Introduction
According to a recent statistical analysis in 2018,
colorectal cancer (CRC) ranked third in regards to cancer incidence
(10.2% of total cancer numbers) and second in terms of cancer
mortality (9.2% of total cancer deaths) (1). Although significant progress has been
made both in conventional treatment options, such as surgery,
radiotherapy, and chemotherapy, and in targeted drugs for CRC
patients, distant metastasis is still the leading cause of
CRC-related death (2). Thus,
identification of novel prognostic biomarkers associated with
metastasis could improve the outcome for patients with CRC.
The E3 ubiquitin-protein ligase RAD18 plays a vital
role in DNA damage bypass and post-replication repair (PRR) through
the promotion of proliferating cell nuclear antigen (PCNA)
mono-ubiquitination at stalled replication forks (3). Recent studies have shown that high
expression of RAD18 in cancerous tissues is associated with cancer
metastasis and tumor progression in a variety of cancers (4–6). In
melanoma, studies demonstrated that RAD18 participates in the
regulation of cell proliferation, and its high expression is
associated with poor five-year patient survival (7). In glioma, RAD18 was found to suppress
apoptosis and accelerate cell proliferation (8). In cervical cancer, RAD18 was found to
promote the migration and metastasis of cancer cells through the
interleukin (IL)-1β pathway (9). In
esophageal squamous cell carcinoma, RAD18 exhibited the
characteristics of an oncogene and promoted tumor metastasis
through the JNK-MMP pathway (10).
Our previous work demonstrated that RAD18 expression increases the
resistance to radiotherapy and chemotherapy in CRC cells (11). However, the association between the
expression of RAD18 and metastasis in CRC remains unclear.
In the present research study, we analyzed
differences in the expression of RAD18 in CRC tissues and found
that overexpression of RAD18 was closely related to the strength of
metastatic and invasive tumor phenotype in CRC. However, the
possible cellular mechanisms and molecular signal regulation were
not well understood. Epithelial-mesenchymal transition (EMT) is
undoubtedly one of the important mechanisms of tumor metastasis
(12). EMT is a process considered
to be one of the initial steps in the invasion and metastasis
cascade, during which tumor epithelial cells dedifferentiate into
mesenchymal cells, separating from the original site to new
transfer sites (13). EMT changes
the polarity of the cells, deconstructs the cell connections,
adjusts the motility of cells, and modifies the cytoskeleton,
changes that together may contribute to the promotion of tumor cell
metastasis (14). To evaluate
whether EMT occurs requires the detection of relevant molecular
markers such as E-cadherin, N-cadherin, and vimentin (15). Hence, we further examined the role
of EMT-related molecular markers in the context of RAD18-mediated
invasion and migration of CRC cells.
Materials and methods
Clinical data and pathological
specimen collection
We collected samples from 93 patients with CRC who
were treated at the Nanjing Medical University Affiliated Suzhou
Hospital from November 2009 to May 2010, and we obtained adjacent
normal tissue samples from 87 of them. The mean age ± standard
deviation was 66.76±14.01 years (range, 22–93 years). Among the
patients, 52 were males and 41 were females. None of the patients
received any treatment (such as chemotherapy, radiotherapy, or
biotherapy) before surgery. All specimens were confirmed to be
adenocarcinoma by pathology. The Ethics Committee of Nanjing
Medical University (Suzhou, Jiangsu, China) approved this research,
and all patients signed an informed consent before surgery. The
follow-up deadline for all patients was June 6, 2016.
Immunohistochemical staining and
pathological evaluation criteria
All tissue samples were immunohistochemically
stained using 10% formalin-fixed for >24 h at room temperature,
and paraffin-embedded [sections (4-µm)] using conventional labeling
with horseradish peroxidase (HRP). The nuclei were counterstained
with hematoxylin. The primary antibodies used were rabbit
polyclonal anti-human RAD18 antibody (dilution 1:100, cat. no.
ab188235; Abcam Biotechnology), and rabbit polyclonal anti-human
E-cadherin (cat. no. ab32741), N-cadherin (cat. no. ab34241), and
vimentin (cat. no. ab36067) antibodies (dilution 1:50;
MultiSciences Biotech). The primary antibody was incubated at 37°C
for 2 h, and horseradish peroxidase (HRP)-conjugated anti-rabbit
antibody (cat. no. ab6721; Abcam Biotechnology) was incubated at
37°C for 1 h. The stained sections were observed by a Leica
microscope (magnification, ×200, Leica). The results were assessed
as follows: The intensity of staining was scored 0, 1, 2, and 3
according to the degree of color (no color, weak, moderate, and
strong color, respectively). The area of staining was scored 0, 1,
2, 3, and 4 according to the following: 0–10, 11–25, 26–50, 51–75
and >75%, respectively. The final score was equal to the product
of the above two scores. Two pathologists examined all specimens in
a blinded manner. When the scoring results differed between the
scorers, a final conclusion was reached through discussion.
Specimens with a final score of ≤6 were taken to have low
expression, and scores >6 were deemed to have high
expression.
Nucleic acid extraction and
quantitative RT-PCR
TRIzol (Invitrogen Life Technologies; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from frozen CRC
tissue samples according to the manufacturers instructions. The RNA
concentration was determined using a NanoDrop2000 (NanoDrop; Thermo
Fisher Scientific, Inc.). Total RNA (1 µg) was reverse transcribed
to cDNA, which was then used as a template for RT-qPCR to determine
the cycle threshold (Cq) of each tissue. The experimental data were
analyzed using the 2−ΔΔCq method (16) to calculate mRNA expression of RAD18,
E-cadherin, N-cadherin, vimentin, and GAPDH. All tests were
repeated three times and normalized to GAPDH. The primers
for RT-PCR were RAD18 F (forward), 5′-GTCCTTTCATCCTCTACTCTCGT-3′
and R (reverse), 5′-TAGCCTTCTCTATGTTGTCTATCCC-3′; E-cadherin F,
5′-CGAGAGCTACACGTTCACGG-3′ and R, 5′-GGGTGTCGAGGGAAAAATAGG-3′;
N-cadherin F, 5′-TGCGGTACAGTGTAACTGGG-3′ and R,
5′-GAAACCGGGCTATCTGCTCG-3′; vimentin F, 5′-CCAGGCAAAGCAGGAGTC-3′
and R, 5′-GGGTATCAACCAGAGGGAGT-3′; GAPDH F,
5′-CGACCACTTTGTCAAGCTCA-3′ and R, 5′-AGGGGAGATTCAGTGTGGTG-3′. The
PCR reactions were performed in duplicate at 95°C for 2 min and
subjected to 40 cycles of 95°C for 5 sec and 60°C for 35 sec.
Cell culture and pharmaceutical
reagents
Human CRC cell lines HCT116, DLD-1, and SW480 were
purchased from the Shanghai Cell Bank (Shanghai, China) and
identified using short tandem repeat profiling. Dulbecco's modified
Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased
from Hyclone. All cells were cultured in DMEM with 10% FBS, 100
U/ml penicillin, and streptomycin in a humidified atmosphere, with
5% CO2 at 37°C in an incubator. Subculturing was carried
out when the cells reached 85% confluence or more, and experiments
were carried out in cells that were passaged three times.
Establishment of stably transfected
cell strains with upregulated and downregulated expression of
RAD18
HCT116, DLD-1, and SW480 cells were seeded in
24-well plates at 5.0×104 cells/well, and confluency was
allowed to reach 70–80% by the next day. A lentivirus-based short
hairpin RNA (shRNA) vector targeting the RAD18 gene and a
lentivirus-based cDNA (GenePharm) were added separately to the
above cells using Lipofectamine 3000 (Invitrogen) according to the
instructions of the manufacturer. The final stably transfected
lines, which were selected with puromycin for 7 days and genotyped
by PCR, were HCT116 LV3, HCT116 RAD18sh, DLD-1 LV3, DLD-1 RAD18sh,
SW480 LV5, and SW480 RAD18.
Extraction of proteins and western
blot analysis of protein expression
The cells were trypsinized, harvested, centrifuged,
washed twice with PBS, and dissolved in RIPA buffer (Beyotime
Biotechnology) on ice, followed by centrifugation at 15,000 × g for
15 min. The supernatant was collected, and the protein
concentration was measured with the Bicinchoninic Acid (BCA)
Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal
aliquots (20 µg) from the samples were loaded and run into each
lane of 10% SDS-PAGE gels (Amresco), and then transferred to a PVDF
membrane (Millipore). After blocking with 5% non-fat milk in
Tween-20 (TBST) in Tris-buffered saline for 1 h at room
temperature, the membrane was incubated overnight at 4°C with the
appropriate concentration of primary antibody. We used the
following antibodies: Rabbit polyclonal anti-human RAD18 antibody
(dilution 1:1,000, cat. no. ab188235; Abcam Biotechnology);
E-cadherin (cat. no. ab32741), N-cadherin (cat. no. ab34241),
vimentin (cat. no. ab36067) polyclonal rabbit anti-human antibodies
(dilution 1:500; MultiSciences Biotech); and β-actin monoclonal
mice anti-human antibody (dilution 1:1,000, cat.no. sc-47778; Santa
Cruz Biotechnology). β-actin antibody was used as a loading control
to ensure equal protein loading. After washing three times with
TBST, the membrane was incubated with HRP-conjugated anti-rabbit or
anti-mouse secondary antibody for 2 h. The protein was visualized
by enhanced chemiluminescence (ECL; Beyotime Institute of
Biotechnology). The western blotting results were quantified by
ImageJ software version 1.52p [National Institutes of Health
(NIH)].
Wound-healing assay and Matrigel
Transwell chamber experiment
Cell migration was examined using the wound-healing
assay. CRC cells were seeded in a 6-well culture plate at
5.0×105 cells/well. In cell cultures that had grown to
confluence, typically 24–48 h later, scratches were made with a
200-µl pipette tip. The detached cells were washed three times with
PBS, and the remaining cells were incubated in culture medium
without serum. Images were captured at 0, 24, 48, 72 and 96 h with
an optical microscope (magnification, ×200) used to assess the
distance covered by the movement of the cells.
Cell invasion was determined by the Matrigel
Transwell chamber experiment following previous descriptions
(17,18). The Transwell chamber (Corning, Inc.)
was pre-coated with 60 µl Matrigel (1:6 dilution; BD Biosciences);
200 ml serum-free medium was added to the upper chamber, and 600 ml
of 10% serum medium was added to the lower chamber, as a chemical
attractant. The upper chamber, which was separated from the lower
one by an 8.0-µm polycarbonate membrane, was inoculated with the
same number of CRC cells (HCT116 and DLD-1, 1×105;
SW480, 5×104), cultured for 48 h, fixed with 3.7%
paraformaldehyde, and stained with Giemsa for 5 min at room
temperature. The cells in the upper chamber were wiped with a
cotton swab, and cells that had passed through the membrane were
photographed under a light microscope (magnification, ×200).
Rescue and recovery experiments
Mutated RAD18-encoding plasmids obtained from
Ribobio were used in the rescue experiment. Transient plasmid
transfection was performed using Lipofectamine 3000 DNA
transfection reagents (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Western blot analysis,
wound-healing assay and Matrigel Transwell chamber experiments were
performed after transfection with the above-described methods.
Statistical methods and data
analysis
We used the SPSS v.18.0 software (SPSS Inc.) for
statistical evaluation of the data, Graphpad PRISM v.5.0 (GraphPad
Software, Inc.) for graphing, and ImageJ software v.1.52p [National
Institutes of Health (NIH)] for analyzing western blot data and for
counting the numbers of Transwell cells. All experiments were
repeated at least three times and are expressed as mean ± standard
error. A Chi-square test was used to analyze the correlation
between RAD18 expression and the clinicopathologic data of
patients. The overall survival rate of patients was calculated
using the Kaplan-Meier method. Univariate and multivariate Cox
proportional hazard regression analysis was used to calculate the
hazard ratio (HR) of each variable to the 95% confidence interval
(CI) for overall patient survival. Student's t-test was used to
compare the western blotting protein expression levels, RT-PCR gene
expression levels, open wound areas, and Transwell cell numbers
between the two groups. For comparison of the three sets of data in
the rescue experiment, we used Newman-Keuls method in one-way
ANOVA. Pearson's correlation was used to analyze the correlation
between the expression of two genes. A two-sided test with a
significance level of α=0.05 (P<0.05) was used.
Results
Immunohistochemistry shows high
expression of RAD18 in cancer tissues
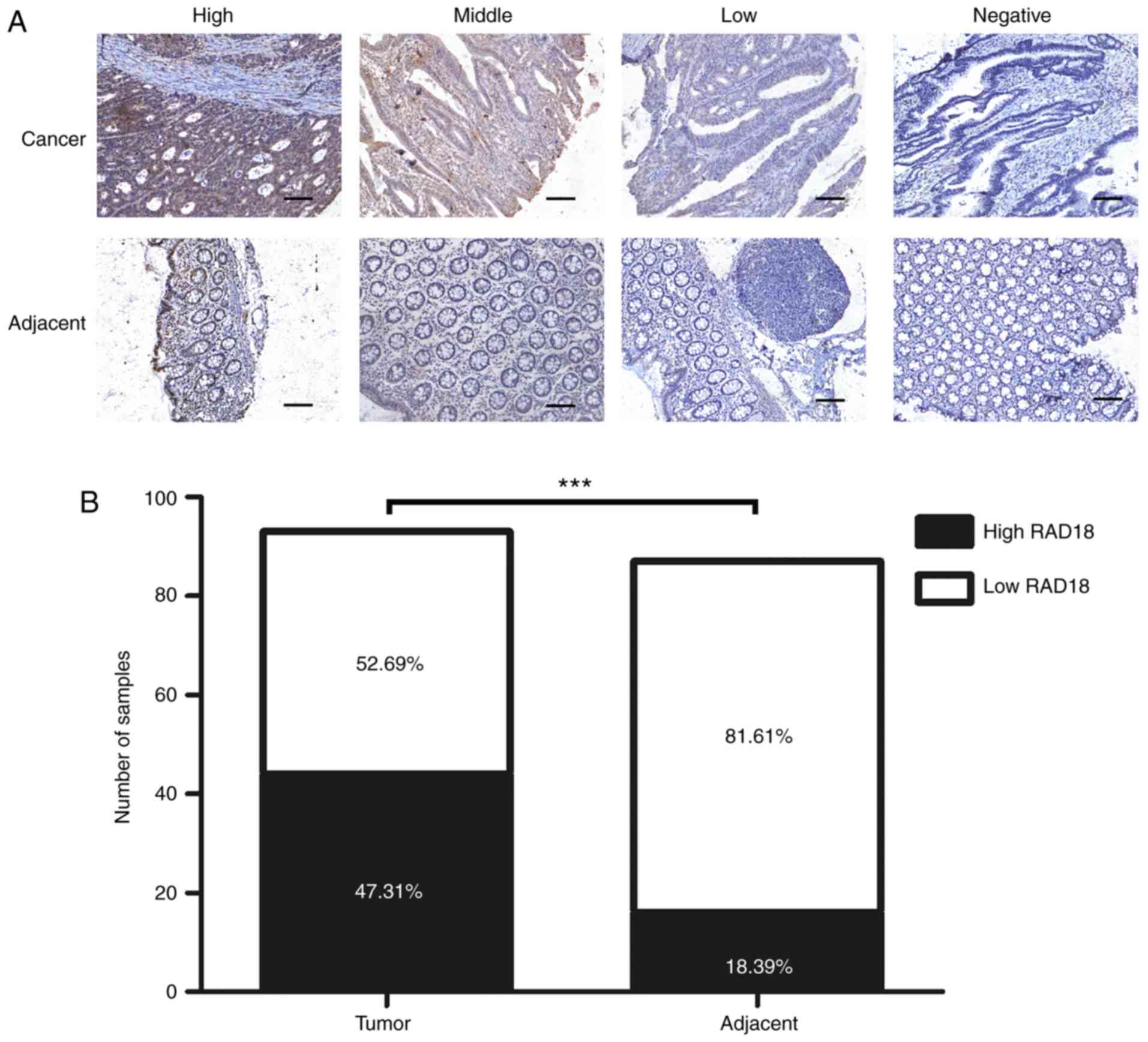
The immunohistochemical staining data of 93 cancer
tissues and 87 corresponding adjacent tissues with the RAD18
antibody are shown in Fig. 1A. Most
of the tumor tissues displayed deep brown staining, according to
the criteria described in Materials and methods. RAD18 was highly
expressed in 44 tissue specimens. Most of the adjacent normal
tissues were stained lightly or appeared unstained, with only 16
tissue samples showing a high expression of RAD18. High expression
of RAD18 was found in 47.31% (44/93) of tumor tissues and 18.39%
(16/87) of normal adjacent tissues. A histogram of RAD18 expression
in tumor and normal tissues (Fig.
1B) shows that RAD18 expression in tumor tissues was
significantly higher than that noted in the normal tissues
(P<0.001).
Clinical data of patients reveals that
RAD18 is associated with pathological stage and lymphatic
metastasis
The association of RAD18 with clinical data and
pathological characteristics of 93 CRC patients is shown in
Table I. Through the Chi-square
test, we demonstrated that the degree of tumor differentiation,
lymph node metastasis, tumor stage, and expression of MSH2 and
RAD18 were positively correlated, and the results were
statistically significant (P<0.05). RAD18 had no significant
correlation with other clinical and pathological factors
(P>0.05). The specific details are shown in Table I. The lymph nodes and pathological
staging findings suggest that RAD18 is closely related to tumor
metastasis.
 | Table I.Association of RAD18 expression with
clinicopathological characteristics of the CRC patients (N=93). |
Table I.
Association of RAD18 expression with
clinicopathological characteristics of the CRC patients (N=93).
|
|
| RAD18 expression, n
(%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Variables | N | Low | High | χ2 | P-value |
|---|
| Sex |
|
|
| 0.591 | 0.442 |
|
Male | 52 | 25 (48.1) | 27 (51.9) |
|
|
|
Female | 41 | 23 (56.1) | 18 (43.9) |
|
|
| Age (years) |
|
|
| 0.056 | 0.813 |
|
≤60 | 34 | 17 (50.0) | 17 (50.0) |
|
|
|
>60 | 59 | 31 (52.5) | 28 (47.5) |
|
|
| Degree of
differentiation |
|
|
| 5.551 | 0.018a |
|
Well/Moderate | 12 | 10 (83.3) | 2 (16.7) |
|
|
|
Poor | 81 | 43 (53.1) | 38 (46.9) |
|
|
| pT |
|
|
| 0.253 | 0.615 |
|
T1-3 | 69 | 34 (55.1) | 35 (44.9) |
|
|
| T4 | 16 | 9 (62.5) | 7 (37.5) |
|
|
| pN |
|
|
| 4.669 | 0.031a |
| N0 | 56 | 34 (60.7) | 22 (39.3) |
|
|
|
N1-3 | 37 | 23 (62.2) | 14 (37.8) |
|
|
| TNM stage |
|
|
| 4.652 | 0.031a |
| II | 54 | 33 (61.1) | 21 (38.9) |
|
|
|
III–IV | 39 | 15 (38.5) | 24 (61.5) |
|
|
| MSH2 |
|
|
| 4.321 | 0.038a |
|
Low | 56 | 24 (42.9) | 32 (57.1) |
|
|
|
High | 37 | 24 (64.9) | 13 (35.1) |
|
|
| MSH6 |
|
|
| 0.014 | 0.905 |
|
Low | 81 | 42 (51.9) | 39 (48.1) |
|
|
|
High | 12 | 6 (50.0) | 6 (50.0) |
|
|
Overall survival analysis illustrates
that RAD18 is an independent prognostic factor
The results of univariate and multivariate Cox
regression analyses are shown in Table
II. Lymph node metastasis, tumor stage, and RAD18 expression
were statistically significant in the univariate survival analysis
(P<0.01). Tumor staging and expression of RAD18 were still
statistically significant (P<0.05) when all significant factors
were analyzed by multivariate analysis. We have reasons to believe
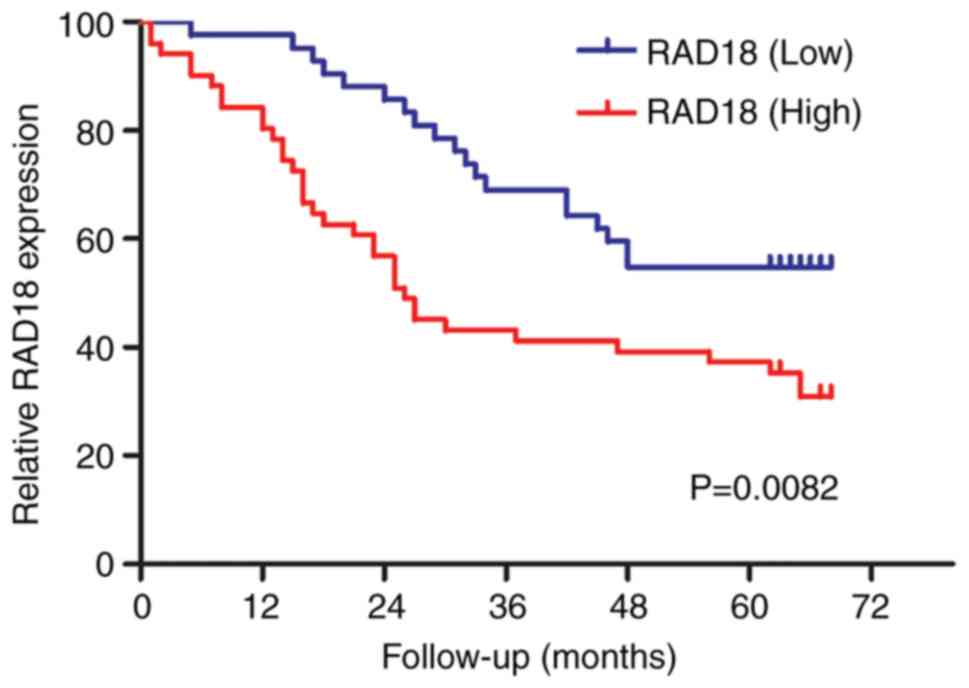
that RAD18 is an independent prognostic factor. As shown in
Fig. 2, we performed Kaplan-Meier
analysis of patients with high and low expression of RAD18. The
results showed that the survival curves were clearly separated and
that the difference was significantly different (P=0.0082).
 | Table II.Univariate and multivariate Cox
regression analysis for OS in CRC patients. |
Table II.
Univariate and multivariate Cox
regression analysis for OS in CRC patients.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variates | Categories | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex | Female vs.
Male | 1.212
(0.706–2.081) | 0.485 |
|
|
| Age | >60 vs. ≤60
years | 1.504
(0.826–2.736) | 0.182 | 1,810
(0.954–3.435) | 0.069 |
|
Differentiation | Poor vs.
Well/Moderate | 1.904
(0.758–4.787) | 0.171 | 1.150
(0.433–3.053) | 0.779 |
| pT | T4 vs. T1-3 | 1.557
(0.899–2.697) | 0.114 | 1.489
(0.823–2.694) | 0.189 |
| pN | N1-3 vs. N0 | 2.782
(1.610–4.809) |
<0.001b | 0.300
(0.061–1.463) | 0.136 |
| TNM stage | III–IV vs. II | 3.092
(1.776–5.383) |
<0.001b | 9.067
(1.781–46.152) | 0.008b |
| MSH2
expression | High vs. Low | 0.635
(0.359–1.122) | 0.118 | 0.760
(0.412–1.402) | 0.760 |
| MSH6
expression | High vs. Low | 0.554
(0.220–1.392) | 0.209 |
|
|
| RAD18
expression | High vs. Low | 2.365
(1.361–4.110) | 0.002b | 1.809
(1.005–3.256) | 0.048a |
Experiments with transfected cell
lines confirm that RAD18 promotes the migration and metastasis of
CRC cells
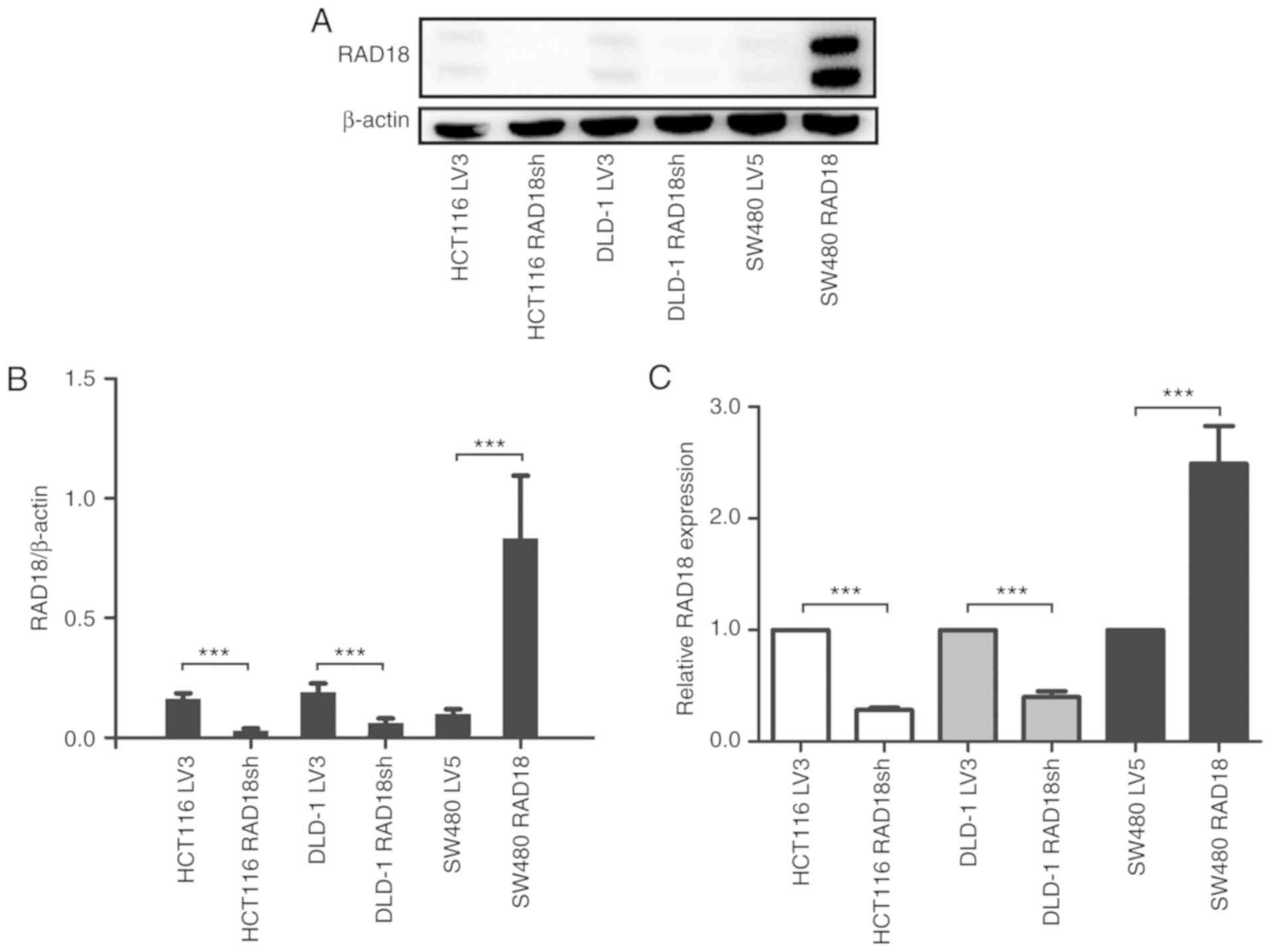
We established stably transfected clones with low
expression of RAD18 by introducing small interfering RNA into CRC
cell lines HCT116 and DLD-1. At the same time, stably transfected
SW480 strains with high expression of RAD18 were obtained by
transfection with a lentivirus vector containing a RAD18
cDNA. The expression of the protein was verified by western blot
analysis (Fig. 3A and B) and RT-PCR
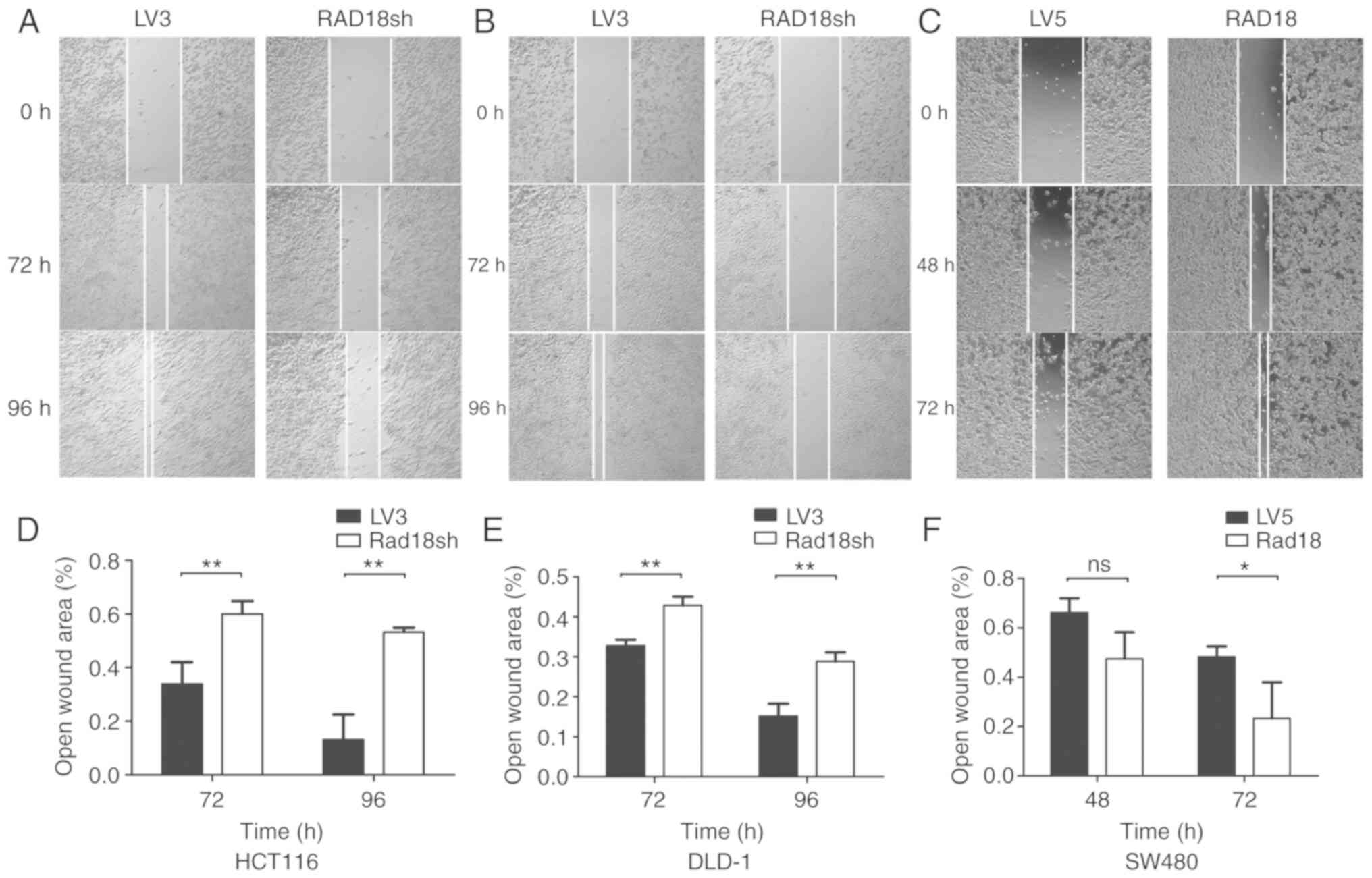
(Fig. 3C). Next, we carried out a
scratch test and Matrigel Transwell experiment on the three pairs
of transgenic cell lines (HCT116, DLD-1, and SW480) knocked down
for RAD18 or overexpressing RAD18. Wound-healing
assays demonstrated the migratory ability of the cells (Fig. 4). We found that the mobility of the
cell lines with low expression of RAD18 was markedly reduced and
the mobility of the cell lines showing high expression of RAD18 was
increased, and the results were statistically significant
(P<0.01 and P<0.05). The Matrigel Transwell experiment
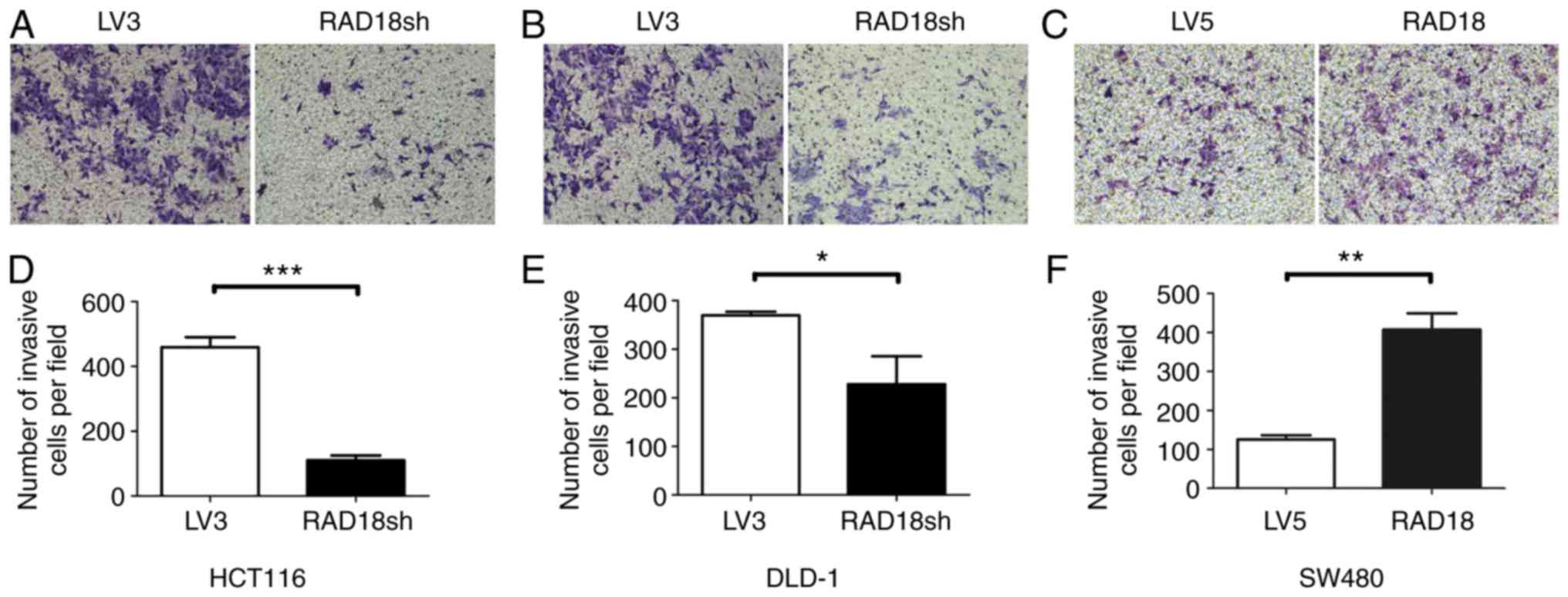
demonstrated the invasiveness of the cells (Fig. 5). Knockdown of RAD18 significantly
reduced the invasive ability of the HCT116 (P<0.001) and DLD-1
cells (P<0.05; Fig. 5A, B, D and
E). In contrast, overexpression of RAD18 significantly
(P<0.01) increased the invasive ability of the SW480 cells
(Fig. 5C and F).
Clinical specimens and cell
experiments indicate that RAD18 promotes CRC metastasis through the
EMT signaling pathway
The proteins from the three CRC cell lines were
subjected to western blot assay to examine the expression of RAD18
and the EMT-related proteins E-cadherin, N-cadherin, and vimentin.
We found that RAD18 expression was negatively correlated with
E-cadherin and positively correlated with N-cadherin and vimentin
(Fig. 6A). These findings were
confirmed by the analysis of the protein levels in clinical samples
using immunohistochemical staining. Images of representative tissue
samples stained for the above EMT-related proteins and RAD18 are
shown in Fig. 6B. Interestingly,
the results of the tissue samples were strikingly consistent with
those obtained with the cell lines. To further explore the
association of EMT-related proteins with RAD18 expression in CRC
tissues at the organizational level, we used RT-PCR to detect the
expression levels of E-cadherin, N-cadherin, vimentin, and
RAD18 mRNAs in 93 CRC clinical samples. The correlation
between the RAD18 and EMT-related protein levels is shown in
Fig. 6C. High expression of RAD18
was accompanied by a high expression of N-cadherin and vimentin
(P<0.001) and low expression of E-cadherin (P<0.001). These
findings indicate that the activation of EMT could be a vital
pathway by which RAD18 promotes migration and metastasis of CRC
cells.
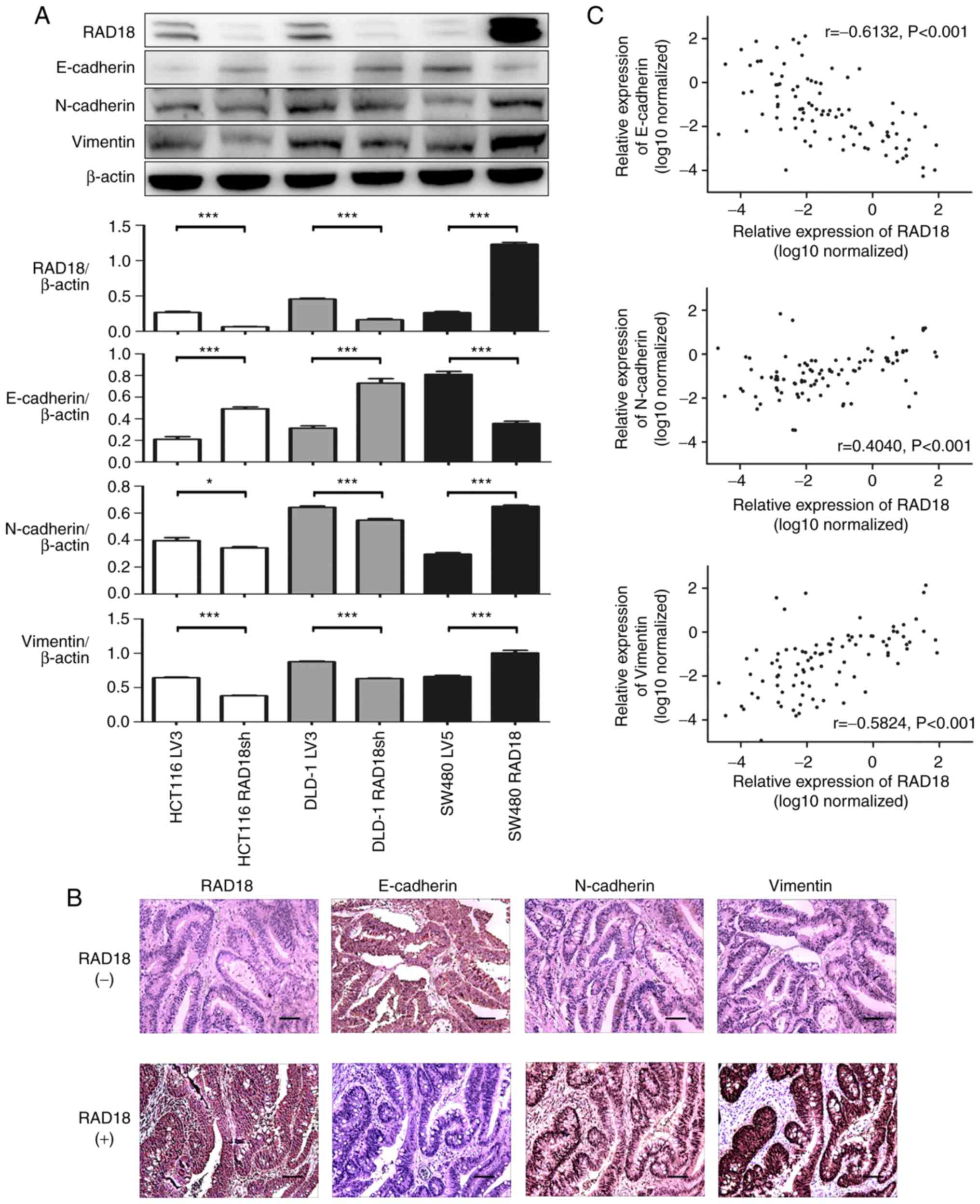 | Figure 6.Overexpression of RAD18 increases the
metastatic potential of CRC cells via the EMT pathway. (A) EMT
biomarkers, including E-cadherin, N-cadherin, vimentin and RAD18
were detected by western blot analysis in HCT116 LV3 cells, HCT116
RAD18sh (RAD18 knockdown), DLD-1 LV3, DLD-1 RAD18sh (RAD18
knockdown), SW480 LV5 and SW480 RAD18 (RAD18-overexpressing) cell
lines. All the experiments were repeated three to four times with
similar findings. Band intensity was quantified by ImageJ software
and are shown by a histogram. *P<0.01, ***P<0.001, Student's
t-test. (B) Immunohistochemical analysis of RAD18, E-cadherin,
N-cadherin and vimentin in CRC tissues. Representative patients,
RAD18 (+) and RAD18 (−) were selected from 93 patients with CRC
(Scale bar, 100 µm; magnification, ×200). (C) E-cadherin,
N-cadherin, vimentin and RAD18 expression was detected in 93 CRC
tissues by PCR. RAD18 expression was positively correlated with
N-cadherin and vimentin expression (P<0.001), but negatively
correlated with E-cadherin expression (P<0.001). EMT,
epithelial-mesenchymal transition; RAD18, E3 ubiquitin protein
ligase; CRC, colorectal cancer. |
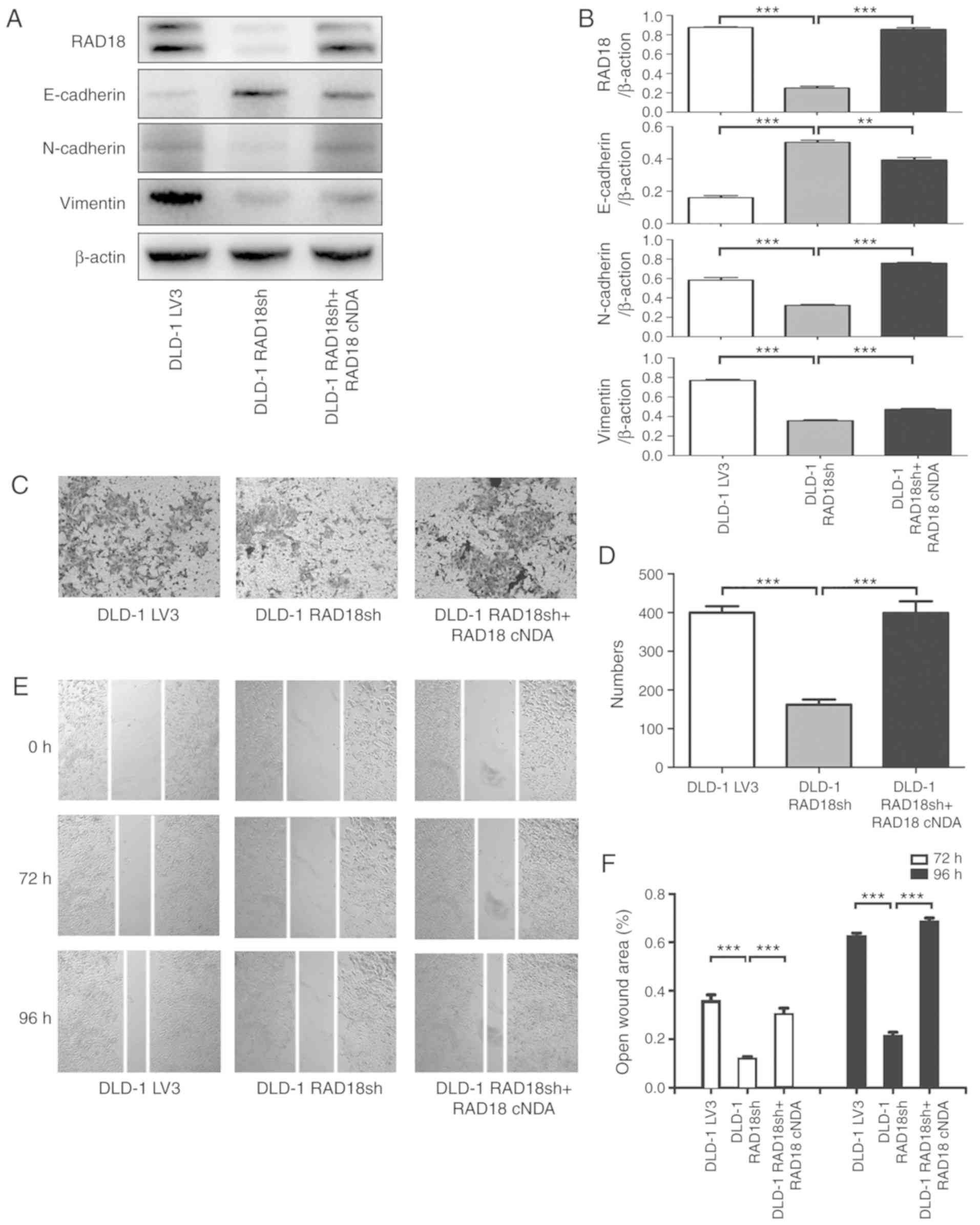
Rescue experiments suggest that the
mutant RAD18 reverses the genetic phenotype of the RAD18-knockdown
cells
After RAD18-silenced DLD-1 RAD18sh cells were
retransfected with the mutated RAD18 gene, the protein expression
of RAD18 was restored, and the expression of EMT-related proteins
was correspondingly reversed (Fig. 7A
and B). Transwell experiment found that the invasion ability of
DLD-1 RAD18sh cells was restored (Fig.
7C and D), and the migration ability of DLD-1 RAD18sh cells was
also restored in the scratch experiment (Fig. 7E and F).
Discussion
Metastasis is one of the main characteristics of
tumors, and most patients with colorectal cancer (CRC) succumb to
the disease due to distant metastasis (19). The transfer of tumor cells from the
primary tumor site to a non-adjacent organ to form a secondary
tumor is a complex multi-step process (20). The first step in tumor cell
metastasis is the infiltration of normal tissues surrounding the
tumor (20). Cancer migration and
invasion are the major factors that determine metastasis (21). Therefore, effective inhibition of
migration and invasion of tumor cells is crucial to the control of
metastasis in CRC. Although a series of intracellular and
extracellular protein biomarkers for CRC have been identified as
potential prognostic and predictive markers by various methods
(22–29), the conversion of the growing
differential proteome data into a database that could be used as a
clinical tool to predict the patient prognosis is still lacking
(30). Thus, it is imperative to
identify more effective biomarkers that can be used for the
reliable prediction of metastasis in CRC.
RAD18 is an E3 ubiquitin ligase that plays a key
role in promoting PCNA mono-ubiquitination. It was reported to be
involved in carcinogenesis and tumor progression owing to its role
in error-prone DNA synthesis. High expression of RAD18 promotes
melanoma cell proliferation (7).
Low expression of RAD18 inhibits glioblastoma development (8). Previously, our data demonstrated that
RAD18 is a cancer-promoting gene for metastatic esophageal squamous
cell cancer (10). In the present
study, we found that RAD18 expression levels were significantly
increased in CRC tissues compared with that noted in the adjacent
non-cancerous normal tissues. The level of RAD18 expression was
also found to be positively associated with lymph node metastasis
and poor prognosis in patients with CRC. Consistent with these
findings, we additionally found that RAD18 promotes mobility and
invasiveness of CRC cells in well-established cell model systems.
However, to elucidate whether this result is due to RAD18, we
further carried out a rescue experiment. The experimental results
showed that the invasion and migration ability of DLD-1 cells were
weakened after downregulation, and the invasion and migration
ability of the cells were restored after the RAD18-c plasmid
was again transfected into the cells. Meanwhile, the expected
synchronous changes were also found in EMT-related proteins. Hence,
RAD18 may play a crucial role in the migration and invasion of CRC
cells.
Epithelial-mesenchymal transition (EMT) is generally
considered to be the first step in cancer metastasis because it
promotes the migration of the tumor cells from the original site to
the tumor stroma. One of the hallmarks of EMT, which is essential
for this process to occur, is the cadherin switch, where E-cadherin
is downregulated, and N-cadherin is upregulated (31–33).
Current studies have shown that EMT in CRC cells is a key factor in
distant metastasis of colorectal cancer (12). In the CRC cell line, we observed
that an increase in RAD18 expression was associated with reduced
E-cadherin expression and increased N-cadherin and vimentin
expression. Consistent with this observation, our clinical data
also demonstrated that the expression of RAD18 affected the
expression of the EMT markers. Therefore, the EMT signaling pathway
could be the molecular mechanism by which RAD18-mediated CRC cell
metastasis takes place.
RAD18 has been found to actively promote migration
and invasion of CRC cells by activating the EMT signaling pathway,
but the exact mechanism remains unclear. Therefore, in future
studies, we will further establish a nude mouse model of CRC
metastasis through caudal vein injection and demonstrate that RAD18
promotes metastasis of colorectal cancer cells to liver, lung and
other organs. At the same time, the collected blood and tissue from
mice will be used to explore and identify the molecular mechanisms
linking RAD18 and the EMT pathway to other signaling pathways that
may contribute to the migration and invasion of CRC cells. Based on
available data, we believe that RAD18 could play a crucial role in
a subset of patients with metastatic tumors and advanced stage
disease. RAD18 may also be a potential therapeutic target for
treating CRC patients.
This research is the first to report that RAD18
promotes the metastasis of CRC. We also demonstrated that the EMT
signaling pathway plays a vital role in RAD18-mediated metastasis.
These conclusions suggest that RAD18 is an essential biomarker for
distant metastasis of CRC, and further studies should aim at
exploring its use for the diagnosis and treatment of metastatic
CRC.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (81672975), and the Six Talent Peaks
Project of Jiangsu Province of China (WSN095).
Availability of data and materials
The data used to support the results of this study
are included in this article.
Authors' contributions
PL conducted most of the experiments and drafted the
manuscript. CH and AG provided guidance and assistance with
conduction of the experiments. XY analyzed the data and plotted the
charts. XX performed the statistical analysis. JZ and JW designed
the research, and reviewed the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Nanjing Medical University, and all patients signed an informed
consent before surgery. The research was conducted following the
Helsinki Declaration of the World Medical Association.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu R, Li J, Xie K, Zhang T, Lei Y, Chen
Y, Zhang L, Huang K, Wang K, Wu H, et al: FGFR4 promotes
stroma-induced epithelial-to-mesenchymal transition in colorectal
cancer. Cancer Res. 73:5926–5935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ting L, Jun H and Junjie C: RAD18 lives a
double life: Its implication in DNA double-strand break repair. DNA
Repair (Amst). 9:1241–1248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu B, Wang H, Zhang L, Sun C, Li H, Jiang
C and Liu X: High expression of RAD18 in glioma induces
radiotherapy resistance via down-regulating P53 expression. Biomed
Pharmacother. 112:1085552019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Y, Gao Y, Zlatanou A, Tateishi S,
Yurchenko V, Rogozin IB and Vaziri C: Diverse roles of RAD18 and
Y-family DNA polymerases in tumorigenesis. Cell Cycle. 17:833–843.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li M, Larsen L and Hedglin M: Rad6/Rad18
competes with DNA polymerases η and δ for PCNA encircling DNA.
Biochemistry. 59:407–416. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wong RP, Aguissa-Tourè AH, Wani AA,
Khosravi S, Martinka M, Martinka M and Li G: Elevated expression of
Rad18 regulates melanoma cell proliferation. Pigment Cell Melanoma
Res. 25:213–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie C, Lu D, Xu M, Qu Z, Zhang W and Wang
H: Knockdown of RAD18 inhibits glioblastoma development. J Cell
Physiol. 234:21100–21112. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lou P, Zou S, Shang Z, He C, Gao A, Hou S
and Zhou J: RAD18 contributes to the migration and invasion of
human cervical cancer cells via the interleukin-1β pathway. Mol Med
Rep. 20:3415–3423. 2019.PubMed/NCBI
|
|
10
|
Zou S, Yang J, Guo J, Su Y, He C, Wu J, Yu
L, Ding WQ and Zhou J: RAD18 promotes the migration and invasion of
esophageal squamous cell cancer via the JNK-MMPs pathway. Cancer
Lett. 417:65–74. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan X, Chen J, Meng Y, He C, Zou S, Li P,
Chen M, Wu J, Ding WQ and Zhou J: RAD18 may function as a predictor
of response to preoperative concurrent chemoradiotherapy in
patients with locally advanced rectal cancer through
caspase-9-caspase-3-dependent apoptotic pathway. Cancer Med.
8:3094–3104. 2019.PubMed/NCBI
|
|
12
|
Loboda A, Nebozhyn MV, Watters JW, Buser
CA, Shaw PM, Huang PS, Van't Veer L, Tollenaar RA, Jackson DB,
Agrawal D, et al: EMT is the dominant program in human colon
cancer. BMC Med Genomics. 4:92011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu J, Li D, Zeng Y, Wang H, Feng W, Qi S
and Yu L: IDH1 mutation promotes proliferation and migration of
glioma cells via EMT induction. J BUON. 24:2458–2464.
2019.PubMed/NCBI
|
|
14
|
Wu K, Li L, Li L and Wang D: Long
non-coding RNA HAL suppresses the migration and invasion of serous
ovarian cancer by inhibiting EMT signaling pathway. Biosci Rep.
40(pii): BSR201944962020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schulz A, Gorodetska I, Behrendt R,
Fuessel S, Erdmann K, Foerster S, Datta K, Mayr T, Dubrovska A and
Muders MH: Linking NRP2 With EMT and Chemoradioresistance in
bladder cancer. Front Oncol. 9:14612020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou J, Zhang S, Xie L, Liu P, Xie F, Wu
J, Cao J and Ding WQ: Overexpression of DNA polymerase iota (Polι)
in esophageal squamous cell carcinoma. Cancer Sci. 103:1574–1579.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou S, Shang ZF, Liu B, Zhang S, Wu J,
Huang M, Ding WQ and Zhou J: DNA polymerase iota (Pol ι) promotes
invasion and metastasis of esophageal squamous cell carcinoma.
Oncotarget. 7:32274–32285. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kho DH, Bae JA, Lee JH, Cho HJ, Cho SH,
Lee JH, Seo YW, Ahn KY, Chung IJ and Kim KK: KITENIN recruits
Dishevelled/PKC delta to form a functional complex and controls the
migration and invasiveness of colorectal cancer cells. Gut.
58:509–519. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Williams CS, Zhang B, Smith JJ, Jayagopal
A, Barrett CW, Pino C, Russ P, Presley SH, Peng D, Rosenblatt DO,
et al: BVES regulates EMT in human corneal and colon cancer cells
and is silenced via promoter methylation in human colorectal
carcinoma. J Clin Invest. 121:4056–4069. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jackstadt R, Röh S, Neumann J, Jung P,
Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A
and Hermeking H: AP4 is a mediator of epithelial-mesenchymal
transition and metastasis in colorectal cancer. J Exp Med.
210:1331–1350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Libanje F, Raingeaud J, Luan R, Thomas Z,
Zajac O, Veiga J, Marisa L, Adam J, Boige V, Malka D, et al: ROCK2
inhibition triggers the collective invasion of colorectal
adenocarcinomas. EMBO J. 38:e992992019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stiegelbauer V, Vychytilova-Faltejskova P,
Karbiener M, Pehserl AM, Reicher A, Resel M, Heitzer E, Ivan C
Bullock M, Ling H, et al: miR-196b-5p regulates colorectal cancer
cell migration and metastases through interaction with HOXB7 and
GALNT5. Clin Cancer Res. 23:5255–5266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kahlert C, Lahes S, Radhakrishnan P, Dutta
S, Mogler C, Herpel E, Brand K, Steinert G, Schneider M,
Mollenhauer M, et al: Overexpression of ZEB2 at the invasion front
of colorectal cancer is an independent prognostic marker and
regulates tumor invasion in vitro. Clin Cancer Res. 17:7654–7663.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang AD, Fan F, Camp ER, van Buren G, Liu
W, Somcio R, Gray MJ, Cheng H, Hoff PM and Ellis LM: Chronic
oxaliplatin resistance induces epithelial-to-mesenchymal transition
in colorectal cancer cell lines. Clin Cancer Res. 12:4147–4153.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kumara HM, Feingold D, Kalady M, Dujovny
N, Senagore A, Hyman N, Cekic V and Whelan RL: Colorectal resection
is associated with persistent proangiogenic plasma protein changes:
Postoperative plasma stimulates in vitro endothelial cell growth,
migration, and invasion. Ann Surg. 249:973–977. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Belov L, Zhou J and Christopherson RI:
Cell surface markers in colorectal cancer prognosis. Int J Mol Sci.
12:78–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Castagnola P and Giaretti W: Mutant KRAS,
chromosomal instability and prognosis in colorectal cancer. Biochim
Biophys Acta. 1756:115–125. 2005.PubMed/NCBI
|
|
32
|
Tokarz P and Blasiak J: The role of
microRNA in metastatic colorectal cancer and its significance in
cancer prognosis and treatment. Acta Biochim Pol. 59:467–674. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|