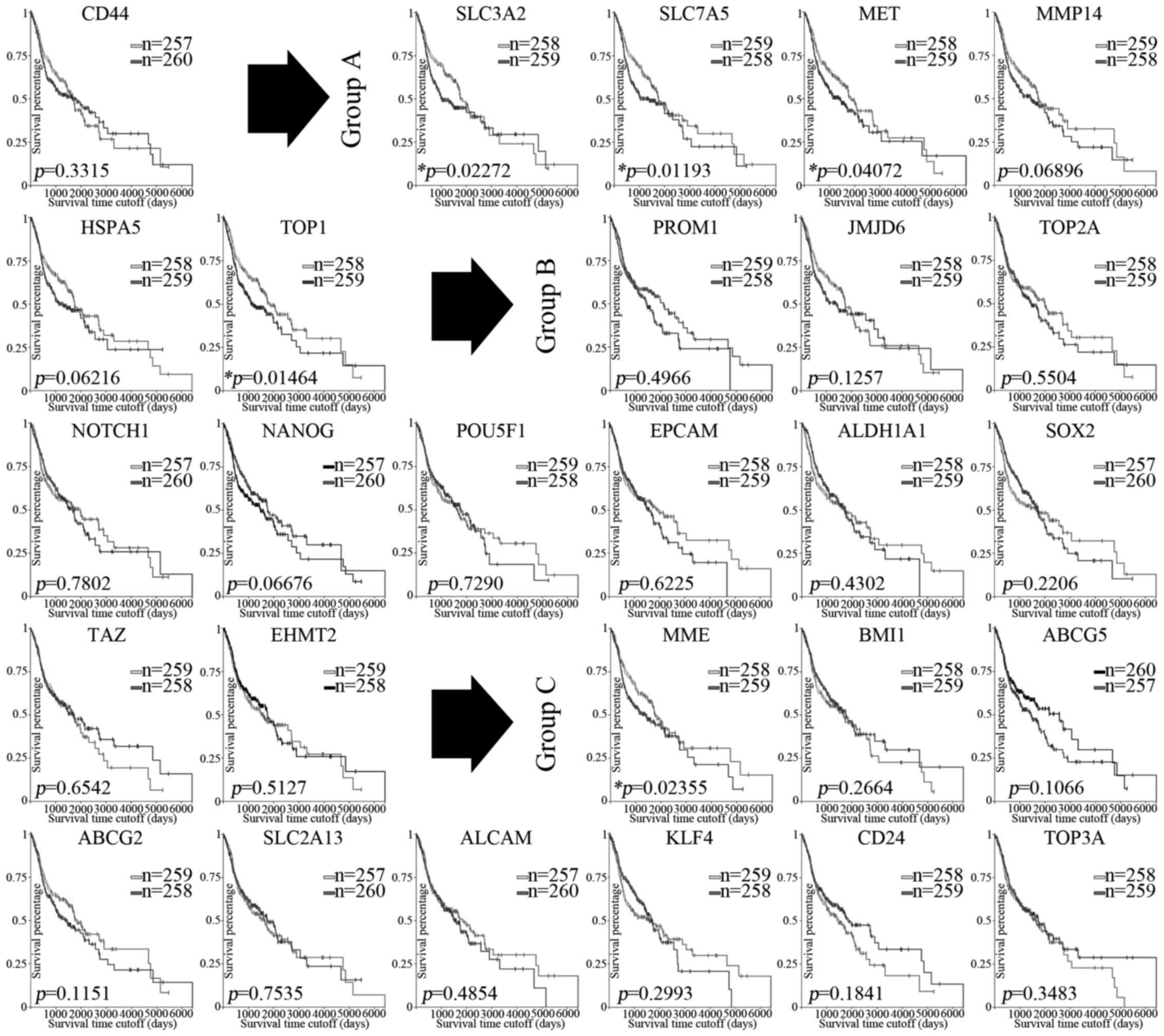
significant. A series of Kaplan-Meier curves were generated to
evaluate the prognostic significance for each reported CSC
biomarker (Fig. 3). Accordingly,
the higher expression of CD98, topoisomerase I, and c-Met (Group
A), and CD10 (Group C) indicate significantly poorer overall
survival (OS) for patients with HNSCC. Unexpectedly, we did not
observe any significant correlations between the expression of
biomarkers in Group B and the overall survival (OS) of the HNSCC
patients.
By analyzing the clinical significance for each
underlying subgroup, we proposed that biomarkers in Group A were
mainly tightly related to clinical outcomes for HNSCC patients, and
biomarkers in Group B were mainly tightly concerned with the
malignant progression in HNSCC. The above data strongly indicate
that the validated biomarkers might regulate CSC properties and
affect the clinicopathological features in HNSCC through different
mechanisms, which warrant further attention to demonstrate the
underlying heterogeneity.
Targeted treatment strategies for
these CSC biomarkers in cancer areas
Currently, there are no targeted therapies for the
CSCs in HNSCC. Despite the fact that HNSCC is a highly prevalent
and deadly cancer, the survival rate for HNSCC patients has not
shown any improvements for years. CSCs are responsible for relapse,
chemoresistance and poor OS, and offer an attractive therapeutic
target. Herein, we searched and summarized the reported targeted
therapies for these molecules in pan-cancer areas (Table III). To date, various agents have
been tested, including compounds, antibodies, and others. Moreover,
some of these agents have been well developed and used in other
types of cancer clinically. No wonder, targeted therapies based on
the validated CSC biomarkers would benefit more patients with
HNSCC. Accordingly, we demonstrate that targeted therapies against
c-Met, topoisomerase I, and GRP78 may improve the survival rate for
HNSCC patients, and targeted therapies against topoisomerase IIα,
EpCAM, and EHMT2 might greatly suppress the malignant progression
of HNSCC.
 | Table III.Targeted therapies for the reported
CSC markers of HNSCC in cancer areas. |
Table III.
Targeted therapies for the reported
CSC markers of HNSCC in cancer areas.
| CSC molecule | Targeted
compound | Targeted
antibody | Others |
|---|
| CD44 | Hyaluronic
acid-based drug delivery | RG7356 |
|
| Subgroup with
significantly positive correlation to the expression pattern of
CD44 (Group A) |
|
c-Met | Cabozantinib,
crizotinib, tepotinib, tivantinib, other small-molecule
inhibitors | BsAbs, mAb |
|
|
Topoisomerase I | Camptothecin, DXd,
organic non-camptothecin compounds, topotecan, LMP-400, NSC724998,
Iirinotecan, betulinic acid, SN-38 |
| Metal complexes,
OSI-211 |
|
GRP78 | Medicarpin,
isoliquiritigenin, HA15 |
| Fusion protein,
GMBP1, KP1339/IT-139 |
|
MT1-MMP |
|
|
Peptide-inhibitor |
| Subgroup with
significantly negative correlation to the expression pattern of
CD44 (Group B) |
|
Topoisomerase IIα | Pixantrone,
glycyrrhetinic acid, halogenated triterpenoid,
2α-bromo-dihydrobetulonic acid, CS1 |
| D11 |
|
Notch1 | PF-03084014 | Brontictuzumab |
|
|
EpCAM | EpCAM
aptamer-mediated delivery | mAbs,
catumaxomab | Immunotoxin,
adoptive T-cell therapy, cytolytic fusion protein |
|
ALDH1 |
Diethylaminobenzaldehyde |
|
|
|
Oct4 | Metformin |
|
|
|
CD133 | CD133
aptamer-mediated delivery |
| Immunotoxin |
|
EHMT2 | UNC0638,
BIX-01294 |
|
|
| Subgroup without
significant correlation to the expression patter of CD44 (Group
C) |
|
CD24 | Anti-CD24 based
drug delivery | mAb |
|
| CD166,
CD10 |
| mAbs |
|
|
Bmi-1 | PTC-209, PTC-028,
PTC596 |
|
|
|
ABCG2 | Anti-ABCG2 based
drug delivery, Ko143, PZ-39, MBL-II-141, YHO-13351, glafenine | Ko143 |
|
Discussion
Emerging studies suggest that cancer stem cells are
responsible for tumor initiation, cancer progression, metastasis
and treatment resistance in HNSCC (4,13).
Several molecules have been identified to isolate and characterize
CSCs in HNSCC, and almost all the CSC biomarkers established to
date with a special emphasis on their impact on malignant
progression and their potentially clinical significance in HNSCC
(12,14,21).
However, none of these biomarkers or their combinations have been
well acknowledged or systematically validated. Consequently, there
are no approved targeted strategies in regards to CSCs for the
treatment of HNSCC patients (22).
Thus, there is a critical need for comprehensive evidence to
evaluate the reported CSC biomarkers for HNSCC.
To date, a total of 27 molecules have been reported
as potential CSC biomarkers for HNSCC. Nevertheless, the validity
and underlying relationship among these molecules have not been
demonstrated. Furthermore, several studies have reported the
limitations and pitfalls underlying the isolation of CSCs with a
single biomarker (23). Thus, we
must analyze and discover the underlying heterogeneity among all
the reported CSC biomarkers (12,13,24–40).
Primarily, we cannot deny the potential heterogeneity from the
inconsistent experimental conditions, the power of experimental
evidence, and the limited sample size for each study reporting the
CSC biomarkers. What's more, it is of paramount importance to
identify a reliable strategy to realize the essential heterogeneity
derived from the CSCs of HNSCC. Recently, different CSC phenotypes
have been implicated in breast cancer (41,42).
In HNSCC, it has been reported that Oct4, Sox2 and CD133 are not
consistently expressed in isolated CSCs (43). Herein, we proposed that the
essential heterogeneity may result from the possible CSC
subpopulations existing in HNSCC. Besides, understanding the
underlying relationship among these CSC-related molecules is
vitally important for demonstrating the biological roles for CSCs
in HNSCC.
Recently, large-scale bioinformatic analyses based
on the TCGA cohort have shown great priority for cancer research
(17,18), which could greatly avoid the
potential heterogeneity from the experimental results and limited
clinical sample sizes. In this study, we conducted a comprehensive
analysis for the expression files of the reported CSC biomarkers in
a large number of primary HNSCC patients from TCGA. By data mining,
we managed to discover the relationship among these molecules and
validate the significance for each molecule in clinicopathological
features and OS for HNSCC. Consequently, the reported CSC
biomarkers were clustered into 3 groups according to their
expression pattern, indicating that there might be subgrouping
clusters existing for all CSCs in HNSCC. Accordingly, we might
propose 2 molecular signatures for the possible CSC clusters
existing in HNSCC, 3 validated biomarkers in group A (CD98, GRP78
and topoisomerase I), and 5 validated biomarkers in group B (JMJD6,
Nanog, EpCAM, topoisomerase IIα and EHMT2).
Previous studies have reported that CSCs are
responsible for cancer initiation and progression, and are
especially resistant to conventional therapy (1,12,30).
In this study, filtered biomarkers belonging to group A were
observed without significant correlation to the malignant
progression of HNSCC, but significantly indicating worse OS for
HNSCC patients. On the contrary, filtered biomarkers belonging to
group B were shown to be significantly correlated to the malignant
characteristics of HNSCC, but without significant correlation to
the OS rates of HNSCC patients. As we know, the clinical outcomes
for HNSCC are determined by malignant phenotypes and treatment
responses of cancer cells. Thus, we may conclude that some CSCs are
responsible for malignant phenotypes, but poorer responses to
treatment strategies, and some CSCs may be responsible for worse
malignant phenotypes, but better responses to treatment. Further
studies for the underlying heterogeneity among all the CSCs in
HNSCC are critically necessary in the future.
Treatment decisions for HNSCC are complex, and
according to the US guidelines, a multidisciplinary approach is
recommended (2). However, the
prognosis of HNSCC remains very poor. Besides, there are still no
approved targeted strategies for CSCs in HNSCC. Targeted strategies
based on the validated CSC biomarkers may effectively supplement
conventional therapies, and benefit HNSCC patients. In this study,
we proposed that targeted strategies against c-Met, topoisomerase
I, and GRP78 show great possible to improve the prognosis of HNSCC
patients, and targeted strategies against topoisomerase IIα, EpCAM
and EHMT2 may potentially suppress the malignant progression of
HNSCC.
In conclusion, we comprehensively evaluated the 27
reported CSC biomarkers for HNSCC based on the TCGA primary HNSCC
cohort. Accordingly, we managed to illustrate the underlying
subgroup clusters among all the CSCs in HNSCC. We proposed that
precisely targeted strategies based on the CSC subgroup clusters
may well supplement conventional therapies, and benefit HNSCC
patients. There is no doubt that numerous studies have improved and
greatly furthered our understanding of the CSCs of HNSCC. However,
more laboratory research and well-designed retrospective or
prospective large-scale studies are still necessary to validate the
conclusions derived from our study for eventually clinical
translation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81371164 and 81602367),
the Science and Technology Commission of Shanghai Municipality (no.
15411950300).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XY and YW conceived and designed the experiments. MX
and LL performed the experiments. MX, LL and SZ summarized and
analyzed the data. MX, LL and XY contributed to writing and
revising the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Medical Ethics
Committee of the Ninth People's Hospital, Shanghai Jiao Tong
University School of Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sun S and Wang Z: Head neck squamous cell
carcinoma c-Met+ cells display cancer stem cell
properties and are responsible for cisplatin-resistance and
metastasis. Int J Cancer. 129:2337–2348. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pearson AT, Jackson TL and Nör JE:
Modeling head and neck cancer stem cell-mediated tumorigenesis.
Cell Mol Life Sci. 73:3279–3289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leemans CR, Snijders PJF and Brakenhoff
RH: The molecular landscape of head and neck cancer. Nat Rev
Cancer. 18:269–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yu CC, Lo WL, Chen YW, Huang PI, Hsu HS,
Tseng LM, Hung SC, Kao SY, Chang CJ and Chiou SH: Bmi-1 regulates
snail expression and promotes metastasis ability in head and neck
squamous cancer-derived ALDH1 positive cells. J Oncol. 2011:1–16.
2011. View Article : Google Scholar
|
|
5
|
Ringash J: Survivorship and quality of
life in head and neck cancer. J Clin Oncol. 33:3322–3327. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prince ME and Ailles LE: Cancer stem cells
in head and neck squamous cell cancer. J Clin Oncol. 26:2871–2875.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Plaks V, Kong N and Werb Z: The cancer
stem cell niche: How essential is the niche in regulating stemness
of tumor cells? Cell Stem Cell. 16:225–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sayed SI, Dwivedi RC, Katna R, Garg A,
Pathak KA, Nutting CM, Rhys-Evans P, Harrington KJ and Kazi R:
Implications of understanding cancer stem cell (CSC) biology in
head and neck squamous cell cancer. Oral Oncol. 47:237–243. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abbaszadegan MR, Bagheri V, Razavi MS,
Momtazi AA, Sahebkar A and Gholamin M: Isolation, identification,
and characterization of cancer stem cells (Review). J Cell Physiol.
232:2008–2018. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wolmarans E, Boy SC, Nel S, Mercier AE and
Pepper MS: Cancer stem cells in head and neck carcinomas:
Identification and possible therapeutic implications. Adv Exp Med
Biol. Nov 15–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shaikh MV, Kala M and Nivsarkar M: CD90 a
potential cancer stem cell marker and a therapeutic target. Cancer
Biomark. 16:301–307. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Q, Shi S, Yen Y, Brown J, Ta JQ and
Le AD: A subpopulation of CD133+ cancer stem-like cells
characterized in human oral squamous cell carcinoma confer
resistance to chemotherapy. Cancer Lett. 289:151–160. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prince ME, Sivanandan R, Kaczorowski A,
Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF and Ailles
LE: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou C and Sun B: The prognostic role of
the cancer stem cell marker aldehyde dehydrogenase 1 in head and
neck squamous cell carcinomas: A meta-analysis. Oral Oncol.
50:1144–1148. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pozzi V, Sartini D, Rocchetti R,
Santarelli A, Rubini C, Morganti S, Giuliante R, Calabrese S, Di
Ruscio G, Orlando F, et al: Identification and characterization of
cancer stem cells from head and neck squamous cell carcinoma cell
lines. Cell Physiol Biochem. 36:784–798. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun W, Bunn P, Jin C, Little P,
Zhabotynsky V, Perou CM, Hayes DN, Chen M and Lin DY: The
association between copy number aberration, DNA methylation and
gene expression in tumor samples. Nucleic Acids Res. 46:3009–3018.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marzouka NA, Eriksson P, Rovira C,
Liedberg F, Sjödahl G and Höglund M: A validation and extended
description of the Lund taxonomy for urothelial carcinoma using the
TCGA cohort. Sci Rep. 8:37372018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F,
Kuhn RM, Karolchik D, Archie J, Lenburg ME, Esserman LJ, et al: The
UCSC cancer genomics browser. Nat Methods. 6:239–240. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kokko LL, Hurme S, Maula SM, Alanen K,
Grénman R, Kinnunen I and Ventelä S: Significance of site-specific
prognosis of cancer stem cell marker CD44 in head and neck
squamous-cell carcinoma. Oral Oncol. 47:510–516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Judd NP, Winkler AE, Murillo-Sauca O,
Brotman JJ, Law JH, Lewis JS Jr, Dunn GP, Bui JD, Sunwoo JB and
Uppaluri R: ERK1/2 regulation of CD44 modulates oral cancer
aggressiveness. Cancer Res. 72:365–374. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Major AG, Pitty LP and Farah CS: Cancer
stem cell markers in head and neck squamous cell carcinoma. Stem
Cells Int. 2013:3194892013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kiang A, Yu MA and Ongkeko WM: Progress
and pitfalls in the identification of cancer stem cell-targeting
therapies in head and neck squamous cell carcinoma. Curr Med Chem.
19:6056–6064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gilormini M, Wozny AS, Battiston-Montagne
P, Ardail D, Alphonse G and Rodriguez-Lafrasse C: Isolation and
characterization of a head and neck squamous cell carcinoma
subpopulation having stem cell characteristics. J Vis Exp.
2016:e539582016.
|
|
24
|
Zimmerer RM, Ludwig N, Kampmann A,
Bittermann G, Spalthoff S, Jungheim M, Gellrich NC and Tavassol F:
CD24+ tumor-initiating cells from oral squamous cell
carcinoma induce initial angiogenesis in vivo. Microvasc Res.
112:101–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Martens-de Kemp SR, Brink A, Stigter-Van
Walsum M, Damen JM, Rustenburg F, Wu T, Van Wieringen WN,
Schuurhuis GJ, Braakhuis BJ, Slijper M, et al: CD98 marks a
subpopulation of head and neck squamous cell carcinoma cells with
stem cell properties. Stem Cell Res. 10:477–488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Waldron NN, Barsky SH, Dougherty PR and
Vallera DA: A bispecific EpCAM/CD133-targeted toxin is effective
against carcinoma. Target Oncol. 9:239–249. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lim YC, Kang HJ and Moon JH: c-Met pathway
promotes self-renewal and tumorigenecity of head and neck squamous
cell carcinoma stem-like cell. Oral Oncol. 50:633–639. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan M, Yang X, Wang L, Clark D, Zuo H, Ye
D, Chen W and Zhang P: Plasma membrane proteomics of tumor spheres
identify CD166 as a novel marker for cancer stem-like cells in head
and neck squamous cell carcinoma. Mol Cell Proteomics.
12:3271–3284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee SH, Do SI, Lee HJ, Kang HJ, Koo BS and
Lim YC: Notch1 signaling contributes to stemness in head and neck
squamous cell carcinoma. Lab Invest. 96:508–516. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fukusumi T, Ishii H, Konno M, Yasui T,
Nakahara S, Takenaka Y, Yamamoto Y, Nishikawa S, Kano Y, Ogawa H,
et al: CD10 as a novel marker of therapeutic resistance and cancer
stem cells in head and neck squamous cell carcinoma. Br J Cancer.
111:506–514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang CC, Zhu LF, Xu XH, Ning TY, Ye JH and
Liu LK: Membrane Type 1 Matrix metalloproteinase induces an
epithelial to mesenchymal transition and cancer stem cell-like
properties in SCC9 cells. BMC Cancer. 13:1712013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chou MY, Hu FW, Yu CH and Yu CC: Sox2
expression involvement in the oncogenicity and radiochemoresistance
of oral cancer stem cells. Oral Oncol. 51:31–39. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct-4 and Nanog in oral cancer stem-like cells and high-grade
oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen D, Wu M, Li Y, Chang I, Yuan Q,
Ekimyan-Salvo M, Deng P, Yu B, Yu Y, Dong J, et al: Targeting
BMI1+ cancer stem cells overcomes chemoresistance and
inhibits metastases in squamous cell carcinoma. Cell Stem Cell.
20:621–634.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Oliveira-Costa JP, Oliveira LR, da
Silveira GG, Soave DF, Soares FA and Ribeiro-Silva A: Topoisomerase
expression in oral squamous cell carcinoma: Relationship with
cancer stem cells profiles and lymph node metastasis. J Oral Pathol
Med. 41:762–768. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Z, Wang Y, Zhu Y, Yuan C, Wang D, Zhang
W, Qi B, Qiu J, Song X, Ye J, et al: The Hippo transducer TAZ
promotes epithelial to mesenchymal transition and cancer stem cell
maintenance in oral cancer. Mol Oncol. 9:1091–1105. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee CR, Lee SH, Rigas NK, Kim RH, Kang MK,
Park NH and Shin KH: Elevated expression of JMJD6 is associated
with oral carcinogenesis and maintains cancer stemness properties.
Carcinogenesis. 37:119–128. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen B, Dong P, Li D and Gao S: Expression
and function of ABCG2 in head and neck squamous cell carcinoma and
cell lines. Exp Ther Med. 2:1151–1157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee DG, Lee JH, Choi BK, Kim MJ, Kim SM,
Kim KS, Chang K, Park SH, Bae YS and Kwon BS:
H+-myo-inositol transporter SLC2A13 as a potential
marker for cancer stem cells in an oral squamous cell carcinoma.
Curr Cancer Drug Targets. 11:966–975. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu MJ, Jan CI, Tsay YG, Yu YH, Huang CY,
Lin SC, Liu CJ, Chen YS, Lo JF and Yu CC: Elimination of head and
neck cancer initiating cells through targeting glucose regulated
protein78 signaling. Mol Cancer. 9:2832010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Da Cruz Paula A and Lopes C: Implications
of different cancer stem cell phenotypes in breast cancer.
Anticancer Res. 37:2173–2183. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Eun K, Ham SW and Kim H: Cancer stem cell
heterogeneity: Origin and new perspectives on CSC targeting. BMB
Rep. 50:117–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Han J, Fujisawa T, Husain SR and Puri RK:
Identification and characterization of cancer stem cells in human
head and neck squamous cell carcinoma. BMC Cancer. 14:1732014.
View Article : Google Scholar : PubMed/NCBI
|