Introduction
Colorectal cancer is one of the most common
malignancies in the world. It is the third most common cancer in
men and the second most common cancer in women worldwide according
to the Global Cancer Statistics 2018 (1). In addition, the incidence rates of
colorectal cancer are ~3-fold higher in developed countries
compared with developing countries (1). To understand the molecular and cellular
mechanism of colorectal cancer, previous studies have tried to
establish several reliable in vitro cancer models (2–5). For
decades, primary patient-derived tumor xenografts have been one of
the most widely used cancer models (6). However, the use of animals can be
challenging, laborious and time-consuming. In vitro culture
systems for patient-derived intestinal epithelial cells have many
advantages and may facilitate development of novel therapies for
colorectal cancer (7,8). Long-term expansion of patient-derived
tumor cells is challenging; however, conditional reprogramming
methodologies for cell culture with a combination of irradiated
feeder cells and a rho-associated coiled-coil containing protein
kinase (ROCK) inhibitor, Y-27632, have been proposed to overcome
the limitations of traditional methods, including the short
lifespan of cancer cells and the low success rates of long-term
expansion without the use of viral infection or gene transduction
(9–12). Moreover, Clevers (13) have successfully obtained in
vitro expansion of epithelial cells in a three-dimensional
matrix by designing an organoid culture that can faithfully
recapitulate the physiology and functionality of intestinal
epithelia, although these cells are not readily applicable to
high-throughput drug screening for evaluating drug responses. In
the present study, it was observed that tumor cells isolated from
colorectal cancer patient-derived xenograft (PDX) could be
efficiently immortalized in conditioned medium from irradiated
feeder cells containing a ROCK inhibitor Y-27632.
The present results suggested that genes related to
the ROCK signaling pathway playing a major role in the mechanism of
conditional reprogramming were also associated with the
immortalization of colorectal cancer cells from human-derived
xenograft tumors. Conditional reprogramming models could be
effectively used to evaluate drug response for personalized
therapeutic approaches, and the present study performed a screening
for anticancer drugs approved by the Food and Drug Administration.
Finally, the potential applications of this established model for
detecting gene-drug interactions and making clinical decisions were
investigated.
Materials and methods
Establishment of PDXs
PDXs were established and analysis of mutation
profiles was performed as previously described (11). Briefly, fresh tumor specimens were
obtained from male and female patients (female to male ratio, 1:1)
with colorectal cancer treated with surgical resection in Samsung
Medical Center from December 2011 to February 2013. Clinical
characteristics are presented in Table
I. The specimens were maintained in RPMI medium (Gibco; Thermo
Fisher Scientific, Inc.) containing 3% penicillin streptomycin, cut
into 2–3 mm3 sized pieces, and embedded in
high-concentrated Matrigel (BD Biosciences). Tumor fragments
embedded in Matrigel were implanted into subcutaneous pockets of
female BALB/c nude mice (age, 6–8 weeks old; weight, 16–18 g;
Orient Bio, Inc.), which were made in each side of the lower back
(n=5–6 for each tumor sample). Mice were maintained under standard
conditions of temperature (20–26°C), relative humidity (30–70%)
under a 12-h light/dark cycle with free access to food and water.
When tumors reached 1,000 mm3 in volume, tumor tissues
were collected for primary cell culture. All animal experiments
were performed according to the protocols approved by the
Institutional Animal Care and Use Committee of Samsung Biomedical
Research Institute. The present study performed in patients and
animals was approved by the Institutional Review Board of Samsung
Medical Center.
 | Table I.Clinical information of PDXs used for
cell culture. |
Table I.
Clinical information of PDXs used for
cell culture.
| Sample | Age | Sex | Tumor | Cell type | Stagea | Preoperative
chemotherapy |
|---|
| PDX1 | 51 | F | Adenocarcinoma | MD | IV | No |
| PDX2 | 57 | M | Adenocarcinoma | MD | IV | No |
| PDX3 | 74 | F | Adenocarcinoma | WD | IV | No |
| PDX4 | 46 | F | Adenocarcinoma | MD | IV | Yes |
Cell isolation from xenograft
tumors
Tumor tissue from xenografts was cut into small
pieces (~1 mm3), washed with 70% ethanol and ice-cold
PBS, mechanically dissociated using gentleMACS Dissociator
(Miltenyi Biotec) for 45 sec at room temperature, and subsequently
incubated at 37°C in S/F M199 media (Gibco; Thermo Fisher
Scientific, Inc.) containing 200 U/ml collagenase (Gibco; Thermo
Fisher Scientific, Inc.) and 0.1 mg/ml DNase (Roche Diagnostics)
for 90 min at 37°C on a tube rotator. The dissociated cell
suspension was passed through a cell strainer (Falcon; BD
Biosciences; cat. no. 352340; pore size, 40 µm), washed with RPMI
(Gibco; Thermo Fisher Scientific, Inc.) containing 20% FBS (Biowest
LLC), and centrifuged at 1,000 rpm for 3 min. Viable cells were
counted using a hemocytometer and a light microscope
(magnification, ×10) and subjected to cell culture.
Cell cultures
Dissociated cells were cultured in a mixture of
conditioned medium from irradiated 3T3-J2 fibroblasts and fresh F
medium (3:1 ratio), which contained three parts of DMEM (Thermo
Fisher Scientific, Inc.), one part of Ham's F-12 Nutrient Mixture
(Gibco; Thermo Fisher Scientific, Inc.) and the following
supplements: 5% FBS (Biowest LLC), 0.4 µg/ml hydrocortisone
(Sigma-Aldrich; Merck KGaA), 5 µg/ml insulin (Sigma-Aldrich; Merck
KGaA), 8.4 ng/ml cholera toxin (Sigma-Aldrich; Merck KGaA), 10
ng/ml epidermal growth factor (EGF; Thermo Fisher Scientific,
Inc.), 10 µmol/l Y-27632 (Enzo Life Sciences), 1%
penicillin-streptomycin (Thermo Fisher Scientific, Inc.), 20 µg/ml
gentamicin and 500 ng/ml Fungizone. For 3D culture, cells were
embedded in Matrigel (BD Bioscience) on ice and plated into 24-well
plates (1×104 cells with 50 µl of Matrigel per well).
Matrigel was polymerized at 37°C for 15 min. In each well, 1 ml of
a mixture of conditioned medium and fresh F media (3:1 ratio)
supplemented with 5 µmol/l Y-27632 was added. The medium was
replaced every other day. All cells were maintained at 37°C in a
humidified incubator with 5% CO2 and passaged at a ratio
of 1:4 when they reached 80–90% confluency. The number of cells was
counted every 3 days using the C-Chip hemocytometer (NanoEnTek,
Inc.).
Differential trypsinization
Cells at 95% confluence were washed with PBS and
treated with 1 ml trypsin/EDTA (0.25%; Gibco; Thermo Fisher
Scientific, Inc.) at 37°C for 1 min. Fibroblasts were efficiently
detached from the bottom of the flask, whereas epithelial cells
remained attached the flask. Trypsin digestion was terminated by
the addition of complete culture media, and the fibroblasts were
removed using PBS.
Epithelial cell enrichment
Dissociated cells were incubated with
anti-epithelial cell adhesion molecule (EpCAM) microbeads (cat. no.
130-061-101; Miltenyi Biotec, Inc.) for 30 min at 4°C.
Subsequently, labeled cells were collected on a magnetic separation
column. After removal of the column from the magnetic field,
EpCAM+ cells were eluted with PBS for cell culture.
Dissociated cells or EpCAM+ cells were cultured in
conditioned medium.
Conditioned medium from irradiated
3T3-J2 fibroblast
The fibroblast cell line 3T3-J2 was obtained from
the American Type Culture Collection. Cells were maintained at 37°C
with 5% CO2 in DMEM containing 10% bovine calf serum
(Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin-streptomycin
(Thermo Fisher Scientific, Inc.). To prepare conditioned medium
from irradiated 3T3-J2 fibroblast, suspension cells were irradiated
at 30 Gy and plated at a density of ~70% in T-175 flasks with 30 ml
F medium. After 3 days, the medium was collected, and fresh F
medium was replaced for an additional 3 days. The collected medium
was passed through a Stericup filter unit (pore size, 0.22 µm; EMD
Millipore) and stored at −80°C. Conditioned medium was mixed with
fresh F medium at a ratio of 3:1 and supplemented with 5 µmol/l
Y-27632.
Immunohistochemistry and hematoxylin
and eosin (HE) staining
Cells were fixed with 4% paraformaldehyde for 1 h at
room temperature and embedded in paraffin. Paraffin-embedded
sections (thickness, 4 µm) were dewaxed in xylene for 30 min at
room temperature and rehydrated in a graded alcohol series.
Endogenous peroxidase was blocked with 3%
H2O2 for 30 min at room temperature. Before
incubating the samples with the primary antibodies, sections were
immersed in 10 mM citrate buffer (pH 6.0), rinsed in TBS. The
following primary antibodies were incubated with the samples for 1
h at room temperature: Anti-cytokeratin 20 (CK20; 1:100; Dako; cat.
no. M7019, Agilent Technologies, Inc.) and Ki67 (1:100; Dako; cat.
no. M7240, Agilent Technologies, Inc.). Sections were subsequently
treated with a biotin-labeled secondary antibody (cat. no. B2763;
Thermo Fisher Scientific, Inc.) for 30 min at room temperature and
incubated with an avidin-biotin-peroxidase complex for 30 min at
room temperature. 3,3′-Diaminobenzidine was used as the chromogen
followed by HE counterstaining. The sections were stained with HE
for 5 min at room temperature to examine cell morphology. Stained
sections were visualized and captured using a slide scanner (Aperio
ScanScope slide scanner; Leica Microsystems, Inc.) with a 20X
objective (final magnification, ×200).
Immunofluorescence staining
Cells were grown on poly-L-lysine coated coverslips.
After reaching 50–60% confluence, cells were fixed in 4%
paraformaldehyde for 10 min at room temperature, permeabilized with
0.2% Triton X-100 in PBS for 5 min at room temperature and
incubated in blocking buffer (2% BSA and 0.2% Triton X-100 in PBS)
for 1 h at room temperature. These cells were labeled with primary
antibodies against EpCAM (Biolegend; cat. no. 324210; 1:20
dilution), Vimentin (Santa Cruz Biotechnology, Inc.; cat. no.
SC6260; 1:100 dilution) and CK20 (Dako; Agilent Technologies, Inc.;
cat. no. M7019; 1:200 dilution) overnight at 4°C. Samples were
incubated for 1 h at room temperature with secondary antibody
(Alexa Fluor 546 goat anti-mouse IgG; Gibco; Thermo Fisher
Scientific, Inc.; cat. no. MOP-A-11003; 1:40 dilution). Coverslips
were mounted onto glass slides using VECTASHIELD mounting media
(Vector Laboratories), sealed with nail polish to prevent drying,
and stored at 4°C. Slides were analyzed with a laser scanning
confocal microscope (magnification, ×20). DAPI and protein signals
were detected at excitation wavelengths of 633 and 488 nm,
respectively.
Short tandem repeat (STR)
analysis
STR analysis was performed using PowerPlex 18D
system (Promega Corporation) according to the manufacturer's
instructions. DNA extracted from tumor tissues and cell pellets was
amplified by multiplex PCR for 18 loci, including 17 STR loci
(D3S1358, tyrosine hydroxylase 1, D21S11, D18S51, Penta E, D5S818,
D13S317, D7S820, D16S539, CSF1PO, Penta D, von Willebrand factor
type A, D8S1179, TPOX, fibrinogen α chain, D19S433 and D2S1338) and
amelogenin. Primers and DNA polymerase were included in the kit
(cat. no. DC1802; Promega Corporation). The internal lane standard
was labeled with the dye WEN (included in the kit). PCR products
were electrophoresed on an ABI 3130×L Genetic Analyzer (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and analyzed with
GeneMapper 4.0 software (Thermo Fisher Scientific, Inc.) using
allelic ladders supplied by Applied Biosystems (Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from patient-derived cancer
cells (PDCs) and tumor tissues from xenografts using a RNAprep Mini
kit (Qiagen). In total, 500 ng RNA was reverse transcribed using
murine leukemia virus reverse transcriptase (cat. no. M0253S; New
England Biolabs, Inc.) primers and reaction buffer were included in
the kit (New England Biolabs, Inc.) for 1 h at 42°C. The RT-qPCR
amplification was performed using a SYBR Green Master Mix (Roche
Diagnostics) in a real-time system under the following conditions:
Initial denaturation at 95°C for 60 sec, followed by 45 cycles of
95°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec, with a final
elongation step at 72°C for 2 min. Human-specific PCR primers
(Roche Diagnostics) were used to analyze expression levels of the
following genes: Human telomerase reverse transcriptase (hTERT),
C-Myc, p16, CDK4 and GAPDH. The primers used for RT-qPCR analysis
were the following: hTERT forward, 5′-CTACTCCTCAGGCGACAAGG-3′ and
reverse 5′-TGGAACCCAGAAAGATGGTC-3′; C-Myc forward,
5′-TCAAGAGGCGAACACACAAC-3′ and reverse, 5′-GGCCTTTTCATTGTTTTCCA-3′;
p16 forward, 5′-GCACCAGAGGCAGTAACCAT-3′ and reverse,
5′-GGAATCCCGTAGCTTCCCTA-3′; CDK4 forward,
5′-GAAACTCTGAAGCCGACCAG-3′ and reverse, 5′-CCTGGGTTCAGCAGAAAGAG-3′;
GAPDH forward, 5′-CTCAGACACCATGGGGAAGGTGA-3′ and reverse,
5′-ATGATCTTGAGGCTGTTGTCATA-3′. mRNA levels of specific genes were
calculated using the 2−∆∆Cq method (14) and normalized to GAPDH.
Mutation profiling
Mutational analysis of PDXs was performed as
previously described (15).
Anticancer drug screening
PDCs grown in conditioned medium were collected and
seeded into 384-well plates at a density of 500 cells/well. After
plating, cells were treated with 65 different drugs under clinical
and preclinical investigation in seven serial 4-fold dilutions (n=2
for each condition) using a Janus Automated Workstation
(PerkinElmer, Inc.). The same drug library was used in a previous
study (16). After 6 days of
incubation at 37°C in a humidified incubator, cell viability was
analyzed using an ATP monitoring system based on Firefly luciferase
enzymatic activity (ATPLite 1step; PerkinElmer, Inc.). Viable cells
were measured using an EnVision Multilabel Reader (PerkinElmer,
Inc.). Drug sensitivity was analyzed by assessing the
IC50, slope of the dose-response curve and area under
the dose-response curve (AUC). Drugs were stored and diluted
according to the manufacturer's instructions (Selleck
Chemicals).
Statistical analysis
All experiments were repeated three times for each
sample. Data are presented as the mean ± SEM. To compare gene
expression levels between the two samples in the PCR analysis,
expression levels derived from the PDX samples were set to zero,
and relative quantification was calculated. Data were analyzed
using Student's t-test. In the drug screening analysis, the mean
and SD of the AUC for every drug were calculated using a reference
samples containing 462 patient-derived tumor cells across 14 cancer
types (16), and Z-scores or standard
scores of AUC were calculated for each drug. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using Graphpad Prism (version
6; GraphPad Software, Inc.).
Results
Primary culture of epithelial cells
from colon cancer PDXs
In our previous studies, a total of 97 PDXs were
successfully established from 143 colorectal cancer specimens
(15,17). Among 18 ×enografts, 10 (55.5%) led to
the establishment of PDCs. In the present study, four PDCs were
selected and characterized since their genomic profiles was
previously identified (Table I). In
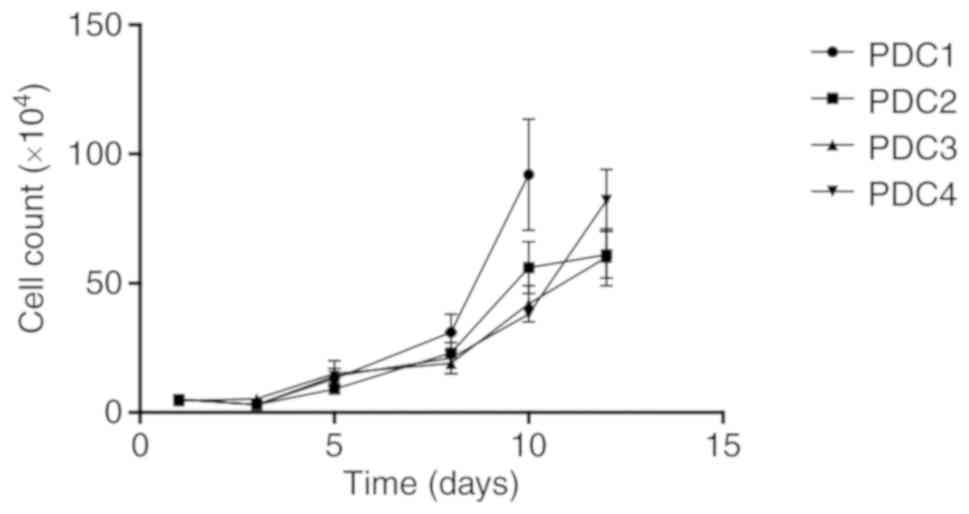
addition, PDCs could grow over 10 passages. Tumor tissues derived
from xenografts were dissociated mechanically and enzymatically.
Dissociated cells were cultured for >5 passages (>30 days) at
37°C in a conditioned medium collected from cultures of irradiated
3T3-J2 cells with 5 µmol/l of Y-27632. PDCs were grown in
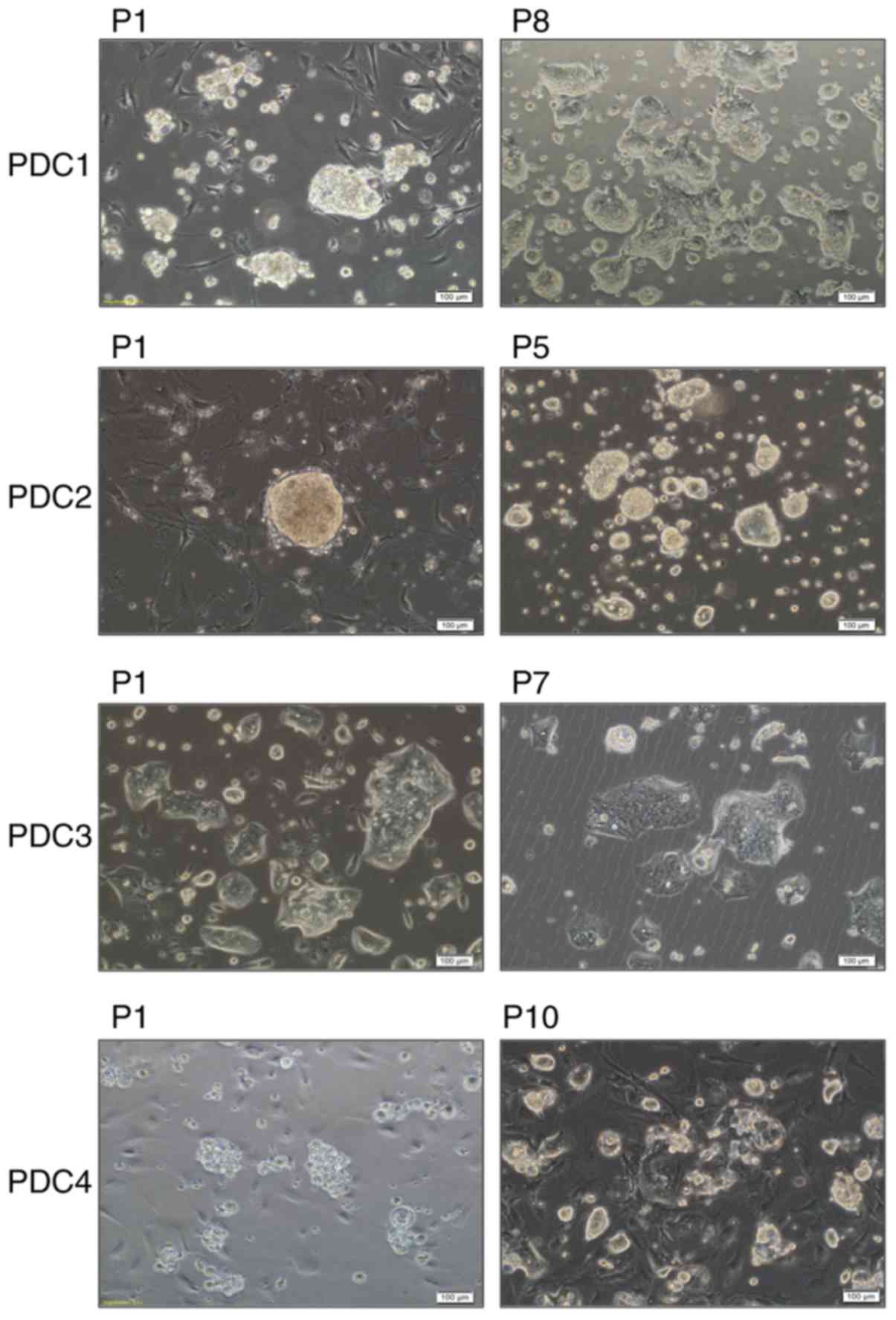
epithelial cell colonies (Fig. 1) and
rapidly proliferated to reach 90% confluence after ~6 days
(Fig. 2). Primary cultures of human
tumor fragments are frequently contaminated by
rapidly-proliferating tumor-associated stromal fibroblasts
(18–21). After 3 days, small clusters of
epithelial cells began to be attached to the Petri dish, and large
colonies of epithelial cells surrounded by stromal fibroblasts were
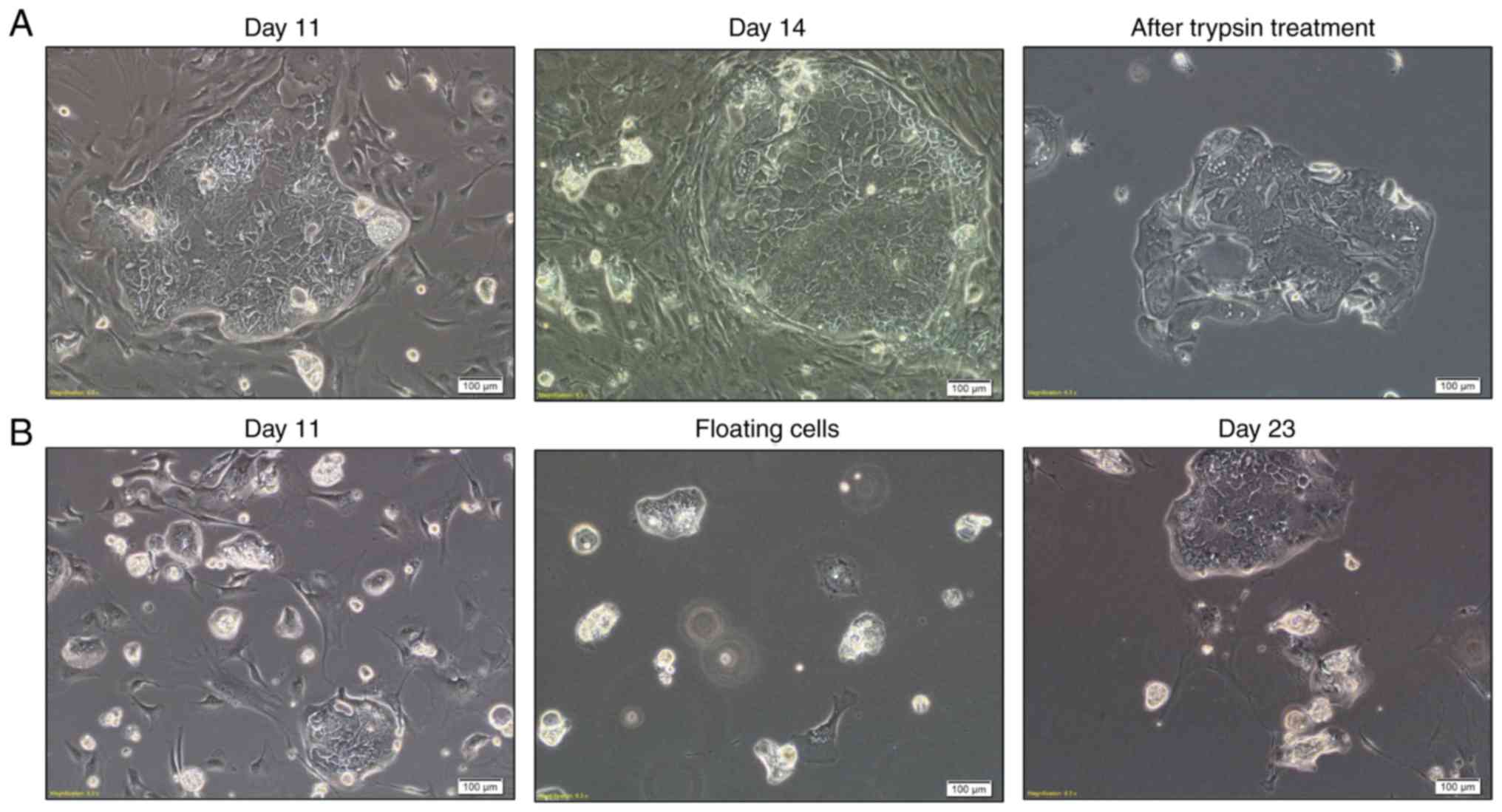
detected after 14 days. For a pure culture of PDCs, differential
trypsinization methods were used to remove stromal fibroblasts
(Fig. 3A). PDC3 floated as small
spheroid cultures for 3 days during initial plating. However, they
adhered when floating cells were seeded into new flasks (Fig. 3B).
Characterization of immortalized
PDCs
PDCs grew in monolayers and displayed epithelial
morphology. Immunocytochemical analysis revealed that epithelial
colonies expressed high levels of membrane-localized EpCAM
(Fig. 4). Colon epithelial
cell-specific marker CK20 was observed in the cytoplasm and cell
membranes of epithelial colonies (Fig.
4). In contrast, the mesenchymal marker vimentin was shown to
specifically stain fibroblasts (22),
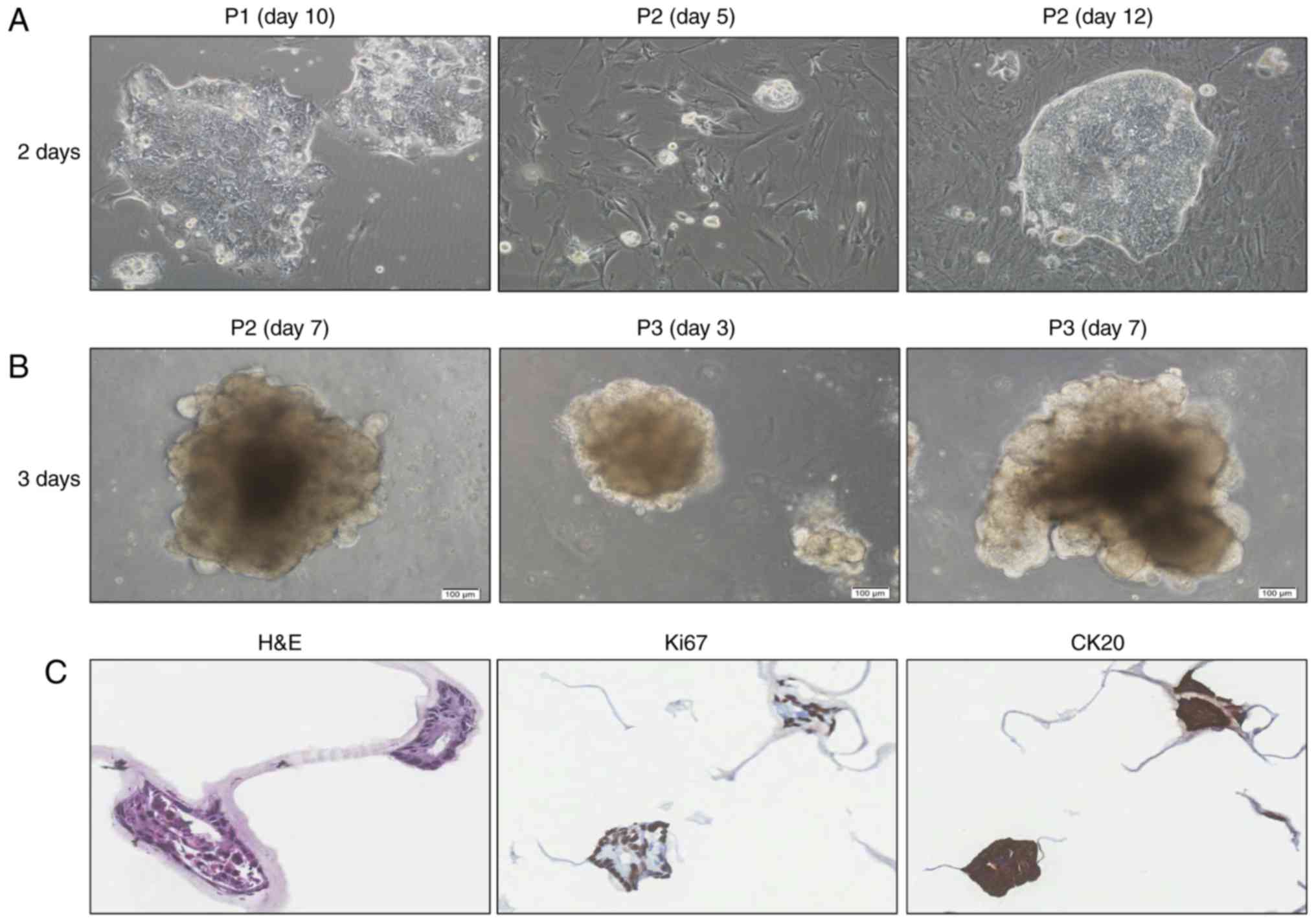
and showed negative staining for all four PDCs (Fig. 4). Epithelial cells were grown in 2D
cultures on stromal cells (Fig. 5A).
When they were transferred to Matrigel, they formed well-defined
spheres without stromal fibroblasts (Fig.
5B). Lumen formation was observed in PDC spheres in cells
stained with proliferation marker protein Ki67 and CK20 (Fig. 5C). STR analysis was performed for 18
loci on different chromosomes to verify that these PDCs were
derived from PDXs without contaminations from other cells during
passaging (Table II).
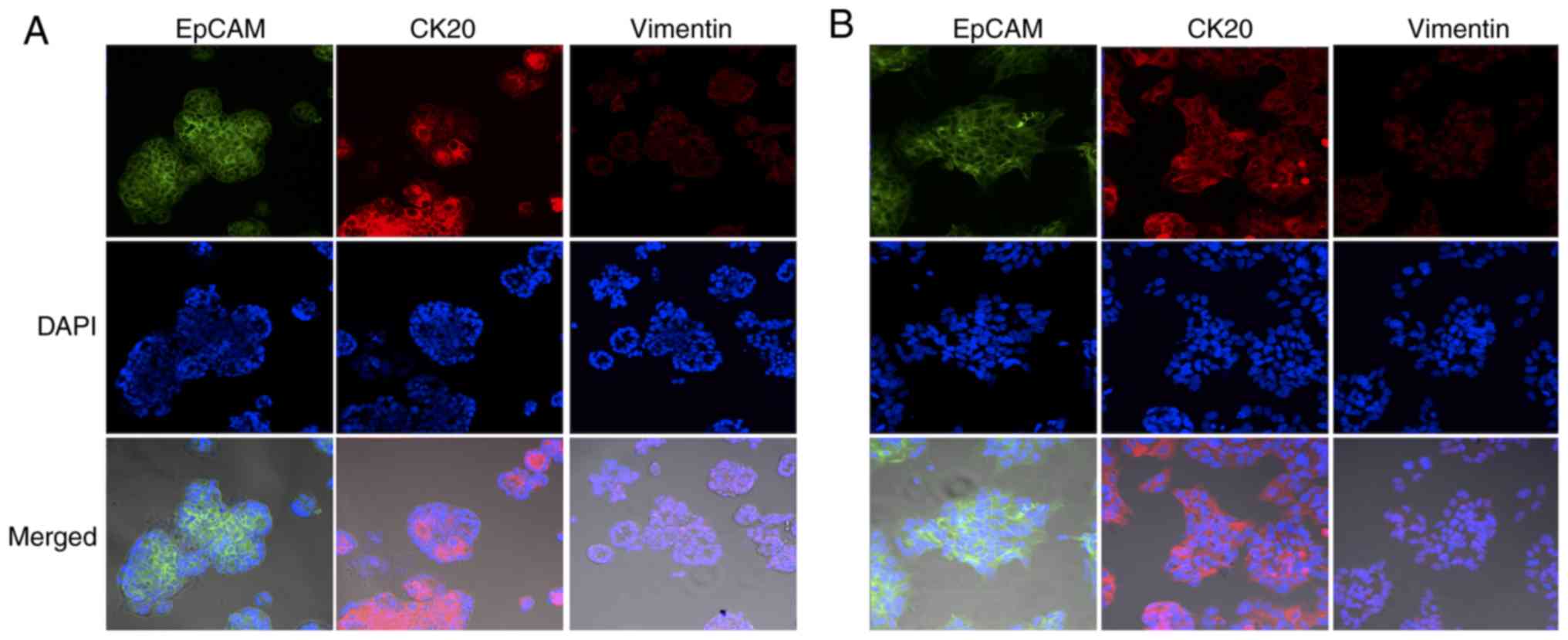 | Figure 4.Immunofluorescence staining of
epithelial cells isolated from (A) PDX2 and (B) PDX3. Cells were
stained by anti-EpCAM, an epithelial cell marker, in green, by
anti-CK20, a colon cancer marker, in red, and by anti-vimentin a
fibroblastic/mesenchymal marker, in red. Nuclei, in blue, were
stained by DAPI. Scale bar, 100 µm; magnification, ×10. EpCAM,
epithelial cell adhesion molecule; PDX, patient-derived xenograft;
CK20, cytokeratin 20. |
 | Table II.Short tandem repeat profiling of PDX
tissues and cultured cells. |
Table II.
Short tandem repeat profiling of PDX
tissues and cultured cells.
|
| PDX1 | PDX2 | PDX3 | PDX4 |
|---|
|
|
|
|
|
|
|---|
| Locus | Tissue | Cell | Tissue | Cell | Tissue | Cell | Tissue | Cell |
|---|
| AMEL | X | X | X,Y | X,Y | X | X | X | X |
| CSF1PO | 11,12 | 11,12 | 11 | 11 | 10,12 | 10,12 | 11,13 | 11,13 |
| D13S317 | 8,9 | 8,9 | 8,9 | 8,9 | 8,10 | 8,10 | 11,12 | 11,12 |
| D16S539 | 11 | 11 | 9,10 | 9,10 | 11,13 | 11,13 | 9 | 9 |
| D18S51 | 14 | 14 | 14 | 14 | 16 | 16 | 18 | 18 |
| D19S433 | 14 | 14 | 13,15 | 13,15 | 13.2,16 | 13.2,16 | 13 | 13 |
| D21S11 | 30 | 30 | 29,30 | 29,30 | 30 | 30 | 22,28.2 | 22,28.2 |
| D2S1338 | 18 | 18 | 23 | 23 | 19 | 19 | 17,18 | 17,18 |
| D3S1358 | 15 | 15 | 16 | 16 | 15,17 | 15,17 | 15 | 15 |
| D5S818 | 10 | 10 | 12 | 12 | 10,12 | 10,12 | 10,13 | 10,13 |
| D7S820 | 11,12 | 11,12 | 10,12 | 10,12 | 8,12 | 8,12 | 12 | 12 |
| D8S1179 | 10,13 | 10,13 | 10,12 | 10,12 | 11,13 | 11 | 13,14 | 13,14 |
| FGA | 23 | 23 | 22 | 22 | 26 | 26 | 22,23 | 22,23 |
| Penta D | 10,12 | 10,12 | 9,13 | 9,13 | 8,11 | 8,11 | 9,10 | 9,10 |
| Penta E | 11 | 11 | 17 | 17 | 11,8 | 11,8 | 24 | 24 |
| TH01 | 9,9.3 | 9,9.3 | 9 | 9 | 9 | 9 | 8 | 8 |
| TPOX | 8 | 8 | 8 | 8 | 8 | 8 | 8,11 | 8,11 |
| vWA | 16 | 16 | 16,18 | 16,18 | 17,18,19 | 17,18 | 17 | 17,18 |
Cooperative effect of feeder cells and
ROCK inhibitor on primary culture
Previous studies have shown that a combination of
feeder cells and ROCK inhibitor is important for primary cell
immortalization (11). Irradiated
feeder cells can induce pronounced telomerase activity and hTERT
expression in keratinocytes (12). To
determine whether PDCs exhibited a similar phenomenon, RT-qPCR was
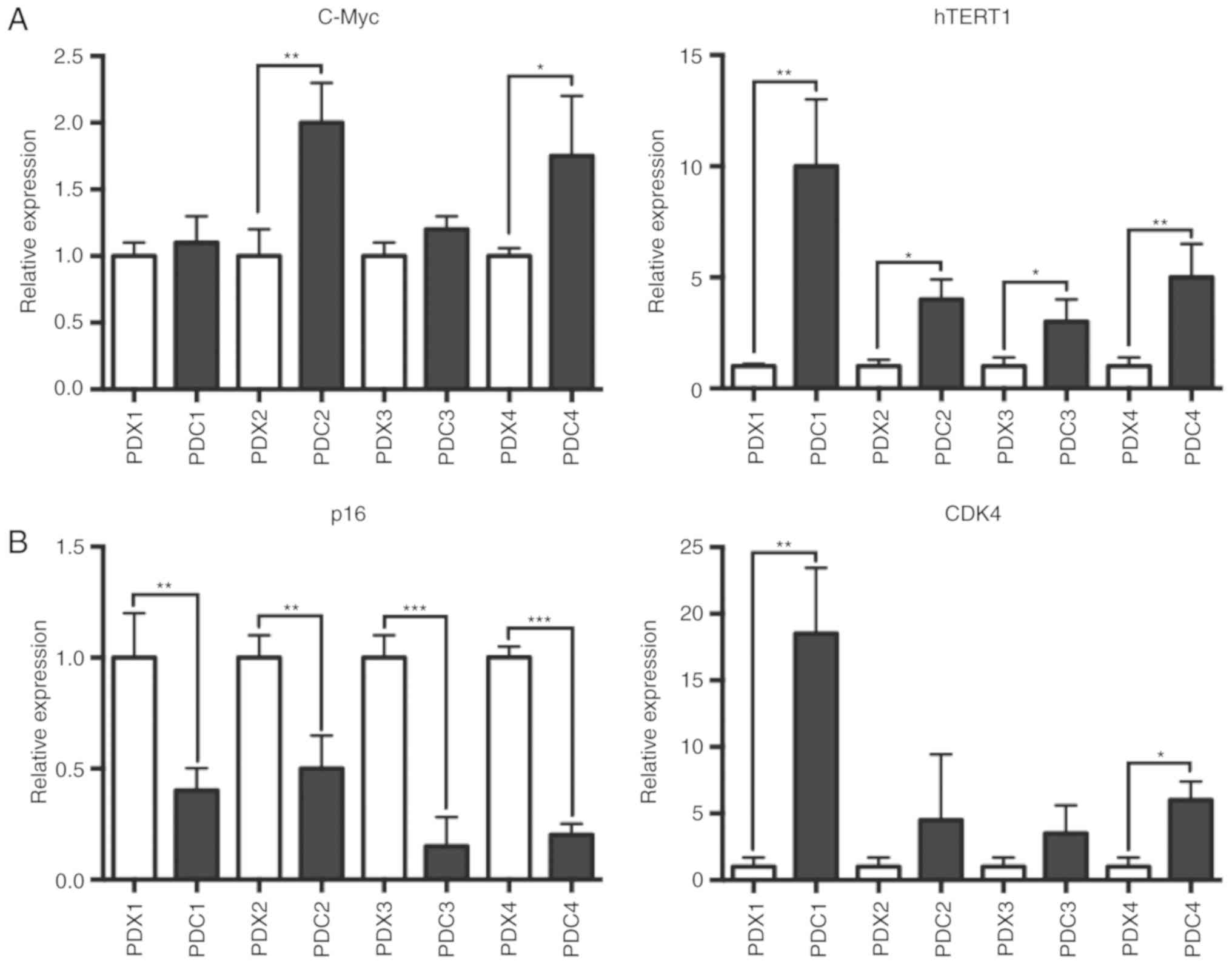
used to measure the expression levels of hTERT. The present RT-qPCR
analysis suggested that hTERT and CDK4 mRNA expression was
increased in PDCs grown in conditioned medium on irradiated feeder
cells with Y-27632 (Fig. 6A). It has
been shown that ROCK inhibitor induces C-Myc mRNA expression in
human keratinocytes and that long-term increase in C-Myc expression
is associated with increased TERT expression (23). C-Myc mRNA level was also increased in
PDCs. In immortalized human keratinocyte, the RB transcriptional
corepressor (RB)/p16 signaling pathway is inactivated (12). The present results suggested that, in
PDCs, p16 mRNA level was also decreased (Fig. 6B).
Screening of 65 FDA-approved
anticancer drugs
To validate the applicability of PDCs for
preclinical drug screening of personalized anticancer therapeutics,
65 FDA-approved anticancer drugs were screened using seven
different concentrations, and cell viability was measured after 6
days of drug exposure. The drug library consisted of agents
targeting 20 molecular targets, including compounds that are
undergoing clinical trial and preclinical investigations (16). The library also included three first
line chemotherapeutics for the treatment of colorectal cancer
(24), such as fluorouracil (5FU),
oxaliplatin and irinotecan. In the present study, drug screening
was performed for all PDCs established, and the results showed a
diverse range of drug sensitivities (Figs. S1–S4).
Following quantitative analysis, various effective drugs showing
statistically significant response scores were selected for each
PDC (Table SI). A small Z-score
indicates the effectiveness of a drug, as previously described
(16). PDC3, being derived from PDX3,
was the most sensitive cell line to various EGF receptor (EGFR)
inhibitors such as afatinib, lapatinib, CI-1033, dacomitinib and
gefitinib, in line with the mutation profiling results of PDXs in
Table III indicating that PDX3 was
the only cell line carrying wild-type KRAS and EGFR. The present
results suggested that PDCs could be used to identify effective
drugs against colorectal cancer cells. In addition, the PDC in
vitro model established in the present study may be suitable
for personalized medicine approaches.
 | Table III.Mutation profiling of PDXs. |
Table III.
Mutation profiling of PDXs.
|
|
| Mutation |
|---|
|
|
|
|
|---|
| Sample | Site | APC | PI3KCA | p53 | KRAS | EGFR |
|---|
| PDX1 | Metastasis | R858X | WT | FS_DEL | G12C | R521K |
| PDX2 | Primary | WT | WT | P72R | G12D | R521K |
| PDX3 | Primary | WT | WT | Y31C | WT | WT |
| PDX4 | Metastasis | R223X | WT | E285K | G13D | R199C |
Discussion
In the past, the propagation of adult neoplastic
epithelial cells required the use of specific media that led to
early onset senescence (25,26). To bypass senescence, the
overexpression of viral oncogenes such as simian virus 40 large T
antigen or E6/E7 proteins of oncogenic human papillomaviruses were
necessary, resulting in genomic alterations and antigenicity
(27–32). In the present study, colorectal cancer
cells derived from xenograft tissue could be conditionally
immortalized by combining the ROCK inhibitor Y-27632 and irradiated
fibroblast feeder cells. Based on STR analysis, cultured cells were
identified to be genetically identical to tumor tissues, suggesting
that continuous proliferation was not due to genomic instability,
but caused by the immortalization of cells. Previous studies have
shown that cells from various human tissues including prostate,
breast and colon could be conditionally immortalized with a ROCK
inhibitor and irradiated fibroblast feeder cells (11). Similarly to prostasphere or
mammosphere, PDCs formed well-defined spheres without contamination
of fibroblasts following administration of Matrigel. In a previous
study, contamination of fibroblasts has been suggested to limit
conditional reprogramming (9). In the
present study, EpCAM staining was used to selectively identify
epithelial cells and it was confirmed that EpCAM-positive cells
could be isolated and cultured without stromal cells. Differential
trypsinization methods were also used to separate epithelial cells
and stromal cells to avoid contaminations.
Although the mechanism of conditional reprogramming
has not been fully elucidated, previous studies have suggested that
increased activity of telomerase and dysregulation of the
cytoskeleton and p16/RB signaling pathway are associated with
conditional immortalization of epithelial cells (33–39). Liu
et al (11) demonstrated that
hTERT expression is increased in conditionally-reprogrammed
prostate and breast cancer cells. In the present study, the
expression levels of hTERT and C-Myc, genes involved in telomerase
activity, were higher in cultured cells compared with tumor
tissues. The present RT-qPCR results suggested that p16 expression
in cultured cells was decreased, whereas CDK4 expression was
increased compared with tumor tissues, suggesting that inactivation
of the p16/RB signaling pathway and dysregulation of the
cytoskeleton may be important mechanisms underlying conditional
reprogramming.
Notably, this efficient primary culture model can be
used for personalized treatment approaches. To assess the
applicability of this model for preclinical screening of
personalized anticancer therapeutics, 65 FDA-approved
chemotherapeutic agents were screened in the present study. The
drugs tested in the established in vitro model included
compounds generally used in the first-line treatment of colorectal
cancer, such as 5FU, oxaliplatin and irinotecan, and monoclonal
antibodies prescribed for targeted therapies, such as lapatinib or
imatinib.
In clinical settings, mutation profiles of KRAS and
EGFR are reliable and validated predictors for the effect of EGFR
inhibitors such as cetuximab (40,41). In
the present study, all PDXs, with the exception of PDX3, were
identified to carry mutations in KRAS and EGFR. In total, five EGFR
inhibitors were identified as effective drugs for PDC3, which were
originated from PDX3 cells expressing wild-type KRAS and EGFR.
Notably, EGFR inhibitors did not affect the growth of PDC1 and
PDC4. However, two EGFR inhibitors were identified as effective
drugs for PDC2, although PDX2, derived from PDC2, was identified to
carry mutations in KRAS and EGFR genes, suggesting potential
resistance to EGFR inhibitors. The present results may be explained
by intra-tumor heterogeneity (ITH), which lead to different
genotypes and phenotype of individual cancer cells within the same
tumor (42). ITH is one of the major
causes causing discordance between the results of drug screening
assays using patient-derived cancer cells and chemotherapy response
(43). Using multiple PDCs from a
single tumor at the same time may help overcoming this limitation.
Performing drug screening assays using the PDC platform established
in the present paper, clinicians could not only predict the
response to individual chemotherapeutic agents, but also consider a
combination of targeted drugs effective in cancer cells isolated
from patients in the first-line treatment (40,41).
The conditional reprogramming method described in
the present study may have several advantages, including lack of
genotypic drift, rapid proliferation, efficient immortalization and
high success rate (~50%) compared with conventional traditional
methods (1–10%) (44), and may allow
experiments such as screening assays of anticancer drug. Recently,
PDX and 3D organoid cultures have emerged as novel in vitro
models to study cancer (45).
Although PDXs present molecular and cellular features that reflect
tumor heterogeneity, they are difficult and expensive to develop
(6,15). By contrast, 3D organoid cultures are
easy to transfect and can be useful for assessing drug responses;
however, since 3D structures can be difficult to analyze by light
microscopy and organoid culture medium contains many growth factors
or small molecular inhibitors that can affect the response to the
tested drugs, using organoid models in automated high-throughput
drug screening is challenging. Considering that the timing for the
analysis of cancer tissues and therapeutic decisions is key in
clinical settings, conditional reprogramming could be the most
appropriate model for the application of personalized medicine
strategies. The high efficiency and robustness of conditionally
reprogrammed cells may increase the importance of biobanking and
facilitate the investigation of cancer cells for genetic and
molecular analysis allowing clinicians to make precise and prompt
decisions for treating patients using personalized treatment
approaches.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Yoo La Lee
(Samsung Advanced Institute for Health Sciences and Technology) and
Ms. Mi Na Hwang (Samsung Advanced Institute for Health Sciences and
Technology) for PDC culture and PDX generation.
Funding
The present study was supported by grants of The
Korea Health Technology R&D Project of The Korea Health
Industry Development Institute, funded by The Ministry of Health
and Welfare, Republic of Korea (grant. nos. HI14C3418 and
HI15C1593), and by The Basic Science Research Program of the
National Research Foundation of Korea, funded by The Ministry of
Education (grant. no. NRF-2017R1D1A1B03034309). In addition, the
present study was supported by The Samsung Medical Center (grant.
no. OTC1190571).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HKH and YBC designed the experiments. DHP and HKH
wrote the manuscript. HKH, DHP, TWK, NHY, YSL and SJS performed
experiments. WYL collected the clinical data and reviewed the
literature. YBC conceived the study and supervised the experiments
and writing of the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work is appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in the present study were
in accordance with the ethical principles of the care and use of
laboratory animals guidelines approved by the Institutional Animal
Care and Use Committee of Samsung Biomedical Research Institute.
The experiments conducted on animal and patient samples were
approved by the Institutional Review Board of Samsung Medical
Center. Written informed consents were obtained from all
participating patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeppesen M, Hagel G, Glenthoj A, Vainer B,
Ibsen P, Harling H, Thastrup O, Jørgensen LN and Thastrup J:
Short-term spheroid culture of primary colorectal cancer cells as
an in vitro model for personalizing cancer medicine. PLoS One.
12:e01830742017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Riedl A, Schlederer M, Pudelko K, Stadler
M, Walter S, Unterleuthner D, Unger C, Kramer N, Hengstschläger M,
Kenner L, et al: Comparison of cancer cells in 2D vs 3D culture
reveals differences in AKT-mTOR-S6K signaling and drug responses. J
Cell Sci. 130:203–218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dolznig H, Rupp C, Puri C, Haslinger C,
Schweifer N, Wieser E, Kerjaschki D and Garin-Chesa P: Modeling
colon adenocarcinomas in vitro a 3D co-culture system induces
cancer-relevant pathways upon tumor cell and stromal fibroblast
interaction. Am J Pathol. 179:487–501. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katt ME, Placone AL, Wong AD, Xu ZS and
Searson PC: In vitro tumor models: Advantages, disadvantages,
variables, and selecting the right platform. Front Bioeng
Biotechnol. 4:122016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hidalgo M, Amant F, Biankin AV, Budinská
E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo
GM, et al: Patient-derived xenograft models: An emerging platform
for translational cancer research. Cancer Discov. 4:998–1013. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gillet JP, Varma S and Gottesman MM: The
clinical relevance of cancer cell lines. J Natl Cancer Inst.
105:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Voskoglou-Nomikos T, Pater JL and Seymour
L: Clinical predictive value of the in vitro cell line, human
xenograft, and mouse allograft preclinical cancer models. Clin
Cancer Res. 9:4227–4239. 2003.PubMed/NCBI
|
|
9
|
Liu X, Krawczyk E, Suprynowicz FA,
Palechor-Ceron N, Yuan H, Dakic A, Simic V, Zheng YL, Sripadhan P,
Chen C, et al: Conditional reprogramming and long-term expansion of
normal and tumor cells from human biospecimens. Nat Protoc.
12:439–451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suprynowicz FA, Upadhyay G, Krawczyk E,
Kramer SC, Hebert JD, Liu X, Yuan H, Cheluvaraju C, Clapp PW,
Boucher RC Jr, et al: Conditionally reprogrammed cells represent a
stem-like state of adult epithelial cells. Proc Natl Acad Sci USA.
109:20035–20040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Ory V, Chapman S, Yuan H, Albanese
C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al:
ROCK inhibitor and feeder cells induce the conditional
reprogramming of epithelial cells. Am J Pathol. 180:599–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chapman S, Liu X, Meyers C, Schlegel R and
McBride AA: Human keratinocytes are efficiently immortalized by a
Rho kinase inhibitor. J Clin Invest. 120:2619–2626. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Clevers H: Modeling development and
disease with organoids. Cell. 165:1586–1597. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho YB, Hong HK, Choi YL, Oh E, Joo KM,
Jin J, Nam DH, Ko YH and Lee WY: Colorectal cancer patient-derived
xenografted tumors maintain characteristic features of the original
tumors. J Surg Res. 187:502–509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JK, Liu Z, Sa JK, Shin S, Wang J,
Bordyuh M, Cho HJ, Elliott O, Chu T, Choi SW, et al:
Pharmacogenomic landscape of patient-derived tumor cells informs
precision oncology therapy. Nat Genet. 50:1399–1411. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oh BY, Lee WY, Jung S, Hong HK, Nam DH,
Park YA, Huh JW, Yun SH, Kim HC, Chun HK and Cho YB: Correlation
between tumor engraftment in patient-derived xenograft models and
clinical outcomes in colorectal cancer patients. Oncotarget.
6:16059–16068. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chopra DP, Yeh K and Brockman RW:
Isolation and characterization of epithelial cell types from the
normal rat colon. Cancer Res. 41:168–175. 1981.PubMed/NCBI
|
|
19
|
McCallum HM and Lowther GW: Long-term
culture of primary breast cancer in defined medium. Breast Cancer
Res Treat. 39:247–259. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Castell JV and Gómez-Lechón MJ: Liver cell
culture techniques. Methods Mol Biol. 481:35–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mitra A, Mishra L and Li S: Technologies
for deriving primary tumor cells for use in personalized cancer
therapy. Trends Biotechnol. 31:347–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugimoto H, Mundel TM, Kieran MW and
Kalluri R: Identification of fibroblast heterogeneity in the tumor
microenvironment. Cancer Biol Ther. 5:1640–1646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dakic A, DiVito K, Fang S, Suprynowicz F,
Gaur A, Li X, Palechor-Ceron N, Simic V, Choudhury S, Yu S, et al:
ROCK inhibitor reduces Myc-induced apoptosis and mediates
immortalization of human keratinocytes. Oncotarget. 7:66740–66753.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
André T, Boni C, Navarro M, Tabernero J,
Hickish T, Topham C, Bonetti A, Clingan P, Bridgewater J, Rivera F
and de Gramont A: Improved overall survival with oxaliplatin,
fluorouracil, and leucovorin as adjuvant treatment in stage II or
III colon cancer in the MOSAIC trial. J Clin Oncol. 27:3109–3116.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Janik K, Popeda M, Peciak J, Rosiak K,
Smolarz M, Treda C, Rieske P, Stoczynska-Fidelus E and Ksiazkiewicz
M: Efficient and simple approach to in vitro culture of primary
epithelial cancer cells. Biosci Rep. 36(pii): e004232016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ali MY, Anand SV, Tangella K, Ramkumar D
and Saif TA: Isolation of primary human colon tumor cells from
surgical tissues and culturing them directly on soft elastic
substrates for traction cytometry. J Vis Exp. 2015:e525322015.
|
|
27
|
Liu X, Dakic A, Chen R, Disbrow GL, Zhang
Y, Dai Y and Schlegel R: Cell-restricted immortalization by human
papillomavirus correlates with telomerase activation and engagement
of the hTERT promoter by Myc. J Virol. 82:11568–11576. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Van der Haegen BA and Shay JW:
Immortalization of human mammary epithelial cells by SV40 large
T-antigen involves a two step mechanism. In Vitro Cell Dev Biol
29A. 180–182. 1993. View Article : Google Scholar
|
|
29
|
Hudson JB, Bedell MA, McCance DJ and
Laiminis LA: Immortalization and altered differentiation of human
keratinocytes in vitro by the E6 and E7 open reading frames of
human papillomavirus type 18. J Virol. 64:519–526. 1990.PubMed/NCBI
|
|
30
|
Münger K, Phelps WC, Bubb V, Howley PM and
Schlegel R: The E6 and E7 genes of the human papillomavirus type 16
together are necessary and sufficient for transformation of primary
human keratinocytes. J Virol. 63:4417–4421. 1989.PubMed/NCBI
|
|
31
|
Hawley-Nelson P, Vousden KH, Hubbert NL,
Lowy DR and Schiller JT: HPV16 E6 and E7 proteins cooperate to
immortalize human foreskin keratinocytes. EMBO J. 8:3905–3910.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schlegel R, Phelps WC, Zhang YL and
Barbosa M: Quantitative keratinocyte assay detects two biological
activities of human papillomavirus DNA and identifies viral types
associated with cervical carcinoma. EMBO J. 7:3181–3187. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang L, Valdez JM, Zhang B, Wei L, Chang
J and Xin L: ROCK inhibitor Y-27632 suppresses dissociation-induced
apoptosis of murine prostate stem/progenitor cells and increases
their cloning efficiency. PLoS One. 6:e182712011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Terunuma A, Limgala RP, Park CJ, Choudhary
I and Vogel JC: Efficient procurement of epithelial stem cells from
human tissue specimens using a Rho-associated protein kinase
inhibitor Y-27632. Tissue Eng Part A. 16:1363–1368. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hanna JH, Saha K and Jaenisch R:
Pluripotency and cellular reprogramming: Facts, hypotheses,
unresolved issues. Cell. 143:508–525. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Takehara T, Teramura T, Onodera Y,
Kakegawa R, Fukunaga N, Takenoshita M, Sagawa N, Fukuda K and Hosoi
Y: Rho-associated kinase inhibitor Y-27632 promotes survival of
cynomolgus monkey embryonic stem cells. Mol Hum Reprod. 14:627–634.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li X, Meng G, Krawetz R, Liu S and
Rancourt DE: The ROCK inhibitor Y-27632 enhances the survival rate
of human embryonic stem cells following cryopreservation. Stem
Cells Dev. 17:1079–1085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Krawetz R, Liu S, Meng G and
Rancourt DE: ROCK inhibitor improves survival of cryopreserved
serum/feeder-free single human embryonic stem cells. Hum Reprod.
24:580–589. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Watanabe K, Ueno M, Kamiya D, Nishiyama A,
Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S,
Muguruma K and Sasai Y: A ROCK inhibitor permits survival of
dissociated human embryonic stem cells. Nat Biotechnol. 25:681–686.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lièvre A, Bachet JB, Le Corre D, Boige V,
Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bardelli A and Jänne PA: The road to
resistance: EGFR mutation and cetuximab. Nat Med. 18:199–200. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schütte M, Risch T, Abdavi-Azar N, Boehnke
K, Schumacher D, Keil M, Yildiriman R, Jandrasits C, Borodina T,
Amstislavskiy V, et al: Molecular dissection of colorectal cancer
in pre-clinical models identifies biomarkers predicting sensitivity
to EGFR inhibitors. Nat Commun. 8:142622017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McGranahan N and Swanton C: Clonal
heterogeneity and tumor evolution: Past, present, and the future.
Cell. 168:613–628. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dangles-Marie V, Pocard M, Richon S,
Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N,
Validire P, et al: Establishment of human colon cancer cell lines
from fresh tumors versus xenografts: Comparison of success rate and
cell line features. Cancer Res. 67:398–407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sachs N and Clevers H: Organoid cultures
for the analysis of cancer phenotypes. Curr Opin Genet Dev.
24:68–73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Edge SB and Compton CC: The American Joint
Committee On Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|