Introduction
Deubiquitinating enzymes (DUBs) play a critical
function through regulation of protein-protein interactions,
protein subcellular localization and protein stability (1,2). Many
DUBs, such as USP11, USP22 and USP37 have been found to play
important functions in several types of cancers (3–7).
Recently, several reports have found that USP11
plays an important role in modulating a number of signaling
cascades, including NF-κB, TGF-β and Notch signaling pathways, and
the DNA damage response (5,8–11).
USP11 exhibits differential functions through interacting with
multiple substrates, such as promyelocytic leukemia protein (PML),
BRCA2 and IκBα (5,8–11). For
example, USP11 interacts with and deubiquitylates IκB, thereby
inhibiting NFκB activation (10). A
recent study showed that USP11 is upregulated in breast cancer, and
its abnormal accumulation predicts a poor patient prognosis
(6). However, the function of USP11
in ovarian cancer remains unknown.
Cancer metastasis is one of the important causes of
the poor outcomes of tumor patients and is a complex, multistep
process (12). Increasing motility
and invasiveness of cancer cells results in the initiation of
metastasis. To increase their invasive abilities, cancer cells
undergo physiological changes, including an
epithelial-to-mesenchymal transition (EMT) (13). EMT is a complex process in which
epithelial cells convert into a mesenchymal phenotype. In addition,
EMT plays crucial functions in tumor progression and metastasis.
Several signaling pathways, such as the tyrosine kinase receptors,
Wnt/β-catenin, TGF-β, PI3K/Akt pathways and Notch regulate and
induce EMT. One characteristic of EMT is the downregulation of
E-cadherin and upregulation of N-cadherin. Moreover, there are
multiple transcription factors that have been found to respond to
EMT, including Slug, Twist and Snail (14–16).
In this study, we verified that USP11 was
significantly upregulated in ovarian cancer tissues and cell lines.
In addition, high expression of USP11 was closely correlated with
tumor size, TNM stage, lymph node metastasis, and predicted a poor
prognosis of ovarian cancer patients. USP11 is a deubiquitinase,
and we found that it interacted with Snail, a key EMT-associated
transcription factor, and its deubiquitinase activity was dependent
on a stabilized Snail. Our research revealed the detailed mechanism
of USP11 in ovarian cancer, and suggests that it may be a novel
molecular therapy target for ovarian cancer.
Materials and methods
Tissue specimens
A total of 143 pairs of ovarian cancer tissues and
adjacent normal tissues were used for RT-qPCR and western blotting
analysis. These patients included 143 females aged between 25–84
years, with a mean of 40.3 years. None of the patients had received
chemotherapy or radiotherapy before surgery. The specimens were
immediately stored at −80°C before using. The Ethical Committees of
Linyi Central Hospital approved all human tissue experiments and
all patients provided written informed consent prior to
participation in the study.
Cell culture
Human ovarian cancer cell lines, OVCAR-3 and SKOV3
and human normal ovarian cell line IOSE80 were obtained from the
National Infrastructure of Cell Line Resource (Beijing, China). All
cells were cultured in HyClone™ Dulbecco's modified Eagle's medium
(DMEM; GE Healthcare Life Sciences, Logan, UT, USA) supplemented
with 100 U/ml penicillin, 100 g/ml streptomycin (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) (17). All cells were cultured at 37°C in a
humidified atmosphere containing 5% CO2.
Cell transfection
The vector (pcDNA3.1) and pcDNA3.1-USP11 were
purchased from Vigene Biosciences (Shandong, China), and
small-interfering RNA (siRNA) targeting USP11 and scramble siRNA
(SCR) were chemically synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The cells were grown to 70–80% confluence prior
to transfection in 6-well plates. For cell transfection, cells were
transfected with 2.5 µg plasmid or 50 nM siRNA using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. After transfection for 48 h, the
expression of USP11 was determined by qRT-PCR. USP11 siRNA-1:
5′-UUAUCUCAUCUUGAAAGAGUG-3′; USP11 siRNA-2:
5′-UGUUGUUGUUCAUGACAUGCA-3′; scramble siRNA (SCR):
5′-GAACCGTGTCTTCCTCAGTATC-3′.
Western blotting
After transfection for 48 h, cells were collected
and washed with cold-PBS three times, and lysed with RIPA buffer
(1% NP-40, 50 mM Tris pH 7.4, 0.1% SDS, 1 mM EDTA, 0.5% DOC and 50
mM NaCl) containing the protease inhibitor mixture (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) at 4°C for 45 min. Lysates were
centrifuged at 13,000 × g for 15 min at 4°C, and the supernatants
were removed to a new tube. The protein concentration was measured
by the Bradford assay (Thermo Fisher Scientific, Inc.). Proteins
(40 µg) were separated by 10% SDS-PAGE, and then transferred to
polyvinylidene fluoride membranes (PVDF; GE Healthcare,
Marlborough, MA, USA). Subsequently, the membranes were blocked
with 5% skimmed milk at room temperature for 1 h, followed by
incubation with specific antibodies at 4°C overnight. The
antibodies were as followed: USP11 (1:1,000; cat. no. ab109232;
Abcam, Cambridge, MA, USA), EMT Kit (1:1,000; cat. no. 9782; Cell
Signaling Technology, Inc., Danvers, MA, USA), β-actin (1:4,000;
cat. no. ab8226; Abcam). The membranes were washed with PBST three
times and incubated with HPR-conjugated second antibodies (1:5,000;
cat. nos. ab6721 and and 97023; Abcam) at room temperature for 1 h
and washed with PBST three times. Finally, the specific blots were
detected using ECL Western Blotting Detection Reagents (GE
Healthcare, Marlborough, MA, USA). The densitometry of blots were
analysized using ImageJ software (version 4.7; National Institutes
of Health, Bethesda, MD, USA).
Quantitative real-time PCR
(RT-qPCR)
Total mRNA was extracted from cells or tissue
samples using Invitrogen™ TRIzol reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions, and reverse
transcription reaction was conducted using the TransScript
First-Strand cDNA Synthesis SuperMix Kit (Transgen Biotech, Co.,
Ltd., Beijing, China). Subsequently, 2 µg cDNA samples were used to
perform RT-qPCR using TransStart Top Green qPCR SuperMix (Transgen
Biotech). Primers were designed as follows: E-cadherin forward,
5′-AAACATCATTGATGCAGACC-3′ and reverse,
5′-GATAGATTCTTGGGTTGGGTC-3′; fibronectin forward,
5′-AATGTGAACGACACATTCCA-3′ and reverse, 5′-ACCACTTGAGCTTGGATAGG-3′;
N-cadherin forward, 5′-CAAAGCCTGGAACATATGTG-3′; and reverse,
5′-GTTTGAAAGGCCATATGTGG-3′; Slug forward,
5′-ACACATACAGTGATTATTTCCC-3′ and reverse,
5′-ACTGTAGTCTTTCCTCTTCAT-3′; Snail forward,
5′-TCTAATCCAGAGTTTACCTTCCAG-3′ and reverse,
5′-TGAAGTAGAGGAGAAGGACGA-3′; Twist forward,
5′-GTACATCGACTTCCTCTACC-3′ and reverse,
5′-GAAACAATGACATCTAGGTCTC-3′; GAPDH forward,
5′-ATTTCCTGGTATGACAACGA-3′ and reverse,
5′-TTGATGGTACATGACAAGGTG-3′. GAPDH was used as an internal control.
The RT-qPCR conditions were as follows: 4 min at 98°C, denaturation
at 98°C for 30 sec, annealing at 56°C for 30 sec and extension at
72°C for 35 sec, performed for 32 cycles. The relative expression
of gene was analyzed by the 2−ΔΔC(q) method (18).
Co-immunoprecipitation (co-IP)
assay
Cells were collected and washed with PBS three
times, and the cells were subsequently lysed using 0.3% IP buffer
(1 mM EDTA, 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.3% NP-40) and
protease inhibitor cocktail at 4°C for 30 min. Subsequently, the
cell lysates were centrifuged at 13,000 × g for 30 min and the
supernatants were collected. Next, 2 µg USP11 antibody (cat. no.
ab109232; Abcam) and 2 µg IgG antibody (cat. no. ab190492; Abcam)
were added into the cell lysates, respectively. After rotation at
4°C overnight, 50 µl protein A beads (Thermo Fisher Scientific,
Inc.) was added. After rotation at 4°C for 3 h, the beads were
collected by centrifugation at 800 × g for 5 min. The beads were
next washed with 0.1% IP buffer (1 mM EDTA, 25 mM Tris-HCl pH 7.5,
150 mM NaCl, 0.1% NP-40) and a protease inhibitor cocktail three
times. After denaturing at 95°C for 10 min, the supernatants were
collected and used for western blot analysis.
Colony formation assay
After transfection for 48 h, ~5,000 cells were
placed in 6-well plates and cultured with serum-free media. Cells
were incubated at 37°C in a humidified atmosphere containing 5%
CO2 for 10 days. Cells were washed with PBS three times
and fixed with 10% methanol at room temperature for 15 min, and
stained with 1% crystal violet at room temperature for 10 min, then
the cells were washed with PBS. Finally, the number of colonies
(cells >50) were counted under an Olympus CKX41 light microscope
(Olympus Corp., Tokyo, Japan). Each experiment was repeated three
times.
CCK-8 assay
After transfection for 48 h, ~2,000 cells were
placed in 96-well plates and cultured with 200 µl media. Every 12
h, 20 µl CCK-8 regent was added and incubated with cells at 37°C in
a humidified atmosphere containing 5% CO2 for 1 h. The
OD 450 was measured. The value of OD 450 indicated the cell
viability. Each experiment was repeated three times.
In vivo deubiquitination analysis
FLAG-Snail and HA-ubiquitin were transfected in
OVCAR-3/siUSP11 cells. After transfection for 48 h, cells were
treated with 10 µM MG132 for 6 h and then lysed with RIPA buffer.
The ubiquitinated status of Snail was detected through western
blotting analysis with the HA antibody.
In vitro deubiquitination
analysis
Briefly, HA-ubiquitin and FLAG-Snail were
co-expressed in OVCAR-3 cells. After transfection for 48 h, cells
were treated with 10 µM MG132 for 6 h and FLAG antibody was used to
isolated ubiquitinated Snail by IP analysis. In a parallel
experiment, vector or Myc-wt-USP11 or Myc-USP11/C318S were
expressed in OVCAR-3 cells, and purified by IP with anti-Myc
Affinity Matrix (Roche Diagnostics, Mannheim, Germany). Next,
Myc-USP11 was eluded with Myc peptide and dialyzed. Finally, the
purified USP11 was incubated with ubiquitinated Snail in a
deubiquitination reaction buffer [1 mM ATP, 5 mM MgCl2,
50 mM HEPES (pH 7.5), 5% glycerol, 1 mM DTT and 100 mM NaCl] at
30°C for 2 h. The ubiquitinated status of Snail was determined by
western blot analysis with an anti-HA antibody.
Transwell migration and invasion
assays
After transfection of the SKOV3 cells with the
plasmid or siRNA for 48 h, ~2×105 cells in serum-free
medium were placed into a chamber with Matrigel (8-µm pore; BD
Biosciences, San Diego, CA, USA) for invasion analysis or into the
chamber without Matrigel (8-µm pore; BD Biosciences) for migration
analysis. Then the chambers were put in the 24-well plates with
DMEM/F-12 supplemented with 10% FBS. The cells were incubated at
37°C with 5% CO2 for 18 h. The cells on the topside were
removed by a cotton swab and the cells on the underside were fixed
with 10% methanol at room temperature for 15 min, and stained with
1% crystal violet at room temperature for 10 min, and then washed
with water. Finally, the number of cells were counted under an
Olympus CKX41 light microscope (Olympus Corp.). Each experiment was
repeated three times.
Statistical analysis
Statistical analyses were performed using SPSS
software 18.0 (SPSS, Inc., Chicago, IL, USA). All data are
represented as the mean ± SD. The significant associations between
the expression of USP11 and clinicopathological parameters of
ovarian cancer patients were determined by two-tailed χ2
tests. Survival curve analysis was assessed using the Kaplan-Meier
method. Analysis of variance followed by Tukey's post hoc test was
used to assess the differences between multiple groups. The
differences between two groups were determined using two-tailed
Student's t-test. P<0.05 was considered to be a statistically
significant difference.
Results
Association between USP11 expression
and clinicopathological variables in the ovarian cancer cases
Previous research has found that USP11 is
upregulated in breast cancer (6,19). To
verify the expression of USP11 in ovarian cancer, we first used
RT-qPCR and western blot analysis to analyze the expression of
USP11 in 143 pairs of ovarian cancer tissues from patients who
received tumor resection at the Linyi Central Hospital during May
2013 and October 2016, and their paired adjacent normal tissues
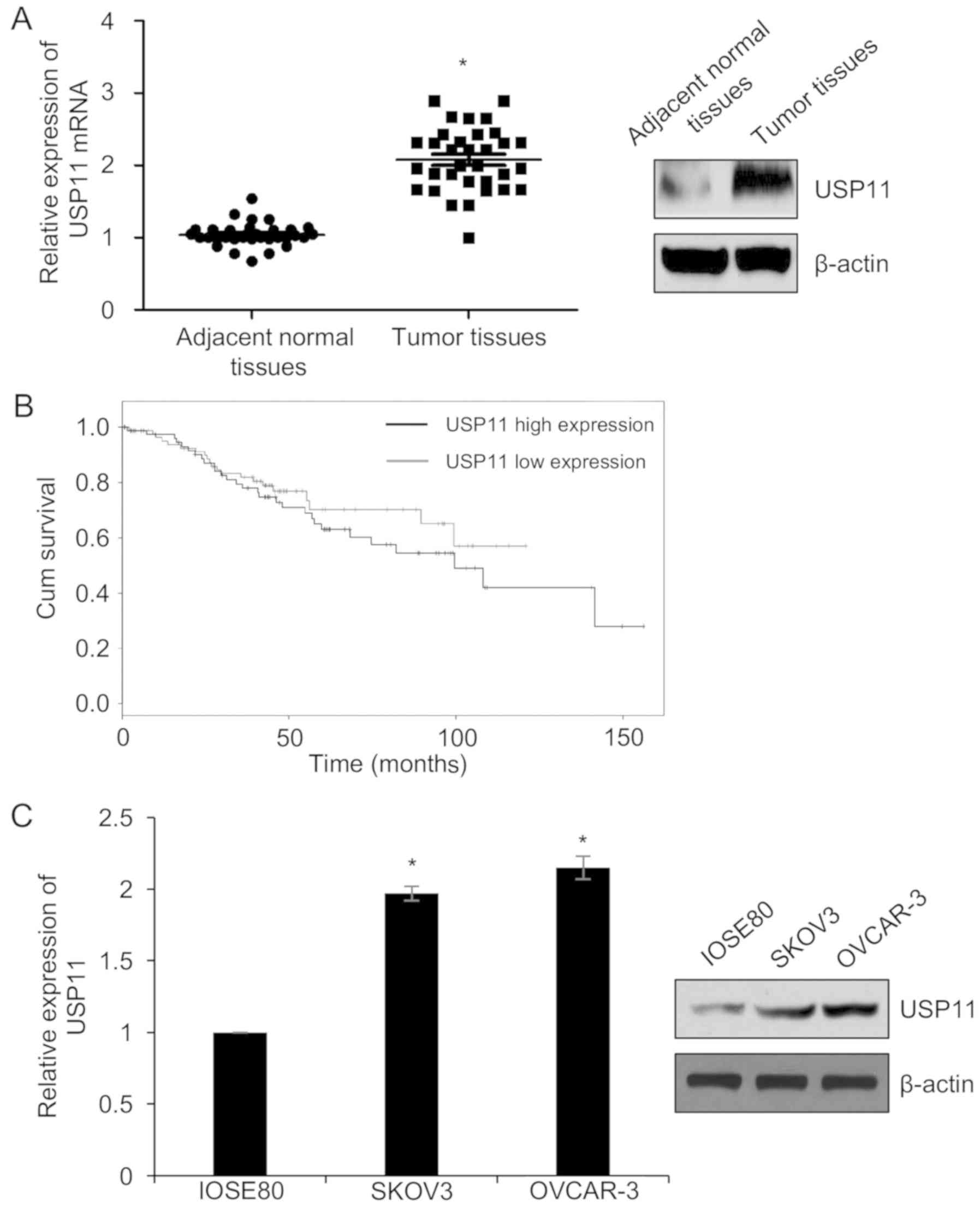
were used as the control group. As shown in Fig. 1A, we found that the mRNA and protein
levels of USP11 were clearly higher in tumor tissues than that in
adjacent normal tissues. Meanwhile, we next analyzed the
association between the expression of USP11 and the
clinico-pathological features of the ovarian cancer patients. The
results demonstrated that higher expression of USP11 was
significantly associated with tumor size, TNM stage, and the
presence of lymph node metastasis (Table I). However, no significant
associations were observed between the USP11 expression level and
patient age (Table I).
Subsequently, we analyzed the correlation between the expression of
USP11 and the survival curve of the ovarian cancer patients. As
shown in Fig. 1B, high expression
of USP11 predicted a poor prognosis of the patients. To investigate
the function of USP11 in ovarian cancer, we next detected the
expression of USP11 in ovarian cancer cell lines, including OVCAR-3
and SKOV3, and the human normal ovarian cell line IOSE80 was used
as a control. We found that USP11 was significantly upregulated in
OVCAR-3 and SKOV3 cells compared to that in the IOSE80 cells
(Fig. 1C). Together, our findings
showed that USP11 is upregulated in ovarian cancer.
 | Table I.Relationship between the expression of
USP11 and the clinicopathological features of the ovarian cancer
cases (n=143). |
Table I.
Relationship between the expression of
USP11 and the clinicopathological features of the ovarian cancer
cases (n=143).
|
|
| USP11 protein
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. of cases | Low (n=56) | High (n=87) | P-value |
|---|
| Age (years) |
|
|
| 0.367 |
|
<40 | 68 | 24 | 44 |
|
| ≥40 | 75 | 32 | 43 |
|
| Tumor size (cm)
(diameter) |
|
|
| 0.002 |
| Small
(≤3) | 69 | 36 | 33 |
|
| Large
(≥3) | 74 | 20 | 54 |
|
| TNM stage |
|
I–II | 76 | 36 | 40 | 0.032 |
|
III–IV | 67 | 20 | 47 |
|
| pT status |
|
|
| 0.014 |
|
pT1 | 71 | 35 | 36 |
|
|
pT2-4 | 72 | 21 | 51 |
|
| pN status |
|
|
| 0.011 |
|
pN0 | 63 | 32 | 31 |
|
|
pN1-2 | 80 | 24 | 56 |
|
| Metastasis |
|
|
| 0.010 |
|
Yes | 65 | 18 | 47 |
|
| No | 78 | 38 | 40 |
|
Knockdown of USP11 suppresses the
migration and invasion of ovarian cancer cell lines
Because high USP11 expression in ovarian cancer
tissues was associated with lymph node metastasis, we assumed that
USP11 may also regulate the migration and invasion of ovarian
cancer cells. To investigate the effect of USP11 on cell migration
and invasion in ovarian cancer, we first over-expressed or knocked
down USP11 in SKOV3 cells and RT-qPCR and western blotting were
used to determine the expression of USP11. As shown in Fig. 2A and B, USP11 was significantly
over-expressed or knocked down in the SKOV3/USP11 group or the
SKOV3/siUSP11 cell group, respectively. The USP11 siRNA#1
(siUSP11#1) showed more silencing efficiency than siRNA#2
(siUSP11#2); thus, siUSP11#1 was used for further experiments.
Subsequently, Transwell migration and invasion analyses were
performed to determine the effect of USP11 on migration and
invasion. The results of the Transwell migration assay showed that
ectopic expression of USP11 increased the number of migrating
cells, compared with the vector group, whereas knockdown of USP11
significantly decreased the number of migrating cells, compared
with the scramble siRNA (SCR) group (Fig. 2C). Similar results were observed for
the Transwell invasion assay. Ectopic expression of USP11 promoted
cell invasion, compared with the vector group, whereas knockdown of
USP11 significantly suppressed cell invasion, compared with the
scramble siRNA (SCR) group (Fig.
2D). The above experiments suggest that USP11 promotes cell
migration and invasion in ovarian cancer cells.
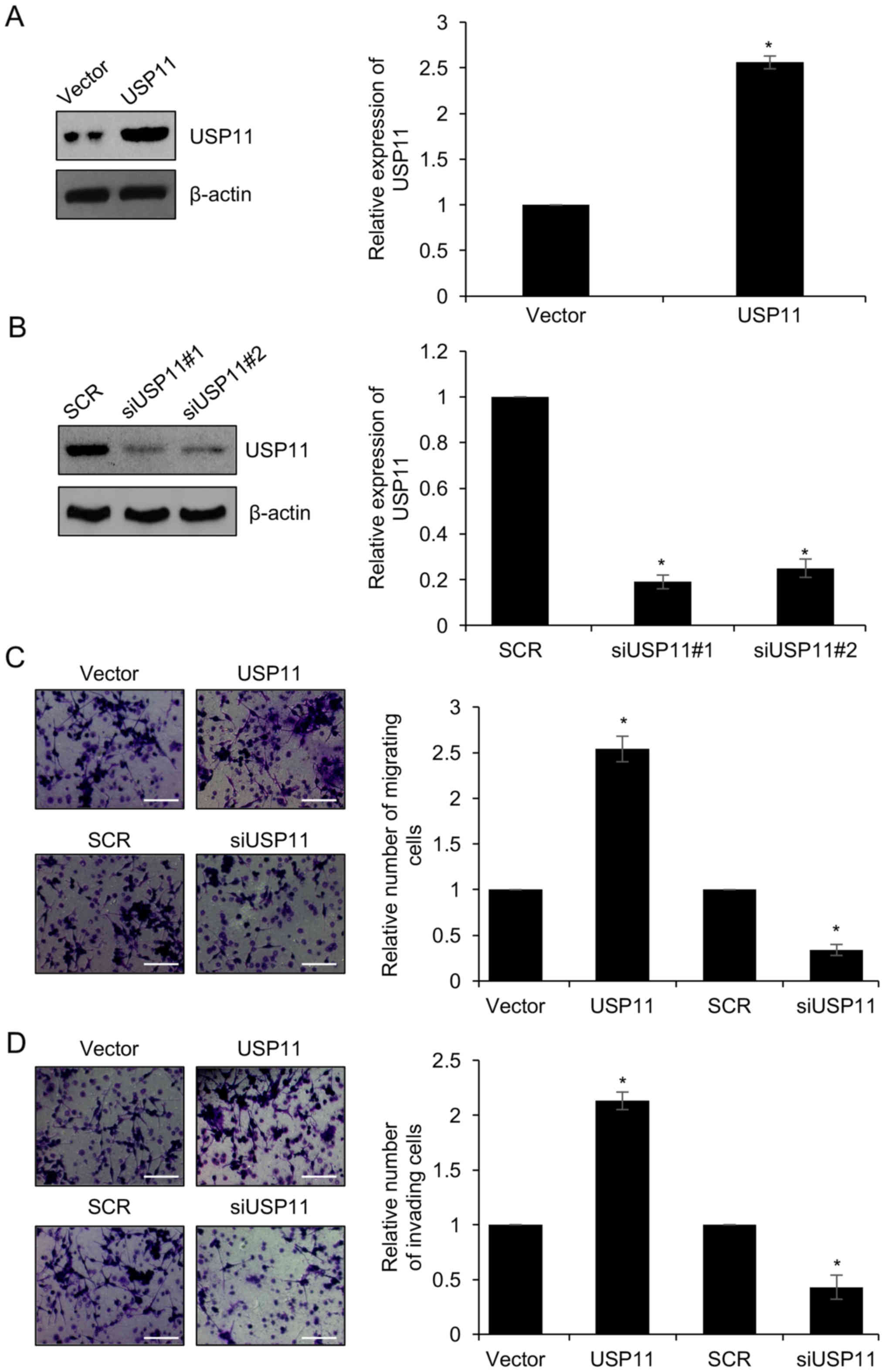 | Figure 2.Knockdown of USP11 suppresses the
migration and invasion of ovarian cell lines. (A) SKOV3 cells were
transfected with empty vector or USP11 expression plasmid for 48 h,
and the mRNA and protein levels of USP11 were examined by RT-qPCR
and western blotting. *P<0.05, USP11 vs. Vector. (B) SKOV3 cells
were transfected with scramble siRNA (SCR) or USP11 siRNA#1
(siUSP11#1) and USP11 siRNA#2 (siUSP11#2) for 48 h, and the mRNA
and protein levels of USP11 were examined by RT-qPCR and western
blotting. *P<0.05, siUSP11 vs. SCR. (C) USP11 was overexpressed
or knocked down in SKOV3 cells. Representative images showing the
migration of SKOV3 cells. The relative number of tumor cells is
quantified on the right. Magnification, ×20; scale bar, 100 µm.
*P<0.05, USP11 vs. Vector, siUSP11 vs. SCR. (D) USP11 was
overexpressed or knocked down in SKOV3 cells. Representative images
showing the invasion of SKOV3 cells. The relative number of tumor
cells is quantified on the right. Magnification, ×20; scale bar,
100 µm. *P<0.05, USP11 vs. Vector, siUSP11 vs. SCR. USP11,
ubiquitin specific peptidase 11. |
USP11 induces an
epithelial-to-mesenchymal transition (EMT) in ovarian cancer
cells
To further determine whether USP11 regulates EMT in
ovarian cancer cells, we overexpressed or knocked down USP11 in
OVCAR-3 cells, and RT-qPCR and western blotting were used to
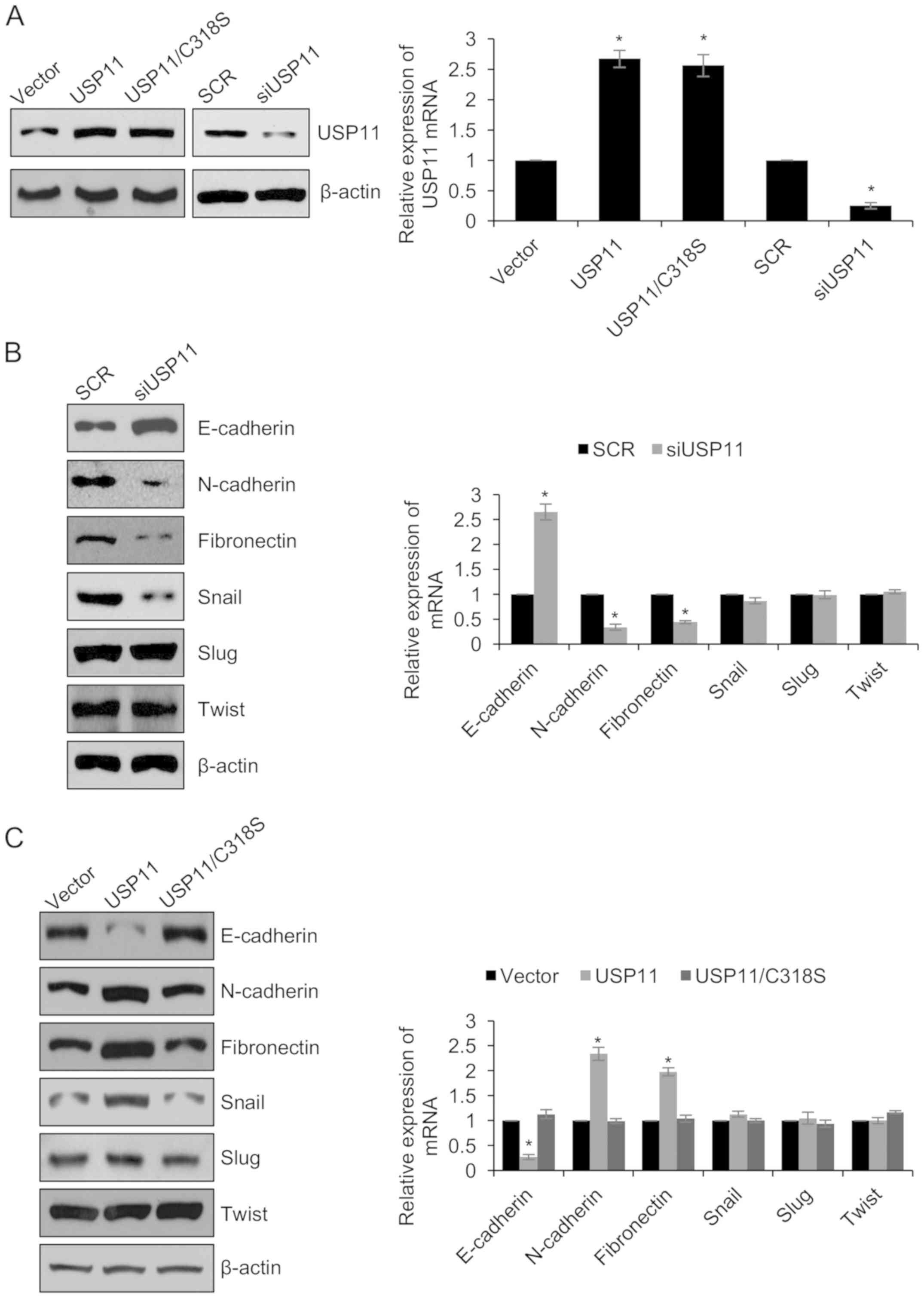
determine the expression of USP11. As shown in Fig. 3A, USP11 was significantly
overexpressed or knocked down in the OVCAR-3/USP11 and
OVCAR-3/siUSP11 group, respectively. Next, we examined the
expression of epithelial marker, E-cadherin, and the expression of
the mesenchymal markers, N-cadherin and fibronectin in OVCAR-3
cells. As shown in Fig. 3B,
E-cadherin expression was increased, while N-cadherin and
fibronectin were decreased in the OVCAR-3/siUSP11 cells compared to
that of the SCR control cells (Fig.
3B). Meanwhile, E-cadherin expression was decreased, while
N-cadherin and fibronectin expression were increased in the
OVCAR-3/USP11 cells compared to that of the vector control cells
(Fig. 3C). To investigate whether
USP11 regulates EMT through its deubiquitinase activity, we
overexpressed USP11/C318S which has no catalytic activity in
OVCAR-3 cells (Fig. 3A). Notably,
USP11/C318S failed to regulate EMT in OVCAR-3 cells (Fig. 3C). That indicates that USP11
regulated EMT is dependent on its deubiquitinase activity.
EMT-associated transcription factors, including the
helix-loop-helix transcription factor Twist, and the zinc-finger
containing proteins Slug and Snail, repress E-cadherin expression
and induce EMT (20). To further
explore the mechanism involving the regulation of EMT by USP11, we
next assessed whether USP11 also regulates these transcription
factors. As shown in Fig. 3B and C,
the protein level of Snail was decreased when USP11 was knocked
down, whereas ectopic expression of wild-type-USP11 (USP11)
increased the expression of Snail at the protein level and
USP11/C318S failed to increase the expression of Snail. At the mRNA
level, both USP11 and USP11/C318S had no effect on the expression
of Snail (Fig. 3B and C). In
addition, Slug and Twist showed little change when USP11 was
overexpressed or knocked down (Fig. 3B
and C). The above results indicate that USP11 regulates Snail
through a post-transcriptional mechanism.
USP11 interacts with Snail and
stabilizes Snail through its deubiquitinase activity
To further decipher the detailed mechanism of USP11
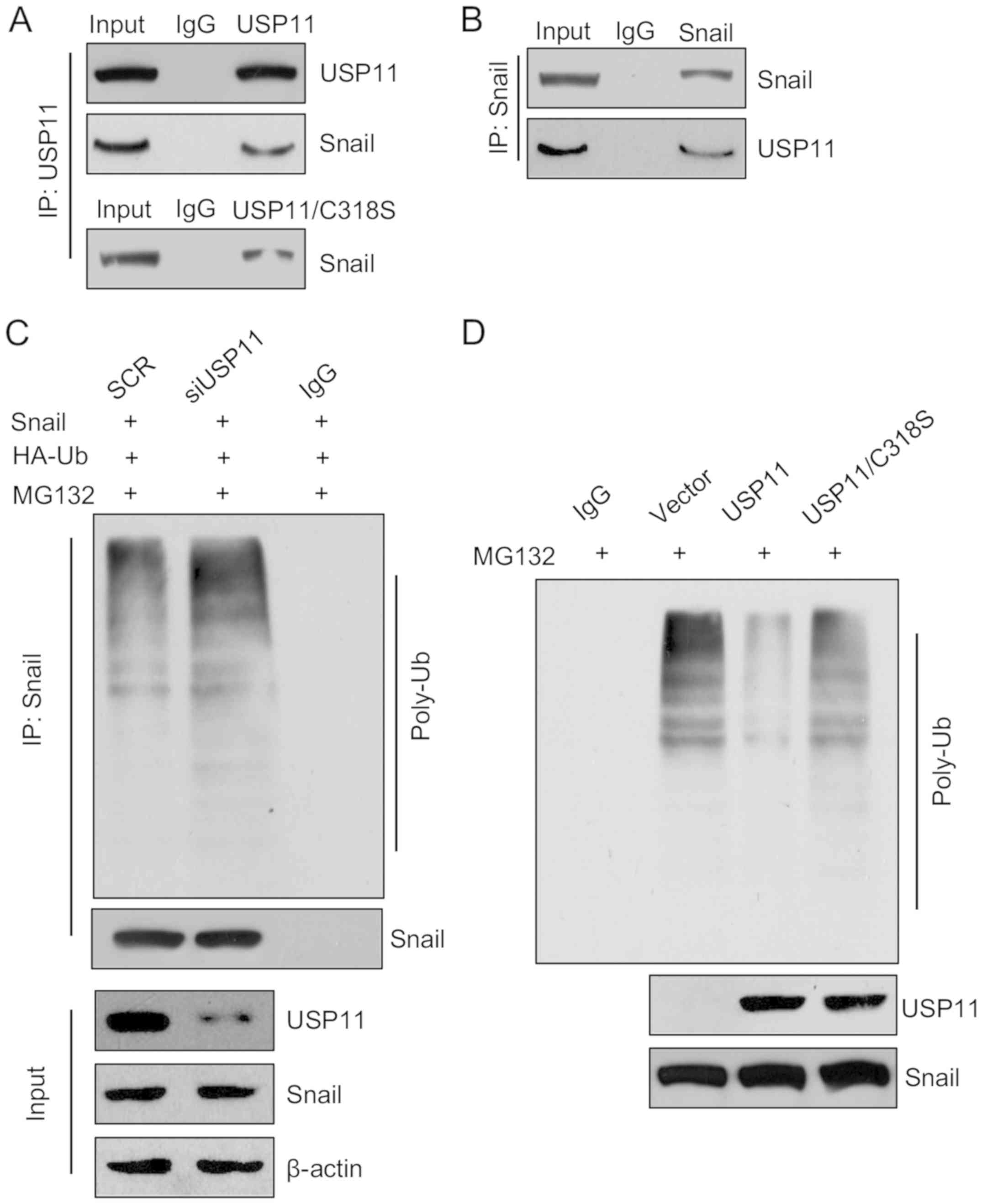
on the EMT, we performed co-immunoprecipitation (co-IP) analysis to
assess whether Snail interacted with USP11 or USP11/C318S. The
results indicated that both USP11 and USP11/C318S could interact
with Snail (Fig. 4A). Meanwhile,
reciprocal immunoprecipitation with anti-Snail and immunoblotting
with anti-USP11 also revealed that Snail could interact with USP11
(Fig. 4B). In brief, our research
suggests USP11 is physically associated with Snail. USP11 as a
deubiquitinase has been found to deubiquitinate promyelocytic
leukemia protein (PML), BRCA2 and IκBα (5,8–11);
thus, we hypothesized USP11 may regulate the expression of Snail
through its deubiquitinase activity. Subsequently, we performed
in vitro and in vivo deubiquitination assays. The
results revealed that Snail was deubiquitinated by USP11, the
ubiquitination of Snail was increased following inhibition of
USP11; however, ectopic expression of USP11 decreased the
ubiquitination of Snail, and USP11/C318S had no effect (Fig. 4C and D). Together, the above results
indicate that USP11 interacts with Snail and stabilizes Snail
through its deubiquitinase activity.
USP11 promotes cell proliferation in
ovarian cancer cells
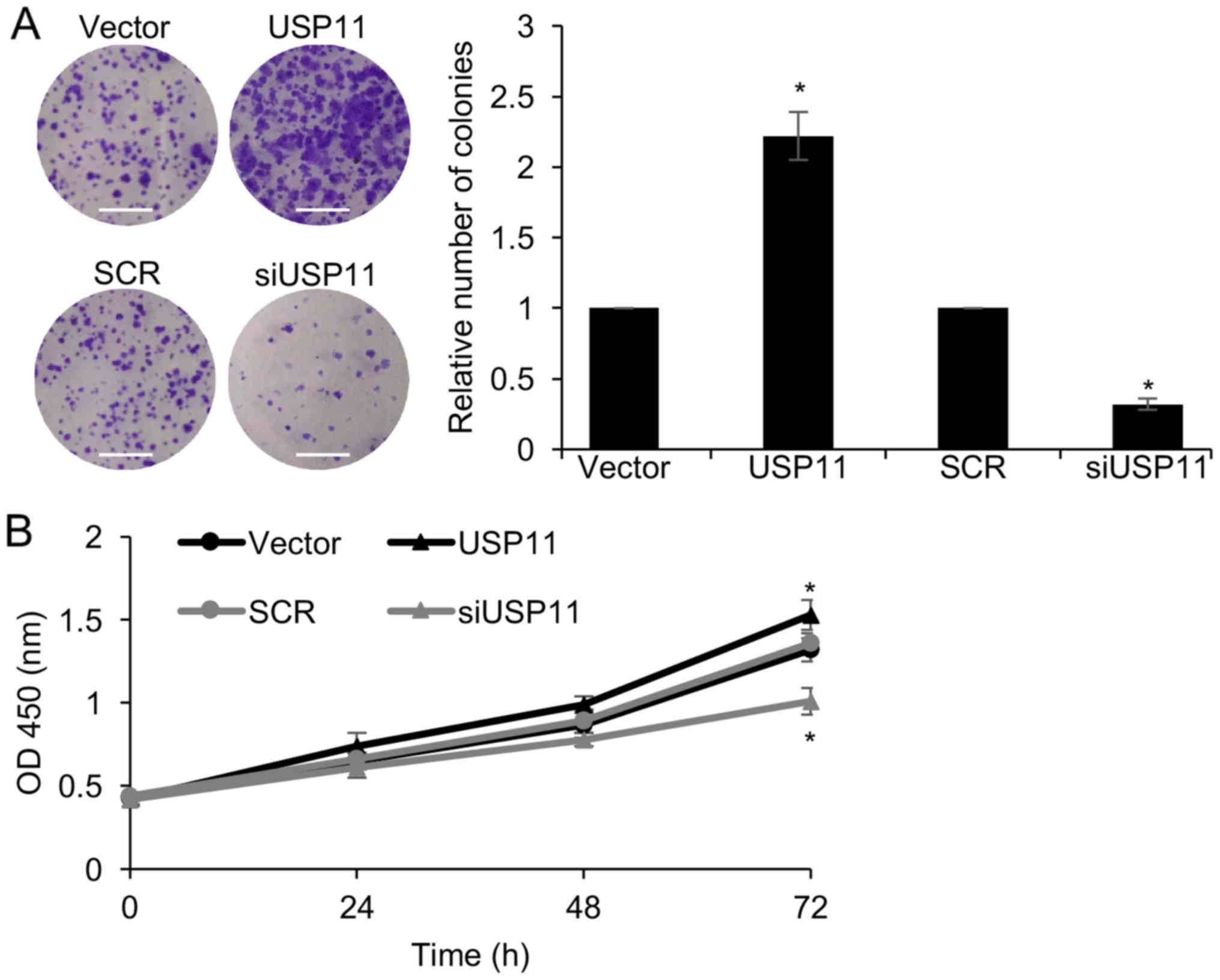
Since the expression of USP11 was associated with
tumor size (Table I), we next
explored the function of USP11 on cell proliferation. Colony
formation and CCK-8 assays were performed. The results of the
colony formation assays indicated that inhibition of USP11
significantly reduced the number of colonies, compared with the SCR
group; however, ectopic expression of USP11 significantly increased
the number of colonies, compared with the vector group (Fig. 5A). In addition, the similar results
were observed for the CCK-8 assay. Knockdown of USP11 inhibited
cellular proliferation and ectopic expression of USP11 facilitated
cellular proliferation (Fig. 5B).
In conclusion, USP11 promotes cell proliferation in ovarian cancer
cells.
Discussion
USP11 has been found to be upregulated in breast
cancer, and its abnormal accumulation predicts a poor prognosis of
breast cancer (6). Yet, the
expression and function of USP11 in ovarian cancer remain
unknown.
The results of RT-qPCR and western blotting analyses
revealed that USP11 was upregulated in ovarian cancer tissues and
cell lines. This finding was similar with previous research about
USP11 in breast cancer (6). In
addition, we also found that high expression of USP11 was closely
associated with tumor size, TNM stage and lymph node metastasis.
The present findings indicate that USP11 plays an important role in
ovarian cancer, thus it is necessary to investigate the detailed
function of USP11 in ovarian cancer.
The epithelial-to-mesenchymal transition (EMT) is a
complex process that is closely related to tumor metastasis. One
characteristic of EMT is the gain of mesenchymal markers, such as
N-cadherin, and the loss of epithelial markers, such as E-cadherin.
This changes the morphology of the cancer cells and improves the
invasive and migratory capabilities of cancer cells. Several
reports have shown that multiple transcription factors play key
role in EMT, such as Snail (21–25).
Abnormal expression of Snail is found in different tumor types, and
its expression correlates with tumor aggressiveness (14,26),
and facilitates tumor recurrence (25). Here, we found that USP11 promoted
the EMT in ovarian cancer cells through regulation of Snail. USP11
as a deubiquitinase has been found to interact with many proteins
and stabilize them through its deubiquitinase activity. IP analysis
showed that USP11 interacted with Snail and in vitro and
in vivo deubiquitination assays revealed that USP11
stabilized Snail by deubiquitinating it. Meanwhile, Transwell
migration and invasion assays demonstrated that USP11 enhanced the
invasive and migratory capabilities of ovarian cancer cells
dependent on its deubiquitinase activity.
Notably, previous research indicated that USP11
suppressed cell proliferation through regulation of Mgl-1 protein
(27). However, we found the
expression of USP11 was associated with a larger tumor size, and
knockdown of USP11 inhibited cell proliferation in ovarian cancer
cells. This suggests that USP11 may play a different role in
different tumors through interacting with different proteins.
Snail has been found to be strongly regulated at the
post-translational level, and it may be degraded rapidly after its
initial synthesis (28). Like some
other growth regulatory proteins such as cyclin D1, Myc as well as
β-catenin, the degradation of Snail is associated with
phosphorylation and poly-ubiquitination (28). Moreover, previous research indicates
that Snail is acetylated by CREB-binding protein (CBP) (29). In particular, poly-ubiquitination
can both suppress EMT by destabilizing Snail and, at the same time,
enhance EMT through auto-ubiquitination of the E3 ubiquitin ligase
tumor necrosis factor receptor-associated factor 6 (TRAF6)
(30). Here, we found that USP11
could stabilize Snail through its deubiquitinase activity.
In conclusion, our research revealed that USP11
promoted EMT in ovarian cancer by stabilizing the EMT-associated
transcription factor Snail, thereby facilitating cancer cell
migration and invasion. Our results suggest USP11 as a novel
molecular therapy target for ovarian cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WW and YL conceived and designed the study. WW, JW,
KZ and HY performed the experiments and analyzed the data. WW and
KZ wrote the paper. WW and YL reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The Ethical Committees of Linyi Central Hospital
approved all human tissue experiments and all patients provided
written informed consent prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Skaar JR, Pagan JK and Pagano M: SCF
ubiquitin ligase-targeted therapies. Nat Rev Drug Discov.
13:889–903. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lipkowitz S and Weissman AM: RINGs of good
and evil: RING finger ubiquitin ligases at the crossroads of tumour
suppression and oncogenesis. Nat Rev Cancer. 11:629–643. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim D, Hong A, Park HI, Shin WH, Yoo L,
Jeon SJ and Chung KC: Deubiquitinating enzyme USP22 positively
regulates c-Myc stability and tumorigenic activity in mammalian and
breast cancer cells. J Cell Physiol. 232:3664–3676. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang QX, Wang XC, Chen SP and Qin XT:
Predictive value of deubiquitination enzymes USP37 in the prognosis
of breast cancer. Zhonghua Yi Xue Za Zhi. 96:944–948. 2016.(In
Chinese). PubMed/NCBI
|
|
5
|
Schoenfeld AR, Apgar S, Dolios G, Wang R
and Aaronson SA: BRCA2 is ubiquitinated in vivo and interacts with
USP11, a deubiquitinating enzyme that exhibits prosurvival function
in the cellular response to DNA damage. Mol Cell Biol.
24:7444–7455. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bayraktar S, Gutierrez Barrera AM, Liu D,
Pusztai L, Litton J, Valero V, Hunt K, Hortobagyi GN, Wu Y, Symmans
F, et al: USP-11 as a predictive and prognostic factor following
neoadjuvant therapy in women with breast cancer. Cancer J.
19:10–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim MS, Yoo KJ, Kang I, Chung HM and Baek
KH: A novel cysteine protease HeLa DUB-1 responsible for cleaving
the ubiquitin in human ovarian cancer cells. Int J Oncol.
25:373–379. 2004.PubMed/NCBI
|
|
8
|
Ramakrishna S, Suresh B and Baek KH: The
role of deubiquitinating enzymes in apoptosis. Cell Mol Life Sci.
68:15–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu HC, Lin YC, Liu CH, Chung HC, Wang YT,
Lin YW, Ma HI, Tu PH, Lawler SE and Chen RH: USP11 regulates PML
stability to control Notch-induced malignancy in brain tumours. Nat
Commun. 5:32142014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun W, Tan X, Shi Y, Xu G, Mao R, Gu X,
Fan Y, Yu Y, Burlingame S, Zhang H, et al: USP11 negatively
regulates TNFalpha-induced NF-kappaB activation by targeting on
IkappaBalpha. Cell Signal. 22:386–394. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Al-Salihi MA, Herhaus L, Macartney T and
Sapkota GP: USP11 augments TGFbeta signalling by deubiquitylating
ALK5. Open Biol. 2:1200632012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gupta GP and Massague J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagaishi M, Nakata S, Ono Y, Hirata K,
Tanaka Y, Suzuki K, Yokoo H and Hyodo A: Tumoral and stromal
expression of Slug, ZEB1, and ZEB2 in brain metastasis. J Clin
Neurosci. 46:124–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grzegrzolka J, Biala M, Wojtyra P,
Kobierzycki C, Olbromski M, Gomulkiewicz A, Piotrowska A, Rys J,
Podhorska-Okolow M and Dziegiel P: Expression of EMT markers SLUG
and TWIST in breast cancer. Anticancer Res. 35:3961–3968.
2015.PubMed/NCBI
|
|
17
|
Soule HD, Vazguez J, Long A, Albert S and
Brennan M: A human cell line from a pleural effusion derived from a
breast carcinoma. J Natl Cancer Inst. 51:1409–1416. 1973.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Z, Luo A, Shrivastava I, He M, Huang
Y, Bahar I, Liu Z and Wan Y: Regulation of XIAP turnover reveals a
role for USP11 in promotion of tumorigenesis. EBioMedicine.
15:48–61. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng H and Kang Y: Multilayer control of
the EMT master regulators. Oncogene. 33:1755–1763. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vetter G, Le Bechec A, Muller J, Muller A,
Moes M, Yatskou M, Al Tanoury Z, Poch O, Vallar L and Friederich E:
Time-resolved analysis of transcriptional events during
SNAI1-triggered epithelial to mesenchymal transition. Biochem
Biophys Res Commun. 385:485–491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Naber HP, Drabsch Y, Snaar-Jagalska BE,
ten Dijke P, van Laar T, Snail and Slug: key regulators of
TGF-β-induced EMT, are sufficient for the induction of single-cell
invasion. Biochem Biophys Res Commun. 435:58–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Moody SE, Perez D, Pan TC, Sarkisian CJ,
Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD and
Chodosh LA: The transcriptional repressor Snail promotes mammary
tumor recurrence. Cancer Cell. 8:197–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blanco MJ, Moreno-Bueno G, Sarrio D,
Locascio A, Cano A, Palacios J and Nieto MA: Correlation of snail
expression with histological grade and lymph node status in breast
carcinomas. Oncogene. 21:3241–3246. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lim KH, Suresh B, Park JH, Kim YS,
Ramakrishna S and Baek KH: Ubiquitin-specific protease 11 functions
as a tumor suppressor by modulating Mgl-1 protein to regulate
cancer cell growth. Oncotarget. 7:14441–14457. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou BP, Deng J, Xia W, Xu J, Li YM,
Gunduz M and Hung MC: Dual regulation of Snail by
GSK-3beta-mediated phosphorylation in control of
epithelial-mesenchymal transition. Nat Cell Biol. 6:931–940. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hsu DS, Wang HJ, Tai SK, Chou CH, Hsieh
CH, Chiu PH, Chen NJ and Yang MH: Acetylation of snail modulates
the cytokinome of cancer cells to enhance the recruitment of
macrophages. Cancer Cell. 26:534–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gudey SK, Sundar R, Mu Y, Wallenius A,
Zang G, Bergh A, Heldin CH and Landström M: TRAF6 stimulates the
tumor-promoting effects of TGFβ type I receptor through
polyubiquitination and activation of presenilin 1. Sci Signal.
7:ra22014. View Article : Google Scholar : PubMed/NCBI
|