Introduction
Zinc finger protein 36 (ZFP36), is a member of the
TISS11 family of RNA-binding proteins that serves a role in
post-transcriptional gene regulation by promoting the decay of
AU-rich element (ARE)-containing mRNAs (1). Inhibition of ZFP36-mediated gene
regulation can induce pathological consequences, and is implicated
in chronic inflammation and cancer (2). Additionally, overexpression of human
ZFP36 in cancer cell lines suppresses the cell cycle (3) and epithelial-mesenchymal transition
(4). Recently, evidence has emerged
from human liver and colorectal cancer cells demonstrating that
ZFP36 expression is downregulated by epigenetic modification
(5,6). Additionally, treatments with
demethylating agent 5-Aza-2′-deoxycytidine (Aza) or histone
deacetylase inhibitors, including trichostatin A, SAHA and sodium
butyrate, increase ZFP36 expression (5–8). These
pieces of evidence indicate that regulation of ZFP36 expression is
affected by epigenetic modification.
Epigenetics is the study of heritable gene
expression regulation that occurs without changes to the DNA
sequence (9). The primary
epigenetic modifications include DNA methylation and a complex set
of histone modifications (10). DNA
methylation is mediated by DNA methyltransferase (DNMTs), of which
humans have three: DNA (cytosine-5)-methyltransferase 1 (DNMT1),
DNMT3A and DNMT3B (11). The
function of DNMT1 is to maintain DNA methylation patterns, whereas
DNMT3A and 3B are responsible for methylation of new CpG sites
(12). Notably, accumulating
evidence demonstrates that abnormal DNA methylation is involved in
cancer progression, including sporadic gastric cancer, and renal
and colorectal carcinoma (13–15).
Resveratrol (Res) is a secondary metabolite produced
by plants, including grapevines, berries and peanuts (16). Multiple studies to date have
investigated the effects of Res on ZFP36 expression in cancer cells
(17–19). These studies demonstrated that Res
can upregulate ZFP36 expression, resulting in the induction of
apoptosis in glioma cells (17). It
was also indicated recently that Res induces ZFP36 expression and
suppresses the production of cytokines in MCF-7 breast cancer
cells, including cyclooxygenase-2 (COX-2) and vascular endothelial
growth factor (VEGF) (20).
However, the molecular mechanism for the transcriptional regulation
of ZFP36 remains poorly understood.
In the present study, it was indicated that Res
induces ZFP36 expression in A549 human lung cancer cells, which
results in the downregulation of ARE-containing genes. Further
analysis indicated that Res inhibits methylation at the ZFP36
promoter region. The present study demonstrated that regulation of
ZFP36 expression in A549 lung cancer cells occurs through an
epigenetic mechanism and posits a potential therapeutic approach
through Res-induced ZFP36 expression in non-small cell lung
cancer.
Materials and methods
Reagents
Res was purchased from Tocris Bioscience (Bristol,
UK). High-glucose Dulbecco's modified Eagle's medium (DMEM),
RPMI-1640 medium, penicillin/streptomycin, Trypsin-EDTA and fetal
bovine serum (FBS) were purchased from Gibco; Thermo Fisher
Scientific, Inc. (Waltham, MA, USA). Dimethyl sulfoxide (DMSO; cat.
no. D2650), Aza (cat. no. A3656), and antibodies against ZFP36
(cat. no. T5327) and β-actin (cat. no. A5441) were purchased from
Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Antibodies against
DNMT1 (cat. no. Ab19905), VEGFA (cat. no. Ab46154) and MYC (cat.
no. Ab32072) were purchased from Abcam (Cambridge, MA, USA). The
antibody against Cyclin D1 (CCND1; cat. no. 2978S) was purchased
from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Cell culture
A549 and H23 human lung cancer cells line was
obtained from American Type Culture Collection (Manassas, VA, USA).
A549 cells were grown in high-glucose DMEM and H23 cells were grown
in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin, and
100 µg/ml streptomycin. Cells were cultured at 37°C in a 5%
CO2 humidified incubator.
Aza treatment
A549 and H23 cells were seeded in 100-mm dished at
1×106 cells/dishes. Cells were allowed to adhere
overnight at 37°C and then were treated with the appropriate
concentration of control [0.1% DMSO (0 µM)] or Aza (5 µM) for 72 h
at 37°C.
Res treatment
A549 cells were seeded in 100-mm dished at
1×106 cells/dishes. Cells were allowed to adherent
overnight at 37°C and then treated with the appropriate
concentration of control [0.1% DMSO (0 µM)] or Res (20, 50 and 100
µM) for 72 h at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was added to cell lysates, according to
the manufacturer's protocols. The RT-PCR was performed using a
Moloney murine leukemia virus reverse transcriptase kit (cat. no.
30201; Beams Biotechnology Co., Ltd., Seognam, Korea).
Complementary DNA synthesis using an iCycler thermocycler (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was performed by incubating
at 37°C for 90 min. RT-qPCR analysis was performed using TOPreal™
SYBR®-Green (Enzynomics, Daejeon, Korea) and the
following program in a Real-Time PCR Instrument Light
Cycle® 480 (Roche Applied Science, Madison, WI, USA):
95°C for 5 min, followed by 45 cycles of 95°C for 30 sec, 60°C for
30 sec and 72°C for 30 sec. The 2−ΔΔCq method was used
to calculate the relative levels of target mRNAs and GAPDH
was used as a reference gene (21).
The primer sequences were used as follows: ZFP36, forward,
5′-TGGGATCCGACCCTGATGAA-3′ and reverse, 5′-AAAACTCCCGCCTCGAAGAC-3′;
MYC, forward, 5′-AGAGTTTCATCTGCGACCCG-3′ and reverse,
5′-AAGCCGCTCCACATACAGTC-3′; CCND1, forward,
5′-TGCCAACCTCCTCAACGAC-3′ and reverse, 5′-TTTGAAGTAGGACACCGAGGG-3′;
VEGFA, forward, 5′-AGGGAAAGGGGCAAAAACGA-3′ and reverse,
5′-GAGGCTCCAGGGCATTAGAC-3′; DNMT1, forward,
5′-TACCTGGACGACCCTGACCTC-3′ and reverse,
5′-TACCTGGACGACCCTGACCTC-3′; GAPDH, forward,
5′-ACGCCACAGTTTCCCGGAGG-3′ and reverse,
5′-GCACCCCTGGCCAAGGTCAT-3′.
Western blotting
Cells were harvested, centrifuged at 890 × g for 1
min at 4°C and lysed with radioimmunoprecipitation buffer [25 mM
Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate
and 0.1% SDS; Thermo Fisher Scientific, Inc.] containing a protease
inhibitor cocktail [4-(2-aminoethyl) benzene sulfonyl fluoride
hydrochloride, aprotinin, bestatin hydrochloride-aminopeptidases,
N-(trans-Epoxysuccinyl)-L-leucine 4-guanidinobutylamide, EDTA and
leupeptin hemisulfate salt; Sigma-Aldrich; Merck KGaA]. Total
protein concentration was determined using a Pierce Bicinchoninic
Acid Assay kit (Thermo Fisher Scientific, Inc.). Equivalent
quantities of total protein (20–30 µg) were separated by SDS-PAGE
on 8–10% polyacrylamide gels, and then transferred to
nitrocellulose membranes (GE Healthcare Life Sciences, Little
Chalfont, UK) using a Trans-Blot® SD Semi-Dry
Electrophoretic Transfer Cell (cat. no. 170-3940; Bio-Rad
Laboratories, Inc.) submerged in transfer buffer (25 mM Tris, pH
8.3, 192 mM glycine and 20% methanol). The membrane was blocked
with 5% skimmed milk in 0.1% Tween-20/Tris-buffered saline (TBST)
for 2 h at room temperature, and then incubated with primary
antibodies for ZFP36, DNMT1, VEGFA, MYC, CCND1 (1:1,000) and mouse
monoclonal anti-β-actin (1:10,000) at 4°C overnight. Subsequently,
the blots were washed three times in TBST and incubated with goat
anti-rabbit horseradish peroxidase (HRP) conjugate (cat. no. 31430;
Thermo Fisher Scientific, Inc.) and goat anti-mouse HRP conjugate
(cat. no. 31460; Thermo Fisher Scientific, Inc.) IgG secondary
antibodies (1:10,000; Thermo Fisher Scientific, Inc.) for 50 min at
room temperature. The specific antibody signals were detected by
chemiluminescence (Advansta, Inc., San Jose, CA, USA) using a LAS
3000 instrument (Fujifilm, Tokyo, Japan).
Methylation assay
Cells were seeded in 100-mm dishes at
1×106 cells/dishes. Cells were allowed to adhere
overnight at 37°C and were then treated with the appropriate
concentration of control [0.1% DMSO (0 µM)], Res (100 µM) or Aza (5
µM) for 72 h 37°C. Following treatment, cells were lysed in buffer
containing 10 mM Tris-HCl (pH 8), 100 mM NaCl, 10 mM EDTA (pH 8),
10% Triton X-100 and proteinase K overnight at 56°C. Subsequently,
NaCl (6 M) was added to the cells, and then lysates were cleared by
centrifugation at 4,700 × g for 10 min at 4°C. A total of 600 µl
isopropanol was used for extraction of DNA from the supernatant. To
analyze CpG methylation of the ZFP36 promoter, the region
(+63 to −894) was selected due to this region containing numerous
transcription factor binding sites (5,6). Among
46 CpG islands, MS1-3 and MS4 were selected for Acc II and Hap II
restriction enzymes, respectively. Subsequently, 1 µg genomic DNA
was digested with 10 U Acc II (cat. no. 1002A, Takara Bio, Inc.,
Osaka, Japan) or Hap II (cat. no. 1053A, Takara Bio, Inc.)
overnight at 37°C prior to qPCR (21). The primer sequences used were as
follows: Methylation Site 1 (MS1), forward,
5′-GAAGGGAACCAGTCCAGGG-3′ and reverse, 5′-CCGAGAGCCGGCTACTTATAG-3′;
MS2, forward, 5′-CTCGGTCACGGCTGTCC-3′ and reverse,
5′-CTGGACTGGTTCCCTTCCG-3′; MS3, forward, 5′-CCCCATCCGTCTGTGTCG-3′
and reverse, 5′-TGTAGAAGGAAACTGGGGCG-3′; and MS4, forward,
5′-CTCAGGTAATCCACCTGCCTC-3′ and reverse,
5′-TACAGGTGTGAGCCACCAAG-3′.
Migration assay
The migration assay was conducted using a culture
insert (IBIDI, LLC, Verona, WI, USA), according to the
manufacturer's protocols. Briefly, cells were seeded in
culture-insert 4 well at 7×104 cells/well. Cells were
allowed to adhere for 24 h at 37°C and then the culture insert was
removed, leaving gaps in the sheets of cells. Different
concentrations of control [0.1% DMSO (0 µM)] and Res (20, 50, and
100 µM) were used to treat cells for 48 h 37°C. After 48 h, the
area destitute of cells was analyzed using a Carl Zeiss microscope
(Olympus TH4 200; Carl Zeiss AG, Oberkochen, Germany).
MTS assay
For the MTS assay, cells were seeded in 96-well
plates at 3×103 cells/well and then cells were allowed
to adhere overnight at 37°C. Different concentrations of control
[0.1% DMSO (0 µM)] and Res (20, 50 and 100 µM) were used to treat
cells for 48 h at 37°C. Following the indicated treatments, a total
of 20 µl CellTiter 96® AQueous One Solution cell
proliferation assay reagent (Promega Corporation, Madison, WI, USA)
was added to each well, according to the manufacturer's protocols,
and absorbance was measured at 490 nm using an Infinite M200 Pro
microplate reader (Tecan Group, Ltd., Mannedorf, Switzerland).
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Statistical significance was determined using the
Student's t-test or one-way analysis of variance was performed
using Dunnett's post test (GraphPad Prism; GraphPad Software, Inc.,
La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
ZFP36 expression is epigenetically
regulated in non-small cell lung cancer cells
To examine the methylation of ZFP36 promoter
regions, methylated CpG island site were predicted and MS1-4
regions were designated up to restriction enzyme availabilities
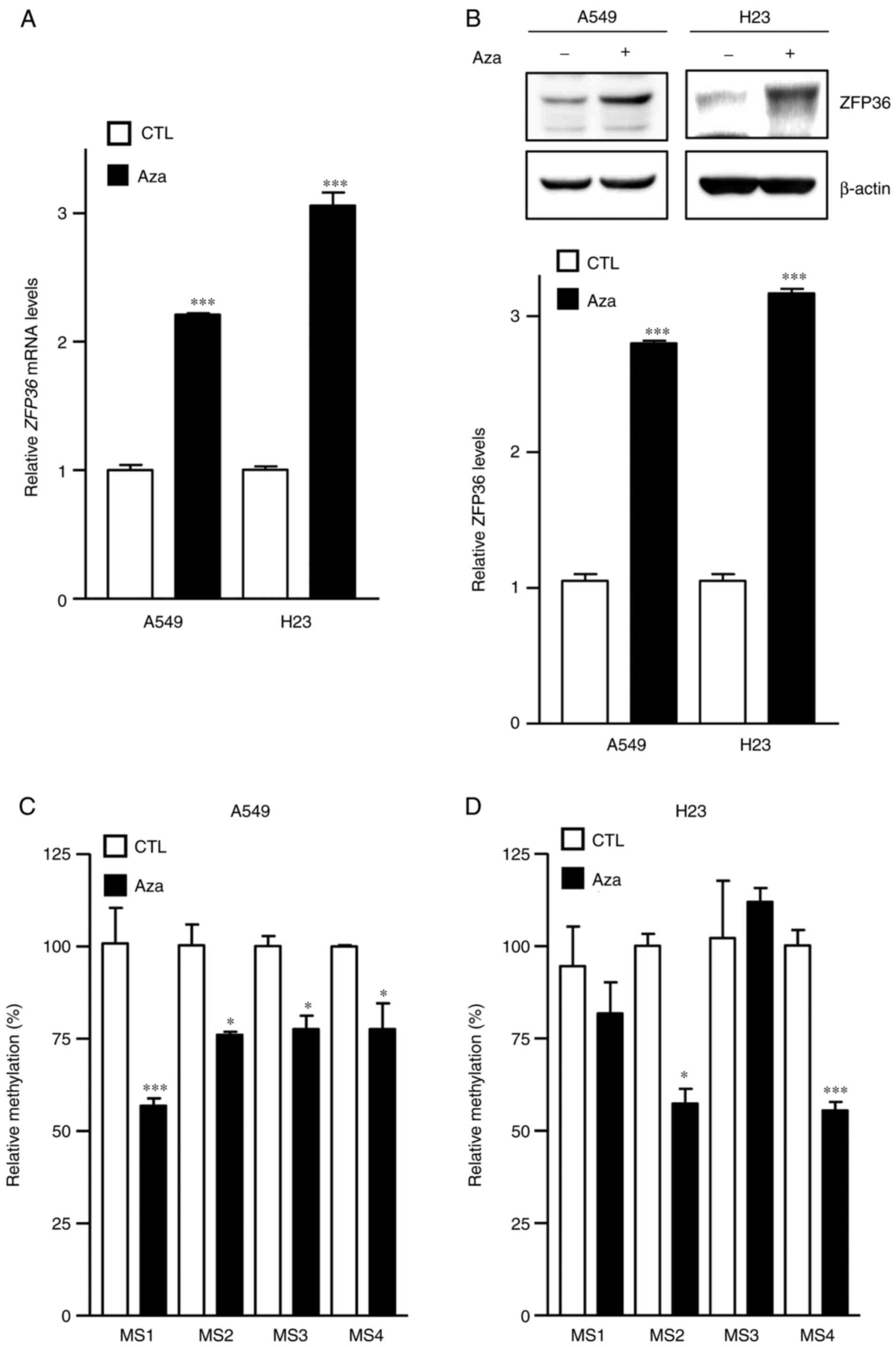
(Fig. 1). To determine the effects
of ZFP36 demethylation, A549 and H23 cells were firstly treated
with Aza, and it was determined that Aza significantly increased
ZFP36 mRNA and protein, compared with the control group
(P<0.001; Fig. 2A and B).
Increased ZFP36 expression following Aza treatment indicated that
ZFP36 expression is epigenetically regulated in A549 and H23 lung
cancer cells. Using enzyme restriction sensitive methylation
analysis with Acc II and Hap II, which recognizes and cleaves CGCG
and CCGG sequences, respectively, ZFP36 promoter methylation was
examined. Epigenetic analysis demonstrated that Aza significantly
reduces methylation at MS1-4 in A549 cells, and MS2 and 4 in H23
cells, compared with the control group (P<0.05 and P<0.001,
respectively; Fig. 2C and D).
Res demethylates ZFP36 promoter CpG
islands
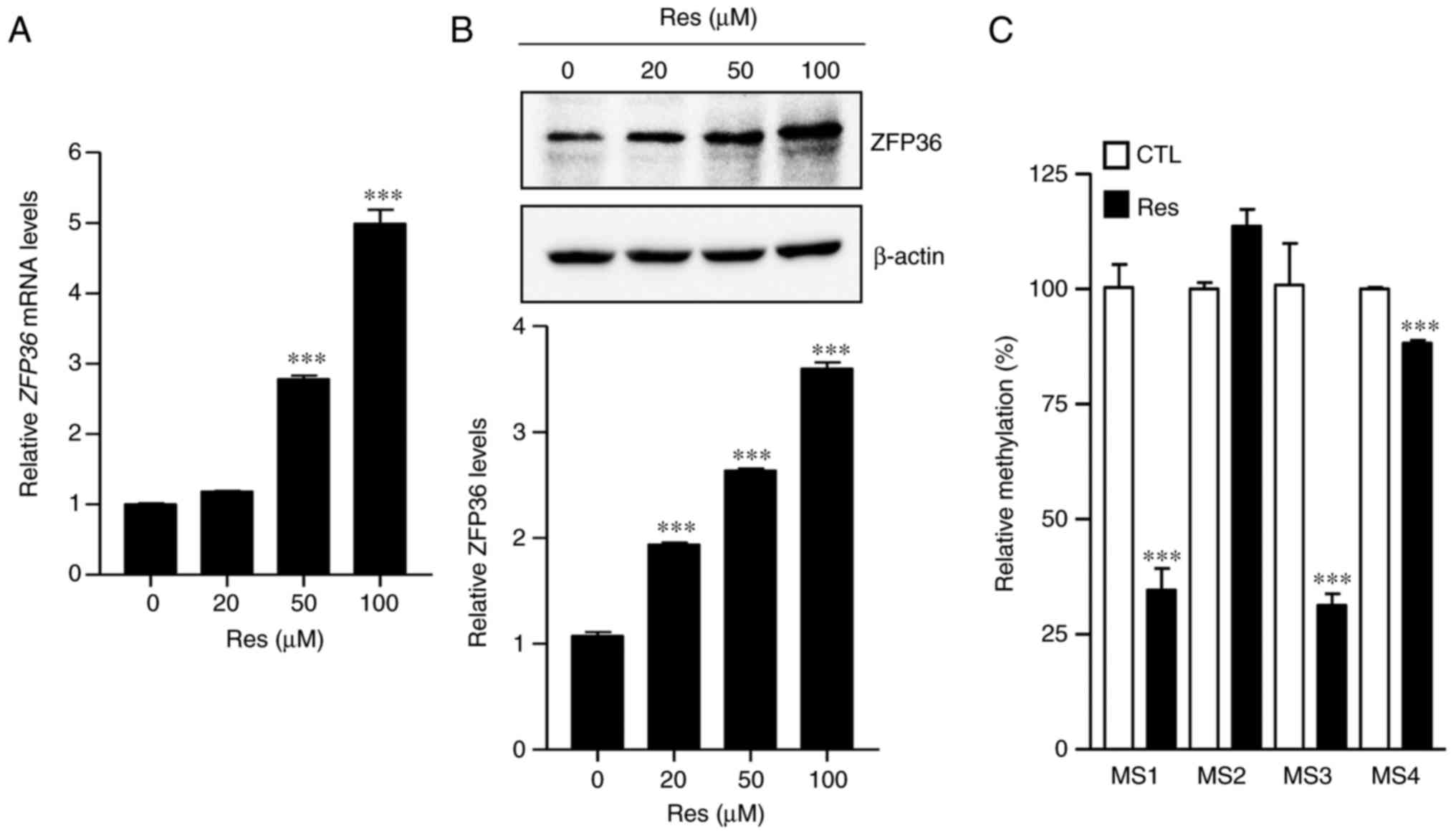
Recently, Res was demonstrated to induce ZFP36
expression in U87MG human glioblastoma cells (17). To determine whether these effects
also occur in lung cancer cells, ZFP36 expression was analyzed in
A549 cells following treatment with Res, and it was determined that
ZFP36 mRNA levels and protein expression were significantly
increased, compared with the control group (P<0.001; Fig. 3A and B). To analyze whether Res
demethylates CpG islands in the ZFP36 promoter, A549 lung
cancer cells were treated with Res and genomic DNA was isolated to
determine its methylation status. Notably, it was determined that
Res significantly reduced methylation at MS1, 3 and 4 of the ZFP36
promoter region, compared with the control group (P<0.001;
Fig. 3C). These data indicate that
Res activates ZFP36 mRNA expression by demethylating CpG
islands in the promoter region.
Res reduces ZFP36 target gene
expression in A549 lung cancer cells
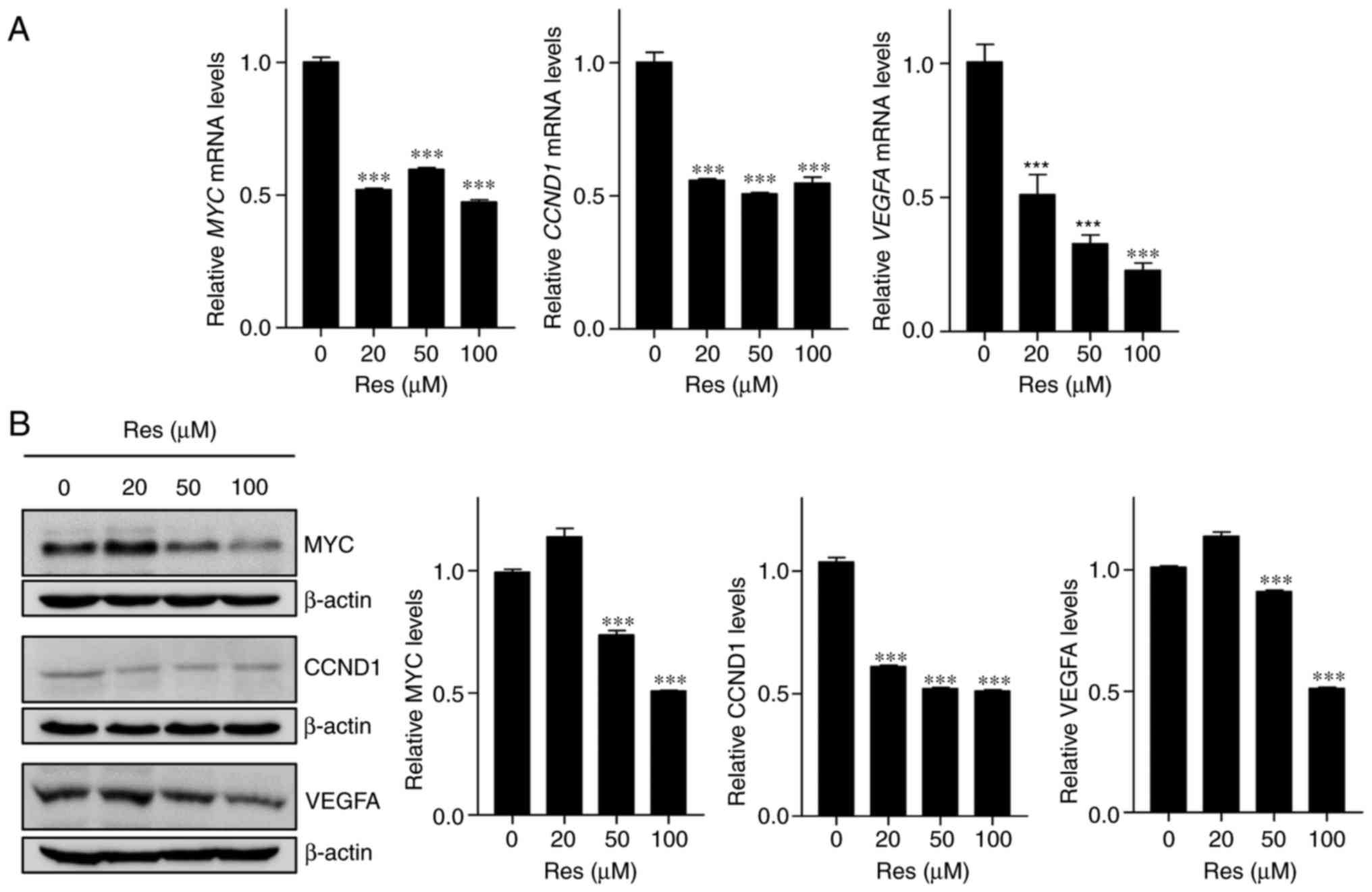
To investigate whether Res-induced ZFP36 expression
affects downstream ARE-mRNA decay, the expression of ZFP36 target
gene was examined following Res treatment. A549 lung cancer cells
were treated with Res, and the transcript and protein expression of
CCND1, MYC and VEGFA were determined by RT-qPCR and
western blotting, respectively. As expected, it was determined that
Res treatment significantly decreased mRNA and protein expression
of these ZFP36 targets, compared with the control group
(P<0.001; Fig. 4A and B).
Res inhibits DNMT1 expression in A549
lung cancer cells
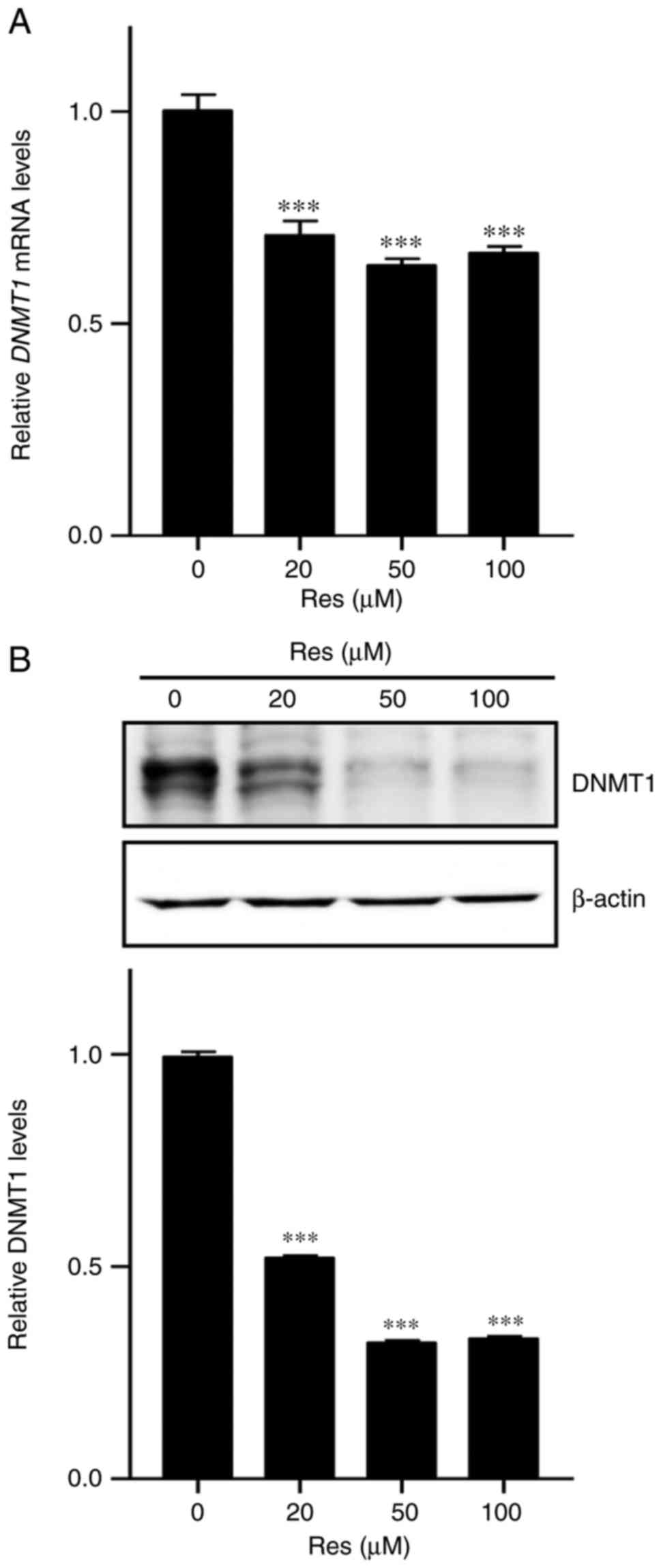
To investigate whether DNMT1 is involved in
regulation of ZFP36 methylation by Res, RT-qPCR and western
blotting were performed to detect DNMT1 mRNA and protein
levels in A549 lung cancer cells, respectively. It was determined
that Res treatment significantly reduced mRNA and protein levels,
compared with the control group (P<0.001; Fig. 5A and B) of DNMT1, indicating that
Res regulates the downregulation of DNMT1 expression in A549 lung
cancer cells.
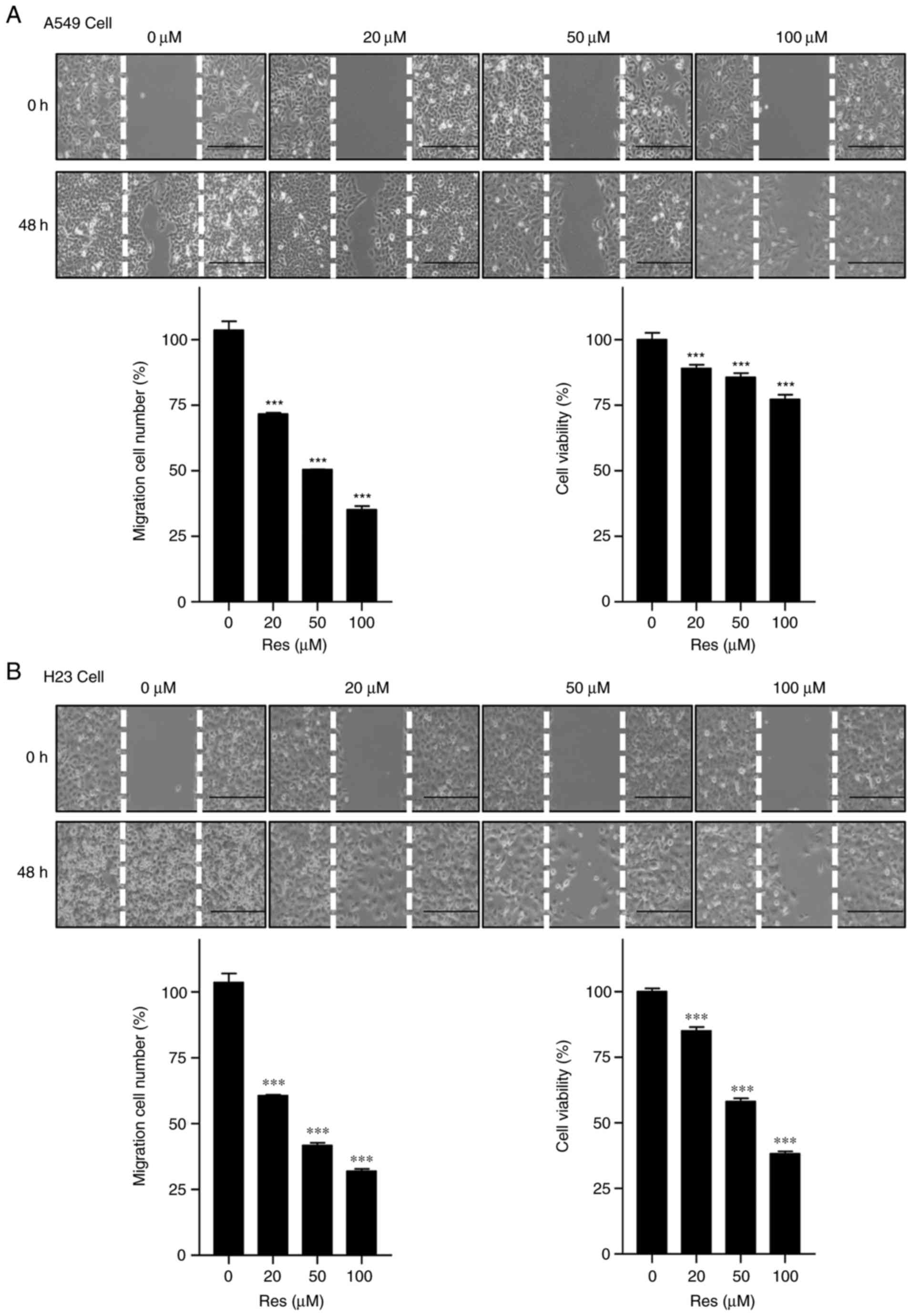
Res inhibits migration and cell
proliferation in non-small cell lung cancer cells
To investigate the migration and anticancer activity
of Res on A549 and H23 lung cancer cells, cell migration and MTS
assays were performed following Res treatment. It was determined
that Res significantly reduced the migration of A549 and H23 cells,
compared with the control group (P<0.001), as assessed with a
wound-healing assay. Additionally, further examination demonstrated
Res significantly inhibited the proliferation of A549 and H23 lung
cancer cells in a concentration-dependent, compared with the
control group (P<0.001; Fig. 6A and
B).
Discussion
ZFP36 is a member of the TISS11 family of
RNA-binding proteins characterized by the presence of two tandem
zinc finger domains (22,23). ZFP36 regulates the expression of
numerous genes through binding ARE regions in the 3′ untranslated
regions of its targets (24), which
include regulators of oncogenic growth, including cyclins, growth
factors and proto-oncogenes (25,26).
Previous studies demonstrated that ZFP36 expression inhibits the
growth of tumor cells through degradation of transcripts involved
in oncogenic growth, including c-FOS, c-MYC, CCND1, VEGFA
and COX-2 (27–32). ZFP36 is also known to destabilize
ARE-containing cytokine mRNAs, including interleukin-1β
(IL-1ß), IL-2, IL-3, IL-6, IL-10, tumor necrosis
factor and granulocyte-macrophage-colony-stimulating factor
(33). Notably, ZFP36 expression is
reduced in human cancer lines, compared with normal cells,
indicating that its loss may convey a pro-oncogenic advantage
(5,34).
Epigenetic regulation primarily occurs by changes in
DNA methylation, histone modification and miRNA expression. The aim
of the present study was to determine how epigenetics regulate
ZFP36 in A549 lung cancer cells. Epigenetic silencing of
ZFP36 may occur through direct modulation of the
ZFP36 gene or by silencing transcription factors that
regulate ZFP36 transcription. Previously, it has been
demonstrated that DNA methylation of a single CpG site in the
ZFP36 promoter in hepatocellular carcinoma is responsible
for downregulating ZFP36 expression (5). Similarly, the present study indicated
that demethylation by Aza treatment increased ZFP36
expression in A549 and H23 lung cancer cells (Fig. 2A and B). As one of the best methods
to analyze promoter methylation, methylation-sensitive restriction
analysis was used to dissect the effect of Aza on the methylation
status of the ZFP36 promoter region (Fig. 2C and D), demonstrating that Aza
effectively demethylated the ZFP36 promoter region.
Collectively, these results indicated that demethylation of the
ZFP36 promoter region is associated with increased
ZFP36 expression.
Aberrant expression of DNMTs is associated with a
variety of human diseases, including cancer (35), neurological disorders (36), immunological diseases (37) and genetic disorders (38). Therefore, DNMTs are promising
therapeutic targets (39). A number
of nutritional compounds, including apigenin, genistein, curcumin,
sulforaphane, epigallocatechin gallate and Res, regulate cellular
epigenetic events, including DNMT activity, in breast, colon, lung
and prostate cancer cells (40–43).
Res decreases DNMT enzymatic activity and mRNA levels of DNMT1,
DNMT3A and DNMT3B in HCC1806 breast cancer cells
(44). Additionally, Res reduces
transcript levels of three DNMT family members in MCF-7 and MDA MB
231 human breast cancer cell lines (45). In vivo studies also
demonstrated that Res suppresses tumor growth (46–51)
and decreases DNMT3B expression in an estrogen-dependent mammary
carcinoma rodent model (52).
However, the majority of aforementioned in vivo studies
demonstrated only the suppression of tumor growth or a DNMT subunit
expression by Res, and further studies are required to verify the
Res-induced epigenetic changes in tumor tissues in an in
vivo model. Furthermore, Res also reverses CpG methylation of
the promoter region of estrogen receptor-α in human breast cancer
MDA-MB468 cells (53), restores the
expression of phosphatase and tensin homolog via promoter CpG
demethylation in MCF-7 breast cancer cells (54), and inhibits DNMT1 expression and
prevents recruitment of DNMT1 to the BRCA-1 promoter in MCF-7 cells
(55). Considering this data,
indicating the possible epigenetic effects of Res, the present
study investigated whether Res treatment could rescue ZFP36
gene expression through promoter demethylation in A549 lung cancer
cells. As depicted in Fig. 3A and
B, Res induces ZFP36 expression in A549 lung cancer
cells. The methylation of the ZFP36 promoter region was then
further verified by Res using methylation-sensitive restriction
analysis (Fig. 3C). Notably, it was
determined that Res also decreased DNMT1 transcript and protein
expression (Fig. 5A and B).
Res was investigated as a natural anticancer agent
in various lung cancer cells. Previous studies have reported that
Res induces autophagy (56),
apoptosis and G1 cell cycle arrest (57). Res also inhibits the
phosphatidylinositol-3-kinase pathway, decreases mammalian target
of rapamycin phosphorylation (58),
and inhibits transforming growth factor β1-induced epithelial to
mesenchymal transition and suppression of cell adhesion (59). Other studies have examined the
chemistry behind the activity of Res. For example, Res contains a
resorcin moiety that chelates metal ions and Keap1 is a nuclear
factor erythroid 2-related factor 2 (Nfr2) inhibitory protein
(60). A549 cells treated with
Res-loaded nanoparticles activated Nrf2-Keap1 signaling, reduced
hydrogen peroxide-induced reactive oxygen species levels, increased
Res uptake and accumulated Nrf2 (61). Considering the anticancer effects of
Res, this polyphenol may be beneficial for the development of
therapeutics.
In conclusion, RT-qPCR and western blot analysis
confirmed that in A549 lung cancer cells, Res induces ZFP36
expression, demethylates the ZFP36 promoter and also reduces
DNMT1 expression. Collectively, Res-induced ZFP36 expression
through promoter demethylation serves an important role in lung
cancer cell proliferation and migration.
Acknowledgements
The authors would like to thank Dr Jeong Woo Park
(University of Ulsan, Ulsan, Korea) for their supports of ZFP36
associated materials.
Funding
The present study was supported by the National
Research Foundation of Korea and by a grant funded by the Korean
Government (Ministry of Education, Science and Technology) (grant
no. 2013R1A2A2A01068964).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SSK, DHL and AF conceived and designed the study. AF
and NAY performed the experiments. AF collected, analyzed the data
and wrote the paper. SK, JR, JYJ, AF and SSK designed the
experiments, interpreted the data, edited and reviewed the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sanduja S, Blanco FF, Young LE, Kaza V and
Dixon DA: The role of tristetraprolin in cancer and inflammation.
Front Biosci. 17:174–88. 2012. View
Article : Google Scholar :
|
|
2
|
Sanduja S, Blanco FF and Dixon DA: The
roles of TTP and BRF proteins in regulated mRNA decay. Wiley
Interdiscip Rev RNA. 2:42–57. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu L, Ning H, Gu L, Wang Q, Lu W, Peng H,
Cui W, Ying B, Ross CR, Wilson GM, et al: Tristetraprolin induces
cell cylces arrest in breast tumor cell through targeting
AP-1/c-Jun and NF-κB pathway. Oncotarget. 6:41679–41691. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yoon NA, Jo HG, Lee UH, Park JH, Yoon JE,
Ryu JY, Kang SS, Min YJ, Ju SA, Seo EH, et al: Tristetraprolin
suppresses the EMT through the down-regulation of Twist1 and Snail1
in cancer cells. Oncotarget. 7:8931–8943. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sohn BH, Park IY, Lee JJ, Yang SJ, Jang
YJ, Park KC, Kim DJ, Lee DC, Sohn HA, Kim TW, et al: Functional
switching of TGF-beta1 signaling in liver cancer via epigenetic
modulation of a single CpG Site in TTP promoter. Gastroenterology.
138:1898–1908. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sobolewski C, Sanduja S, Blanco FF, Hu L
and Dixon DA: Histone deacetylase inhibitors activate
tristetraprolin expression through induction of early growth
response protein 1 (EGR1) in Colorectal Cancer Cells. Biomolecules.
5:2035–2055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng XT, Xiao XQ and Dai JJ: Sodium
butyrate down-regulates tristetraprolin-mediated cyclin B1
expression independent of the formation of processing bodies. Int J
Biochem Cell Biol. 69:241–248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tran DD, Koch A, Allister A, Saran S,
Ewald F, Kock M, Nashan B and Tamura T: Treatment with MAPKAP2
(MK2) inhibitor and DNA methylation inhibitor, 5-aza dC,
synergistically triggers apoptosis in hepatocellular carcinoma
(HCC) via tristetraprolin (TTP). Cell Signal. 28:1872–1880. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Laird PW: Cancer epigenetics. Hum Mol
Genet. 14:R65–R76. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su LJ, Mahabir S, Ellison GL, McGuinn LA
and Reid BC: Epigenetic contributions to the relationship between
cancer and dietary intake of nutrients, bioactive food components,
and environmental toxicants. Front Genet. 2:912012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ferguson-Smith AC and Greally JM:
Epigenetics: Perceptive enzymes. Nature. 449:148–149. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meeran SM, Ahmed A and Tollefsbol TO:
Epigenetic targets of bioactive dietary components for cancer
prevention and therapy. Clin Epigenetics. 1:101–116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suzuki H, Itoh F, Toyota M, Kikuchi T,
Kakiuchi H, Hinoda Y and Imai K: Distinct methylation pattern and
microsatellite instability in sporadic gastric cancer. Int J
Cancer. 83:309–313. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herman JG, Latif F, Weng Y, Lerman MI,
Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, et al:
Silencing of the VHL tumor-suppressor gene by DNA methylation in
renal carcinoma. Proc Natl Acad Sci USA. 91:9700–9704. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herman JG, Umar A, Polyak K, Graff JR,
Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW,
et al: Incidence and functional consequences of hMLH1
promoter hypermethylation in colorectal carcinoma. Proc Natl Acad
Sci USA. 95:6870–6875. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Delmas D, Lancon A, Colin D, Jannin B and
Latruffe N: Resveratrol as a chemopreventive agent: A promising
molecule for fighting cancer. Curr Drug Targets. 7:423–442. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ryu J, Yoon NA, Seong H, Jeong JY, Kang S,
Park NM, Choi JI, Lee DH, Roh GS, Kim HJ, et al: Resveratrol
induces glioma cell apoptosis through activation of
tristetraprolin. Mol Cells. 38:991–997. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee SR, Jin H, Kim WT, Kim WJ, Kim SZ,
Leem SH and Kim SM: Tristetraprolin activation by resveratrol
inhibits the proliferation and metastasis of colorectal cancer
cells. Int J Oncol. 53:1269–1278. 2018.PubMed/NCBI
|
|
19
|
Kim WT, Jin H, Lee SR, Kim SZ, Leem SH and
Kim SM: Mediation of the anticancer effect of resveratrol via the
upregulation tristetraprolin in gastric cancer cell. Med Chem.
8:29–37. 2018.
|
|
20
|
Li C, Tang C and He G: Tristetraprolin: A
novel mediator of the anticancer properties of resveratrol. Genet
Mol Res. 15:2016.doi: 10.4238/gmr.15027213.
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
DuBois RN, McLane MW, Ryder K, Lau LF and
Nathans D: A growth factor-inducible nuclear protein with a novel
cysteine/histidine repetitive sequence. J Biol Chem.
265:19185–19191. 1990.PubMed/NCBI
|
|
23
|
Varnum BC, Ma QF, Chi TH, Fletcher B and
Herschman HR: The TIS11 primary response gene is a member of a gene
family that encodes proteins with a highly conserved sequence
containing an unusual Cys-His repeat. Mol Cell Biol. 11:1754–1758.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao H, Deterding LJ and Blackshear PJ:
Phosphorylation site analysis of the anti-inflammatory and
mRNA-destabilizing protein tristetraprolin. Expert Rev Proteomics.
4:711–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bakheet T, Frevel M, Williams BR, Greer W
and Khabar KS: Ared (Human au-rich element-containing mRNA database
reveals an unexpectedly diverse functional repertoire of encoded
proteins). Nucleic Acids Res. 29:246–254. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CY and Shyu AB: AU-rich elements:
Characterization and importance in mRNA degradation. Trends Biochem
Sci. 20:465–470. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hau HH, Walsh RJ, Ogilvie RL, Williams DA,
Reilly CS and Bohjanen PR: Tristetraprolin recruits functional mRNA
decay complexes to ARE sequences. J Cell Biochem. 100:1477–1492.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee HH, Son YJ, Lee WH, Park YW, Chae SW,
Cho WJ, Kim YM, Choi HJ, Choi DH, Jung SW, et al: Tristetraprolin
regulates expression of VEGF and tumorigenesis in human colon
cancer. Int J Cancer. 126:1817–1827. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marderosian M, Sharma A, Funk AP,
Vartanian R, Masri J, Jo OD and Gera JF: Tristetraprolin regulates
Cyclin D1 and c-Myc mRNA stability in response to rapamycin
in an Akt-dependent manner via p38 MAPK signaling. Oncogene.
25:6277–6290. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Essafi-Benkhadir K, Onesto C, Stebe E,
Monori C and Pagès G: Tristetraprolin inhibits Ras-dependent tumor
vascularization by inducing vascular endothelial growth factor mRNA
degradation. Mol Biol Cell. 18:4648–4658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suswam E, Li Y, Zhang X, Gillespie GY, Li
X, Shacka JJ, Lu L, Zheng L and King PH: Tristetraprolin
downregulates interleukin 8 and vascular endothelial growth factor
in malignant glioma cells. Cancer Res. 68:674–682. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Young LE, Sanduja S, Bemis-Standoli K,
Pena EA, Price RL and Dixon DA: The mRNA binding proteins HuR and
tristetraprolin regulate cyclooxygenase 2 expression during colon
carcinogenesis. Gastroenterology. 136:1669–1679. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anderson P: Post-transcriptional control
of cytokine production. Nat Immunol. 9:353–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Carrick DM and Balckshear PJ: Comparative
expression of tristetraprolin (TTP) family member transcripts in
normal human tissues and cancer cell lines. Arch Biochem Biophys.
462:278–285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gaudet F, Hodgson G, Eden A,
Jackson-Grusby L, Dausman J, Gray JW, Leohardt H and Jaenisch R:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fuso A, Nicolia V, Cavallaro RA and Scarpa
S: DNA methylase and demethylase activities are modulated by
one-carbon metabolism in Alzheimer's disease models. J Nutr
Biochem. 22:242–251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
López-Pedrera C, Pérez-Sánchez C,
Ramos-Casals M, Santos-Gonzalez M, Rodriguez-Ariza A and Cuadrado
MJ: Cardiovascular risk in systemic autoimmune diseases. Epigenetic
mechanisms of immune regulatory functions. Clin Dev Immunol.
2012:9746482012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sacconi S, Camaño P, de Greef JC, Lemmers
RJ, Salviati L, Boileau P, Lopez de Munain Arregui A, van der
Maarel SM and Desnuelle C: Patients with a phenotype consistent
with facioscapulohumeral muscular dystrophy display genetic and
epigenetic heterogeneity. J Med Genet. 49:41–46. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chik F and Szyf M: Effects of specific
DNMT gene depletion on cancer cell transformation and breast cancer
cell invasion; Toward selective DNMT inhibitors. Carcinogenesis.
32:224–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hardy TM and Tollefsbol TO: Epigenetic
diet: Impact on the epigenome and cancer. Epigenomics. 3:503–518.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khan SI, Aumsuwan P, Khan IA, Walker LA
and Dasmahapatra AK: Epigenetic events associated with breast
cancer and their prevention by dietary components targeting the
epigenome. Chem Res Toxicol. 25:61–73. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gerhauser C: Cancer chemoprevention and
nutriepigenetics: State of the art and future challenges. Top Curr
Chem. 329:73–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang Z, Huang Q, Ji L, Wang Y, Qi X, Liu
L, Liu Z and Lu L: Epigenetic regulation of active Chinese herbal
components for cancer prevention and treatment. A follow-up review.
Pharmacol Res. 114:1–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kala R, Shah HN, Martin SL and Tollefsbol
TO: Epigenetic-based combinatorial resveratrol and pterostilbene
alters DNA damage response by affecting SIRT1 and DNMT enzyme
expression, including SIRT1-dependent γ-H2AX and telomerase
regulation in triple-negative breast cancer. BMC Cancer.
15:6722015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mirza S, Sharma G, Parshad R, Gupta SD,
Pandya P and Ralhan R: Expression of DNA methyltransferases in
breast cancer patients and to analyze the effect of natural
compounds on DNA methyltransferases and associated proteins. J
Breast Cancer. 16:23–31. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Garvina S, Ollinger K and Dabrosin C:
Resveratrol induces apoptosis and inhibits angiogenesis in human
breast cancer xenografts in vivo. Cancer Lett. 231:113–122. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu YZ, Wu K, Huang J, Liu Y, Wang X, Meng
ZJ, Yuan SX, Wang DX, Luo JY, Zuo GW, et al: The PTEN/PI3K/Aktand
Wnt/β-catenin signaling pathways are involved in the inhibitory
effect of resveratrol on human colon cancer cell proliferation. Int
J Oncol. 45:104–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee M, Choi B, Kundu JK, Shin YK, Na HK
and Surh YJ: Resveratrol suppresses growth of human ovarian cancer
cells in culture and in a murine xenograft model: Eukaryotic
elongation factor 1A2 as a potential target. Cancer Res.
69:7449–7458. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiao Y, Li H, Liu Y, Guo A, Xu X, Qu X,
Wang S, Zhao J, Li Y and Cao Y: Resveratrol inhibits the invasion
of glioblastoma-initiating cells via down-regulation of the
PI3K/Akt/NF-κB signaling pathway. Nutrients. 7:4383–4402. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang T, Zhang J, Zhou J, Zhu M, Wang L and
Yan L: Resveratrol inhibits interleukin-6 induced invasion of human
gastric cancer cells. Biomed Pharmacother. 99:766–773. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Oi N, Jeong CH, Nadas J, Cho YY, Pugliese
A, Bode A and Dong Z: Resveratrol, a red Wine polyphenol,
suppresses pancreatic cancer by inhibiting leukotriene A4
hydrolase. Cancer Res. 70:9755–9763. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qin W, Zhang K, Clarke K, Weiland T and
Sauter ER: Methylation and miRNA effects of resveratrol on mammary
tumors vs. normal tissue. Nutr Cancer. 66:270–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lee H, Zhang P, Herrmann A, Yang C, Xin H,
Wang Z, Hoon DS, Forman SJ, Jove R, Riggs AD, et al: Acetylated
STAT3 is crucial for methylation of tumor-suppressor gene promoter
and inhibition by resveratrol results in demethylation. Proc Natl
Acad Sci USA. 109:7765–7769. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stefanska B, Salamé P, Bednarek A and
Fabianowska-Majewska K: Comparative effects of retinoic acid,
vitamin D and resveratrol alone and in combination with adenosine
analogues on methylation and expression of phosphatase and tensin
homologue tumour suppressor gene in breast cancer cells. Br J Nutr.
107:781–790. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Papoutsis AJ, Borg JL, Selmin OI and
Romagnolo DF: BRCA-1 promoter hypermethylation and silencing
induced by the aromatic hydrocarbon receptor-ligand TCDD are
prevented by resveratrol in MCF-7 cells. J Nurt Biochem.
23:1324–1332. 2012. View Article : Google Scholar
|
|
56
|
Ohshiro K, Rayala SK, Kondo S, Gaur A,
Vadlamudi RK, El-Naggar AK and Kumar R: Identifying the estrogen
receptor coactivator PELP1 in autophagosomes. Cancer Res.
67:8164–8171. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Whyte L, Huang YY, Torres K and Mehta RG:
Molecular mechanisms of resveratrol action in lung cancer cells
using dual protein and microarray analyses. Cancer Res.
67:12007–12017. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ebi H, Tomida S, Takeuchi T, Arima C, Sato
T, Mitsudomi T, Yatabe Y, Osada H and Takahashi T: Relationship of
deregulated signaling converging onto mTOR with prognosis and
classification of lung adenocarcinoma shown by two independent in
silico analyses. Cancer Res. 69:4027–4035. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wu JY, Tsai KW, Shee JJ, Li YZ, Chen CH,
Chuang JJ and Liu YW: 4-Chloro-3,5-dihydroxystilbene, a resveratrol
derivative, induces lung cancer cell death. Acta Pharmacol Sin.
31:81–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dinkova-Kostova AT, Holtzclaw WD and
Wakabayashi N: Keap1, the sensor for electrophiles and oxidants
that regulates the phase 2 response, is a zinc metalloprotein.
Biochemistry. 44:6889–6899. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim JH, Park EY, Ha HK, Jo CM, Lee WJ, Lee
SS and Kim JW: Resveratrol-loaded nanoparticles induce antioxidant
activity against oxidative stress. Asian Australas J Anim Sci.
29:288–298. 2016. View Article : Google Scholar : PubMed/NCBI
|