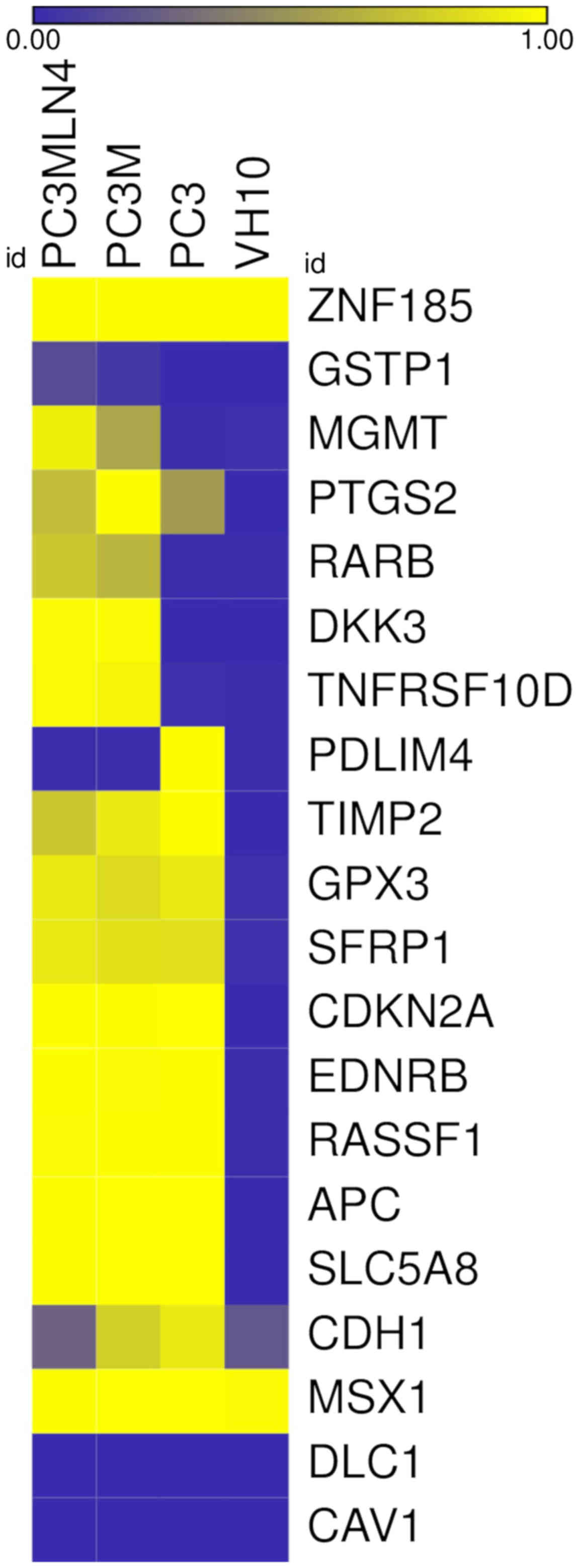
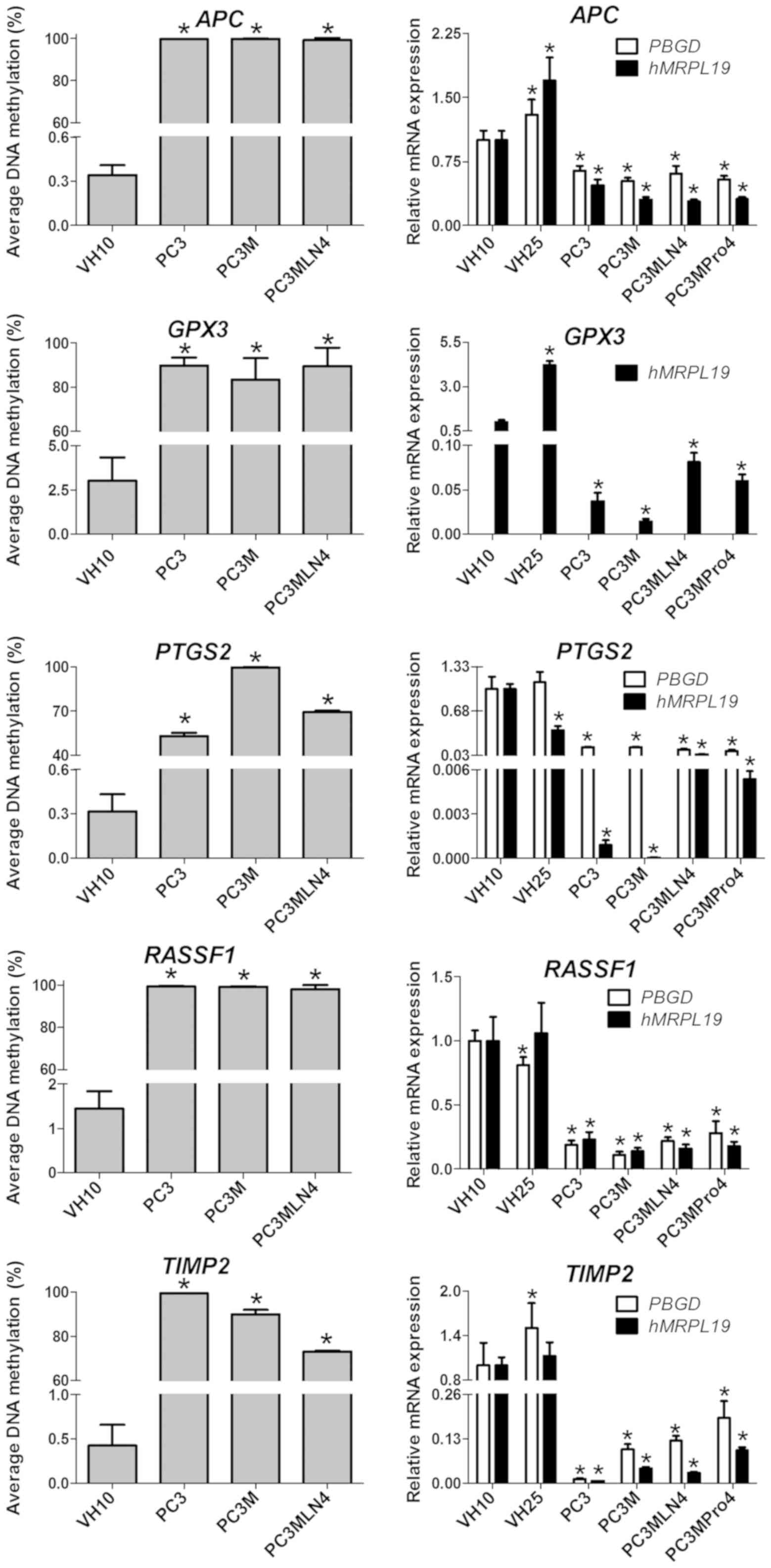
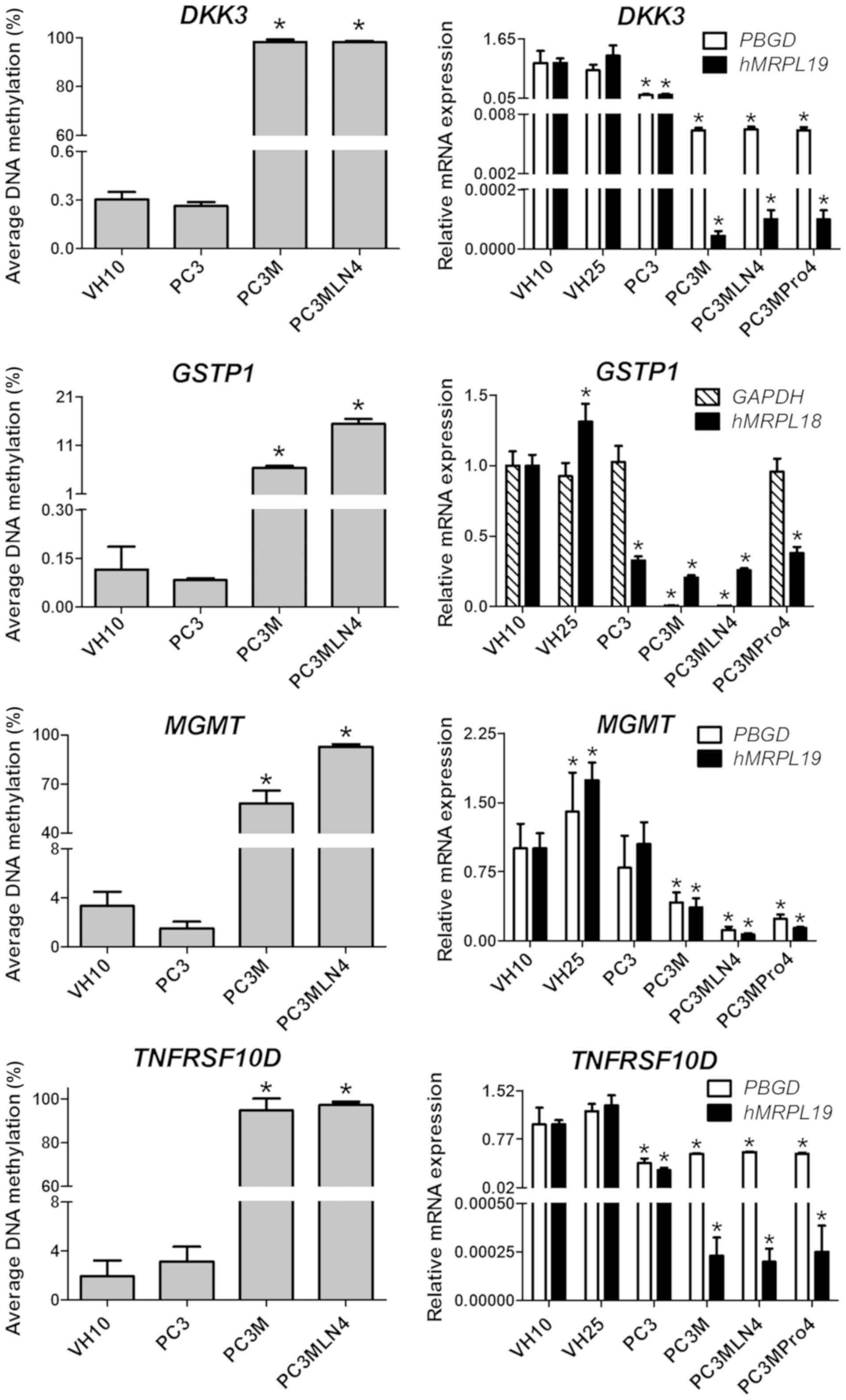
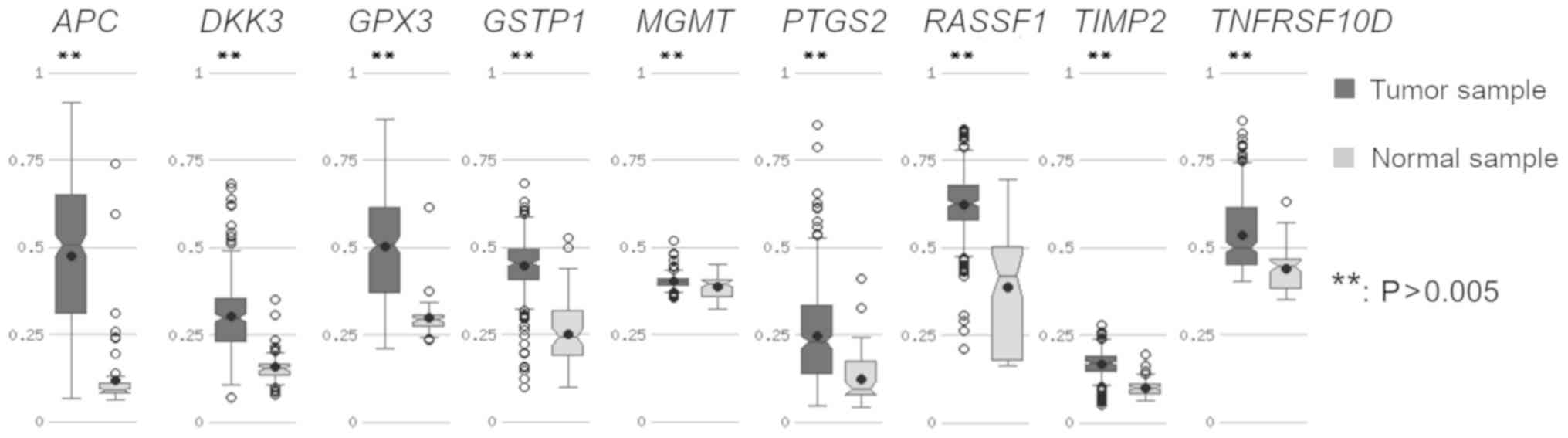
|
1
|
The Cancer Genome Atlas-National Cancer
Institute. https://cancergenome.nih.gov/2018 10 222011
|
|
2
|
Baylin SB and Herman JG: DNA
hypermethylation in tumorigenesis: Epigenetics joins genetics.
Trends Genet. 16:168–174. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang M and Park JY: DNA methylation in
promoter region as biomarkers in prostate cancer. Methods Mol Biol.
863:67–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Golub TR, Slonim DK, Tamayo P, Huard C,
Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri
MA, et al: Molecular classification of cancer: Class discovery and
class prediction by gene expression monitoring. Science.
286:531–537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van't Veer LJ, Dai H, van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salazar R, Roepman P, Capella G, Moreno V,
Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga
J, et al: Gene expression signature to improve prognosis prediction
of stage II and III colorectal cancer. J Clin Oncol. 29:17–24.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo L, Ma Y, Ward R, Castranova V, Shi X
and Qian Y: Constructing molecular classifiers for the accurate
prognosis of lung adenocarcinoma. Clin Cancer Res. 12:3344–3354.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toyota M, Ahuja N, Ohe-Toyota M, Herman
JG, Baylin SB and Issa JP: CpG island methylator phenotype in
colorectal cancer. Proc Natl Acad Sci USA. 96:8681–8686. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi H, Chen J, Li Y, Li G, Zhong R, Du D,
Meng R, Kong W and Lu M: Identification of a six microRNA signature
as a novel potential prognostic biomarker in patients with head and
neck squamous cell carcinoma. Oncotarget. 7:21579–21590.
2016.PubMed/NCBI
|
|
10
|
Voronkov A and Krauss S: Wnt/beta-catenin
signaling and small molecule inhibitors. Curr Pharm Des.
19:634–664. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu P, Rudick M and Anderson RG: Multiple
functions of caveolin-1. J Biol Chem. 277:41295–41298. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rayess H, Wang MB and Srivatsan ES:
Cellular senescence and tumor suppressor gene p16. Int J Cancer.
130:1715–1725. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Angst BD, Marcozzi C and Magee AI: The
cadherin superfamily: Diversity in form and function. J Cell Sci.
114:629–641. 2001.PubMed/NCBI
|
|
14
|
Kim TY, Vigil D, Der CJ and Juliano RL:
Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in
regulation of the cytoskeleton and cell motility. Cancer Metastasis
Rev. 28:77–83. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Niehrs C: Function and biological roles of
the Dickkopf family of Wnt modulators. Oncogene. 25:7469–7481.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arai H, Nakao K, Takaya K, Hosoda K, Ogawa
Y, Nakanishi S and Imura H: The human endothelin-B receptor gene.
Structural organization and chromosomal assignment. J Biol Chem.
268:3463–3470. 1993.PubMed/NCBI
|
|
17
|
Chen B, Rao X, House MG, Nephew KP, Cullen
KJ and Guo Z: GPx3 promoter hypermethylation is a frequent event in
human cancer and is associated with tumorigenesis and chemotherapy
response. Cancer Lett. 309:37–45. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayes JD and Pulford DJ: The glutathione
S-transferase supergene family: Regulation of GST and the
contribution of the isoenzymes to cancer chemoprotection and drug
resistance. Crit Rev Biochem Mol Biol. 30:445–600. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kaina B, Christmann M, Naumann S and Roos
WP: MGMT: Key node in the battle against genotoxicity,
carcinogenicity and apoptosis induced by alkylating agents. DNA
Repair. 6:1079–1099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bonito NA, Borley J, Wilhelm-Benartzi CS,
Ghaem-Maghami S and Brown R: Epigenetic regulation of the homeobox
gene MSX1 associates with platinum-resistant disease in high-grade
serous epithelial ovarian cancer. Clin Cancer Res. 22:3097–3104.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vanaja DK, Grossmann ME, Cheville JC, Gazi
MH, Gong A, Zhang JS, Ajtai K, Burghardt TP and Young CY: PDLIM4,
an actin binding protein, suppresses prostate cancer cell growth.
Cancer Invest. 27:264–272. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chandrasekharan NV and Simmons DL: The
cyclooxygenases. Genome Biol. 5:2412004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang D, Kryvenko ON, Mitrache N, Do KC,
Jankowski M, Chitale DA, Trudeau S, Rundle A, Belinsky SA and
Rybicki BA: Methylation of the RARB gene increases prostate cancer
risk in black Americans. J Urol. 190:317–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Volodko N, Gordon M, Salla M, Ghazaleh HA
and Baksh S: RASSF tumor suppressor gene family: Biological
functions and regulation. FEBS Lett. 588:2671–2684. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mo S, Su Z, Heng B, Chen W, Shi L, Du X
and Lai C: SFRP1 promoter methylation and renal carcinoma risk: A
systematic review and meta-analysis. J Nippon Med Sch. 85:78–86.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ganapathy V, Thangaraju M, Gopal E, Martin
PM, Itagaki S, Miyauchi S and Prasad PD: Sodium-coupled
monocarboxylate transporters in normal tissues and in cancer. AAPS
J. 10:193–199. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brew K and Nagase H: The tissue inhibitors
of metalloproteinases (TIMPs): An ancient family with structural
and functional diversity. Biochim Biophys Acta. 1803:55–71. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan G, Ni J, Wei YF, Yu G, Gentz R and
Dixit VM: An antagonist decoy receptor and a death
domain-containing receptor for TRAIL. Science. 277:815–818. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang JS, Gong A and Young CY: ZNF185, an
actin-cytoskeleton-associated growth inhibitory LIM protein in
prostate cancer. Oncogene. 26:111–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pettaway CA, Pathak S, Greene G, Ramirez
E, Wilson MR, Killion JJ and Fidler IJ: Selection of highly
metastatic variants of different human prostatic carcinomas using
orthotopic implantation in nude mice. Clin Cancer Res. 2:1627–1636.
1996.PubMed/NCBI
|
|
31
|
Wang C, Norton JT, Ghosh S, Kim J, Fushimi
K, Wu JY, Stack MS and Huang S: Polypyrimidine tract-binding
protein (PTB) differentially affects malignancy in a cell
line-dependent manner. J Biol Chem. 283:20277–20287. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Godthelp BC, van Buul PP, Jaspers NG,
Elghalbzouri-Maghrani E, van Duijn-Goedhart A, Arwert F, Joenje H
and Zdzienicka MZ: Cellular characterization of cells from the
Fanconi anemia complementation group, FA-D1/BRCA2. Mutat Res.
601:191–201. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wiegant WW, Meyers M, Verkaik NS, van der
Burg M, Darroudi F, Romeijn R, Bernatowska E, Wolska-Kusnierz B,
Mikoluc B, Jaspers NG, et al: A novel radiosensitive SCID patient
with a pronounced G2/M sensitivity. DNA Repair.
9:365–373. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bastian PJ, Ellinger J, Wellmann A,
Wernert N, Heukamp LC, Müller SC and von Ruecker A: Diagnostic and
prognostic information in prostate cancer with the help of a small
set of hypermethylated gene loci. Clin Cancer Res. 11:4097–4106.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu YP, Yu G, Tseng G, Cieply K, Nelson J,
Defrances M, Zarnegar R, Michalopoulos G and Luo JH: Glutathione
peroxidase 3, deleted or methylated in prostate cancer, suppresses
prostate cancer growth and metastasis. Cancer Res. 67:8043–8050.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Norton JT, Pollock CB, Wang C, Schink JC,
Kim JJ and Huang S: Perinucleolar Compartment prevalence is a
phenotypic pancancer marker of malignancy. Cancer. 113:861–869.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Frycz B, Pinczewska A and Jagodziński PP:
Maślan sodu obniża ekspresję dehydrogenazy 17β-hydroksysteroidowej
typu 1-szego W linii komórkowej raka gruczołu krokowego LNCaP.
Nowiny Lekarskie. 80:283–287. 2011.
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:D856–D861.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
MethHC, . A database of DNA methylation
and gene expression in human cancers. http://methhc.mbc.nctu.edu.tw/php/index.phpOctober
23–2018
|
|
41
|
van de Vijver MJ, He YD, van't Veer LJ,
Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C,
Marton MJ, et al: A gene-expression signature as a predictor of
survival in breast cancer. N Engl J Med. 347:1999–2009. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
O'Connell MJ, Lavery I, Yothers G, Paik S,
Clark-Langone KM, Lopatin M, Watson D, Baehner FL, Shak S, Baker J,
et al: Relationship between tumor gene expression and recurrence in
four independent studies of patients with stage II/III colon cancer
treated with surgery alone or surgery plus adjuvant fluorouracil
plus leucovorin. J Clin Oncol. 28:3937–3944. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Roszkowski K, Furtak J, Zurawski B,
Szylberg T and Lewandowska MA: Potential role of methylation marker
in glioma supporting clinical decisions. Int J Mol Sci. 17(pii):
E18762016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Song L, Peng X, Li Y, Xiao W, Jia J, Dong
C, Gong Y, Zhou G and Han X: The SEPT9 gene methylation assay is
capable of detecting colorectal adenoma in opportunistic screening.
Epigenomics. 9:599–610. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang Q, Liu CX, Gu X, Wilt G, Shaffer J,
Zhang Y and Devgan V: EpiTect Methyl II PCR Array System: A simple
tool for screening regional DNA methylation of a large number of
genes or samples without bisulfite conversion. Qiagen. https://www.qiagen.com/ch/resources/resourcedetail?id=39ec06aa-ec53-4acd-aa15-67b5882efbb6&lang=en(cited
2018-11-09).
|
|
46
|
Kang GH, Lee S, Lee HJ and Hwang KS:
Aberrant CpG island hypermethylation of multiple genes in prostate
cancer and prostatic intraepithelial neoplasia. J Pathol.
202:233–240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Richiardi L, Fiano V, Vizzini L, De Marco
L, Delsedime L, Akre O, Tos AG and Merletti F: Promoter methylation
in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients.
J Clin Oncol. 27:3161–3168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Llorca-Cardeñosa MJ, Fleitas T,
Ibarrola-Villava M, Peña-Chilet M, Mongort C, Martinez-Ciarpaglini
C, Navarro L, Gambardella V, Castillo J, Roselló S, et al:
Epigenetic changes in localized gastric cancer: The role of RUNX3
in tumor progression and the immune microenvironment. Oncotarget.
7:63424–63436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gu YM, Ma YH, Zhao WG and Chen J:
Dickkopf3 overexpression inhibits pancreatic cancer cell growth in
vitro. World J Gastroenterol. 17:3810–3817. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Veeck J, Wild PJ, Fuchs T, Schüffler PJ,
Hartmann A, Knüchel R and Dahl E: Prognostic relevance of
Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter
methylation in human breast cancer. BMC Cancer. 9:2172009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Romero D and Kypta R: Dickkopf-3 function
in the prostate: Implications for epithelial homeostasis and tumor
progression. Bioarchitecture. 3:42–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Romero D, Kawano Y, Bengoa N, Walker MM,
Maltry N, Niehrs C, Waxman J and Kypta R: Downregulation of
Dickkopf-3 disrupts prostate acinar morphogenesis through
TGF-β/Smad signalling. J Cell Sci. 126:1858–1867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lodygin D, Epanchintsev A, Menssen A,
Diebold J and Hermeking H: Functional epigenomics identifies genes
frequently silenced in prostate cancer. Cancer Res. 65:4218–4227.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yu YP, Paranjpe S, Nelson J, Finkelstein
S, Ren B, Kokkinakis D, Michalopoulos G and Luo JH: High throughput
screening of methylation status of genes in prostate cancer using
an oligonucleotide methylation array. Carcinogenesis. 26:471–479.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Falck E, Karlsson S, Carlsson J, Helenius
G, Karlsson M and Klinga-Levan K: Loss of glutathione peroxidase 3
expression is correlated with epigenetic mechanisms in endometrial
adenocarcinoma. Cancer Cell Int. 10:462010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yegnasubramanian S, Kowalski J, Gonzalgo
ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs
WB and Nelson WG: Hypermethylation of CpG islands in primary and
metastatic human prostate cancer. Cancer Res. 64:1975–1986. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Strand SH, Orntoft TF and Sorensen KD:
Prognostic DNA methylation markers for prostate cancer. Int J Mol
Sci. 15:16544–16576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rosenbaum E, Hoque MO, Cohen Y, Zahurak M,
Eisenberger MA, Epstein JI, Partin AW and Sidransky D: Promoter
hypermethylation as an independent prognostic factor for relapse in
patients with prostate cancer following radical prostatectomy. Clin
Cancer Res. 11:8321–8325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Richiardi L, Fiano V, Grasso C, Zugna D,
Delsedime L, Gillio-Tos A and Merletti F: Methylation of APC and
GSTP1 in non-neoplastic tissue adjacent to prostate tumour and
mortality from prostate cancer. PLoS One. 8:e681622013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bastian PJ, Palapattu GS, Lin X,
Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW
and Nelson WG: Preoperative serum DNA GSTP1 CpG island
hypermethylation and the risk of early prostate-specific antigen
recurrence following radical prostatectomy. Clin Cancer Res.
11:4037–4043. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Konishi N, Nakamura M, Kishi M, Nishimine
M, Ishida E and Shimada K: DNA hypermethylation status of multiple
genes in prostate adenocarcinomas. Jpn J Cancer Res. 93:767–773.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ellinger J, Bastian PJ, Haan KI, Heukamp
LC, Buettner R, Fimmers R, Mueller SC and von Ruecker A:
Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of
prostate cancer patients qualify as diagnostic and prognostic
indicators. Int J Cancer. 122:138–143. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Vasiljević N, Wu K, Brentnall AR, Kim DC,
Thorat MA, Kudahetti SC, Mao X, Xue L, Yu Y, Shaw GL, et al:
Absolute quantitation of DNA methylation of 28 candidate genes in
prostate cancer using pyrosequencing. Dis Markers. 30:151–161.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Morris MR, Ricketts C, Gentle D,
Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F and
Maher ER: Identification of candidate tumour suppressor genes
frequently methylated in renal cell carcinoma. Oncogene.
29:2104–2117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Boumber YA, Kondo Y, Chen X, Shen L,
Gharibyan V, Konishi K, Estey E, Kantarjian H, Garcia-Manero G and
Issa JP: RIL, a LIM gene on 5q31, is silenced by methylation in
cancer and sensitizes cancer cells to apoptosis. Cancer Res.
67:1997–2005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu L, Yoon JH, Dammann R and Pfeifer GP:
Frequent hypermethylation of the RASSF1A gene in prostate cancer.
Oncogene. 21:6835–6840. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Maruyama R, Toyooka S, Toyooka KO, Virmani
AK, Zöchbauer- Müller S, Farinas AJ, Minna JD, McConnell J, Frenkel
EP and Gazdar AF: Aberrant promoter methylation profile of prostate
cancers and its relationship to clinicopathological features. Clin
Cancer Res. 8:514–519. 2002.PubMed/NCBI
|
|
68
|
Imren S, Kohn DB, Shimada H, Blavier L and
DeClerck YA: Overexpression of tissue inhibitor of
metalloproteinases-2 retroviral-mediated gene transfer in vivo
inhibits tumor growth and invasion. Cancer Res. 56:2891–2895.
1996.PubMed/NCBI
|
|
69
|
Pulukuri SM, Patibandla S, Patel J, Estes
N and Rao JS: Epigenetic inactivation of the tissue inhibitor of
metalloproteinase-2 (TIMP-2) gene in human prostate tumors.
Oncogene. 26:5229–5237. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ross JS, Kaur P, Sheehan CE, Fisher HA,
Kaufman RA Jr and Kallakury BV: Prognostic significance of matrix
metalloproteinase 2 and tissue inhibitor of metalloproteinase 2
expression in prostate cancer. Mod Pathol. 16:198–205. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Ratzinger G, Mitteregger S, Wolf B, Berger
R, Zelger B, Weinlich G, Fritsch P, Goebel G and Fiegl H:
Association of TNFRSF10D DNA-methylation with the survival of
melanoma patients. Int J Mol Sci. 15:11984–11995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hornstein M, Hoffmann MJ, Alexa A,
Yamanaka M, Müller M, Jung V, Rahnenführer J and Schulz WA: Protein
phosphatase and TRAIL receptor genes as new candidate tumor genes
on chromosome 8p in prostate cancer. Cancer Genomics Proteomics.
5:123–136. 2008.PubMed/NCBI
|
|
73
|
Catalanotto C, Cogoni C and Zardo G:
MicroRNA in control of gene expression: An overview of nuclear
functions. Int J Mol Sci. 17(pii): E17122016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kubiak M and Lewandowska MA: Can chromatin
conformation technologies bring light into human molecular
pathology? Acta Biochim Pol. 62:483–489. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Audia JE and Campbell RM: Histone
modifications and cancer. Cold Spring Harb Perspect Biol.
8:a0195212016. View Article : Google Scholar : PubMed/NCBI
|