Introduction
Osteosarcomas (OS) are uncommon tumors. Despite
their rarity, however, OS is the most common primary malignancy
tumor of the bone affecting children and adolescents (1). OS comprises 56% of all bone cancers in
individuals under 20 years of age (2). In children, the peak incidence occurs
between 13 and 16 years of age (3),
and the most common site of OS is the metaphysis of long bones,
especially the distal femur (2).
Chemotherapy is now considered as a standard component of OS
treatment, not only in children, but also in adults. The long-term
survival rate following the incidence of a local tumor without
chemotherapy has been reported as being only 16%, whereas treatment
with ≥3 types of chemotherapy has been shown to improve the
survival rate to 70% (4).
Clinically, the traditional first-line chemotherapy regimen for OS
is a combination of doxorubicin (DOX), cisplatin (CDDP), and
methotrexate (MTX) (5). However,
the development of drug resistance leads to a decrease in the
therapeutic efficacy of the drugs, and an increasing number of
studies are focusing on this issue.
MicroRNAs (miRNAs) have been studied and reported on
in numerous fields. miRNAs bind to the 3′-untranslated region
(3′-UTR), either perfectly or imperfectly, to contribute to the
translational suppression, or the degradation, of different target
mRNAs (6). In addition, they have
been demonstrated to be endogenous small RNA molecules that are
able to regulate diverse biological functions, including
tumorigenesis, progression and chemosensitivity of different cancer
types (7,8). Previously published studies have
revealed that miRNAs have a crucial role in the drug responsiveness
of OS (9,10). That the regulation of autophagy is
associated with OS has been confirmed (11,12).
In addition, activating autophagy has been shown to cause cytotoxic
drugs to develop drug resistance (13,14).
It is now generally well understood that miRNAs exert a role in
autophagy to regulate drug resistance in OS treatment. It was
reported that miR-101 suppressed the expression of
autophagy-related genes including LC3 and Atg5 in U2OS cells, and
promoted cell sensitivity to DOX (15). Furthermore, miR-410 was shown to
markedly inhibit autophagy by regulating autophagy-related gene
16L1 expression, thereby enhancing chemosensitivity (16).
The main focus of the present study was on
microRNA-22 (miR-22), which is located on chromosome 17p13 and
fulfills crucial roles as a tumor suppressor in certain types of
malignancies (17,18). It has been reported that
overexpression of miR-22 significantly decreased cell proliferation
and survival, and induced cell apoptosis in p53-mutated colon
cancer cells (19). In addition,
miR-22 was shown to target the truncated neurokinin-1 receptor and
estrogen receptor-α to suppress the proliferation, invasion and
metastasis of breast cancer cells (20). miR-22 was subsequently shown to
enhance the radiosensitivity of small cell lung cancer cells
through targeting the ATPase, WRN helicase interacting protein 1
(WRNIP1) (21). With regard to
studies of OS, the level of miR-22 was found to be significantly
decreased in patients with OS, and miR-22 could target S100A11 to
increase the sensitivity of CDDP by preventing MG63 cells from
proliferating and metastasizing (22). miR-22 was also shown to suppress
high-mobility-group box 1 (HMGB1)-regulated autophagy in OS
cells treated with DOX and CDDP (23,24).
These studies have suggested that miR-22 is associated with
autophagy, and moreover, that it is able to regulate autophagy by
targeting certain autophagy-related genes, such that miR-22 can
affect the drug sensitivity of OS treatments. In our previous
study, we mainly argued that CDDP could increase the autophagy of
MG63 cells and the expression of autophagy-related genes including
microtubule-associated protein 1 light chain 3 (LC3),
autophagy related 5 (ATG5) and Beclin-1 (BECN1) and
miR-22 could inhibit the upregulation of those genes induced by
CDDP. However, the specific pathway associated with miR-22 and the
effect of miR-22 on MG63 and CDDP-resistant cells still need to be
investigated (25). In addition,
only a few studies have focused attention on the pathway that is
influenced by miR-22 in OS. It has been reported that miR-22-3p
enhanced the chemosensitivity of gastrointestinal stromal tumor
cell lines to CDDP via the phosphatase and tensin homolog deleted
on chromosome 10 (PTEN)/phosphoinositide 3-kinase (PI3K)/Akt
pathway (26). The
PI3K/AKT/mammalian target of rapamycin (mTOR) pathway is known to
be associated with autophagy; however; whether miR-22 is able to
regulate the chemosensitivity of OS cell lines (or even
drug-resistant cell lines) via the PI3K/AKT/mTOR pathway has yet to
be fully elucidated. The present study aimed to further investigate
the role of miR-22 in the PI3K/AKT/mTOR pathway in order to explore
the effect of miR-22 in CDDP-resistance of OS both in vivo
and in vitro.
Materials and methods
Selection of the cell line
Human osteosarcoma cell lines, including MG63, U2OS,
Saos2 and OS9901 (27–30), were obtained from the Cell Bank of
the Chinese Academy of Sciences (Shanghai, China). The cells were
allowed to proliferate in Hyclone® DMEM medium (HyClone;
GE Healthcare Life Sciences) containing 10% Hyclone®
FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin, and incubated
at a temperature of 37°C in an atmosphere of 5% CO2.
CDDP (2 µM) was added to the cell lines for 24 h. Subsequently,
reverse transcription-PCR (RT-PCR) assay was performed to
investigate the expression levels of BECN1 and ATG5
in each cell line.
Cell culture and transfection
The drug-resistant cell line (MG63/CDDP) was
obtained by plating MG63 cells (1×106 cells per well)
onto a 6-well plate and adding 2 µM CDDP for 24 h. Subsequently,
the dead cells were removed by PBS (Nanjing Dongji, China). After
the cells had reached 80% confluency, 2 µM CDDP was added again for
24 h. This procedure was repeated until the subsequent addition of
CDDP did not lead to any further cell death. The cells that were
finally obtained were of the drug-resistant cell line
(MG63/CDDP).
Invitrogen® Lipofectamine™ 3000
(Lipo3000; Life Technologies; Thermo Fisher Scientific, Inc.) was
used for all transfection assays, according to the manufacturer's
protocol. The MG63 and the MG63/CDDP cells were transiently
transfected using negative control (NC) or miR-22 mimic at room
temperature. Aliquots (50 µl) of Gibco® Opti-MEM (Thermo
Fisher Scientific, Inc.) were used to dilute 50 nM NC or mimic;
subsequently, the mixture was added to 3 µl diluted Lipo3000, prior
to further mixing and incubating the mixture for 20 min at room
temperature. Subsequently, the cells were added to a 6-well plate
which contained 100 µl liposome transfection mixture (Invitrogen;
Thermo Fisher Scientific, Inc.). After incubation for 6 h, the
medium was replaced by Hyclone® DMEM medium containing
10% FBS. After 48 h of incubation, the cells were harvested after a
centrifugation step (1,000 rpm, 5 min, room temperature).
The sequences of the NC and miR-22 mimic constructs
were as follows. NC: Sense, UUCUCCGAACGUGUCACGUTT and antisense,
ACGUGACACGUUCGGAGAATT; miR-22: Sense, AAGCUGCCAGUUGAAGAACUGU and
antisense, AGUUCUUCAACUGGCAGCUUUU. The MG63 and MG63/CDDP cell
lines stably expressed miR-22 with lentivirus particles.
Cell proliferation assay
The MG63 and MG63/CDDP cell lines, respectively,
were cultured in Hyclone® DMEM Complete™ culture medium
containing 10% FBS in a cell incubator containing 5% CO2
at 37°C. The plates were inoculated with 100 µl of cells
(5×105 cells/ml was added per well), with the cells
being added to each well of a 12-well plate. Cells were adherent to
the wall of the plate, and transfection with miR-22 and CDDP was
allowed to take place for 48 h. After transfection, 2 µM CDDP was
added. A solution of bromodeoxyuridine (BrdU) (Sigma-Aldrich; Merck
KGaA) was made up to a final concentration of 0.03 mg/ml, and BrdU
was subsequently added to the cells at 6 and 12 h after
transfection. The cells were then incubated at 37°C for 3 h. The
culture solution was removed, and the cells were washed 3 times
with PBS (5 min each wash). Paraformaldehyde (4%, v/v) was used to
fix the cells at room temperature for 10 min. The paraformaldehyde
was subsequently removed, and the cells were washed 3 times with
PBS (5 min each wash). PBS containing 0.5% Triton X-100 (Alladin)
was added, and the membrane was placed on the ice for 10 min.
PBS/Triton X-100 was removed, and the membrane was washed 3 times
with precooled PBS (5 min each wash). PBS/3% BSA (Sigma-Aldrich;
Merck KGaA) was added to seal the membrane at room temperature for
30 min. The BrdU antibody (cat. no. ab8955, Abcam) was diluted in
PBS/1% BSA solution at the ratio of 1:100. After removal of the
sealing solution, the primary antibody was added and either
incubated at room temperature for 2 h, or at 4°C overnight. The
secondary antibody [Alexa Fluor 488 donkey anti-mouse IgG(H+L)l;
cat. no. ab150105; Abcam] for fluorescence was diluted in PBS-1%
BSA at a ratio of 1:400, and after the PBS-T was removed, the
secondary antibody was added to the membrane and incubated for 1 h
in the dark at room temperature. Subsequently, the secondary
antibody was removed by washing with PBS. DAPI (100 ng/ml) was
added, and subsequently incubated with the membrane at room
temperature in the dark for 10 min. The DAPI was then removed,
anti-fluorescence quenching solution (cat. no. P0126; Beyotime
Institute of Biotechnology) was added, and the membrane was either
placed in the dark at 4°C, or photographs were directly captured
under a fluorescence microscope (magnification, ×200; Ts2R;
Nikon).
Monodansylcadaverine (MDC)
staining
MDC (cat. no. G0170; purchased from Beijing Solarbio
Science & Technology Co., Ltd.), is a marker for autolysosomes,
and is used as a tracer for autophagic vesicles. Cells were fixed
and incubated with MDC for 30 min at 37°C, and subsequently stained
with DAPI. An anti-fluorescence quenching slide (to avoid light)
was used for the fluorescence study using a confocal fluorescence
microscope (LSM710; Zeiss GmbH). Cells were stained with MDC for 30
min at 37°C, and subsequently the fluorescence integrated optical
density (IOD) values of MDC in each group were measured.
Flow cytometric assay
Detection of changes in the expression levels of the
autophagy-related protein, microtubule-associated proteins 1A/1B
light chain 3B (LC3), were qualitatively evaluated by flow
cytometry (using a BD Calibur flow cytometer; BD Biosciences).
Cells were fixed with 4% paraformaldehyde for a duration of 10 min,
followed by three washes with PBS (5 min each wash). PBS/1% BSA
solution was used for preparing a 1:500 dilution of the LC3
antibody. Subsequently, the cells were incubated for 2 h with the
primary antibody (cat. no. ab229327; Abcam) at room temperature.
The Alexa Fluor-488 conjugated secondary antibody (cat. no. A11008;
Invitrogen; Thermo Fisher Scientific, Inc.) was dissolved in PBS/1%
BSA solution to achieve a 1:400 dilution, and then added to the
fixed cells in the dark at room temperature (for 1 h). The treated
cells were analyzed by flow cytometry on a BD FACS Calibur
platform. Changes in cellular fluorescence were detected in the FL1
channel (BD Caliber, FACSCalibur; BD Biosciences).
Inoculation of tumor cells
To obtained the miR-22 overexpressed vector, the
pre-miR-22 hairpin was constructed in the pLV6-EF-1Af/puro
lentivirus plasmid (Genepharma). The plasmid was co-transfected
into 293T cells using Lipofectamine 2000 with pG-P1-VSVG,
pG-P2-REV, pG-P3-RRE plasmids. After culture for 48 h, the
lentivirus particle was harvested and concentrated with lenti-X
concentrator according to the manufacturer's procedure (Clontech
Laboratories). MG63 and MG63/CDDP cells were plated in a 6-well
plate. After being cultured for 8 h, the lentivirus particles were
poured in the plate to transduce cells with a multiplicity of
infection (MOI) of 1:50. The stable clones were selected using 1
µg/ml puromycin for about 1 week.
After 7 days of adaptive feeding (26°C, 12 h of
light and 12 h of darkness every day, 200 ml water per day), 40
male nude mice aged 8 weeks (SCXK2015-0001; Model Animal Research
Center of Nanjing University, Nanjing, China) were randomly divided
into 8 treatment groups, with 5 mice in each group: i) MG63; ii)
MG63+miR-22, iii) MG63+CDDP; iv) MG63+CDDP+miR-22; v) MG63/CDDP;
vi) MG63/CDDP+miR-22; vii) MG63/CDDP+CDDP; and viii)
MG63/CDDP+CDDP+miR-22. Subcutaneous inoculation of 5×106
(100 µl) cells in the right flank was performed. The nude mice were
fed normally, and after the appearance of small nodules (following
7 days of inoculation), the MG63, MG63+miR-22, MG63/CDDP and
MG63/CDDP+miR-22 groups were administered a normal saline injection
of 100 µl each time, twice per week for 5 weeks. For the MG63+CDDP,
MG63+CDDP+miR-22, MG63/CDDP+CDDP and MG63/CDDP+CDDP+miR-22
treatment groups, CDDP was administered at a dose of 2 mg/kg each
time, twice per week for 5 weeks. Animal health and behavior were
monitored every day. The dose of pentobarbital administration was
100 mg/kg and the manner of euthanasia was intraperitoneal
injection. Tumor samples were collected after 6 weeks of normal
feeding. Tumor volume (V) was calculated according to the formula:
V = length × (width)2/2.
Treatment with a specific inhibitor of
PI3K
The MG63 cells with or without miR-22 transfection
were divided into two groups: A control group and an inhibitor
group. DMSO at a concentration of 10 µM (C6295; Sigma-Aldrich;
Merck KGaA) was added to the cells of the control group. Wortmannin
at a concentration of 10 µM (S2758; Selleck Chemicals) was added to
the cells of the inhibitor group. Each group had four treatment
subgroups, comprising treatments of MG63, MG63+miR-22, MG63+CDDP
and MG63+CDDP+miR-22. Subsequently, RT-quantitative (q)PCR and
western blot analysis were performed.
RT-qPCR
Invitrogen TRIzol® reagent (Thermo Fisher
Scientific, Inc.) was used to isolate total RNA from cells or
tissues, according to the manufacturer's protocol. The PrimeScript
RT reagent kit (Takara Biotechnology Co., Ltd.) was used for
conversion of RNA into cDNA; the miRNA extraction kit of Guangzhou
RiboBio Co., Ltd. was used to achieve the same objective with
respect to miRNA. Quantification of transcripts was accomplished by
performing RT-qPCR using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.)., and the ABI StepOne Plus Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to perform
qPCR using the SYBR-Green PCR Kit (Takara Biotechnology Co., Ltd.).
The qPCR protocol consisted of the following steps: a) initial
denaturation for 5 min at 95°C; and b) 40 cycles involving
denaturation at a temperature of 95°C for 10 sec, followed by
annealing and extension at 60°C for 34 sec. For miR-22 expression
in RT-qPCR, U6 was used as the reference gene. These experiments
were repeated three times. miR-22 and U6 primers were amplified by
a Bulgeloop miRNA RT-qPCR kit (Guangzhou RiboBio Co., Ltd.).
Detailed information of the primers employed for the qPCR
experiments is shown in Table I,
whereas the thermocycling conditions used for the qPCR experiments
are shown in Table II.
 | Table I.Detailed information of primers for
the qPCR experiments. |
Table I.
Detailed information of primers for
the qPCR experiments.
| Gene | Sense | Antisense | bp |
|---|
| GAPDH |
CTTTGGTATCGTGGAAGGACTC |
AGTAGAGGCAGGGATGATGT | 133 |
| PI3K |
GTATCCCGAGAAGCAGGATTTAG |
CAGAGAGAGGATCTCGTGTAGAA | 127 |
| Akt |
CTTCTATGGCGCTGAGATTGT |
GCCCGAAGTCTGTGATCTTAAT | 131 |
| mTOR |
GTGGAAACAGGACCCATGAA |
CCATTCCAGCCAGTCATCTT | 102 |
 | Table II.Thermal conditions used for the qPCR
experiments. |
Table II.
Thermal conditions used for the qPCR
experiments.
| Temperature | Time (sec) |
|
|---|
| 95°C | 600 |
|
| 95°C | 5 | 40 cycles |
| 60°C | 60 |
|
| 95°C | 15 | Melting curve |
| 60°C | 60 |
|
| 95°C | 15 |
|
Western blot analysis
The cells were bathed in pre-chilled PBS and
subsequently lysed in 200 µl RIPA buffer (Beyotime Institute of
Biotechnology) for 30 min duration by keeping the vials on ice. The
cell solution was then centrifuged at 13,500 × g at 4°C for 5 min.
SDS-PAGE (Bio-Rad Laboratories, Inc.) was used to separate the
cellular proteins (8% SDS-PAGE for PI3K, mTOR, p-mTOR and 10%
SDS-PAGE for Akt, p-Akt, GAPDH), followed by blot transfer to a
PVDF membrane (EMD Millipore). Each protein sample (30 µg aliquots)
was loaded onto the SDS-PAGE gel. Post-blotting, 5% skimmed milk
was used for incubation of the membrane at 4°C for 1 h. 5%
BSA-containing TBS/Tween-20 (TBST) solution was used to prepare
1:1,000 dilutions of the primary antibody (see the next paragraph
for further details), and this was subsequently incubated with the
membrane overnight at 4°C. The membranes were washed thrice with
TBST at room temperature (10 min each wash). The secondary antibody
was diluted with TBST (1:5,000 dilution), and the membranes were
then incubated with the secondary antibody at room temperature for
1 h, followed by three washes with TBST at room temperature (10 min
each wash). The chemiluminescence reaction was carried out using
the Tanon chemiluminescence sensor system (Tanon Science and
Technology Co., Ltd.). GAPDH was used as the loading control.
The primary antibodies used for western blotting
were as follows: Anti-Akt (cat. no. 10176-2-AP; ProteinTech Group,
Inc.); anti-phosphorylated (p)-Akt (cat. no. ab81283; Abcam);
anti-PI3K (cat. no. ab151549; Abcam); anti-mTOR (cat. no.
20657-1-AP; ProteinTech Group, Inc.); anti-p-mTOR (cat. no.
ab84400; Abcam,); and anti-GAPDH (cat. no. 200306-7E4; ZenBio,
Inc.). The secondary antibodies used were as follows: HRP goat
anti-mouse IgG(H+L) (cat. no. A0216, Beyotime Institute of
Biotechnology); HRP goat anti-rabbit IgG(H+L) (cat. no. A0208;
Beyotime Institute of Biotechnology).
Immunohistochemistry
Paraffin sections were placed in an oven at 65°C for
2 h, dewaxed to water, and washed with PBS three times (5 min each
wash). The slices were placed in EDTA buffer, and put into the
microwave for repair. Power was cut off after medium fire to
boiling, and low fire to boiling at intervals of 10 min. The slices
were subsequently placed in 3% hydrogen peroxide solution
(Sinopharm Chemical Reagent Co., Ltd.), and incubated at room
temperature for 10 min to block endogenous peroxidase. The slides
were then washed three times in PBS (5 min each wash), prior to
being treated with 5% BSA for 20 min after drying. Subsequently,
the BSA was removed, and 50 µl diluted primary antibody (1:100) was
applied to cover the tissue of each slice, followed by an
incubation overnight at 4°C. Subsequently, the PBS solution was
removed, and 50–100 µl of the secondary antibody was added to each
section, also followed by an incubation at 4°C for 50 min.
Subsequently, 50–100 µl fresh DAB (Sinopharm Chemical Reagent Co.,
Ltd.) was added to each slice, and the extent of the color
development was assessed under a microscope. After the process of
color development was complete, the slides were rinsed with
distilled water, redyed with hematoxylin, differentiated with 1%
hydrochloric acid alcohol (1 sec), rinsed with tap water, turned
back into a blue color upon addition of ammonia, and rinsed with
running water. The sections were subjected to a gradient of alcohol
(70–100%) for 10 min, dehydrated and dried; subsequently, xylene
(Sinopharm Chemical Reagent Co., Ltd.) was added, and neutral gum
(Sinopharm Chemical Reagent Co., Ltd.) was used for sealing.
The antibodies used in the immunohistochemical
analyses were as follows: Anti-p-Akt (cat. no. ab81283; Abcam);
anti-PI3K (cat. no. ab151549; Abcam); and anti-p-mTOR (cat. no.
ab84400; Abcam).
Statistical analysis
Each experiment was performed three times, and the
results are shown as the mean ± standard deviation. SPSS 17.0
software (SPSS, Inc.) was used for statistical analysis.
Differences among three or more groups were compared by one-way
analysis of variance (ANOVA); Tukey's test was used for comparisons
of data between two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Transfection of miR-22 is
successful
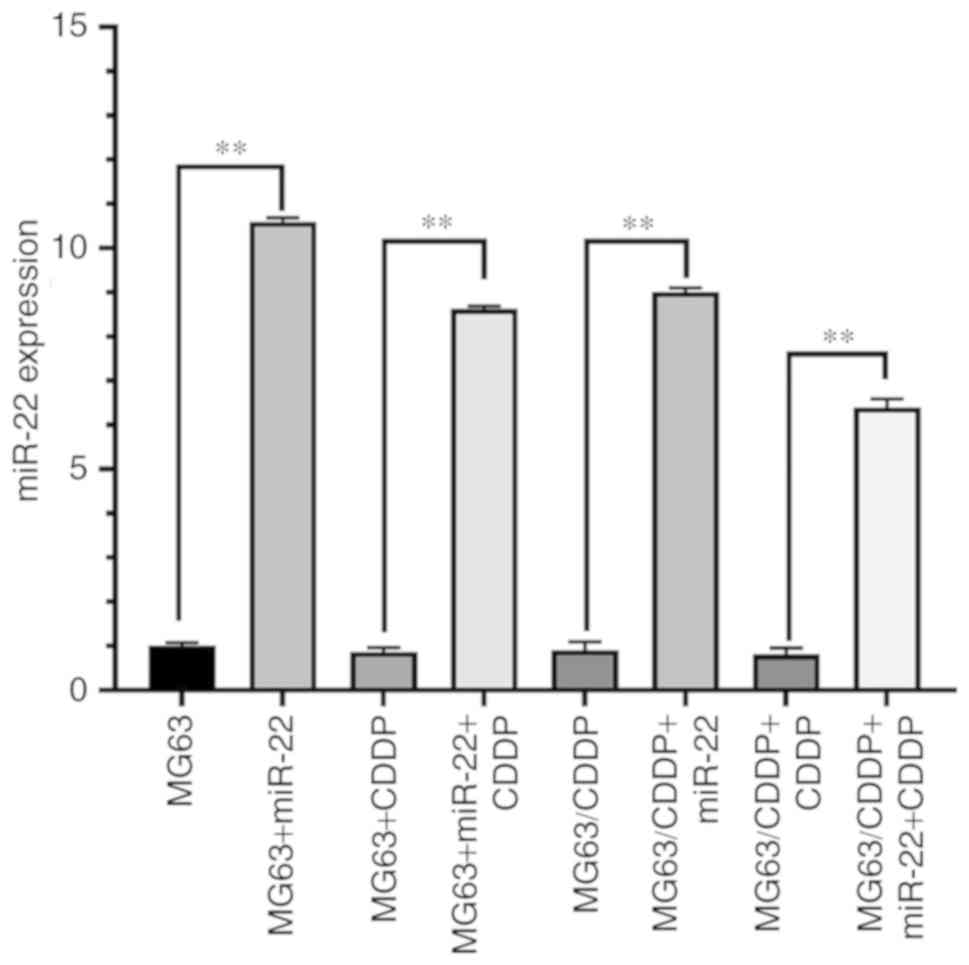
Fig. 1 shows the
mRNA expression of miR-22 in each group. The expression of miR-22
in the transfection group was higher than the group without miR-22
transfection (P<0.01) for all cell lines used. Thus,
transfection was successful.
Expression of the autophagy-related
genes is highest in the MG63 cells compared with other cell
lines
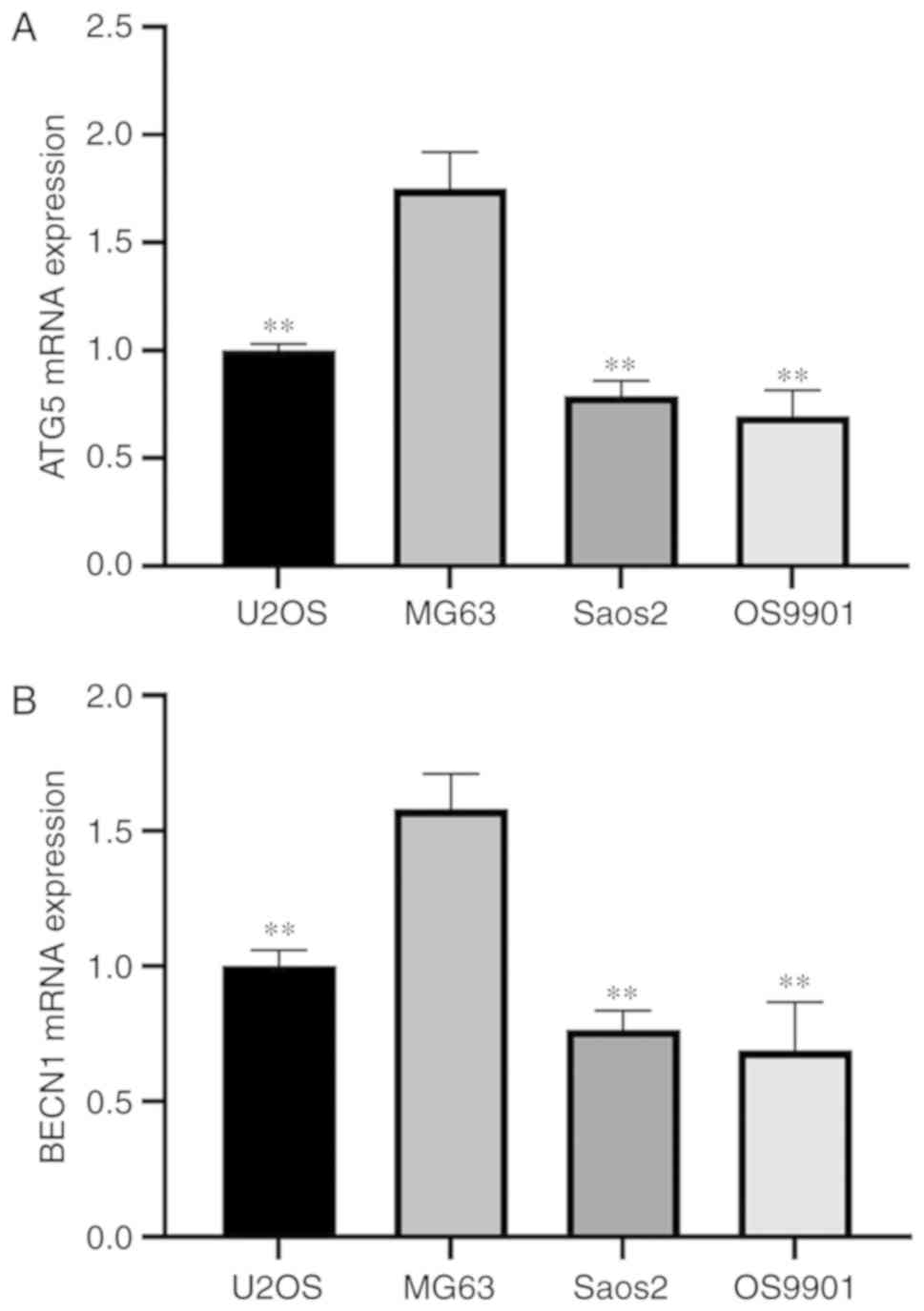
Fig. 2 shows that
the expression levels of the autophagy-related genes BECN1 and ATG5
in MG63 cells were significantly higher compared with the other
cell lines, including U2OS, Saos2 and OS9901 (P<0.01). Thus, we
selected the MG63 cell line as our experimental cell line.
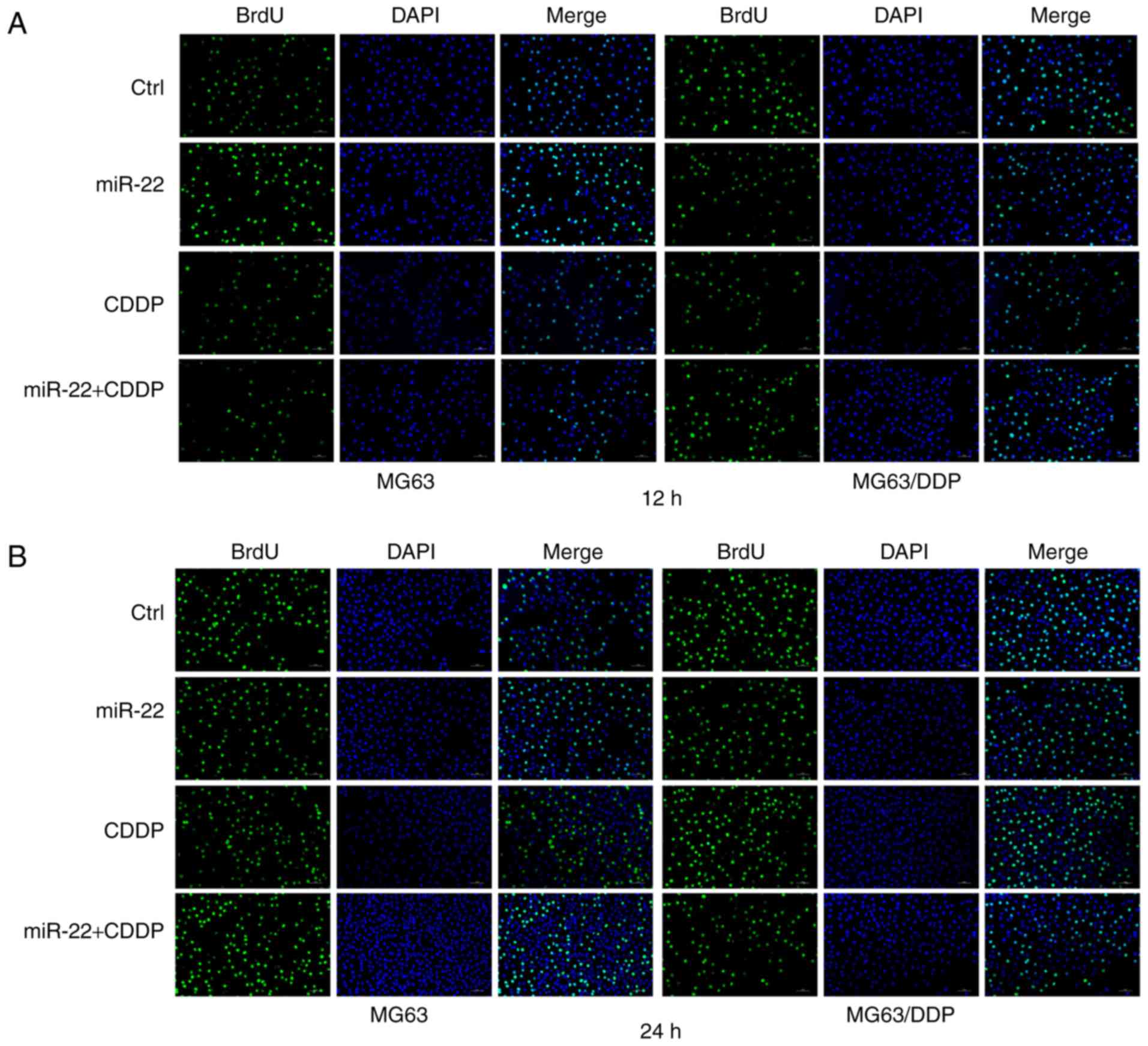
miR-22 inhibits proliferation of the
MG63 and MG63/CDDP cell lines
The results of the cell proliferation experiments
are shown in Fig. 3A-C. The BrdU
ratio values in the miR-22 group in both the MG63 and MG63/CDDP
cell lines were lower than each control group (P<0.05).
Moreover, the values for the miR-22+CDDP treatment group in the two
cell lines were lower compared with the respective control groups
at the 12 and 24 h time-points (P<0.01).
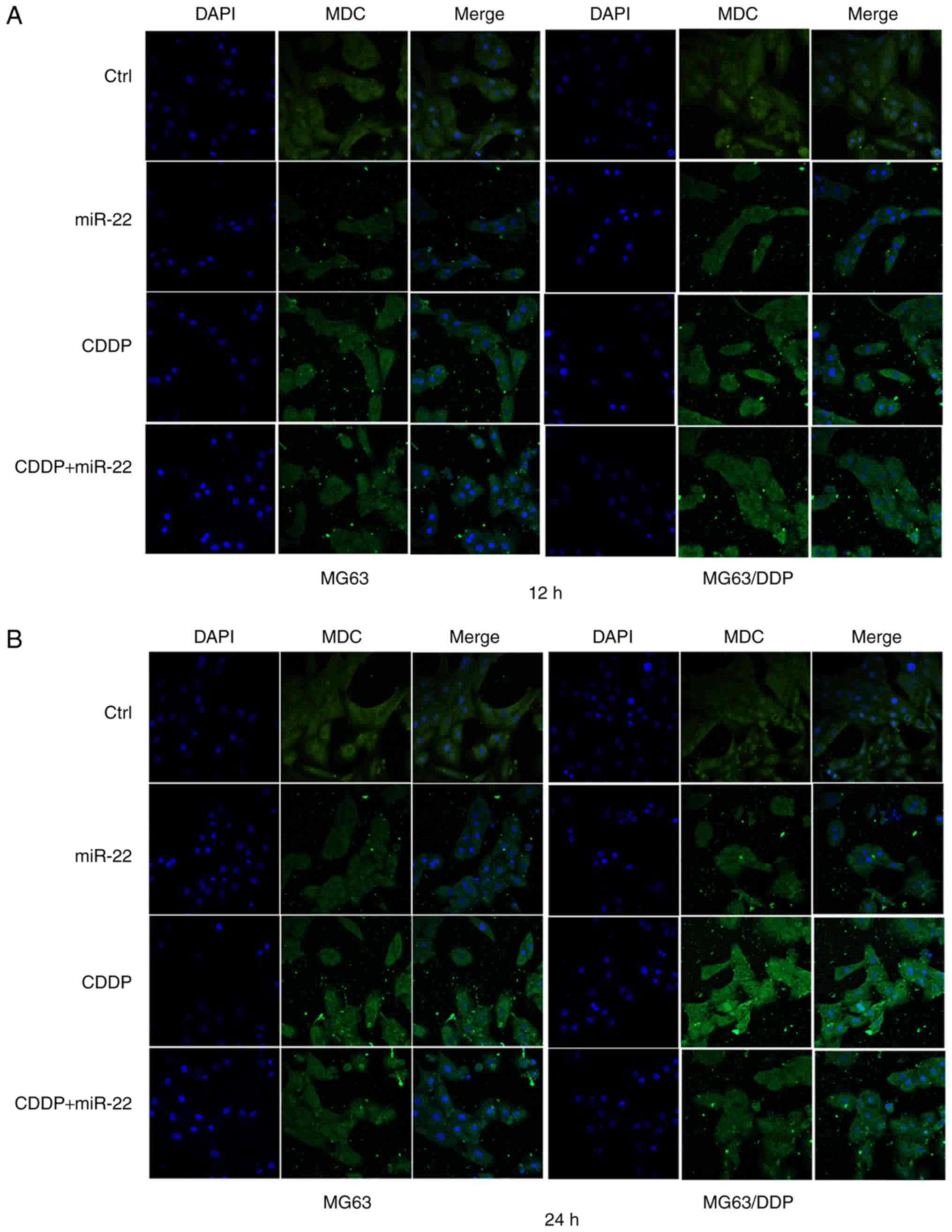
miR-22 suppresses autophagy of the
MG63 and MG63/CDDP cell lines
The results of MDC staining are shown in Fig. 4. The autophagy rates were calculated
according to the IOD of MDC (Fig.
4C). A higher value of IOD corresponds to a higher autophagy
rate. The MG63 cell control group exhibited a lower IOD value
compared with the control group of MG63/CDDP cells at the 12 and 24
h time-point (P<0.05). The IODs of groups that were transfected
by miR-22 for 12 and 24 h in the MG63 and MG63/CDDP cells were
lower when compared with each control group (P<0.01).
Furthermore, the group with combined treatment of miR-22 and CDDP
exhibited smaller IOD values compared with the CDDP treatment alone
groups of the MG63 and MG63/CDDP cell lines at the times of 12 and
24 h (P<0.01).
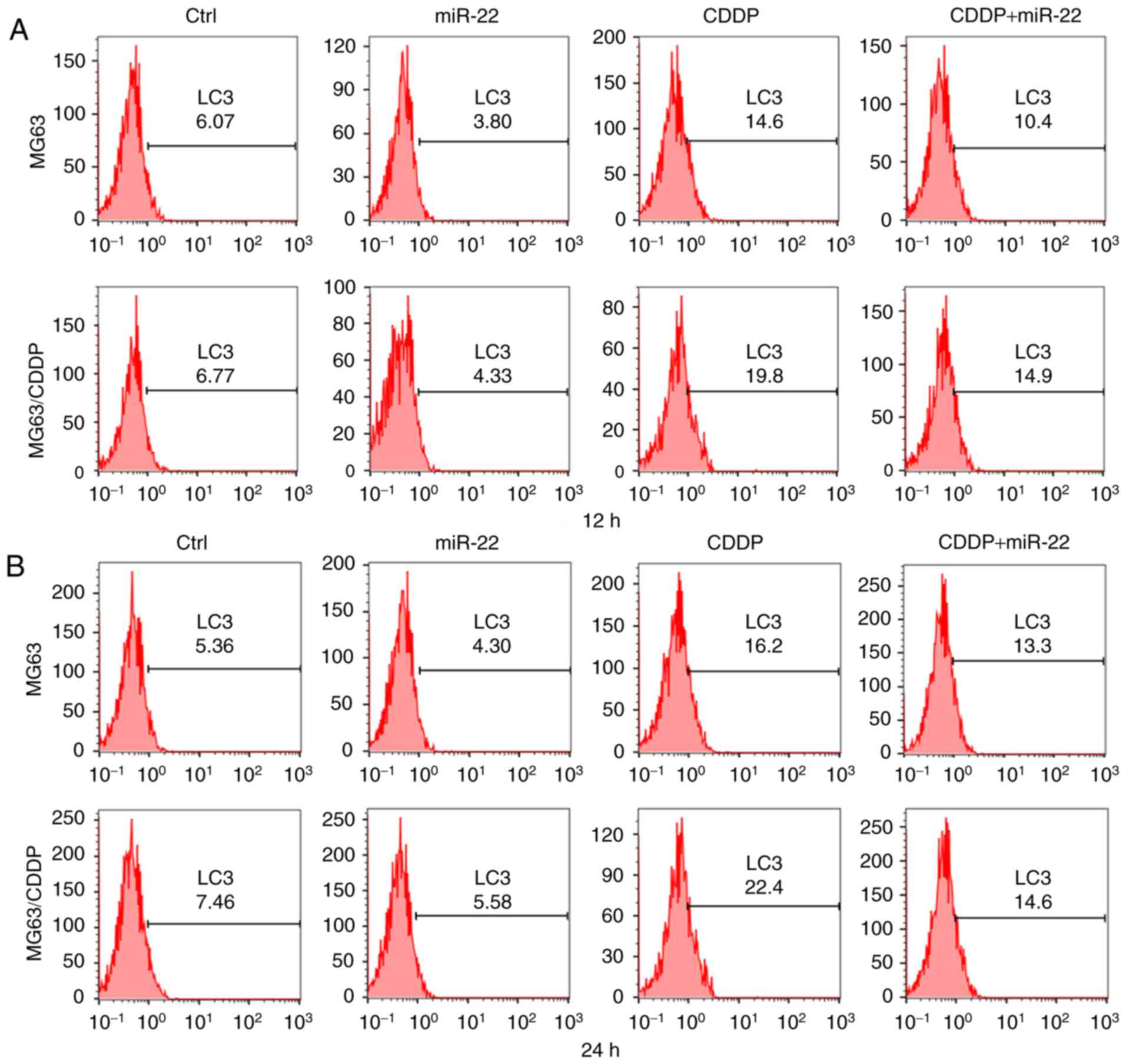
Flow cytometric assay is used to detect expression
of genes in cells by using fluorescent antibodies. In addition,
expression of LC3 is associated with autophagy. Moreover, the
results consolidated to findings which were obtained by measuring
LC3B II/LC3B I, MDC staining and electron microscopy observation
(25,31). Thus, we performed LC3 flow cytometry
to verify autophagy. Fig. 5A-C
revealed that the fluorescence values of LC3 in the miR-22 group of
the MG63 and MG63/CDDP cell lines were lower compared with each
respective control group, whereas the value was higher in the CDDP
treatment groups of the two cell lines compared with the control,
which revealed that miR-22 inhibited autophagy, whereas CDDP
induced autophagy (P<0.05). Furthermore, miR-22 decreased the
level of autophagy that was induced by CDDP in the MG63 and
MG63/CDDP cells (P<0.01). The results obtained from the LC3 flow
cytometric assays at the 12 and 24 h time-points were similar.
CDDP induces autophagy of the MG63 and
MG63/CDDP cell lines
As shown in Fig. 4C,
the IOD values in the CDDP treatment group at times of 12 and 24 h
were higher compared with the control groups for each cell line
(P<0.01). As shown in Fig. 6A-C,
the mRNA expression levels of PI3K, Akt and mTOR, and the protein
expression levels of PI3K, p-Akt and p-mTOR, were upregulated in
MG63/CDDP cells; it was also observed that CDDP could downregulate
the expression of these genes in the MG63 and MG63/CDDP cells
compared with each control group (P<0.05). The results obtained
following treatment with CDDP for 12 and 24 h were similar.
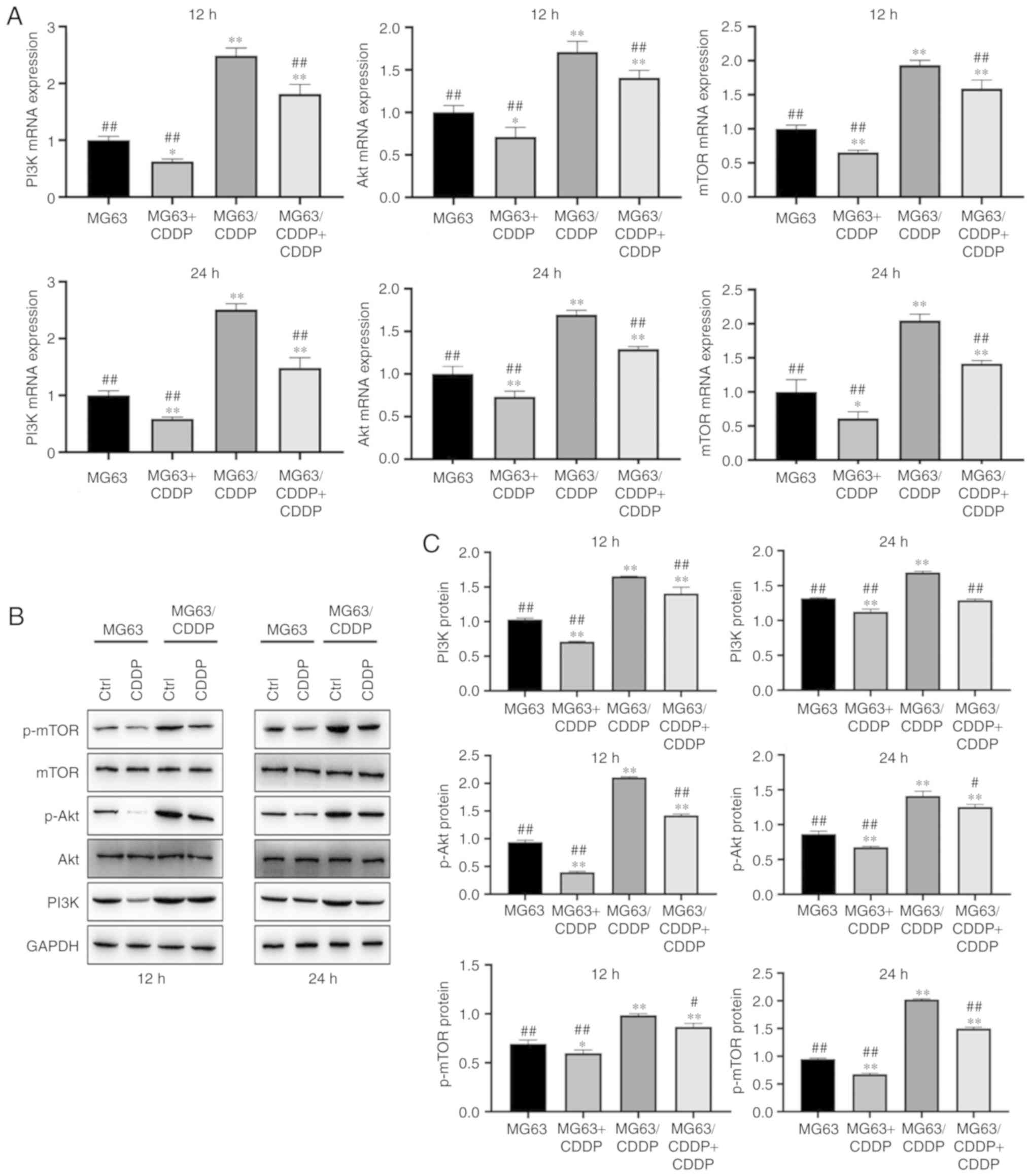 | Figure 6.(A) mRNA expression of PI3K, Akt and
mTOR following treatment with CDDP for 12 and 24 h. (B) Protein
expression of mTOR, p-mTOR, Akt, p-Akt and PI3K after treatment
with CDDP for 12 and 24 h, followed by western blot analysis. (C)
Protein expression of PI3K, p-Akt and p-mTOR after treatment with
CDDP for 12 and 24 h. *P<0.05 and **P<0.01, compared
with the control group of MG63; #P<0.05 and
##P<0.01, compared with the control group of
MG63/CDDP. To determine the extent of phosphorylation of Akt, the
value shown for p-Akt is the ratio of the IODs of p-Akt and Akt;
for p-mTOR, the extent of phosphorylation of mTOR is shown as the
ratio of the IODs of p-mTOR and mTOR. PI3K, phosphoinositide
3-kinase; mTOR, mammalian target of rapamycin; p-, phosphorylated;
CDDP, cisplatin; IOD, integrated optical density. |
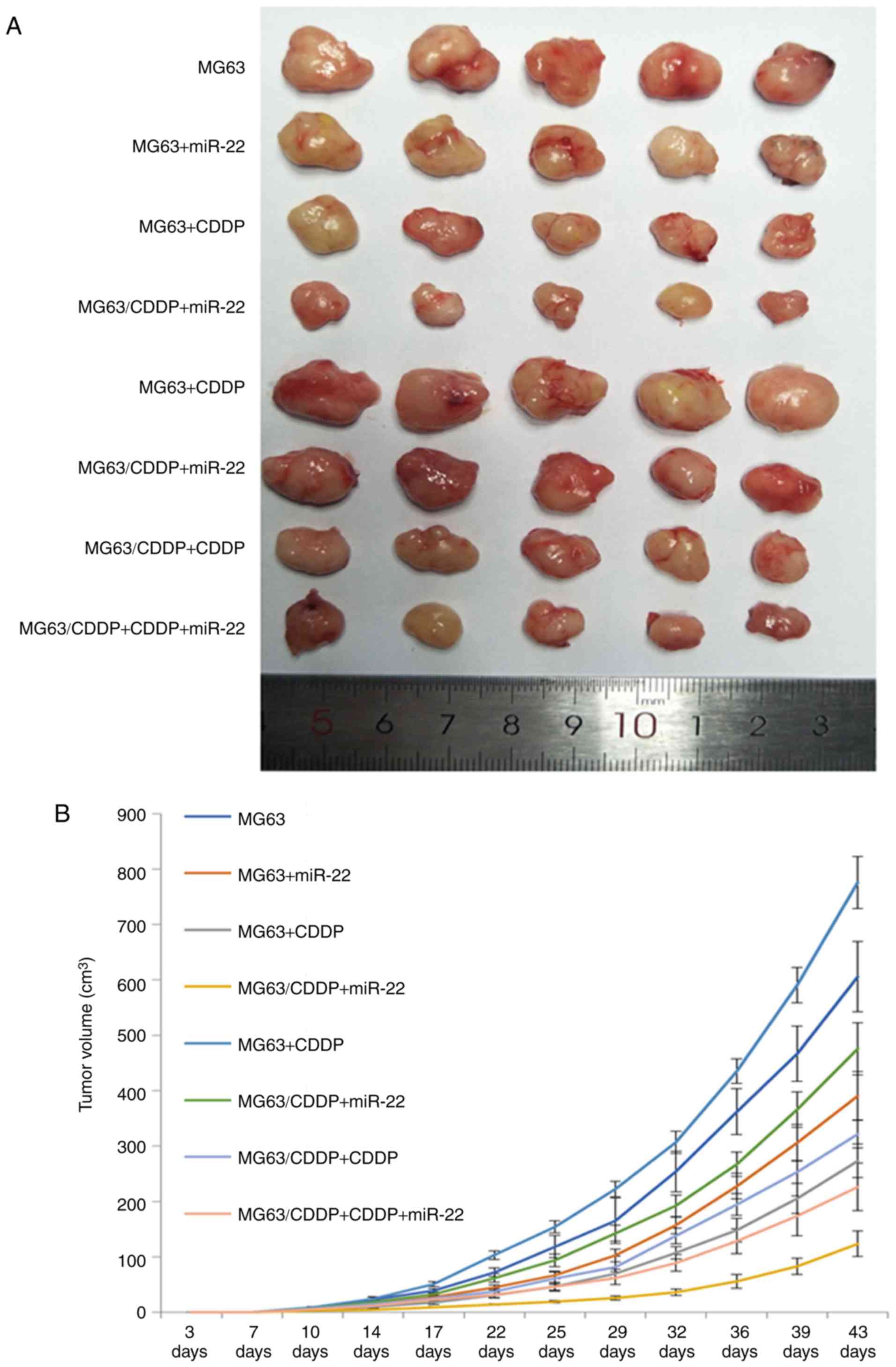
miR-22 inhibits tumor growth and
increases the anti-proliferative effect of CDDP
Images of the tumors are shown in Fig. 7A. The growth curves obtained after
inoculating the mice with MG63 cells or MG63/CDDP cells are shown
in Fig. 6B. The volume and weight
of the tumors are shown in Fig. 7C.
The tumors resulting from inoculation of either MG63 cells or
MG63/CDDP cells combined with miR-22 and CDDP treatments were shown
to have the smallest volume and weight compared with each of the
control groups (P<0.01). Both miR-22 and CDDP were able to
inhibit growth of the tumors (P<0.01). The tumor volumes and
weights resulting after inoculation of the mice with the MG63/CDDP
cells were larger compared with the tumors resulting from the
inoculation of MG63 cells (P<0.01).
miR-22 downregulates the PI3K/Akt/mTOR
pathway to suppress autophagy
As shown in Fig. 8A and
B, miR-22 was able to downregulate the expression levels of
PI3K, Akt and mTOR in the MG63 and MG63/CDDP cell lines compared
with each control group at the mRNA level (P<0.05). miR-22 also
downregulated the expression of PI3K, p-Akt and p-mTOR in the MG63
and MG63/CDDP cell lines compared with each control group at the
protein level (P<0.05). The MG63/CDDP cell line revealed an
upregulation of PI3K, p-Akt and p-mTOR compared with the MG63
control group, whereas both miR-22 and CDDP were able to inhibit
the upregulation of these genes (P<0.05).
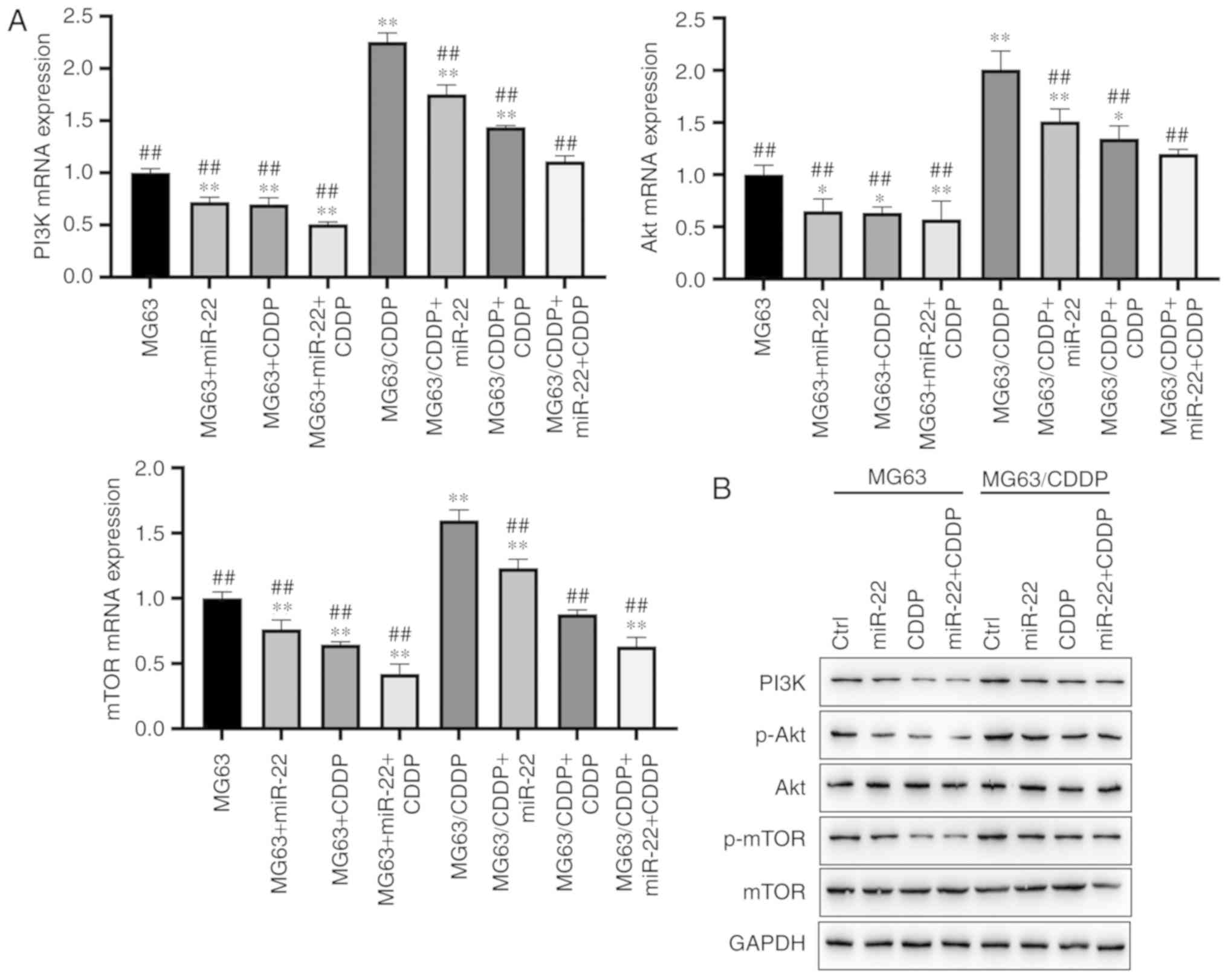 | Figure 8.Protein expression levels of PI3K,
Akt and mTOR in the MG63 and MG63/CDDP cell lines. (A) mRNA
expression of PI3K, Akt and mTOR. (B and C) Protein expression of
PI3K, p-Akt and p-mTOR as determined by western blot analysis.
*P<0.05 and **P<0.01, compared with the control group of
MG63; ##P<0.01, compared with the control group of
MG63/CDDP. To determine the extent of phosphorylation of Akt, the
value shown for p-Akt is the ratio of the IODs of p-Akt and Akt;
for p-mTOR, the extent of phosphorylation of mTOR is shown as the
ratio of the IODs of p-mTOR and mTOR. PI3K, phosphoinositide
3-kinase; mTOR, mammalian target of rapamycin; p-, phosphorylated;
CDDP, cisplatin; IOD, integrated optical density. |
The immunohistochemical results are shown in
Fig. 9A-D. The IOD values of each
gene are shown in Fig. 9D. The
control group of the MG63/CDDP cells was found to have a higher
PI3K IOD value compared with that of control group of the MG63 cell
line (P<0.05). The expression levels identified in the CDDP and
the miR-22 group were low for both of the both cell lines compared
with each control group (P<0.05). The control group of the
MG63/CDDP cells exhibited a higher p-Akt IOD value compared with
that of the MG63 cells (P<0.05). The expression of p-Akt in the
CDDP and miR-22 groups was downregulated in both the cell lines
compared with each control group (P<0.05). In addition, the
expression of the other groups compared with each control group was
downregulated (P<0.05). The control group of the MG63/CDDP cells
was shown to have a higher p-mTOR IOD value compared with that of
the MG63 cells (P<0.05). Treatment with CDDP and miR-22 in the
MG63 and MG63/CDDP cells led to lower IOD values compared with each
control group (P<0.05). In addition, combined treatment with
CDDP and miR-22 led to lower values compared with each control
group (P<0.01).
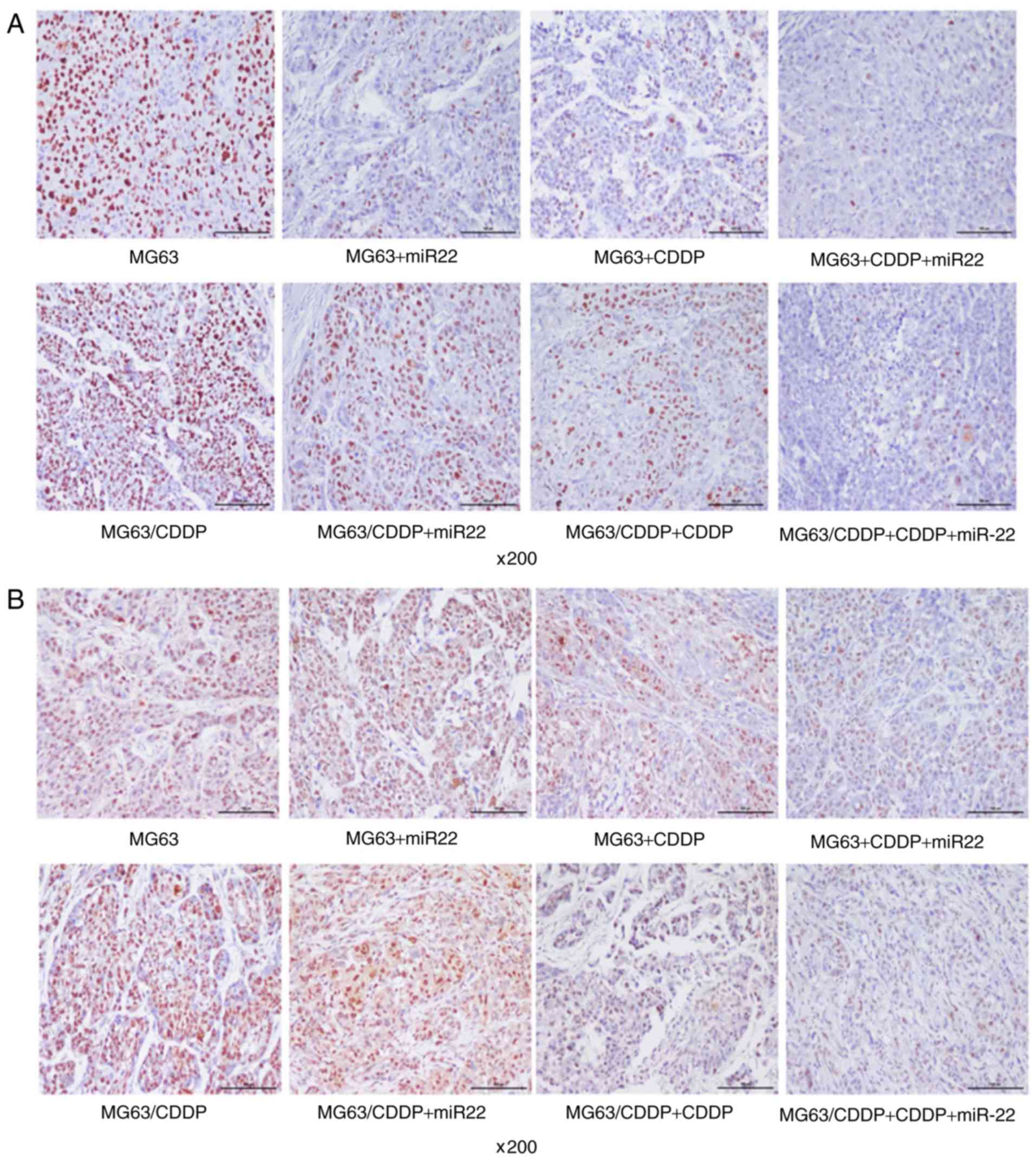 | Figure 9.Immunohistochemistry results.
Immunohistochemical results of (A) PI3K and (B) p-Akt are shown.
PI3K, phosphoinositide 3-kinase; mTOR, mammalian target of
rapamycin; p-, phosphorylated; CDDP, cisplatin; IOD, integrated
optical density. Immunohistochemistry results. Immunohistochemical
results of (C) p-mTOR are shown. (D) The immunohistochemical IOD
values of PI3K, p-Akt, and p-mTOR after inoculation with MG63 and
MG63/CDDP cells. *P<0.05 and **P<0.01, compared with the
control group of MG63; #P<0.05 and
##P<0.01, compared with the control group of
MG63/CDDP. To determine the extent of phosphorylation of Akt, the
value shown for p-Akt is the ratio of the IODs of p-Akt and Akt;
for p-mTOR, the extent of phosphorylation of mTOR is shown as the
ratio of the IODs of p-mTOR and mTOR. PI3K, phosphoinositide
3-kinase; mTOR, mammalian target of rapamycin; p-, phosphorylated;
CDDP, cisplatin; IOD, integrated optical density. |
Role exerted by miR-22 is similar to
that of wortmannin, a specific inhibitor of PI3K, in terms of
regulating the PI3K/Akt/mTOR pathway
Fig. 10A and B show
that the expression levels of LC3 (LC3II/I for protein expression),
PI3K, Akt and mTOR in the MG63 cells were downregulated in the
wortmannin-treated group compared with the DMSO group at both the
mRNA and protein level (P<0.05) while the expression of p62 was
upregulated (P<0.01). These results suggested that wortmannin
could suppress the PI3K/Akt/mTOR pathway and autophagy. In
addition, in the CDDP-treated cells, the expression levels of PI3K,
Akt and mTOR were downregulated in the wortmannin group compared
with the DMSO group at both the mRNA and the protein level
(P<0.05). In addition, the expression of p62 in both mRNA and
protein level was upregulated in the MG63+CDDP+wortmannin group
compared with the MG63+ CDDP+DMSO group while LC3 was downregulated
(P<0.01). These results suggested that, in MG63 cells that has
been treated with CDDP, the role of miR-22 was similar to that of
wortmannin in terms of regulating the PI3K/Akt/mTOR pathway and
autophagy. In addition, when applied to the miR-22+MG63 group,
wortmannin upregulated the expression of p62 and downregulated the
expression of LC3, PI3K, Akt and mTOR at both the mRNA and protein
level compared with the DMSO-treatment group (P<0.05), which
suggest that inhibitors of PI3K are able to enhance the effects
elicited by miR-22.
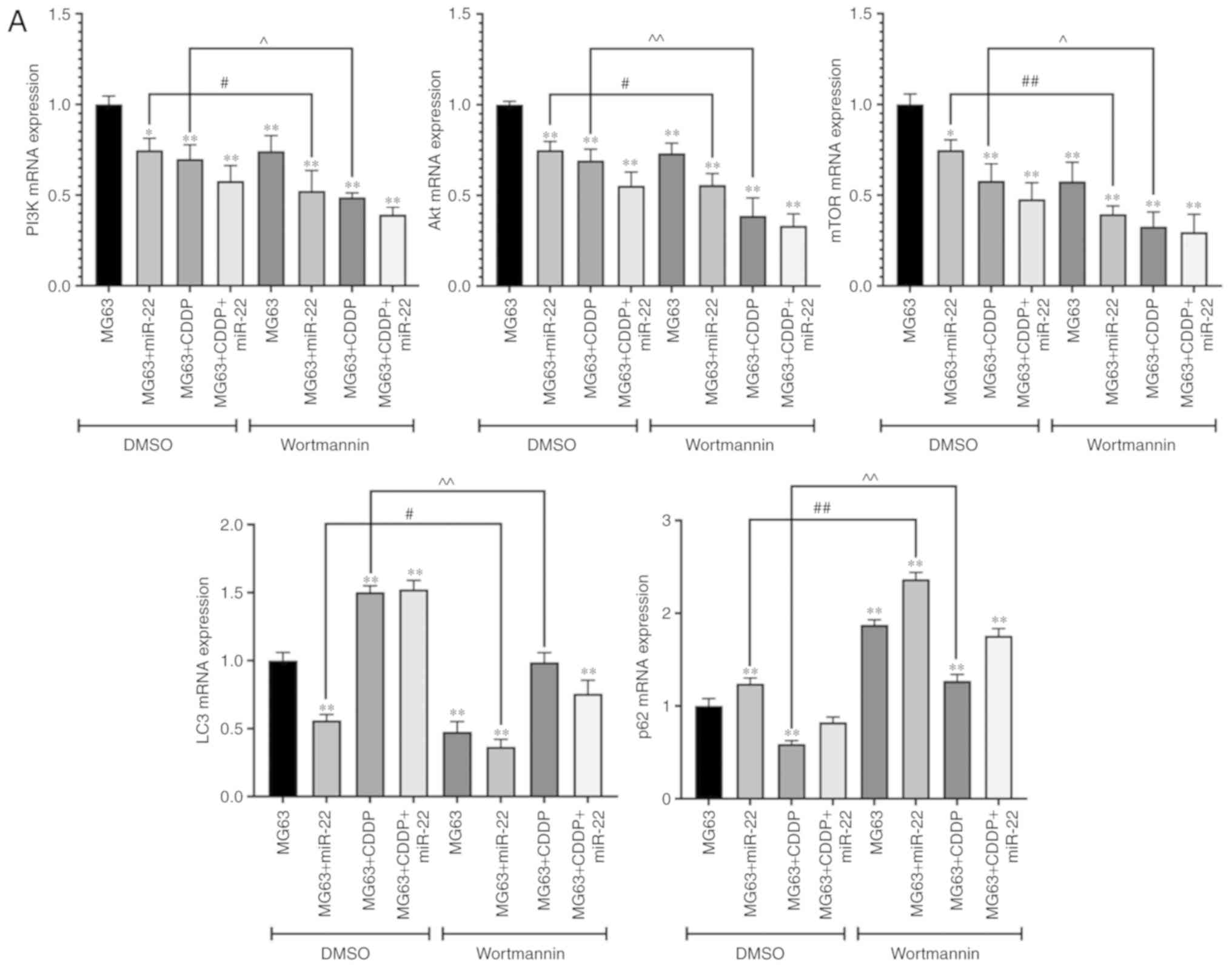 | Figure 10.Effect of wortmannin on the mRNA and
protein expression levels of PI3K, Akt and mTOR. (A) mRNA
expression levels of PI3K, Akt and mTOR in the DMSO (control) group
and the wortmannin-treated group are shown. *P<0.05 and
**P<0.01, compared with group MG63+DMSO. Further comparisons
between two group are indicated with solid lines. The labels
including # and ^ on the top of solid lines means they are
statistically significant: #P<0.05 and
##P<0.01, the wortmannin group of MG63+CDDP compared
with the DMSO group of MG63+CDDP; ^P<0.05 and
^^P<0.01, the wortmannin group of MG63+miR-22
compared with the DMSO group of MG63+miR-22. To determine the
extent of phosphorylation of Akt, the value shown for p-Akt is the
ratio of the IODs of p-Akt and Akt; for p-mTOR, the extent of
phosphorylation of mTOR is shown as the ratio of the IODs of p-mTOR
and mTOR. PI3K, phosphoinositide 3-kinase; mTOR, mammalian target
of rapamycin; p-, phosphorylated; CDDP, cisplatin; IOD, integrated
optical density. Effect of wortmannin on the mRNA and protein
expression levels of PI3K, Akt and mTOR. (B) The protein expression
levels of PI3K, p-Akt and p-mTOR are shown, as determined by
western blot analysis. *P<0.05 and **P<0.01, compared with
group MG63+DMSO. Further comparisons between two group are
indicated with solid lines. The labels including # and ^ on the top
of solid lines means they are statistically significant:
#P<0.05 and ##P<0.01, the wortmannin
group of MG63+CDDP compared with the DMSO group of MG63+CDDP;
^P<0.05 and ^^P<0.01, the wortmannin
group of MG63+miR-22 compared with the DMSO group of MG63+miR-22.
To determine the extent of phosphorylation of Akt, the value shown
for p-Akt is the ratio of the IODs of p-Akt and Akt; for p-mTOR,
the extent of phosphorylation of mTOR is shown as the ratio of the
IODs of p-mTOR and mTOR. PI3K, phosphoinositide 3-kinase; mTOR,
mammalian target of rapamycin; p-, phosphorylated; CDDP, cisplatin;
IOD, integrated optical density. |
Discussion
Chemoresistance may be regulated in numerous ways,
mediated via drug export transporters, DNA repair mechanisms,
cancer stem cells, resistance to apoptosis, self-sufficiency for
growth factor signaling, angiogenic switch, and immunological
pathways (32). Autophagy has been
found to be closely associated with chemoresistance. On one hand,
autophagy directs damaged components of the cells within
autophagosomes and ensures their removal, helping to maintain
cellular homeostasis. In addition, autophagy is able to protect
cells by maintaining a balance among the synthesis, degradation,
and subsequent recycling of essential molecules under the condition
of nutrient deprivation (33,34).
On the other hand, autophagy may induce cell death under the
condition of an excessive loss of proteins (35). Therefore, autophagy is considered to
be a ‘double-edged sword’. It has been shown that overexpression of
miR-155 promoted autophagy induced by anticancer drugs, and also
increases cell viability to modulate drug resistance in OS cells
(36). However, in the majority of
the studies that have been published on chemoresistance associated
with OS treatment, inhibition of autophagy led to a reduction in
drug resistance and an improvement in cell sensitivity (37,15).
In the present study, it was shown that miR-22 inhibited the
proliferation of cells, including the MG63 and MG63/CDDP cell
lines. Furthermore, miR-22 was able to corroborate the effect of
CDDP in terms of fulfilling its anti-proliferative role. In
addition, the results of the present study confirmed that miR-22
regulates chemoresistance by suppressing autophagy, and suppression
of autophagy, in turn, reduces the resistance to CDDP in both MG63
and MG63/CDDP cells.
In the in vivo study, the results obtained
followed the identical trend to those of the in vitro study.
Following inoculation of the tumor cells, the tumor volumes were
observed to be comparatively small in the MG63+CDDP+miR-22 group,
which indicates that miR-22 was able to increase the cell
sensitivity to CDDP. In the MG63/CDDP+miR-22 group, the tumor
volumes were smaller compared with the MG63/CDDP treatment group,
which suggested that miR-22 decreased the resistance associated
with CDDP. In the present study, it was shown that CDDP upregulated
autophagy, which, in turn, may have increased the resistance of
CDDP. However, miR-22 not only inhibited autophagy in tumor cells,
but it also inhibited the CDDP-induced autophagy. It has been
reported that autophagy can contribute to increased levels of
chemoresistance in cancer; however, it also contributes to the
inhibition of chemoresistance in certain types of cancer (32,38).
In the present study, our results showed that CDDP could induce
autophagy, and therefore it could be hypothesized that CDDP
resistance in OS could be acquired by activating autophagy. It was
also possible to surmise that resistant tumor cells (of the
MG63/CDDP cell line) could acquire chemoresistance through
autophagy to protect the cells from the toxic effects of CDDP, even
though CDDP could induce autophagy, and autophagy was upregulated
in the resistant cells. The present study further confirmed that
miR-22 inhibited autophagy in the resistant OS cells, and therefore
miR-22 could fulfill roles in decreasing the proliferation, and in
inhibiting the growth, of tumor cells.
Previous studies have investigated the role of
miR-22 in chemoresistance in OS treatment. It was previously
reported that miR-22 targets the gene HMGB1 to suppress
autophagy, leading to a reduction in the levels of adriamycin and
CDDP resistance (23,24). Another recently published study
demonstrated that miR-22 suppressed the proliferation, and promote
the sensitivity, of OS cells via metadherin (MTDH)-mediated
autophagy (25). The PI3K/Akt/mTOR
pathway is an important pathway that regulates autophagy. It has
been reported that the PI3K pathway contributes to the growth of
cancer cells by providing basic metabolites through glycolysis and
lipogenesis (39). In the present
study, we found that wortmannin as a specific inhibitor of PI3K
inhibited autophagy by regulating the PI3K/Akt/mTOR pathway.
Moreover, the role of miR-22 was similar to wortmannin in the MG63
cells and inhibitors of PI3K were able to enhance the effects
elicited by miR-22. In addition, a recent study proposed the
hypothesis that inhibition of autophagy by suppressing the
PI3K/Akt/mTOR pathway by overexpression of HSP90AA1 could decrease
drug resistance in OS (40). It has
been shown that the proliferation and survival of melanoma are
associated with PI3K/Akt (41). In
addition, the growth of tumor cells, such as breast cancer cells,
could be promoted by mTOR-mediated mitochondrial biogenesis and
function (42). Therefore, the
downregulation of PI3K, Akt and mTOR is advantageous to anticancer
treatment. It has been reported that PI3K is the key component of
the pathway in terms of activating mTOR-induced autophagy (43). In addition, CDDP treatment
inactivated the PI3K/AKT/mTOR signaling pathway to induce autophagy
in endometrial cancer cells (44).
In the present study, it was shown that the expression of PI3K,
Akt, and mTOR was downregulated by miR-22 and CDDP, whereas these
proteins were upregulated in the resistant cells. We surmised that
drug resistance was obtained by activating autophagy via the
PI3K/Akt/mTOR pathway. It could be hypothesized that miR-22 is able
to inhibit autophagy induced by CDDP and decrease drug resistance
via the PI3K/Akt/mTOR pathway both in vivo and in
vitro. The results of the present study coincided with the
results of a previous study, which revealed which PI3K could be
downregulated to inhibit autophagy, leading to suppression of MG63
cell proliferation activity and increasing the sensitivity to CDDP
(45). A recent study argued that
miR-22-3p enhances the chemosensitivity of gastrointestinal stromal
tumor cell lines to CDDP via the phosphatase and tensin homolog
deleted on chromosome 10 (PTEN)/PI3K/Akt pathway (26). Interestingly, in the present study,
CDDP downregulated the PI3K/Akt/mTOR pathway, whereas it was able
to induce autophagy. However, the upregulation of the PI3K/Akt/mTOR
pathway occurred concomitantly with an increase in chemoresistance.
This result suggested that CDDP may downregulate the PI3K/Akt/mTOR
pathway to exert an antitumor effect, although excessive autophagy
induced by CDDP may lead to upregulation of the PI3K/Akt/mTOR
pathway to further increase the chemoresistance of CDDP. In
addition, downregulation of the PI3K/Akt/mTOR pathway was
significant in terms of preventing chemoresistance. The present
study provides new insight that miR-22 could play a role in
enhancing the chemosensitivity of OS to CDDP by inhibiting the
PI3K/Akt/mTOR pathway (26).
In conclusion, the present study demonstrated that
miR-22 was able to both inhibit tumor cell proliferative activity
and decrease CDDP resistance by inhibiting autophagy via the
PI3K/Akt/mTOR pathway in vivo and in vitro. Although
this is significant and novel information, the internal mechanism
of the pathway, more target genes and associated pathways, and the
precise details of the biological metabolism have yet to be
elucidated, and require further study. Our study provides new
insight into the anti-chemoresistance in OS, and may contribute to
the development of novel therapeutic drugs.
Acknowledgements
No funding was received.
Funding
The present study is supported by the National
Natural Science Foundation of China (grant no. 81660440) and the
Natural Science Foundation of Inner Mongolia (grant no.
2018MS08031).
Availability of data and materials
The analyzed datasets generated and/or analyzed
during the current study are available from the corresponding
author on reasonable request.
Authors' contributions
CYM, ZQZ, SBG, WF, CS, YXW and HQX participated in
the design of the study and drafted the manuscript. RB, LS and WZ
collected the data and performed the statistical analyses. CYM,
ZQZ, SBG and WF were major contributors to the design of this study
and revised the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All animal experiments were performed following the
approval of the Inner Mongolia Medical University Animal Ethics
Committee and according to the Guidelines for the Care and Use of
Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
OS
|
osteosarcoma
|
|
miRNA
|
microRNA
|
|
miR-22
|
microRNA-22
|
|
CDDP
|
cisplatin
|
|
LC3
|
microtubule-associated protein 1 light
chain 3
|
|
MTDH
|
metadherin
|
|
PTEN
|
phosphatase and tensin homolog deleted
on chromosome 10
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
mTOR
|
mammalian target of rapamycin
|
|
MDC
|
monodansylcadaverine
|
|
ANOVA
|
analysis of variance
|
|
IOD
|
integral optical density
|
References
|
1
|
Ries LAG, Smith MA, Gurney JG, et al:
Cancer incidence and survival among children and adolescents:
United States SEER Program 1975–1995. 1999.
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004.
Cancer. 115:1531–1543. 2010. View Article : Google Scholar
|
|
3
|
Bleyer A, O'Leary M, Barr R, et al: Cancer
epidemiology in older adolescents and young adults 15 to 29 years
of age, including SEER incidence and survival: 1975–2000. 2006.
|
|
4
|
Anninga JK, Gelderblom H, Fiocco M, Kroep
JR, Taminiau AH, Hogendoorn PC and Egeler RM: Chemotherapeutic
adjuvant treatment for osteosarcoma: Where do we stand? Eur J
Cancer. 47:2431–2445. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kager L, Tamamyan G and Bielack S: Novel
insights and therapeutic interventions for pediatric osteosarcoma.
Future Oncol. 13:357–368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: MiRNAs in human cancer. J Pathol. 223:102–115. 2015.
View Article : Google Scholar
|
|
7
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gougelet A, Pissaloux D, Besse A, Perez J,
Duc A, Dutour A, Blay JY and Alberti L: Micro-RNA profiles in
osteosarcoma as a predictive tool for ifosfamide response. Int J
Cancer. 129:680–690. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Torreggiani E, Roncuzzi L, Perut F, Zini N
and Baldini N: Multimodal transfer of MDR by exosomes in human
osteosarcoma. Int J Oncol. 49:189–196. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan MC and Jian W: Role of autophagy in
cancer. Medical Recapitulate. 7:9612010.
|
|
12
|
Sheng C, Jiang YZ, Huang L, Zhou RJ, Yu
KD, Liu Y and Shao ZM: The residual tumor autophagy marker LC3B
serves as a prognostic marker in local advanced breast cancer after
neoadjuvant chemotherapy. Clin Cancer Res. 19:6853–6862. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: Therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu
Y, Xie M, Yin X, Livesey KM, Lotze MT, et al: HMGB1-Induced
autophagy promotes chemotherapy resistance in leukemia cells.
Leukemia. 25:23–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang Z, Huo L, Li K, Wu Y and Hu Z:
Blocked autophagy by miR-101 enhances osteosarcoma cell
chemosensitivity in vitro. ScientificWorldJournal. 2014:7947562014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen R, Li X, He B and Hu W: MicroRNA-410
regulates autophagy-related gene ATG16L1 expression and enhances
chemosensitivity via autophagy inhibition in osteosarcoma. Mol Med
Rep. 15:1326–1334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Damavandi Z, Torkashvand S, Vasei M,
Soltani BM, Tavallaei M and Mowla SJ: Aberrant expression of breast
development-related microRNAs, miR-22, miR-132, and miR-212, in
breast tumor tissues. J Breast Cancer. 19:148–155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Zhang Y, Zhao J, Kong F and Chen Y:
Overexpression of miR-22 reverses paclitaxel-induced
chemoresistance through activation of PTEN signaling in p53-mutated
colon cancer cells. Mol Cell Biochem. 357:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Zhang L, Tong Y, Yu M, Wang M, Dong
D, Shao J, Zhang F, Niu R and Zhou Y: MicroRNA-22 inhibits
proliferation, invasion and metastasis of breast cancer cells
through targeting truncated neurokinin-1 receptor and ERalpha. Life
Sci. 217:57–69. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang W, Han X, Wang J, Wang L, Xu Z, Wei
Q, Zhang W and Wang H: MiR-22 enhances the radiosensitivity of
small-cell lung cancer by targeting the WRNIP1. J Cell Biochem.
120:17650–17661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou X, Natino D, Zhai X, Gao Z and He X:
MicroRNA22 inhibits the proliferation and migration, and increases
the cisplatin sensitivity, of osteosarcoma cells. Mol Med Rep.
17:7209–7217. 2018.PubMed/NCBI
|
|
23
|
Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang
Y, Wang Y, Zhao W and Wang W: MiR-22 inhibits osteosarcoma cell
proliferation and migration by targeting HMGB1 and inhibiting
HMGB1-mediated autophagy. Tumor Biol. 35:7025–7034. 2014.
View Article : Google Scholar
|
|
24
|
Li X, Wang S, Chen Y, Liu G and Yang X:
MiR-22 targets the 3′ UTR of HMGB1 and inhibits the
HMGB1-associated autophagy in osteosarcoma cells during
chemotherapy. Tumor Biol. 35:6021–6028. 2014. View Article : Google Scholar
|
|
25
|
Wang P, Zhao ZQ, Guo SB, Yang TY, Chang
ZQ, Li DH, Zhao W, Wang YX, Sun C, Wang Y and Feng W: Roles of
microRNA-22 in suppressing proliferation and promoting sensitivity
of osteosarcoma cells via metadherin-mediated autophagy. Orthop
Surg. 11:285–293. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Cheng M, Mi L, Qiu Y, Hao W and Li
L: Mir-22-3p enhances the chemosensitivity of gastrointestinal
stromal tumor cell lines to cisplatin through PTEN/PI3K/Akt
Pathway. Iran J Allergy Asthma Immunol. 17:318–325. 2018.PubMed/NCBI
|
|
27
|
Li Y, Geng P, Jiang W, Wang Y, Yao J, Lin
X, Liu J, Huang L, Su B and Chen H: Enhancement of radiosensitivity
by 5-Aza-CdR through activation of G2/M checkpoint response and
apoptosis in osteosarcoma cells. Tumour Biol. 35:4831–4839. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang TM, Guo SF, Chen CR, Zhang XY and Li
WG: Anti-Osteosarcoma effects and mechanisms of
4-O-amino-phenol-4′-demethylepipodophyllotoxin ether. J Pharm
Pharmacol. 60:179–188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang J, Shen L, Yang Q and Zhang C:
Overexpression of metadherin mediates metastasis of osteosarcoma by
regulating epithelial-mesenchymal transition. Cell Prolif.
47:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye H, Lin J, Yao X, Li Y, Lin X and Lu H:
Overexpression of long non-coding RNA NNT-AS1 correlates with tumor
progression and poor prognosis in osteosarcoma. Cell Physiol
Biochem. 45:1904–1914. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de la Calle C, Joubert PE, Law HK, Hasan M
and Albert ML: Simultaneous assessment of autophagy and apoptosis
using multispectral imaging cytometry. Autophagy. 7:1045–1051.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sui X, Chen R, Wang Z, Huang Z, Kong N,
Zhang M, Han W, Lou F, Yang J, Zhang Q, et al: Autophagy and
chemotherapy resistance: A promising therapeutic target for cancer
treatment. Cell Death Dis. 4:e8382013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh R, Kaushik S, Wang Y, Xiang Y, Novak
I, Komatsu M, Tanaka K, Cuervo AM and Czaja MJ: Autophagy regulates
lipid metabolism. Nature. 458:1131–1135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen L, Jiang K, Jiang H and Wei P:
MiR-155 mediates drug resistance in osteosarcoma cells via inducing
autophagy. Exp Ther Med. 8:527–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu R, Liu S, Chen H and Lao L:
MicroRNA-30a downregulation contributes to chemoresistance of
osteosarcoma cells through activating beclin-1-mediated autophagy.
Oncol Rep. 35:1757–1763. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Degenhardt K, Mathew R, Beaudoin B, Bray
K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al:
Autophagy promotes tumor cell survival and restricts necrosis,
inflammation, and tumorigenesis. Cancer Cell. 10:51–64. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hussain A, Qazi AK, Mupparapu N, Guru SK,
Kumar A, Sharma PR, Singh SK, Singh P, Dar MJ, Bharate SB, et al:
Modulation of glycolysis and lipogenesis by novel PI3K selective
molecule represses tumor angiogenesis and decreases colorectal
cancer growth. Cancer Lett. 374:250–260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiao X, Wang W, Li Y, Yang D, Li X, Shen
C, Liu Y, Ke X, Guo S and Guo Z: HSP90AA1-Mediated autophagy
promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res.
37:2012018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leonardi GC, Falzone L, Salemi R, Zanghì
A, Spandidos DA, Mccubrey JA, Candido S and Libra M: Cutaneous
melanoma: From pathogenesis to therapy (Review). Int J Oncol.
52:1071–1080. 2018.PubMed/NCBI
|
|
42
|
Zhang L and Han J: Branched-Chain amino
acid transaminase 1 (BCAT1) promotes the growth of breast cancer
cells through improving mTOR-mediated mitochondrial biogenesis and
function. Biochem Biophys Res Commun. 486:224–231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kumar S, Guru SK, Pathania AS, Manda S,
Kumar A, Bharate SB, Vishwakarma RA, Malik F and Bhushan S:
Fascaplysin induces caspase mediated crosstalk between apoptosis
and autophagy through the inhibition of PI3K/AKT/mTOR signaling
cascade in human leukemia HL-60 cells. J Cell Biochem. 116:985–997.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin Q, Wang Y, Chen D, Sheng X, Liu J and
Xiong H: Cisplatin regulates cell autophagy in endometrial cancer
cells via the PI3K/AKT/mTOR signalling pathway. Oncol Lett.
13:3567–3571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miao XD, Cao L, Zhang Q, Hu XY and Zhang
Y: Effect of PI3K-mediated autophagy in human osteosarcoma MG63
cells on sensitivity to chemotherapy with cisplatin. Asian Pac J
Trop Med. 8:731–738. 2015. View Article : Google Scholar : PubMed/NCBI
|