Introduction
Despite advances in screening, diagnosis and
curative treatment, hepatocellular carcinoma (HCC) remains a
leading cause of cancer-associated morbidity and mortality
worldwide (1). As a complex and
heterogeneous disease, HCC carries different molecular profiles,
distinct clinical responses to therapeutic agents and thus an
unfavorable prognosis, with a mortality rate of ~500,000 worldwide
per year (2,3). The recurrence rate is high, even for
patients who have undergone curative resection with early-stage
HCC, rendering the 5 year survival probability of <30% in
patients suffered from HCC (4,5). Current
risk assessment, treatment decisions and prognostic prediction of
HCC mainly rely on features restricted to a cancer cell-centric
focus, such as the tumor-node-metastasis (TNM) stage and Child-Pugh
scoring system (6,7). However, conventional clinicopathological
characteristics do not enable the precise predicting of
individualized prognoses in patients with HCC. Such poor outcomes
and high heterogeneity highlight challenging issues and the urgent
need to develop novel predictive biomarkers with potential links to
patient prognosis and therapeutics options.
Recent advances in high-throughput technologies have
unexpectedly provided an opportunity to define the genome-wide
landscape of HCC, and contributed to the rapid development of
molecular signatures for prognostic prediction and the further
personalization and precision of treatment paradigms (8–10). With
the pervasive application of microarray and next-generation
sequencing technology, studies involving mutations, copy number
variation, gene expression, non-coding RNA expression and immune
infiltration have been performed to identify potential HCC-related
loci in oncogenic pathways and interpret complex scenarios in the
malignant progression of HCC (11–15). These
studies, although with promising outcomes, have primarily focused
on transcriptional levels and driver mutations, whereas the
systematic analysis of variations in transcript architecture has
received less attention.
Alternative splicing (AS), as a post-transcriptional
modification process, holds the potential to generate varied
isoforms and reprogram protein diversity among ~90% of human
multi-exon genes (16). Therein, the
selective inclusion or exclusion of specific exon regions from
precursor mRNAs, followed by multiple permutations and combinations
of spliced exons are taken together to produce mature mRNA
(17). As the most important
mechanism for expanding the coding capacity and increasing the
proteome complexity, AS has been experimentally validated as having
a decisive role in controlling growth and development (18,19). The
evidence accumulated in previous decades has demonstrated that
specific splicing variants may be involved in several hallmarks of
carcinogenesis, including anti-apoptotic mechanisms, angiogenesis,
immune evasion, epithelial-mesenchymal transition and metastasis,
further emphasizing the significance of AS for determining clinical
outcomes in cancer, particularly HCC (19–25).
However, few studies have systematically investigated the AS
landscape within the HCC microenvironment and its association with
prognosis. Further comprehensive understanding of the shifts in the
splicing pattern involved in the regulation of HCC may be a vital
step in developing targeted therapy and helping to predict
treatment response and patient prognosis.
By contrast, splicing factors (SFs), as the executor
of splicing behaviors, function in alternative exon usage and
splicing site selection by recognizing cis-regulatory
elements within the alternative exons or flanking introns (26–28).
Substantial evidence implicates somatic mutations and the
differential expression of SFs as the dominant mechanism in the
initial steps of mRNA splicing in normal and cancer cells (29,30).
Notably, aberrant SFs can give rise to the oncogenic splicing
isoforms that confer various advantages to cancer cells (31,32). As AS
events and their clinical relevance in malignancies are only
superficially understood, it is imperative to elucidate the
intricate interwoven relationships between particular SFs and AS
events, and the intrinsic regulatory mechanism in HCC.
The Cancer Genome Atlas (TCGA) project supplements
abundant resources for the investigation of genome-wide AS patterns
in cancer. In the present study, to elucidate the global portrait
of aberrant AS events in HCC, the integrated splicing variant data
of 290 patients with HCC and corresponding clinical information
were profiled based on the TCGA database. In addition, prognostic
signatures were constructed with high clinical efficacy. Further
assessment of the splicing regulatory network between SFs and their
potential targets may shed new light on the mechanisms of genetic
variants in tumorigenesis and development.
Materials and methods
Data curation process
Third-level mRNA sequencing data and corresponding
clinical information of the HCC cohort were obtained from TCGA data
portal (http://tcga-data.nci.nih.gov/),
containing 374 HCC tissues and 50 adjacent non-tumor tissues
(33). To generate the AS profiles
for each patient with HCC, SpliceSeq (version 2.1), a java
application that unambiguously quantifies the inclusion level of
each exon and splice junction, was used to evaluate the mRNA
splicing patterns for the patients with HCC (34). A total of 290 patients who met the
following criteria were included: i) A definite histological
diagnosis of HCC; ii) patients were alive at least 30 days
following initial pathological diagnosis; iii) patients had
corresponding mRNA splicing data. The percent spliced in (PSI)
score, ranging from zero to one, was commonly used to evaluate the
transcript ratio of a specific gene to a particular splicing
pattern. In order to generate the most reliable set of AS events
possible, a series of stringent filters were implemented
(percentage of samples with PSI score ≥75 and average PSI score
≥0.05) and missing PSI values were input using the k-nearest
neighbor algorithm with the impute R package (35).
To describe an AS event precisely, each AS event was
assigned a unique annotation, which consisted of the particular
splicing type, the ID number in the TCGASpliceSeq database and the
matched gene symbol. For example, in ‘RI_C9orf9_ID_87994’, the
retained intron (RI) represents the splicing type, C9orf9 is the
counterpart gene symbol and ID_87994 represents the specific order
number in the TCGASpliceSeq database.
Survival analysis and construction of
a prognostic signature for patients with HCC
Following rigorous screening, a total of 290
patients with HCC with aberrant AS profiles and survival
information were subjected to subsequent analyses. For each
specific AS event, the PSI scores were dichotomized based on the
median cut among patients with HCC. Univariate Cox regression
analyses were then performed to identify the association between AS
events and overall survival (OS), with a threshold of P<0.05.
UpSet plot, generated using the UpSetR R package (version 1.3.2;
http://github.com/hms-dbmi/UpSetR),
was used to qualitatively visualize the intersecting sets among
seven types of survival-associated AS event (36). For high-dimension data, the
traditional Cox regression model cannot be applied directly. To
further reduce redundancy and render the model more practical and
parsimonious, forward stepwise selection with the Akaike
Information Criterion (AIC) was used, which starts with a null
model and gradually adds the variable whose inclusion offers the
most statistically significant improvement to the fitness of the
model until balancing the AIC score of the model to a minimum
(37). Therefore, among the top 20
(if available) most significant AS events within each AS type in
univariate Cox regression analysis, the key AS events were
sub-selected by the forward stepwise procedure to construct the AS
signature, respectively. The AS events included in each AS
signature were then combined to construct the final AS signature
through a secondary implementation of forward stepwise selection.
Finally, the AS-related risk score of each signature was calculated
utilizing the regression coefficients derived from multivariate Cox
regression analysis to multiply the PSI score of each key feature,
respectively. Based on the median cut-off value of the risk score,
the log-rank test and Kaplan-Meier survival analysis were used to
validate the statistical difference between the low-risk and
high-risk subgroups. Furthermore, the clinical predictive efficacy
of each prognostic signature was quantitatively evaluated by
fitting the time-dependent receiver-operator characteristic (ROC)
curve with the area under the curve (AUC) calculated. Therein, the
model selection by AIC in a stepwise algorithm was based on the
stepAIC function in the MASS R package, whereas the dynamic AUC
values of the time-dependent ROC curve were calculated with the
timeROC R package (38).
Independence of the final AS signature
from clinicopathological features
To investigate the independent prognostic value of
the final AS signature from the available conventional
clinicopathological characteristics (including age, gender, alcohol
consumption history, hepatitis B status, hepatitis C status, family
cancer history, serum α-fetoprotein (AFP) level, Child-Pugh
classification, residual tumor status, vascular invasion degree,
histologic grade and pathologic stage) in patients with HCC,
univariate and subsequent multivariate Cox regression analyses were
performed. To confirm whether the final AS signature was of high
applicability and robust in various subgroups, stratification Cox
analyses were also conducted.
Functional enrichment and interaction
analysis of survival-associated AS events
The parent genes of the survival-associated AS
events determined by univariate Cox regression analysis were
subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analyses by setting the false discovery rate
(FDR) <0.05. The above analyses were performed with
clusterProfiler R package (version 3.7) (39). Additionally, the gene interaction
network was visualized and analyzed using the Reactome FIPlugIn
(version 7.0.2; http://apps.cytoscape.org/apps/reactomefiplugin) in
Cytoscape (version 3.6.1) with the purpose of searching important
hub genes.
Construction of the potential SF-AS
regulatory network in the HCC cohort
A list of 88 human SFs was created by integrating
the SpliceAid 2 database (www.introni.it/spliceaid.html) and the data reported
by Xiong et al, who collected experimentally validated SFs
through hand-curated screening of literature and databases
(40). First, the corresponding
expression levels of SFs in the TCGA-HCC dataset were integrated
and normalized with the variance stabilizing transformation
function of the DESeq2 package (41,42). The
HCC samples and adjacent normal samples were compared to identify
differentially expressed SFs using Student's t-test. The
correlation between the normalized expression value of SFs and OS
was then assessed through fitting univariate regression analysis in
the entire cohort, for which SFs with P<0.05 were selected as
prognostic SFs for further analysis. The ‘surv_cutpoint’ function
of the ‘survminer’ R package was used to iteratively determine the
optimal cutpoints of prognostic SFs achieving the maximally
selected rank statistics. Furthermore, Spearman's correlation
analyses were performed between the expression values of prognostic
SFs and PSI scores of the most significant AS events in each AS
type. P-values were adjusted by the Benjamini-Hochberg procedure
and the significance threshold was set at adjusted P<0.05.
Gene set variation analysis (GSVA) for
SF-AS correlation pairs
GSVA is a differential functional gene set
enrichment analysis, which can detect subtle pathway activity
changes over heterogeneous samples by calculating sample-wise gene
set enrichment scores. GSVA was performed using the GSVA package
from Bioconductor (release version 3.8; www.bioconductor.org) to further search for a
significantly enriched set of GO and canonical pathways (KEGG,
Reactome and BioCarta pathway databases) in HCC tissues, downloaded
from the Molecular Signatures Database (www.broadinstitute.org) (43). Differential gene set analysis between
the tumor and adjacent normal samples was then performed using the
limma package (44). |logFC| >0.4
and FDR <0.05 for GO terms and |logFC| >0.2 and FDR <0.05
for pathway sets were considered statistically significant,
respectively.
Results
Overview of AS events in the TCGA HCC
cohort
The integrated mRNA sequencing data of 290 patients
with HCC with AS events profiles were included in the present
study. The median follow-up period was 15.75 months (range 1–122.5
months). Details of the study design are presented as a flowchart
in Fig. 1A. Overall, a total of
34,163 AS events from 8,986 parent genes were detected based on the
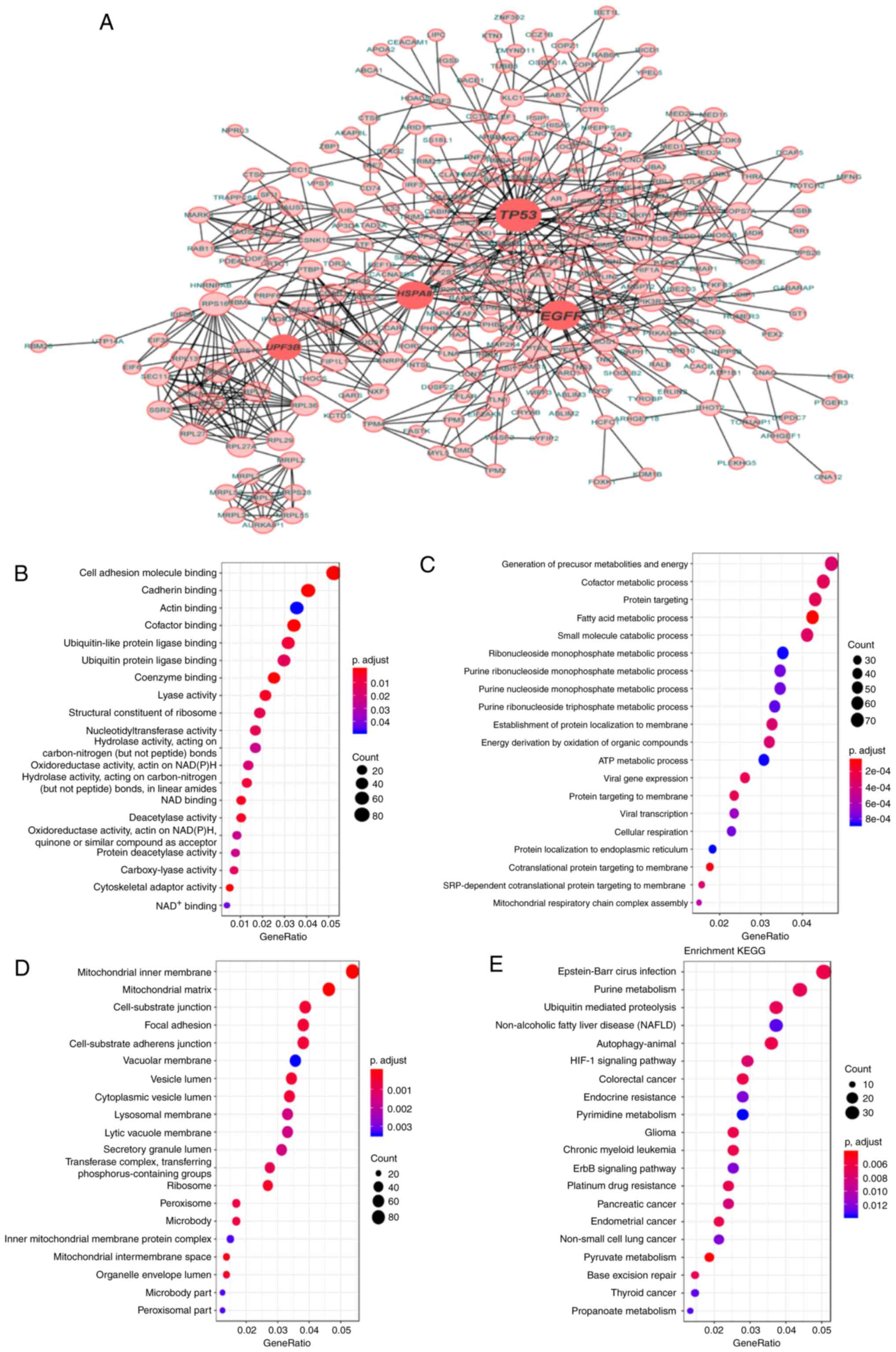
results of SpliceSeq analysis. As shown in Fig. 1C, seven types of AS event were
generated via different mechanisms. Furthermore, the AS events and
genes in each AS type were separated into a survival-related group
(P<0.05) or a survival-irrelevant group (P≥0.05) (Fig. 1B). In total, 1,805 survival-associated
AS events were identified from 1,314 parent genes, which contained
565 exon skips (ESs) in 479 genes, 541 alternate terminators (ATs)
in 356 genes, 341 alternate promoters (APs) in 258 genes, 137
alternate acceptor sites (AAs) in 128 genes, 112 RIs in 104 genes,
100 alternate donor sites (ADs) in 95 genes and nine mutually
exclusive exons (MEs) in nine genes. It was noted that one gene may
have two or more AS events associated with OS, therefore, an UpSet
plot was generated to quantitatively analyze the interactive sets
between the seven AS types (Fig. 1D).
Accordingly, the survival-associated AS events mostly belonged to
one parent gene, whereas several genes had up to four types of AS
event which were all significantly related to prognosis. For
example, RI, AA, AP and ES in the CIRBP gene (red dotted line) were
significantly associated with OS in HCC, and AD, AP and ES in the
BSCL2 gene (green dotted line) were significantly associated with
OS in HCC.
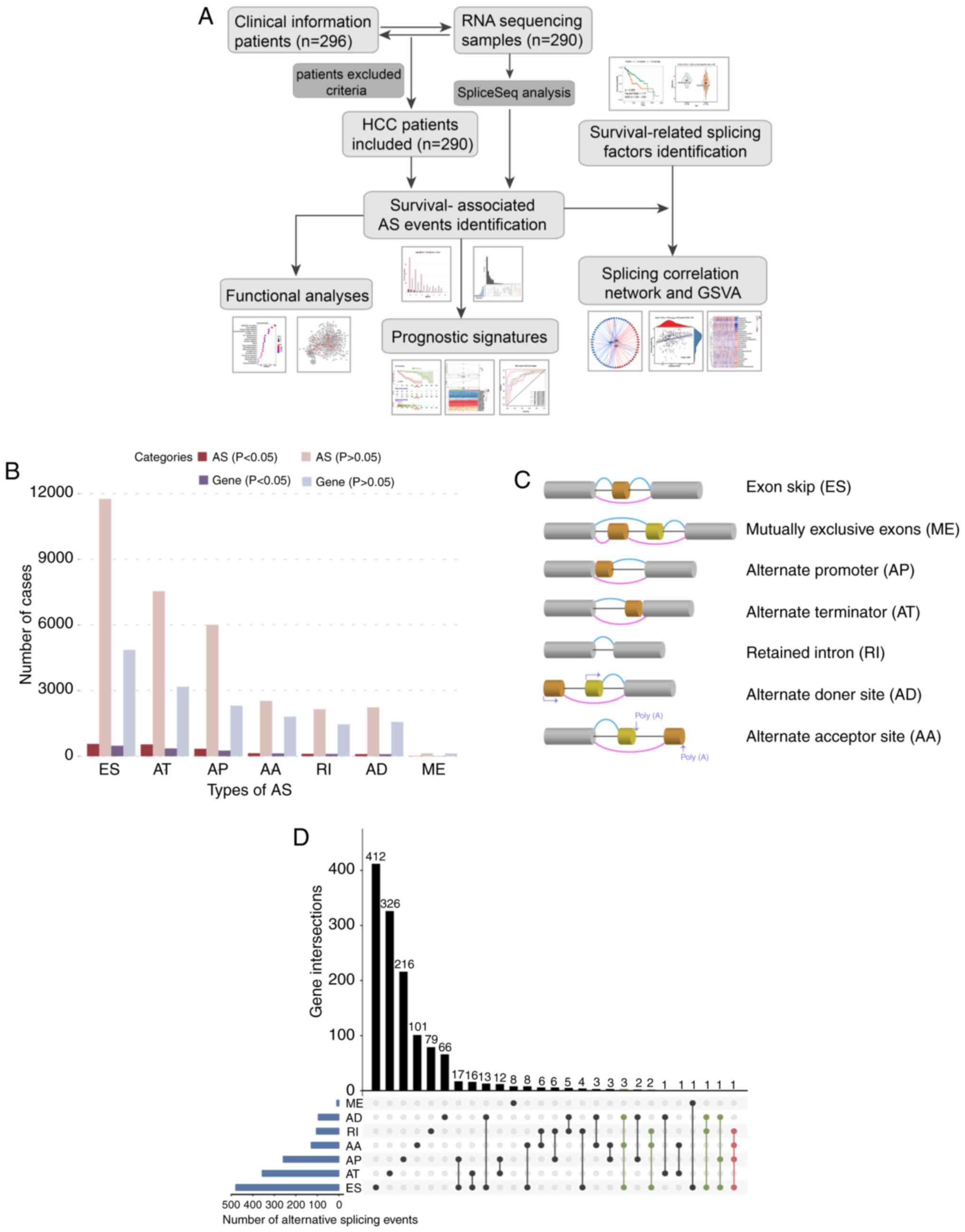 | Figure 1.Overview of AS events profiling in
HCC. (A) Flowchart for profiling the AS events of patients with HCC
in large-scale sequencing data. (B) Number of AS events and parent
genes from 290 patients with HCC. (C) Illustrations for seven types
of AS events, including ES, ME, RI, AP, AT, AD and AA. (D) UpSet
plot of parent gene interactions between the seven types of
survival-associated AS events in HCC. AS, alternative splicing;
HCC, hepatocellular carcinoma; GSVA, gene set variation analysis;
ES, exon skip; ME, mutually exclusive exons; RI, retained intron;
AP, alternate promoter; AT, alternate terminator; AD, alternate
donor site; AA, alternate acceptor site. |
Survival analysis and construction of
a prognostic signature for patients with HCC
To identify independent prognostic factors,
univariate Cox survival analysis was performed to assess the
association between clinical OS and each type of AS event. In each
AS event, the patients with HCC were divided into two subgroups
based on the median PSI score. The top 20 significant
survival-associated events of the seven types of AS, with the
exception of ME with only nine AS events, are presented as forest
plots in Fig. 2. As shown in the
forest plots, it appeared that the survival-associated AS events
were almost equally distributed in the favorable prognostic and
reverse subgroups, regardless of the AS type. Notably, AS
signatures constructed with 11 ES events, six AT events, 10 AD
events, 10 AA events, seven AP events, six RI events or seven ME
events all showed significant predictive power in distinguishing
poor or good clinical outcomes for the patients with HCC
(P<0.0001), and AA, AP, AT and ES had the highest predictive
power among the seven types with a median survival of >3,000
days in the low-risk subgroup (Fig.
3A-G). A final prognostic model, which included only 26 AS
events to minimize the AIC, was then constructed (high vs. low: 827
vs. 3,125 days, P<2e-16, Fig. 5A).
Furthermore, the ROC curves of 1, 3 and 5 years were applied to
compare the predictive efficiency among different AS models. As
shown in Fig. 5C, it was confirmed
that the final prognostic model incorporated with all types of AS
event exhibited a higher prognostic efficiency compared with the
others, and the AUCs for 1, 3 and 5 years were 0.937, 0.902 and
0.985, respectively. Additionally, the distribution of the survival
status and risk scores of patients, and the splicing pattern of
specific AS events included in each signature were visualized
(Figs. 4 and 5B). Detailed information of the specific AS
events involved in each AS signature and final prognostic model are
listed in Table I. Additionally, the
relative PSI scores of particular AS events comprised in the final
AS signature between the low-risk and high-risk subgroups are
depicted in Fig. S1.
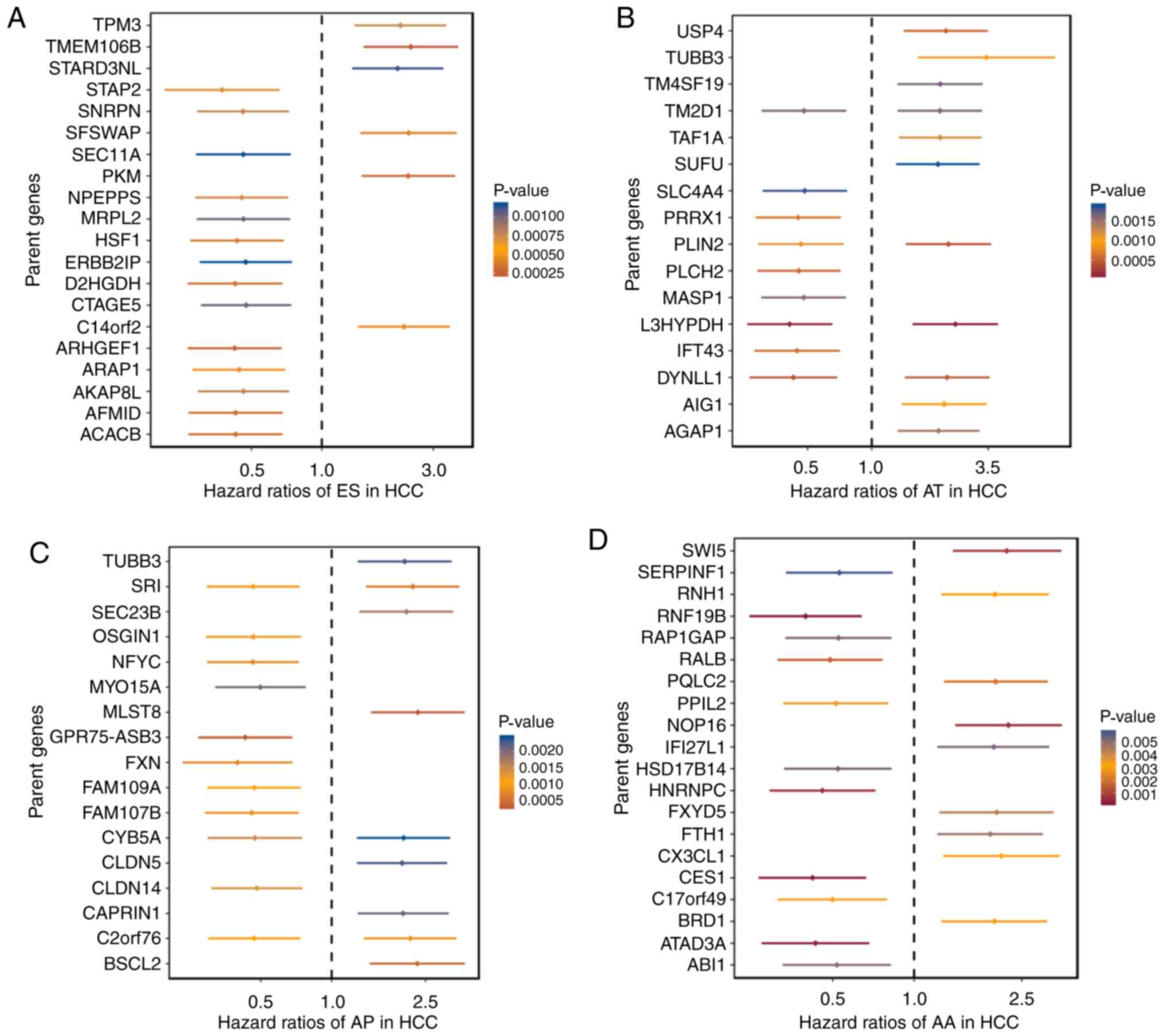 | Figure 2.Forrest plots of hazard ratios of
survival-associated alternative splicing events in HCC. Hazard
ratios of top 20 most significant survival-associated (A) ES, (B)
AT, (C) AP, (D) AA. P-values are indicated by the color scale of
the legend. HCC, hepatocellular carcinoma; ES, exon skip; AT,
alternate terminator; AP, alternate promoter; AA, alternate
acceptor site; RI, retained intron; AD, alternate donor site; ME,
mutually exclusive exons. Forrest plots of hazard ratios of
survival-associated alternative splicing events in HCC. Hazard
ratios of top 20 most significant survival-associated (E) RI and
(F) AD events. (G) Hazard ratios of significant survival-associated
ME events (only nine available). P-values are indicated by the
color scale of the legend. HCC, hepatocellular carcinoma; ES, exon
skip; AT, alternate terminator; AP, alternate promoter; AA,
alternate acceptor site; RI, retained intron; AD, alternate donor
site; ME, mutually exclusive exons. |
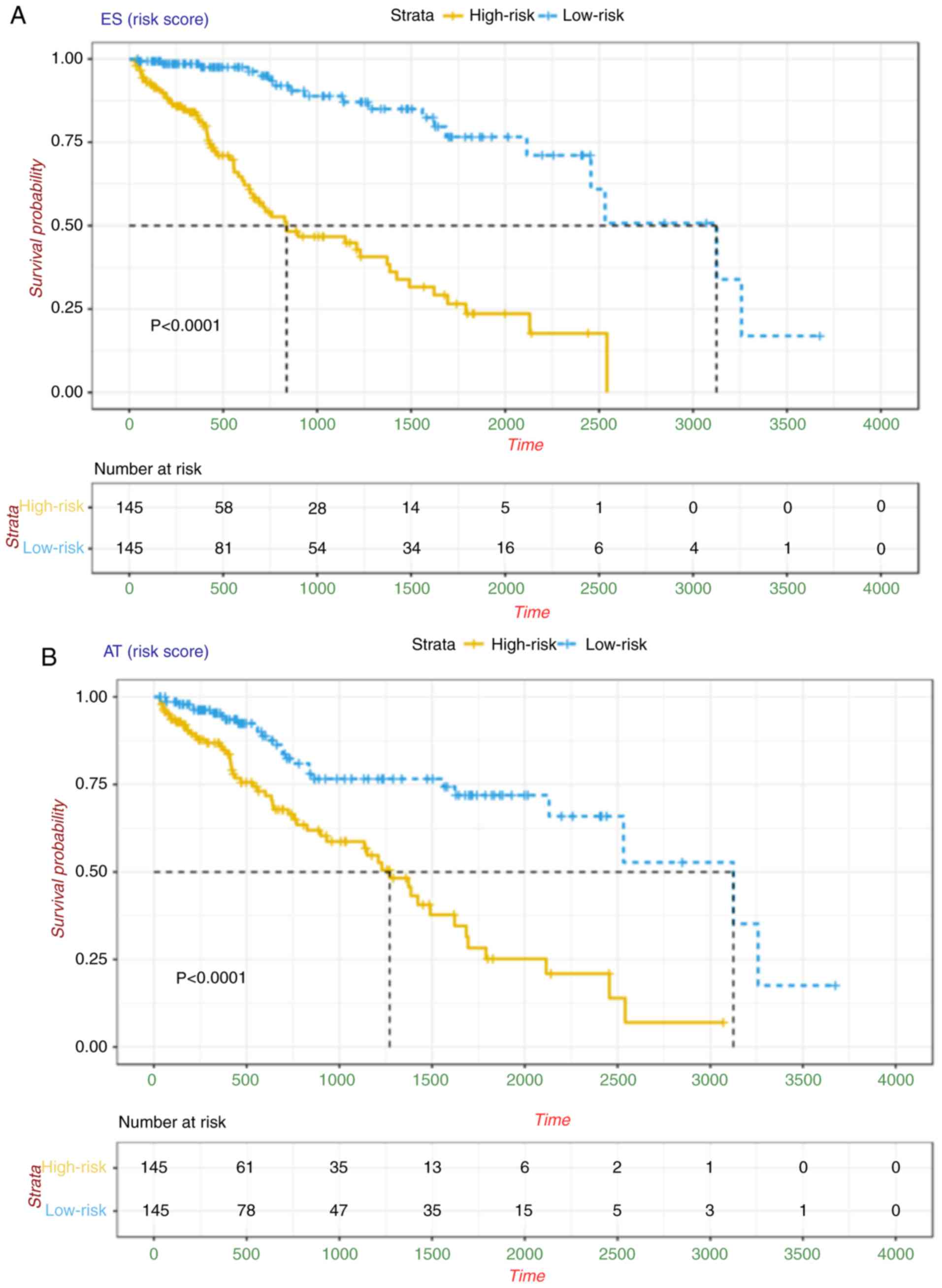 | Figure 3.Kaplan-Meier plots of AS signatures
built with seven types of AS events in HCC. Kaplan-Meier curves of
prognostic predictors built with candidate (A) ES and (B) AT. The
yellow line indicates the high-risk subgroup and the blue line
indicates the low-risk subgroup. P-values and median survival times
(dotted line) are shown in each signature. HCC, hepatocellular
carcinoma; AS, alternative splicing; ES, exon skip; AT, alternate
terminator; AP, alternate promoter; AA, alternate acceptor site;
RI, retained intron; AD, alternate donor site; ME, mutually
exclusive exons. Kaplan-Meier plots of AS signatures built with
seven types of AS events in HCC. Kaplan-Meier curves of prognostic
predictors built with candidate (C) AP and (D) AA. The yellow line
indicates the high-risk subgroup and the blue line indicates the
low-risk subgroup. P-values and median survival times (dotted line)
are shown in each signature. HCC, hepatocellular carcinoma; AS,
alternative splicing; ES, exon skip; AT, alternate terminator; AP,
alternate promoter; AA, alternate acceptor site; RI, retained
intron; AD, alternate donor site; ME, mutually exclusive exons.
Kaplan-Meier plots of AS signatures built with seven types of AS
events in HCC. Kaplan-Meier curves of prognostic predictors built
with candidate (E) RI and (F) AD. The yellow line indicates the
high-risk subgroup and the blue line indicates the low-risk
subgroup. P-values and median survival times (dotted line) are
shown in each signature. HCC, hepatocellular carcinoma; AS,
alternative splicing; ES, exon skip; AT, alternate terminator; AP,
alternate promoter; AA, alternate acceptor site; RI, retained
intron; AD, alternate donor site; ME, mutually exclusive exons.
Kaplan-Meier plots of AS signatures built with seven types of AS
events in HCC. Kaplan-Meier curves of prognostic predictors built
with candidate (G) ME events in HCC, respectively. The yellow line
indicates the high-risk subgroup and the blue line indicates the
low-risk subgroup. P-values and median survival times (dotted line)
are shown in each signature. HCC, hepatocellular carcinoma; AS,
alternative splicing; ES, exon skip; AT, alternate terminator; AP,
alternate promoter; AA, alternate acceptor site; RI, retained
intron; AD, alternate donor site; ME, mutually exclusive exons. |
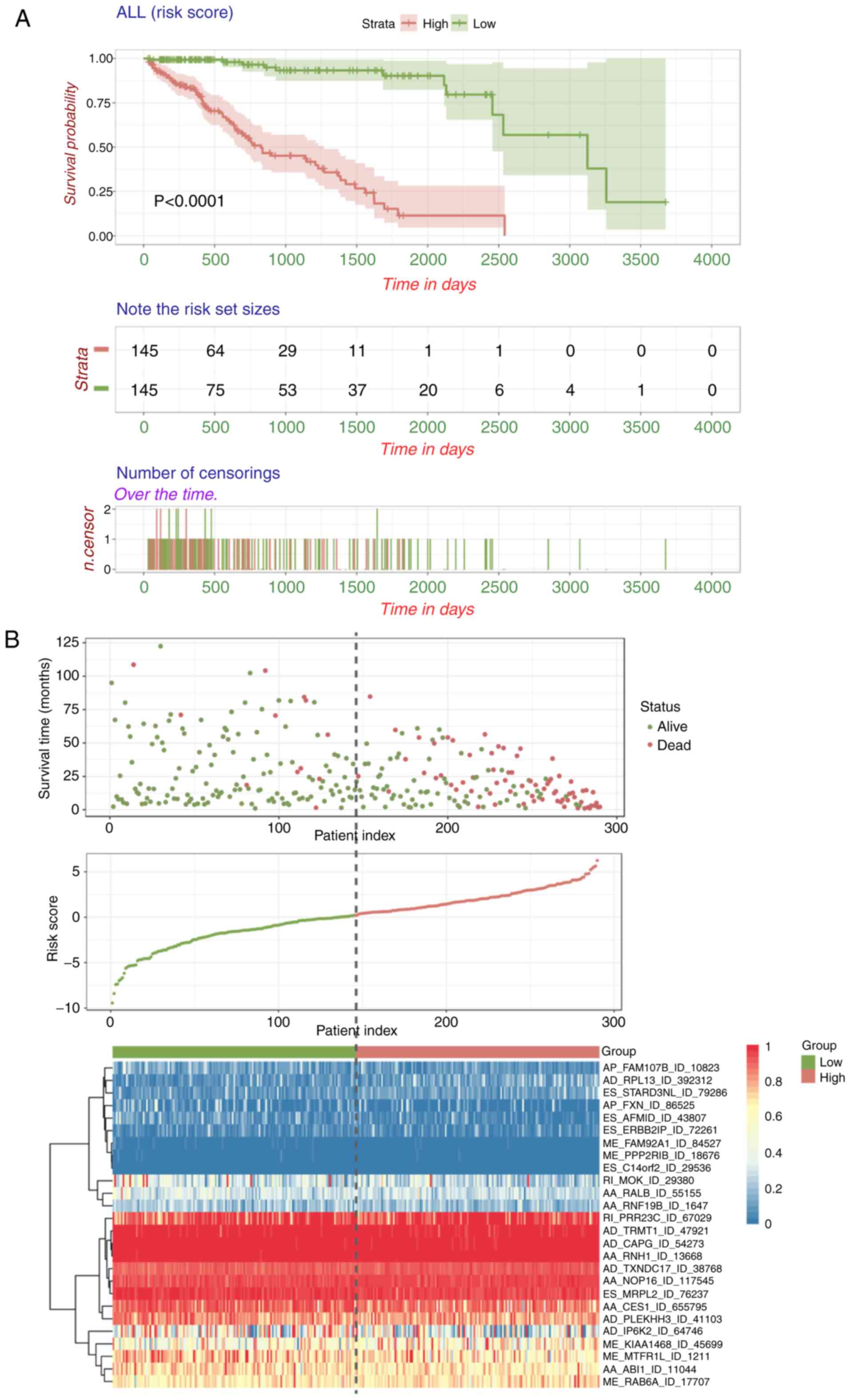 | Figure 5.Construction and validation of final
signature combining all types of prognostic AS event. (A)
Kaplan-Meier curves of prognostic predictors built with all types
of survival-associated splicing event. (B) Distribution of the
survival status and risk scores of patients, and the splicing
pattern of AS events included in the final AS signature. The color
transition from blue to red indicates the increasing percent
spliced in score of particular AS event from low to high. The
colors of the ROC curves represent the different AS types and the
respective AUC values are shown. HCC, hepatocellular carcinoma AS,
alternative splicing; ROC, receiver-operator characteristic; AUC,
area under the curve; ES, exon skip; AT, alternate terminator; AP,
alternate promoter; AA, alternate acceptor site; RI, retained
intron; AD, alternate donor site; ME, mutually exclusive exons. (C)
ROC curves of all prognostic signatures for 1, 3 and 5 years in
HCC. The colors of the ROC curves represent the different AS types
and the respective AUC values are shown. HCC, hepatocellular
carcinoma AS, alternative splicing; ROC, receiver-operator
characteristic; AUC, area under the curve; ES, exon skip; AT,
alternate terminator; AP, alternate promoter; AA, alternate
acceptor site; RI, retained intron; AD, alternate donor site; ME,
mutually exclusive exons. |
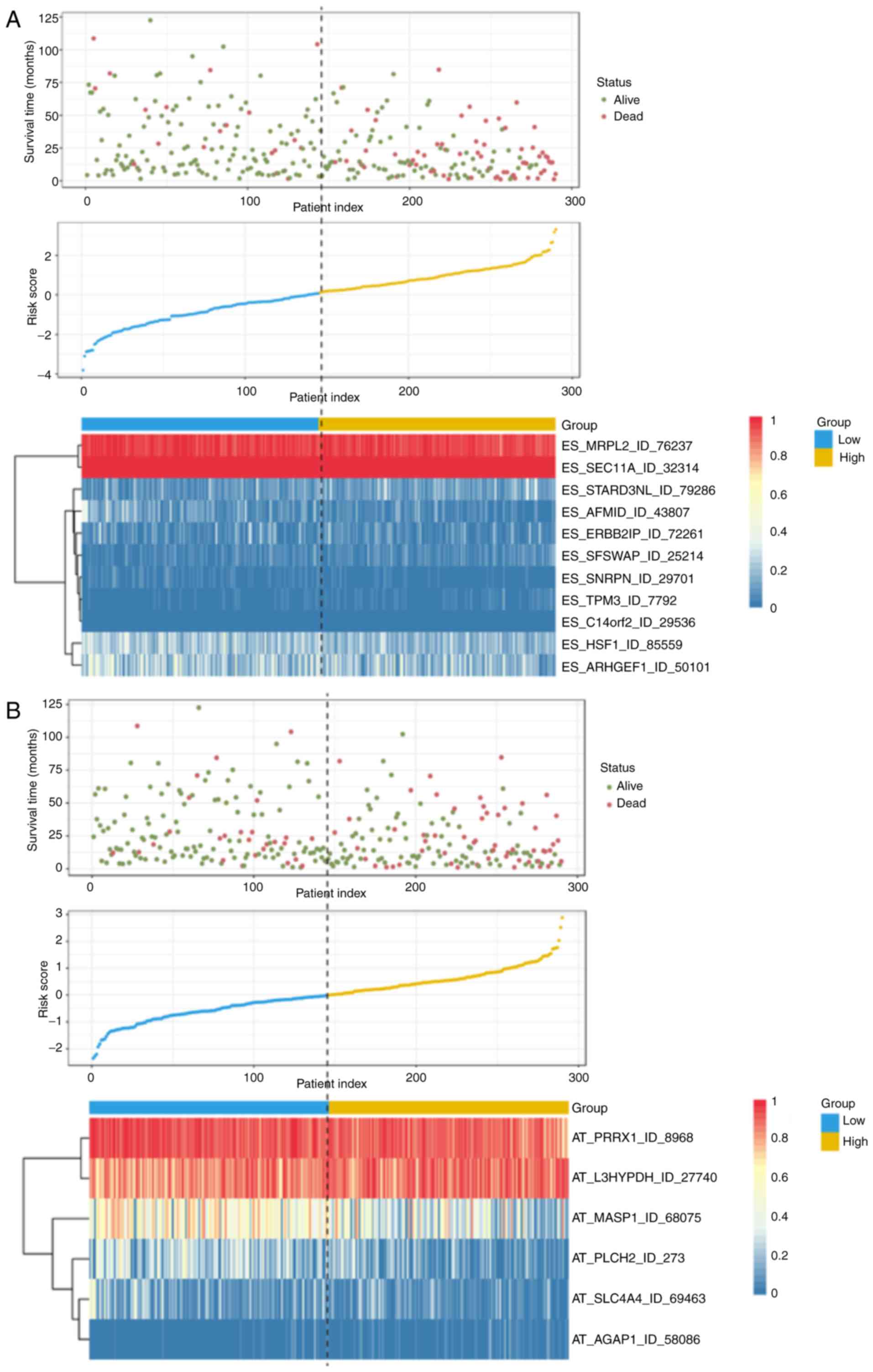 | Figure 4.Determination and analysis of
prognostic signatures based on each AS type in the HCC cohort.
Distribution of the survival status and risk scores of patients,
and the splicing pattern of AS events included in each AS signature
are shown. Patients with HCC were divided into high- and low-risk
subgroups based on the median cut-off risk score calculated
separately. The upper part of each assembly indicates the
distribution of the survival status and survival times of patients
ranked by risk score, the middle part represents the risk score
curve, and the heatmap below displays splicing pattern of the AS
signature from each AS type. The color transition from blue to red
indicates the increasing percent spliced in score of the
corresponding AS event from low to high. For (A) ES and (B) AT risk
scores (corresponding to each AS type) were calculated and AS
signatures were constructed using each type of prognostic splicing
event. HCC, hepatocellular carcinoma; AS, alternative splicing; ES,
exon skip; AT, alternate terminator; AP, alternate promoter; AA,
alternate acceptor site; RI, retained intron; AD, alternate donor
site; ME, mutually exclusive exons. The color transition from blue
to red indicates the increasing percent spliced in score of the
corresponding AS event from low to high. For (C) AP and (D) AA,
risk scores (corresponding to each AS type) were calculated and AS
signatures were constructed using each type of prognostic splicing
event. HCC, hepatocellular carcinoma; AS, alternative splicing; ES,
exon skip; AT, alternate terminator; AP, alternate promoter; AA,
alternate acceptor site; RI, retained intron; AD, alternate donor
site; ME, mutually exclusive exons. The color transition from blue
to red indicates the increasing percent spliced in score of the
corresponding AS event from low to high. For (E) RI and (F) AD risk
scores (corresponding to each AS type) were calculated and AS
signatures were constructed using each type of prognostic splicing
event. HCC, hepatocellular carcinoma; AS, alternative splicing; ES,
exon skip; AT, alternate terminator; AP, alternate promoter; AA,
alternate acceptor site; RI, retained intron; AD, alternate donor
site; ME, mutually exclusive exons. The color transition from blue
to red indicates the increasing percent spliced in score of the
corresponding AS event from low to high. For (G) ME, risk scores
(corresponding to each AS type) were calculated and AS signatures
were constructed using each type of prognostic splicing event. HCC,
hepatocellular carcinoma; AS, alternative splicing; ES, exon skip;
AT, alternate terminator; AP, alternate promoter; AA, alternate
acceptor site; RI, retained intron; AD, alternate donor site; ME,
mutually exclusive exons. |
 | Table I.Detailed information of specific AS
events involved in each AS signature and final prognostic
model. |
Table I.
Detailed information of specific AS
events involved in each AS signature and final prognostic
model.
| Gene symbol | AS ID | AS type | Exons | From exon | To exon | In final
signature |
|---|
| ABI1 | ID_11044 | AA | 11.1 | 9 | 11.2 | Yes |
| SWI5 | ID_87732 | AA | 3.1 | 2 | 3.2 | No |
| CES1 | ID_655795 | AA | 10.1 | 7 | 10.2 | Yes |
| NOP16 | ID_117545 | AA | 5.1 | 4.1 | 5.2 | Yes |
| PPIL2 | ID_61245 | AA | 22.4:22.5:22.6 | 22.2 | 22.7 | No |
| RNH1 | ID_13668 | AA | 6.1 | 5.3 | 6.2 | Yes |
| HSD17B14 | ID_50819 | AA | 4.1 | 3 | 4.2 | No |
| RALB | ID_55155 | AA | 3.1 | 2 | 3.2 | Yes |
| FXYD5 | ID_49079 | AA | 1.5 | 1.1 | 1.6 | No |
| RNF19B | ID_1647 | AA | 3.1 | 2 | 3.2 | Yes |
| IP6K2 | ID_64746 | AD | 11.6:11.7 | 11.5 | 11.9 | Yes |
| TXNDC17 | ID_38768 | AD | 1.2 | 1.1 | 2.1 | Yes |
| RCHY1 | ID_69521 | AD | 7.2 | 7.1 | 8.1 | No |
| AURKAIP1 | ID_152 | AD | 1.2 | 1.1 | 1.5 | No |
| RPL13 | ID_392312 | AD | 1.3:1.4 | 1.2 | 2 | Yes |
| PLEKHH3 | ID_41103 | AD | 11.2 | 11.1 | 12.2 | Yes |
| CAPG | ID_54273 | AD | 7.2 | 7.1 | 8 | Yes |
| TRMT1 | ID_47921 | AD | 2.4 | 2.3 | 3 | Yes |
| ATG9A | ID_57639 | AD | 10.2 | 10.1 | 11 | No |
| ECI2 | ID_75224 | AD | 1.2:1.3 | 1.1 | 2.1 | No |
| MLST8 | ID_33211 | AP | 2 |
|
| No |
| GPR75-ASB3 | ID_53555 | AP |
|
|
| No |
| FAM107B | ID_10823 | AP | 4 |
|
| Yes |
| FXN | ID_86525 | AP | 2 |
|
| Yes |
| CLDN5 | ID_61069 | AP |
|
|
| No |
| TUBB3 | ID_38167 | AP | 4 |
|
| No |
| NFYC | ID_2015 | AP | 4 |
|
| No |
| MTFR1L | ID_1211 | ME | 5|6 | 4.2 | 7.2 | Yes |
| CMC2 | ID_37707 | ME | 6|7 | 5 | 9 | No |
| PPP2R1B | ID_18676 | ME | 4|5 | 3 | 6 | Yes |
| SLC39A14 | ID_140283 | ME | 5|6 | 4 | 7 | No |
| RAB6A | ID_17707 | ME | 5|6 | 4 | 7 | Yes |
| FAM92A1 | ID_84527 | ME | 7|8 | 5.1 | 9.2 | Yes |
| KIAA1468 | ID_45699 | ME | 24|25 | 23 | 26 | Yes |
| MOK | ID_29380 | RI | 15.2:15.3:15.4 | 15.1 | 15.5 | Yes |
| MBD6 | ID_22634 | RI | 13.4 | 13.3 | 13.5 | No |
| C9orf9 | ID_87994 | RI | 5.2 | 5.1 | 5.3 | No |
| PSMC5 | ID_43009 | RI | 2.2 | 2.1 | 2.3 | No |
| NUDT22 | ID_16589 | RI | 1.3 | 1.2 | 1.4 | No |
| PRR23C | ID_67029 | RI | 1.2 | 1.1 | 1.3 | Yes |
| HSF1 | ID_85559 | ES | 10 | 9 | 11 | No |
| MRPL2 | ID_76237 | ES | 6 | 5 | 7 | Yes |
| SNRPN | ID_29701 | ES | 11 | 10.4 | 12 | No |
| SEC11A | ID_32314 | ES | 8 | 5 | 9 | No |
| TPM3 | ID_7792 | ES | 12 | 11 | 15 | No |
| STARD3NL | ID_79286 | ES | 2 | 1 | 3 | Yes |
| C14orf2 | ID_29536 | ES | 3 | 2 | 5 | Yes |
| ARHGEF1 | ID_50101 | ES | 15 | 14 | 16 | No |
| SFSWAP | ID_25214 | ES | 15 | 14 | 16.1 | No |
| AFMID | ID_43807 | ES | 7:8:9:10 | 6 | 12 | Yes |
| ERBB2IP | ID_72261 | ES | 22 | 21 | 24.1 | Yes |
| PRRX1 | ID_8968 | AT | 5 |
|
| No |
| MASP1 | ID_68075 | AT | 19 |
|
| No |
| SLC4A4 | ID_69463 | AT | 14.2 |
|
| No |
| PLCH2 | ID_273 | AT | 22.3 |
|
| No |
| L3HYPDH | ID_27740 | AT | 7 |
|
| No |
| AGAP1 | ID_58086 | AT | 11 |
|
| No |
Independent predictive power of the
final AS signature for patients with HCC
Univariate and multivariate Cox hazard regression
analyses of data in the TCGA HCC cohort were performed in order to
further investigate whether the final AS-based signature was an
independent prognostic factor, with the AS signature treated as a
binary variable. The results of comprehensive univariate analysis
suggested that age, hepatitis B status, family cancer history,
serum AFP level, degree of vascular invasion, pathologic stage and
AS signature were all correlated with the OS of patients with HCC
(Fig. 6A). Therefore, these
significant risk factors were included in a multivariate analysis,
which showed that the AS signature (HR: 12.573; 95% CI:
4.957–31.893; P=9.79e-08), AFP level and vascular invasion were
three independent prognostic factors when adjusted by those factors
(Fig. 6B). In addition, in order to
investigate the prognostic value of the AS signature in a
stratified cohort, the patients were classified into various
subgroups based on relative complete clinical features and
stratification analysis was performed. As shown in Fig. S2, the prognostic signature identified
patients with distinct prognoses in all cohorts analyzed, thus
confirming its robustness for independently predicting HCC
prognosis.
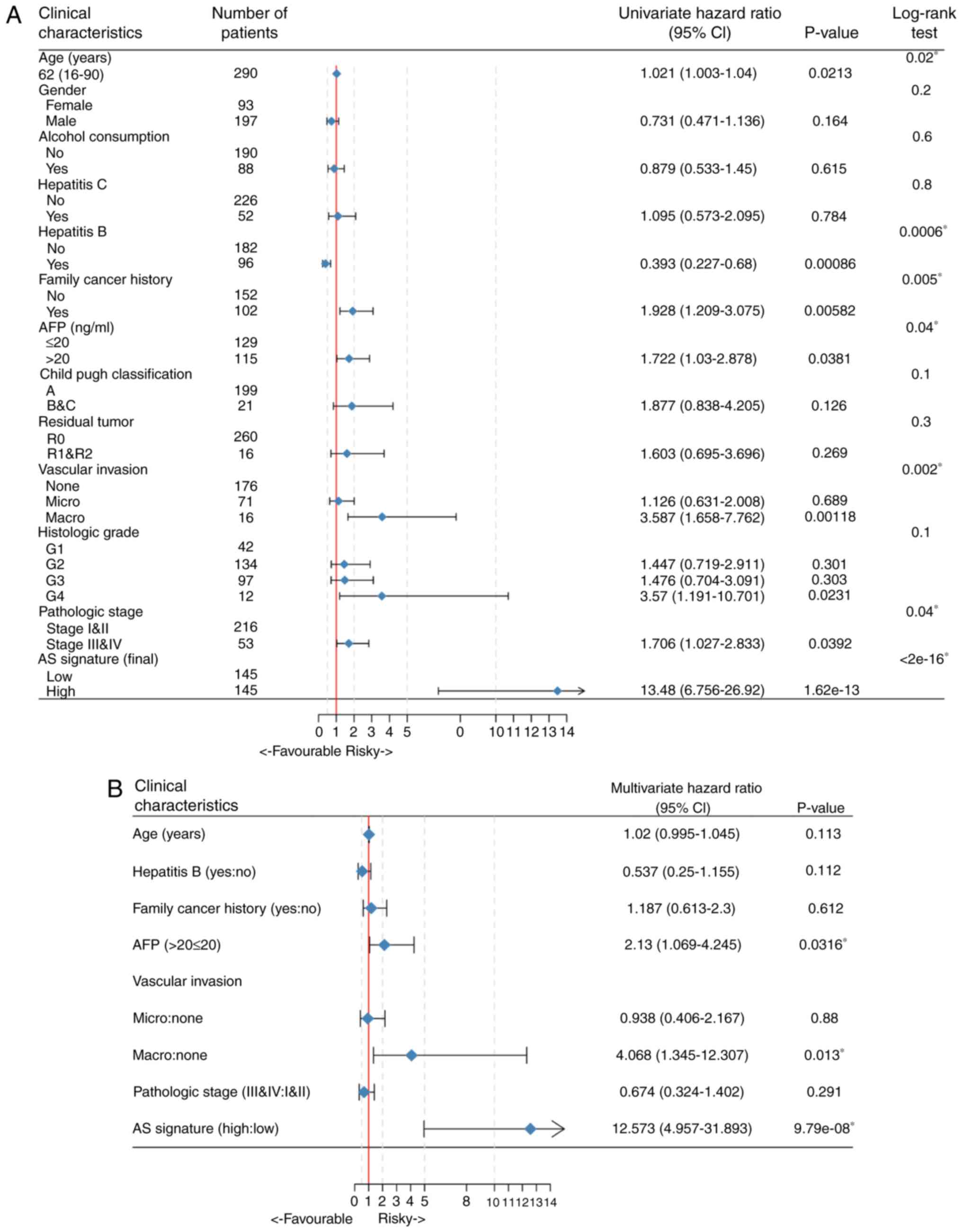 | Figure 6.Forest plots of the associations of
clinicopathological characteristics with OS in the hepatocellular
carcinoma cohort. (A) Univariate Cox regression analysis of the
relation between clinicopathological features and final AS
signature regarding prognostic value. (B) Multivariate Cox analysis
of the associations between clinical risk factors, including age,
hepatitis B, family cancer history, AFP level, vascular invasion,
pathologic stage and AS signature, and OS. The hazard ratios (blue
diamonds) and 95% CI (horizontal lines) are depicted, respectively.
AS, alternative splicing; OS, overall survival; CI, confidence
intervals; AFP, α-fetoprotein. |
Functional enrichment and interaction
analysis of survival-associated AS events
To shed light on the potential impact of prognostic
AS events to their corresponding proteins, GO and KEGG enrichment
analyses were conducted based on those parent genes that generated
prognostic AS events. A total of 20 GO terms were identified in the
aspect of molecular function (MF), including cell adhesion molecule
binding and cadherin binding pathways (Fig. 7B). A total of 176 biological process
(BP) terms (Fig. 7C) and 71 cellular
component (CC) terms (Fig. 7D) were
also enriched significantly, indicating obvious changes in the
purine-related metabolic process, protein targeting, focal adhesion
and cell-surface junction pathways. Additionally, the KEGG
enrichment analysis revealed a total of 48 enriched pathways, the
majority of which were relevant to the liver cancer trilogy,
invasiveness and distant metastasis of hepatoma cells, cancer
pathway and nucleotide metabolism (Fig.
7E). Of note, certain KEGG pathways that are known to be
involved in tumorigenesis and the progression of HCC were also
enriched, including ubiquitin-mediated proteolysis (FDR
<0.0067), p53 signaling pathway (FDR <0.028), AMPK signaling
pathway (FDR <0.017), HIF-1 signaling pathway (FDR <0.0059)
and EGFR tyrosine kinase inhibitor resistance (FDR <0.017).
Coincidentally, cancer-related pathways were also identified,
including colorectal cancer, glioma, pancreatic cancer, endometrial
cancer and non-small cell lung cancer (Fig. 7E). Taken together, the parent genes
associated with prognostic AS events may serve an important role in
the carcinogenesis, progression and metastasis of HCC. Therefore,
these specific dysregulated AS events may orchestrate the
post-transcriptional modification of parent genes and further
modify protein features in patients with HCC. Furthermore, to
demonstrate the interactive relationship between the prognostic AS
events from a biological systems point of view, all of the
identified parent genes in HCC were input into Cytoscape to
generate a gene interaction network. The results revealed several
important hub genes in the entire network, including TP53, EGFR,
HSPA8 and UPF3B (Fig. 7A).
Potential cancer-specific SF-AS
regulatory network in the HCC cohort
It has been shown that the global prognosis-related
AS events can be orchestrated by a limited number of SFs,
particularly in HCC. With access to normalized level-three RNA-seq
profiles of TCGA-HCC samples, the present study identified 53 SFs
whose expression levels differed significantly between tumor
tissues and adjacent normal tissues, among which 27 SFs were
upregulated and 26 were downregulated (Fig. 8A). To systematically analyze the
cancer-specific splicing regulatory connections between SFs and AS
events in HCC, the prognostic SFs were identified and a
cancer-specific SF-AS correlation network was constructed. In this
network, three SFs, including PCBP2 (P=0.009, HR: 1.77, 95% CI:
1.09–2.85), SFPQ (P<0.001, HR: 2.02, 95% CI: 1.25–3.27) and
SRSF10 (P=0.019, HR: 1.74, 95% CI: 1.12–2.7) were screened as being
prognostic (Fig. 8C), and their
corresponding expression levels are presented in Fig. 8B. In addition, a total of 62
survival-associated AS events were used for constructing an SF-AS
regulatory network (Fig. 8D), among
which were 26 risky AS events (HR >1) and 36 favorable events
(HR <1). Of note, the majority of risky prognostic AS events
(red dots) were positively correlated (red lines) with the
expression of SFs (green diamonds), whereas favorable prognostic AS
events (blue dots) were negatively correlated (blue lines) with
these SFs. It appears that aberrant AS events can be co-regulated
by the synergistic or competitive influence of distinct prognostic
SFs. These results further demonstrated the underlying mechanisms
that indicate AS offer potential in expanding the coding capacity
of transcripts. In addition, representative correlation pairs
within this regulatory network are shown in scatter plots (Fig. 8E). For example, the expression of SFPQ
was positively correlated with the RI event of RASSF7 but
negatively correlated with the ES event of AFMID.
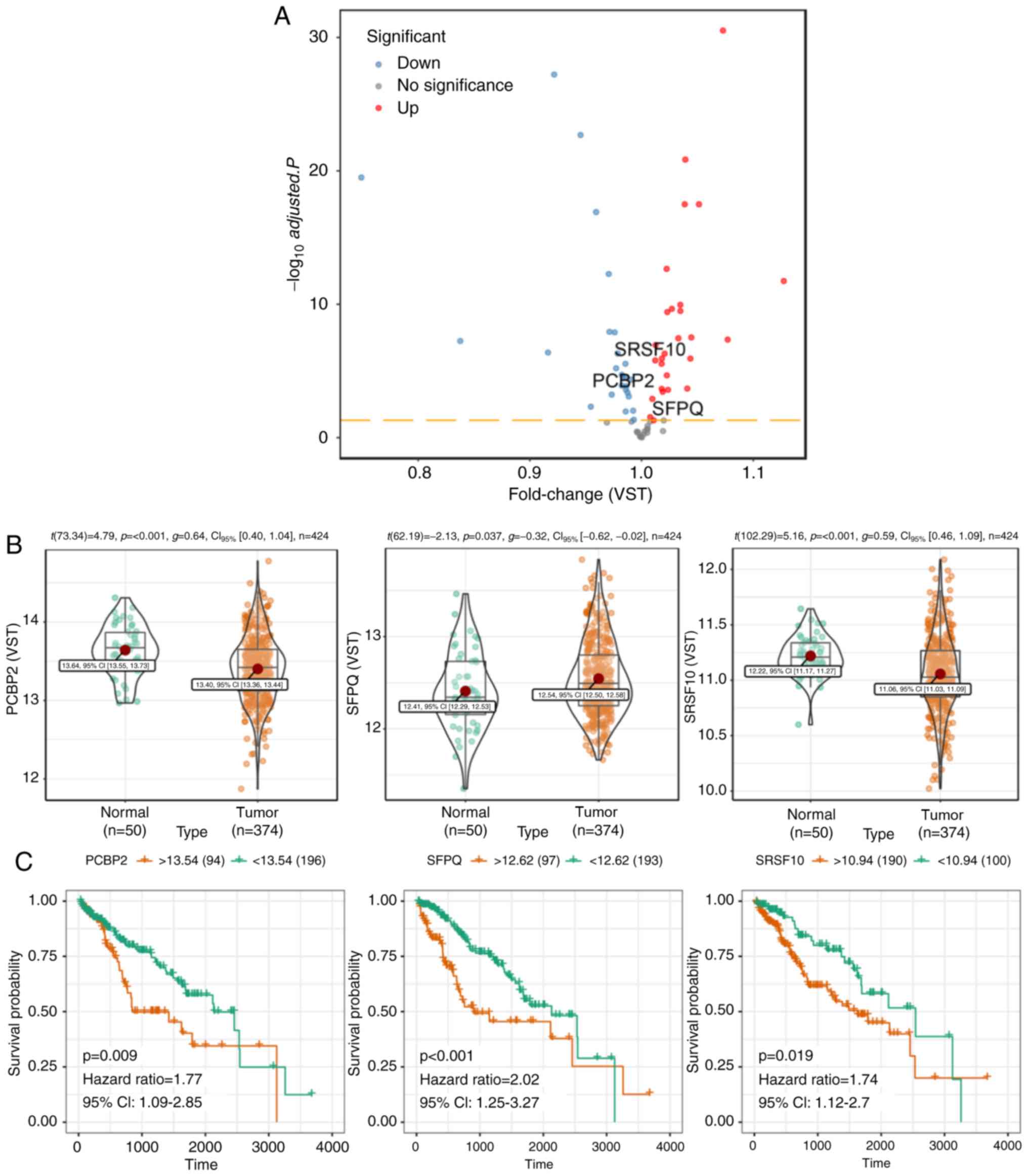 | Figure 8.Construction of a potential SF-AS
regulatory network in HCC. (A) Volcano plot visualizing
differential expressed SFs in The Cancer Genome Atlas dataset of
HCC. The red and blue dots in the plot represent the differentially
expressed SFs with statistical significance (adjusted P<0.05).
And the prognostic SFs are marked in the diagram. (B) Relative
expression levels (VST) of screened prognostic SFs (PCBP2, SFPQ and
SRSF10) between primary HCC and para-cancerous tissues. (C)
Kaplan-Meier survival curves of PCBP2, SFPQ and SRSF10 in the HCC
cohort with optimal cut-off values shown. HCC, hepatocellular
carcinoma; AS, alternative splicing; SF, splicing factor; VST,
variance stabilizing transformation; PSI, percent spliced in; CI,
confidence intervals; RI, retained intron; AD, alternate donor
site; ES, exon skip; AT, alternate terminator. (D) Splicing
correlation network built among the significant correlation pairs.
Three prognostic SFs (green diamonds) were positively (red line) or
negatively (blue line) correlated with 36 favorable AS events (blue
dots) and 26 adverse AS events (red dots). (E) Representative
scatter plots between the normalized expression (VST) of the
particular SF and PSI score of the AS event. HCC, hepatocellular
carcinoma; AS, alternative splicing; SF, splicing factor; VST,
variance stabilizing transformation; PSI, percent spliced in; CI,
confidence intervals; RI, retained intron; AD, alternate donor
site; ES, exon skip; AT, alternate terminator. |
GSVA for SF-AS correlation pairs
The potential mechanism underlying the SF-AS
regulatory relationships, either in a positive or negative
correlation, was investigated. Spearman's correlation analyses were
conducted between the identified SFs described above and
survival-associated AS events, P-values were adjusted using the
Benjamini-Hochberg correction. The significant SF-AS correlation
pairs were filtered with an adjusted P-value of <0.05. The
parent genes of these correlation pairs were then sent for GSVA and
differential enrichment analysis between tumor and adjacent normal
samples. The results were visualized as volcano plots and heatmaps,
respectively (Figs. S3 and S4), which showed that the tumor tissue
exhibited increased activities in cell proliferation and cell
cycle, and decreased activities in immune response and cell
adhesion.
Discussion
HCC is heterogeneous tumor from a molecular point of
view (45). Over the last decade,
significant efforts have been made to reveal the molecular changes
in genomic profiles involved in the development of HCC (46). Such studies have contributed to the
determination of prognostic genetic signatures, including genes,
microRNAs and non-coding RNAs, which has promoted the
identification of relevant prognostic markers and even therapeutic
targets (47–49). In addition, as a major
post-transcriptional biological behavior to expand genomic coding
capacity and increase protein diversity, AS has been shown to have
more potential significance in cancer biology.
Preliminary investigations of AS in HCC have
demonstrated the crucial cancer-associated phenotypes can be
converted by specific AS events and changes through inducing cell
proliferation, promoting angiogenesis or avoiding apoptosis
(20). For example, PKM2, resulting
from ME events of exons 9 and 10 of the Pkm gene, is closely linked
to the tumorigenesis of HCC by controlling cancer metabolic
homeostasis and inflammation (50).
Similarly, KIA1 is regarded as a metastatic suppressor gene in HCC;
in-depth investigation revealed that KITENIN, as a spliced variant
of KAI1 lacking exon 7 at the COOH-terminal region, enhances
distant metastasis by facilitating cell invasion and antagonizing
the expression of KIA1 and other metastasis-suppressing genes
(51). There has been much success in
research into the diversity of HCC-specific splicing variants
owning to the advancement of high-throughput technology. Hui et
al analyzed eight paired tumor-normal HCC specimens using SMRT
sequencing, and reported 233 novel AS events occurred in 223 known
genes (52). More recently, Li et
al identified 243 differential AS events in the development and
progression of HCC, and these were closely involved in
metabolism-related pathways and cancer hallmarks (53). Accordingly, aberrant AS events,
considered as another hallmark of cancer, may serve as promising
diagnostic, predictive and prognostic biomarkers for patients with
HCC. However, due to the lack of corresponding clinical information
and limited sample size, few have annotated the detected AS events
with clinical meaning in a systematic manner, particularly for
patients with HCC.
To the best of our knowledge, the present study is
the first attempt at a comprehensive and integrated computational
investigation of the AS event characteristics of HCC, further
broadening the novel field of prognostic and molecule-targeted
implications. Based on the splicing pattern and bioinformatics
algorithm of SpliceSeq, AS events can be roughly divided into seven
types, which are depicted in Fig. 1C.
Following strict filtering and screening, the preliminarily
analysis detected a total of 34,163 AS events from 8,986 parent
genes, among which the majority of detectable AS events belonged to
the ES type (36.08%). Recently, the prognostic values of splicing
variants have been widely identified in various types of cancer,
including non-small cell lung cancer, pan-gastrointestinal
adenocarcinomas, ovarian cancer and esophageal carcinoma (54–57). Such
studies have developed a series of AS-based prognostic signatures
that have performed fairly well in distinguishing between poor and
good outcomes in relevant patients. Similarly, a total of 1,805 AS
events from 1,314 parent genes were identified to be associated
with OS in patients with HCC, using univariate Cox regression
analyses. It is noteworthy that one gene can hold differential AS
events that have a significantly opposite influence on survival
according to the forest plots, which would have been undetectable
if focus had been on transcriptional expression levels only,
further highlighting the indispensable role of AS in oncogenesis.
Compared with previous studies that used multivariate Cox
regression analyses to screen key features in a prognostic panel,
the present study applied the stepwise variable selection approach
with the AIC criterion, which is more scientific and stringent for
high-dimensional data mining. Encouragingly, the time-dependent ROC
curves confirmed that the final AS signature, comprised of 26 AS
events, had a noteworthy ability to distinguish between the
distinct prognoses for patients with HCC, which also indicated that
splicing events may be preeminent and ideal indicators for
predicting cancer prognoses. In addition, the proposed AS signature
was independent of traditional clinical risk factors and of high
clinical applicability in stratified patients.
Furthermore, the present study attempted to
investigate the potential mechanism of prognostic AS events.
Notably, the functional enrichment analysis revealed several
significant interfered pathways, including ubiquitin-mediated
proteolysis, ribosome and p53 signaling pathway, which were in
accordance with previous studies concerning the genome-wide
investigation of AS in gastrointestinal adenocarcinoma and
colorectal cancer, respectively (40,56).
Therefore, the cancer-related outcome resulting from AS alteration
may be disturbed via certain shared cancer pathways. Additionally,
it was revealed that the HIF-1 pathway and AMPK signaling pathway
were significantly enriched biological processes, which have been
gradually recognized to be associated with HCC-specific prognosis.
For example, Jiang et al revealed that HIF-1 directly bound
to the Rpn10 promoter and further promoted HCC cell proliferation
(58). A cohort study (n=419) by Wang
et al also suggested that HIF-1 was significantly associated
with TNM stage, hepatitis B virus infection, tumor size, portal
vein tumor thrombus and vascular invasion and further served as an
independent adverse prognostic factor for patients with HCC with
liver cirrhosis (59). AMP-activated
protein kinase (AMPK), as a conserved heterotrimeric protein kinase
complex, serves a vital role in cancer development and linking
metabolism through mediating multiple mechanisms related to cell
cycle, apoptosis and autophagy (60).
In vitro, the phosphorylation status of AMPK has an
anticancer effect via inhibition of the NF-κB signaling pathway in
HCC (61). Previous investigations on
HCC support the reliability and accuracy of the bioinformatics
analyses performed in the present study. The results suggest that
the poor outcomes of patients with HCC resulted from aberrant AS
events that may be disturbed by a certain vital oncogenic
biological process.
As widespread AS alterations within the tumor
microenvironment may be extensively orchestrated by a limited
number of SFs, the present study focused on the altered prognostic
SFs and the SF-AS splicing correlation network. Through
differential expression analysis combined with survival analysis,
three SFs (SRSF10, PCBP2 and SFPQ) were filtered as candidates and
all three were recognized as adverse prognostic factors. The
function of splicing regulator SRSF10 may be partly mediated by the
generation of BCLAF1-L through the alternative inclusion of exon
5a; the overexpression of the BCLAF1-L isoform has been shown to be
associated with increased tumorigenic potential and a higher tumor
grade in colon cancer (62). Liu
et al revealed that SRSF10 can modulate the AP events of
IL-1 towards mIL1RAP in cervical cancer oncogenesis, which promoted
NF-κB activation and inhibited macrophage phagocytosis (63). Poly(C)-binding protein 2 (PCBP2), as a
multifunctional adapter that contributes to mRNA stabilization,
translational silencing and enhancement via the poly(C)-binding
motif, has been widely reported to mediate ambiguous functions in
various types of cancer, including gastric cancer, breast cancer,
pancreatic ductal adenocarcinoma, esophageal squamous cell
carcinoma and glioblastoma (37,64–68). A
particular AS event was also implicated in the presence of the
binding site of PCBP2 within the exon 6 splicing acceptor (69). PCBP2 is pivotal in attenuating the
innate immune response against hepatitis C infection and promoting
alcoholic liver fibrosis, both of which are recognized as clinical
carcinogenic factors for HCC (70,71). As a
member of the Drosophila behavior Human Splicing family,
SFPQ, containing both RNA- and DNA-binding motifs, is linked to
multiple biological behaviors, including pre-mRNA splicing,
transcriptional regulation and DNA mismatch repair (21,72,73).
Previous studies have also revealed that SFPQ is correlated with
the development of cisplatin resistance in liver cancer and
regarded as an adverse prognostic biomarker (74). However, until now, few studies have
addressed the involvement of SFs in AS events in HCC, and the
regulatory roles of SFs in generating varied transcript isoforms
require further validation in HCC samples. The splicing correlation
network showed distinguished interactions between the identified
SFs and the significant prognostic AS events. Of note, SFs can have
opposite effects in the regulation of AS events, even from the same
gene, and one particular AS event can be synergistically or
antagonistically regulated by different SFs. This demonstrates that
these complicated underlying connections offer potential clues to
further understanding the crucial process that participates in
regulation of HCC. Another noteworthy finding of the present study
was that the majority of the favorable prognostic AS events were
negatively regulated by SFs, whereas the risky AS events were
mainly positively regulated by SFs. This indicates the multiple
altered SFs may promote the invasive and metastatic potential of
HCC via co-regulating the AS events of genes, which may offer novel
insights for elucidating the mechanisms underlying the biogenesis
and progression of HCC.
However, there were several limitations in the
present study that require clarification. Firstly, the patients
enrolled were exclusively from a single cohort and the sample size
of the HCC cohort was limited. Secondly, due to the lack of other
independent cohort concerning AS events among HCC patients, further
validation and reproducible analysis could not be performed at
present. Thirdly, owning to the retrospective nature of the present
study, prospective cohorts with a larger sample size are warranted.
Therefore, further verification of the in silico analysis
performed is required in the future.
In conclusion, the present study provided a
comprehensive picture of the global changes in mRNA splicing
signatures in HCC, developed a robust prognostic model and
constructed a splicing correlation network, were valuable in
elucidating the underlying mechanisms of AS and contributed to the
identification of novel prognostic markers and therapeutic targets
for further validation.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science
Foundation of Liaoning Province (grant no. 20180550781).
Availability of data and materials
The RNA-seq data, splicing variant data and clinical
information of HCC used in this study were acquired and integrated
from the TCGA and TCGA SpliceSeq.
Authors' contributions
DZ and JL contributed to the conception and design
of the study. YD and ZW also participated in the conception of the
study. DZ performed the statistical analysis. DZ and YD were
involved in the preparation of the figures and tables. DZ, ZW and
YD reviewed the results and participated in the discussion of the
data. DZ and YD prepared and wrote the manuscript. ZW and JL
revised the manuscript. All authors read and approved the final
version of this manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AA
|
alternate acceptor site
|
|
AD
|
alternate donor site
|
|
AFP
|
α-fetoprotein
|
|
AP
|
alternate promoter
|
|
AS
|
alternative splicing
|
|
AT
|
alternate terminator
|
|
AUC
|
area under the curve
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
CI
|
confidence intervals
|
|
ES
|
exon skip
|
|
FDR
|
false discovery rate
|
|
GO
|
Gene Ontology
|
|
GSVA
|
gene set variation analysis
|
|
HCC
|
hepatocellular carcinoma
|
|
HR
|
hazard ratio
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ME
|
mutually exclusive exons
|
|
MF
|
molecular function
|
|
OS
|
overall survival
|
|
PSI
|
percent spliced in
|
|
RI
|
retained intron
|
|
ROC
|
receiver-operator characteristic
|
|
SF
|
splicing factor
|
|
TCGA
|
The Cancer Genome Atlas
|
|
VST
|
variance stabilizing
transformation
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vitale A, Peck-Radosavljevic M, Giannini
EG, Vibert E, Sieghart W, Van Poucke S and Pawlik TM: Personalized
treatment of patients with very early hepatocellular carcinoma. J
Hepatol. 66:412–423. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ulahannan SV, Duffy AG, McNeel TS, Kish
JK, Dickie LA, Rahma OE, McGlynn KA, Greten TF and Altekruse SF:
Earlier presentation and application of curative treatments in
hepatocellular carcinoma. Hepatology. 60:1637–1644. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tabrizian P, Jibara G, Shrager B, Schwartz
M and Roayaie S: Recurrence of hepatocellular cancer after
resection: Patterns, treatments, and prognosis. Ann Surg.
261:947–955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang JH, Wang CC, Hung CH, Chen CL and Lu
SN: Survival comparison between surgical resection and
radiofrequency ablation for patients in BCLC very early/early stage
hepatocellular carcinoma. J Hepatol. 56:412–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ueno M, Hayami S, Shigekawa Y, Kawai M,
Hirono S, Okada K, Tamai H, Shingaki N, Mori Y, Ichinose M and
Yamaue H: Prognostic impact of surgery and radiofrequency ablation
on single nodular HCC 5 cm: Cohort study based on serum HCC
markers. J Hepatol. 63:1352–1359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hiraoka A, Kumada T, Kudo M, Hirooka M,
Tsuji K, Itobayashi E, Kariyama K, Ishikawa T, Tajiri K, Ochi H, et
al: Albumin-bilirubin (ALBI) grade as part of the evidence-based
clinical practice guideline for HCC of the japan society of
hepatology: A comparison with the liver damage and child-pugh
classifications. Liver Cancer. 6:204–215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li S and Mao M: Next generation sequencing
reveals genetic landscape of hepatocellular carcinomas. Cancer
Lett. 340:247–253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang W, Skanderup AJ and Lee CG: Advances
in genomic hepatocellular carcinoma research. Gigascience.
7:112018. View Article : Google Scholar
|
|
10
|
Zhang YC, Zhou Q and Wu YL: The emerging
roles of NGS-based liquid biopsy in non-small cell lung cancer. J
Hematol Oncol. 10:1672017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cancer Genome Atlas Research Network.
Electronic address, . simplewheeler@bcm.edu; Cancer
Genome Atlas Research Network: Comprehensive and integrative
genomic characterization of hepatocellular carcinoma. Cell.
169:1327–1341.e23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villanueva A, Portela A, Sayols S,
Battiston C, Hoshida Y, Mendez-Gonzalez J, Imbeaud S, Letouzé E,
Hernandez-Gea V, Cornella H, et al: DNA methylation-based prognosis
and epidrivers in hepatocellular carcinoma. Hepatology.
61:1945–1956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiu J, Peng B, Tang Y, Qian Y, Guo P, Li
M, Luo J, Chen B, Tang H, Lu C, et al: CpG Methylation signature
predicts recurrence in early-stage hepatocellular carcinoma:
Results from a multicenter study. J Clin Oncol. 35:734–742. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Long J, Wang A, Bai Y, Lin J, Yang X, Wang
D, Yang X, Jiang Y and Zhao H: Development and validation of a
TP53-associated immune prognostic model for hepatocellular
carcinoma. EBioMedicine. 42:363–374. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Long J, Bai Y, Yang X, Lin J, Yang X, Wang
D, He L, Zheng Y and Zhao H: Construction and comprehensive
analysis of a ceRNA network to reveal potential prognostic
biomarkers for hepatocellular carcinoma. Cancer Cell Int.
19:902019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baralle FE and Giudice J: Alternative
splicing as a regulator of development and tissue identity. Nat Rev
Mol Cell Biol. 18:437–451. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gamazon ER and Stranger BE: Genomics of
alternative splicing: Evolution, development and pathophysiology.
Hum Genet. 133:679–687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang ET, Sandberg R, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Montes M, Sanford BL, Comiskey DF and
Chandler DS: RNA Splicing and disease: Animal models to therapies.
Trends Genet. 35:68–87. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Climente-Gonzalez H, Porta-Pardo E, Godzik
A and Eyras E: The functional impact of alternative splicing in
cancer. Cell Rep. 20:2215–2226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tao Y, Ma C, Fan Q, Wang Y, Han T and Sun
C: MicroRNA-1296 facilitates proliferation, migration and invasion
of colorectal cancer cells by targeting SFPQ. J Cancer.
9:2317–2326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
David CJ and Manley JL: Alternative
pre-mRNA splicing regulation in cancer: Pathways and programs
unhinged. Genes Dev. 24:2343–2364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song X, Zeng Z, Wei H and Wang Z:
Alternative splicing in cancers: From aberrant regulation to new
therapeutics. Semin Cell Dev Biol. 75:13–22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Y, Xu W, Ji J, Feng D, Sourbier C, Yang
Y, Qu J, Zeng Z, Wang C, Chang X, et al: Alternative splicing of
the cell fate determinant numb in hepatocellular carcinoma.
Hepatology. 62:1122–1131. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Berasain C, Goni S, Castillo J, Latasa MU,
Prieto J and Avila MA: Impairment of pre-mRNA splicing in liver
disease: Mechanisms and consequences. World J Gastroenterol.
16:3091–3102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ratnadiwakara M, Mohenska M and Anko ML:
Splicing factors as regulators of miRNA biogenesis-links to human
disease. Semin Cell Dev Biol. 79:113–122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen M and Manley JL: Mechanisms of
alternative splicing regulation: Insights from molecular and
genomics approaches. Nat Rev Mol Cell Biol. 10:741–754. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wahl MC, Will CL and Luhrmann R: The
spliceosome: Design principles of a dynamic RNP machine. Cell.
136:701–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prochazka L, Tesarik R and Turanek J:
Regulation of alternative splicing of CD44 in cancer. Cell Signal.
26:2234–2239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshida K and Ogawa S: Splicing factor
mutations and cancer. Wiley Interdiscip Rev RNA. 5:445–459. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
El Marabti E and Younis I: The cancer
spliceome: Reprograming of alternative splicing in cancer. Front
Mol Biosci. 5:802018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anczukow O and Krainer AR: Splicing-factor
alterations in cancers. RNA. 22:1285–1301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tomczak K, Czerwinska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
34
|
Ryan MC, Cleland J, Kim R, Wong WC and
Weinstein JN: SpliceSeq: A resource for analysis and visualization
of RNA-Seq data on alternative splicing and its functional impacts.
Bioinformatics. 28:2385–2387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liao SG, Lin Y, Kang DD, Chandra D, Bon J,
Kaminski N, Sciurba FC and Tseng GC: Missing value imputation in
high-dimensional phenomic data: Imputable or not, and how? BMC
Bioinformatics. 15:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lex A, Gehlenborg N, Strobelt H, Vuillemot
R and Pfister H: UpSet: Visualization of intersecting sets. IEEE
Trans Vis Comput Graph. 20:1983–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ye J, Zhou G, Zhang Z, Sun L, He X and
Zhou J: Poly (C)-binding protein 2 (PCBP2) promotes the progression
of esophageal squamous cell carcinoma (ESCC) through regulating
cellular proliferation and apoptosis. Pathol Res Pract.
212:717–725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kamarudin AN, Cox T and Kolamunnage-Dona
R: Time-dependent ROC curve analysis in medical research: Current
methods and applications. BMC Med Res Methodol. 17:532017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiong Y, Deng Y, Wang K, Zhou H, Zheng X,
Si L and Fu Z: Profiles of alternative splicing in colorectal
cancer and their clinical significance: A study based on
large-scale sequencing data. EBioMedicine. 36:183–195. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Piva F, Giulietti M, Burini AB and
Principato G: SpliceAid 2: A database of human splicing factors
expression data and RNA target motifs. Hum Mutat. 33:81–85. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-Seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin DC, Mayakonda A, Dinh HQ, Huang P, Lin
L, Liu X, Ding LW, Wang J, Berman BP, Song EW, et al: Genomic and
epigenomic heterogeneity of hepatocellular carcinoma. Cancer Res.
77:2255–2265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang BD and Lee NH: Aberrant RNA splicing
in cancer and drug resistance. Cancers (Basel). 10:112018.
View Article : Google Scholar :
|
|
47
|
Shi YM, Li YY, Lin JY, Zheng L, Zhu YM and
Huang J: The discovery of a novel eight-mRNA-lncRNA signature
predicting survival of hepatocellular carcinoma patients. J Cell
Biochem. Nov 28–2018.(Epub ahead of print). doi:
10.1002/jcb.28028.
|
|
48
|
Liu G, Wang H, Fu JD, Liu JY, Yan AG and
Guan YY: A five-miRNA expression signature predicts survival in
hepatocellular carcinoma. APMIS. 125:614–622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gu JX, Zhang X, Miao RC, Xiang XH, Fu YN,
Zhang JY, Zhang JY, Liu C and Qu K: Six-long non-coding RNA
signature predicts recurrence-free survival in hepatocellular
carcinoma. World J Gastroenterol. 25:220–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dayton TL, Gocheva V, Miller KM, Israelsen
WJ, Bhutkar A, Clish CB, Davidson SM, Luengo A, Bronson RT, Jacks T
and Vander Heiden MG: Germline loss of PKM2 promotes metabolic
distress and hepatocellular carcinoma. Genes Dev. 30:1020–1033.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee JH, Park SR, Chay KO, Seo YW, Kook H,
Ahn KY, Kim YJ and Kim KK: KAI1 COOH-terminal interacting
tetraspanin (KITENIN), a member of the tetraspanin family,
interacts with KAI1, a tumor metastasis suppressor, and enhances
metastasis of cancer. Cancer Res. 64:4235–4243. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen H, Gao F, He M, Ding XF, Wong AM, Sze
SC, Yu AC, Sun T, Chan AW, Wang X and Wong N: Long-read RNA
sequencing identifies Alternative splice variants in hepatocellular
carcinoma and tumor-specific isoforms. Hepatology. Jan
13–2019.(Epub ahead of print). doi: 10.1002/hep.30500.
|
|
53
|
Li S, Hu Z, Zhao Y, Huang S and He X:
Transcriptome-wide analysis reveals the landscape of aberrant
alternative splicing events in liver cancer. Hepatology.
69:359–375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z
and He J: Prognostic alternative mRNA splicing signature in
non-small cell lung cancer. Cancer Lett. 393:40–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu J, Chen Z and Yong L: Systematic
profiling of alternative splicing signature reveals prognostic
predictor for ovarian cancer. Gynecol Oncol. 148:368–374. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lin P, He RQ, Ma FC, Liang L, He Y, Yang
H, Dang YW and Chen G: Systematic analysis of survival-associated
alternative splicing signatures in gastrointestinal
pan-adenocarcinomas. EBioMedicine. 34:46–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mao S, Li Y, Lu Z, Che Y, Sun S, Huang J,
Lei Y, Wang X, Liu C, Zheng S, et al: Survival-associated
alternative splicing signatures in esophageal carcinoma.
Carcinogenesis. 40:121–130. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jiang Z, Zhou Q, Ge C, Yang J, Li H, Chen
T, Xie H, Cui Y, Shao M, Li J and Tian H: Rpn10 promotes tumor
progression by regulating hypoxia-inducible factor 1 alpha through
the PTEN/Akt signaling pathway in hepatocellular carcinoma. Cancer
Lett. 447:1–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang D, Zhang X, Lu Y, Wang X and Zhu L:
Hypoxia inducible factor 1α in hepatocellular carcinoma with
cirrhosis: Association with prognosis. Pathol Res Pract.
214:1987–1992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jones RG and Thompson CB: Tumor
suppressors and cell metabolism: A recipe for cancer growth. Genes
Dev. 23:537–548. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zheng L, Yang W, Wu F, Wang C, Yu L, Tang
L, Qiu B, Li Y, Guo L, Wu M, et al: Prognostic significance of AMPK
activation and therapeutic effects of metformin in hepatocellular
carcinoma. Clin Cancer Res. 19:5372–5380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou X, Li X, Cheng Y, Wu W, Xie Z, Xi Q,
Han J, Wu G, Fang J and Feng Y: BCLAF1 and its splicing regulator
SRSF10 regulate the tumorigenic potential of colon cancer cells.
Nat Commun. 5:45812014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu F, Dai M, Xu Q, Zhu X, Zhou Y, Jiang
S, Wang Y, Ai Z, Ma L, Zhang Y, et al: SRSF10-mediated IL1RAP
alternative splicing regulates cervical cancer oncogenesis via
mIL1RAP- NF-κB-CD47 axis. Oncogene. 37:2394–2409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chen C, Lei J, Zheng Q, Tan S, Ding K and
Yu C: Poly(rC) binding protein 2 (PCBP2) promotes the viability of
human gastric cancer cells by regulating CDK2. FEBS Open Bio.
8:764–773. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Luo K and Zhuang K: High expression of
PCBP2 is associated with progression and poor prognosis in patients
with glioblastoma. Biomed Pharmacother. 94:659–665. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li F, Bullough KZ, Vashisht AA,
Wohlschlegel JA and Philpott CC: Poly(rC)-binding protein 2
regulates hippo signaling to control growth in breast epithelial
cells. Mol Cell Biol. 36:2121–2131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hu CE, Liu YC, Zhang HD and Huang GJ: The
RNA-binding protein PCBP2 facilitates gastric carcinoma growth by
targeting miR-34a. Biochem Biophys Res Commun. 448:437–442. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wan C, Gong C, Zhang H, Hua L, Li X, Chen
X, Chen Y, Ding X, He S, Cao W, et al: β2-adrenergic receptor
signaling promotes pancreatic ductal adenocarcinoma (PDAC)
progression through facilitating PCBP2-dependent c-myc expression.
Cancer Lett. 373:67–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ghanem LR, Kromer A, Silverman IM, Ji X,
Gazzara M, Nguyen N, Aguilar G, Martinelli M, Barash Y and
Liebhaber SA: Poly(C)-binding protein Pcbp2 enables differentiation
of definitive erythropoiesis by directing functional splicing of
the runx1 transcript. Mol Cell Biol. 38:162018. View Article : Google Scholar
|
|
70
|
Liu H, Chen Z, Jin W, Barve A, Wan YY and
Cheng K: Silencing of α-complex protein-2 reverses alcohol- and
cytokine-induced fibrogenesis in hepatic stellate cells. Liver Res.
1:70–79. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Qin Y, Xue B, Liu C, Wang X, Tian R, Xie
Q, Guo M, Li G, Yang D and Zhu H: NLRX1 mediates MAVS degradation
to attenuate the hepatitis C virus-induced innate immune response
through PCBP2. J Virol. 91:232017. View Article : Google Scholar
|
|
72
|
Mora Gallardo C, Sánchez de Diego A,
Gutiérrez Hernández J, Talavera-Gutiérrez A, Fischer T, Martínez-A
C and van Wely KHM: Dido3-dependent SFPQ recruitment maintains
efficiency in mammalian alternative splicing. Nucleic Acids Res.
47:5381–5394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ray D, Kazan H, Cook KB, Weirauch MT,
Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al:
A compendium of RNA-binding motifs for decoding gene regulation.
Nature. 499:172–177. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ru Y, Chen XJ, Guo WZ, Gao SG, Qi YJ, Chen
P, Feng XS and Zhang SJ: NEAT1_2-SFPQ axis mediates cisplatin
resistance in liver cancer cells in vitro. Onco Targets Ther.
11:5695–5702. 2018. View Article : Google Scholar : PubMed/NCBI
|