Introduction
Lung cancer is a malignant tumor with the highest
incidence and mortality in the world (1,2). The
5-year survival rate has not improved significantly in the past 25
years, and is approximately 18% (3), even with specialized treatment
combination of surgery, chemotherapy, and radiation therapy
(4). For lung cancer patients, one
of the most important causes for such a low survival rate is the
occurrence of drug resistance that develops during the
chemotherapeutic procedure (5).
Therefore, in order to obtain better results in lung cancer
therapy, it is crucial to find effective ways to counter the drug
resistance by exploring the underlying mechanisms of
chemoresistance. Cisplatin is widely used in the clinical treatment
of lung cancer due to its marked anticancer effect, and broad
anticancer spectrum (6), and its
main function is through inhibition of DNA synthesis to induce
tumor cell apoptosis (7,8). Resistance to cisplatin seriously
affects the clinical efficacy of lung cancer, and its main
mechanism has not been elucidated clearly. Recently, the effects of
miRNAs on the development of cancer drug resistance has gained
attention.
MicroRNAs (miRNAs) are small non-coding RNA
molecules (approximately 20–25 nucleotides) which are known to
negatively modulate the expression of targeted genes by completely
or partially binding with the 3′-untranslated region (3′-UTR) of
the mRNA (9–11). This function of miRNAs plays an
important role in the development of various malignancies (12,13).
Aberrant miRNA expression has been observed in both physiological
and pathological processes, such as apoptosis and chemotherapy
resistance, in multiple human cancers (14).
Nuclear factor erythroid 2-like 2 (NFE2L2; commonly
known as Nrf2) is a critical transcription factor in the regulation
of antioxidant and cytoprotective genes, by binding to and
activating the expression of promoters containing the antioxidant
response element (ARE). Nrf2 has been reported to be targeted by
many miRNAs. miR-27a-5p, miR-142-5p, miR-28 and miR-93-5p
suppressed cancer chemoprevention activity by downregulating the
Nrf2 mRNA and protein levels (15).
miR-93 was able to regulate the oncogenic process in mammaries
through regulation of its target gene NRF2 (16). MicroRNA-140-5p aggravated
doxorubicin-induced cardiotoxicity by promoting myocardial
oxidative stress by targeting Nrf2 and Sirt2 (17). Nevertheless, the biological role of
miR-144-3p in modulating lung cancer drug resistance by targeting
Nrf2 is not well understood.
In the present study, we investigated the expression
levels of miR-144-3p in tumor and adjacent tissues of lung cancer
patients and in lung cancer cell lines, in order to identify the
functional role of miR-144-3p in lung cancer biology. In addition,
we elucidated the regulatory cisplatin resistance involving
miR-144-3p and Nrf2 in lung cancer cell multidrug resistance.
Furthermore, in the course of the experiment, we unexpectedly found
that there was a regulation loop between miR-144-3p and Nrf2 in
regulating the cisplatin resistance of lung cancer cells.
Materials and methods
Cell culture
A549, H1299, HepG2, MCF-7, HeLa and Cos-7 cell lines
were used in the present study. The A549 and H1299 cell lines were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA) (nos. CCL-185 and CRL-5803, respectively). These
cells were seeded in RPMI-1640 medium and fetal bovine serum (FBS)
to a final concentration of 10%. HepG2, MCF-7, HeLa and Cos-7 cells
used in this study were purchased from ATCC (nos. HB-8065, HTB-22,
CCL-2 and CRL-1651, respectively). These cells were seeded in
Dulbecco's modified Eagle's medium-high glucose (DMEM-HG; Hyclone;
GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
FBS) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All
of the aforementioned cell lines were cultured at 37°C in a
humidified air with 5% CO2.
Plasmid constructs and cell
transfection
Human Nrf2 (Gene ID: 4780) was inserted into
expression vector (pcDNA3.1-). ShNrf2 and its corresponding control
were kindly provided by Professor Jian Dong (North Carolina State
University, Raleigh, NC, USA). miR-144-3p mimic, miR-144-3p
inhibitor or the appropriate negative controls (NC) of miRNA mimic
(mimic NC) and miRNA inhibitor (inhibitor NC) were obtained from
Guanzhou RiboBio Co., Ltd. (Guanzhou, China). For transfection
experiments, the cells were cultured in growth medium without
antibiotics at 60% confluence for 2 days, and then transfected with
transfection reagent (FuGENE® HD; Roche Diagnostics,
Basel, Switzerland) according to the manufacturer's instructions.
After incubation for 6 h, the medium was removed and replaced with
normal culture medium for 48 h.
MTT assay
Cell growth was estimated by a modified MTT assay.
As a measurement of cell growth, the cells were seeded onto a
96-well dish and grown in medium containing 10% FBS. After the
cells were treated daily with cisplatin (1, 2, 4, 8, 16, 32 and 64
µM) for 24 h, the MTT reagent (2.5 mg/ml) was added and the optical
density (570 nm) was measured by ELISA reader.
Quantitative real-time RT-PCR
Total RNA, including miRNA, was extracted by using a
e.Z.N.A miRNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA)
according to the manufacturer's protocol, and the sample was
reverse-transcribed using M-MLV reverse transcriptase (Promega
Corporation, Madison, WI, USA). Real-time PCR was performed using
Applied Biosystems Step One™ Real-Time PCR System. Fast
SYBR® Green Master Mix was obtained from Applied
Biosystems; Thermo Fisher Scientific, Inc. Data presented as the
relative expression levels of Nrf2, HO-1, NQO-1, Bcl-2 and
caspase-3 were normalized by GAPDH. The primers for the PCR
analysis are listed in Table I.
Amplification of U6 small nuclear RNA served as an endogenous
control used to normalize miR-144-3p expression data. Thermocycling
conditions were as follows: 95°C for 5 min followed by 40 cycles at
95°C for 10 sec and 60°C for 30 sec, then a melting curve analysis
from 60 to 95°C every 0.2°C for 1.5 min was obtained. Each sample
was analyzed in triplicate, and quantified using the
2−ΔΔCq method (18).
 | Table I.Primers used for RT-PCR. |
Table I.
Primers used for RT-PCR.
| Nrf2 | F:
TCAGCGACGGAAAGAGTA |
|
| R:
GGGAGTAGTTGGCAGATC |
| HO-1 | F:
GTGAAGCGGCTCCACGAG |
|
| R:
GGCAATGTTGGGGAAGGT |
| Caspase-3 | F:
GAGTTCGGTGGGGTCATG |
|
| R:
GGAGAAATCAAACAGAGGC |
| Bcl-2 | F:
GAGTTCGGTGGGGTCATG |
|
| R:
GGAGAAATCAAACAGAGGC |
| NQO-1 | F:
GAAAGGACATCACAGGTAA |
|
| R:
GGGAACTGGAATATCACAA |
| GAPDH | F:
CTCCTCCACCTTTGACGC |
|
| R:
CCACCACCCTGTTGCTGT |
Protein extraction and western
blotting
For the western blot analysis, protein samples were
extracted from the cells with Protein Extraction Reagent (Pierce;
Thermo Fisher Scientific, Inc.). The concentrations of the proteins
were determined using the BCA Quantification kit (Beyotime
Institute of Biotechnology, Beijing, China) for subsequent sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The
proteins (20 µg) were separated by SDS PAGE (10%) and transferred
onto a PVDF membrane. The membrane was blocked using 5% non-fat
milk at 25°C for 1 h, and then incubated with primary antibodies
overnight at 4°C. The antibodies used were as follows: Anti-human
GAPDH antibody (dilution 1:2,000; cat. no. 97166; Cell Signaling
Technology, Inc., Danvers, MA, USA), anti-human Nrf2 antibody
(dilution 1:1,000; cat. no. sc-81342; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), anti-human HO-1 antibody (dilution 1:1,000;
cat. no. 5853; Cell Signaling Technology, Inc.), anti-human NQO-1
antibody (dilution 1:1,000; cat. no. ab28947; Abcam, Cambridge,
UK), anti-human Bcl-2 antibody (dilution 1:1,000; cat. no. sc-509)
and anti-human caspase-3 antibody (dilution 1:1,000; cat. no.
sc-271759) (both from Santa Cruz Biotechnology, Inc.). Then, the
membrane was incubated with IRDyeTM-800 conjugated anti-mouse or
anti-rabbit secondary antibodies (dilution 1:5,000; cat. nos.
115-005-146 and 115-005-144; Jackson ImmunoResearch Laboratories,
Inc., West Grove, PA, USA) for 1 h at room temperature. The protein
signals were visualized with the Odyssey Infrared Imaging System
(LI-COR Biosciences, Lincoln, NE, USA). GAPDH expression was used
as an internal control.
Luciferase constructs, site-mutation,
and luciferase assay
The luciferase constructs were as follows: i)
Nrf2-3′-UTR-WT: The Nrf2 3'-UTR region was fused to the pmirGLO
reporter vector; ii) Nrf2-3′-UTR-MUT: The same as the
Nrf2-3′-UTR-WT, except that the miR-144-3p binding site was
mutated; iii) miR-144-3p-luc: The miR-144-3p promoter region (about
2000 bp) was fused to the pGL3-Basic reporter vector; iv)
Mut-miR-144-3p-luc: The same as miR-144-3p-luc, except that the
Nrf2 binding site was mutated. The cells (2×105/well)
were plated in 24-well plates. To ascertain the effect of
miR-144-3p on Nrf2, Cos-7 cells were co-transfected with miR-144-3p
mimic or mimic NC in combination with Nrf2-3′-UTR-WT or
Nrf2-3′-UTR-MUT. In order to explore the mechanism of Nrf2 acting
on miR-144-3p, Cos-7 cells were co-transfected with Nrf2 or control
(pcDNA3.1-) in combination with miR-144-3p-luc, or
mut-miR-144-3p-luc. Cells were harvested 48 h after transfection
and luciferase activity was assessed using the Dual Luciferase
Assay System (Promega Corp.). The results were expressed as a fold
induction relative to the cells transfected with the control after
normalization to Renilla activity. In the dual luciferase assay
results, all columns represented the mean result of three
independent experiments and the error bars represented the standard
deviation.
Chromatin immunoprecipitation (ChIP)
assay
We used a commercial Chromatin Immunoprecipitation
(ChIP) Assay kit (Merck KGaA, Darmstadt, Germany) by following the
manufacturer's instructions. After the treatment, each test group
was incubated with 1% formaldehyde to cross-link DNA-protein
complexes. After washing with ice-cold PBS three times, the cells
were lysed in SDS lysis buffer. Then lysates were sonicated to
shear DNA to ~200-1,000 bp fragments. We then used an anti-Nrf2
antibody to immunoprecipitate the cross-linked protein at 4°C
overnight. IgG acted as the negative control. The DNA was used as a
template for PCR and utilized the Nrf2 binding site. The PCR
products were separated on 1% agarose gel.
Statistical analysis
High throughput sequencing data and prognostic data
were derived from the TCGA database (GSE56036) and LinkedOmics
database (ID-3650). The software of TargetScan was used to predict
gene targets. Data were expressed as the mean ± SE, accompanied by
the number of experiments performed independently, and analyzed by
t-tests. Differences at P<0.05 were considered to be
statistically significant.
Results
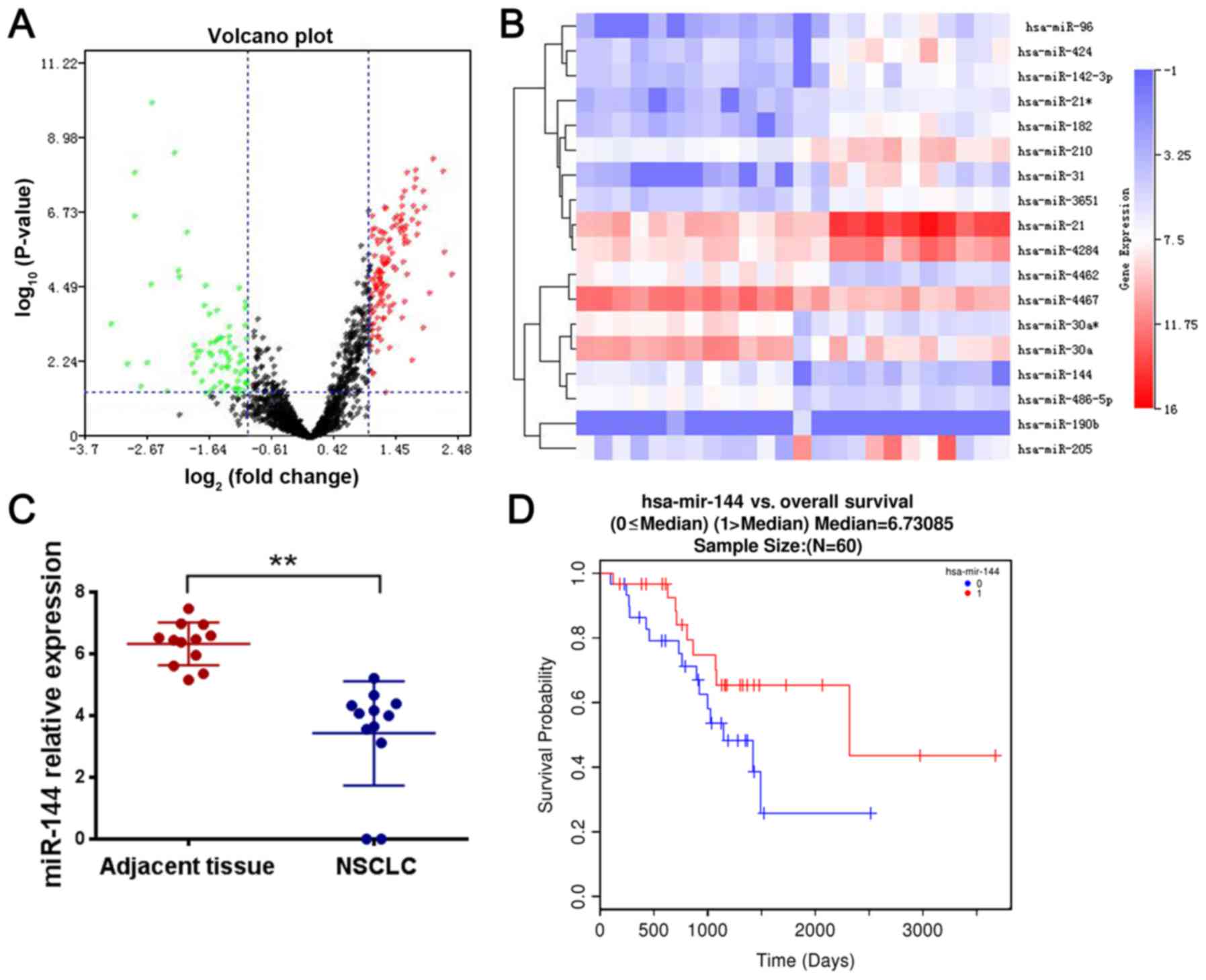
miR-144-3p expression is downregulated
in lung cancer tissues and associated with poor prognosis
There was a significant difference of miR-144-3p
expression in tumor tissues and normal tissues in patients with
platinum insensitivity drugs by analyzing high-throughput
sequencing data from the TCGA database (GSE56036). The results
revealed that miR-144-3p expression was significantly decreased in
lung cancer tissues compared with adjacent non-tumor tissues
(Fig. 1A-C). The emergence of drug
resistance often indicates a poor prognosis. To assess the clinical
significance of miR-144-3p overexpression in lung cancer, we
evaluated the association between miR-144-3p levels and patient
clinicopathological characteristics based on the TCGA database
(GSE56036) and LinkedOmics database (ID-3650). Kaplan-Meier
survival analysis revealed that patients with higher miR-144-3p
levels had longer overall survival and progression-free survival
times than those who had lower levels of miR-144-3p (Fig. 1D). These findings indicated that the
expression of miR-144-3p was decreased in lung cancer tissues and
associated with poor prognosis. Thus, we performed experimental
research on whether miR-144-3p was related to cisplatin
resistance.
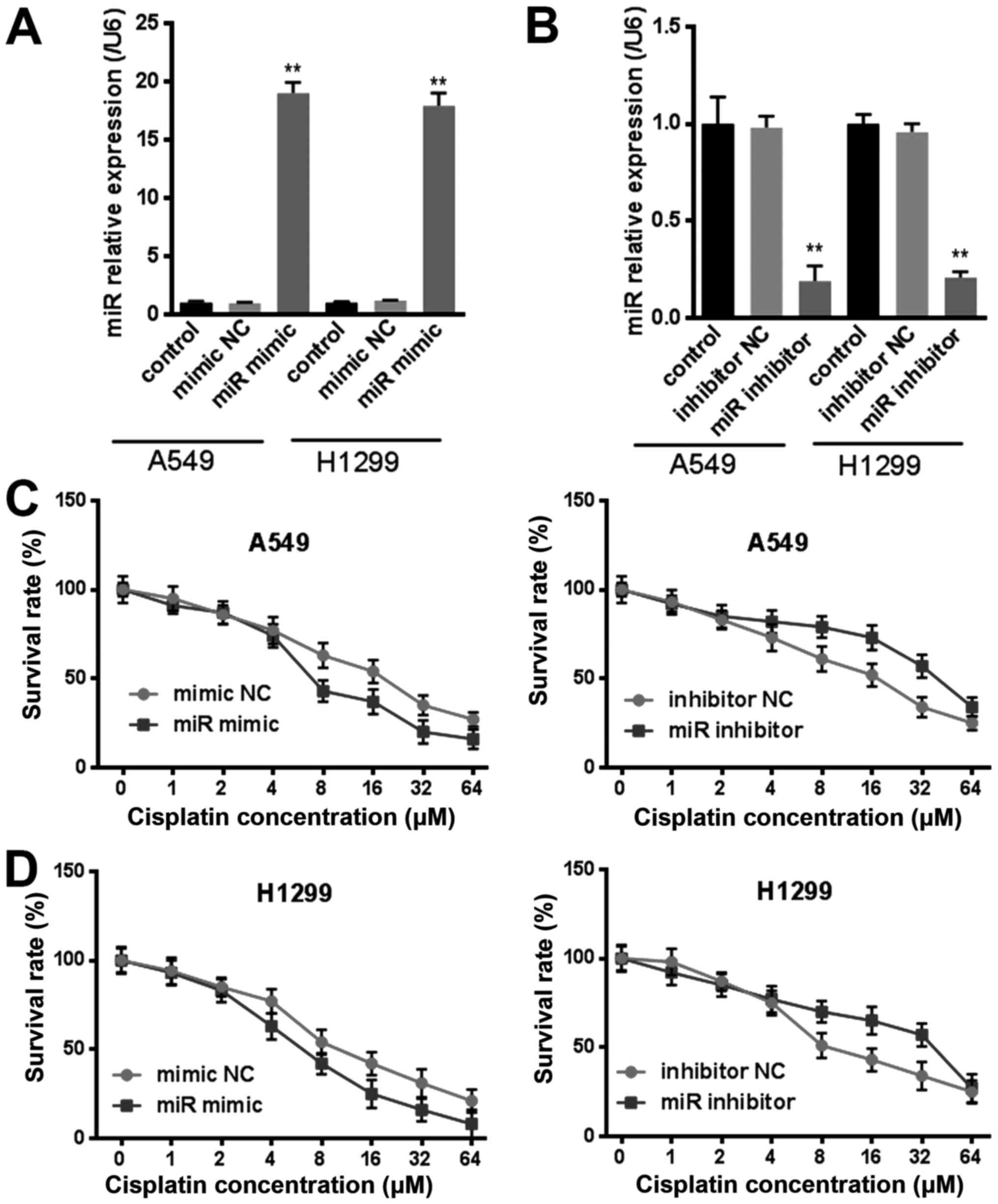
miR-144-3p inhibits cisplatin
resistance in lung cancer cells
To further investigate whether miR-144-3p was
involved in drug resistance in lung cancer, we transfected
miR-144-3p mimic or miR-144-3p inhibitor to lung cancer cells
(Fig. 2A and B) and then exposed
the cells to cisplatin with rising levels of concentration for 24
h. The viability of lung cancer cells that were transfected with
miR-144-3p were more sensitive to cisplatin in comparison with the
mimic NC group (Fig. 2C and D).
However, the miR-144-3p inhibitor did the opposite, the resistance
of cells was significantly enhanced to cisplatin (Fig. 2C and D). These results indicated
that miR-144-3p inhibited cisplatin resistance in lung cancer
cells. Thus, targeting miR-144-3p could reverse the cisplatin
resistant behavior in lung tumor cells.
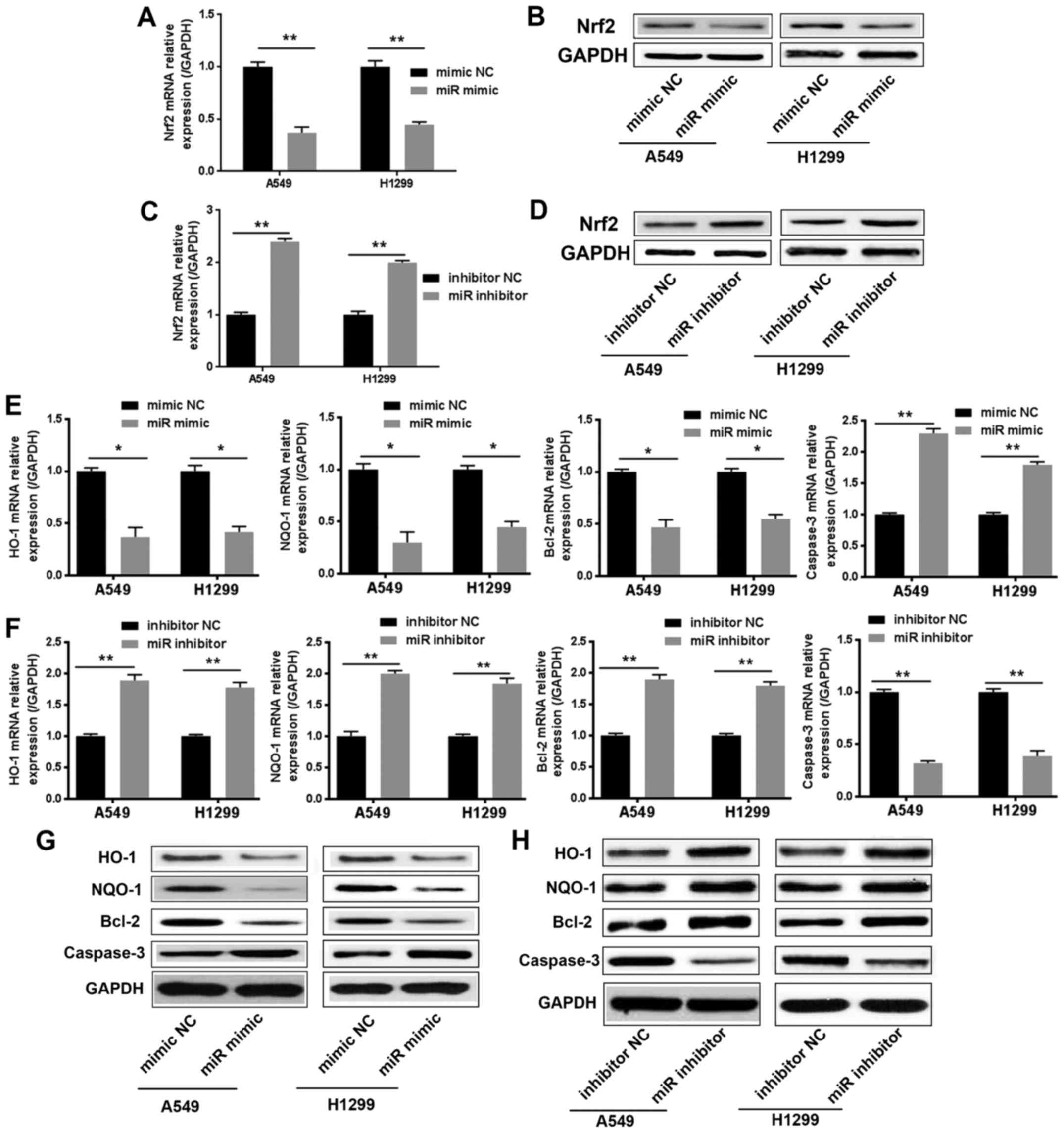
miR-144-3p inhibits the Nrf2 pathway
during the cisplatin resistance process in lung cancer cells
In order to further explore how miR-144-3p regulated
drug resistance, we used TargetScan to predict potential miR-144-3p
target genes. Nrf2 was identified during the scanning process.
Nrf2, as a nuclear transcription factor, which binds to the ARE
sequences and activates the molecules downstream, was considered to
render cancer cells resistant to drugs including cisplatin. We next
investigated whether miR-144-3p could regulate the Nrf2 pathway
during the cisplatin resistance process in lung cancer cells.
Within 24 h of cisplatin treatment, we examined the
mRNA and protein levels of Nrf2 in lung cancer cells after
transfection with the miR-144-3p mimic or miR-144-3p inhibitor.
Data revealed that overexpressed miR-144-3p effectively suppressed
the mRNA and protein levels of Nrf2 in lung cancer cells (Fig. 3A and B). However, the mRNA and
protein levels of Nrf2 were upregulated in the miR-144-3p
inhibitor-transfected lung cancer cells (Fig. 3C and D). Concurrently, the
expression of Nrf2 downstream target genes, which are involved in
drug resistance, were examined. As revealed in Fig. 3E-H, the expression of HO-1, NQO1 and
Bcl-2 was downregulated significantly in the miR-144-3p
mimic-transfected group compared to the miR-144-3p mimic-NC group.
However, the miR-144-3p inhibitor-transfected group exhibited the
opposite results. Then, we investigated the expression of caspase-3
by Real-time PCR and western blot assays. The results confirmed
that the expression of caspase-3 and miR-144-3p exhibited a
positive association trend (Fig.
3E-H). These results revealed that miR-144-3p could inhibit the
expression of Nrf2 against the drug resistance of lung cancer
cells.
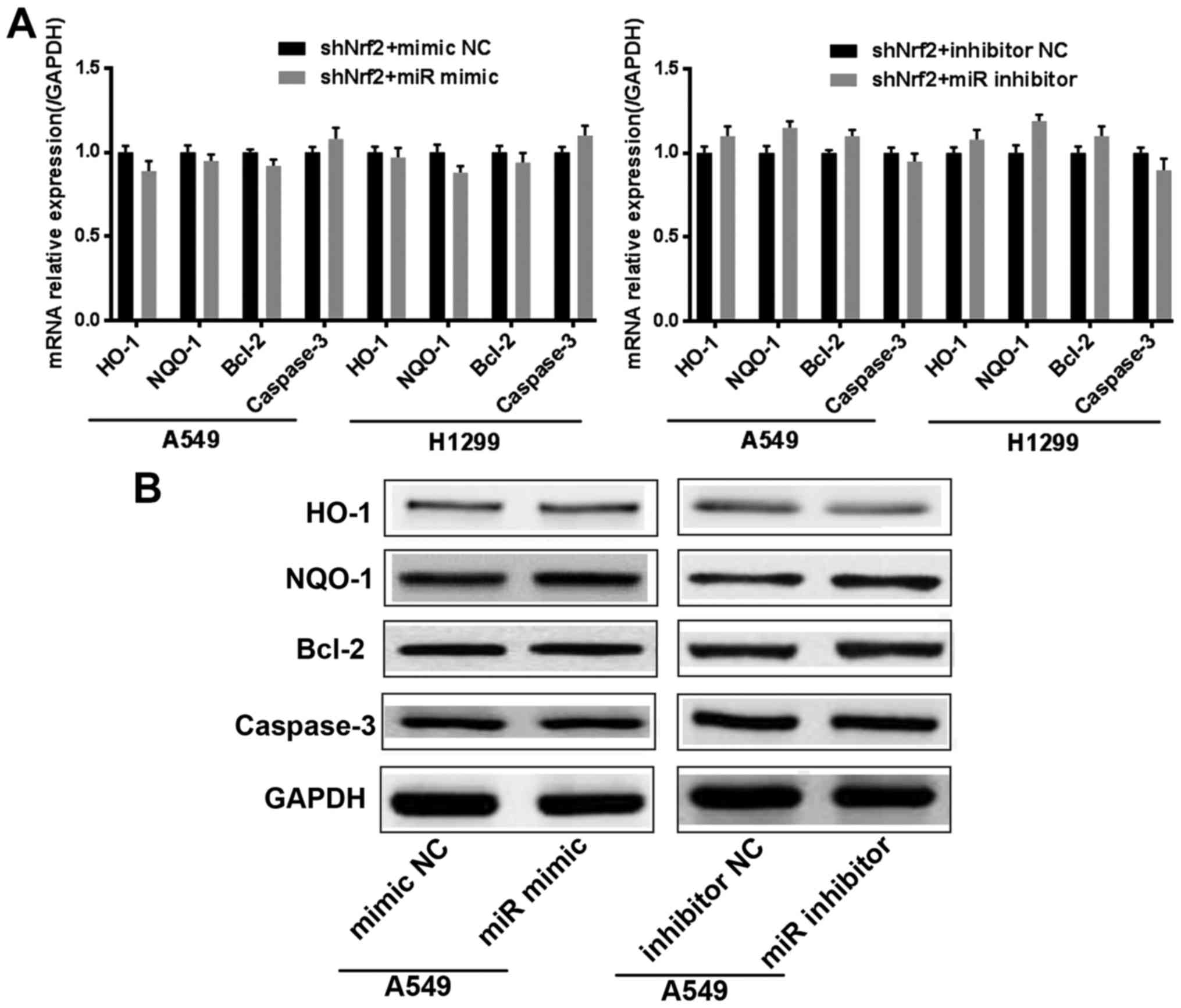
miR-144-3p promotes cisplatin
sensitivity by adjusting Nrf2 in lung cancer cells
To further determine that the effect of miR-144-3p
in cisplatin resistance was achieved by regulating Nrf2, we
generated two stable lung cancer cell lines with Nrf2 knocked down
(shNrf2 group). Then miR-144-3p was overexpressed or inhibited in
the shNrf2 stable lung cancer cells. We used Real-time PCR to
assess the mRNA levels of HO-1, NQO1, Bcl-2 and caspase-3.
Concurrently, western blotting was also used to detect the protein
levels of HO-1, NQO1, Bcl-2 and caspase-3. The results confirmed
that, when Nrf2 was knocked down, miR-144-3p lost the ability to
regulate the resistance in cisplatin-treated lung cancer cells
(Fig. 4A and B). All of the
aforementioned results indicated that miR-144-3p required Nrf2 to
promote cisplatin sensitivity in lung cancer cells.
miR-144-3p mediates Nrf2 expression by
targeting the 3′-UTR in lung cancer cells
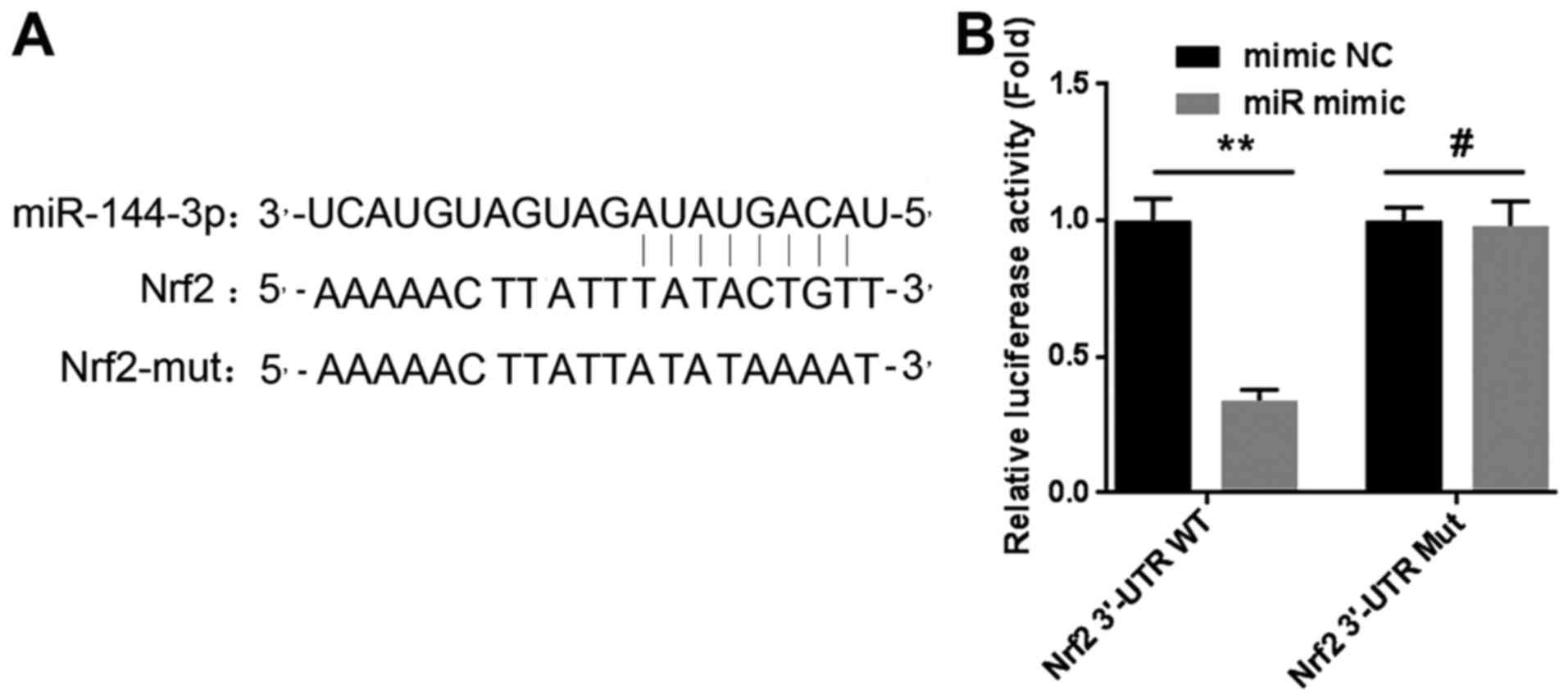
As reported in research, miRNAs are known to
negatively modulate the expression of targeted genes by completely
or partially binding with the 3′-untranslated regions (3′-UTR).
Thus, we constructed two plasmids of luciferase reporter gene
vectors named Nrf2-3′-UTR-WT and Nrf2-3′-UTR-MUT respectively. We
then examined the effect of miR-144-3p on the Nrf2 3′-UTR by
luciferase assay. The results confirmed that miR-144-3p could bind
to the 3′-UTR region of Nrf2 directly, reducing the mRNA level of
Nrf2 (Fig. 5A and B). The
experimental results revealed that miR-144-3p mediated Nrf2
expression by targeting the 3′-UTR in lung cancer cells.
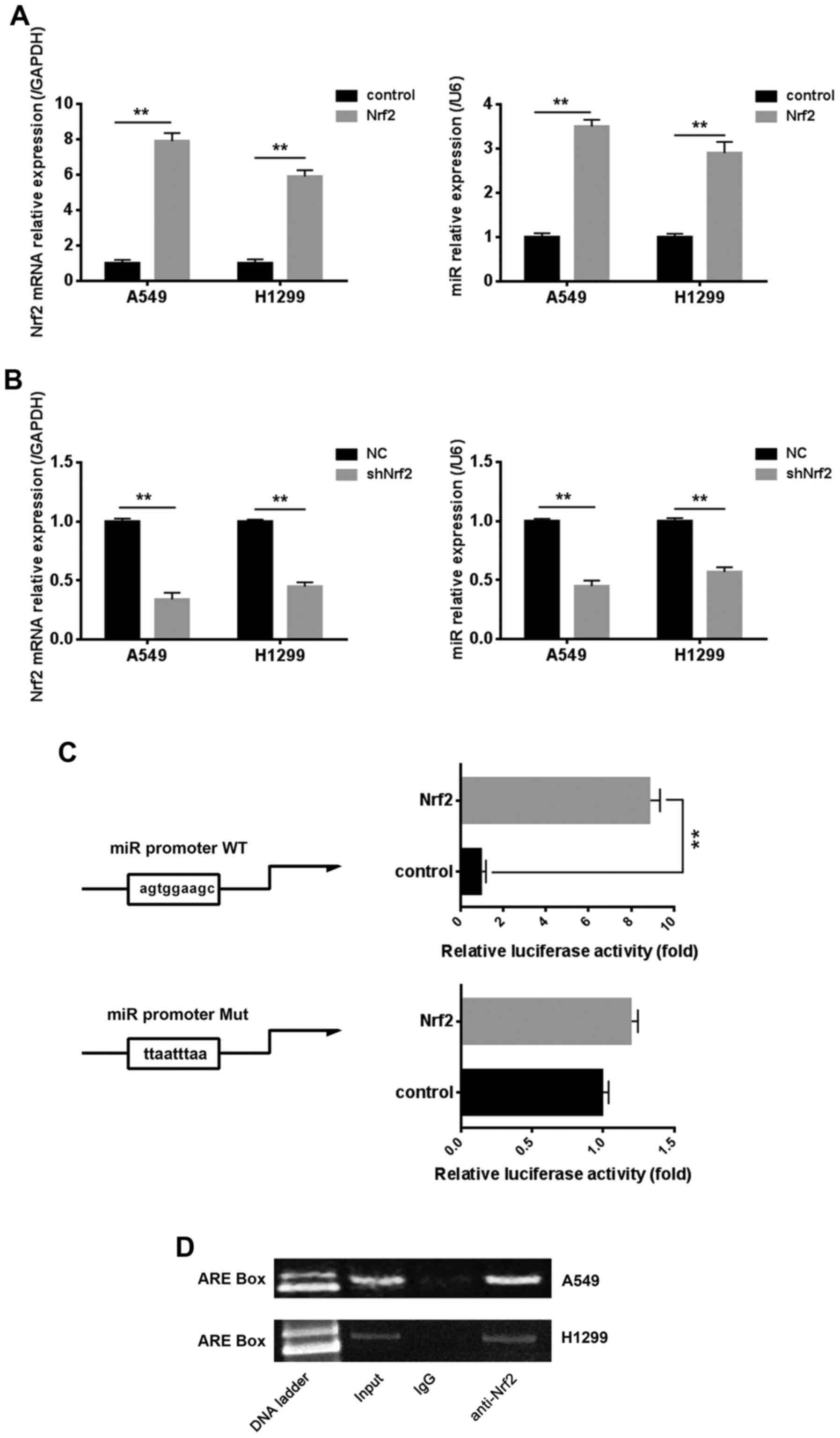
Nrf2 also affects the expression of
miR-144-3p by binding to the ARE box in the miR-144-3p
promoter
Previous research confirmed that miR-144-3p could
inhibit the cisplatin resistance of lung cancer cells via Nrf2, and
we revealed the possible molecular mechanisms between miR-144-3p
and Nrf2 in this process. Notably, we found that Nrf2 could reverse
regulate the expression of miR-144-3p. There was one potential ARE
box on the miR-144-3p promoter region. As revealed in Fig. 6A and B, the results of Real-time PCR
indicated that the mRNA level of miR-144-3p was positively
associated with the mRNA level of Nrf2, whether Nrf2 was
overexpressed or knocked down. Nrf2 regulated target genes by
binding to the ARE box existing in the promoter region. There was
one potential ARE box on the miR-144-3p promoter region. The
results of the luciferase assay indicated that the transcriptional
activity of miR-144-3p promoter could be upregulated by Nrf2.
However, the miR-144-3p transcriptional activity was not affected
when the ARE box was mutated (Fig.
6C). These results may indicate that Nrf2 affected the
miR-144-3p transcriptional activity by binding to the ARE box. To
further assess the mechanism of this regulation, we used chromatin
immunoprecipitation (ChIP) to investigate in lung cancer cells.
Data revealed that Nrf2 could be combined with the ARE box,
consistent with results of the luciferase assay (Fig. 6D).
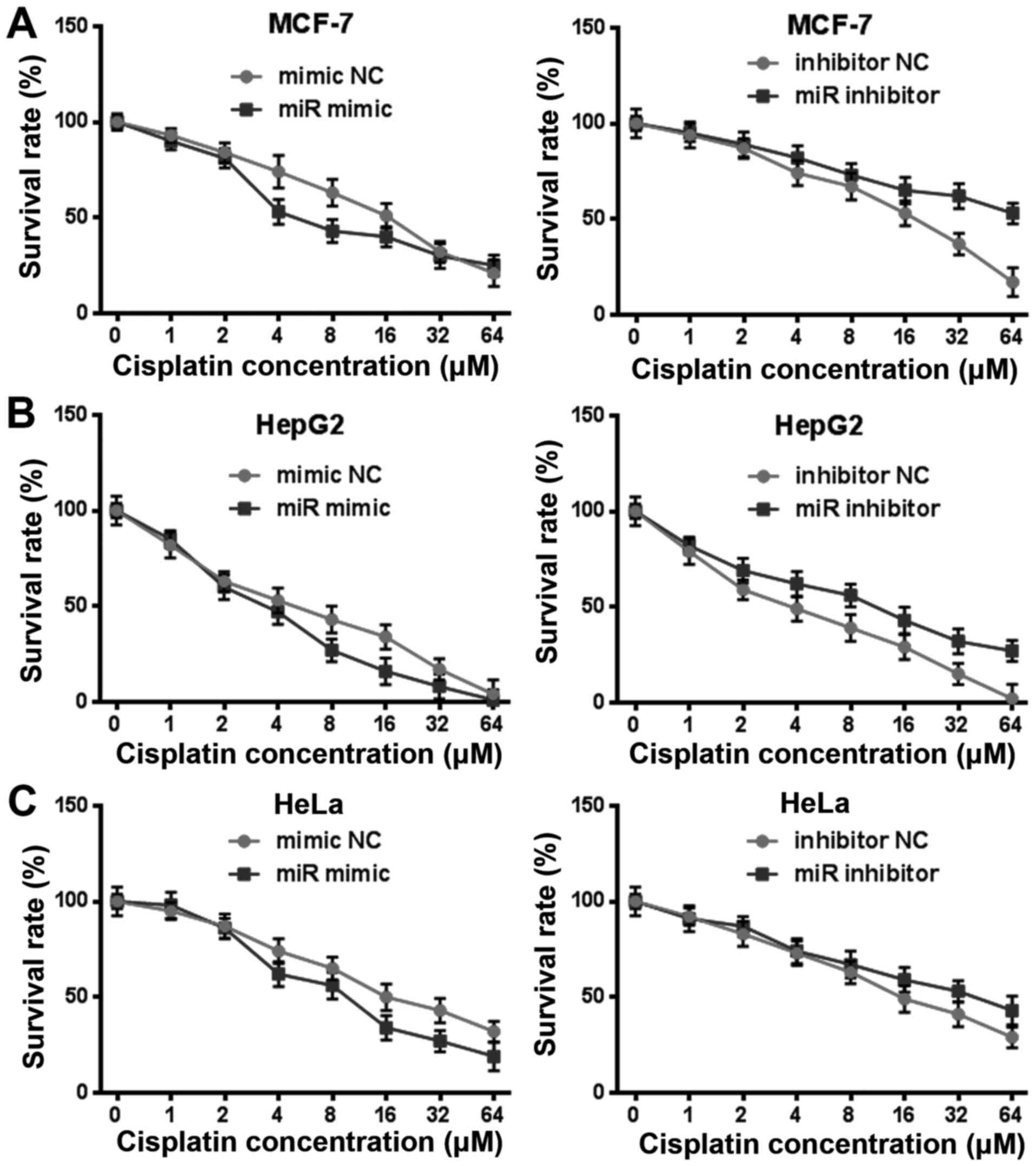
miR-144-3p also regulates cisplatin
resistance in other tumor cells
In order to clarify whether miR-144-3p played a
crucial role to cisplatin resistance in other tumor cells, we
exposed HepG2, HeLa, MCF-7 to cisplatin with rising levels of
concentration for 24 h after transfection miR-144-3p mimic or
miR-144-3p inhibitor (Fig. 7A-C).
The viability of these tumor cells that were transfected with
miR-144-3p were more sensitive to cisplatin in comparison with the
mimic NC group. Conversely, the resistance of cells after
transfection the miR-144-3p inhibitor was significantly enhanced to
cisplatin. These results indicated that miR-144-3p could also
regulate cisplatin resistance in other tumor cells.
Discussion
Chemotherapy is one of the main treatments for
malignant tumors. However, congenital or acquired resistance,
especially multidrug resistance, often leads to chemotherapy
failure which has become an unavoidable problem in the process of
cancer treatment in clinics. However, there have been limited
effective solutions to solve the resistance problem in cancer
treatment over the years (19–21).
Searching for new chemical resistance mechanisms as well as new
molecular targets are urgent in order to solve drug resistance.
In recent years, research has revealed that the
aberrant expression of microRNAs (miRNAs) is closely related to the
occurrence, development, prognosis and drug resistance of cancer.
Although its underlying mechanism still remains uncertain, current
experiments and observations have revealed that miRNAs are involved
in multidrug resistance processes and play a crucial role in a
variety of signaling pathways through negative regulation. For
instance, miR-181a, miR-302a and miR-487a can increase the
sensitivity of breast cancer to mitoxantrone by inhibiting BCP
expression (22–24). In addition, microRNA-130b targets
PTEN to induce resistance to cisplatin in lung cancer cells by
activating the Wnt/β-catenin pathway (25). These discoveries suggest that miRNAs
may be used as a target for predicting chemosensitivity and
reversing resistance to tumor therapy. In the present study, we
determined that the expression of miR-144-3p was significantly
downregulated in lung cancer tissue compared to matched adjacent
normal lung tissue and associated with poor prognosis, especially
in advanced patients with lung cancer. Notably, we found that the
change of miR-144-3p levels was related to cisplatin resistance. To
further demonstrate whether miR-144-3p was involved in drug
resistance in lung cancer, we transfected miR-144-3p mimic or
miR-144-3p inhibitor to lung cancer cells and then exposed these
cells to cisplatin with increasing levels of concentration. The
viability of lung cancer cells that were transfected with
miR-144-3p were more sensitive to cisplatin in comparison with the
mimic NC group, while with the miR-144-3p inhibitor the opposite
was observed. These findings indicated that miR-144-3p inhibited
cisplatin resistance in lung cancer cells. Furthermore, miR-144-3p
played a crucial role in cisplatin resistance in other tumor cells,
such as HepG2, HeLa and MCF-7.
Nrf2 is an important molecule associated with the
resistance of anticancer drugs and activates the molecules
downstream, such as antioxidant molecules, detoxification proteins
and inhibits cell apoptosis (Bcl-2) and multidrug resistant
drug-associated proteins (MRPs), rendering cancer cells resistant
to drugs (26–28). Nrf2 regulates downstream target gene
expression mainly by binding with ARE elements in the target gene
promoter region. When the activated Nrf2 enters the nucleus, and
combines with the ARE sequence, the ARE-regulated genes involving
metabolism, intracellular redox balance, apoptosis and drug
resistance begin to transcribe and induce the expression of target
genes and play the role of cell protection (29). Nrf2 has been revealed to be closely
connected with cisplatin resistance (30,31),
however we know little about the relationship between miR-144-3p
and Nrf2. Our research findings revealed that targeting Nrf2 at a
post-transcriptional stage via miR-144-3p could regulate its
downregulation process, thus decreasing the expression of target
genes related to drug resistance.
In addition, we revealed that the overexpression of
Nrf2 can activate the expression of miR-144-3p by binding the
antioxidant response element (ARE) in the promoter region. Perhaps
this is a self-protection mechanism of the body when drug
resistance occurs during the chemotherapy procedure.
In conclusion, we determined that miR-144-3p
expression was related with the survival rate and cisplatin
resistance of lung cancer patients. Furthermore, there was a
positive association between the levels of miR-144-3p and the
sensitivity of lung cancer cells to cisplatin. Moreover, we
demonstrated that miR-144-3p reacted to cisplatin resistance in
lung cancer cells by altering its target Nrf2. Furthermore,
overexpression of Nrf2, in turn, could regulate the expression of
miR-144-3p by binding to ARE in the miR-144-3p promoter region.
This may be a self-protection mechanism of the body. In addition,
we also found that miR-144-3p could also regulate cisplatin
resistance in other tumor cells. Cisplatin is an early-used and
mature drug among antitumor drugs, but intrinsic and acquired
resistance limits the clinical application of cisplatin. The
present study, may provide some theoretical reference for the
clinical inhibition of cisplatin resistance.
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the
Science and Technology Fund of Tianjin Municipal Health Bureau
(2015KZ013).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
YY, HL and JQ designed the experiments. YY, HL, JX,
DS, LZ, BL, LW and GL performed the experiments, analyzed and
interpreted the data. YY and HL were major contributors in writing
the manuscript. All authors read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jones GS and Baldwin DR: Recent advances
in the management of lung cancer. Clin Med (Lond). 18 Suppl
2:S41–S46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Waqar SN and Morgensztern D: Treatment
advances in small cell lung cancer (SCLC). Pharmacol Ther.
180:16–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsvetkova E and Goss GD: Drug resistance
and its significance for treatment decisions in non-small cell lung
cancer. Curr Oncol. 19 Suppl 1:S45–S51. 2012.PubMed/NCBI
|
|
6
|
Barr MP, Gray SG, Hoffmann AC, Hilger RA,
Thomale J, O'Flaherty JD, Fennell DA, Richard D, O'Leary JJ and
O'Byrne KJ: Generation and characterisation of cisplatin-resistant
non-small cell lung cancer cell lines displaying a stem-like
signature. PLoS One. 8:e541932013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rudolph C, Melau C, Nielsen JE, Jensen
Vile K, Liu D, Pena-Diaz J, Rajpert-De Meyts E, Rasmussen LJ and
Jørgensen A: Involvement of the DNA mismatch repair system in
cisplatin sensitivity of testicular germ cell tumours. Cell Oncol
(Dordr). 40:341–355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan M, Liaw CS, Ji SM, Tan HH, Wong CY,
Thike AA, Tan PH, Ho GH and Lee AS: Identification of circulating
microRNA signatures for breast cancer detection. Clin Cancer Res.
19:4477–4487. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Natalie G, Walker RC, Hee KC, Winter S and
Hunter KW: Inherited variation in miR-290 expression suppresses
breast cancer progression by targeting the metastasis
susceptibility gene Arid4b. Cancer Res. 73:2671–2681. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guarnieri DJ and Dileone RJ: MicroRNAs: A
new class of gene regulators. Ann Med. 40:197–208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wahid F, Shehzad AT, Khan T and Kim YY:
MicroRNAs: Synthesis, mechanism, function, and recent clinical
trials. Biochim Biophys Acta. 1803:1231–1243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoelscher SC, Doppler SA, Dreßen M, Lahm
H, Lange R and Krane M: MicroRNAs: Pleiotropic players in
congenital heart disease and regeneration. J Thorac Dis. 9 Suppl
1:S64–S81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kastl L, Brown I and Schofield AC:
miRNA-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dongoran RA and Wu TY: Abstract 5269:
Cryptotanshinone activate Nrf2 expression through microRNA
regulations. Cancer Res. 77 Suppl 13:52692017. View Article : Google Scholar
|
|
16
|
Singh B, Ronghe AM, Chatterjee A, Bhat NK
and Bhat HK: MicroRNA-93 regulates NRF2 expression and is
associated with breast carcinogenesis. Carcinogenesis.
34:1165–1172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao L, Qi Y, Xu L, Tao X, Han X, Yin L
and Peng J: MicroRNA-140-5p aggravates doxorubicin-induced
cardiotoxicity by promoting myocardial oxidative stress via
targeting Nrf2 and Sirt2. Redox Biol. 15:284–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Holohan C, Van Schaeybroeck S, Longley DB
and Johnston PG: Cancer drug resistance: An evolving paradigm. Nat
Rev Cancer. 13:714–726. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Housman G, Byler S, Heerboth S, Lapinska
K, Longacre M, Snyder N and Sarkar S: Drug resistance in cancer: An
overview. Cancers (Basel). 6:1769–1792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Periti P and Mini E: Drug resistance in
cancer: An overview of the clinical aspects. J Chemother. 1:5–9.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiao X, Zhao L, Ma M, Bai X, He M, Yan Y,
Wang Y, Chen Q, Zhao X, Zhou M, et al: MiR-181a enhances drug
sensitivity in mitoxantone-resistant breast cancer cells by
targeting breast cancer resistance protein (BCRP/ABCG2). Breast
Cancer Res Treat. 139:717–730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma MT, He M, Wang Y, Jiao XY, Zhao L, Bai
XF, Yu ZJ, Wu HZ, Sun ML, Song ZG and Wei MJ: miR-487a resensitizes
mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by
targeting breast cancer resistance protein (BCRP/ABCG2). Cancer
Lett. 339:107–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwabedissen Meyer zu HE and Kroemer HK:
In vitro and in vivo evidence for the importance of breast cancer
resistance protein transporters (BCRP/MXR/ABCP/ABCG2). Handb Exp
Pharmacol. 1–371. 2011.
|
|
25
|
Zhang Q, Zhang B, Sun L, Yan Q, Zhang Y,
Zhang Z, Su Y and Wang C: MicroRNA-130b targets PTEN to induce
resistance to cisplatin in lung cancer cells by activating
Wnt/β-catenin pathway. Cell Biochem Funct. 36:194–202. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Niture SK and Jaiswal AK: Nrf2-induced
antiapoptotic Bcl-xL protein enhances cell survival and drug
resistance. Free Radic Biol Med. 57:119–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Niture SK and Jaiswal AK: Nrf2 protein
up-regulates antiapoptotic protein Bcl-2 and prevents cellular
apoptosis. J Biol Chem. 287:9873–9886. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Young LC, Campling BG, Cole SP, Deeley RG
and Gerlach JH: Multidrug resistance proteins MRP3, MRP1, and MRP2
in lung cancer: Correlation of protein levels with drug response
and messenger RNA levels. Clin Cancer Res. 7:1798–1804.
2001.PubMed/NCBI
|
|
29
|
Hayes JD, McMahon M, Chowdhry S and
Dinkova-Kostova AT: Cancer chemoprevention mechanisms mediated
through the Keap1-Nrf2 pathway. Antioxid Redox Signal.
13:1713–1748. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Solis LM, Behrens C, Dong W, Suraokar M,
Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN,
et al: Nrf2 and Keap1 abnormalities in non-small cell lung
carcinoma and association with clinicopathologic features. Clin
Cancer Res. 16:3743–3753. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Homma S, Ishii Y, Morishima Y, Yamadori T,
Matsuno Y, Haraguchi N, Kikuchi N, Satoh H, Sakamoto T, Hizawa N,
et al: Nrf2 enhances cell proliferation and resistance to
anticancer drugs in human lung cancer. Clin Cancer Res.
15:3423–3432. 2009. View Article : Google Scholar : PubMed/NCBI
|