Introduction
Several studies have reported that mouse or human
fibroblasts can be directly reprogrammed into neurons, neural stem
cells, or glial cells by the introduction of cell-specific
transcription factors (1–9). Malignant gliomas can also be converted
into functional neurons with the aid of neural-cell-specific
transcription factors (10,11). These studies suggest the possibility
that direct reprogramming technology can change the fate of a cell,
irrespective of the cell type.
Direct reprogramming studies have mostly used the
lentivirus system to introduce cell-specific transcription factors
into donor cells (8); this may,
however, lead to the insertion of the host chromosome. However, in
a previous study, a direct reprogramming technology was developed
based on small-molecule compounds to overcome the critical problem
of the virus platform (12,13). This new technology enabled the
conversion of mouse and human somatic cells into neurons, neural
stem cells, or glial cells, without inserting the host chromosome.
Therefore, these small-molecule compounds can replace the
transcription factors in direct reprogramming.
Previous studies have reported that gliomas are
derived from glial precursor cells (14), and a significant portion of gliomas
strongly react with glial precursor cell-specific markers (15). However, whether or not gliomas can
be transformed into glial cells has not been explicitly
investigated. Therefore, we hypothesized that regulating the
glial-specific endogenous gene expression of gliomas by a
combination of small-molecule compounds could affect glial
reprogramming.
Materials and methods
Cell culture
In the present study, rat C6 and human U87MG glioma
cells (of unknown origin) (HTB-14™; The American Type Culture
Collection, Manassas, VA, USA) were used. The cells were cultured
in Dulbecco's modified Eagle's medium (DMEM; Life Technologies;
Thermo Fisher Scientific, Inc., Waltham, A, USA) containing 10%
fetal bovine serum (FBS; HyClone Laboratories; GE Healthcare,
Chicago, IL, USA), 0.1% β-mercaptoethanol (Life Technologies;
Thermo Fisher Scientific, Inc.), 1% penicillin/streptomycin (P/S)
(Life Technologies; Thermo Fisher Scientific, Inc.), 1%
non-essential amino acids (NEAA; Life Technologies; Thermo Fisher
Scientific, Inc.) and 1% sodium pyruvate (Life Technologies; Thermo
Fisher Scientific, Inc.), and incubated at 37°C in a humidified
atmosphere containing 5% CO2.
Differentiation and maturation of
glial cells
Glioma cells were seeded on 1% basement membrane
matrix-coated plates (BD Biosciences, San Jose, CA, USA) at a
density of 1×103 cells/cm2. After incubating
the cells at 37°C for 24 h, we replaced the medium with glial
differentiation medium, which consisted of neurobasal
medium:advanced DMEM/F12 (1X) (1:1), 1% N2 supplement (100X), 2%
B27 supplement (50X), 0.05% bovine serum albumin (BSA), 1% P/S, 1%
glutamax-I (100X), 0.1% β-mercaptoethanol (1000X) (all of which
were from Life Technologies; Thermo Fisher Scientific, Inc.), BDNF
(10 ng/ml), GDNF (10 ng/ml) and NT-3 (10 ng/ml) (all from
PeproTech, Inc., Rocky Hill, NJ, USA). We then added a combination
of small molecules, composed of either forskolin (Tocris
Bioscience, Bristol, UK) and rapamycin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) or forskolin, rapamycin and T3 (Sigma-Aldrich;
Merck KGaA). After 7 days, we replaced the medium with only
I-BET151 (Tocris Bioscience) for maturation of the differentiated
cells. The cells were then incubated at 37°C for 7 additional days.
We replaced the media every 2–3 days. In some experiments, we used
temozolomide (TMZ; 50 µM) to compare the growth inhibition effect
of the small-molecule combination.
The small molecules were dissolved in dimethyl
sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA) and diluted in glial
differentiation medium to the following final concentrations:
forskolin, 100 µM; rapamycin, 100 nM; T3, 100 nM; I-BET151, 1 µM;
and TMZ, 50 µM. We tested the glial cell differentiation into
glioma in three independent experiments.
MTT assay
For measuring cell proliferation, we used the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Sigma-Aldrich; Merck KGaA). Cells from each group,
containing the same volume of DMSO as that in the differentiation
media, were differentiated for 7 or 14 days in 24-well plates.
Then, we added 5 mg/ml MTT solution to the media and incubated them
at 37°C for 3 h. The media were removed, and formazan crystals were
dissolved using DMSO. The samples were then incubated at 37°C for
10 min and transferred to 96-well plates, and the absorbance was
measured at 490 nm using VersaMax ELISA microplate reader
(Molecular Devices, Sunnyvale, CA, USA). Results were acquired from
three independent experiments.
Immunofluorescence and cell
quantification
Media were removed and the cells were washed with
phosphate-buffered saline (PBS; HyClone Laboratories; GE
Healthcare). The cells were then fixed with 4% paraformaldehyde
(Millipore, Temecula, CA, USA), pH 7.2, for 10 min at room
temperature. Subsequently, they were washed thrice with 0.3%
Tween-20 (Life Technologies; Thermo Fisher Scientific, Inc.) in PBS
for 3 min. The blocking procedure was performed in PBS with 10%
normal donkey serum and 0.3% Triton-X 100 for 30 min at room
temperature. Primary antibodies, diluted in the blocking buffer,
namely, mouse monoclonal anti-Nestin (dilution 1:1,000; cat. no.
ab6142; Abcam, Cambridge, MA, USA), rabbit polyclonal anti-NG2
(dilution 1:250; cat. no. ab5320; Millipore), rabbit polyclonal
anti-Olig2 (dilution 1:100; cat. no. ab42453; Abcam), rabbit
polyclonal anti-PDGFRa (dilution 1:500; cat. no. sc-338; Santa Cruz
Biotechnology, Santa Cruz, CA, USA;) mouse monoclonal anti-MBP
(dilution 1:1,000; cat. no. ab2404; Abcam), mouse monoclonal
anti-CNP (dilution 1:1,000; cat. no. NE1020; Millipore), mouse
monoclonal anti-oligodendrocyte (RIP; dilution 1:50,000; cat. no.
MAB1580; Millipore), rabbit polyclonal anti-GFAP (dilution 1:1,000;
cat. no. ab7260; Abcam), mouse monoclonal anti-GFAP (dilution
1:1,000; cat. no. G3893; Sigma-Aldrich; Merck KGaA), mouse
monoclonal anti-O4 (dilution 1:100; cat. no. MAB345; Millipore) and
rabbit polyclonal anti-Ki-67 (dilution 1:250; cat. no. ab15580;
Abcam), were allowed to react with the samples for 60 min at room
temperature. After washing with PBS with 0.3% Tween-20, secondary
antibodies diluted in the blocking buffer were added and allowed to
react for 30 min at room temperature: FITC donkey anti-rabbit (cat.
no. 711-095-152) or anti-mouse (cat. no. 715-095-151) and Cy3
donkey anti-rabbit (cat. no. 711-165-152) or anti-mouse (cat. no.
715-165-151) (dilution 1:500; all of them are from Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA) were used
as secondary antibodies. The samples were then washed thrice and
stained using 4′,6′-diamino-2-phenylindole (DAPI; Vector
Laboratories, Burlingame, CA, USA). The samples were then covered
with glass coverslips and examined using a confocal laser scanning
microscope (LSM 700; Carl Zeiss, Oberkochen, Germany).
Flow cytometry
To determine the number of positive cells stained by
each antibody (GFAP, CNP, RIP and Ki-67), we performed flow
cytometric analyses. Briefly, the cells were harvested using 0.25%
trypsin/EDTA (Life Technologies; Thermo Fisher Scientific, Inc.).
For fixation, 4% paraformaldehyde was added to the cell pellet and
incubated for 10 min. After centrifugation at 400 × g for 5 min,
the supernatant was discarded and the cell pellet was washed twice
with 0.3% Tween-20 in PBS for 3 min. Next, the primary antibodies
(GFAP, 1:1,000; CNP, 1:1,000; RIP, 1:50,000; and Ki-67, 1:250) were
added to the pellet and allowed to react for 60 min at room
temperature. Following a second wash using the same procedure,
secondary antibodies (FITC donkey anti-rabbit or anti-mouse and Cy3
donkey anti-rabbit or anti-mouse; dilution 1:500) were added to the
cell pellet and allowed to react for 30 min at room temperature.
Finally, the pellet was washed and re-suspended in PBS. Samples
were analyzed using a FACSCalibur (Becton-Dickinson; BD
Biosciences, San Jose, CA, USA) and CellQuest software
(Becton-Dickinson; BD Biosciences). Results were acquired from
three independent experiments.
Microarray
Microarray analysis was performed according to the
manufacturer's instructions. After total RNA isolation, cDNA was
synthesized using the GeneChip WT (Whole Transcript) Amplification
kit (Thermo Fisher Scientific, Inc.). Labeled DNA target was
hybridized to the Affymetrix GeneChip Array (Affymetrix; Thermo
Fisher Scientific, Inc.). Hybridized arrays were washed and stained
on a GeneChip Fluidics Station 450 and scanned on a GCS3000 Scanner
(Thermo Fisher Scientific, Inc.). Analysis was performed using
Affymetrix® GeneChip Command Console®
Software (AGCC; Affymetrix; Thermo Fisher Scientific, Inc.).
Microarray data were deposited in a public database (https://www.ncbi.nlm.nih.gov/geo/), GSE101337
(For undifferentiated C6 and SMiG), and 101338 (For single clone
no. 4 and single clone no. 8).
Cell counting
To determine the proliferation rate of untreated
glioma cells, i.e., those without the small molecules, we seeded
them at a density of 1×103 cells/cm2 on a
24-well plate. When the cells reached confluence, they were
harvested using 0.25% trypsin/EDTA. After being centrifuged at 268
× g for 3 min and re-suspended in 1 ml culture medium, the cells
were diluted to half concentration in trypan blue (Life
Technologies; Thermo Fisher Scientific, Inc.) and counted using the
manual cell counting method. All the cells were then seeded on
12-well plates (Corning Inc., Corning, NY, USA). The cells were
counted when they reached confluence, and were re-plated on wider
plates, using the same method. Results were acquired from three
independent experiments.
Single colony selection
We performed a serial dilution of glioma cells for
isolation of a single colony. Briefly, we added 100 µl culture
media into all the wells of a 96-well plate. Approximately 100
cells were mixed with 100 µl culture medium and added into the
first well of a 96-well plate. Then, 100 µl was taken from the
first well, and then serially diluted to the next well, and so on.
After confirming the single cell isolation, cells were incubated
until a colony was formed. After 14 days, each colony was treated
with trypsin for 5 min, and maintained in the culture media.
We tested the glial differentiation efficiency with
a few isolated glioma. We used the clone no. 4 and 8 glioma for the
present study.
Statistical analysis
A two-tailed Student's t-test was performed to
assess differences between two groups. Two-way analysis of variance
(ANOVA) followed by Tukey's post hoc test was performed to assess
differences among three groups or more. Data are presented as mean
± standard error of the mean (SEM) and P-values <0.05 were
considered to indicate a statistically significant result.
Results
Direct conversion of malignant glioma
cells into non-proliferating glial cells by treatment with a
combination of small molecules
A schematic diagram was provided to facilitate
understanding of the present study (Fig. 1A). First, we confirmed that
undifferentiated glioma cells were positive for specific markers of
glial progenitor cells, namely, Nestin, NG2, Olig2 and PDGFRa
(Fig. 1B). However, the glioma
cells were negative for other markers of glial cells, namely, GFAP,
CNP, RIP and O4 (Fig. 1B).
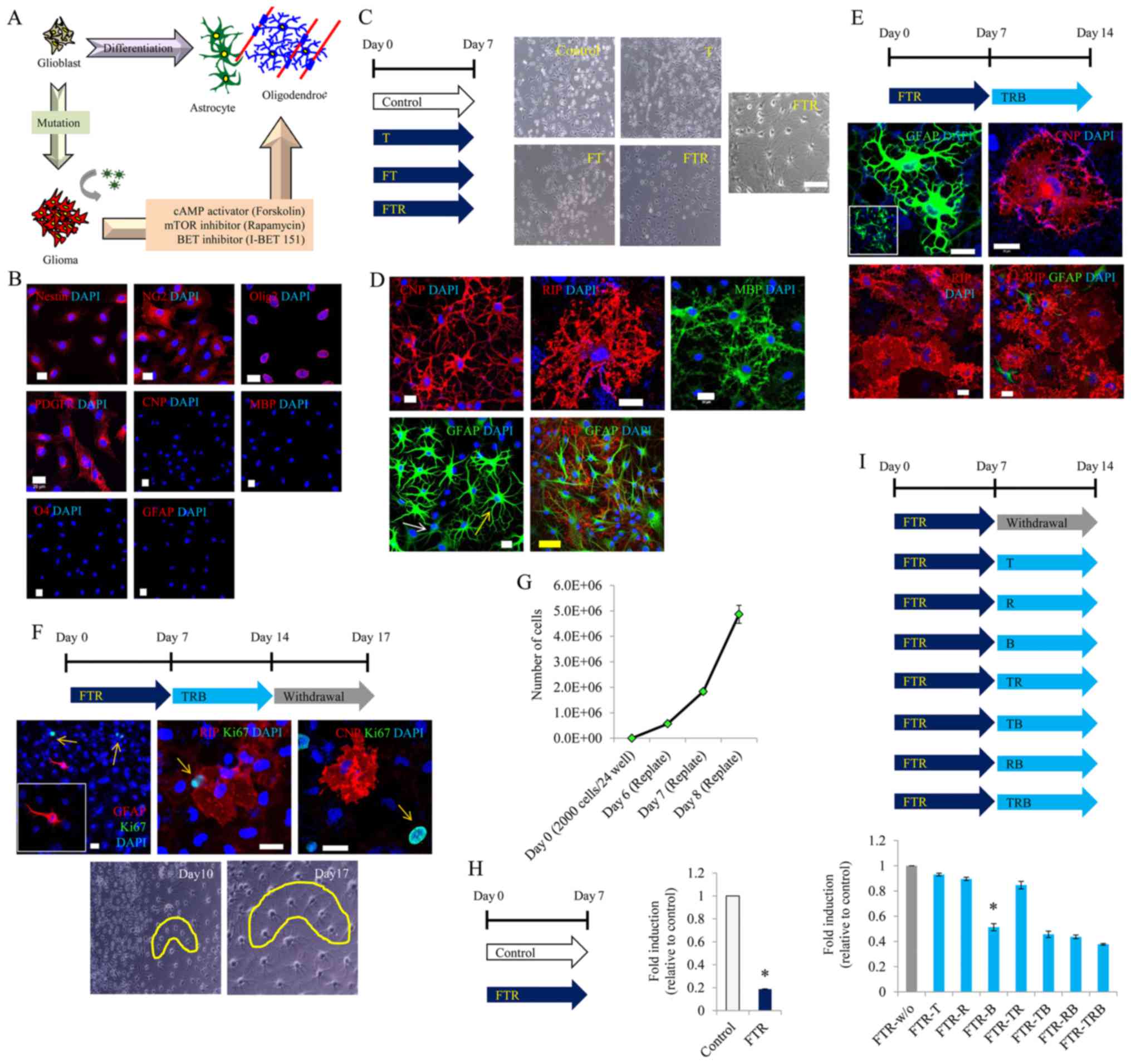 | Figure 1.Glial differentiation of malignant
glioma cells by treatment with small molecules. (A) Schematic
diagram. (B) Representative image of undifferentiated glioma cells
stained with Nestin, NG2, PDGFR, Olig2, GFAP, CNP, RIP and O4.
Scale bar, 20 µm. (C) Representative image 7 days after induction
of glial differentiation. T (T3), FT (forskolin/T3), FTR
(forskolin/T3/rapamycin). Original magnification, ×100; Scale bar,
100 µm. (D) Representative image of GFAP, CNP and RIP staining 7
days after induction of glial differentiation. Scale bars, 20 µm
(white) and 50 µm (yellow). (E) Representative image of GFAP, CNP
and RIP staining 14 days after induction of glial differentiation.
Scale bars, 20 µm. TRB, T3/rapamycin/I-BET151. (F) Representative
image of Ki-67, GFAP, CNP and RIP staining after 3 days of small
molecule withdrawal following 14 days of glial differentiation.
Scale bar, 20 µm. (G) Quantitative result of cell proliferation in
basal media. (H) Quantitative result of cell density 7 days after
induction of glial differentiation. (I) Quantitative result of cell
density 14 days after induction of glial differentiation.
*P<0.05. Data are presented as the mean ± SEM. |
Therefore, we aimed to ascertain whether T3, which
is typically used to differentiate glial progenitor cells into
oligodendrocytes, could convert gliomas into glial cells (i.e.,
oligodendrocytes). However, the T3-treated glioma cells were not
transformed into a glial-specific morphology (Fig. 1C). Next, we added T3 and forskolin
to the glioma cells, but could not confirm glial-specific
morphology until 7 days of incubation (Fig. 1C). Nevertheless, we persisted in our
efforts to convert C6 glioma cells into glial cells by adding
rapamycin to T3 and forskolin, and incubating the cells for 7 days.
With this, we were finally able to observe glial-specific
morphology (Fig. 1C), as well as
glial-specific markers such as GFAP, CNP, and RIP (Fig. 1D).
After several experiments, we confirmed that this
glial-specific morphology could be maintained using medium
containing rapamycin, T3 and I-BET151 (Fig. 1E). To further confirm whether the
converted cells were fully differentiated into glial cells, they
were cultured in the absence of small molecules for 3 days.
Consequently, we observed that the glial-specific morphology was
retained; the GFAP-, CNP- and RIP-positive cells did not react with
Ki-67 (Fig. 1F).
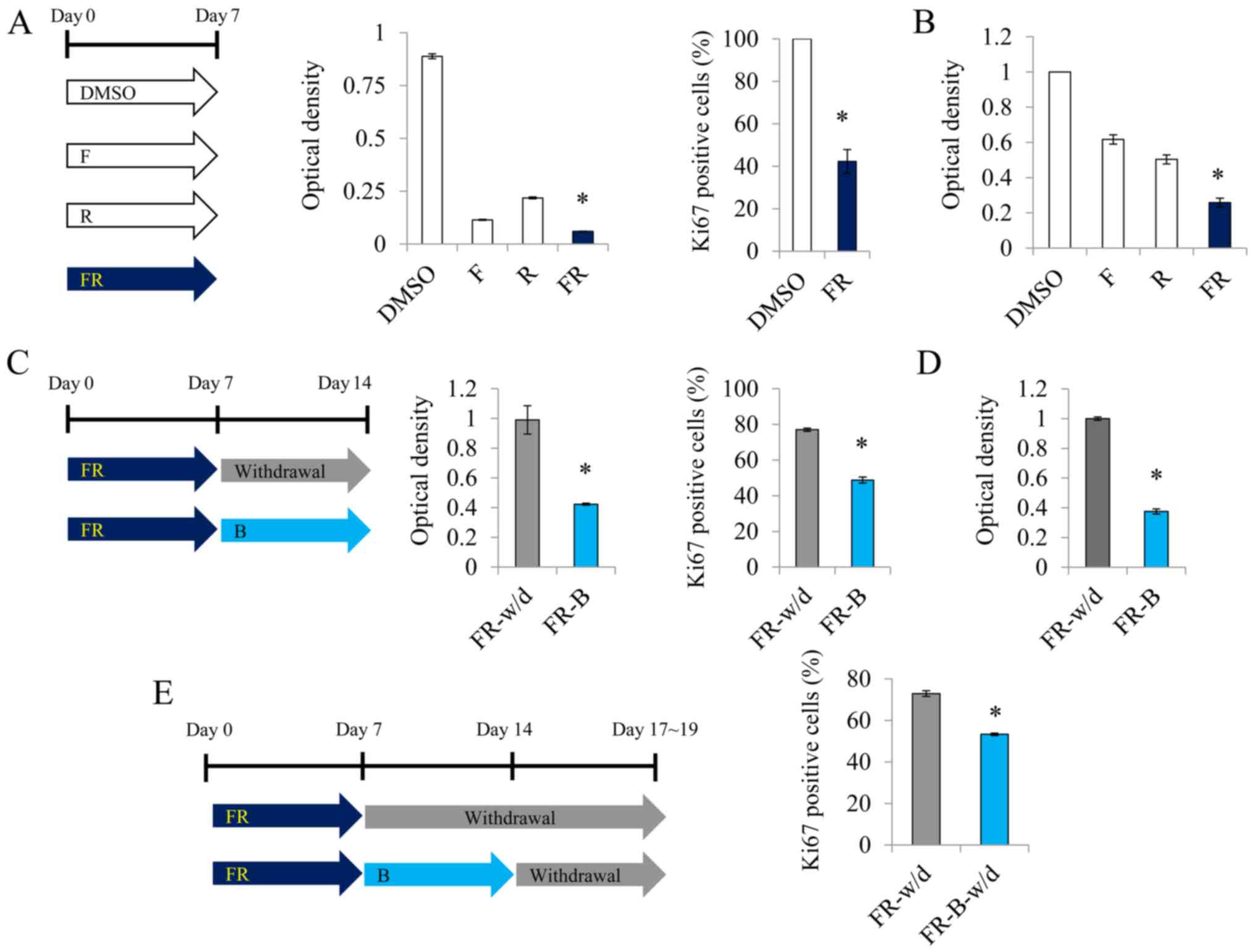
Next, we investigated the growth inhibitory effect
of the small molecules. The undifferentiated glioma cells grew
rapidly, despite a very low cell density (Fig. 1G). In contrast, treatment with a
combination of forskolin, T3 and rapamycin conspicuously decreased
the cell proliferation (Fig. 1H).
Further experimentation confirmed that I-BET151 played a major role
in the growth inhibition after glial induction (Fig. 1I).
Glial differentiation of malignant
glioma cells through optimal treatment with small molecules
To determine the major compound among forskolin, T3,
rapamycin and I-BET151, responsible for glial induction, we
performed further experiments. We confirmed that glioma cells were
converted into GFAP-, CNP-, RIP- and O4-positive cells with
glial-specific morphology, even in the absence of T3 (Fig. 2A and B). These converted cells did
not react with Ki-67 (marker for dividing cells) (Fig. 2B). Thus, T3 did not affect the glial
conversion of glioma cells.
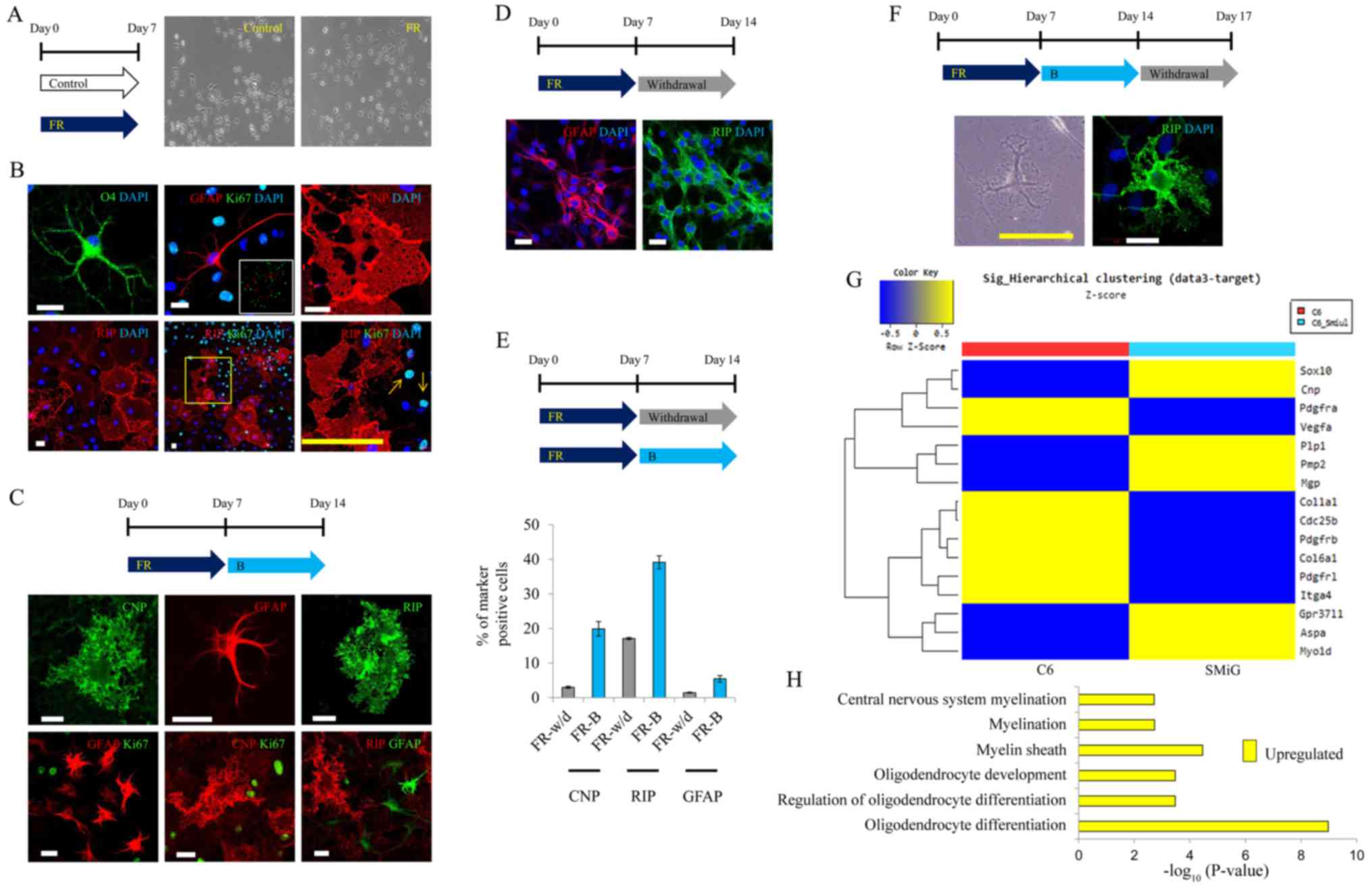 | Figure 2.Glial differentiation of malignant
glioma cells through optimal treatment with small molecules. (A)
Representative image 7 days after induction of glial
differentiation. (original magnification, ×100). FR,
forskolin/rapamycin). (B) Representative image of GFAP, CNP, RIP,
O4 and Ki-67 staining 7 days after induction of glial
differentiation. Scale bars, 20 µm (white) and 100 µm (yellow). B,
I-BET151. (C) Representative image of GFAP, CNP, RIP and Ki-67
staining 14 days after induction of glial differentiation. Scale
bars, 20 µm. (D) Representative image of GFAP and RIP staining
after 7 days of small molecule withdrawal following 7 days of glial
differentiation. Scale bars, 50 µm. (E) Quantitative result of the
glial-specific marker-positive cells after glial differentiation
for 14 days. (F) Representative image of RIP staining after three
days of small molecule withdrawal following 14 days of glial
differentiation. Scale bars, 20 µm (white) and 200 µm (yellow). (G)
Lists of upregulated or downregulated genes in
small-molecule-induced gliomas (SMiGs). (H) Upregulated Gene
Ontology terms related to glial differentiation in SMiGs. |
We further confirmed that glial-specific morphology
was maintained in the presence of I-BET151 after induction of glial
differentiation using forskolin and rapamycin, for 7 days (Fig. 2C). In contrast, it was difficult to
maintain glial-specific morphology in the absence of I-BET151
(Fig. 2D). As a quantitative
result, the number of CNP-, RIP- and GFAP-positive cells clearly
increased in the SMiGs converted using forskolin, rapamycin and
I-BET151 (Fig. 2E). We also
observed that glial-specific morphology was retained even when
small molecules were withdrawn for 3 days (Fig. 2F).
Gene expression profile in
undifferentiated glioma cells and SMiGs
We performed microarray analysis to compare the gene
expression pattern between glioma cells and SMiGs. In a microarray
analysis, gene expression profile related to oligodendrocyte
differentiation and myelination was significantly induced in the
SMiGs. In contrast, gene expression profiles relating to cell
division and mitosis, or extracellular matrix (ECM) and vessel
development were markedly upregulated in glioma cells (Fig. 2G). Gene Ontology related to glial
differentiation, including oligodendrocyte differentiation and
myelination, was significantly different between undifferentiated
glioma cells and SMiGs (Fig. 2H).
These results revealed that specific characteristics of malignant
gliomas can be converted into those of glial cells by treatment
with a combination of forskolin, rapamycin and I-BET151.
Proliferation of malignant glioma
cells is abrogated by a combination of small molecules
We investigated whether the strong proliferation
potency of malignant gliomas can be inhibited by glial conversion.
On day 7 after treatment with a combination of forskolin and
rapamycin, we confirmed that the proliferation of glioma cells was
significantly reduced compared to that of the single-molecule
treatment group, and the rate of Ki-67-positive cells was
significantly decreased compared to that in the DMSO group
(Fig. 3A). We also confirmed that
the proliferation of U87MG glioma cells was significantly reduced
compared to that of the single-molecule treatment group (Fig. 3B). Further treatment with I-BET151
was more effective in inhibiting cell proliferation (Fig. 3C and D). This pattern was similar
even in the absence of the small molecules (Fig. 3E). These results indicated that the
strong proliferation capacity of malignant glioma cell can be
controlled by glial conversion.
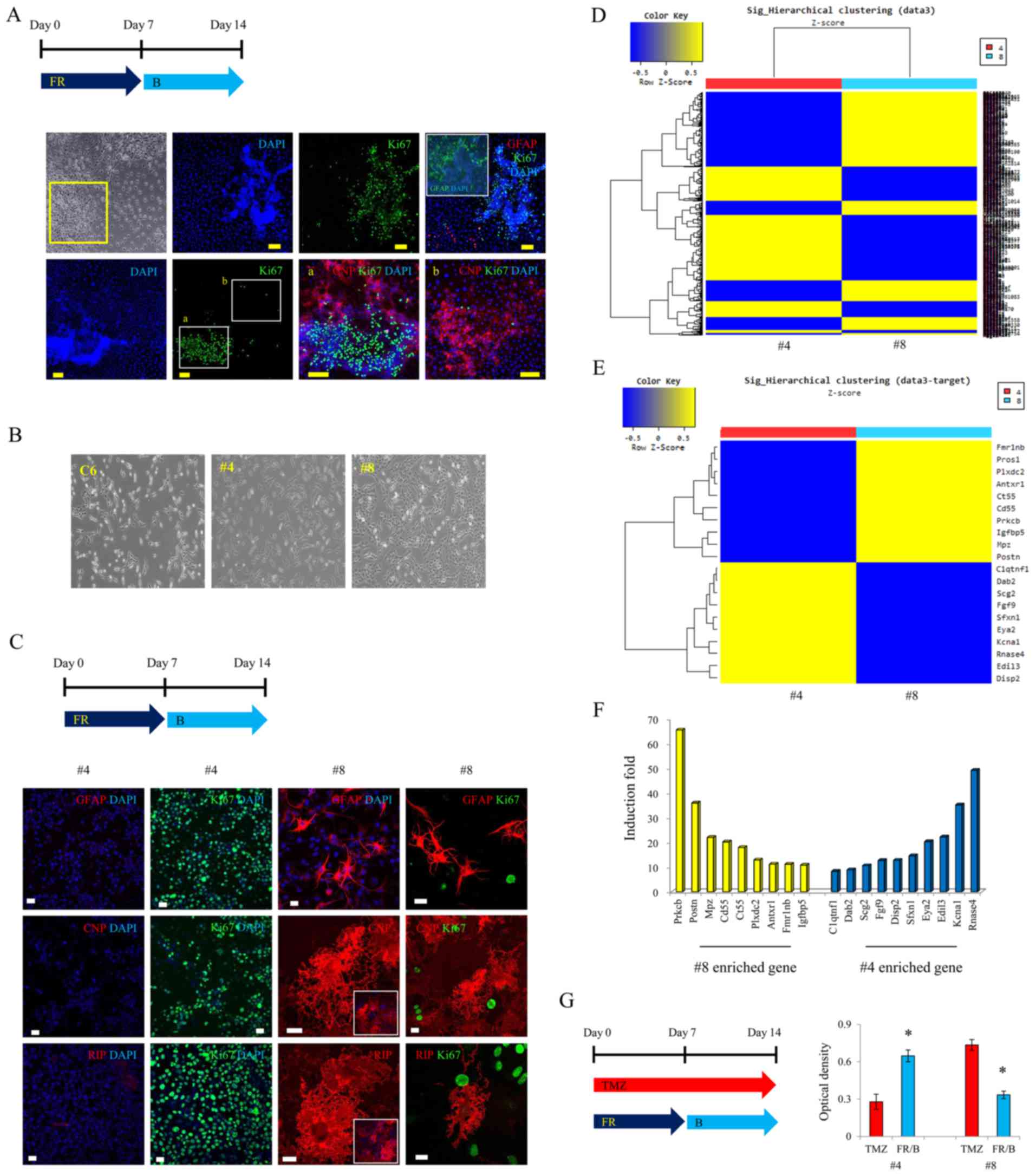
Specific cell type responds to the
small-molecule combination for glial conversion
Some cells were not positive for GFAP, CNP, or RIP
after glial induction by small molecules (Fig. 4A). We, therefore, selected each
single colony considering the glioma's heterogeneity (Fig. 4B). Of the many colonies we isolated,
colony no. 4 did not react with GFAP, CNP or RIP after glial
induction with forskolin, rapamycin and I-BET151; it was only
positive for Ki-67 (Fig. 4C).
However, most of colony no. 8 was positive for GFAP, CNP and RIP
after glial induction with forskolin, rapamycin and I-BET151
(Fig. 4C). GFAP-, CNP- and
RIP-positive cells did not react with Ki-67 (Fig. 4C). We, then, analyzed the gene
expression patterns between colony no. 4 and 8, using a microarray
analysis (Fig. 4D). The top 10
most-expressed genes in colony no. 8 were prkcb, postn, mpz,
pros1, cd55, ct55, plxdc2, antxr1, fmr1nb and igfbp5
(Fig. 4E and F). We then compared
the proliferation-inhibition effect of the small-molecule
combination with that of a standard medication such as temozolomide
(TMZ). For colony no. 4, cell proliferation was reduced by TMZ, but
not by the small-molecule combination (Fig. 4G), although, our small molecules
inhibited proliferation more effectively than TMZ in colony no. 8
(Fig. 4G).
Discussion
In our previous study, we demonstrated that
malignant gliomas can be reprogrammed into functional neurons,
using a combination treatment of forskolin and CHIR99021 (GSK3
inhibitor). It is very exciting to report that the fate of gliomas
can be changed diversely, into non-proliferating neurons or glial
cells, depending on the small molecule combinations chosen.
In this preliminary study, we demonstrated the
conversion of malignant glioma cells to glial cells, using
small-molecule compounds that have been widely used for glial
differentiation. Unexpectedly, the malignant glioma cells did not
acquire the glial-specific morphology in the presence of T3 and
forskolin, which are known to induce glial differentiation
(16). After various experiments,
we determined that the glioma cells can be transformed to
glial-specific morphology using only forskolin and rapamycin,
without T3. We assume that the characteristics and differentiation
mechanism of malignant gliomas are not the same as those of glial
precursor cells; however, gliomas are derived from glial precursor
cells, and a significant portion of gliomas still strongly react
with glial precursor cell-specific markers (14,15). A
previous study reported that mTOR inhibition prevents
oligodendrocyte differentiation (17). mTOR inhibition was shown to prevent
the conversion of glial precursor cells into gliomas (14). However, the role of mTOR inhibition
in glioma reprogramming to glial cells remains unknown. This also
indicates that the differentiation mechanism of malignant gliomas
may not be the same as those of glial precursor cells, and the
synergistic effect of cAMP activation and mTOR inhibition may play
a major role in the conversion of glioma into glial cells.
BET inhibitor was classified as an anticancer agent
after clinical trials in the United States and Europe (18,19).
In this study, a BET (bromodomain and extra-terminal motif)
inhibitor, I-BET151, was added to the media 7 days after the
induction of glial differentiation, using a combination of
forskolin and rapamycin. Treatment with I-BET151 not only helped to
maintain the glial-specific morphology, but also strongly inhibited
cell proliferation (Fig. 2C-F).
According to a previous study, the BET inhibitor accelerates the
differentiation of mouse primary glial progenitors into
oligodendrocytes (20). To the best
of our knowledge, this is the first study that reports the effect
of I-BET151 on the differentiation of a malignant glioma into glial
cells.
The heterogeneity of glioblastomas has been
previously reported to affect their drug response, with each single
clone responding differently (21).
In this study, we observed that some cell populations did not
convert to a glial-like morphology even after treatment with
forskolin, rapamycin and I-BET151 while most of the population
remained positive to Ki-67. Thus, we compared the gene expression
profiles in single clones such as no. 4 or 8. In the microarray
analysis, the malignancy- or metastasis-related genes were found to
be highly expressed in no. 8 clone (Fig. 4E and F) (22–29).
In addition, expression of pdgfrb and
sox2 was higher in no. 8 than in no. 4 (data not shown). The
expression and amplification of gene in glioma have been reported
(30). In brain tumors, sox2
expression was found to be positively correlated with the grade of
malignancy (31,32). pdgfrb expression has been
reported to be correlated with the metastatic behavior in cancer
(33); pdgfrb is
preferentially expressed in glioma stem cells and its activation
promotes glioma stem cell self-renewal (34).
The genes mpz and plp1 were found to
be highly overexpressed in no. 8. These genes are reported to be
closely related to myelin and oligodendroglioma (35,36).
Given that no. 8 clone showed a good response to our drug, this
indicates that the above genes may be specific markers for our
small-molecule combination. Future studies may need to investigate
whether the expression levels of the above genes are implicated in
glial induction through a combination treatment of our small
molecules.
Moreover, pdgfra, pdgfrb, pdgfrl, met, vegfa
and colla1 have been implicated in cancer invasion and
proliferation (37,38). Our results showed that the
expression of these genes was downregulated by treatment with the
specific small molecules (Fig. 2G),
thereby indicating that the glial differentiation may influence
cancer growth.
The existence of differentiation-resistant cells
suggests that this combination of small molecules is neither
sufficient nor robust to convert all the glioma cells. However, the
standard drug for glioblastoma, temozolomide (TMZ), is also not
effective for all heterogeneous glioblastoma; in analogy, our
combination of small molecules was found to be quite effective on
some types of glioma. It is noteworthy that our small-molecule
combination inhibited the proliferation of glioma cells more
efficiently than standard anticancer drugs such as TMZ. We
demonstrated that the combination treatment inhibited the
proliferation of glioma no. 8 more efficiently than TMZ, thereby
suggesting that this combination may be effective on TMZ-resistant
cells. Thus, a combined therapy, using both TMZ and our molecules,
can be applied to TMZ-resistant cells for effective treatment.
Thus, extensive research using patient-derived gliomas would be
required to confirm this hypothesis.
Our findings suggest that malignant gliomas, derived
from mutations in glial precursor cells, can be converted to
non-proliferating glial cells with glial-specific characteristics.
Moreover, the proliferation of malignant cells can be highly
suppressed by the combination treatment of a cAMP activator, mTOR
inhibitor and BET inhibitor. In future, we will continue to
investigate the genes that are specifically involved in glial
differentiation of glioma.
Acknowledgements
Not applicable.
Funding
The present study was also supported by the Basic
Science Research Program through the NRF, funded by the Ministry of
Education (no. 2015R1D1A1A02059821, 2016R1D1A1A02937027 and
2018R1D1A1B07044055).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JO and YK designed the study, performed the
experiments and wrote the paper. YH was involved in the conception
of the study, wrote, reviewed and edited the manuscript. DB
performed the data analysis, reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SMiGs
|
small-molecule-induced gliomas
|
|
TMZ
|
temozolomide
|
|
cAMP
|
cyclic adenosine monophosphate
|
|
mTOR
|
mammalian target of rapamycin
|
|
BET
|
bromodomain and extra-terminal
motif
|
References
|
1
|
Thier M, Wörsdörfer P, Lakes YB, Gorris R,
Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nöthen MM,
et al: Direct conversion of fibroblasts into stably expandable
neural stem cells. Cell Stem Cell. 10:473–479. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tian E, Sun G, Sun G, Chao J, Ye P, Warden
C, Riggs AD and Shi Y: Small-molecule-based lineage reprogramming
creates functional astrocytes. Cell Rep. 16:781–792. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang M, Lin YH, Sun YJ, Zhu S, Zheng J,
Liu K, Cao N, Li K, Huang Y and Ding S: Pharmacological
reprogramming of fibroblasts into neural stem cells by
signaling-directed transcriptional activation. Cell Stem Cell.
18:653–667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caiazzo M, Giannelli S, Valente P, Lignani
G, Carissimo A, Sessa A, Colasante G, Bartolomeo R, Massimino L,
Ferroni S, et al: Direct conversion of fibroblasts into functional
astrocytes by defined transcription factors. Stem Cell Reports.
4:pp. 25–36. 2015, View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan
W, Wang M, Yang W and Pei G: Generation of neural progenitor cells
by chemical cocktails and hypoxia. Cell Res. 25:645–646. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang
H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al: Direct conversion of
fibroblasts to neurons by reprogramming PTB-regulated microRNA
circuits. Cell. 152:82–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ladewig J, Mertens J, Kesavan J, Doerr J,
Poppe D, Glaue F, Herms S, Wernet P, Kögler G, Müller FJ, et al:
Small molecules enable highly efficient neuronal conversion of
human fibroblasts. Nat Methods. 9:575–578. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pang ZP, Yang N, Vierbuchen T, Ostermeier
A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC
and Wernig M: Induction of human neuronal cells by defined
transcription factors. Nature. 476:220–223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambasudhan R, Talantova M, Coleman R, Yuan
X, Zhu S, Lipton SA and Ding S: Direct reprogramming of adult human
fibroblasts to functional neurons under defined conditions. Cell
Stem Cell. 9:113–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su Z, Zang T, Liu ML, Wang LL, Niu W and
Zhang CL: Reprogramming the fate of human glioma cells to impede
brain tumor development. Cell Death Dis. 5:e14632014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guichet PO, Bieche I, Teigell M, Serguera
C, Rothhut B, Rigau V, Scamps F, Ripoll C, Vacher S, Taviaux S, et
al: Cell death and neuronal differentiation of glioblastoma
stem-like cells induced by neurogenic transcription factors. Glia.
61:225–239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu W, Qiu B, Guan W, Wang Q, Wang M, Li W,
Gao L, Shen L, Huang Y, Xie G, et al: Direct conversion of normal
and Alzheimer's disease human fibroblasts into neuronal cells by
small molecules. Cell Stem Cell. 17:204–212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Zuo X, Jing J, Ma Y, Wang J, Liu D,
Zhu J, Du X, Xiong L, Du Y, et al: Small-molecule-driven direct
reprogramming of mouse fibroblasts into functional neurons. Cell
Stem Cell. 17:195–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Galvao RP, Kasina A, McNeill RS, Harbin
JE, Foreman O, Verhaak RG, Nishiyama A, Miller CR and Zong H:
Transformation of quiescent adult oligodendrocyte precursor cells
into malignant glioma through a multistep reactivation process.
Proc Natl Acad Sci USA. 111:E4214–E4223. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hayes J, Thygesen H, Droop A, Hughes TA,
Westhead D, Lawler SE, Wurdak H and Short SC: Prognostic microRNAs
in high-grade glioma reveal a link to oligodendrocyte precursor
differentiation. Oncoscience. 2:252–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim JB, Lee H, Araúzo-Bravo MJ, Hwang K,
Nam D, Park MR, Zaehres H, Park KI and Lee SJ: Oct4-induced
oligodendrocyte progenitor cells enhance functional recovery in
spinal cord injury model. EMBO J. 34:2971–2983. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tyler WA, Gangoli N, Gokina P, Kim HA,
Covey M, Levison SW and Wood TL: Activation of the mammalian target
of rapamycin (mTOR) is essential for oligodendrocyte
differentiation. J Neurosci. 29:6367–6378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garnier JM, Sharp PP and Burns CJ: BET
bromodomain inhibitors: A patent review. Expert Opin Ther Pat.
24:185–199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi J and Vakoc CR: The mechanisms behind
the therapeutic activity of BET bromodomain inhibition. Mol Cell.
54:728–736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gacias M, Gerona-Navarro G, Plotnikov AN,
Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen
B, et al: Selective chemical modulation of gene transcription
favors oligodendrocyte lineage progression. Chem Biol. 21:841–854.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meyer M, Reimand J, Lan X, Head R, Zhu X,
Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, et al:
Single cell-derived clonal analysis of human glioblastoma links
functional and genomic heterogeneity. Proc Natl Acad Sci USA.
112:851–856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang Y, He W, Wei Y, Qu Z, Zeng J and Qin
C: Screening key genes and pathways in glioma based on gene set
enrichment analysis and meta-analysis. J Mol Neurosci. 50:324–332.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nanda A, Buckhaults P, Seaman S, Agrawal
N, Boutin P, Shankara S, Nacht M, Teicher B, Stampfl J, Singh S, et
al: Identification of a binding partner for the endothelial cell
surface proteins TEM7 and TEM7R. Cancer Res. 64:8507–8511. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Davies G, Rmali KA, Watkins G, Mansel RE,
Mason MD and Jiang WG: Elevated levels of tumour endothelial
marker-8 in human breast cancer and its clinical significance. Int
J Oncol. 29:1311–1317. 2006.PubMed/NCBI
|
|
25
|
Song MH, Kim YR, Lee JW, Lee CH and Lee
SY: Cancer/testis antigen NY-SAR-35 enhances cell proliferation,
migration, and invasion. Int J Oncol. 48:569–576. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tomiyoshi G, Nakanishi A, Takenaka K,
Yoshida K and Miki Y: Novel BRCA2-interacting protein BJ-HCC-20A
inhibits the induction of apoptosis in response to DNA damage.
Cancer Sci. 99:747–754. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ikeda J, Morii E, Liu Y, Qiu Y, Nakamichi
N, Jokoji R, Miyoshi Y, Noguchi S and Aozasa K: Prognostic
significance of CD55 expression in breast cancer. Clin Cancer Res.
14:4780–4786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park SY, Piao Y, Jeong KJ, Dong J and de
Groot JF: Periostin (POSTN) regulates tumor resistance to
antiangiogenic therapy in glioma models. Mol Cancer Ther.
15:2187–2197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Che Mat MF, Abdul Murad NA, Ibrahim K,
Mohd Mokhtar N, Wan Ngah WZ, Harun R and Jamal R: Silencing of
PROS1 induces apoptosis and inhibits migration and invasion of
glioblastoma multiforme cells. Int J Oncol. 49:2359–2366. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Annovazzi L, Mellai M, Caldera V, Valente
G and Schiffer D: SOX2 expression and amplification in gliomas and
glioma cell lines. Cancer Genomics Proteomics. 8:139–147.
2011.PubMed/NCBI
|
|
31
|
Schmitz M, Temme A, Senner V, Ebner R,
Schwind S, Stevanovic S, Wehner R, Schackert G, Schackert HK,
Fussel M, et al: Identification of SOX2 as a novel
glioma-associated antigen and potential target for T cell-based
immunotherapy. Br J Cancer. 96:1293–1301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phi JH, Park SH, Kim SK, Paek SH, Kim JH,
Lee YJ, Cho BK, Park CK, Lee DH and Wang KC: Sox2 expression in
brain tumors: A reflection of the neuroglial differentiation
pathway. Am J Surg Pathol. 32:103–112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wehler TC, Frerichs K, Graf C, Drescher D,
Schimanski K, Biesterfeld S, Berger MR, Kanzler S, Junginger T,
Galle PR, et al: PDGFRalpha/beta expression correlates with the
metastatic behavior of human colorectal cancer: A possible
rationale for a molecular targeting strategy. Oncol Rep.
19:697–704. 2008.PubMed/NCBI
|
|
34
|
Heldin CH: Targeting the PDGF signaling
pathway in tumor treatment. Cell Commun Signal. 11:972013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Golfinos JG, Norman SA, Coons SW, Norman
RA, Ballecer C and Scheck AC: Expression of the genes encoding
myelin basic protein and proteolipid protein in human malignant
gliomas. Clin Cancer Res. 3:799–804. 1997.PubMed/NCBI
|
|
36
|
Popko B, Pearl DK, Walker DM, Comas TC,
Baerwald KD, Burger PC, Scheithauer BW and Yates AJ: Molecular
markers that identify human astrocytomas and oligodendrogliomas. J
Neuropathol Exp Neurol. 61:329–338. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakada M, Kita D, Watanabe T, Hayashi Y,
Teng L, Pyko IV and Hamada J: Aberrant signaling pathways in
glioma. Cancers. 3:3242–3278. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Motegi H, Kamoshima Y, Terasaka S,
Kobayashi H and Houkin K: Type 1 collagen as a potential niche
component for CD133-positive glioblastoma cells. Neuropathology.
34:378–385. 2014.PubMed/NCBI
|