Introduction
Dysregulation of the ubiquitin-proteasome pathway
serves an important role in tumor development and progression
(1). As the largest family of
multiunit E3 ubiquitin ligases (crucial control points in the
ubiquitin-proteasome pathway), cullin-RING ligases (CRLs) target
numerous ubiquitinated protein substrates for proteasome-targeted
degradation (2,3). Importantly, CRL activation requires
cullin neddylation (4,5), an ubiquitination modification process in
which the ubiquitin-like protein neural precursor cell expressed
developmentally downregulated 8 (NEDD8) is conjugated to cullins
(4–8).
The process involves NEDD8-activating enzyme E1 (NAE1),
NEDD8-conjugating enzyme E2M (UBE2M) and E3 NEDD8 ligases (6,9). UBE2M
interacts with a specific E3 ubiquitin ligase to promote the
neddylation of cullins (10), thereby
regulating cell cycle checkpoint control, DNA damage response,
apoptosis and senescence via diverse pathways. As an essential
component of neddylation, UBE2M is the critical factor in the
progression of various human carcinomas, including human renal cell
carcinoma and lung cancers, and intrahepatic cholangiocarcinoma
(ICC) (11–14).
According to the Surveillance, Epidemiology and End
Results program, ICC is one of the most common liver cancers in the
US, accounting for ~15% of primary liver cancers (15). The incidence and mortality of ICC are
gradually increasing worldwide (16).
Additionally, due to its frequently asymptomatic nature, patients
with ICC are usually diagnosed with late-stage disease, preventing
curative treatment and resulting in worse prognosis (17,18).
Therefore, effective therapies are urgently required to improve
outcomes. MLN4924 is an NAE inhibitor that blocks NAE activity and
efficiently suppresses cullin neddylation, inhibiting the growth of
ICC cells in vitro and in vivo (14). However, UBE2M, another critical
neddylation regulator is poorly understood. In the present study,
the roles and underlying mechanisms of UBE2M in ICC were
investigated for the first time, to the best of our knowledge.
Materials and methods
Patients and tissue specimens
ICC tissue samples were obtained from 81 patients
with a pathological diagnosis of primary ICC that underwent radical
resection for their tumor in Lanzhou University Second Hospital
between March 2009 and August 2017. Adjacent matched non-cancerous
livers were obtained from an incision >5 cm away from the
carcinoma sites. A total of 29 female and 52 male patients ranging
46–75 years old, who had not received distant metastases nor any
preoperative anticancer therapy. Furthermore, for reverse
transcription-quantitative PCR (RT-qPCR) analysis, normal
intrahepatic bile duct were obtained from 12 patients (5 female and
7 male individuals ranging 43–74 years old), and ICC tissues were
obtained from 46 patients (26 male and 20 female patients ranging
33–75 years old) at Lanzhou University Second Hospital between June
2016 and July 2018. A protocol for use of human surgical samples
was approved by the Lanzhou University Second Hospital Ethics
Committee, and informed consent was obtained from each patient.
The clinicopathological characteristics of patients
with ICC are presented in Table I.
Outpatient follow-up procedures were performed via letter and
telephone, and are presented in Table
II. The median follow-up period of the 81 patients was 18.5
months (range, 3.5–90.5 months; SD, 19.3 months). The interval
between the date of surgery and the date of first relapse was
defined as the time to relapse (TTR); for patients without relapse,
the interval between the date of surgery and the end of the
follow-up period was recorded. Patients without relapse were
censored at the date of the last follow-up. Systemic chemotherapy
was given to patients with recurrence; the main recurrence sites
were the hilar and bile duct.
 | Table I.Associations between UBE2M expression
and clinicopathologic characteristics in patients with intrahepatic
cholangiocarcinoma. |
Table I.
Associations between UBE2M expression
and clinicopathologic characteristics in patients with intrahepatic
cholangiocarcinoma.
|
| UBE2M
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
factors | Low | High | P-value |
|---|
| Age (years) |
|
| 0.586 |
|
<61 | 16 | 25 |
|
|
≥61 | 18 | 22 |
|
| Sex |
|
| 0.698 |
|
Female | 13 | 16 |
|
|
Male | 21 | 31 |
|
| HBsAg |
|
| 0.184 |
|
Negative | 19 | 33 |
|
|
Positive | 15 | 14 |
|
| Serum AFP
(ng/ml) |
|
| 0.130a |
|
<20 | 33 | 40 |
|
|
≥20 | 1 | 7 |
|
| Serum CA 19-9
(ng/ml) |
|
| 0.888 |
|
<36 | 15 | 20 |
|
|
≥36 | 19 | 27 |
|
| Serum ALT
(U/l) |
|
| 0.306 |
|
<40 | 26 | 31 |
|
|
≥40 | 8 | 16 |
|
| Serum ALP
(U/l) |
|
| 0.072a |
|
<145 | 29 | 31 |
|
|
≥145 | 5 | 16 |
|
| Serum CEA
(µg/l) |
|
| 0.901 |
|
<5 | 20 | 27 |
|
| ≥5 | 14 | 20 |
|
| gGGT (U/l) |
|
| 0.191 |
|
<60 | 18 | 18 |
|
|
≥60 | 16 | 29 |
|
| Child-Pugh
score |
|
| 0.130a |
| A | 33 | 40 |
|
| B | 1 | 7 |
|
| Liver
cirrhosis |
|
| 0.342a |
| No | 27 | 42 |
|
|
Yes | 7 | 5 |
|
| Tumor size
(cm) |
|
| 0.599 |
|
<5 | 15 | 18 |
|
| ≥5 | 19 | 29 |
|
| Tumor number |
|
| 0.530 |
|
Single | 28 | 36 |
|
|
Multiple | 6 | 11 |
|
| Microvascular/bile
duct invasion |
|
| 0.532 |
| No | 26 | 33 |
|
|
Yes | 8 | 14 |
|
| Lymphatic
metastasis |
|
| 0.300 |
| No | 22 | 25 |
|
|
Yes | 12 | 22 |
|
| Tumor
encapsulation |
|
| 0.108a |
|
Complete | 2 | 9 |
|
|
None | 32 | 38 |
|
| Tumor
differentiation |
|
| 0.124 |
|
Poor | 19 | 34 |
|
|
Moderate/well | 15 | 13 |
|
 | Table II.Univariate and multivariate analyses
of prognostic factors in patients with intrahepatic
cholangiocarcinoma. |
Table II.
Univariate and multivariate analyses
of prognostic factors in patients with intrahepatic
cholangiocarcinoma.
| A, Univariate
analysis |
|---|
|
|---|
|
| RFS | OS |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≥61 years vs.
<61 years) | 1.208
(0.735–1.985) | 0.457 | 1.131
(0.560–1.966) | 0.664 |
| Sex (male vs.
female) | 0.737
(0.442–1.229) | 0.243 | 0.623
(0.355–1.094) | 0.099 |
| HBsAg (positive vs.
negative) | 0.346
(0.192–0.622) |
<0.001a | 0.258
(0.130–0.512) |
<0.001a |
| Serum AFP (≥20
ng/ml vs. <20 ng/ml) | 1.182
(0.822–3.995) | 0.141 | 1.859
(0.834–4.143) | 0.129 |
| Serum CA 19-9 (≥36
ng/ml vs. <36 ng/ml) | 1.589
(0.954–2.646) | 0.075 | 2.308
(1.261–4.226) | 0.007a |
| Serum ALT (≥40 U/l
vs. <40 U/l) | 1.073
(0.626–1.840) | 0.798 | 1.184
(0.655–2.141) | 0.557 |
| Serum ALP (≥145 U/l
vs. <145 U/l) | 1.430
(0.826–2.475) | 0.202 | 1.806
(1.006–3.244) | 0.048a |
| Serum CEA (≥5 µg/l
vs. <5 µg/l) | 1.885
(1.143–3.109) | 0.013a | 2.824
(1.605–4.969) |
<0.001a |
| γGGT (≥60 U/l vs.
<60 U/l) | 1.304
(0.789–2.155) | 0.301 | 1.760
(0.989–3.133) | 0.054 |
| Child-Pugh score (A
vs. B) | 1.200
(0.546–2.638) | 0.649 | 1.294
(0.551–3.038) | 0.554 |
| Liver cirrhosis (no
vs. yes) | 0.571
(0.271–1.203) | 0.140 | 0.539
(0.229–1.266) | 0.156 |
| Tumor size (≥5 cm
vs. <5 cm) | 1.190
(0.716–1.975) | 0.502 | 1.722
(0.958–3.096) | 0.069 |
| Tumor number
(multiple vs. single) | 1.344
(0.741–2.438) | 0.330 | 1.736
(0.922–3.266) | 0.087 |
| Vascular invasion
(yes vs. no) | 1.181
(0.688–2.027) | 0.546 | 1.060
(0.579–1.941) | 0.850 |
| Lymphatic
metastasis (yes vs. no) | 1.848
(1.119–3.053) | 0.016a | 2.838
(1.614–4.990) |
<0.001a |
| Tumor encapsulation
(complete vs. none) | 0.805
(0.383–1.694) | 0.568 | 0.589
(0.233–1.486) | 0.262 |
| Tumor
differentiation (poor vs. moderate/well) | 0.677
(0.395–1.161) | 0.155 | 0.604
(0.326–1.121) | 0.110 |
| UBE2M (high vs.
low) | 1.185
(1.084–3.038) | 0.023a | 2.470
(1.372–4.445) | 0.003a |
|
| B, Multivariate
analysis |
|
|
| RFS | OS |
|
|
|
|
|
Variable | HR (95%
CI) | P-value | HR (95%
CI) | P-value |
|
| HBsAg (positive vs.
negative) | 0.365
(0.198–0.672) | 0.001a | 0.280
(0.135–0.578) | 0.001a |
| Serum CEA (≥5 µg/l
vs. <5 µg/l) | 1.348
(0.788–2.034) | 0.275 | 2.051
(1.108–3.797) | 0.022a |
| Lymphatic
metastasis (yes vs. no) | 1.518
(0.895–2.577) | 0.122 | 1.891
(1.038–3.442) | 0.037a |
| UBE2M (high vs.
low) | 1.892
(1.11–3.214) | 0.018a | 2.842
(1.151–5.332) | 0.001a |
Immunohistochemistry (IHC)
ICC tissue samples obtained from representative
regions and noncancerous liver tissue were stained via IHC. An
arraying machine (Beecher Instruments, Inc.) was used to construct
tissue microarrays. The tissues were fixed in 10% formalin neutral
buffer solution for 24 h at room temperature. Following
deparaffinization in xylene and rehydration in a graded ethanol
series, ICC tissue array sections (4–5 µm thick) were treated with
1% H2O2 to block endogenous peroxidase
activity and antigen retrieval was performed with 0.01% citrate
buffer (pH 6.0) for 10 min in a microwave oven. Rabbit anti-UBE2M
monoclonal primary antibodies (1:100; cat. no. ab109507; Abcam)
were added and incubated for 30 min at room temperature. Then, the
tissues washed with TBS and incubated with the secondary antibodies
(1:100; cat. no. SPN-9001; OriGene Technologies, Inc.) for 2 h at
room temperature. Following visualization of protein expression
with 3,3′-diaminobenzidine (DAB), sections were counterstained with
hematoxylin at room temperature for 5 min, and comparisons were
performed between tumor/normal pairs. Photos were acquired under a
Nikon Optiphot microscope equipped with an Optronics
charge-coupled-device camera (magnification, ×200; Nikon
Corporation); five random fields per sample were analyzed.
Immunostaining intensity was processed with Image-Pro Plus version
4.1 software (Media Cybernetics, Inc.) and scored as follows: No
staining (0), light brown (1), brown
(2), and dark brown (3), and scores of 2 and 3 were classified as
high expression, whereas scores of 0 and 1 were classified as low
expression in prognostic analyses.
RT-qPCR analysis
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with RNase-free DNase was
used to extract total RNA from 46 ICC tissues and 12 normal
intrahepatic bile duct tissues, as well as HUCCT1 and QBC939 cells.
A PrimeScript™ RT reagent kit (Takara Bio, Inc.) was used to
perform RT according to the manufacturer's protocols. Then, qPCR
was performed on an ABI 7500 thermocycler (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using Power SYBR Green PCR
MasterMix (Applied Biosystems; Thermo Fisher Scientific, Inc.) as
follows: Initial step at 94°C for 5 min, followed by 40 cycles of
denaturation at 94°C for 30 sec, annealing at 55–60°C for 15 sec
and extension at 72°C for 60 sec, with a final extension at 72°C
for 7 min. β-actin was used as endogenous control, and the
2−ΔΔCq method was performed to measure the expression of
target genes (19). The sequences of
the primers used are as follows: Human β-actin, forward
5′-TGACGTGGACATCCGCAAAG-3′, reverse 5′-CTGGAAGGTGGACAGCGAGG-3′;
human UBE2M, forward 5′-ATGAGGGCTTCTACAAGAGTGG-3′, reverse
5′-ATTGTCTCACACTTCACCTTGG-3′.
Cell lines and culture
Human cholangiocarcinoma cell lines QBC939 and
HUCCT1 were obtained from the Third Military Medical University
(Chongqing, China) and the Cell Resource Center of Tohoku
University, respectively. 293T cells were obtained from the
American Type Culture Collection. Cells were cultured in Dulbecco's
modified Eagle's medium (HyClone; GE Healthcare Life Sciences)
supplemented with 10% fetal bovine serum (FBS; Biochrom AG; Merck
KGaA), 1% penicillin and streptomycin (Corning Inc.) at 37°C and
95% humidity with 5% CO2.
UBE2M short hairpin (sh)RNA and
transfection
The target sequence for the UBE2M shRNA was
5′-GGGCUUCUACAAGAGUGGGAAGUUU-3′ (Invitrogen; Thermo Fisher
Scientific, Inc). The sequence of shUBE2M was designed according to
a previous study (11). pGC-fu-EGFP
(Shanghai GeneChem Co., Ltd.) double-digested by the restriction
enzymes EcoRI (cat. no. R0101S; New England BioLabs, Inc.)
and BamHI (cat. no. R0136S; New England BioLabs, Inc.) was
used as a vector. Positive lentiviral pGC-fu-EGFP vectors with
shUBE2M were packaged with pHelper1.0 and pHelper 2.0 vectors
(Shanghai GeneChem Co., Ltd.) into 293T cells (4×107
cells/dish) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), and the virus were collected and
centrifuged at 4,000 × g for 10 min at 4°C after transfection for
48 h. Then the virus was filtrated with a 0.45-µm filter and 500 µl
virus was infected into QBC939 or HUCCT1 cells (5×105
cells/well) in DMEM supplemented with 10% FBS and polybrene at a
titer of 3×108 TU/ml, using Lipofectamine 2000, whereas
empty vector pGC-fu-EGFP was used as an empty control (shNC).
Following infection for 96 h, puromycin was added to the medium at
4 µg/ml and cultured for 48 h after transfection, and then images
were captured.
Cell viability and colony formation
assays
Cells transfected with shUBE2M or shNC at a density
of 1,500 cells/well were plated into 96-well plates in triplicate.
A Cell Counting Kit-8 assay (CCK-8; Beyotime Institute of
Biotechnology) was used to determine the cell viability at 48 h
following the aforementioned 96-h transfection period according to
the manufacturer's protocols. Following incubation with 10 µl CCK-8
reagent for 4 h, the absorbance was measured at 450 nm using an
automatic multi-well spectrophotometer (Bio-Rad Laboratories,
Inc.).
For clonogenic assays, 200 cells/well were seeded in
60-mm petri dishes in triplicate. Following incubation for 12 days
at 37°C, colonies were fixed with 4% paraformaldehyde for 15 min
and stained with 0.5% crystal violet for 15 min (both at room
temperature). The number of colonies with >50 cells was counted
using a fluorescence microscope (magnification, ×40; Olympus
Corporation).
Apoptosis analysis
A fluorescence microscope (magnification, ×400;
Olympus Corporation) was used to analyze apoptosis in at least five
fields per sample at 48 h after cell seeding in 6-well plates at
2.0×106 cells/well.
Propidium iodide (PI) staining and
analysis
Harvested cells were fixed with 70% ethanol at −20°C
overnight. Following two washes with ice-cold PBS, cells were
stained with PI (36 µg/ml; Sigma-Aldrich; Merck KGaA) containing
RNase (10 µg/ml; Sigma-Aldrich; Merck KGaA) in the dark at 37°C for
15 min, then a CyAn™ ADP flow cytometer (Beckman Coulter, Inc.) was
used to analyze apoptosis. Apoptosis was determined by the
population percentage of cells in sub-G1. Data analysis was
performed using ModFit LT 5.0 software (Verity Software House).
Protein extraction and western
blotting
Cellular proteins were extracted using cell lysis
buffer (Beyotime Institute of Biotechnology), then the
concentration was measured with a Pierce™ BCA protein assay kit
(Thermo Fisher Scientific, Inc.). Protein (50 µg) was separated via
12% SDS-PAGE and then transferred to PVDF membranes (EMD
Millipore). Membranes were blocked with 5% skim milk at room
temperature for 60 min, and then incubated at 4°C overnight with
the following primary antibodies: NEDD8 (1:1,000; cat. no. ab81264;
Abcam); UBE2M (1:1,000; cat. no. ab109507; Abcam); origin
recognition complex subunit 1 (ORC1; 1:1,000; cat. no. ab85830;
Abcam); chromatin-licensing and DNA replication factor 1 (CDT1;
1:1,000; cat. no. ab202067; Abcam); poly (ADP-ribose) polymerase
(PARP; 1:1,000; cat. no. ab74290; Abcam); cleaved (c)PARP (1:1,000;
cat. no. ab4830; Abcam; caspase-3 (1:1,000; cat. no. ab197202;
Abcam); caspase-9 (1:1,000; cat. no. ab219590; Abcam); cullin 1
(1:1,000; cat. no. ab75817; Abcam); and γ-H2A histone family member
X (γ-H2AX; 1:1,000; cat. no. ab26350; Abcam). Secondary horseradish
peroxidase-conjugated antibodies against rabbit immunoglobulin G
(IgG; 1:5,000; cat. no. sc-2004) or mouse IgG (1:5,000; cat. no.
sc-2005; both Santa Cruz Biotechnology, Inc.) were then used at
room temperature for 2 h. Bands were visualized using Pierce™ ECL
Western Blotting Substrate (Thermo Fisher Scientific, Inc.).
In vivo mouse xenograft study
The present study followed animal handling and
experimental procedures, and was approved by the Animal Care and
Use Committee of Lanzhou University Second Hospital (permit no.
2019A-019). Following anesthesia via 1.9% ether inhalation for 5
min (anesthesia was monitored based on the respiratory rate),
5-week-old male athymic nude mice (n=14, 20–35 g; housed in an
animal room maintained at 20–22°C and 30–70% relative humidity
under a 12:12-h light/dark cycle) were subcutaneously injected with
QBC939 cells in the groin (5×106 cells/injection). All
animals (7 mice/group) had free access to standard food and
drinking water. After 3 days, the tumor-bearing mice were randomly
divided into shNC and shUBE2M groups. Lentivirus was intratumorally
injected in mice (5×108 copies per injection). Vernier
calipers were used to measure the tumor size once every other day.
At 7 weeks later, the mice were sacrificed via cervical dislocation
(confirmed by the cessation of breathing and heartbeat, and loss of
nerve reflexes), and tumor xenografts were collected, weighed and
photographed. Tumor tissues fixed with 10% formalin neutral buffer
solution for 24 h in room temperature were paraffin-embedded and
sectioned (4–5 µm thickness) for IHC. Paraffin sections were
deparaffinized in 100% xylene and rehydrated in a graded ethanol
series, and then peroxidase blocking with 1%
H2O2 for 30 min at room temperature and
antigen retrieval were performed. Sections were incubated overnight
at 4°C with primary antibody against Ki-67 (1:100; cat. no.
ab15580; Abcam), followed by incubation for 15 min at room
temperautere with a biotin-labeled secondary antibody (1:5,000;
Product # 65-6140; Invitrogen) and subsequently with
streptavidin-conjugated horseradish peroxidase (Fuzhou Maixin
Biotech Co., Ltd.). DAB was applied to develop the peroxidase
reaction. Then the sections were counterstained with hematoxylin
(Fisher CS401-D) for 20 sec at room temperature, mounted in neutral
gum and analyzed using a Optiphot microscope (magnification, ×200;
Nikon Corporation) in five random fields per sample.
GSE dataset analyses
Public data for ICC [GSE26566 (20), containing 104 cases of ICC, 59 cases
of matched non-cancerous livers and 6 cases of normal intrahepatic
bile duct] were obtained from Gene Expression Omnibus (GEO)
database (21).
Protein-protein interaction (PPI)
network analyses
The STRING database (version 11; http://string-db.org/cgi/input.pl) was used to
construct the PPI network of UBE2M (22).
Statistical analyses
Data were presented as the mean ± standard deviation
of three or more independent repeats. Comparisons between two
groups were performed using Student's t-test, Fisher's exact test
or χ2 test. Kaplan-Meier analysis and log-rank test, or
univariate and multivariate analyses were used to analyze
associations between TTR and overall survival and prognostic
factors as appropriate. All statistical analysis were performed
using SPSS software 18.0 (SPSS, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
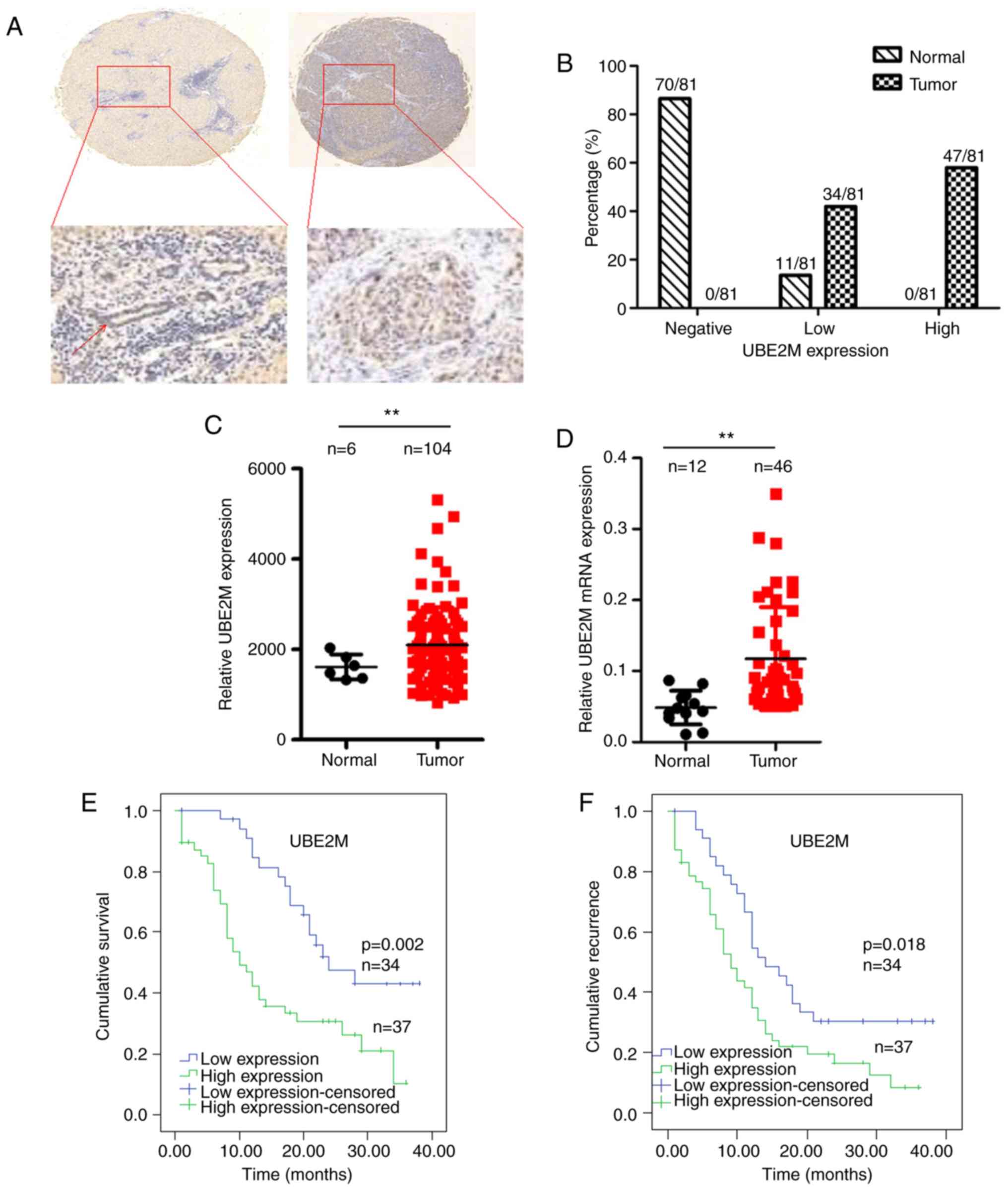
UBE2M protein is overexpressed in ICC
tissue specimens and associated with clinical outcomes
To evaluate the expression of UBE2M in ICC tissues,
UBE2M expression was measured via IHC staining in tissue specimens
derived from 81 pairs of primary ICC tissues and adjacent normal
tissues. It was revealed that UBE2M was overexpressed in ICC tumor
tissue compared with matched non-tumor tissue (Fig. 1A and B).
To further investigate the effects of UBE2M on ICC
progression, UBE2M protein expression in ICC tumors from the
GSE26566 dataset was analyzed; as presented in Fig. 1C, UBE2M protein was significantly
upregulated in ICC tumor tissues compared with normal intrahepatic
bile duct in the dataset (P<0.01). RT-qPCR analysis revealed
that UBE2M mRNA was significantly overexpressed in ICC tumor tissue
compared with normal intrahepatic bile duct tissues (Fig. 1D). These findings indicated that UBE2M
was overexpressed in ICC tumor tissues compared with in normal
tissues.
To demonstrate the prognostic relevance of UBE2M
expression in ICC, patients were separated into high (strong or
moderate intensities) or low (weak or negative intensities)
expression groups according to IHC staining of ICC tumor tissues.
The expression of UBE2M was not significantly associated with any
of the assessed clinicopathological variables (Table I). Kaplan-Meier analysis revealed that
the overall survival of patients with high expression of UBE2M was
significantly reduced compared with those with low expression
(P=0.002; Fig. 1E). The TTR of
patients with high UBE2M expression (median, 9 months) was
significantly shorter than those with low expression (median, 14
months; P=0.018; Fig. 1F). Univariate
analysis revealed that high expression of UBE2M was clearly
associated with worse prognosis and reduced TTR in patients with
ICC (Table II). Furthermore,
multivariate Cox proportional hazards regression analysis was
performed for several clinicopathological factors (Table II). It was demonstrated that that
high expression of UBE2M (P<0.001), negative hepatitis B surface
antigen (HBsAg; P<0.001), high expression of serum
carcinoembryonic antigen (CEA; P<0.05) and lymphatic metastasis
(P<0.05) were independent prognostic factors for poor overall
survival. Additionally, multivariate regression analysis showed
that UBE2M overexpression and positive HBsAg were independent
factors for recurrence-free survival (P=0.018 and P<0.001,
respectively).
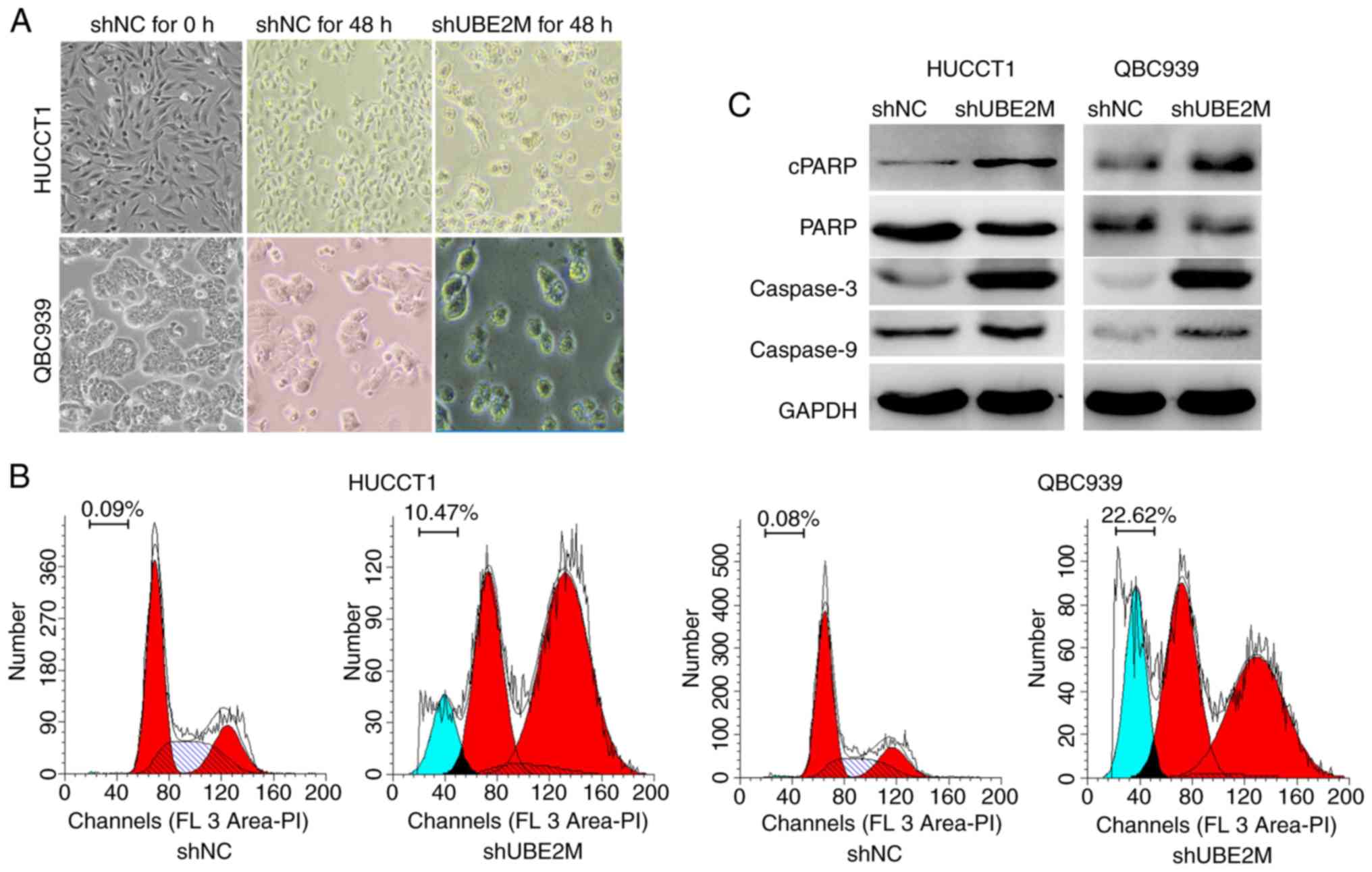
UBE2M knockdown inhibits the growth of
QBC939 cells and HUCCT cells
To further determine the role of UBE2M in ICC tumor
cell lines, shUBE2M was used to knock down UBE2M expression in the
ICC cell lines QBC939 and HUCCT1. As shown in Fig. 2C, western blotting revealed that UBE2M
shRNA markedly downregulated expression of UBE2M protein in QBC939
and HUCCT1 cells (Fig. 2C).
Subsequently, the effects of UBE2M knockdown on the viability and
colony formation of these ICC cells were evaluated; CCK-8 analysis
revealed that the viability of shUBE2M cells was significantly
inhibited compared with that of shNC cells (P<0.001; Fig. 2A). Similarly, colony formation was
significantly decreased in shUBE2M cells (P<0.001; Fig. 2B). These data indicated that the UBE2M
suppression resulted in the inhibition of ICC cell viability.
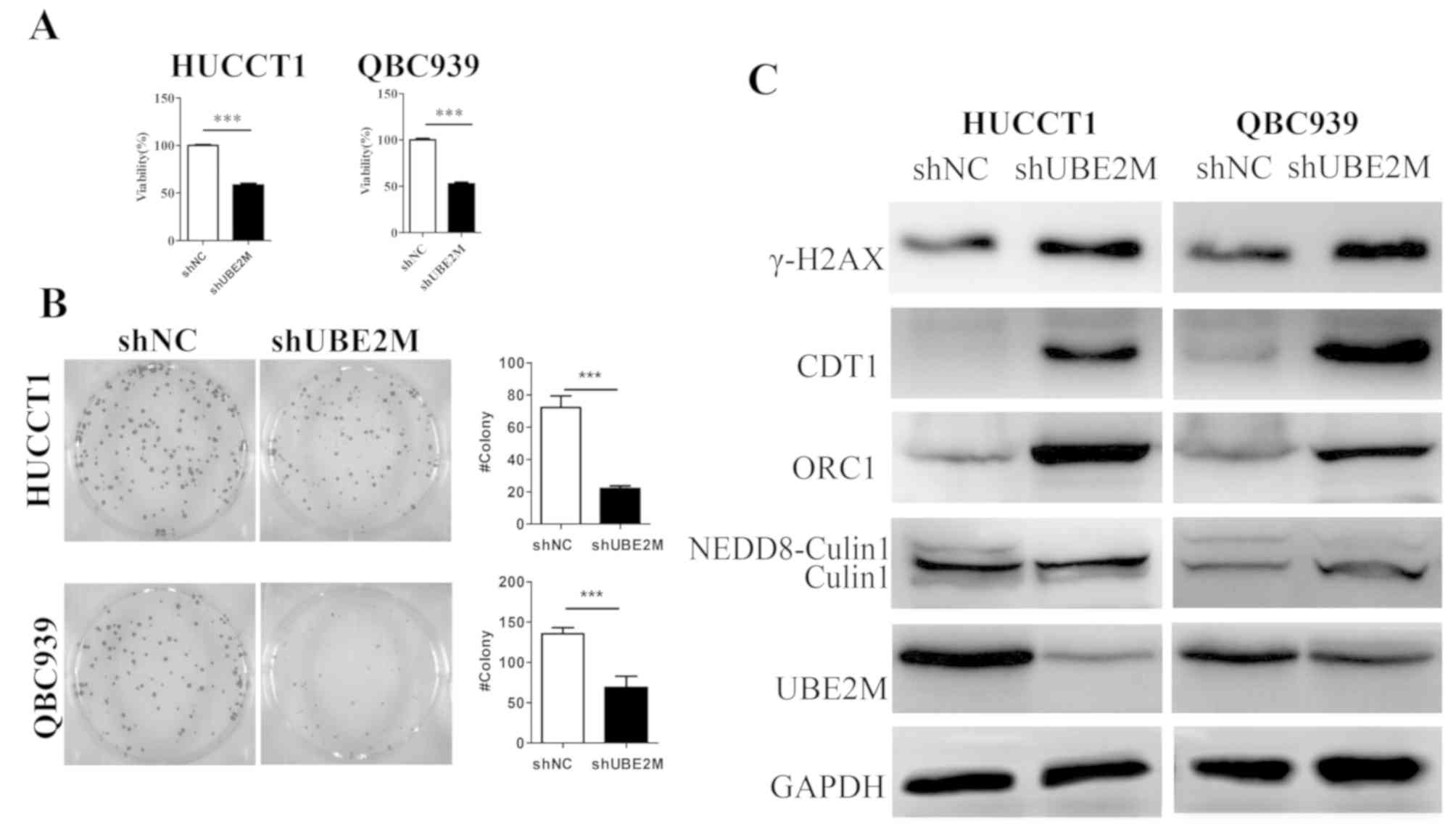 | Figure 2.Effects of UBE2M knockdown on the
viability and colony formation of intrahepatic cholangiocarcinoma
cells. (A) Effects of UBE2M knockdown on the viability of HUCCT1
and QBC939 cells. Cells were transfected with shUBE2M for 48 h, and
the viability was assessed by Cell Counting Kit-8 assays (n=3). (B)
Effects of UBE2M knockdown on clonogenic survival. HUCCT1 and
QBC939 cells were transfected with shUBE2M for 48 h (n=3). (C)
Effects of UBE2M knockdown on the neddylation of cullin 1, and the
expression of its substrates in HUCCT1 and QBC939 cells. Knockdown
of UBE2M induced accumulation of the cullin-RING ligase substrates
ORC1, CDT1, γ-H2AX. Data are presented as the mean ± SD.
***P<0.001. CDT1, chromatin-licensing and DNA replication factor
1; γ-H2AX, γ-H2A histone family member X; NC, negative control;
NEDD8, neural precursor cell expressed developmentally
downregulated 8; ORC1, origin recognition complex subunit 1; sh,
short hairpin (RNA); UBE2M, NEDD8-conjugating enzyme E2M. |
As UBE2M is an important component of neddylation,
the expression of the cullin substrates ORC1 and CDT1 were
subsequently investigated. Immunoblotting revealed that the
expression levels of cullin 1 in ICC cell lines were not notably
changed, whereas NEDD8-cullin1 expression was notably reduced
following UBE2M knockdown. The cullin substrates ORC1 and CDT1,
whose overexpression induce DNA damage responses, were clearly
upregulated in shUBE2M cells. Additionally, upregulation of γ-H2AX,
a marker of DNA double strand breaks (DSBs), was observed in the
shUBE2M groups (Fig. 2C).
UBE2M knockdown induces ICC tumor cell
apoptosis
The previous findings indicated that UBE2M silencing
suppressed the viability and proliferation of ICC tumor cells. To
further investigate the potential mechanisms involved, the
expression of apoptosis-associated proteins was evaluated,
including PARP, cPARP, caspase-3, and caspase-9. UBE2M knockdown
appeared to induce apoptosis as demonstrated by a shrunk cellular
morphology (Fig. 3A), the
upregulation of cPARP, and caspase-3 and −9 (Fig. 3C), and the appearance of a sub-G1 peak
(Fig. 3B).
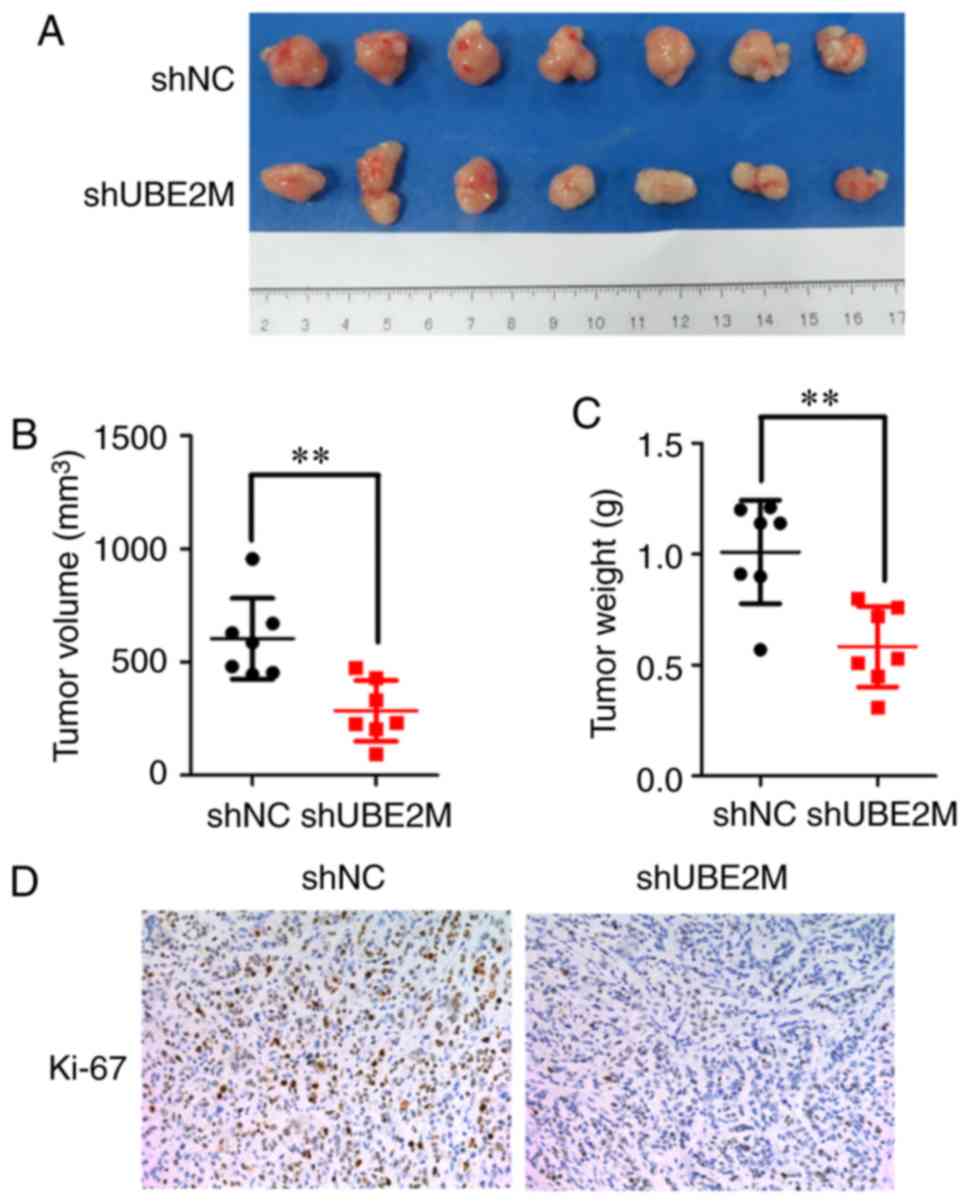
Effects of UBE2M knockdown on mouse
xenograft growth
Due to the antitumor effects of UBE2M knockdown on
ICC cells in vitro, its effects were further examined in
nude mouse xenografts. Tumor-bearing nude mice were intratumorally
injected with lenti-shUBE2M, and then tumor growth was monitored.
UBE2M knockdown in reduced growth of tumors, with significantly
decreased tumor volume (P<0.01; Fig.
4A) and weight (P<0.01; Fig.
4B) compared with the control group injected with lenti-shNC.
IHC staining demonstrated that injection of lenti-shUBE2M led to a
marked reduction of Ki67 expression, a marker of proliferation, in
nude mouse xenografts (Fig. 4C).
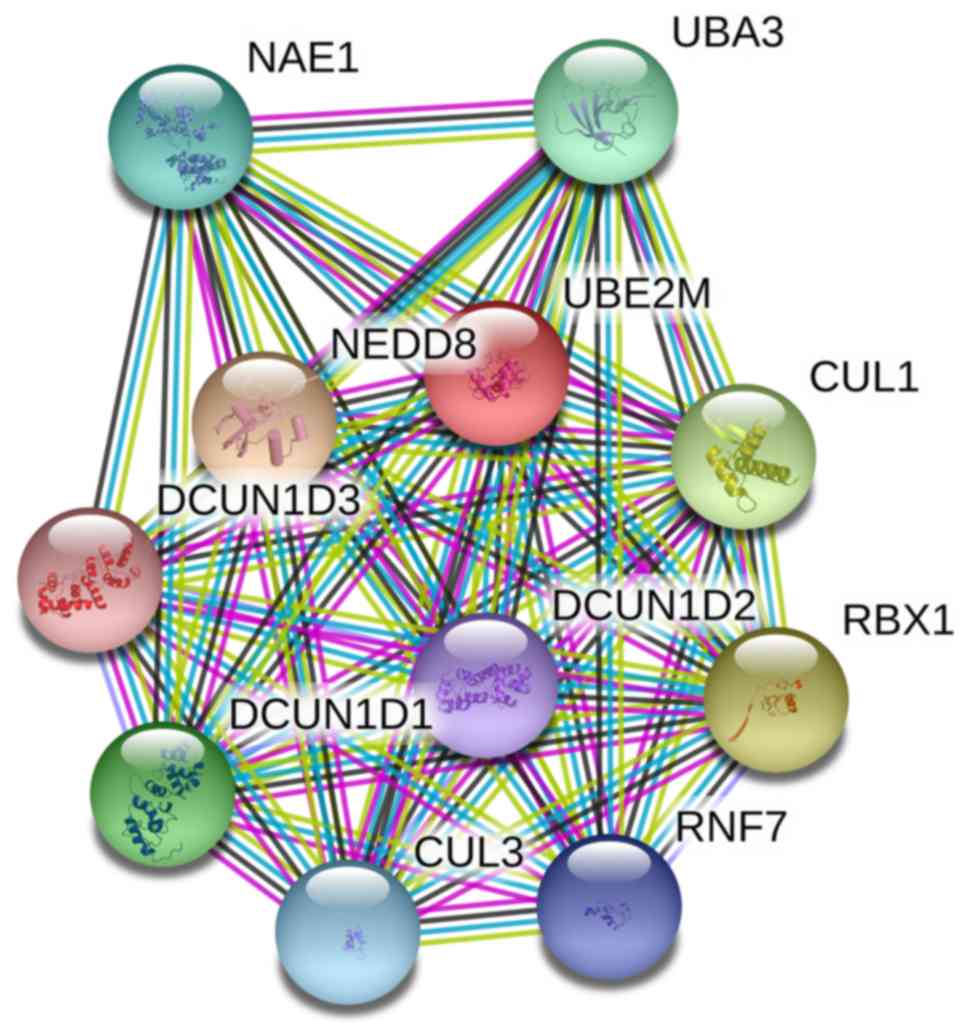
Finally, protein-protein interaction (PPI) network
analysis was performed using the STRING database, based on previous
studies into UBE2M function (13,23–25). As
presented in Fig. 5, UBE2M was
revealed to interact with CUL1, CUL3, defective in cullin
neddylation 1 (DCUN1)D1, DCUN1D2, DCUN1D3, NAE1, NEDD8, RING-box
protein 1, ring finger protein 7 and ubiquitin-like modifier
activating enzyme 3.
Discussion
ICC represents the second most common hepatic
carcinoma, with a majority of patients diagnosed at a nonsurgical
stage; even following early radical resection, the rate of
recurrence after surgery is 60–70% (26). Therefore, postoperative adjuvant
therapy is often considered, and the development of effective
treatment strategies is urgently required. As one of the critical
molecular pathways involved in carcinogenesis and the progression
of various cancers, including lung, breast, bladder, gastric and
prostate cancers (27–31), the neddylation pathway was also
upregulated in ICC (14). Neddylation
modification and CRL E3 ligase are attractive anticancer targets
(32–34). An inhibitor of NEDD8-activating enzyme
E1, MLN4924, was demonstrated to suppress the growth of ICC tumor
cells both in vitro and in vivo (14); however, there has been limited
investigation into the effects of Nedd8-conjugating enzyme
UBE2M.
In the present study, it was reported for the first
time, to the best of our knowledge, that UBE2M protein was
overexpressed in ICC tissue specimens compared with corresponding
normal tissues, consistent to with previous studies in other types
of human cancers, including lung cancer, and head and neck squamous
cell carcinoma (27,35). However, the imbalanced group numbers
in the GSE26566 dataset, including 104 cases of ICC and 6 cases of
normal intrahepatic bile duct, may impact the interpretation of
these results. More normal intrahepatic bile duct samples should be
collected for further analysis in future studies. Overexpression of
UBE2M was previously associated with worse prognosis in lung cancer
(27), suggesting that UBE2M
expression may be a novel prognostic marker for patients with ICC.
In the study, it was demonstrated that the expression of UBE2M was
not significantly associated with any assessed clinicopathologic
features in patients with ICC, which may be due to the small sample
size; a previous study reported that Neuropilin-1 expression was
not significantly associated with clinicopathologic characteristics
in breast cancer tissues as a result of the small number of
patients enrolled in this study (36).
Silencing of UBE2M expression inhibited ICC cell
growth and led to apoptosis. Molecularly, knockdown of UBE2M
expression blocked the second step of neddylation pathway in HUCCT1
cells and QBC939 cells, leading to accumulation of cullin
substrates ORC1 and CDT1, and subsequently resulting in DSBs and
the induction DNA damage responses, as revealed by the upregulated
expression of γ-H2AX. UBE2M silencing results in increased DNA
breakages and sensitivity to DNA-damaging agents (24). Additionally, aberrant DNA repair and
increased apoptosis was observed, as determined by low expression
of PARP, which is involved in DNA repair (37) and apoptosis (38), and increased cPARP, caspase-3 and
caspase-9 following UBE2M knockdown (11). Induction of cellular apoptosis is
regarded as an important antitumor mechanism; various stresses,
including antitumor gene activation (39), telomere dysfunction (40,41) and
DNA damage (42) can all initiate
cellular apoptosis.
Mechanistically, UBE2M knockdown resulted in the
reduced viability and colony formation of ICC cell lines,
potentially due to DNA damage responses and apoptosis. UBE2M
knockdown not only inhibited the colony formation of ICC cells
in vitro, but also suppressed ICC tumor growth in
vivo, as determined by the reduced size of tumor xenografts
infected with the shUBE2M lentivirus.
Additionally, protein-protein interaction (PPI)
network analysis was performed for UBE2M to identify molecules
potentially involved in its effects. In future experiments, gene
expression or RNA sequencing followed by gene set enrichment
analysis on cell lines will be performed pre- and post-shUBE2M
treatment to identify the relevant molecular pathways involved in
the development of ICC.
In conclusion, the present study revealed that UBE2M
served an important role in ICC progression, and that targeting
UBE2M protein is a potential anticancer strategy for ICC. Further
investigation of the underlying mechanisms of UBE2M in ICC is
required.
Acknowledgements
Not applicable.
Funding
This work was supported by Medical Research Project
of Lanzhou Chengguan District, China (grant no. 2018-01-11).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BZ, CG and ZJ conceived the study, carried out the
overall experimental design and data interpretation, and prepared
and revised the manuscript. BZ performed the majority of the
experiments. CG, DS and JM performed immunohistochemical assays. JZ
and LG performed the colony formation assay. JG performed the
western blot analysis. All authors have read and approved the final
version of this manuscript.
Ethics approval and consent to
participate
The human study was approved by the Lanzhou
University Second Hospital Ethics Committee, and informed consent
was obtained from each patient. The animal study was approved by
the Animal Care and Use Committee of Lanzhou University Second
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bedford L, Lowe J, Dick LR, Mayer RJ and
Brownell JE: Ubiquitin-like protein conjugation and the
ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov.
10:29–46. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
O'Connor HF and Huibregtse JM:
Enzyme-substrate relationships in the ubiquitin system: Approaches
for identifying substrates of ubiquitin ligases. Cell Mol Life Sci.
74:3363–3375. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Petroski MD and Deshaies RJ: Function and
regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol.
6:9–20. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deshaies RJ, Emberley ED and Saha A:
Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell
Biochem. 54:41–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ying J, Zhang M, Qiu X and Lu Y: Targeting
the neddylation pathway in cells as a potential therapeutic
approach for diseases. Cancer Chemother Pharmacol. 81:797–808.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xirodimas DP: Novel substrates and
functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans.
36:802–806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zuo W, Huang F, Chiang YJ, Li M, Du J,
Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, et al: C-Cbl-mediated
neddylation antagonizes ubiquitination and degradation of the TGF-β
type II receptor. Mol Cell. 49:499–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao Y, Morgan MA and Sun Y: Targeting
neddylation pathways to inactivate cullin-RING ligases for
anticancer therapy. Antioxid Redox Signal. 21:2383–2400. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watson IR, Irwin MS and Ohh M: NEDD8
pathways in cancer, sine quibus non. Cancer Cell. 19:168–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang DT, Ayrault O, Hunt HW, Taherbhoy
AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF and
Schulman BA: E2-RING expansion of the NEDD8 cascade confers
specificity to cullin modification. Mol Cell. 33:483–495. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cukras S, Morffy N, Ohn T and Kee Y:
Inactivating UBE2M impacts the DNA damage response and genome
integrity involving multiple cullin ligases. PLoS One.
9:e1018442014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu B, Deng Y, Bi R, Guo H, Shu C, Shah NK,
Chang J, Liu G, Du Y, Wei W and Wang C: A first-in-class inhibitor,
MLN4924 (pevonedistat), induces cell-cycle arrest, senescence, and
apoptosis in human renal cell carcinoma by suppressing
UBE2M-dependent neddylation modification. Cancer Chemother
Pharmacol. 81:1083–1093. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou W, Xu J, Tan M, Li H, Li H, Wei W and
Sun Y: UBE2M is a stress-inducible dual E2 for neddylation and
ubiquitylation that promotes targeted degradation of UBE2F. Mol
Cell. 70:1008–1024. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Q, Yu GY, Shi JY, Li LH, Zhang WJ,
Wang ZC, Yang LX, Duan M, Zhao H, Wang XY, et al: Neddylation
pathway is up-regulated in human intrahepatic cholangiocarcinoma
and serves as a potential therapeutic target. Oncotarget.
5:7820–7832. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altekruse SF, Devesa SS, Dickie LA,
McGlynn KA and Kleiner DE: Histological classification of liver and
intrahepatic bile duct cancers in SEER registries. J Registry
Manag. 38:201–205. 2011.PubMed/NCBI
|
|
16
|
Bertuccio P, Bosetti C, Levi F, Decarli A,
Negri E and La Vecchia C: A comparison of trends in mortality from
primary liver cancer and intrahepatic cholangiocarcinoma in Europe.
Ann Oncol. 24:1667–1674. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bridgewater J, Galle PR, Khan SA, Llovet
JM, Park JW, Patel T, Pawlik TM and Gores GJ: Guidelines for the
diagnosis and management of intrahepatic cholangiocarcinoma. J
Hepatol. 60:1268–1289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Poultsides GA, Zhu AX, Choti MA and Pawlik
TM: Intrahepatic cholangiocarcinoma. Surg Clin North Am.
90:817–837. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 14:1021–1031. 2012. View Article : Google Scholar
|
|
21
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Kang J, Zhang W, Cai L, Wang S,
Liang Y, Jiang Y, Liu X, Zhang Y, Ruan H, et al: Validation of
NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung
cancer. EBioMedicine. 45:81–91. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brown JS, Lukashchuk N, Sczaniecka-Clift
M, Britton S, le Sage C, Calsou P, Calsou P, Beli P, Galanty Y and
Jackson SP: Neddylation promotes ubiquitylation and release of ku
from DNA-damage sites. Cell Rep. 11:704–714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jimeno S, Fernandez-Avila MJ, Cruz-Garcia
A, Cepeda-Garcia C, Gomez-Cabello D and Huertas P: Neddylation
inhibits CtIP-mediated resection and regulates DNA double strand
break repair pathway choice. Nucleic Acids Res. 43:987–999. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mavros MN, Economopoulos KP, Alexiou VG
and Pawlik TM: Treatment and prognosis for patients with
intrahepatic cholangiocarcinoma: Systematic review and
meta-analysis. JAMA Surg. 149:565–574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L, Wang M, Yu G, Chen P, Li H, Wei D,
Zhu J, Xie L, Jia H, Shi J, et al: Overactivated neddylation
pathway as a therapeutic target in lung cancer. J Natl Cancer Inst.
106:dju0832014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang W, Liu Z, Qu P, Zhou Z, Zeng Y, Fan
J, Liu Y, Guo Y and Qiu J: Knockdown of regulator of cullins-1
(ROC1) expression induces bladder cancer cell cycle arrest at the
G2 phase and senescence. PLoS One. 8:e627342013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lan H, Tang Z, Jin H and Sun Y:
Neddylation inhibitor MLN4924 suppresses growth and migration of
human gastric cancer cells. Sci Rep. 6:242182016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wen M, Kwon Y, Wang Y, Mao JH and Wei G:
Elevated expression of UBE2T exhibits oncogenic properties in human
prostate cancer. Oncotarget. 6:25226–25239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ueki T, Park JH, Nishidate T, Kijima K,
Hirata K, Nakamura Y and Katagiri T: Ubiquitination and
downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T
overexpression in human breast cancer cells. Cancer Res.
69:8752–8760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu S and Yu L: Targeting cullin-RING
ligases for cancer treatment: Rationales, advances and therapeutic
implications. Cytotechnology. 68:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jia L, Soengas MS and Sun Y: ROC1/RBX1 E3
ubiquitin ligase silencing suppresses tumor cell growth via
sequential induction of G2-M arrest, apoptosis, and senescence.
Cancer Res. 69:4974–4982. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou L, Zhang W, Sun Y and Jia L: Protein
neddylation and its alterations in human cancers for targeted
therapy. Cell Signal. 44:92–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang W, Liang Y, Li L, Wang X, Yan Z,
Dong C, Zeng MS, Zhong Q, Liu XK, Yu J, et al: The nedd8-activating
enzyme inhibitor MLN4924 (TAK-924/Pevonedistat) induces apoptosis
via c-myc-noxa axis in head and neck squamous cell carcinoma. Cell
Prolif. 52:e125362019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Seifi-Alan M, Shams R, Bandehpour M,
Mirfakhraie R and Ghafouri-Fard S: Neuropilin-1 expression is
associated with lymph node metastasis in breast cancer tissues.
Cancer Manag Res. 10:1969–1974. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hosoya N and Miyagawa K: Targeting DNA
damage response in cancer therapy. Cancer Sci. 105:370–388. 2018.
View Article : Google Scholar
|
|
38
|
Affar EB, Germain M, Winstall E,
Vodenicharov M, Shah RG, Salvesen GS and Poirier GG:
Caspase-3-mediated processing of poly(ADP-ribose) glycohydrolase
during apoptosis. J Biol Chem. 276:2935–2942. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pistritto G, Trisciuoglio D, Ceci C,
Garufi A and D'Orazi G: Apoptosis as anticancer mechanism: Function
and dysfunction of its modulators and targeted therapeutic
strategies. Aging (Albany NY). 8:603–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chin L, Artandi SE, Shen Q, Tam A, Lee SL,
Gottlieb GJ, Greider CW and DePinho RA: P53 deficiency rescues the
adverse effects of telomere loss and cooperates with telomere
dysfunction to accelerate carcinogenesis. Cell. 97:527–538. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Titen SW and Golic KG: Telomere loss
provokes multiple pathways to apoptosis and produces genomic
instability in Drosophila melanogaster. Genetics. 180:1821–1832.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kee Y, Huang M, Chang S, Moreau LA, Park
E, Smith PG and D'Andrea AD: Inhibition of the nedd8 system
sensitizes cells to DNA interstrand cross-linking agents. Mol
Cancer Res. 10:369–377. 2012. View Article : Google Scholar : PubMed/NCBI
|