Introduction
Curcumin, also known as diferuloylmethane, is a
natural hydrophobic molecule derived from Curcuma longa
(also known as turmeric). The structure of curcumin is comprised of
three chemical entities: Two aromatic ring systems containing
o-methoxy phenolic groups, connected by a seven-carbon linker
consisting of an α, β-unsaturated β-diketone moiety (1–3). Curcumin
is a common food additive used as a spice and a coloring agent
(4). This compound has a particularly
good safety profile; its toxicity and tolerability have been
tested, and a previous study revealed that it is highly tolerable
at doses up to 12 g/day in patients, with no curcumin-related
toxicities, due to its low bioavailability (5–7).
It has been reported that curcumin is able to block
the nuclear factor (NF)-κB pathway, which is a critical mediator of
intracellular signaling that has been linked to proinflammatory
signals that control the expression of a vast array of genes
involved in immune and stress responses (8). Notably, this suppression is mediated by
numerous inflammatory stimuli through inhibition of IKKα kinase. It
has also been reported that the increased levels of tumor necrosis
factor-α, interleukin (IL)-6 and IL-8 in a model of acute kidney
injury are significantly reduced by curcumin treatment (2). The primary role of curcumin as an
antioxidant is to intercept peroxyl free radicals formed during
lipid peroxidation. This prevents free-radical chain reactions
which induce deterioration of the lipid membrane (9). Furthermore, a previous study indicated
that the enhancement of apoptosis induced by high doses of curcumin
is regulated by antioxidants, such as N-acetyl-l-cysteine (10). Previous studies demonstrated that
curcumin prevents several types of cancer (3–5,11,12). In
addition, curcumin has been reported to possess anti-invasive
properties and is able to interfere with the epithelial-mesenchymal
transition (EMT) (13). During EMT,
cells shift from an epithelial phenotype towards a mesenchymal
phenotype, modifying the expression profile of some adhesion
molecules and allowing cells to adopt invasive, migratory behaviors
(14).
Cervical cancer is the fourth most common type of
cancer in women worldwide and is commonly caused by sexually
acquired infection with high-risk human papillomavirus (HR-HPV)
(15). Two types of HR-HPV (HPV16 and
18) cause 70% of cervical cancer cases and precancerous cervical
lesions (16). HR-HPV E6 and E7
oncoproteins are overexpressed in cervical carcinoma; this is
principally mediated by integration of the virus into the host
genome (17). These oncoproteins are
considered to be the primary carcinogenic factors in cervical
cancer cells. E6 protein binds to tumor suppressor protein p53, and
causes its ubiquitination and proteolysis via the proteasome
pathway, whereas E7 protein inactivates the retinoblastoma protein
by inducing its degradation. Therefore, E6 and E7 oncogenes are
strongly involved in cell cycle progression and reduced apoptosis,
which lead to carcinogenesis (18,19). Our
previous study reported that HPV16 E6 and E7 oncoproteins are
involved in promoting Pirin gene (PIR) upregulation in epithelial
cervical and oral cancer cells. Since the PIR gene encodes for
Pirin, an oxidative stress sensor, these previous findings
suggested that E6 and E7 are involved in cervical cancer
progression through Pirin (20).
Pirin is a highly conserved 32-kDa protein assigned to the cupin
superfamily of proteins (21,22). Its structure comprises two barrel
domains, with an Fe (II) cofactor bound within the cavity of the
N-terminal domain (23). Furthermore,
it has been reported that Pirin may act as an activator of the
NF-κB pathway (24). It has
previously been demonstrated that Pirin is involved in EMT
regulation, as well as cell migration (20,25,26). There
is mounting evidence of a hierarchy that controls the expression of
transcriptional regulators of EMT. During development and cancer
progression, the contribution of different EMT inducers is
dependent on the cellular context (14). Previous findings have suggested that
curcumin is able to decrease the levels of HR-HPV E6/E7 in cervical
cancer cells (6). The present study
revealed that curcumin decreased EMT of cervical cancer cells, at
least in part, through a Pirin-dependent mechanism, which in turn
may be dependent on HPV16 E6/E7 levels.
Materials and methods
Cell culture and curcumin
treatment
The SiHa (HTB-35) cell line was obtained directly
from the American Type Culture Collection. The cells were incubated
in RPMI-1640 basal medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare) and antibiotics (100 U/ml penicillin, 100 g/ml
streptomycin) at 37°C in an atmosphere containing 5%
CO2. Cells were serum-depleted for 24 h and were treated
with 20 µM curcumin (Sigma-Aldrich; Merck KGaA) or DMSO for 72 h at
37°C in an atmosphere containing 5% CO2 in a humidified
incubator.
Cell viability assay
Cells were grown to 80% confluence and were
incubated with serum-free RPMI-1640 medium in 60-mm plates for 24
h. The cells were then trypsinized with EDTA-trypsin mixture
(Invitrogen; Thermo Fisher Scientific, Inc.) and counted using
trypan blue exclusion staining with a Neubauer chamber (27). For the MTS assay, cells were cultured
to 80% confluence in a 96-well flat-bottomed microtiter plate with
RPMI-1640 medium and were treated with 20 µM curcumin for 24–96 h;
DMSO was used as a control. Finally, 20 µl MTS (Promega
Corporation) was added to each well, the plate was incubated for 3
h at 37°C and absorbance was measured at 492 nm using a microplate
reader (Biotek Instruments, Inc.).
Knockdown of PIR and E6/E7
Cells at 80% confluence were serum-depleted for 24 h
and were transfected with 30 µM small interfering RNA (siRNA)
targeting PIR (cat. no. sc-61359; Santa Cruz Biotechnology, Inc.),
HPV16 E6/E7 (cat. no. sc-270423; Santa Cruz Biotechnology, Inc.)
and Silencer™ Select Negative Control No. 1 siRNA (cat. no.
4390843; Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h at
37°C in an atmosphere containing 5% CO2 using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. RNA and
proteins were collected for subsequent analyses.
RNA extraction and cDNA synthesis
RNA was isolated from cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Following chloroform purification and
isopropanol precipitation, RNA was suspended in DEPC-water and
stored at −80°C. The RNA was treated with RQ1 RNase-free DNase
(Promega Corporation) at 37°C for 120 min and was incubated with
RQ1 DNase Stop Solution for 10 min. cDNA was prepared in a 20-ml
reaction volume containing DNAse-treated RNA (1 µg), 1 unit RNAse
inhibitor (Promega Corporation), 0.4 µg/ml random primers (Promega
Corporation), 2 mM dNTP (Promega Corporation) and 10 units Moloney
Murine Leukemia Virus reverse transcriptase (Promega Corporation).
The reaction mixture was incubated for 1 h at 37°C.
Quantitative polymerase chain reaction
(qPCR)
cDNA was subjected to qPCR quantification of gene
expression with specific primers (Table
I) using a Rotor-Gene 6000 system (Qiagen, Inc.) under the
following conditions: 95°C for 10 min followed by 40 cycles of
denaturation at 95°C for 15 sec, annealing at 55°C for 20 sec and
extension at 72°C for 20 sec. To obtain the dissociation curve, the
temperature was increased from 70 to 90°C (0.5°C increase at each
step). The reaction was performed using 2X SYBR Green Master Mix
(Bioline), 0.4 µM primer pair, 10.5 µl RNase-free water and 1 µl
cDNA in a 25-µl final volume. Specific primers and amplification
conditions were adjusted for each RNA sequence. Standard curves for
each gene were generated independently by preparing 10-fold serial
dilutions of DNA amplicons. The relative copy number of each sample
was calculated through the 2−∆∆Cq method using
Rotor-Gene software version 4 (Qiagen, Inc.) (28). Endogenous β-actin mRNA levels were
used for normalization of mRNA expression. All reactions were
performed in triplicate.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Product size
(bp) | Primer
sequence | Annealing
temperature, °C |
|---|
| E6 | 96 | F:
5′-CTGCAAGCAACAGTTACTGCGA-3′ |
|
|
|
|
R:5′-TCACACACTGCATATGGATTCCC-3′ | 58 |
| E7 | 120 | F:
5′-CAATATTGTAATGGGCTCTGTCC-3′ |
|
|
|
| R:
5′-ATTTGCAACCAGAGACAACTGAT-3′ | 58 |
| E-cadherin | 135 | F:
5′-GCACCGGTCGACAAAGGACA-3′ |
|
|
|
| R:
5′-AGTCCCAGGCGTAGACCAAGA-3′ | 58 |
| N-cadherin | 121 | F:
5′-AAGAACGCCAGGCCAAACAA-3′ |
|
|
|
| R:
5′-TGCAGCTGGCTCAAGTCATA-3′ | 58 |
| Vimentin | 131 | F:
5′-GCCCTTGACATTGAGATTGCCA-3′ |
|
|
|
| R:
5′-TCAACCAGAGGGAGTGAATCCA-3′ | 58 |
| Slug | 117 | F:
5′-CTCCATTCCACGCCCAGCTAC-3′ |
|
|
|
| R:
5′-AGCCACTGTGGTCCTTGGAG-3′ | 60 |
| Snail | 84 | F:
5′-AGGCTCGAAAGGCCTTCAACT-3′ |
|
|
|
| R:
5′-TGTGGCTTCGGATGTGCATCT-3′ | 60 |
| Zeb1 | 157 | F:
5′-ACTGCCTGGTGATGCTGAAA-3′ |
|
|
|
| R:
5′-CCCAAACTGCAAGAAACGCT-3′ | 60 |
| PIR | 131 | F:
5′-TCAAATTGGACCCAGGAGCC-3′ |
|
|
|
| R:
5′-TCCAAGCACTGCTGTGTGAT-3′ | 55 |
Western blotting
Total protein was extracted from cells with a lysis
buffer [20 mM Tris (pH 8.0), 1% SDS] containing protease inhibitor
cocktail (Roche Diagnostics). The cells were incubated at 4°C for 1
h, sonicated at 20 kHz for 20 sec on ice and centrifuged at 12,000
× g for 10 min at 4°C. The proteins were quantified using the
Pierce Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher
Scientific, Inc.) and 30 µg protein extract was loaded per well.
Proteins were separated by 12% SDS-PAGE and were then transferred
to Hybond-P ECL membranes (Amersham; GE Healthcare) using 20 mM
Tris and 150 mM glycine (pH 8.3), in 20% methanol with semi-dry
transfer apparatus (Bio-Rad Laboratories, Inc.). Membranes were
then incubated for 2 h at room temperature with the blocking
reagent [5% bovine serum albumin (AppliChem GmbH) in 0.5% Tris
buffered saline/0.1% Tween-20 (TBST, pH 7.6)] and were incubated
overnight at room temperature with the following primary
antibodies: β-actin (cat. no. ab6276; Abcam), N-cadherin (cat. no.
ab98952; Abcam), zinc finger E-box binding homeobox 1 (Zeb1; cat.
no. pa520979; Thermo Fisher Scientific, Inc.), Pirin (cat. no.
ab51360; Abcam), HPV16 E6 (cat. no. sc-460; Santa Cruz
Biotechnology, Inc.), HPV16 E7 (cat. no. sc-6981; Santa Cruz
Biotechnology, Inc.), E-cadherin (cat. no. sc-21791; Santa Cruz
Biotechnology, Inc.), Slug (cat. no. sc-166476; Santa Cruz
Biotechnology, Inc.) and Vimentin (cat. no. sc-6260; Santa Cruz
Biotechnology, Inc.), which were diluted 1:1,000 in TBST. Membranes
were washed three times in TBST and were then incubated for 1 h at
room temperature with peroxidase-labeled secondary IgG antibodies
(anti-mouse cat. no. 554002; BD Pharmingen; BD Biosciences; and
anti-rabbit cat. no. sc-2004; Santa Cruz Biotechnology, Inc.),
which were diluted 1:2,000 in TBST. After washing three times in
TBST, immune complexes were detected using an ECL system (Amersham;
GE Healthcare) according to the manufacturer's protocol. For
semi-quantitative analysis, ImageJ software version 1.52a (National
Institutes of Health) was used.
Cell migration assays
For the three-dimensional migration assay, the
bottom side of the upper chamber of a Transwell system (pore size,
8 µm; Corning, Inc.) was coated with 1 µg/ml fibronectin (cat. no.
PHE0023; Gibco; Thermo Fisher Scientific, Inc.) and incubated
overnight at 4°C. A total of 50,000 SiHa cells in supplement-free
medium were seeded in the upper chamber, whereas 500 µl complete
medium was added per well. Cells were then allowed to migrate for 7
h at 37°C in an atmosphere containing 5% CO2. Migrated
cells were fixed and stained for 15 min at room temperature with
0.5% w/v crystal violet/methanol solution, whereas non-migrated
cells were removed using a cotton tip. Migrated cells were counted
in eight fields for each experiment using an inverted light
microscope.
The wound-healing assay was carried out according to
previously reported protocols (29).
Briefly, SiHa cells were serum-depleted for 24 h. Subsequently,
4×105 cells were seeded in 6-well plates at 90% cell
confluence and exposed to 20 µM curcumin for 72 h at 37°C. A 10-µl
pipette tip was used to scrape the cell monolayer in a straight
line and the cells were washed with PBS and incubated in RPMI
medium containing 10% FBS. Images of the scratches were captured at
0 and 18 h after wound generation, using a polarized light inverted
microscope (magnification, ×20).
Statistical analysis
Comparison of means between two groups was performed
using a Mann Whitney test, whereas comparisons between multiple
groups were performed using one-way ANOVA and Tukey's post hoc
test. Data were analyzed using GraphPad Prism version 7 software
(GraphPad Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
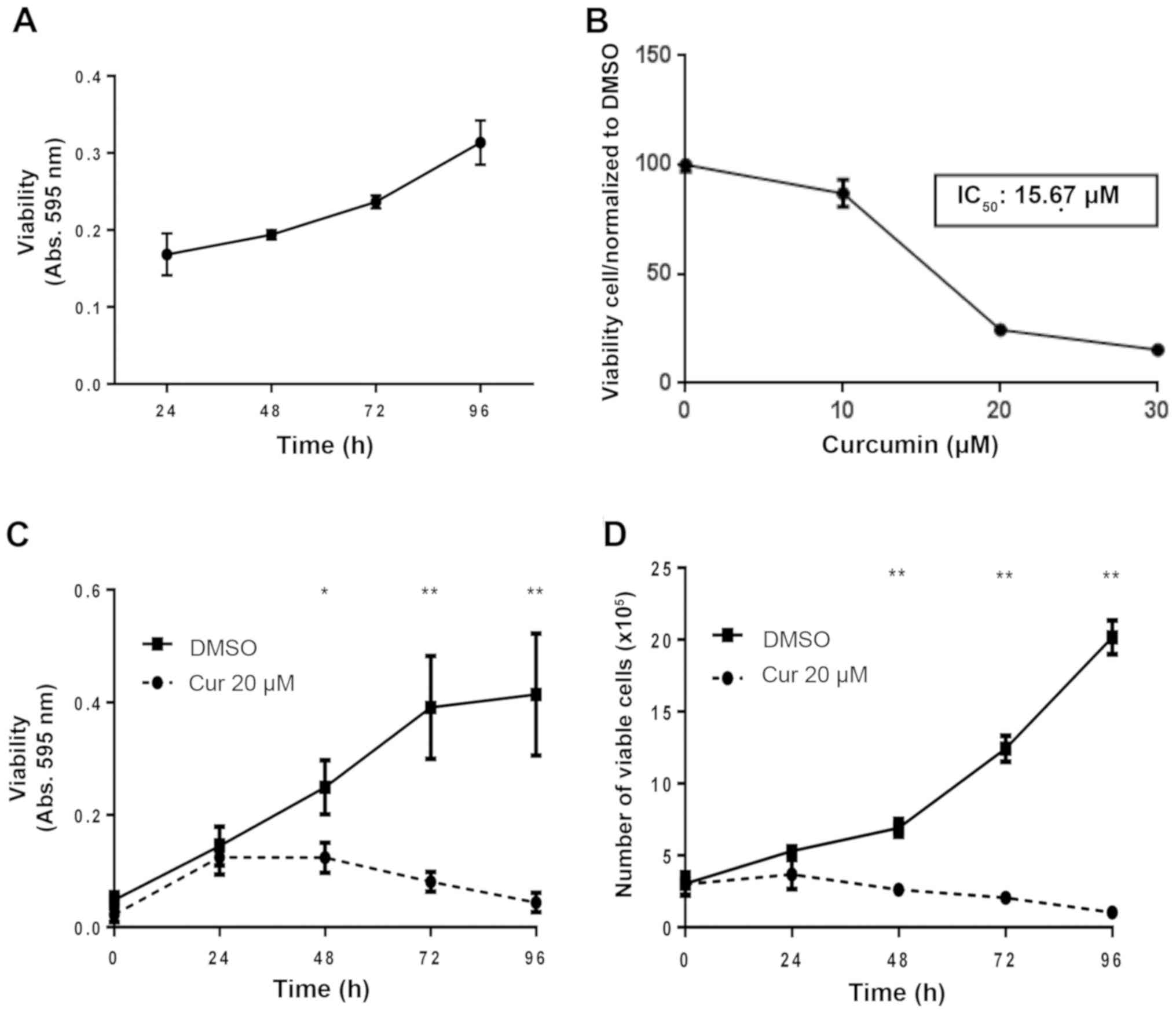
Curcumin decreases viability of
cervical cancer cells
The present study used SiHa cervical carcinoma
cells, which harbor two copies of HPV16 per cell. A cell
viability-time curve is shown in Fig.
1A. In order to determine the appropriate curcumin
concentration to be used in subsequent analyses, the viability of
SiHa cells in response to various curcumin concentrations was
determined by MTS assay (Fig. 1B).
The IC50 value was determined to be 15.67 µM.
Considering this concentration as a reference and according to
previously reported data (30,31), a
concentration of 20 µM curcumin was used for subsequent
experiments. Cell viability in response to this curcumin
concentration was measured by MTS assay (Fig. 1C) and cell counting was conducted
using the Trypan blue exclusion assay (Fig. 1D). Although significant cell death was
found at 48 and 72 h (Fig. 1C and D),
the curcumin exposure time for subsequent analyses was extended to
72 h.
Curcumin alters E6/E7 and Pirin levels
in cervical cancer cells
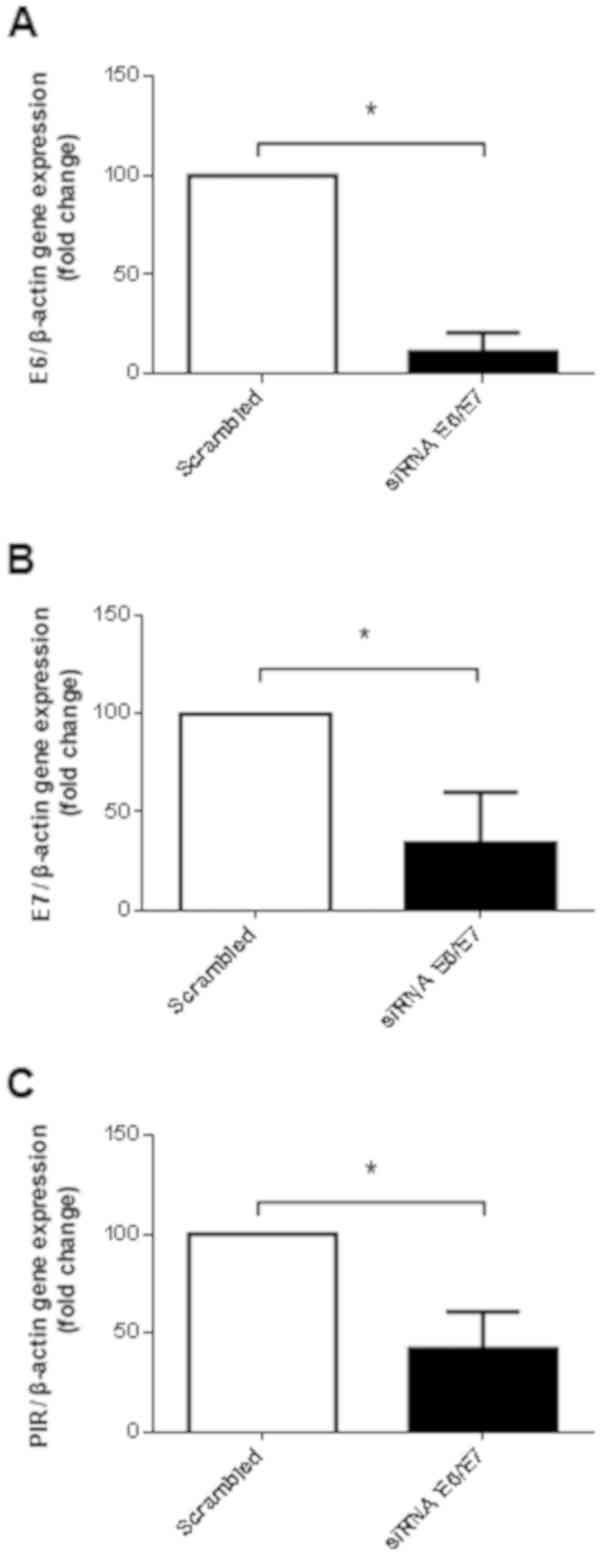
Firstly, this study confirmed that PIR expression is
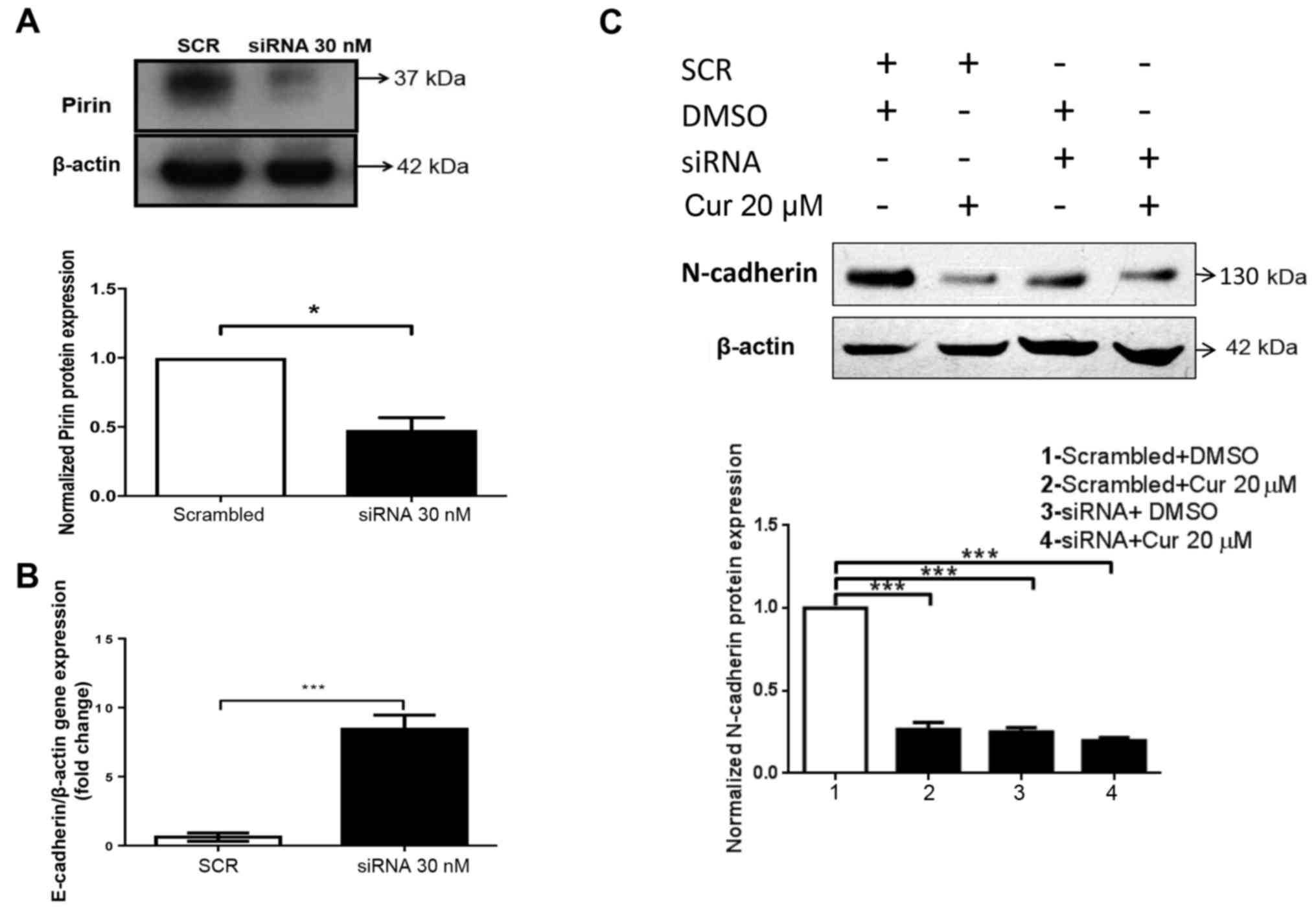
dependent on E6/E7 in SiHa cells. Accordingly, as shown in Fig. 2A and B, siRNA induced a significant
reduction in the mRNA expression levels of E6 and E7. In addition,
the mRNA expression levels of PIR were significantly decreased in
response to E6/E7 knockdown (Fig.
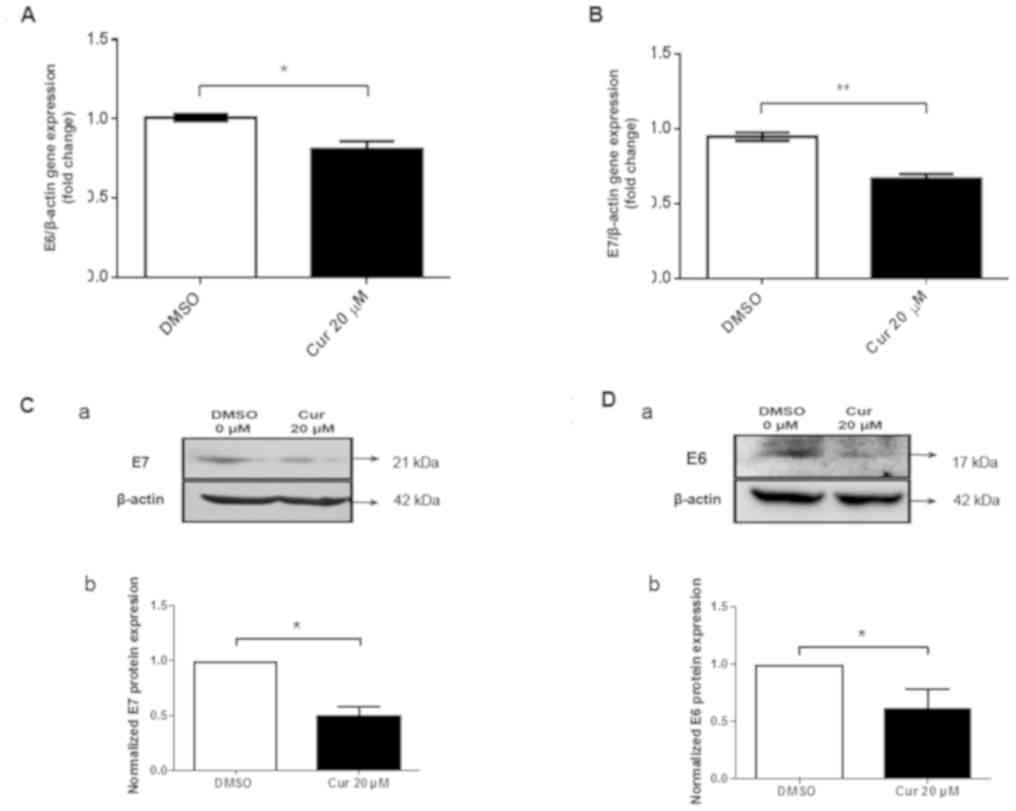
2C). To determine the effects of curcumin on alterations in E6
and E7 gene expression, SiHa cells were exposed to 20 µM curcumin
for 72 h. Firstly, it was demonstrated that curcumin induced a
significant decrease in endogenous E6 and E7 mRNA expression levels
(Fig. 3A and B). A significant
decrease in E6 and E7 protein expression was also observed in
response to curcumin (Fig. 3C and D).
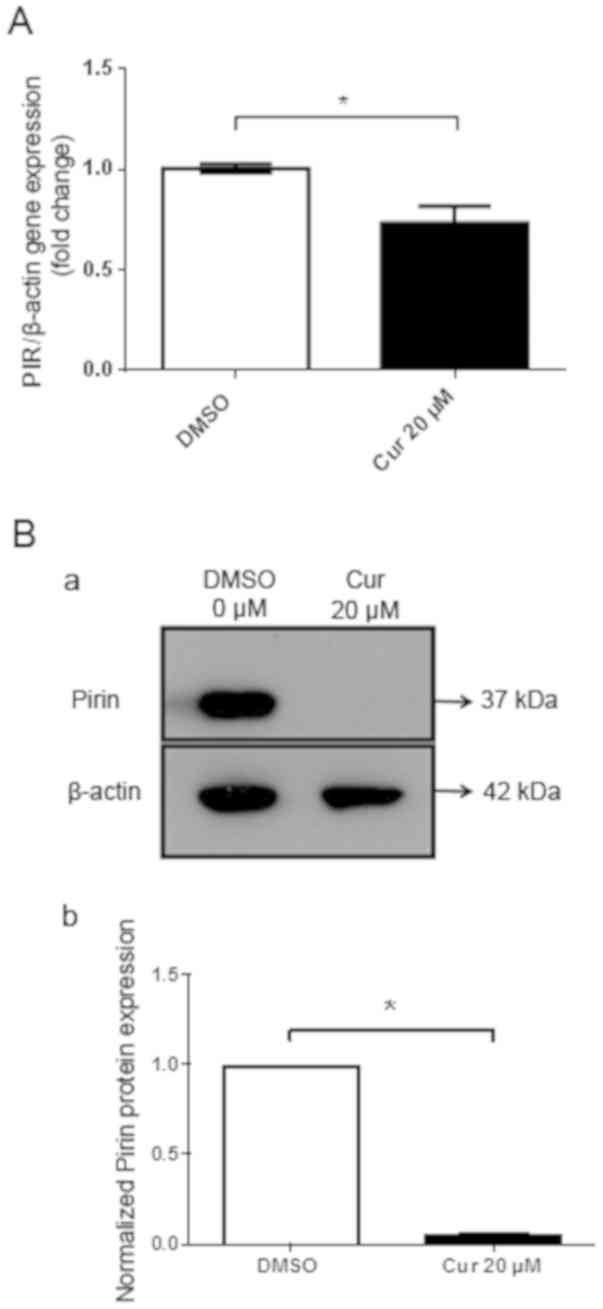
In addition, the effects of curcumin exposure on the mRNA and
protein expression levels of Pirin were analyzed. A significant
decrease in PIR mRNA expression (Fig.
4A) and Pirin protein expression (Fig. 4B) was detected following exposure to
20 µM curcumin for 72 h. Taken together, E6 and E7 may regulate
Pirin in SiHa cells, whereas curcumin may decrease the levels of
E6/E7 and Pirin in these cells.
Pirin promotes EMT in cervical cancer
cells
To determine whether knockdown of PIR affects EMT,
cells were transfected with a siRNA. Western blotting was initially
performed to confirm the efficiency of PIR siRNA. As shown in
Fig. 5A, a significant decrease in
Pirin protein expression was revealed. Subsequently, a significant
increase in the mRNA expression levels of E-cadherin was observed
(Fig. 5B) following PIR knockdown,
although it should be noted that the mRNA expression levels of
N-cadherin and Vimentin were not detected by RT-qPCR (data not
shown). The increase in E-cadherin expression suggested that
dependence may exist between Pirin and EMT. Subsequently, PIR was
knocked down and SiHa cells were exposed to 20 µM curcumin for 72
h. In the PIR siRNA + DMSO group, the protein expression levels of
N-cadherin were significantly decreased (Fig. 5C). Furthermore, N-cadherin expression
was also decreased in the scrambled siRNA + curcumin and PIR siRNA
+ curcumin groups, suggesting that both curcumin and Pirin may be
involved in N-cadherin downregulation.
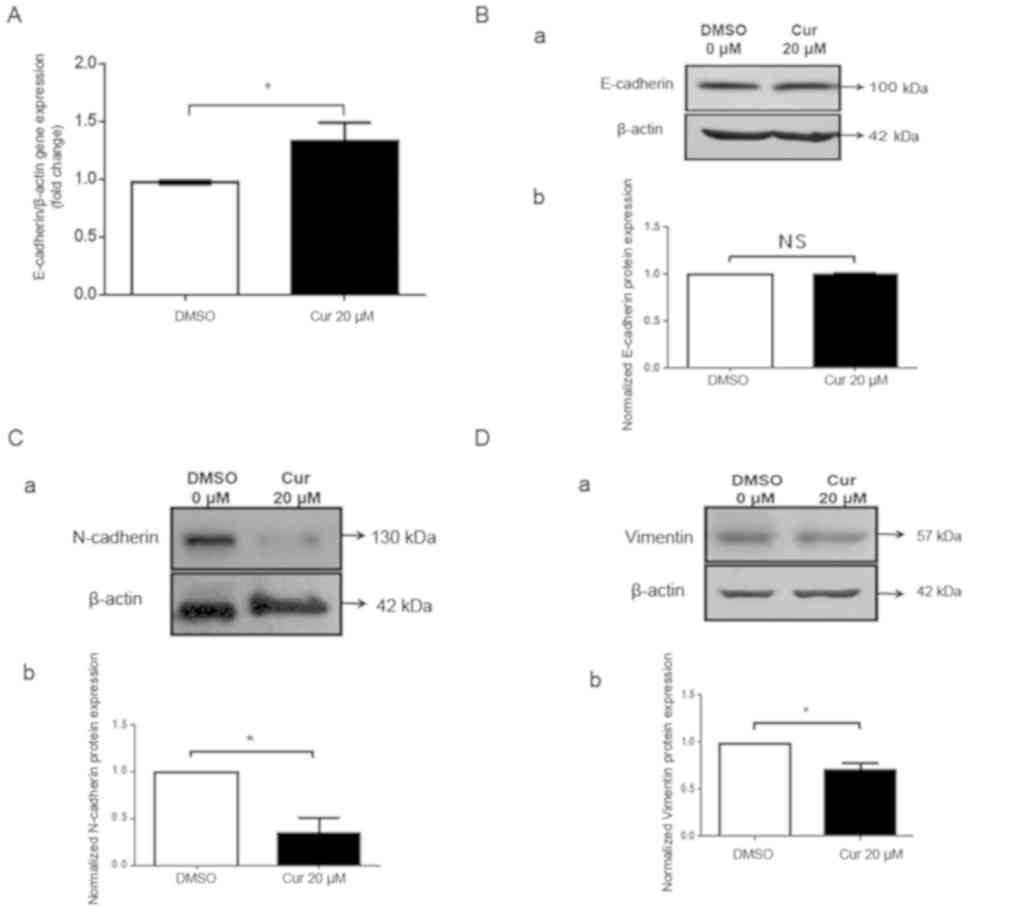
Curcumin decreases EMT
The effects of curcumin on EMT were evaluated; the
epithelial marker E-cadherin was detected. The mRNA expression
levels of E-cadherin were increased (Fig.
6A); however, the protein expression levels were not affected
by 20 µM curcumin (Fig. 6B). In
addition, the expression levels of the mesenchymal markers
N-cadherin (Fig. 6C), Vimentin
(Fig. 6D), Slug (Fig. S1A) and Zeb1 (Fig. S1B) were detected; the protein
expression levels of all of these markers were reduced by ≥40% in
response to curcumin. Finally, the mRNA expression levels of Zeb1
(Fig. S1B) and Snail (Fig. S1C) were increased in response to
curcumin. The discrepancy between the mRNA and protein expression
levels of Zeb1 may be due to changes induced by curcumin at the
post-translational level; notably, mechanisms of protein
degradation may have been induced, which affected EMT-related
proteins. These observations suggested that curcumin may induce a
decrease in SiHa cell EMT.
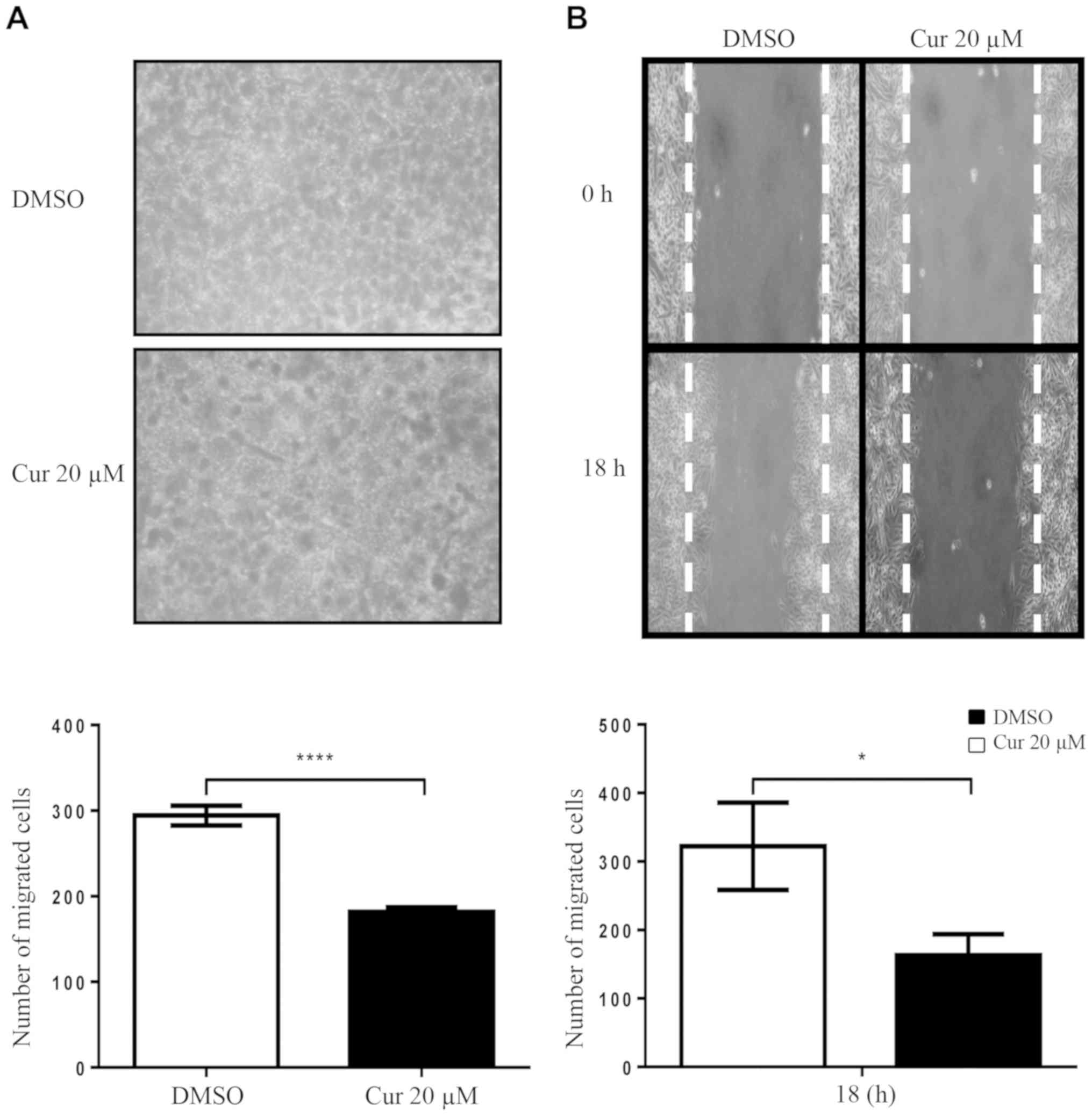
Curcumin exposure decreases cell
migration
Cell migration was evaluated in SiHa cells exposed
to curcumin using Transwell and wound healing assays. As shown in
Fig. 7A, a significant decrease in
cell migration was observed following exposure to 20 µM curcumin
for 72 h. To further confirm these results, a wound-healing assay
was conducted. As shown in Fig. 7B,
it was confirmed that curcumin reduced migration of SiHa cells.
Discussion
Curcumin has previously been described as an
inhibitor of carcinogenesis (13,32) and as
a regulator of EMT (33); however,
the mechanism by which this polyphenolic compound regulates EMT has
not been established. Therefore, this study proposed a mechanism to
explain this issue in cervical cancer cells. The SiHa cell line,
which harbors two integrated copies of HPV16 (34), was exposed to 20 µM curcumin for 72 h;
this curcumin concentration has been supported by previous reports
(30,31). In addition, a significant decrease in
E6 and E7 was observed in cells exposed to curcumin. This finding
is in accordance with the findings of previous studies, in
vitro (6) and in vivo
(12).
Pirin is a well-known oxidative stress sensor;
however, its specific function has not been fully elucidated. It is
known that human PIR is expressed at different levels in various
organs, including the heart, brain, liver, kidney, lung, pancreas,
placenta and skeletal muscle; the highest expression is observed in
the liver and heart, whereas the lowest is in the brain and
pancreas (11,35). To demonstrate the participation of
Pirin in the regulation of EMT, a siRNA against PIR was used;
post-transfection with this siRNA, an increase in E-cadherin mRNA
expression was observed. These data suggested that EMT may be
regulated by Pirin. These results are similar to previously
documented findings (25) in a HeLa
cell model; in this previous study, Pirin was downregulated to
demonstrate that it has the ability to promote EMT. In addition,
these same authors concluded that Pirin regulates EMT independently
to binding to BCL3-Slug (25).
The loss of E-cadherin expression is considered a
crucial step in the progression of low-grade lesions to invasive
carcinoma, and is a fundamental event in EMT (36). In addition, N-cadherin stimulates cell
migration and invasion. It has previously been demonstrated that
aberrant expression of N-cadherin makes cancer cells more motile
and invasive (37). In addition,
Vimentin is regarded as a canonical marker of EMT (38). Snail transcription factors bind to
E-box consensus sequences in the E-cadherin promoter, with the help
of local modifications of chromatin structure, and participate in
downregulation of the transcriptional activation of the E-cadherin
promoter. Slug (also known as SNAI2) is a member of the Snail
family of transcriptional repressors, which is capable of
suppressing E-cadherin expression and thereby triggering EMT, thus
suggesting that it may act as a promoter of migration (39).
It has been reported that Snail and its family
member Slug are capable of suppressing E-cadherin in epithelial
cells via E-box elements in the proximal E-cadherin promoter
(40). Zeb gene activation frequently
occurs upon Snail activation, and Snail1 has been reported to
activate Zeb1; however, Zeb is active in some tumors that lack
Snail expression (41); therefore,
Zeb protein may be an upstream marker of EMT that can be evaluated
together with E-cadherin and N-cadherin. This study demonstrated
that knocking down PIR decreased the expression levels of
N-cadherin, suggesting that Pirin regulates EMT. Furthermore, it
was observed that the combination of curcumin + PIR siRNA decreased
N-cadherin.
These data suggested that curcumin may negatively
regulate the protein expression levels of Pirin, which in turn may
regulate the expression of N-cadherin and affect the EMT. This
evidence provides important information regarding the mechanism by
which curcumin decreases EMT. However, these data are insufficient
to deny the possibility of a Pirin-independent mechanism, by which
curcumin decreases N-cadherin and EMT. Notably, this study
demonstrated that curcumin decreased the protein expression levels
of Pirin and the mRNA expression levels of PIR, confirming the
mechanism proposed in this study. The findings of this study also
suggested that curcumin may participate in the decrease of EMT in
cervical cancer cells; however, there was no alteration in the
protein expression levels of E-cadherin post-exposure to curcumin,
thus suggesting that other mechanisms mediated by curcumin may be
involved in the regulation of the levels of E-cadherin (for
example, microRNAs). In addition, the increase in Snail and Zeb1
mRNA expression contrasted the results obtained by western
blotting. Previously, differences in the response of EMT markers
have been reported in HeLa cells (HPV-18) (30) and in breast cancer cell lines exposed
to curcumin (13).
Finally, to analyze the phenotypic alterations
associated with the curcumin-mediated decrease in EMT, cell
migration was evaluated by two methods. A significant decrease in
migration was observed, which was similar to the findings reported
by previous studies in this cellular model (30), in CaSki cells (HPV16) (6), in mice (12) and in breast cancer (13). In conclusion, curcumin decreased EMT
and migration in cervical cancer cells. In addition, curcumin
decreased Pirin expression and this protein was revealed to
regulate EMT. These results suggested a novel mechanism by which
curcumin may exerts its effect, in which the Pirin protein has an
important role. Therefore, the Pirin protein may be considered an
important pharmacological target in the treatment of cervical
cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mr. Leodán A.
Crispin (Instituto de Alta Investigación, Universidad de Tarapacá,
Arica) for his technical support.
Funding
This study was funded by Fondecyt [grant nos.
1161219 (FA) and 1151214 (HC)] and Conicyt-Fondap (grant no.
15130011).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
VAA designed and performed the majority of the
experiments, analyzed and discussed data, and approved the
manuscript. DCB, JPMB, NG and JCO performed additional experiments,
contributed to the discussion and approved the manuscript. HRC, JT
and OL contributed to the experimental design and manuscript
discussion, and provided reagents. FA and GMC designed experiments,
discussed the results and wrote the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Priyadarsini KI: The chemistry of
curcumin: From extraction to therapeutic agent. Molecules.
19:20091–20112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar P, Sulakhiya K, Barua CC and Mundhe
N: TNF-α, IL-6 and IL-10 expressions, responsible for disparity in
action of curcumin against cisplatin-induced nephrotoxicity in
rats. Mol Cell Biochem. 431:113–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baghbani F, Chegeni M, Moztarzadeh F,
Hadian-Ghazvini S and Raz M: Novel ultrasound-responsive
chitosan/perfluorohexane nanodroplets for image-guided smart
delivery of an anticancer agent: Curcumin. Mater Sci Eng C.
74:186–193. 2017. View Article : Google Scholar
|
|
4
|
Cheng A-L, Hsu C-H, Lin J-K, Hsu M-M, Ho
Y-F, Shen T-S, Ko J-Y, Lin J-T, Lin B-R, Ming-Shiang W, et al:
Phase I clinical trial of curcumin, a chemopreventive agent, in
patients with high-risk or pre-malignant lesions. Anticancer Res
21B. 2895–2900. 2001.
|
|
5
|
Liu Z, Liu J, Zhao L, Geng H, Ma J, Zhang
Z, Yu D and Zhong C: Curcumin reverses benzidine-induced
epithelial-mesenchymal transition via suppression of ERK5/AP-1 in
SV-40 immortalized human urothelial cells. Int J Oncol.
50:1321–1329. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maher DM, Bell MC, O'Donnell EA, Gupta BK,
Jaggi M and Chauhan SC: Curcumin suppresses human papillomavirus
oncoproteins, restores p53, Rb, and PTPN13 proteins and inhibits
benzo[a]pyrene-induced upregulation of HPV E7. Mol Carcinog.
50:47–57. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park W, Amin AR, Chen ZG and Shin DM: New
perspectives of curcumin in cancer prevention. Cancer Prev Res
(Phila). 6:387–400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Panahi Y, Darvishi B, Ghanei M, Jowzi N,
Beiraghdar F and Varnamkhasti BS: Molecular mechanisms of curcumins
suppressing effects on tumorigenesis, angiogenesis and metastasis,
focusing on NF-κB pathway. Cytokine Growth Factor Rev. 28:21–29.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wright JS: Predicting the antioxidant
activity of curcumin and curcuminoids. J Mol Struct. 591:207–217.
2002. View Article : Google Scholar
|
|
10
|
Scharstuhl A, Mutsaers HAM, Pennings SWC,
Szarek WA, Russel FGM and Wagener FADTG: Curcumin-induced
fibroblast apoptosis and in vitro wound contraction are regulated
by antioxidants and heme oxygenase: Implications for scar
formation. J Cell Mol Med. 13:712–725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brzóska K, Stępkowski TM and Kruszewski M:
Basal PIR expression in HeLa cells is driven by NRF2 via
evolutionary conserved antioxidant response element. Mol Cell
Biochem. 389:99–111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zaman MS, Chauhan N, Yallapu MM, Gara RK,
Maher DM, Kumari S, Sikander M, Khan S, Zafar N, Jaggi M, et al:
Curcumin nanoformulation for cervical cancer treatment. Sci Rep.
6:200512016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gallardo M and Calaf GM: Curcumin inhibits
invasive capabilities through epithelial mesenchymal transition in
breast cancer cell lines. Int J Oncol. 49:1019–1027. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
OMS, . Cervical cancer estimated
incidence, Mortality and prevalence worldwide in 2012. http://gco.iarc.fr/today/data/pdf/fact-sheets/cancers/cancer-fact-sheets-16.pdfMarch
1–2019
|
|
16
|
OMS, . Human papillomavirus (HPV) and
cervical cancer. 2016, http://www.who.int/mediacentre/factsheets/fs380/en/March
1–2019
|
|
17
|
zur Hausen H: Human papillomavirus &
cervical cancer. Indian J Med Res. 130:2092009.PubMed/NCBI
|
|
18
|
Senapati R, Senapati NN and Dwibedi B:
Molecular mechanisms of HPV mediated neoplastic progression. Infect
Agent Cancer. 11:592016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Niu G, Wang D, Pei Y and Sun L: Systematic
identification of key genes and pathways in the development of
invasive cervical cancer. Gene. 618:28–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carrillo D, Muñoz JP, Huerta H, Leal G,
Corvalán A, León O, Calaf GM, Urzúa U, Boccardo E, Tapia JC, et al:
Upregulation of PIR gene expression induced by human papillomavirus
E6 and E7 in epithelial oral and cervical cells. Open Biol.
7:1701112017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Licciulli S, Luise C, Zanardi A, Giorgetti
L, Viale G, Lanfrancone L, Carbone R and Alcalay M: Pirin
delocalization in melanoma progression identified by high content
immuno-detection based approaches. BMC Cell Biol. 11:52010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zeng Q, Li X, Bartlam M, Wang G, Pang H
and Rao Z: Purification, crystallization and preliminary X-ray
analysis of human pirin. Acta Crystallogr D Biol Crystallogr.
59:1496–1498. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pang H, Bartlam M, Zeng Q, Miyatake H,
Hisano T, Miki K, Wong L-L, Gao GF and Rao Z: Crystal structure of
human pirin: An iron-binding nuclear protein and transcription
cofactor. J Biol Chem. 279:1491–1498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu F, Rehmani I, Esaki S, Fu R, Chen L,
de Serrano V and Liu A: Pirin is an iron-dependent redox regulator
of NF-κB. Proc Natl Acad Sci USA. 110:9722–9727. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Komai K, Niwa Y, Sasazawa Y and Simizu S:
Pirin regulates epithelial to mesenchymal transition independently
of Bcl3-Slug signaling. FEBS Lett. 589:738–743. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mishra A and Das BC: Curcumin as an
anti-human papillomavirus and anti-cancer compound. Future Oncol.
11:2487–2490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tennant JR: Evaluation of the trypan blue
technique for determination of cell viability. Transplantation.
2:685–694. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rodriguez LG, Wu X and Guan JL:
Wound-healing assay. Methods Mol Biol. 294:23–29. 2005.PubMed/NCBI
|
|
30
|
Thacker PC and Karunagaran D: Curcumin and
emodin down-regulate TGF-β signaling pathway in human cervical
cancer cells. PLoS. 10:e01200452015. View Article : Google Scholar
|
|
31
|
Roy M and Mukherjee S: Reversal of
resistance towards cisplatin by curcumin in cervical cancer cells.
Asian Pac J Cancer Prev. 15:1403–1410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shao Z-M, Shen Z-Z, Liu C-H, Sartippour
MR, Go VL, Heber D and Nguyen M: Curcumin exerts multiple
suppressive effects on human breast carcinoma cells. Int J Cancer.
98:234–240. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Calaf GM, Echiburú-Chau C, Wen G, Balajee
AS and Roy D: Effect of curcumin on irradiated and
estrogen-transformed human breast cell lines. Int J Oncol.
40:436–442. 2012.PubMed/NCBI
|
|
34
|
Meissner JD: Nucleotide sequences and
further characterization of human papillomavirus DNA present in the
CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol.
80:1725–1733. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Licciulli S, Cambiaghi V, Scafetta G,
Gruszka AM and Alcalay M: Pirin downregulation is a feature of AML
and leads to impairment of terminal myeloid differentiation.
Leukemia. 24:429–437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perl AK, Wilgenbus P, Dahl U, Semb H and
Christofori G: A causal role for E-cadherin in the transition from
adenoma to carcinoma. Nature. 392:190–193. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Derycke LD and Bracke ME: N-cadherin in
the spotlight of cell-cell adhesion, differentiation,
embryogenesis, invasion and signalling. Int J Dev Biol. 48:463–476.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bolós V, Peinado H, Pérez-Moreno MA, Fraga
MF, Esteller M and Cano A: The transcription factor Slug represses
E-cadherin expression and induces epithelial to mesenchymal
transitions: A comparison with Snail and E47 repressors. J Cell
Sci. 116:499–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Adhikary A, Chakraborty S, Mazumdar M,
Ghosh S, Mukherjee S, Manna A, Mohanty S, Nakka KK, Joshi S, De A,
et al: Inhibition of epithelial to mesenchymal transition by
E-cadherin up-regulation via repression of slug transcription and
inhibition of E-cadherin degradation: Dual role of scaffold/matrix
attachment region-binding protein 1 (SMAR1) in breast cancer cells.
J Biol Chem. 289:25431–25444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu WS, You RI, Cheng CC, Lee MC, Lin TY
and Hu CT: Snail collaborates with EGR-1 and SP-1 to directly
activate transcription of MMP 9 and ZEB1. Sci Rep. 7:177532017.
View Article : Google Scholar : PubMed/NCBI
|