Introduction
Colorectal cancer (CRC) is the third most frequent
malignancy worldwide and the fourth most common cause of
cancer-associated mortalities (1).
Globally, ~1.36 million people are diagnosed with CRC each year and
approximately one-half will succumb to this disease (1,2). A
number of genes that are mutated in the multistep process of
colorectal carcinogenesis and progression have been demonstrated to
influence the prognosis of patients with CRC and their response to
treatment (3–6). In addition to these somatic genetic
mutations, epigenetic alterations and particularly the aberrant
hypermethylation of gene promoter regions leading to
transcriptional silencing are suggested to be important in CRC
tumorigenesis (7). This mechanism
is responsible for the functional inactivation of numerous tumor
suppressor genes in CRC (7,8), including human MutL homolog 1,
tissue inhibitor of metalloproteinase-3, p14, death-associated
protein kinase, adenomatous polyposis coli, O-6-methylguanine-DNA
methyltransferase and p16INK4 [(p16) or
cyclin-dependent kinase (CDK) inhibitor 2a] (9–11).
In normal cells, p16 and retinoblastoma (Rb)
proteins serve an important role in regulating the cell cycle
pathway (12,13). Rb is phosphorylated by the cyclin
D1-CDK4/6 complex, resulting in its dissociation from transcription
E2 factor (E2F) (13). The
subsequent transcriptional activation of E2F leads to progression
of the cell cycle from G1 to S phase (14). As p16 interferes with cell cycle
progression by inactivating CDK4/6, decreased expression or
inactivation of p16 attenuates the ability of Rb to inhibit cell
proliferation (15). The p16-Rb
pathway is suppressed in a number of cancer types via genetic or
epigenetic alterations in Rb and/or p16, through overexpression of
the cyclin D1/CDK4 complex, in addition to a number of other
mechanisms (12,13,15,16).
In particular, deletion or mutation of the p16 gene is
frequently observed in cancer of the biliary tract, lung, pancreas
and esophagus and in brain tumors (17–21).
Deletion of p16 has been associated with a late clinical
stage in esophageal cancer, and with lymphatic invasion and distant
metastasis in pancreatic cancer (22,23).
In gall bladder and lung cancer, p16 deletions and mutations
are associated with poor prognosis (24,25).
Decreased p16 expression due to hypermethylation of
the p16 promoter was detected in 32–55% CRC cases (26–30).
Although p16 mRNA expression is inversely correlated with tumor
size and lymph node metastasis (31), its influence on the prognosis of
patients with CRC remains unclear (32). A previous Japanese study identified
that p16 promoter hypermethylation in the primary tumors of
patients with CRC was associated with a shorter survival (33). This result was supported by a
subsequent meta-analysis (34). In
the present study, it was investigated whether p16 mRNA expression
was associated with methylation of the p16 gene promoter and
with patient prognosis in CRC.
Materials and methods
Patients with CRC and tissues
The present study included 101 patients with primary
CRC who underwent surgery at the Kanazawa University Hospital
(Kanazawa, Japan) between April 1999 and December 2002. Eligible
patients were aged 20 years or older and had histologically proven
adenocarcinoma of the colon and rectum. Exclusion criteria included
absolute contraindications to general anesthesia and/or surgery.
Their clinicopathological characteristics are presented in Table I. The survival status was determined
for all patients and the median follow-up period was 54.5 months.
In total, 54 patients (53.5%) received postoperative 5-fluorouracil
(5-FU)-based adjuvant chemotherapy.
 | Table I.Association between p16
methylation status or p16 mRNA expression and clinicopathological
features in colorectal cancer. |
Table I.
Association between p16
methylation status or p16 mRNA expression and clinicopathological
features in colorectal cancer.
| Clincopathological
features | n | p16
methylation | P-value | p16 mRNA | P-value |
|---|
| Sex |
|
Male | 57 | 0.455
(0–1.462) | 0.383 | 2.020
(0.960–5.250) | 0.584 |
|
Female | 44 | 0.060
(0–2.105) |
| 1.950
(0.85–3.548) |
|
| Age, years |
|
≥65 | 56 | 0.271
(0–1.955) | 0.900 | 2.040
(1.023–4.558) | 0.494 |
|
<65 | 45 | 0.398
(0–1.220) |
| 1.860
(0.700–4.460) |
|
| Tumor site |
|
Proximal | 29 | 0.455
(0–1.917) | 0.259 | 2.220
(0.780–3.630) | 0.588 |
|
Distal | 48 | 0.127
(0–0.863) |
| 1.825
(1.033–4.520) |
|
| Not
known | 24 | 1.068
(0.055–4.814) |
| 2.585
(0.925–9.550) |
|
| Tumor
histology |
|
Well | 34 | 0.161
(0–1.117) | 0.018 | 2.600
(1.240–4.360) | 0.066 |
|
Moderately | 34 | 0.082
(0–0.575) |
| 1.775
(0.857–4.380) |
|
|
Poorly | 4 | 0.162
(0.551–12.930) |
| 0.335
(0.270–0.760) |
|
|
Mucinous | 4 | 38.990
(15.750–80.514) |
| 1.940
(1.137–2.557) |
|
| Not
known | 25 | 1.028
(0.006–4.002) |
| 3.070
(0.960–9.050) |
|
| T stage |
| T2 | 3 | 0.000
(0–24.523) | 0.438 | 1.710
(1.025–3.085) | 0.929 |
| T3 | 56 | 0.150
(0–0.911) |
| 2.085
(0.932–4.187) |
|
| T4 | 18 | 0.438
(0–1.008) |
| 1.750
(1.045–4.137) |
|
| Not
known | 24 | 1.068
(0.055–4.814) |
| 2.585
(0.925–9.550) |
|
| Stage |
| 2 | 36 | 0.012
(0–0.658) | 0.021 | 1.970
(0.933–4.968) | 0.435 |
| 3 | 41 | 0.455
(0–3.154) |
| 1.860
(0.970–3.380) |
|
| 4 | 0 |
|
|
|
|
| Not
known | 24 | 1.068
(0.055–4.814) |
| 2.585
(0.925–9.550) |
|
| Adjuvant
chemotherapy |
|
Yes | 54 | 0.494
(0–2.585) | 0.169 | 1.915
(0.850–4.623) | 0.833 |
| No | 44 | 0.192
(0–1.000) |
| 1.970
(0.998–4.473) |
|
| Not
known | 3 | 0 (0–0.804) |
| 4.330
(2.845–4.640) |
|
Tumor tissue samples collected from the fresh
surgical specimen were cryopreserved in liquid nitrogen and stored
at −80°C for extraction of DNA and RNA. The remaining surgical
specimen was fixed with 10% neutral-buffered formalin for 1–2 days
at room temperature and embedded in paraffin for histopathological
examination. The tumor stage was determined according to the Union
for International Cancer Control tumor, node and metastasis (TNM)
classification (35). Genomic DNA
and total RNA were extracted from the same tumor tissues using the
QIAmp DNA Mini kit and the RNeasy Mini kit (both from Qiagen GmbH,
Hilden, Germany), respectively, according to the manufacturer's
protocols.
The present study was performed in accordance with
the Declaration of Helsinki. The design and protocol for the
present study were approved by the Kanazawa University Human Genome
and Gene Analysis Research Ethics Committee, and written informed
consent was obtained from the majority of the patients.
Quantification of p16 methylation by
the MethyLight assay
Bisulfite conversion of genomic DNA was performed as
previously described (36). DNA was
denatured using 0.2 M NaOH and subsequently incubated with
bisulfite for 16 h at 50°C. The bisulfite-converted DNA was
purified using the Wizard DNA purification kit (Promega
Corporation, Madison, WI, USA) and precipitated with ethanol. The
DNA sample was resuspended in water and stored at −30°C.
MethyLight, a fluorescence-based real-time PCR assay
was used to measure the level of p16 promoter methylation as
previously described (37). The
sense and antisense primers used for amplifying the
bisulfite-converted p16 promoter were:
5′-TGGAATTTTCGGTTGATTGGTT-3′ and 5′-AACAACGTCCGCACCTCCT-3′,
respectively (37). These primers
were used with the probe 5′-6FAM-ACCCGACCCCGAACGCG-TAMRA-3′ to
measure CpG methylation of the p16 promoter region by
real-time PCR (38). The
specificity for amplification of methylated DNA was confirmed
separately using human sperm DNA (unmethylated) and SssI (New
England BioLabs, Inc., Ipswich, MA, USA)-treated sperm DNA (fully
methylated) in the assay. Actin was amplified as a control for the
total amount of DNA using the sense primer,
5′-TGGTGATGGAGGAGGTTTAGTAAGT-3′ and antisense primer,
5′-AACCAATAAAACCTACTCCTCCCTTAA-3′; and probe,
5′-6FAM-ACCACCACCCAACACACAATAACAAACACA-TAMRA-3′ (38). The percentage of fully methylated
fraction [percentage of methylated reference (PMR)] at a specific
gene locus was calculated by dividing the gene:actin ratio of the
sample DNA by the gene:actin ratio of the SssI-treated sperm DNA
and multiplying by 100 (38). PMR
values obtained using MethyLight were classified into high (PMR ≥4)
and low (PMR <4) methylation categories, according to previous
studies (9,39,40).
Analysis of p16 mRNA expression
Reverse transcription (RT)-PCR was used to measure
p16 mRNA expression, as previously described (41). The relative expression of mRNA was
quantified using the 2−ΔΔCq method (42). The expression of actin was measured
as an internal standard and the level of p16 mRNA expression in
each tumor sample was normalized to the expression of actin. The
cut-off value for high and low levels of p16 mRNA expression was
defined as the median expression level for all tumor samples.
Cut-off values for p16 PMR and mRNA expression were used to
compare p16 promoter methylation and mRNA expression in the
same tumors.
Proliferating cell nuclear antigen (PCNA) mRNA
expression levels were additionally measured, using actin
expression level as the internal standard. p16 mRNA expression
relative to PCNA (p16/PCNA) was calculated as the p16 mRNA:actin
ratio divided by the PCNA mRNA:actin ratio in the same cDNA sample.
The cut-off value for p16/PCNA expression was defined as the median
p16/PCNA level for all tumor samples. This was used to investigate
the influence of p16 mRNA expression on the survival of patients
with CRC.
Statistical analysis
For statistical comparison of tumor p16
methylation and mRNA expression with clinicopathological factors
and tumor stage, the Fisher's exact test was used to study
non-continuous variables, and the Mann-Whitney U test and
Kruskal-Wallis test were used to study continuous variables. The
Mann-Whitney U test was used to compare p16 methylation with
p16 mRNA expression. The survival of patients was evaluated using
Kaplan-Meier analysis. Univariate and multivariate analyses of
survival were conducted using Cox proportional hazards regression.
All statistical analyses were performed with EZR (version 1.29,
Jichi Medical University, Shimotsuke, Japan), which is a graphical
user interface for R (The R Foundation for Statistical Computing,
Vienna, Austria). More precisely, it is a modified version of R
commander designed to add statistical functions frequently used in
biostatistics (43).
Results
p16 promoter methylation and p16 mRNA
expression in CRC
The levels of p16 gene promoter methylation
and p16 mRNA expression in the tumor samples of all patients with
CRC are presented in Table I in
association with clinicopathological features. p16
methylation (PMR >0) was detected in 67 cases (66.3%) and the
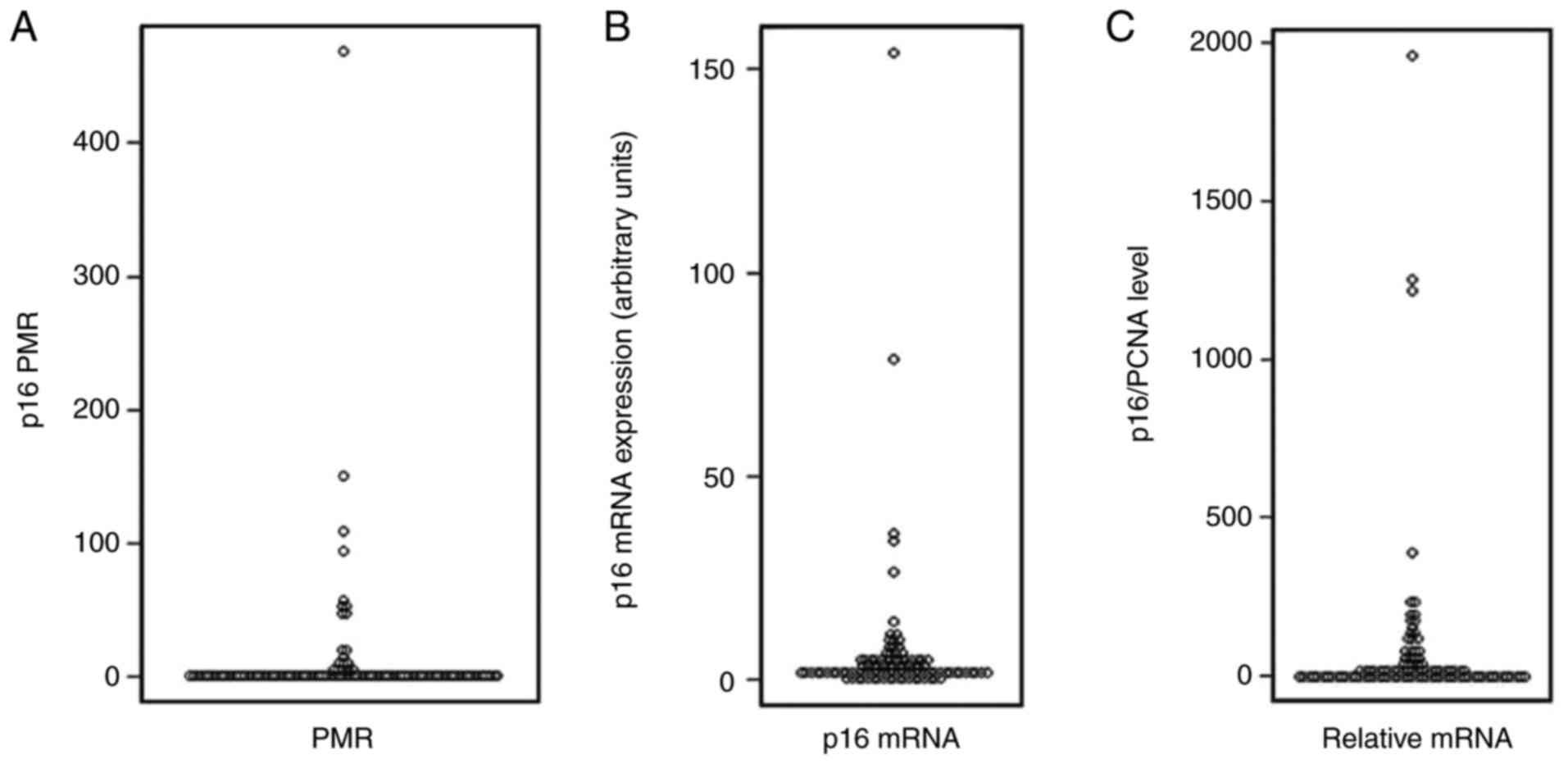
median PMR value was 0.34 (range, 0.00–468.6; Fig. 1A). A PMR cut-off value of ≥4 was
used to define high methylation and PMR <4 for low methylation,
according to previous studies (9,38–40).
Using this definition, the high methylation group comprised 18
(17.8%) of the 101 patients (Table
II), in agreement with previous studies (9,33). The
range of relative p16 mRNA expression levels was between 0 and
154.0, with a median expression level of 1.98 (Fig. 1B). As p16 mRNA expression is likely
to be associated with cell cycle (44), the p16 mRNA expression level was
normalized to that of PCNA expression in the same tumor and
designated as p16/PCNA expression. The relative values of p16/PCNA
ranged between 0 and 1,957.6, with a median value of 11.46
(Fig. 1C). To evaluate the
influence of p16 mRNA expression on survival outcome, patients were
divided into two groups (high and low) according to the median
value for p16 mRNA or p16/PCNA expression.
 | Table II.Association between p16
methylation status or p16/PCNA expression and clinicopathological
features in colorectal cancer. |
Table II.
Association between p16
methylation status or p16/PCNA expression and clinicopathological
features in colorectal cancer.
|
|
| p16
methylation | p16/PCNA mRNA |
|---|
|
|
|
|
|
|---|
| Features | n | High, n=18 | Low, n=83 | P-value | High, n=50 | Low, n=51 | P-value |
|---|
| Sex |
|
Male | 57 | 9 | 48 | 0.605 | 29 | 28 | 0.842 |
|
Female | 44 | 9 | 35 |
| 21 | 23 |
|
| Age, years |
|
≥65 | 56 | 11 | 45 | 0.794 | 29 | 27 | 0.690 |
|
<65 | 45 | 7 | 38 |
| 21 | 24 |
|
| Site |
|
Proximal | 29 | 7 | 22 | 0.090 | 12 | 17 | 0.814 |
|
Distal | 48 | 4 | 44 |
| 22 | 26 |
|
| Not
known | 24 | 7 | 17 |
| 16 | 8 |
|
| Tumor
histology |
|
Well | 34 | 3 | 31 | 0.017 | 18 | 16 | 0.210 |
|
Moderately | 34 | 4 | 30 |
| 13 | 21 |
|
|
Poorly | 4 | 1 | 3 |
| 0 | 4 |
|
|
Mucinous | 4 | 3 | 1 |
| 2 | 2 |
|
| Not
known | 25 | 7 | 18 |
| 17 | 8 |
|
| T stage |
| T2 | 3 | 1 | 2 | 0.353 | 0 | 3 | 0.344 |
| T3 | 56 | 7 | 49 |
| 25 | 31 |
|
| T4 | 18 | 3 | 15 |
| 9 | 9 |
|
| Not
known | 24 | 7 | 17 |
| 16 | 8 |
|
| Stage |
| 2 | 36 | 1 | 35 | 0.008 | 16 | 20 | 1.000 |
| 3 | 41 | 10 | 31 |
| 18 | 23 |
|
| 4 | 0 | 0 | 0 |
| 0 | 0 |
|
| Not
known | 24 | 7 | 17 |
| 16 | 8 |
|
| Adjuvant
chemotherapy |
|
Yes | 54 | 11 | 43 | 0.611 | 29 | 25 | 0.543 |
| No | 44 | 7 | 37 |
| 20 | 24 |
|
| Not
known | 3 | 0 | 3 |
| 1 | 2 |
|
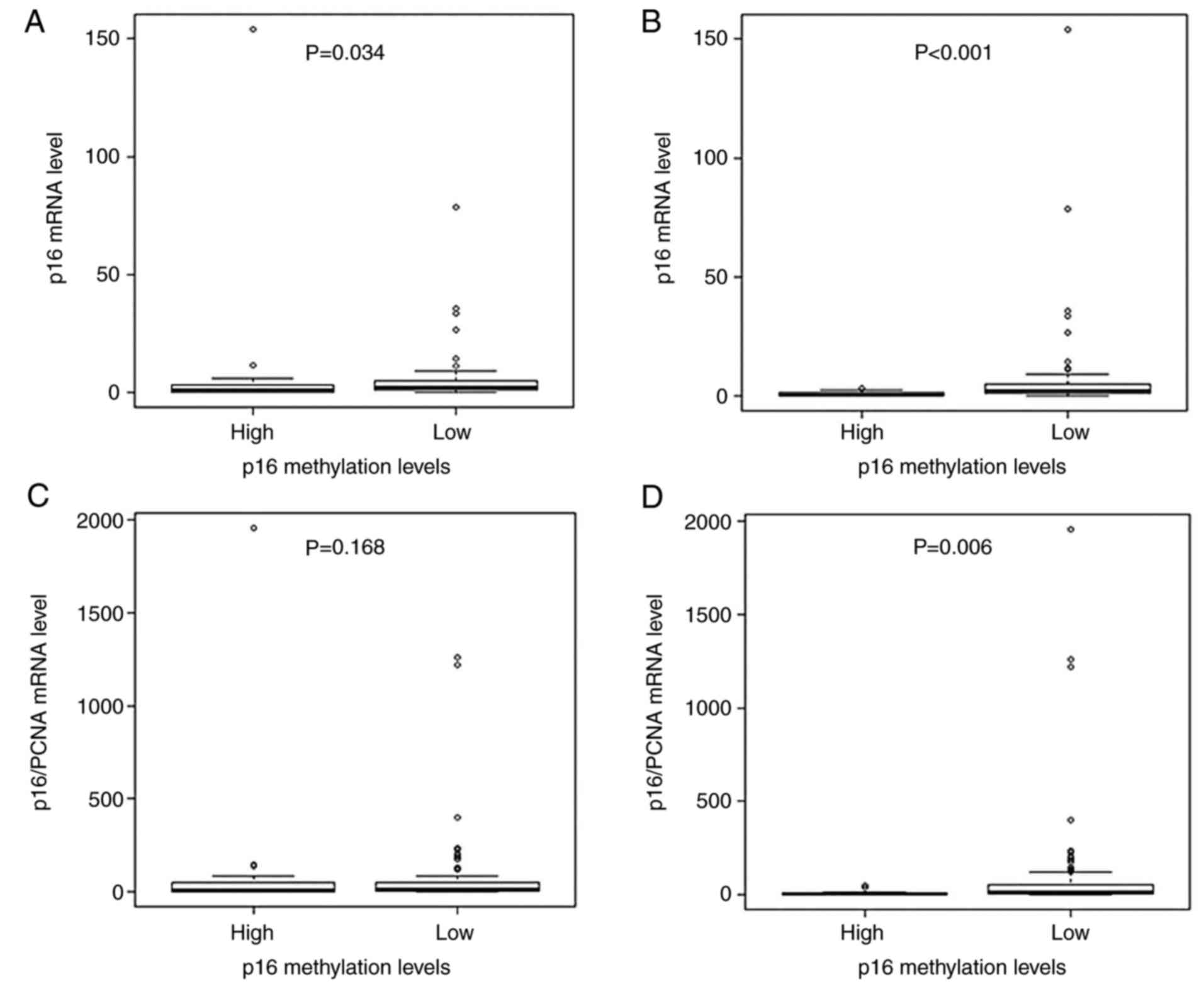
Comparison of p16 mRNA expression between the
p16 high and low methylation groups identified a significant
inverse association (P=0.034; Fig.
2A). When a higher PMR cut-off value of 10 was used rather than
4, a strong inverse association with p16 mRNA expression was
observed between the high (n=12; 11.9%) and low (n=89; 87.1%)
p16 methylation groups (P<0.001; Fig. 2B). Subsequently, p16/PCNA expression
was compared between the p16 high and low methylation
groups. Using a PMR value of 4 to classify p16 methylation,
no significant association was observed between p16
methylation and p16/PCNA expression (P=0.168; Fig. 2C). However, when classified
according to the higher PMR cut-off value of 10, a significant
inverse association was observed between p16 methylation and
p16/PCNA expression (P=0.006; Fig.
2D).
Tumor p16 methylation and prognosis of
patients with CRC
Comparison of tumor p16 methylation levels
with clinical and histopathologic characteristics of patients with
CRC is presented in Table II. High
p16 methylation (PMR >4) level was more frequently
detected in mucinous (P=0.017) compared with the other histological
types and was significantly associated with later clinical stage
(P=0.008); however, not with the sex or age of the patient, tumor
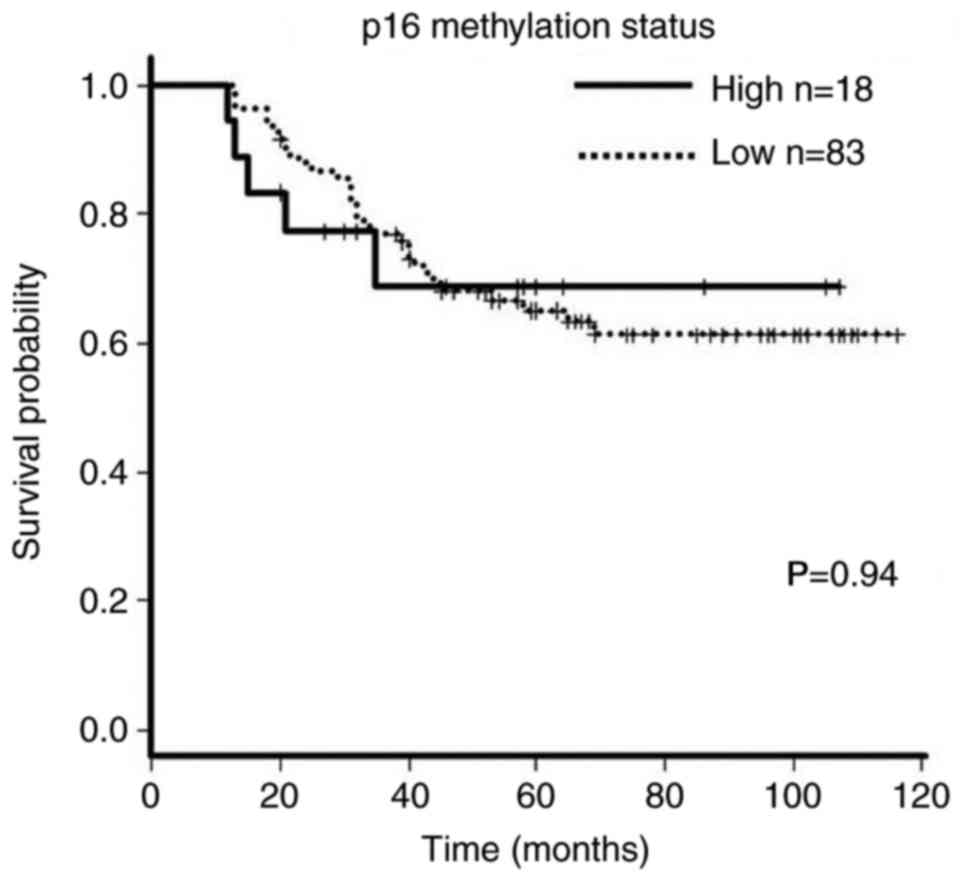
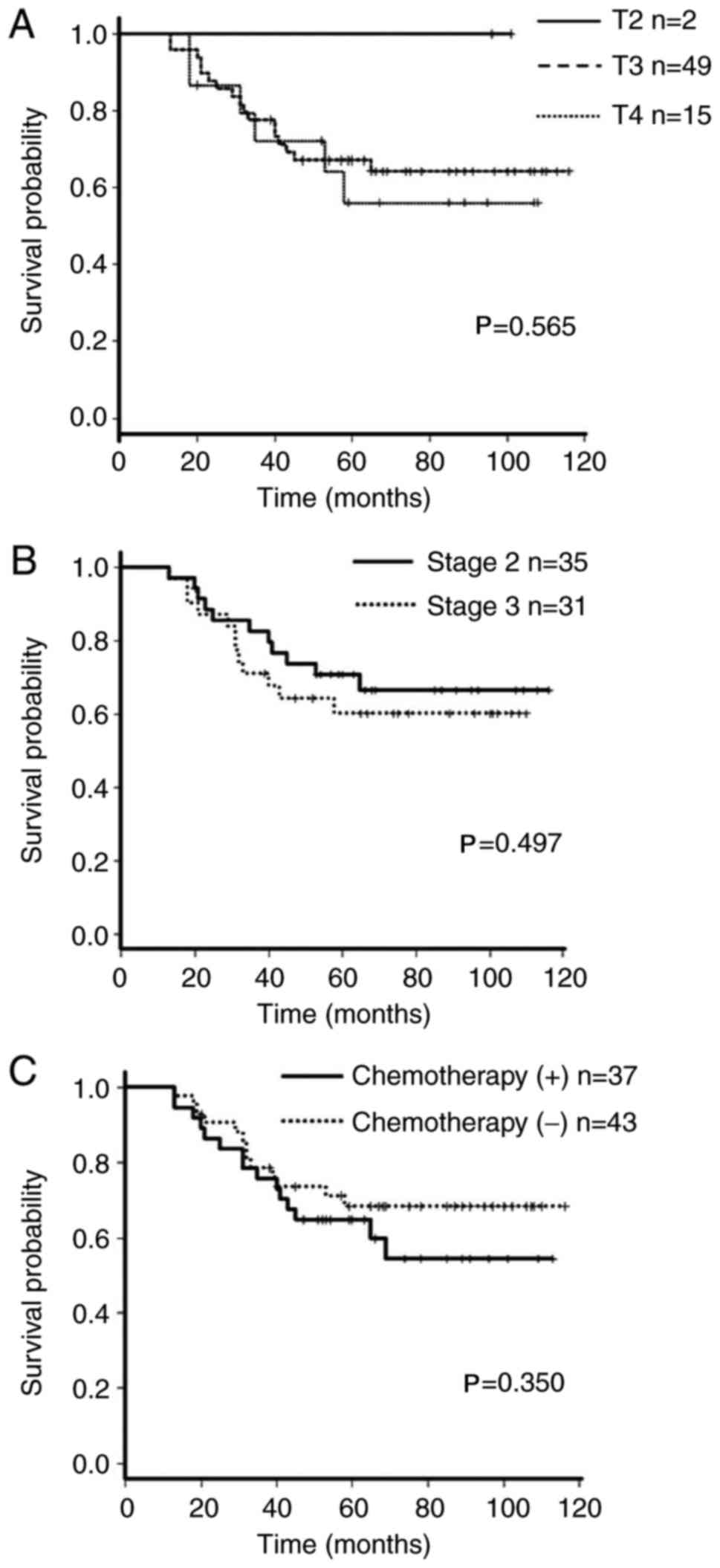
site, T stage or adjuvant chemotherapy. There was no significant
difference in prognosis between the high and low p16
methylation groups (P=0.94; Fig.
3).
Tumor p16 mRNA expression and
prognosis of patients with CRC
No significant differences in p16 mRNA expression
levels (high or low) were observed according to sex or age of the
patient, or with tumor site, histology, T stage, or adjuvant
chemotherapy (Table III). The
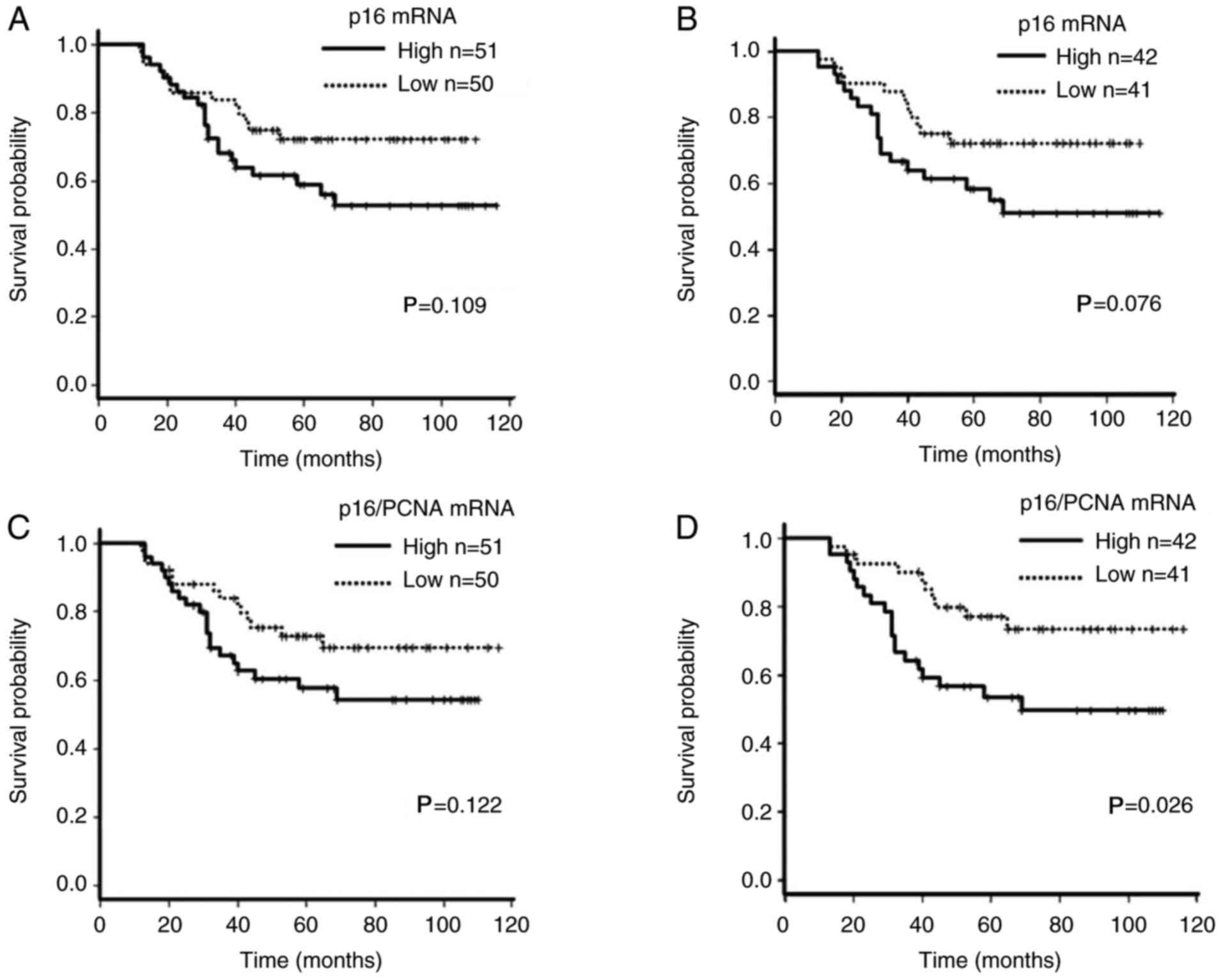
survival of patients with CRC with high p16 mRNA expression was
worse compared with patients with low expression; however, this did
not reach statistical significance (P=0.109; Fig. 4A). The majority (83/101; 82%) of
patients with CRC demonstrated low levels (PMR <4) of p16
methylation (Table II; Fig. 2A). When these patients were divided
into high and low p16 mRNA expression groups defined by a median
level of value (2.15), the high expression group (n=42)
demonstrated a worse outcome compared with the low expression group
(n=41; P=0.076; Fig. 4B).
 | Table III.Association between p16 mRNA
expression and clinicopathological features in colorectal
cancer. |
Table III.
Association between p16 mRNA
expression and clinicopathological features in colorectal
cancer.
|
|
| p16 mRNA |
|
|---|
|
|
|
|
|
|---|
| Features | n | High, n=51 | Low, n=50 | P-value |
|---|
| Sex |
|
Male | 57 | 29 | 28 | 1.000 |
|
Female | 44 | 22 | 22 |
|
| Age, years |
|
|
| 0.842 |
|
≥65 | 56 | 29 | 27 |
|
|
<65 | 45 | 22 | 23 |
|
| Site |
|
Proximal | 29 | 16 | 13 | 0.485 |
|
Distal | 48 | 22 | 26 |
|
| Not
known | 24 | 13 | 11 |
|
| Tumor
histology |
|
Well | 34 | 19 | 15 | 0.668 |
|
Moderately | 34 | 15 | 19 |
|
|
Poorly | 4 | 1 | 3 |
|
|
Mucinous | 4 | 2 | 2 |
|
| Not
known | 25 | 14 | 11 |
|
| T stage |
| T2 | 3 | 1 | 2 | 0.507 |
| T3 | 56 | 30 | 26 |
|
| T4 | 18 | 7 | 11 |
|
| Not
known | 24 | 13 | 11 |
|
| Stage |
| 2 | 36 | 18 | 18 | 1.000 |
| 3 | 41 | 20 | 21 |
|
| 4 | 0 | 0 | 0 |
|
| Not
known | 24 | 13 | 11 |
|
| Adjuvant
chemotherapy |
|
Yes | 54 | 27 | 27 | 1.000 |
| No | 44 | 22 | 22 |
|
| Not
known | 3 | 2 | 1 |
|
Patients with CRC were additionally divided into
p16/PCNA high (n=50) and low (n=51) groups according to the median
value (11.46). No differences in any of the clinical and
histopathologic parameters were observed between these two groups
(Table II), nor was there a
significant difference in patient survival between the high and low
p16/PCNA groups (P=0.122; Fig. 4C).
The 83 patients with low tumor p16 methylation were further
examined by dividing them into groups with high or low p16/PCNA
expression according to the median value (12.49). In this analysis,
patients with high p16/PCNA expression demonstrated a significantly
worse survival (P=0.026; Fig.
4D).
The patient group with a low p16 methylation
(PMR <4) was additionally evaluated using Cox regression
analysis for the prognostic significance of various clinical and
histopathologic features and for p16 mRNA and p16/PCNA expression
levels. p16/PCNA mRNA expression was the only significant
prognostic factor in univariate analysis (Table IV; P=0.026). The influence of T
stage, tumor stage and adjuvant chemotherapy on the overall
survival of patients with CRC was analyzed using the Kaplan-Meier
method. There was no association between any of these factors and
the prognosis of the patients (Fig.
5). Multivariate analysis identified that low p16/PCNA
expression was an independent factor for better survival in
patients with low p16 methylation (hazard ratio 0.287; 95%
confidence interval 0.110–0.747; P=0.011; Table IV).
 | Table IV.Univariate and multivariate analysis
for the prognostic significance of clinicopathological factors and
p16/PCNA mRNA. |
Table IV.
Univariate and multivariate analysis
for the prognostic significance of clinicopathological factors and
p16/PCNA mRNA.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Clinicopathological
factors Variables | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Male | 0.912 | 0.443–1.877 | 0.802 | 0.660 | 0.223–1.947 | 0.451 |
| Age, years | 1.026 | 0.996–1.058 | 0.095 | 1.019 | 0.977–1.063 | 0.379 |
| Proximal vs.
distal | 0.516 | 0.192–1.389 | 0.190 | 0.446 | 0.126–1.574 | 0.209 |
| Well-differentiated
histology vs. others | 0.585 | 0.250–1.370 | 0.217 | 0.643 | 0.231–1.796 | 0.400 |
| T stage | 1.448 | 0.631–3.332 | 0.383 | 1.602 | 0.604–4.245 | 0.343 |
| Stage | 1.326 | 0.584–3.007 | 0.500 | 1.723 | 0.677–4.245 | 0.343 |
| Adjuvant
chemotherapy | 0.704 | 0.664–1.481 | 0.355 | 0.439 | 0.144–1.336 | 0.147 |
| p16/PCNA mRNA | 0.422 | 0.197–0.902 | 0.026 | 0.287 | 0.110–0.747 | 0.011 |
Discussion
Tumor p16 methylation has previously been
identified as a prognostic factor for a worse outcome in CRC
(33,34,41).
However, other previous studies demonstrated that p16
methylation has no impact on prognosis (32) or predicts worse outcome only in
patients with poorly differentiated CRC (45). A possible mechanism for the putative
association between tumor p16 methylation and survival of
patients with CRC is that expression of p16 protein is diminished,
thereby promoting tumor cell proliferation and invasion (46,47).
In the present study, however, no significant association was
observed between tumor p16 methylation and the outcome of
patients with CRC, thus supporting a previous study (32). A number of technical reasons, in
addition to the number of patients examined may account for the
inconsistent results demonstrated by different previous studies and
the present study. The technical reasons include differences in the
tissue samples analyzed (fresh compared with fixed tissues), the
methods used to quantify p16 methylation levels and the
cut-off values used in the analyses (32–34,41,45).
p16 promoter methylation is associated with
decreased expression of p16 mRNA in clinical samples of CRC
(26). In the present study, an
inverse association between tumor p16 methylation and the
expression of its transcript was additionally identified. This was
most pronounced in tumors with high methylation levels (PMR
>10). However, within the high methylation group (PMR ≥4) a
number of cases with relatively increased expression of p16 mRNA
were identified, in agreement with previous studies, which
demonstrated that tumor cells with p16 promoter methylation
may express p16 mRNA (31,41). Therefore, p16 mRNA expression may
not be controlled exclusively through promoter methylation;
however, may additionally be influenced by other factors, including
the Ras signaling pathway (48),
which is frequently activated in CRC due to KRAS
proto-oncogene GTPase and NRAS proto-oncogene GTPase
mutations.
Previous studies demonstrated that low p16 protein
expression in CRC was associated with larger tumor size, lymph node
metastasis and faster tumor proliferation (31,49,50),
and is thus likely to account for an association with worse
prognosis (51). However, little is
known regarding the prognostic impact of p16 mRNA expression in
patients with CRC. Although an inverse association between tumor
p16 methylation and mRNA expression was observed in the
present study, relatively few patients (18/101) demonstrated high
levels of p16 methylation, defined as PMR ≥4. Therefore,
patients with low tumor p16 methylation levels were
investigated, as it was hypothesized that p16 mRNA expression may
affect patient survival, as those patients may exhibit a wide range
of p16 mRNA expression for analysis. An unexpected result of the
present study was that patients with high p16/PCNA mRNA expression
demonstrated significantly worse survival. Furthermore, this was
demonstrated by multivariate analysis to be independent of other
factors that potentially influence patient survival, including
tumor stage and histological types. Despite its well-recognized
tumor suppressor role (52), p16 is
overexpressed at the mRNA and protein expression levels in tumor
tissues compared with adjacent normal mucosa (31,51).
Similar to the present results, patients with breast (53,54)
and prostate cancer (55) with high
p16 expression were additionally identified to have a worse
prognosis. Notably, although p16 is a critical cell cycle regulator
and its mRNA expression is likely to be associated with cell cycle
(44), none of the previous studies
investigating the association of p16 promoter methylation
with prognosis of patients with CRC (32,34)
scored a copy number of p16 mRNA. Therefore, the clinical
implications of p16 mRNA and protein expression in different cancer
types and association with p16 methylation require further
investigation.
Established tumor cell lines with Rb deletion
demonstrated activated p16 transcription and increased p16
protein expression (56). It has
additionally been demonstrated that cell cycle regulation by p16 is
lost in tumor cells with inactivated Rb (57), and that the efficacy of exogenously
expressed p16 in cancer cells depends on Rb function (58). Furthermore, the overexpression of
transcription factor E2F1 promotes p16 transcription
(59), whereas CDK4 overexpression
in sarcoma cells is thought to increase p16 expression through a
feedback loop (60). This putative
feedback regulation in the expression and function of Rb pathway
mediators may explain the paradoxical association observed in the
present study and in others between high p16 expression and worse
patient survival.
In summary, p16 is a CDK4 inhibitor that counteracts
the cell cycle process by sustaining the Rb-mediated pathway.
Accordingly, p16 has been recognized as a tumor suppressor that is
lost or inactivated through gene mutation, deletion or promoter
methylation in various cancer types, including CRC. Previous
studies demonstrated an association between p16 gene
promoter hypermethylation and worse prognosis in patients with CRC,
in addition to an inverse association between p16 expression and
tumor progression. However, the effect of p16 mRNA expression on
the prognosis of patients with cancer is controversial. In the
present study, it was demonstrated that p16 mRNA expression in the
tumors was inversely associated with the levels of p16
promoter methylation. In addition, multivariate analysis determined
that high p16 mRNA expression normalized to PCNA mRNA expression
(p16/PCNA) was an independent prognostic factor for poor survival
of patients with CRC. These results identified a previously
unrecognized and paradoxical association between high expression of
p16 mRNA and worse prognosis of patients with CRC, although a
similar association has been demonstrated in other cancer
types.
Acknowledgements
The authors would like to thank Dr Barry Iacopetta
(University of Western Australia, Crawley, Australia) for
critically reading and editing the manuscript.
Funding
The present study was supported in part by
Grants-in-Aid for Scientific Research from the Japan Society for
the Promotion of Science (grant nos. 20390353 and 23390321).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HK made substantial contributions to the design and
conception of the study, and the acquisition, analysis and
interpretation of the data, and drafted the manuscript. HT and TM
collected the clinical samples and contributed to the
interpretation of the data. TM helped to draft and finalize the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki. As the tissues used in the present
study were from the patients diagnosed between 1999 and 2002,
written informed consent was obtained from the patients prior to
the tissue sample collection. In accordance with Japanese ethical
guidelines and law, the study protocol was reviewed and approved by
the Kanazawa University Human Genome/Gene Analysis Research Ethics
Committee (approval no. 181; Kanazawa, Japan). At the start of the
study, it was not possible to directly contact the specific
patients to explain the present study. According to the Human
Genome/Gene Analysis Research Ethics Committee, the present study
was announced on our website, providing these patients an
opportunity to opt out of the present study. By the date indicated,
none of them refused to be included in the present study. All
samples were anonymized before analysis was performed to guarantee
the protection of privacy.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
5-FU
|
5-fluorouracil
|
|
PCNA
|
proliferating cell nuclear antigen
|
|
PCR
|
polymerase chain reaction
|
|
PMR
|
percentage of methylated reference
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M and Bray F: Cancer incidence and mortality
worldwide: Sources, methods and major patterns in GLOBOCAN 2012.
Int J Cancer. 136:E359–E386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Markowitz SD and Bertagnolli MM: Molecular
basis of colorectal cancer. N Engl J Med. 361:2449–2460. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carethers JM and Jung BH: Genetics and
genetic biomarkers in sporadic colorectal cancer. Gastroenterology.
149:1177–1190.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Issa JP: CpG island methylator phenotype
in cancer. Nat Rev Cancer. 4:988–993. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kondo Y and Issa JPJ: Epigenetic changes
in colorectal cancer. Cancer Metastasis Rev. 23:29–39. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iacopetta B, Grieu F, Li W, Ruszkiewicz A,
Caruso M, Moore J and Kawakami K: APC gene methylation is
inversely correlated with features of the CpG island methylator
phenotype in colorectal cancer. Int J Cancer. 119:2272–2278. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Derks S, Postma C, Moerkerk P, van den
Bosch SM, Carvalho B, Hermsen MA, Giaretti W, Herman JG, Weijenberg
MP, de Bruïne AP, et al: Promoter methylation precedes chromosomal
alterations in colorectal cancer development. Cell Oncol.
28:247–257. 2006.PubMed/NCBI
|
|
11
|
Toyota M, Ahuja N, Ohe-Toyota M, Herman
JG, Baylin SB and Issa JPJ: CpG island methylator phenotype in
colorectal cancer. Proc Natl Acad Sci USA. 96:8681–8686. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Knudsen ES and Wang JY: Targeting the
RB-pathway in cancer therapy. Clin Cancer Res. 16:1094–1099. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Munro S, Carr SM and La Thangue NB:
Diversity within the pRb pathway: Is there a code of conduct?
Oncogene. 31:4343–4352. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Engelmann D and Pützer BM: The dark side
of E2F1: In transit beyond apoptosis. Cancer Res. 72:571–575. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Witkiewicz AK, Knudsen KE, Dicker AP and
Knudsen ES: The meaning of p16ink4a expression in
tumors: Functional significance, clinical associations and future
developments. Cell Cycle. 10:2497–2503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Knudsen KE, Diehl JA, Haiman CA and
Knudsen ES: Cyclin D1: Polymorphism, aberrant splicing and cancer
risk. Oncogene. 25:1620–1628. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Caca K, Feisthammel J, Klee K, Tannapfel
A, Witzigmann H, Wittekind C, Mössner J and Berr F: Inactivation of
the INK4a/ARF locus and p53 in sporadic extrahepatic bile duct
cancers and bile tract cancer cell lines. Int J Cancer. 97:481–488.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Toyooka S, Mitsudomi T, Soh J, Aokage K,
Yamane M, Oto T, Kiura K and Miyoshi S: Molecular oncology of lung
cancer. Gen Thorac Cardiovasc Surg. 59:527–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hong SM, Park JY, Hruban RH and Goggins M:
Molecular signatures of pancreatic cancer. Arch Pathol Lab Med.
135:716–727. 2011.PubMed/NCBI
|
|
20
|
Mori T, Miura K, Aoki T, Nishihira T, Mori
S and Nakamura Y: Frequent somatic mutation of the MTS1/CDK4I
(multiple tumor suppressor/cyclin-dependent kinase 4 inhibitor)
gene in esophageal squamous cell carcinoma. Cancer Res.
54:3396–3397. 1994.PubMed/NCBI
|
|
21
|
He J, Mokhtari K, Sanson M, Marie Y, Kujas
M, Huguet S, Leuraud P, Capelle L, Delattre JY, Poirier J, et al:
Glioblastomas with an oligodendroglial component: A pathological
and molecular study. J Neuropathol Exp Neurol. 60:863–871. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maesawa C, Tamura G, Nishizuka S,
Ogasawara S, Ishida K, Terashima M, Sakata K, Sato N, Saito K and
Satodate R: Inactivation of the CDKN2 gene by homozygous
deletion and de novo methylation is associated with advanced stage
esophageal squamous cell carcinoma. Cancer Res. 56:3875–3878.
1996.PubMed/NCBI
|
|
23
|
Oshima M, Okano K, Muraki S, Haba R, Maeba
T, Suzuki Y and Yachida S: Immunohistochemically detected
expression of 3 major genes (CDKN2A/p16TP53, and
SMAD4/DPC4) strongly predicts survival in patients with
resectable pancreatic cancer. Ann Surg. 258:336–346. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ueki T, Hsing AW, Gao YT, Wang BS, Shen
MC, Cheng J, Deng J, Fraumeni JF Jr and Rashid A: Alterations of
p16 and prognosis in biliary tract cancers from a population-based
study in China. Clin Cancer Res. 10:1717–1725. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Taga S, Osaki T, Ohgami A, Imoto H,
Yoshimatsu T, Yoshino I, Yano K, Nakanishi R, Ichiyoshi Y and
Yasumoto K: Prognostic value of the immunohistochemical detection
of p16INK4 expression in nonsmall cell lung carcinoma. Cancer.
80:389–395. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Herman JG, Merlo A, Mao L, Lapidus RG,
Issa JP, Davidson NE, Sidransky D and Baylin SB: Inactivation of
the CDKN2/p16/MTS1 gene is frequently associated with
aberrant DNA methylation in all common human cancers. Cancer Res.
55:4525–4530. 1995.PubMed/NCBI
|
|
27
|
Guan RJ, Fu Y, Holt PR and Pardee AB:
Association of K-ras mutations with p16 methylation in human colon
cancer. Gastroenterology. 116:1063–1071. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burri N, Shaw P, Bouzourene H, Sordat I,
Sordat B, Gillet M, Schorderet D, Bosman FT and Chaubert P:
Methylation silencing and mutations of the p14ARF and p16INK4a
genes in colon cancer. Lab Invest. 81:217–229. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yi J, Wang ZW, Cang H, Chen YY, Zhao R, Yu
BM and Tang XM: p16 gene methylation in colorectal cancers
associated with Duke's staging. World J Gastroenterol. 7:722–725.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Esteller M, González S, Risques RA,
Marcuello E, Mangues R, Germà JR, Herman JG, Capellà G and Peinado
MA: K-rasp16 aberrations confer poor prognosis in human
colorectal cancer. J Clin Oncol. 19:299–304. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim BN, Yamamoto H, Ikeda K, Damdinsuren
B, Sugita Y, Ngan CY, Fujie Y, Ogawa M, Hata T, Ikeda M, et al:
Methylation and expression of p16INK4 tumor suppressor
gene in primary colorectal cancer tissues. Int J Oncol.
26:1217–1226. 2005.PubMed/NCBI
|
|
32
|
Shima K, Nosho K, Baba Y, Cantor M,
Meyerhardt JA, Giovannucci EL, Fuchs CS and Ogino S: Prognostic
significance of CDKN2A (p16) promoter methylation and loss of
expression in 902 colorectal cancers: Cohort study and literature
review. Int J Cancer. 128:1080–1094. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maeda K, Kawakami K, Ishida Y, Ishiguro K,
Omura K and Watanabe G: Hypermethylation of the CDKN2A gene in
colorectal cancer is associated with shorter survival. Oncol Rep.
10:935–938. 2003.PubMed/NCBI
|
|
34
|
Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J
and Chen M: The prognostic value of CDKN2A hypermethylation
in colorectal cancer: A meta-analysis. Br J Cancer. 108:2542–2548.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union Against Cancer (UICC) TNM Classification of
Malignant Tumors. 7th. Wiley-Blackwell; Oxford: 2009
|
|
36
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D and Laird PW: MethyLight: A high-throughput
assay to measure DNA methylation. Nucleic Acids Res. 28:E322001.
View Article : Google Scholar
|
|
38
|
Eads CA, Lord RV, Wickramasinghe K, Long
TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester
TR, Skinner KA, et al: Epigenetic patterns in the progression of
esophageal adenocarcinoma. Cancer Res. 61:3410–3418.
2001.PubMed/NCBI
|
|
39
|
Ogino S, Kawasaki T, Brahmandam M, Cantor
M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ,
Laird PW, Loda M, et al: Precision and performance characteristics
of bisulfite conversion and real-time PCR (MethyLight) for
quantitative DNA methylation analysis. J Mol Diagn. 8:209–217.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ogino S, Cantor M, Kawasaki T, Brahmandam
M, Kirkner GJ, Weisenberger DJ, Campan M, Laird PW, Loda M and
Fuchs CS: CpG island methylator phenotype (CIMP) of colorectal
cancer is best characterised by quantitative DNA methylation
analysis and prospective cohort studies. Gut. 55:1000–1006. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mitomi H, Fukui N, Tanaka N, Kanazawa H,
Saito T, Matsuoka T and Yao T: Aberrant p16INK4a
methylation is a frequent event in colorectal cancers: Prognostic
value and relation to mRNA expression and immunoreactivity. J
Cancer Res Clin Oncol. 136:323–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li J, Poi MJ and Tsai MD: The regulatory
mechanisms of tumor suppressor P16INK4A and relevance to
cancer. Biochemistry. 50:5566–5582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Veganzones-de-Castro S, Rafael-Fernández
S, Vidaurreta-Lázaro M, de-la-Orden V, Mediero-Valeros B, Fernández
C and Maestro-de las Casas ML: p16 gene methylation in colorectal
cancer patients with long-term follow-up. Rev Esp Enferm Dig.
104:111–117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tada T, Watanabe T, Kazama S, Kanazawa T,
Hata K, Komuro Y and Nagawa H: Reduced p16 expression correlates
with lymphatic invasion in colorectal cancers.
Hepatogastroenterology. 50:1756–1760. 2002.
|
|
47
|
Rayess H, Wang MB and Srivatsan ES:
Cellular senescence and tumor suppressor gene p16. Int J Cancer.
130:1715–1725. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Serrano M, Gómez-Lahoz E, DePinho RA,
Beach D and Bar-Sagi D: Inhibition of ras-induced proliferation and
cellular transformation by p16INK4. Science. 267:249–252. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dai CY, Furth EE, Mick R, Koh J, Takayama
T, Niitsu Y and Enders GH: p16 INK4a expression begins early in
human colon neoplasia and correlates inversely with markers of cell
proliferation. Gastroenterology. 119:929–942. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Palmqvist R, Rutegård JN, Bozoky B,
Landberg G and Stenling R: Human colorectal cancers with an intact
p16/cyclin D1/pRb pathway have up-regulated p16 expression and
decreased proliferation in small invasive tumor clusters. Am J
Pathol. 157:1947–1953. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao P, Hu YC and Talbot IC: Expressing
patterns of p16 and CDK4 correlated to prognosis in colorectal
carcinoma. World J Gastroenterol. 9:2202–2206. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nat Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dublin EA, Patel NK, Gillett CE, Smith P,
Peters G and Barnes DM: Retinoblastoma and p16 proteins in mammary
carcinoma: Their relationship to cyclin D1 and histopathological
parameters. Int J Cancer. 79:71–75. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hui R, Macmillan RD, Kenny FS, Musgrove
EA, Blamey RW, Nicholson RI, Robertson JF and Sutherland RL:
INK4a gene expression and methylation in primary breast
cancer: Overexpression of p16INK4a messenger RNA is a
marker of poor prognosis. Clin Cancer Res. 6:2777–2787.
2000.PubMed/NCBI
|
|
55
|
Quinn DI, Henshall SM and Sutherland RL:
Molecular markers of prostate cancer outcome. Eur J Cancer.
41:858–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Parry D, Bates S, Mann DJ and Peters G:
Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with
high levels of p16INK4/MTS1 tumour suppressor gene product. EMBO J.
14:503–511. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Knudsen ES and Knudsen KE: Tailoring to
RB: Tumour suppressor status and therapeutic response. Nat Rev
Cancer. 8:714–724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Craig C, Kim M, Ohri E, Wersto R, Katayose
D, Li Z, Choi YH, Mudahar B, Srivastava S, Seth P, et al: Effects
of adenovirus-mediated p16INK4A expression on cell cycle
arrest are determined by endogenous p16 and Rb status in human
cancer cells. Oncogene. 16:265–272. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Khleif SN, DeGregori J, Yee CL, Otterson
GA, Kaye FJ, Nevins JR and Howley PM: Inhibition of cyclin
D-CDK4/CDK6 activity is associated with an E2F-mediated induction
of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA.
93:4350–4354. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yao J, Pollock RE, Lang A, Tan M, Pisters
PW, Goodrich D, El-Naggar A and Yu D: Infrequent mutation of the
p16/MTS1 gene and overexpression of cyclin-dependent kinase 4 in
human primary soft-tissue sarcoma. Clin Cancer Res. 4:1065–1070.
1998.PubMed/NCBI
|