Introduction
Pancreatic cancer (PC) is the fourth most aggressive
digestive cancer, and the 5-year overall survival rate is <5%
(1). Although therapeutic
strategies for treatment of PC have improved, patient outcomes
remain unsatisfactory due to a high degree of metastasis (2,3).
Epithelial-mesenchymal transition (EMT) is a key part of the
metastatic process in PC. EMT is associated with significant
changes in the expression of thousands of genes and is
characterized by the downregulation of epithelial markers, such as
E-cadherin, and the upregulation of mesenchymal markers, such as
N-cadherin and vimentin (4). The
process of EMT is regulated by several signaling pathways, such as
the MEK/ERK (5), AKT (6) and Wnt signaling pathways (7). Activation of the Wnt signaling pathway
results in nuclear translocation of β-catenin, where it co-operates
with TCF4 and increases the transcription of mesenchymal markers
(8). Therefore, identification of
upstream regulatory mechanisms of the Wnt signaling pathway and
thus EMT may serve as targets for treatment of patients with
PC.
Epigenetic regulation, including through non-coding
RNAs, are upstream regulatory mechanisms which may regulate the Wnt
signaling pathway and thus EMT (9).
Long noncoding RNAs (lncRNAs) are a type of non-coding RNA >200
nucleotides in length. Competing endogenous RNAs (ceRNAs) sponge
target microRNAs (miRNAs) by binding to the 3′ untranslated regions
(3′UTR) of the target miRNAs and mRNAs and regulate their
expression (10). Various lncRNAs
have been demonstrated to be involved in the metastasis of PC
(11). Yang et al (12) demonstrated that lncRNA DLX6-AS1
expression was increased in PC tissues and cell lines, and
modulated the Wnt/β-catenin pathway to promote proliferation,
migration and invasion of cells by sponging miR-497-5p (12). LincRNA H19 activated the Wnt
signaling pathway and promoted the metastasis of PC cells by
regulating the miR-194/PFTK1 axis (13). The biological functions and
associated mechanisms of Linc00261 have been demonstrated in
various types of cancer (14,15).
Linc00261 expression was reduced in non-small cell lung cancer
cells and had the capacity to inhibit the progression of non-small
cell lung cancer (16). Linc00261
suppressed proliferation and migration of colon cancer by sponging
miR-324-3p (17). However, the role
of Linc00261 in PC is unknown, to the best of our knowledge.
Forkhead box O3 (FOXO3) is member of the forkhead
box O transcription factors, which is involved in EMT by regulating
the Wnt signaling pathway (18).
Recently, numerous studies have revealed that FOXO3 is inactivated
in different types of cancer and may serve as a tumor suppressor
(19). In PC, by inhibiting the Wnt
signaling pathway and thus EMT, FOXO3 also exhibited an inhibitory
effect on metastasis in vitro and in vivo (20). However, the molecular regulatory
mechanism by which FOXO3 results in these effects are not
completely understood.
In the present study, the effect of Linc00261 on the
metastasis of PC cells, and the association between Linc00261, EMT
and metastasis was assessed. Linc00261 overexpression inhibited
metastasis of PC cells, EMT and the Wnt signaling pathway by
regulating the miR-552-5p/FOXO3 axis. Linc00261 may serve as a
suppressive lncRNA in PC and thus may be a potentially useful
biomarker for the diagnosis of PC and effective target for
treatment.
Materials and methods
Clinical specimens
A total of 54 pairs of PC tissues and corresponding
adjacent non-tumor tissues were collected at the Affiliated
Hospital of Guizhou Medical University (Guizhou, China) between
July 2014 and March 2019. The mean age of patients enrolled in the
present was 55±7.4 years, (range, 45–71 years) with 25 males and 29
females. None of patients enrolled in the present study received
neoadjuvant chemotherapy, radiotherapy or immunotherapy prior to
surgery. The present study was approved by the Ethics Committee of
Guizhou Medical University and performed in accordance with the
Declaration of Helsinki (Approval no. 2019LS146). All patients
provided written informed consent for participation.
Bioinformatics method
The expression of Linc00261 in PC tissues and
adjacent tissues was first assessed by online database GEPIA (URL:
http://gepia.cancer-pku.cn/). LogFC>1
and a P-value <0.05 was set as cut-offs. TargetScan (version
7.2; URL: http://www.targetscan.org/vert_72/) was used to
determine the target miRNA of Linc00261, while the target gene of
target miRNA was determined by TargetScan and miRwalk (version 3.0;
URL: http://mirwalk.umm.uni-heidelberg.de/). The pathways
that the target genes were enriched in, were analysed using R
software (version: 3.5.2; The R Foundation; URL: http://www.r-project.org/). P<0.05 was used as a
threshold for a pathway to be considered significantly
enriched.
Cell culture and transfection
A total of 6 PC cell lines, CFPAC-1 (liver
metastasis derivation, metastasis potential), AsPC-1 (ascites
derivation, metastasis potential), MIA-PaCa-2 (primary tumor,
non-metastasis potential), Capan-2 (primary tumor, non-metastasis
potential), BXPC-3 (primary tumor, non-metastasis potential) and
PANC-1 (primary tumor, metastasis potential), and the normal
pancreatic duct epithelial cell line HPDE were all purchased from
American Type Culture Collection. CFPAC-1, MIA-PaCa-2 and PANC-1
cells were cultured in high-glucose DMEM medium (Gibco; Thermo
Fisher Scientific, Inc.), whereas CFPAC-1, AsPC-1, Capan-2 and
BXPC-3 cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.). All cell lines were cultured at 37°C in a
humidified atmosphere containing 5% CO2.
A Linc00261 expression lentivirus was generated by
subcloning the PCR-amplified full-length human Linc00261 cDNA into
the pMSCV retrovirus plasmid (GeneCopoeia, Inc.). Empty pMSCV
retrovirus plasmid was used as a negative control for Linc00261
expression lentivirus. Linc00261-targeting and scramble short
hairpin RNA (shRNA) oligonucleotides were cloned into the
pSuper-retro-puro vector to generate pSuper-retro-Linc00261-RNAi.
The shRNA sequences were: shLinc00261, CAGTCGCTTGGTTTGAGCTCAAATA;
scramble, UUCUCCGAACGUGUCACGUTT. The miR-552-5p mimic and inhibitor
were obtained from GeneCopoeia, Inc. PANC-1 and Mia-PaCa2 cells
were seeded into 6-well plates at a density of
1×105/well. After the cells had adhered, a lentivirus
was added according to manufacturer's protocol (MOI=5 for PANC-1,
MOI=10 for MIA-PaCa2). To obtain stably overexpressing Linc00261
cells or stable knockdown cells, cells were selected for 14 days
using 1 µg/ml puromycin 48 h after transfection.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA of tissues and cells were extracted using
TRIzol reagent (Wuhan Boster Biological Technology, Ltd.). Total
RNA was reverse-transcribed into cDNA using PrimeScript RT reagent
kit (Yeasen Biotechnology, Co., Ltd.). qPCR was performed using
SYBR Green Master mix (Yeasen, Biotechnology, Co., Ltd.). Relative
expression was normalized to GAPDH and fold-change was calculated
using the 2−ΔΔCq method (21). The sequences of the primers used
were: Linc00261 forward, GTCAGAAGGAAAGGCCGTGA and reverse,
TGAGCCGAGATGAACAGGTG; miR-552-5p forward, CCGCACAGGTGACTGGTTAGA and
reverse, GTGCAGGGTCCGAGGT; FOXO3 forward, CGGACAAACGGCTCACTCT and
reverse, GGACCCGCATGAATCGACTAT; E-cadherin forward,
CGAGAGCTACACGTTCACGG and reverse, GGGTGTCGAGGGAAAAATAGG; N-cadherin
forward, GGGTGTCGAGGGAAAAATAGG and reverse,
ATGCACATCCTTCGATAAGACTG; vimentin forward, GACGCCATCAACACCGAGTT and
reverse, CTTTGTCGTTGGTTAGCTGGT; and GAPDH forward,
AGAAGGCTGGGGCTCATTTG and reverse, AGGGGCCATCCACAGTCTTC.
Wound healing assay
Total 5×105 PC cells were seeded into
6-well plates and cultured until they reached a confluence of 90%.
A 200-µl pipette tip was used to scratch the monolayer to create a
wound. After scratching, the cells were washed using PBS to remove
detached cells. Cells were subsequently incubated in the serum-free
DMEM and cultured for 48 h. Phase-contrast microscopy was used to
capture the images in 6 random fields.
Transwell invasion assay
Cells were suspended in medium without FBS and a
total of 1×105 PC cells were then added to the upper
chamber pre-coated with Matrigel (BD Biosciences), and DMEM
supplemented with 20% FBS was added to the lower chamber. Cells
were cultured at 37°C for 48 h, after which, the cells which had
invaded were fixed using 4% paraformaldehyde and stained using 0.5%
crystal violet (Wuhan Boster Biological Technology, Ltd.) at room
temperature for 30 min. After washing with PBS, the chambers were
air-dried and observed under an inverted light microscope (Olympus
Corporation). The number of invaded cells in five random fields
were counted using Image Pro-Plus (version: 6.0; Sevice and
technology, URL: http://www.xrayscan.com/software-image-pro-plus/).
Animal model of liver metastasis
The nude mice used in the present study were
obtained from Huafukang Biotechnology Co., Ltd. Animals were kept
in specific pathogen-free conditions. NC and Linc00261
overexpression of PANC-1 cells (2×106 cells) were
injected into the spleen of nude mice (n=6 per group). After
intravenous anesthesia with pentobarbital at a concentration of 2.5
mg/100 g, the mice were sacrificed via cervival dislocation in 10
weeks after injection, and the liver was harvested, weighed, imaged
and embedded in 10% paraffin. All animal experiments were approved
by the Ethics Committee of Guizhou Medical University (Approval no.
1900069).
H&E staining
The liver tissues were fixed in 4% paraformaldehyde
for 30 min at room temperature and sectioned into 4 µm-thick
sections. After heating to 60°C for 1 h, the specimens were
deparaffinized using xylene at room temperature and rehydrated in a
descending graded series of ethanol (100, 80, 60 and 40%).
Subsequently, the samples were stained with hematoxylin for 5 min
and eosin for 10 min both at room temperature. After washing with
PBS, images were captured using an upright metallurgical microscope
(×200; Olympus Corp.).
Luciferase reporter assay
PC cells were plated in 6-well plates at density of
5×105. Subsequently, the cells were co-transfected with
psiCHECK™-2 vector and miR-552-5p mimics. Then, the cells were
lysed and used to measure firefly and Renilla luciferase
activity according to manufacturer's protocol (Guangzhou RiboBio
Co., Ltd.). Subsequently, the luciferase activity was normalized to
the firefly luciferase internal control.
Western blotting
Total protein was extracted from PC cells using RIPA
lysis buffer (Wuhan Boster Biological Technology, Ltd.), and the
protein concentration was quantified using a bicinchoninic acid
protein assay kit (Wuhan Boster Biological Technology, Ltd.).
Proteins (30 µg per lane) were loaded on a 10% gel and resolved
using SDS-PAGE and transferred to PVDF membranes (EMD Millipore).
Membranes were blocked using 5% skimmed milk, and subsequently
incubated overnight using primary antibodies against target
proteins, FOXO3 (cat. no. 10849-1-AP), β-catenin (cat. no.
51067-2-AP), TCF4 (cat. no. 22337-1-AP), E-cadherin (cat. no.
20874-1-AP), N-cadherin (cat. no. 22018-1-AP), vimentin (cat. no.
10366-1-AP) and GAPDH (cat. no. 60004-1-Ig) (all 1:1,000;
ProteinTech Group, Inc.) at 4°C. Subsequently, the membranes were
washed using TBS-Tween twice, and subsequently the membranes were
incubated with horseradish peroxidase-conjugated anti-mouse (cat.
no. BA1051) and anti-rabbit (cat. no. BA1055) secondary antibody
(all 1:2,500; Wuhan Boster Biological Technology, Ltd.) and signals
were visualized using enhanced chemiluminescent reagent (Wuhan
Boster Biological Technology, Ltd.). Image Pro-Plus software was
used to analyze the expression of protein, while GAPDH was used as
a loading control.
Statistical analysis
Statistical analysis was performed using SPSS
version 21.0 (IBM Corp). Survival curves were plotted using
Kaplan-Meier plotter (kmplot.com).
Comparisons were performed using a one-way ANOVA combined with
LSD-t test or a paired Student's t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Linc00261 expression is decreased in
PC
The expression and association of Linc00261 with
clinical traits were examined. Bioinformatics analysis of data
obtained from GEPIA revealed that Linc00261 was significantly
reduced in PC tissues compared with the matching adjacent tissues
(Fig. 1A; P<0.05). Reduced
expression of Linc00261 in patients with PC predicted less
favorable outcomes (Fig. 1B;
P<0.05). To verify the results obtained from the bioinformatics
analysis, RT-qPCR was performed on the 54 pairs of PC tissues and
adjacent matching tissues, and the results revealed that the
expression of Linc00261 was also decreased in our PC tissues
compared with adjacent non-tumor tissues (Fig. 1C; P<0.05). A χ2 test
was performed to analyze the relationship between the expression of
Linc00261 and various clinicopathological characteristics. The
results revealed that increased expression of Linc00261 was
significantly negatively associated with several clinical traits of
PC, such as tumor size, lymph node metastasis, TNM stage, distant
metastasis, perineural invasion and blood vessel invasion (Table I). Furthermore, it was demonstrated
that the expression of Linc00261 was also reduced in all the PC
cell lines compared with the control pancreatic epithelial cell
line (Fig. 1D; P<0.05).
Collectively, Linc00261 may serve as a tumor-suppressive lncRNA in
PC.
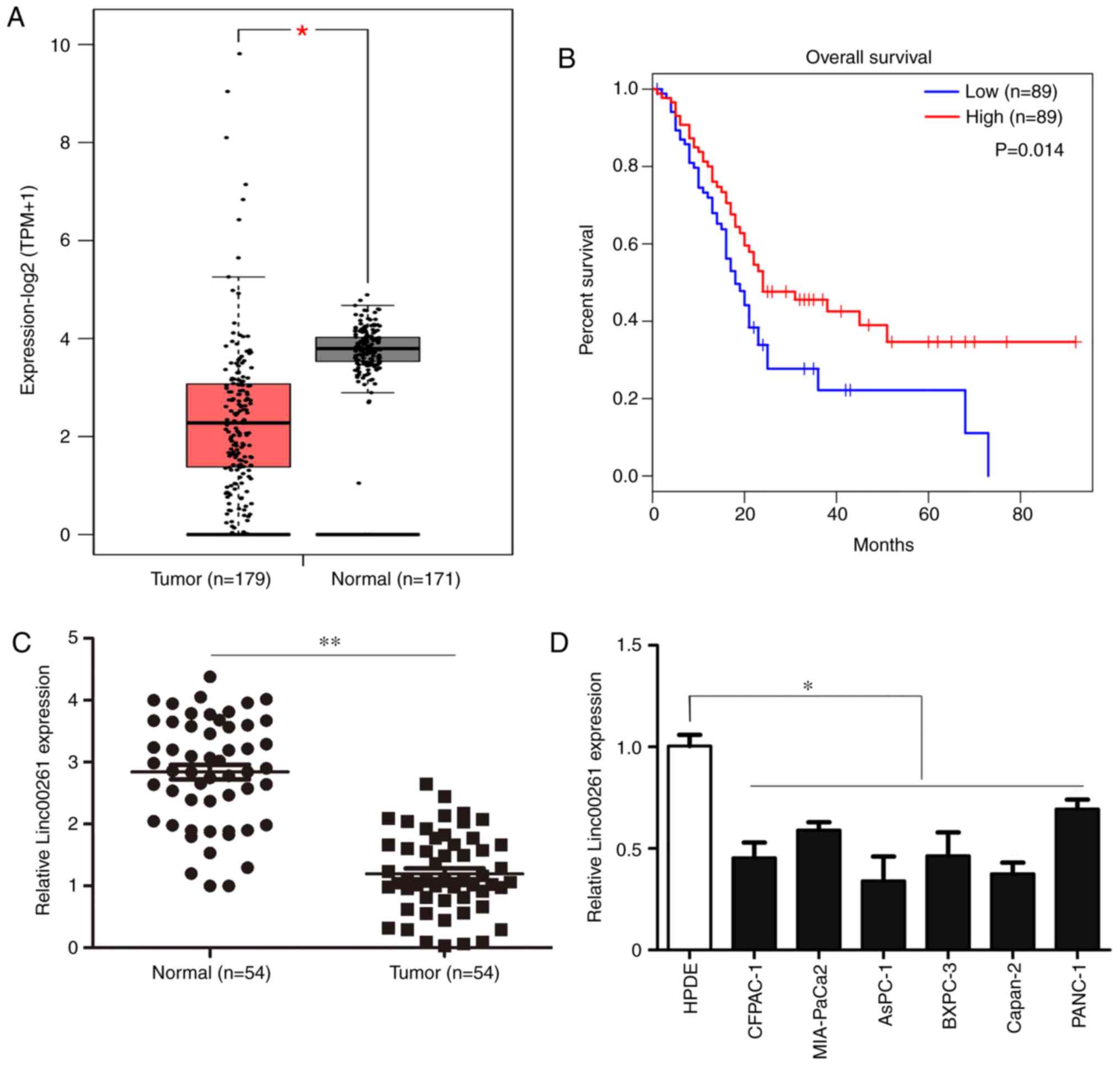 | Figure 1.Linc00261 is significantly
downregulated in PC. (A) The expression of Linc00261 in PC tissues
and adjacent healthy tissues was analyzed using the online
database, GEPIA. (B) Association between the prognosis and the
expression of Linc00261 was analyzed using the online database,
GEPIA. (C) Expression of Linc00261 was measured in 54 PC tissues
and 54 adjacent non-tumor tissues using reverse
transcription-quantitative PCR. (D) The expression of Linc00261 in
the PC cell lines, CFPAC-1, AsPC-1, MIA-PaCa2, Capan-2, BXPC-3 and
PANC-1 was significantly decreased compared with the HPDE cells.
*P<0.05, **P<0.01. PC, pancreatic cancer; HPDE, human
pancreatic epithelium cells. |
 | Table I.The relationship between the
expression of Linc00261 and various clinicopathological
characteristics. |
Table I.
The relationship between the
expression of Linc00261 and various clinicopathological
characteristics.
| Features | n | Low | High | χ2 | P-value |
|---|
| All cases |
| 54 | 27 | 27 |
|
| Age |
|
|
| 1.187 | 0.276 |
|
<60 | 28 | 12 | 16 |
|
|
|
≥60 | 26 | 15 | 11 |
|
|
| Sex |
|
|
| 1.2 | 0.273 |
|
Man | 30 | 13 | 17 |
|
|
|
Female | 24 | 14 | 10 |
|
|
| Tumor size
(cm) |
|
|
| 6.135 | 0.013 |
|
<2 | 23 | 7 | 16 |
|
|
| ≥2 | 31 | 20 | 11 |
|
|
| Lymph node
metastasis |
|
|
| 9.012 | 0.003 |
|
Negative | 25 | 7 | 18 |
|
|
|
Positive | 29 | 20 | 9 |
|
|
| TNM stage |
|
|
| 14.7 | 0.001 |
| I and
II | 24 | 5 | 19 |
|
|
| III and
IV | 30 | 22 | 8 |
|
|
| Distant
metastasis |
|
|
| 9.012 | 0.003 |
|
Negative | 26 | 7 | 18 |
|
|
|
Positive | 29 | 20 | 9 |
|
|
| Perineural
invasion |
|
|
| 6.135 | 0.013 |
|
Negative | 23 | 7 | 16 |
|
|
|
Positive | 31 | 20 | 11 |
|
|
| Blood vessel
invasion |
|
|
| 7.67 | 0.006 |
|
Negative | 22 | 6 | 16 |
|
|
|
Positive | 32 | 21 | 11 |
|
|
Linc00261 overexpression inhibits cell
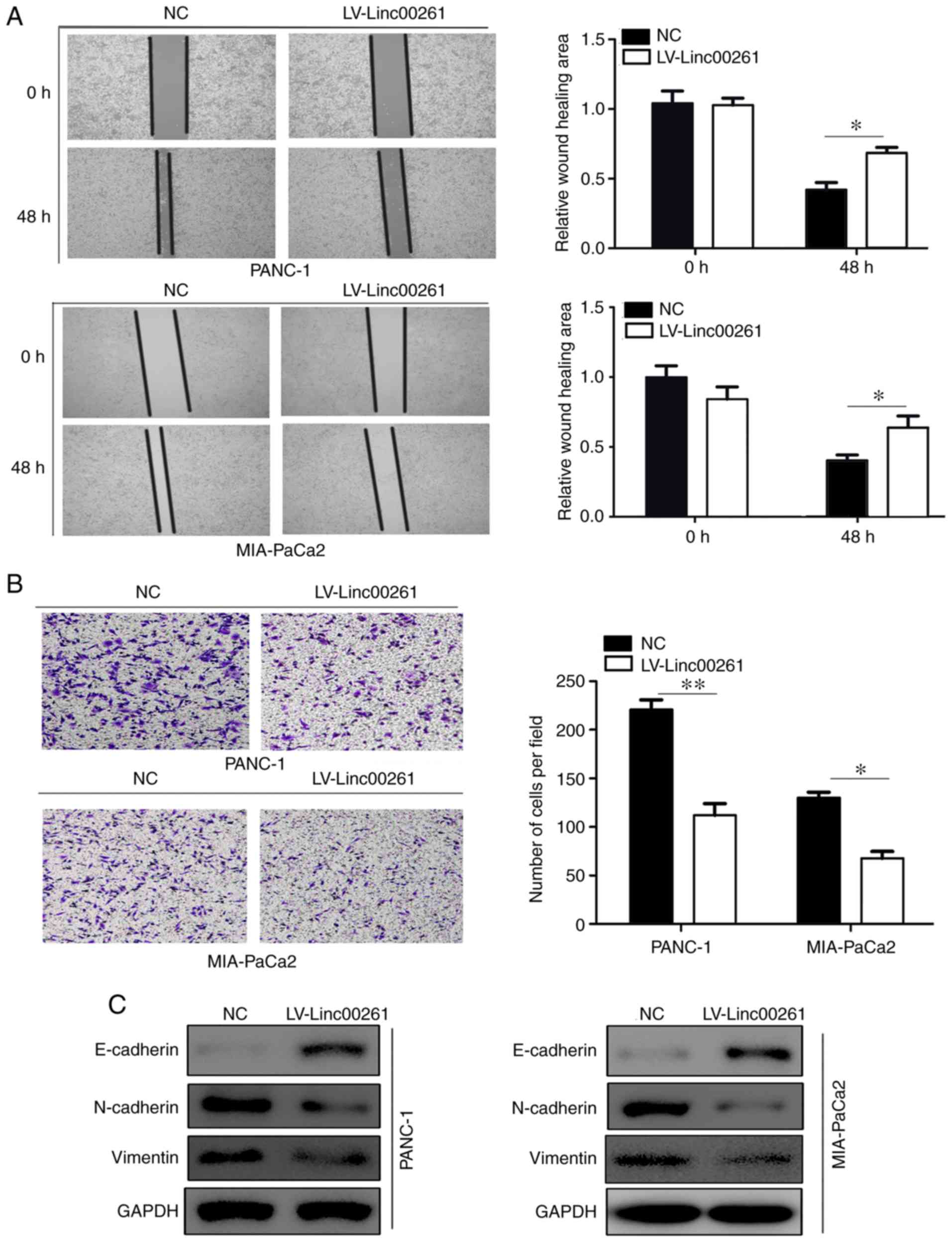
migration and invasion, and EMT in PC cells
Then, PANC-1 and MIA-PaCa2 cell lines which were
derived from primary tumors with different metastatic ability were
employed for our study. The migratory ability of PC cells in
MIA-PaCa2 and PANC-1 cells transfected with empty vector or
Linc00261 overexpression lentivirus was assessed using a wound
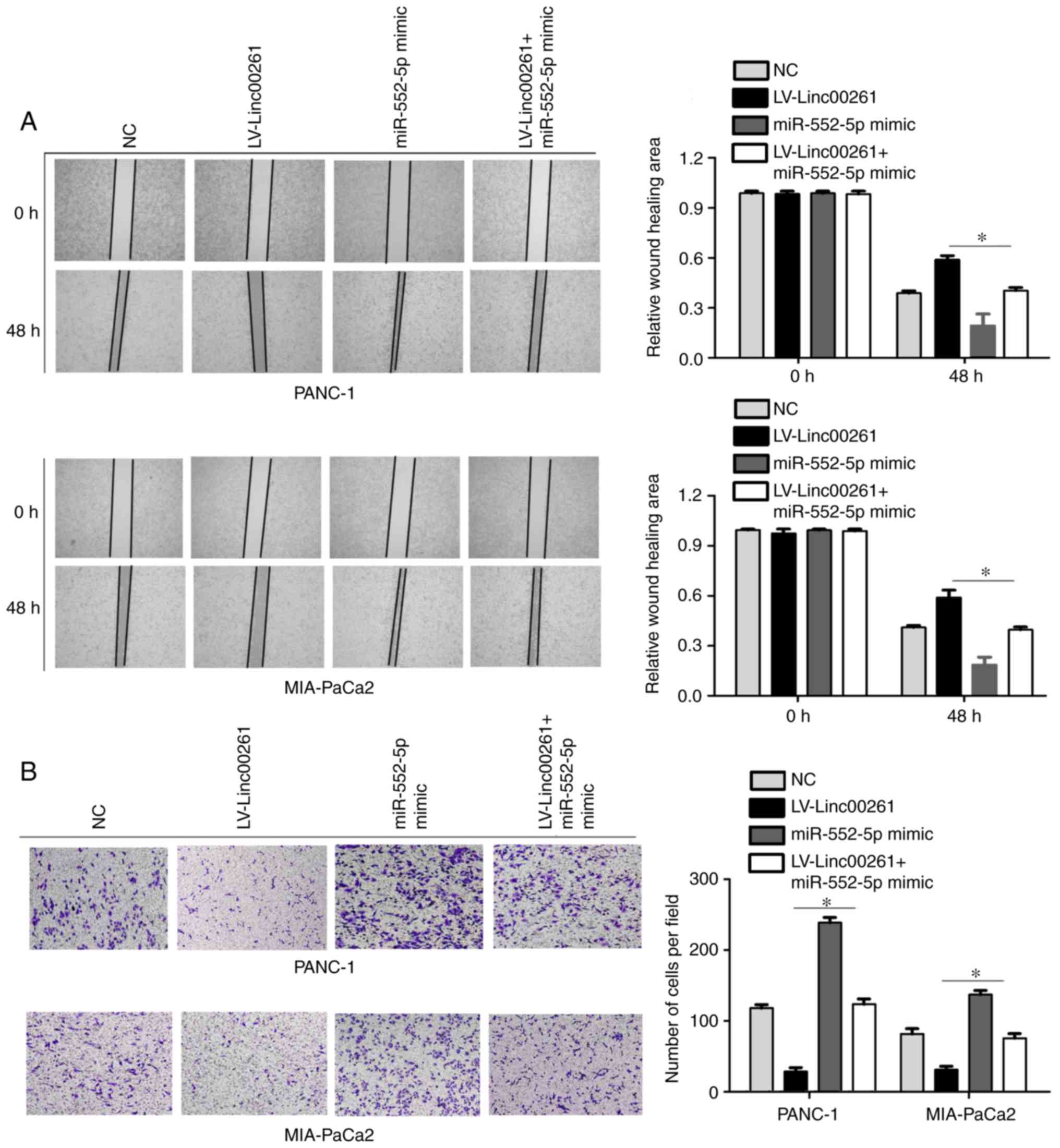
healing assay. Linc00261 overexpression both significantly reduced
migration of PANC-1 and MIA-PaCa2 cells (Fig. 2A; P<0.05). Similarly, Transwell
invasion assays revealed that overexpression of Linc00261 both
significantly suppressed the invasive capacity of PANC-1 and
MIA-PaCa2 cells (Fig. 2B;
P<0.05). Various studies have demonstrated that
epithelial-mesenchymal transition promotes the process of
metastasis of cancer cells (22).
The expression of several EMT markers, including E-cadherin,
N-cadherin and vimentin were assessed in MIA-PaCa2 and PANC-1 cells
transfected with empty vector and Linc00261 overexpression
lentivirus, and the results revealed that the expression of
E-cadherin was increased in the Linc00261 overexpression cells,
whereas the expression of N-cadherin and vimentin were markedly
reduced (Fig. 2C; P<0.05).
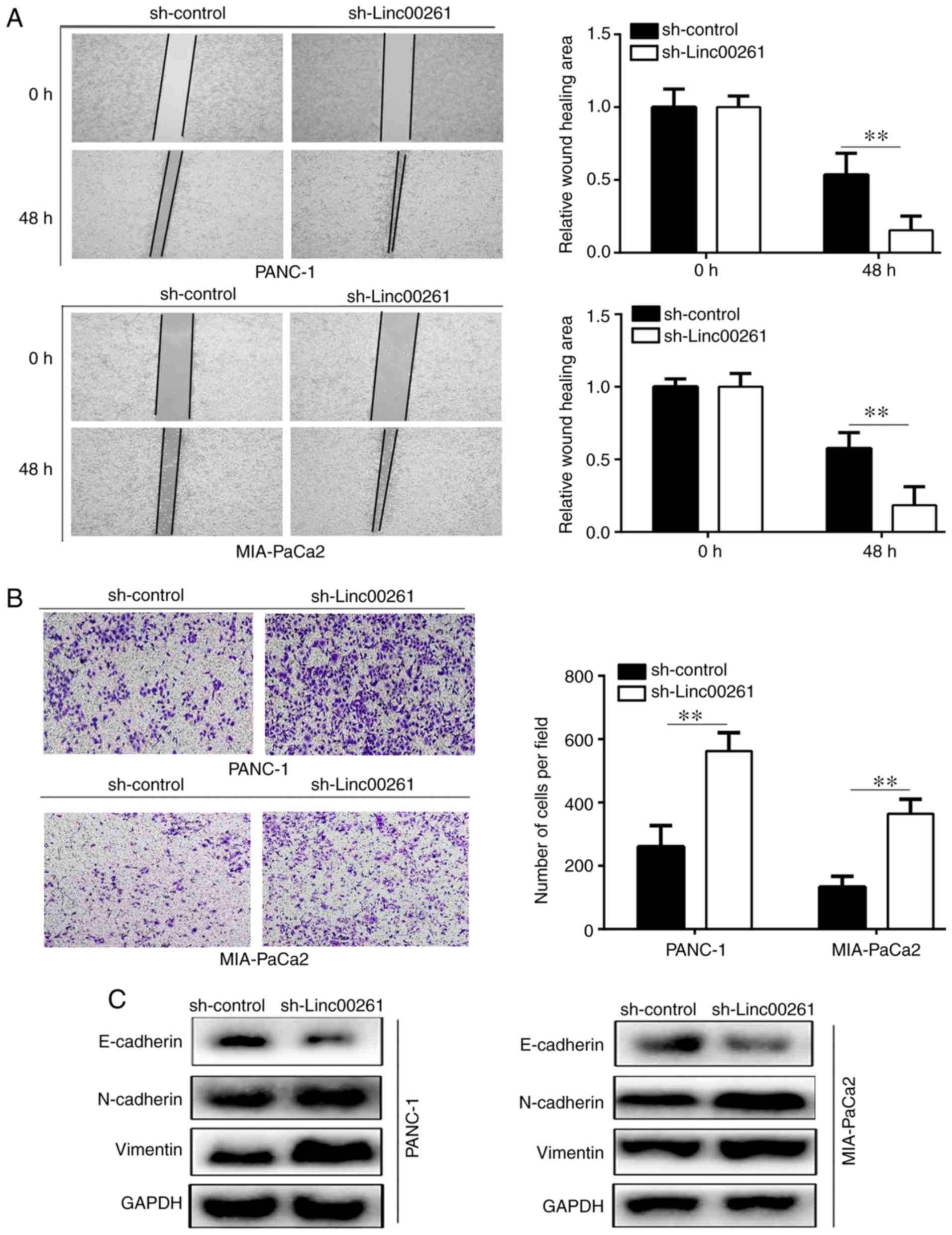
Linc00261 knockdown increases
migration and invasion, and EMT of PC cells
The results of the wound healing assays revealed
that knockdown of Linc00261 significantly increased the migration
of PANC-1 and MIA-PaCa2 cells (Fig.
3A; P<0.05). Similarly, the results of the Transwell
invasion assays revealed that Linc00261 knockdown significantly
increased invasion of PANC-1 and MIA-PaCa2 cells (Fig. 3B; P<0.05). The results of western
blotting revealed that Linc00261 knockdown significantly decreased
the expression of E-cadherin, whereas the expression levels of
N-cadherin and vimentin were increased (Fig. 3C; P<0.05).
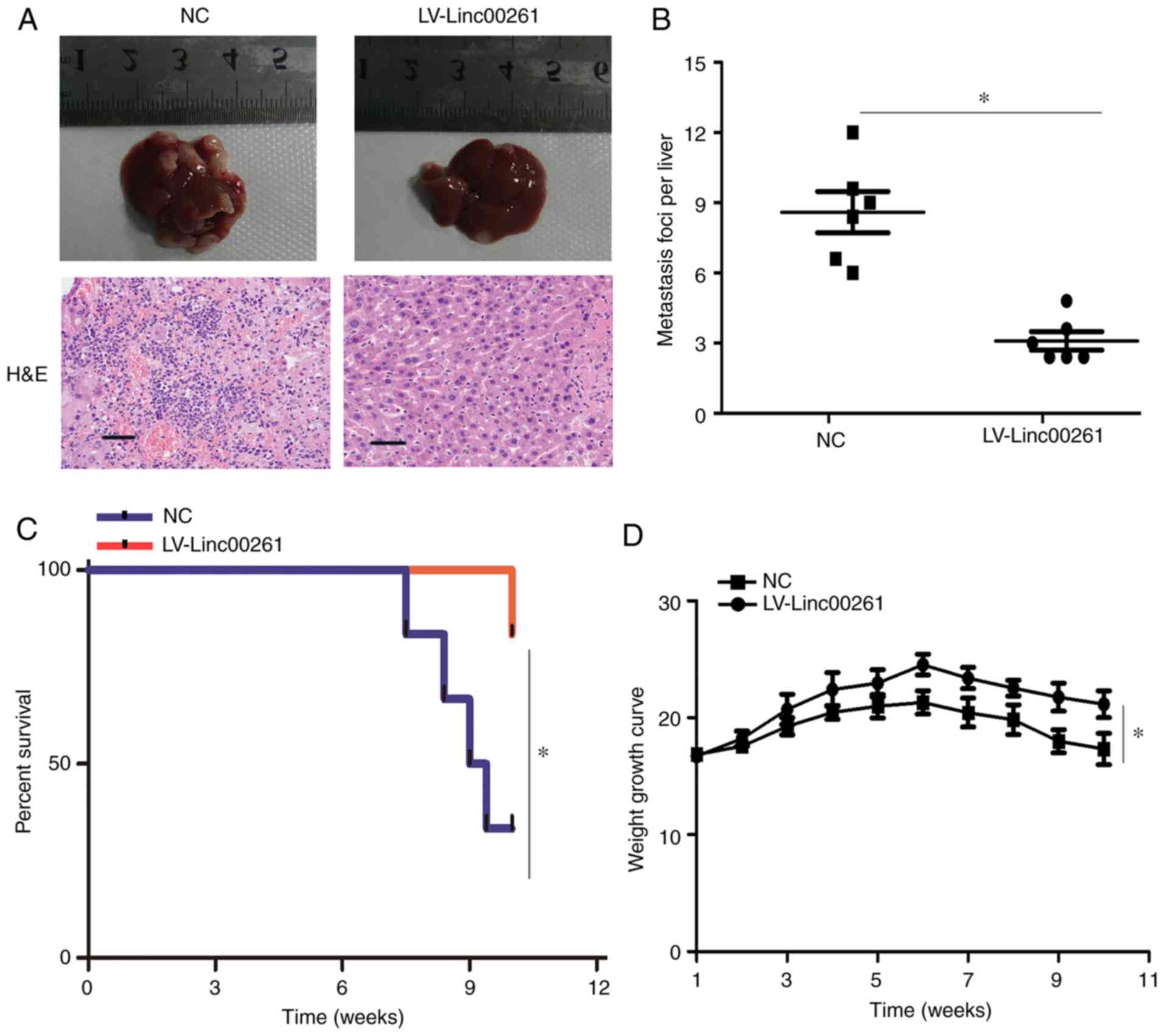
Linc00261 reduces metastasis of PC
cells in vivo
PANC-1 cells transfected with empty vector or
Linc00261 overexpression lentivirus were injected into the spleen
of nude mice. After 10 weeks, metastasis of PANC-1 cells was
assessed, and the results revealed that the number of metastatic
foci was significantly decreased in the mice injected with the
Linc00261 cells (Fig. 4A and B;
P<0.05). Similarly, the rate of death in mice was significantly
lower in the mice injected with Linc00261-overexpressing cells
compared with the mice injected with the empty vector control cells
(Fig. 4C; P<0.05), as well as
weight loss (Fig. 4D, P<0.05).
Collectively, these results indicated that Linc00261 suppressed
metastasis of PC cells in vivo.
miR-552-5p is a target of Linc00261 in
PC
Target miRNAs of Linc00261 were predicted using
TargetScan, and the analysis revealed that miR-552-5p possessed
potential binding sites for Linc00261 (Fig. 5A). A dual luciferase reporter assay
was performed and the results revealed that luciferase activity was
decreased in PANC-1 and MIA-PaCa2 cells co-transfected with
miR-552-5p mimic and Linc00261-WT plasmid compared with cells
co-transfected with NC mimic and the Linc00261-WT plasmid (Fig. 5B; P<0.05). The expression levels
of miR-552-5p were inversely associated with Linc00261 (Fig. 5C; r=−0.3561; P<0.01).
Transfection of miR-552-5p inhibitor into PANC-1 and MIA-PaCa2
cells increased the expression of Linc00261 (Fig. 5D; P<0.05). Similarly, the
expression levels of miR-552-5p were significantly decreased in
Linc00261-overexpressed cells (Fig.
5E; P<0.05). In addition, the expression of miR-552-5p was
increased in PC tissues and PC cell lines (Fig. 5F and G; P<0.05). These results
indicated that miR-552-5p may be an important target of Linc00261
in PC.
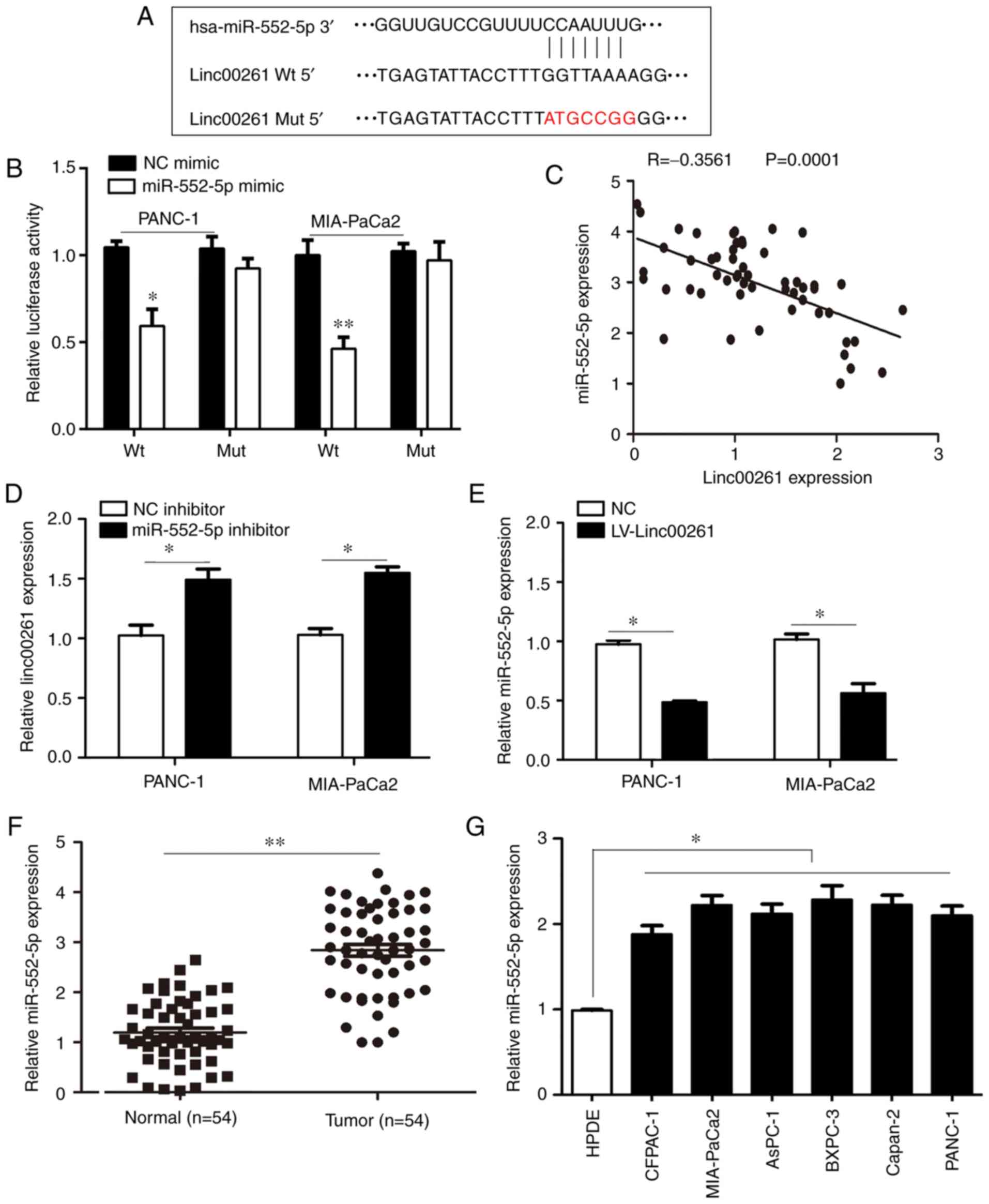 | Figure 5.miR-552-5p is a target of Linc00261
in PC. (A) Linc00261 predicted binding site in the 3′UTR of
miR-552-5p mRNA. (B) miR-552-5p mimics reduced luciferase activity
in PC cells, this was not observed with the NC mimics. (C)
Expression of Linc00261 was inversely associated with expression of
miR-552-5p in PC tissues. (D) miR-552-5p inhibition increased the
mRNA expression levels of Linc00261 in PC cells. (E) Overexpression
of Linc00261 decreases the expression of miR-552-5p. (F) The mRNA
expression levels of miR-552-5p were detected in PC tissues and
adjacent non-tumor tissues using reverse transcription-quantitative
PCR. (G) mRNA expression levels of miR-552-5p in the CFPAC-1,
AsPC-1, MIA-PaCa2, Capan-2, BXPC-3 and PANC-1 PC cells were
significantly higher compared with HPDE cells. *P<0.05,
**P<0.01. PC, pancreatic cancer; UTR, untranslated region; NC,
negative control; miR, microRNA; HPDE, human pancreatic epithelium
cells. |
miR-552-5p upregulates the expression
of FOXO3 in PC
Since miR-552-5p was considered an important target
of Linc00261 in PC, the targets of miR-552-5p were predicted. Using
the online tools, miRWalk and TargetScan, a total of 332 potential
target genes regulated by miR-552-5p were predicted (Fig. 6A). These target genes were primarily
enriched for 5 pathways, including the FOXO signaling pathway
(Fig. 6B). FOXO3, which is involved
in the FOXO signaling pathway was determined to possess binding
sites for miR-552-5p (Fig. 6C). Due
to evidence that FOXO3 serves a key role in PC (20), our focus was on FOXO3. Luciferase
activity was significantly reduced in cells co-transfected with
miR-552-5p mimic and FOXO3-WT plasmid compared with cells
co-transfected with miR-552-5p mimic and FOXO3-Mut plasmid
(Fig. 6D; P<0.05). Furthermore,
the expression of FOXO3 was decreased in tumor samples, and
negatively associated with the expression of miR-552-5p in PC
(Fig. 6E and F; P<0.05).
Similarly, the mRNA levels of FOXO3 in PC cell lines were also
decreased compared with the normal pancreatic epithelial cell line
(Fig. 6G; P<0.05). Collectively,
FOXO3 was considered to be an important target of miR-552-5p in
PC.
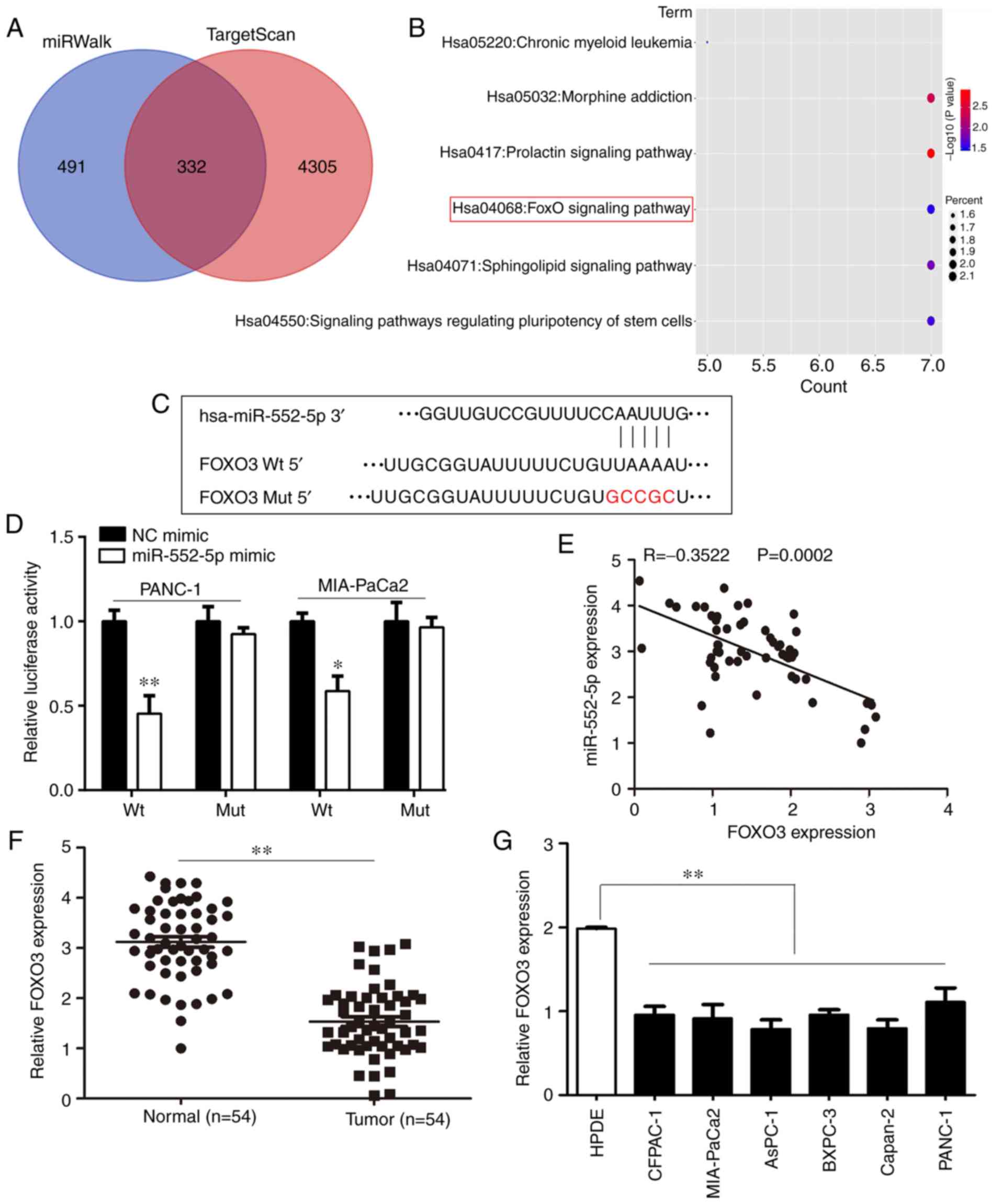 | Figure 6.FOXO3 is a target of miR-552-5p in
PC. (A) Using TargetScan and miRwalk online databases, the target
genes of miR-552-5p were predicted and 332 genes were considered
relevant based on the results from both databases. (B) Bubble chart
showing the pathways in which miR-552-5p targeted genes are
enriched in. (C) miR-552-5p was predicted to bind to the 3′UTR of
FOXO3 mRNA. The mutated site is highlighted. (D) miR-552-5p mimics
reduced the luciferase intensity in PC cells, whereas mutation of
the associated element in 3′-UTR of FOXO3 mRNA abolished the effect
of miR-552-5p mimic on luciferase activity. (E) The expression of
miR-552-5p was inversely associated with expression of FOXO3 in PC
tissues. (F) The mRNA expression levels of FOXO3 in PC tissues and
adjacent non-tumor tissues. (G) The mRNA expression levels of FOXO3
in CFPAC-1, AsPC-1, MIA-Paca2, Capan-2, BXPC-3 and PANC-1 PC cells
were significantly decreased compared with HDPE cells. *P<0.05,
**P<0.01. PC, pancreatic cancer; miR, microRNA; UTR,
untranslated region; HPDE, human pancreatic epithelium cells;
FOXO3, forkhead box O3. |
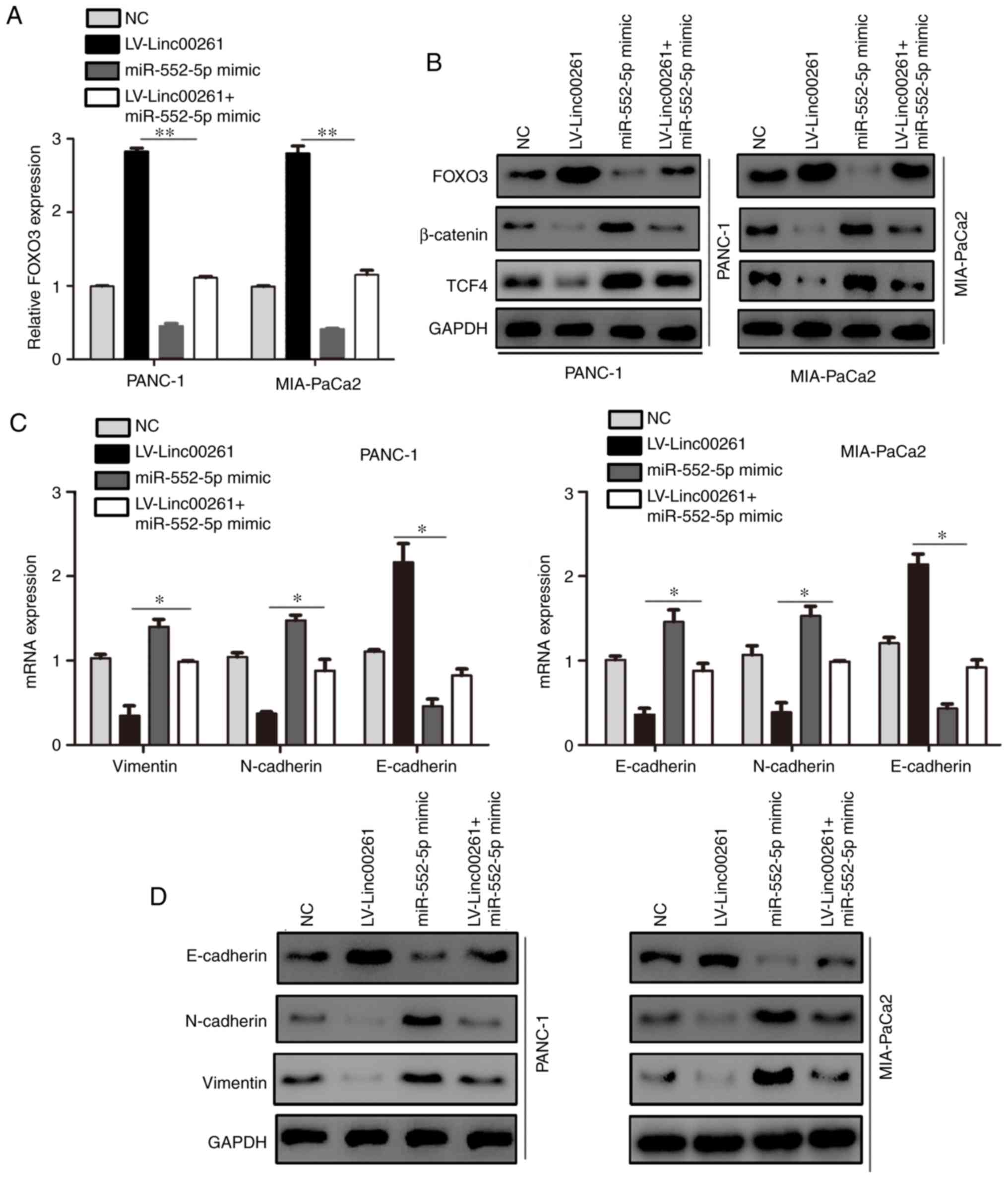
Linc00261 inhibits the activity of the
β-catenin/TCF4 pathway and cell metastasis via a miR-552-5p/FOXO3
axis
To determine whether the effect of Linc00261 on PC
was dependent on miR-552-5p/FOXO3, the expression of miR-552-5p was
restored in Linc00261-overexpressing cells. miR-552-5p
overexpression increased the mRNA and protein expression levels of
FOXO3 (Fig. 7A and B; P<0.05).
Since it has been demonstrated that FOXO3 regulates the Wnt
signaling pathway (23), the
expression of β-catenin and TCF4 was assessed. The results revealed
that Linc00261 overexpression decreased the expression of β-catenin
and TCF4, whereas miR-552-5p prevented the inhibitory effects of
Linc00261 on the Wnt signaling pathway (Fig. 7B; P<0.05). The expression levels
of E-cadherin, N-cadherin and vimentin, which are also downstream
proteins of TCF4, were assessed using RT-qPCR and western blotting.
Restoration of miR-552-5p alleviated the effects of Linc00261
overexpression on the expression of E-cadherin, N-cadherin and
vimentin (Fig. 7C and D;
P<0.05). Similarly, miR-552-5p reversed the inhibitory effects
of Linc00261 on the migration and invasion of PC cells (Fig. 8A and B; P<0.05).
Discussion
PC exhibits a high metastatic capacity, thus any
factor which promotes metastasis of PC may be involved in the
progression of PC. LncRNAs have been revealed to regulate the
metastasis of PC previously (24).
Since Linc00261 has been demonstrated to function as a
tumor-suppressive LincRNA in a range of different types of cancer
(25,26), it was hypothesized that Linc00261
may also participate in the pathogenesis of PC.
Consistent with previous studies, in the present
study, Linc00261 expression was decreased in PC tissues and 6 PC
cell lines compared with the adjacent non-tumor tissues and normal
pancreatic epithelial cells, respectively. Reduced expression of
Linc00261 predicted less favorable outcomes in patients with PC and
Linc00261 overexpression decreased PC cell migration and invasion
in vitro as well as EMT, whereas Linc00261 knockdown
increased cell migration and invasion in vitro. Similarly,
Linc00261 overexpression decreased PC cell metastasis in
vivo. Together these data indicated that Linc00261 is a
potential tumor-suppressive lncRNA in PC. Therefore, determining
the underlying mechanism by which Linc00261 reduced metastasis of
PC cells may assist in understanding the development of PC.
Numerous studies have demonstrated that specific
lncRNAs may act as ceRNAs which are primarily located in the
cytosol (27,28). In the present study, through
bioinformatics analysis and dual luciferase activity assays,
Linc00261 was revealed to bind with miR-552-5p in PC cells. A
previous study revealed that miR-552-5p acted as an oncogenic miRNA
in osteosarcoma and promoted the development of osteosarcoma
(29). Therefore, Linc00261 may
inhibit PC cell metastasis by sponging miR-552-5p. The results of
the present study revealed that the expression of miR-552-5p was
negatively associated with Linc00261 in PC tissues. Linc00261
overexpression significantly decreased the expression of miR-552-5p
in PC cells. Furthermore, the expression of miR-552-5p was
significantly upregulated in PC tissues and cell lines. Therefore,
miR-552-5p was considered a target miRNA of Linc00261 in PC.
Using bioinformatics analysis to predict the target
genes of miR-552-5p, it was demonstrated that the target genes were
significantly enriched in the FOXO signaling pathway and FOXO3 was
a direct target of miR-552-5p. Since the FOXO pathway, and
specifically FOXO3 can regulate the Wnt signaling pathway, which is
involved in the development of PC (18), a focus was placed on FOXO3 and it
was revealed that miR-552-5p overexpression significantly decreased
its expression. Additionally, the expression of FOXO3 was
negatively associated with miR-552-5p. Linc00261 overexpression
significantly decreased the expression of FOXO3, and inhibited the
Wnt signaling pathway and thus EMT. Ectopically restoring
miR-552-5p expression in Linc00261-overexpressing cells
significantly alleviated the inhibitory effect of Linc00261 on the
expression of FOXO3, the Wnt pathway and EMT. Similarly, Linc00261
significantly decreased the migratory and invasive capacities of PC
cells, whereas miR-552-5p restoration in Linc00261-overexpressing
PC cells significantly increased the migration and invasion of PC
cells.
In conclusion, the present study demonstrated that
Linc00261 is a tumor-suppressive lncRNA in PC. Linc00261 inhibited
PC cell metastasis in vitro and in vivo by regulating
the miR-552-5p/FOXO3 axis. These results may assist in improving
our understanding of the molecular mechanisms underlying the
development and metastasis of PC, and indicates that Linc00261 may
be a potential biomarker for diagnosis and target for
treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81060176) and Major
Projects of Applied and Basic Research Program of Guizhou Province
(grant no. J-[2015]2003).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
TC, SL and ZZ collected and interpreted the data.
TC, SL, ZZ, JZ, YX, YS, JL, SX, DM, BG analyzed the data. TC wrote
the manuscript. All authors have read and approved the final
version of the article.
Ethics approval and consent to
participate
All patients who had provided samples provided
informed consent. The present study was approved by the Ethics
Committee of the Guizhou Medicine University (Approval no.
2019LS146) and was performed in accordance with the Declaration of
Helsinki. All animal experiments were approved by the Ethics
Committee of Guizhou Medical University (Approval no. 1900069).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee SH, Chang PH, Chen PT, Lu CH, Hung YS,
Tsang NM, Hung CY, Chen JS, Hsu HC, Chen YY and Chou WC:
Association of time interval between cancer diagnosis and
initiation of palliative chemotherapy with overall survival in
patients with unresectable pancreatic cancer. Cancer Med.
8:3471–3478. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Skau Rasmussen L, Vittrup B, Ladekarl M,
Pfeiffer P, Karen Yilmaz M, Østergaard Poulsen L, Østerlind K,
Palnæs Hansen C, Bau Mortensen M, Viborg Mortensen F, et al: The
effect of postoperative gemcitabine on overall survival in patients
with resected pancreatic cancer: A nationwide population-based
Danish register study. Acta Oncol. 58:864–871. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elaileh A, Saharia A, Potter L, Baio F,
Ghafel A, Abdelrahim M and Heyne K: Promising new treatments for
pancreatic cancer in the era of targeted and immune therapies. Am J
Cancer Res. 9:1871–1888. 2019.PubMed/NCBI
|
|
4
|
Mitschke J, Burk UC and Reinheckel T: The
role of proteases in epithelial-to-mesenchymal cell transitions in
cancer. Cancer Metastasis Rev. 38:431–444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang
J, Qian Y, Ma Y, Wang F, Li H, et al: FAT1 prevents epithelial
mesenchymal transition (EMT) via MAPK/ERK signaling pathway in
esophageal squamous cell cancer. Cancer Lett. 397:83–93. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam
D, Lee J, Lee SG, Yang WM, Um JY, et al: Ginkgolic acid inhibits
invasion and migration and TGF-β-induced EMT of lung cancer cells
through PI3K/Akt/mTOR inactivation. J Cell Physiol. 232:346–354.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang Y, Bai YS and Wang Q: CDGSH Iron
Sulfur domain 2 activates proliferation and EMT of pancreatic
cancer cells via Wnt/β-catenin pathway and has prognostic value in
human pancreatic cancer. Oncol Res. 25:605–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kleszcz R: The canonical Wnt pathway.
Postepy Biochem. 65:183–192. 2019.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin Y, Dong C and Zhou BP: Epigenetic
regulation of EMT: The Snail story. Curr Pharm Des. 20:1698–1705.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang X, Zhi X, Gao Y, Ta N, Jiang H and
Zheng J: LncRNAs in pancreatic cancer. Oncotarget. 7:57379–57390.
2016.PubMed/NCBI
|
|
12
|
Yang J, Ye Z, Mei D, Gu H and Zhang J:
Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating
miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer.
Cancer Manag Res. 11:4209–4221. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Y, Zhu Q, Yang W, Shan Y, Yu Z, Zhang
Q and Wu H: LncRNA H19/miR-194/PFTK1 axis modulates the cell
proliferation and migration of pancreatic cancer. J Cell Biochem.
120:3874–3886. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang ZK, Yang L, Wu LL, Mao H, Zhou YH,
Zhang PF and Dai GH: Long non-coding RNA LINC00261 sensitizes human
colon cancer cells to cisplatin therapy. Braz J Med Biol Res.
51:e67932017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu Y, Li L, Zheng Z, Chen S, Chen E and Hu
Y: Long non-coding RNA linc00261 suppresses gastric cancer
progression via promoting Slug degradation. J Cell Mol Med.
21:955–967. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi J, Ma H, Wang H, Zhu W, Jiang S, Dou R
and Yan B: Overexpression of LINC00261 inhibits non-small cell lung
cancer cells progression by interacting with miR-522-3p and
suppressing Wnt signaling. J Cell Biochem. 120:18378–18387.
2019.PubMed/NCBI
|
|
17
|
Yan D, Liu W, Liu Y and Luo M: LINC00261
suppresses human colon cancer progression via sponging miR-324-3p
and inactivating the Wnt/β-catenin pathway. J Cell Physiol.
234:22648–22656. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu H, Yin J, Wang H, Jiang G, Deng M,
Zhang G, Bu X, Cai S, Du J and He Z: FOXO3a modulates WNT/β-catenin
signaling and suppresses epithelial-to-mesenchymal transition in
prostate cancer cells. Cell Signal. 27:510–518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yao S, Fan LY and Lam EW: The FOXO3-FOXM1
axis: A key cancer drug target and a modulator of cancer drug
resistance. Semin Cancer Biol. 50:77–89. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Yang R, Dong Y, Chen M, Wang Y and
Wang G: Knockdown of FOXO3a induces epithelial-mesenchymal
transition and promotes metastasis of pancreatic ductal
adenocarcinoma by activation of the β-catenin/TCF4 pathway through
SPRY2. J Exp Clin Cancer Res. 38:382019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chaffer CL, San Juan BP, Lim E and
Weinberg RA: EMT, cell plasticity and metastasis. Cancer Metastasis
Rev. 35:645–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iyer S, Ambrogini E, Bartell SM, Han L,
Roberson PK, de Cabo R, Jilka RL, Weinstein RS, O'Brien CA,
Manolagas SC and Almeida M: FOXOs attenuate bone formation by
suppressing Wnt signaling. J Clin Invest. 123:3409–3419. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Previdi MC, Carotenuto P, Zito D, Pandolfo
R and Braconi C: Noncoding RNAs as novel biomarkers in pancreatic
cancer: What do we know? Future Oncol. 13:443–453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Sha L, Huang L, Yang S, Zhou Q,
Luo X and Shi B: LINC00261 functions as a competing endogenous RNA
to regulate BCL2L11 expression by sponging miR-132-3p in
endometriosis. Am J Transl Res. 11:2269–2279. 2019.PubMed/NCBI
|
|
26
|
Cheng D, Jiang S, Chen J, Li J, Ao L and
Zhang Y: Upregulated long noncoding RNA Linc00261 in pre-eclampsia
and its effect on trophoblast invasion and migration via regulating
miR-558/TIMP4 signaling pathway. J Cell Biochem. 120:13243–13253.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin K, Jiang H, Zhuang SS, Qin YS, Qiu GD,
She YQ, Zheng JT, Chen C, Fang L and Zhang SY: Long noncoding RNA
LINC00261 induces chemosensitization to 5-fluorouracil by mediating
methylation-dependent repression of DPYD in human esophageal
cancer. FASEB J. 33:1972–1988. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang Q, Sang L and Du S: Long noncoding
RNA LINC00261 regulates endometrial carcinoma progression by
modulating miRNA/FOXO1 expression. Cell Biochem Funct. 36:323–330.
2018. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai W, Xu Y, Yin J, Zuo W and Su Z:
miR-552-5p facilitates osteosarcoma cell proliferation and
metastasis by targeting WIF1. Exp Ther Med. 17:3781–3788.
2019.PubMed/NCBI
|