Introduction
DNA replication in mammalian cells is accurately
controlled by a number of protein factors. The initiation of DNA
replication that takes place at replication origins is coordinately
controlled by multiple proteins, including ORC, CDC6, CDC45, CDKs,
CTD1, GINS, and MCM2-7 helicase complex (1,2).
Loading of the replicative MCM2-7 helicase complex on the
replication initiation sites is considered to be of primary
importance (3). Recently, a
molecular model for the formation of stable double hexamers at
replication origins has been proposed (4). The replicative helicase is not only
required for DNA unwinding but also for tethering DNA primase to
synthesize short RNA primers for DNA chain elongation on the
lagging strand (5). In yeast cells,
among this helicase complex, minichromosome maintenance 4 (Mcm4),
co-operating with Sld3 and Dbf4, plays an essential role in the
regulation of origin firing and replication fork progression
(6). A mutation in the MCM4
gene has been reported in mammary adenocarcinomas in mice (7). The G486D mutation in the MCM4 protein
affects formation of the MCM2/4/6/7 complex, and that could cause
the generation of human cancer (8).
Moreover, whole genome sequencing of human thymic adenocarcinoma
revealed that a complex chromosomal rearrangement in chromosome 8
caused fusion of the MCM4 and SNTB1 genes (9). These lines of evidence indicate that
dysregulation of the MCM4 function may be deleterious for control
of the initiation of DNA replication.
Recent studies on molecular structure revealed that
the MCM2-7 hexamer physically interacts with ORC-Cdc6 and Ctd1
proteins to be loaded onto the replication initiation site in yeast
(10,11). Moreover, it has been reported that
phosphorylation and SUMOylation of MCM4 regulate the accurate
initiation of replication (12–14).
Although the structure and functions of MCM4 have been studied, its
mechanism of gene expression has not been revealed. Surveillance of
the human genomic DNA database indicated that the MCM4 gene
is head-head bound with the protein kinase, DNA-activated,
catalytic subunit (PRKDC) gene, which encodes DNA-PKcs
(15). In the present study, a
luciferase (Luc) expression plasmid containing 309 bp of the
5′-upstream end of the human MCM4 gene was constructed. The
transfection and Luc reporter assay revealed that the 309-bp
fragment functioned as a promoter and responded to
trans-resveratrol (Rsv) both in HeLa S3 and HL-60 cells. A
natural polyphenolic compound, Rsv, which is known to stimulate
NAD+-dependent deacetylase sirtuin and lengthen the
lifespan of model animals, upregulates the expression of the DNA
repair-associated genes (16). For
example, expression of the TP53 and HELB genes, which
encode tumor suppressor p53 (17)
and RecD-like DNA helicase HDHB (18), respectively, are induced by Rsv in
HeLa S3 cells, and notably, a duplicated GGAA motif is present in
the 5′ upstream end of these two genes (19,20).
In contrast, in the human TERT and WRN gene promoter
regions, a GC-box has been identified as a common Rsv-responsive
element (21).
In the present study, deletion and point mutations
on the GGAA motif and the GC-box markedly decreased MCM4
promoter activity and its response to Rsv both in HeLa S3 and HL-60
cells. Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting revealed that the MCM4 gene
transcripts and its encoding protein accumulated in HeLa S3 cells.
Electrophoretic mobility shift assay (EMSA) with various antibodies
revealed that PU.1 (SPI1) and Sp1 bind to the Rsv-responsive
sequence. Collectively, the findings indicated that the GGAA motif
and the GC-box are essential for the control of MCM4 gene
expression in response to Rsv treatment of HeLa S3 cells.
Materials and methods
Materials
trans-Resveratrol (Rsv) (cat. no.
CAS501-36-0) was purchased from Cayman Chemical (19,20).
Cells and cell culture
Human cervical carcinoma (HeLa S3) cells (19,20)
and human promyelotic leukemia (HL-60) cells (22) were grown in Dulbecco's modified
Eagle's medium (DMEM) and RPMI-1640 medium (Nacalai Tesque, Inc.),
respectively, supplemented with 10% fetal bovine serum (FBS)
(Biosera) and penicillin-streptomycin at 37°C in a humidified
atmosphere with 5% CO2.
Construction of Luc reporter
plasmids
The Luc reporter plasmids, carrying 309 bp, which
contains both transcription start sites (TSSs) of the human
MCM4 and PRKDC genes, were constructed by the slight
modification of a previously described procedure (19–22).
Briefly, polymerase chain reaction (PCR) was performed with the
hPRKDC-0028/AhPRKDC-0336 primer pair (Table I) and genomic DNAs that were
extracted from HeLa S3 cells. The amplified DNA fragment was
treated with HindIII and then ligated into the multi-cloning
site of pGL4.10[luc2] (Promega Corporation). The resultant
plasmids, containing the 309-bp fragment in correct and reverse
orientations, were named pGL4-MCM4-309 and pGL4-PRKDC-309,
respectively. Similarly, other Luc reporter plasmids were
constructed by ligating a PCR-amplified DNA fragment into the
KpnI/XhoI site of pGL4.10[luc2]. The sense and
anti-sense primers used for the amplification of the DNA fragments
are presented in Table I.
Nucleotide sequences were confirmed by a DNA sequencing service
(FASMAC; Greiner Japan, Inc.) with primers Rv
(TAGCAAAATAGGCTGTCCCC) and GL (CTTTATGTTTTTGGCGTCTTCC). The Luc
reporter plasmids pGL4-PIF1, pGL4-TP53-551, pGL4-RB1, and
pGL4-CDKN1A (pGL4-p21) were constructed as previously described
(19,22,23).
 | Table I.Primer pairs used for amplifying 5′
upstream regions of the human MCM4 gene. |
Table I.
Primer pairs used for amplifying 5′
upstream regions of the human MCM4 gene.
| Luc plasmid | Primer | Sequence (5′ to
3′) |
|---|
| pGL4-MCM4-309 | AhPRKDC-0028 |
ATTAAGCTTGATGACCGGCCAGGGCAGCAC |
|
| hPRKDC-0336 |
GGGAAGCTTAGCCACCCAAACTACCTCCGC |
| pGL4-MCM4-d1 | hMCM4-0088 |
ATTGGTACCCAGCAGGGAGCAACGCACACC |
|
| AhMCM4-0336 |
ATTGGTACCCAGCAGGGAGCAACGCACACC |
| pGL4-MCM4-d2 | hMCM4-0159 |
ATTGGTACCTCGGCCCGGACCCGGAAATGC |
|
| AhMCM4-0336 |
ACGCTCGAGTAGCCACCCAAACTACCTCCG |
| pGL4-MCM4-d3 | hMCM4-0217 |
ATTGGTACCAGGAACTTTCCCGGGGACCCC |
|
| AhMCM4-0336 |
ACGCTCGAGTAGCCACCCAAACTACCTCCG |
| pGL4-MCM4-d4 | hMCM4-0269 |
ATGGGTACCGCGCCTCTTTGGCCCGAATCA |
|
| AhMCM4-0336 |
ACGCTCGAGTAGCCACCCAAACTACCTCCG |
| pGL4-MCM4-d5 | hMCM4-0028 |
ATTGGTACCTTGATGACCGGCCAGGGCAGC |
|
| AhMCM4-0181 |
ATTCTCGAGGCATTTCCGGGTCCGGGCCGA |
| pGL4-MCM4-d6 | hMCM4-0028 |
ATTGGTACCTTGATGACCGGCCAGGGCAGC |
|
| AhMCM4-0153 |
ATTCTCGAGCACGCGCGGGAGCGGGACTCG |
| pGL4-MCM4-d7WT | hMCM4-0159 |
ATTGGTACCTCGGCCCGGACCCGGAAATGC |
|
| AhMCM4-0204 |
AATCTCGAGCAGCCCCGCCTCCGCGCGTAGGGGCA |
| pGL4-MCM4-d7M1 | hmMCM4-0159 |
ATTGGTACCTCGGCCCGGACCCTTAAATGC |
|
| AhMCM4-0204 |
AATCTCGAGCAGCCCCGCCTCCGCGCGTAGGGGCA |
| pGL4-MCM4-d7M2 | hMCM4-0159 |
ATTGGTACCTCGGCCCGGACCCGGAAATGC |
|
| AhmMCM4-0204 |
AATCTCGAGCAGCACAGCATCCGCGCGTAGGGGCA |
| pGL4-MCM4-d7MM | hmMCM4-0159 |
ATTGGTACCTCGGCCCGGACCCTTAAATGC |
|
| AhmMCM4-0204 |
AATCTCGAGCAGCACAGCATCCGCGCGTAGGGGCA |
Transcription factor binding sequence
analysis
The nucleotide sequence of the cloned 309-bp DNA
fragment was subjected to analysis of human transcription factor
binding elements by JASPAR 2016 (http://jaspar2016.genereg.net/).
Transient transfection and Luc
assay
Luc reporter plasmids were transfected into HeLa S3
or HL-60 cells by the DEAE-dextran method in 96-well plates
(24), and after 24 h of
transfection, the culture medium was changed to Rsv (20 µM)
containing DMEM or RPMI-1640 medium with 10% FBS, respectively.
After a further 24 h of incubation, cells were collected and lysed
with 100 µl of 1X cell culture lysis reagent, containing 25 mM
Tris-phosphate (pH 7.8), 2 mM DTT, 2 mM
1,2-diaminocyclohexane-N,N,N′,N′,-tetraacetic acid, 10% glycerol,
and 1% Triton X-100 and then mixed and centrifuged at 12,000 × g
for 5 sec. The supernatant was stored at −80°C. The Luc assay was
performed with a Luciferase assay system (Promega Corporation), and
relative Luc activities were calculated as previously described
(20,22–25).
Western blot analysis
Cells were collected after Rsv-treatment. They were
lysed in a RIPA buffer [20 mM Tris-HCl (pH 7.4), 0.1% SDS, 1%
Triton X-100, and 1% sodium deoxychlate]. The amount of Protein
amount was analyzed with a protein assay kit (BioRad Laboratories,
Inc.) according to the manufacturer's protocol. After SDS-PAGE (15%
acrylamide) (15 to 25 µg proteins/lane) and blotting onto a PVDF
(Immobilon-P) membrane as previously described (19,20),
Western blot analysis was carried out with antibodies against MCM4
(cat. no. sc-48407; Santa Cruz Biotechnology, Inc.), and β-actin
(cat. no. A5441; Sigma-Aldrich; Merck KGaA) (1:1,000) at 20°C for 1
h, followed by the incubation with horseradish
peroxidase-conjugated anti-rabbit (cat. no. A0545) or anti-mouse
IgG (cat. no. A9917) secondary antibodies (Sigma-Aldrich; Merck
KGaA) (1:10,000) at 20°C for 1 h in a Blocking reagent TBS
containing 0.05% Tween 20 and 0.5% casein. Signal intensities were
detected with ImmunoStar LD (FUJIFILM Wako Pure Chemical
Corporation) and quantified with a ChemiDoc image analysis system
and ImageLab 6.0 software (BioRad Laboratories, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
First-strand cDNAs were synthesized with ReverTra
Ace (Toyobo Life Science), random primers (Takara Bio, Inc.), and
total RNAs extracted from HeLa S3 cells. Real-time PCR analysis was
carried out using a Mx3000P Real-Time qPCR System (Stratagene;
Agilent Technologies, Inc.) (19,20).
For PCR amplification, cDNAs were amplified by Thunderbird Realtime
PCR Master Mix (Toyobo Life Science) and 0.3 µM of each primer
pair. The primer pairs for amplifying the human MCM4 and
GAPDH transcripts were hMCM4-2097:
AGGACTACATTGCCTACGCG/AhMCM4-2216: AAACCATTCCCCGGCTACTG and
hGAPDH556/hGAPDH642 (19,20), respectively. Amplification was
carried out initially for 1 min at 95°C, followed by 40 cycles at
95°C (15 sec) and 58°C (30 sec). Quantitative PCR analysis for each
sample was carried out in triplicate. Relative gene expression
values were obtained by normalizing Cq (quantification
cycle) values of target genes in comparison with Cq
values of the GAPDH gene using the ΔΔCq method
(26).
EMSA
Nuclear extracts were prepared from either mock- or
Rsv (20 µM)-treated cells as previously described (27). The double-stranded DNA probes d7WT,
d7M1, d7M2, and d7MM were obtained by annealing and treating primer
pairs hMCM4-0159/AhMCM4-0204, hmMCM4- 0159/AhMCM4-0204,
hMCM4-0159/AhmMCM4-0204, and hmMCM4-0159/AhmMCM4-0204,
respectively, with T4 polymerase (Table II). Double-stranded d7WT probe
(approximately 0.1 ng) was labeled with digoxigenin (DIG) (Roche
Applied Science), and binding reactions were carried out in a
buffer containing 0.2 mM EDTA, 20% glycerol, 20 mM Hepes-KOH (pH
7.9), 100 mM KCl, 1 mM DTT, 1 mM PMSF, 50 ng/µl of poly (dI-dC),
and 5 ng/µl of poly-L-Lysine at 20°C for 20 min (27). The resulting reaction mixture was
separated by native TBE-PAGE and transferred to a nylon membrane
(PALL Corporation) in 0.5X TBE buffer and UV cross-linked with a
transilluminator. Detection of labeled DNAs was performed with an
alkaline phosphatase-conjugated anti-DIG antibody and CSPD ECL
substrate (Roche Applied Science). Chemiluminescence was detected
by a ChemiDoc image analysis system (BioRad Laboratories, Inc.).
For competition EMSAs (28), a
molar excess of unlabeled competitor probe was included in the
binding reaction, as indicated in the figure legends. For EMSA
supershift analysis, antibodies (1 µl) anti-PU.1, anti-ETS1,
anti-NF-κB (p50), anti-STAT4, anti-IDH1, and anti-Sp1 (cat. nos.
sc-22805, sc-111, sc-8414, sc-485, sc-49996, and sc-59,
respectively; Santa Cruz Biotechnology, Inc.), anti-ELK1 (cat. no.
E3401; Sigma-Aldrich; Merck KGaA), anti-STAT1 (cat. no. 06–501; EMD
Millipore), and anti-KLF4 (cat. no. GTX101508; GeneTex, Inc.) were
added to the reaction mixture, containing nuclear proteins, poly
(dI-dC), and poly-L-Lysine, then incubated at 20°C for 20 min.
Then, DIG-labeled probe was added to start the binding
reaction.
 | Table II.The double-stranded oligonucleotides
used for EMSA. |
Table II.
The double-stranded oligonucleotides
used for EMSA.
| Name | Sequence |
|---|
| d7WT |
5-attggtacCTCGGCCCGGACCCGGAAATGCCCCTACGCGCGGAGGCGGGGCTGCtcgagatt-3 |
|
|
3-taaccatgGAGCCGGGCCTGGGCCTTTACGGGGATGCGCGCCTCCGCCCCGACGagctctaa-5 |
| d7M1 |
5-attggtacCTCGGCCCGGACCCTTAAATGCCCCTACGCGCGGAGGCGGGGCTGCtcgagatt-3 |
|
|
3-taaccatgGAGCCGGGCCTGGGAATTTACGGGGATGCGCGCCTCCGCCCCGACGagctctaa-5 |
| d7M2 |
5-attggtacCTCGGCCCGGACCCGGAAATGCCCCTACGCGCGGATGCTGTGCTGCtcgagatt-3 |
|
|
3-taaccatgGAGCCGGGCCTGGGCCTTTACGGGGATGCGCGCCTACGACACGACGagctctaa-5 |
| d7MM |
5-attggtacCTCGGCCCGGACCCTTAAATGCCCCTACGCGCGGATGCTGTGCTGCtcgagatt-3 |
|
|
3-taaccatgGAGCCGGGCCTGGGAATTTACGGGGATGCGCGCCTACGACACGACGagctctaa-5 |
Statistical analysis
Standard deviations (SD) for each data were
calculated and results are presented as the means ± SD from three
independent experiments. Statistical analysis for data in Figs. 1 and 3 was performed with the Student's
t-test (*P<0.05 and **P<0.01, as indicated in the
figures and legends, were considered to indicate statistically
significant differences).
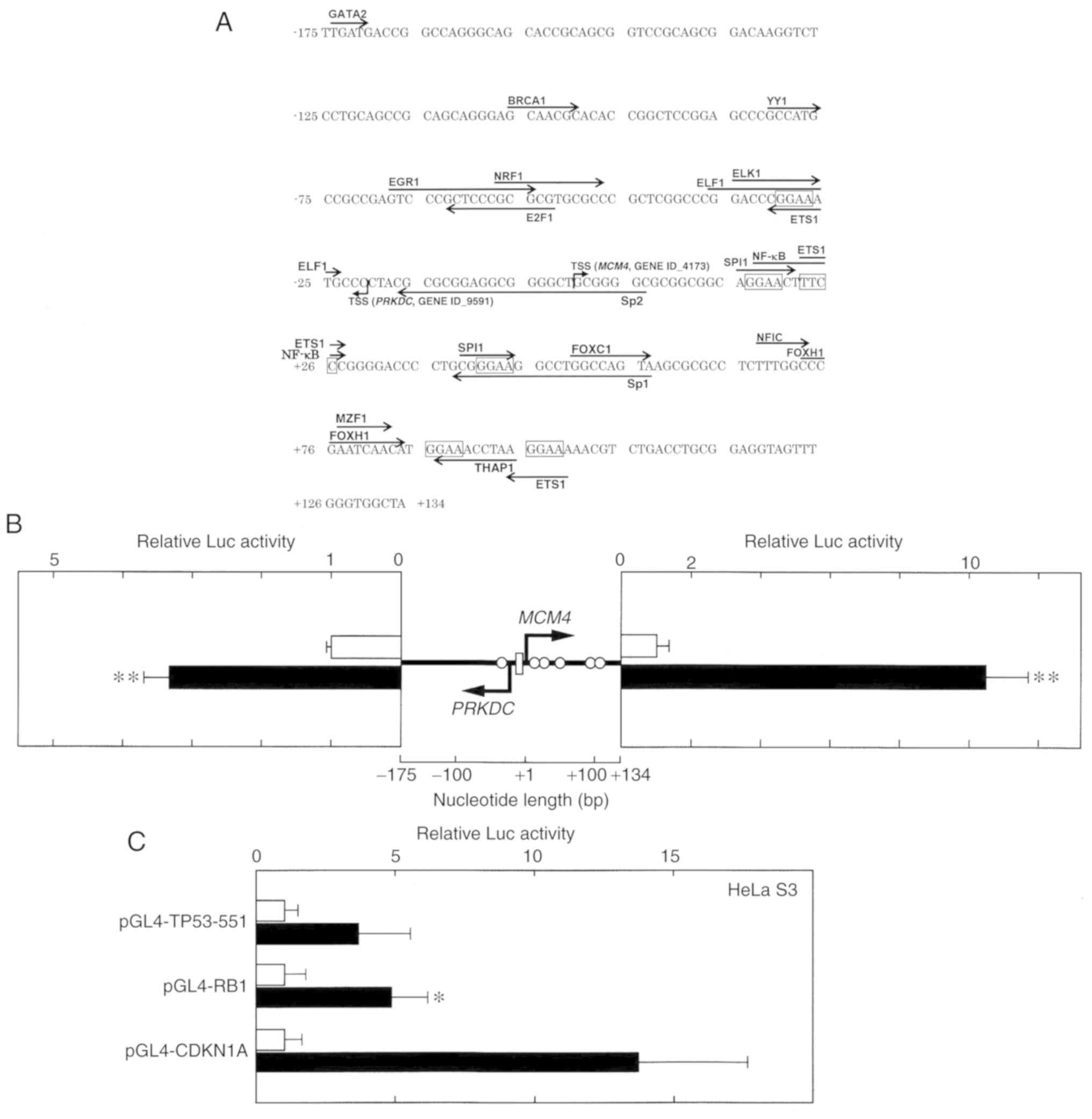 | Figure 1.Characterization of the human
MCM4/PRKDC bi-directional promoter region. (A) The
nucleotide sequence of the 309-bp fragment that was obtained from
PCR is presented. The most upstream 5′ end of the human MCM4
(NM_005914.3 and NM_182746.2) and PRKDC (NM_001081640.1 and
NM_006904.6) cDNAs are designated transcription start sites (TSSs).
Putative transcription factor-binding sites (JASPAR database
program, threshold >90%) are indicated by arrows. (B) The 309-bp
fragment, which contained both TSSs of the MCM4 and the
PRKDC genes, is schematically presented (center). Open
circles and a rectangle represent GGAA (TTCC) motifs and a GC-box,
respectively. The luciferase (Luc) reporter plasmids pGL4-PRKDC-309
(left) or pGL4-MCM4-309 (right) were transfected into HeLa S3
cells, which were treated with (closed columns) or without (open
columns) Rsv (20 µM) for 24 h. Luc activities were normalized to
that of the pGL4-PIF1-transfected cells. Histograms show relative
Luc activities compared with that of the Rsv non-treated cells. (C)
The Luc reporter plasmids pGL4-TP53-551, pGL4-RB1, and pGL4-CDKN1A
were transfected into HeLa S3 cells, which were treated with or
without Rsv (20 µM) for 24 h. Results show fold activation of the
normalized Luc activities compared with that of Rsv-non-treated
cells. (B and C) Results are presented as the means ± SD from at
least three independent experiments. Statistical analysis for the
results between Rsv-treated and non-treated cells was performed
with the Student's t-test. *P<0.05. TSSs, transcription start
sites; Rsv, trans-resveratrol. MCM4, minichromosome
maintenance 4; PRKDC, protein kinase, DNA-activated,
catalytic subunit. |
Results
Isolation and characterization of the
human MCM4/PRKDC bi-directional promoter region
It has been revealed that GGAA duplex-containing
human DNA repair-associated gene promoters, including the
HELB promoter, respond to Rsv, which upregulates the
NAD+/NADPH ratio in HeLa S3 cells (29). HELB associates with CDC45 that
interacts with MCM helicase to construct the CMG (CDC45-MCM-GINS)
complex. On the basis of this background, it was hypothesized that
MCM promoter would respond to Rsv in concert with the
HELB promoter. First, the 309-bp fragment of the
bi-directional MCM4/PRKDC promoter region (30) was amplified and isolated by PCR.
Sequence analysis revealed that the pGL4-MCM4-309 and
pGL4-PRKDC-309 plasmids contain a nucleotide identical to NCBI
Sequence IDs NC_018919.2 (nucleotide from 48924950 to 48925258) and
NC_000008.11 (nucleotide from 47960028 to 47960336) and that it
covers the sequence of the most upstream 5′ end of the cDNA
(Sequence IDs: NM_005914.3 and NM_182746.2 for the variants 1 and 2
of MCM4, respectively; GENE ID, MCM4: 4173). This 309-bp
region also contains a 5′ upstream end of variants 1 and 2 of the
PRDKC mRNA (Sequence ID: NM_006904.6 and NM_001081640.1,
respectively; GENE ID, PRKDC: 5591) in a reverse orientation to
that of the MCM4 gene. The TSS was tentatively set as +1 at
the most upstream 5′ end of the MCM4 transcripts shown in
the human genomic DNA database. The JASPAR 2016 database program
(http://jaspar2016.genereg.net/)
indicated that the characteristic recognition sequences of several
known transcription factors are contained (Fig. 1A). Although no evident sequences
similar to the TATA or CCAAT boxes were found, putative binding
sites for GATA2 (−174 to −171), BRCA1 (−106 to −100), YY1 (−81 to
−76), ERG1 (−68 to −55), E2F1 (−63 to −53), NRF1 (−57 to −47), ELF1
(−36 to −24), ELK1 (−35 to −26), ETS1 (−31 to −26), Sp2 (−16 to
+21), SPI1 (+16 to +21, +39 to +44), NF-κB (+18 to +27), Sp1 (+38
to +57), FOXC1 (+50 to +57), NFIC (+68 to +73), FOXC1 (+73 to +84),
MZF1 (+77 to +82), and THAP1 (+87 to +95) were contained in the
309-bp region. To examine whether the isolated DNA fragment
contains functional promoter activity, Luc reporter plasmids
pGL4-PRKDC-309 and pGL4-MCM4-309 were transiently transfected into
HeLa S3 cells. The relative Luc activities of the pGL4-PRKDC-309-
(Fig. 1B, left panel) and
pGL4-MCM4-309-transfected cells (Fig.
1B, right panel) increased after the addition of Rsv to the
cell culture. It has been observed that HeLa S3 cells are not
killed or not induced to proliferate with 20 µM of the Rsv
treatment, and the activation of the human TP53 gene
promoter was most prominent with the concentration (20). Based on the observation, the
experimental condition for HeLa S3 cells was set as 20 µM. The
upregulation of Luc activities in response to Rsv was significantly
greater in the pGL4-MCM4-309-transfected cells than in the
pGL4-PRKDC-309-transfected cells. The MCM4 gene/protein
expression and promoter activity was further examined. In this
experimental setting, the duplicated GGAA motif containing
promoters of the human RB1 and CDKN1A (p21) genes
responded positively to Rsv (Fig.
1C).
Effects of Rsv on MCM4 gene expression
and its protein amount in HeLa S3 cells
Next, total RNAs were extracted from cells after
adding Rsv to the culture medium, and RT-qPCR was carried out
(Fig. 2A). Since apparent
up-/down-regulation of the expression of the GAPDH in HeLa
S3 cells in response to trans-resveratrol (Rsv) (20) has not been observed, this gene was
used as a control for the RT-qPCR experiment. The relative gene
expression of MCM4 compared with that of GAPDH began
to increase at 2 h after Rsv treatment and then reached a plateau.
Western blot analysis revealed that the amount of MCM4 protein
peaked at 24 h after the treatment (Fig. 2B). The slight decrease at 32 h may
have been caused by degradation of the MCM4 protein, non-coding
regulatory RNAs, or another post-transcriptional regulation in HeLa
S3 cells. However, after a further 12 h of incubation it increased
again.
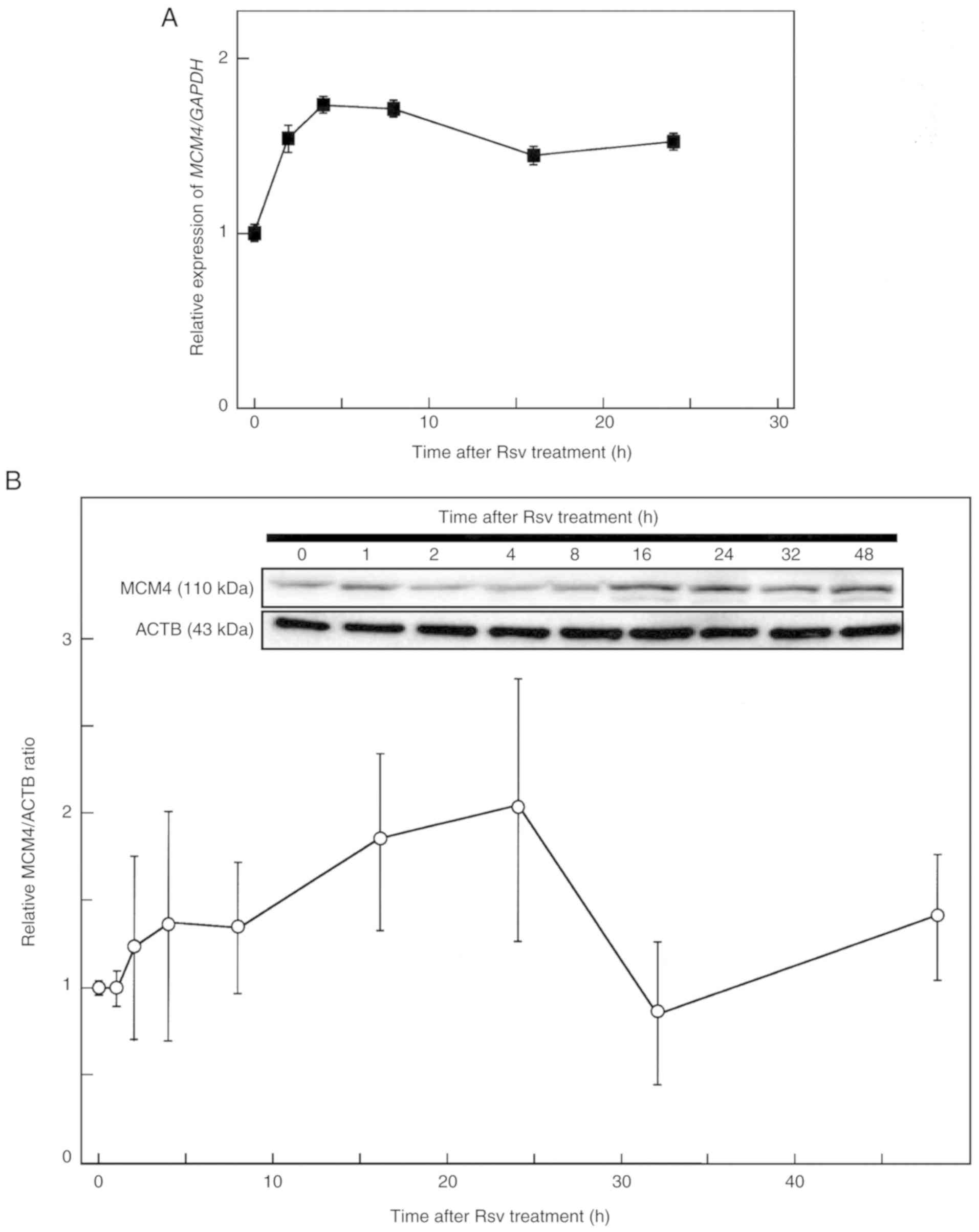 | Figure 2.Effects of Rsv on MCM4 gene
and protein expression. (A) The culture medium of HeLa S3 cells was
changed to DMEM (containing 10% FBS) with 20 µM of Rsv. Cells were
harvested after 0, 2, 4, 8, 16, and 24 h of treatment. Total RNAs
were extracted from cells, and synthesized cDNAs were subjected to
real-time quantitative PCR with primer pairs to amplify MCM4
(upper panel) and GAPDH (lower panel) transcripts. The results
revealed the relative MCM4/GAPDH gene expression ratio
compared with that of Rsv non-treated cells. Results are presented
as the means ± SD from at least three independent experiments. (B)
HeLa S3 cells were collected after 0, 1, 2, 4, 8, 16, 24, 32, and
48 h of Rsv (20 µM) treatment. The extracted proteins were
separated by a 15% SDS-PAGE, and western blotting was performed
with primary antibodies against MCM4 and ACTB (β-actin) (upper and
lower rows, respectively). The signal of each band was quantified,
and the results revealed the relative MCM4/ACTB expression ratio
compared with that of the non-treated control cells (0 h
treatment). Results are presented as the means ± SD from three
independent experiments. Rsv, trans-resveratrol;
MCM4, minichromosome maintenance 4. |
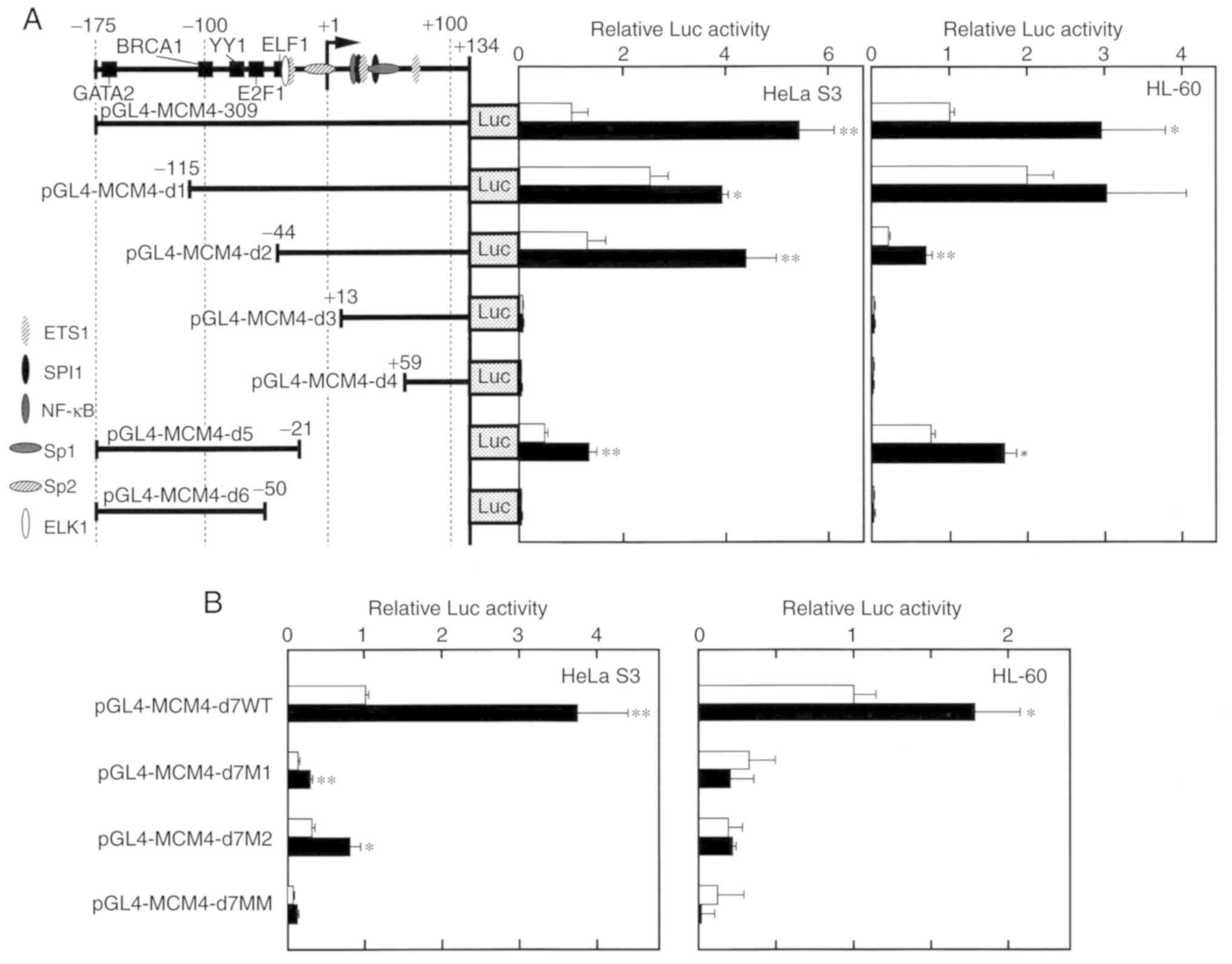
Effect of Rsv on the MCM4 promoter
activity
To narrow the Rsv-responsive sequence, deletion from
the 5′ and 3′ ends of the 309-bp MCM4 promoter region was
introduced into the pGL4-MCM4-309 plasmid (Fig. 3A). The induction by Rsv was observed
in the HeLa S3 and HL-60 cells transfected with pGL4-MCM4-d1 and
d2, but no apparent Luc activity was observed in the cells
transfected with pGL4-MCM4-d3, -d4, and -d6. Comparison of the Luc
activities from the cells transfected with pGL4-MCM4-d2 and -d3
indicated that the 57 nucleotides from −44 to +12 were of primary
importance for MCM4 promoter activity and its positive
response to Rsv. The response was observed in
pGL4-MCM4-d5-transfected cells, indicating that the sequence from
−44 to −21, containing the putative c-ETS binding sequence and
GC-box, was the minimum Rsv responding core element both in HeLa S3
and HL-60 cells. To further examine the contribution of these
cis-elements, point mutations were introduced in the Luc
expression construct pGL4-MCM4-d7WT, containing the nucleotide from
−44 to +2, and a transient transfection experiment was carried out.
Mutations on the c-ETS element and GC-box (in pGL4-MCM4-d7M1 and
-d7M2, respectively) greatly reduced basal promoter activity and
its response to Rsv (Fig. 3B).
Cells that were transfected with pGL4-MCM4-d7MM, carrying double
mutations on both the c-ETS and GC-box elements, also exhibited no
apparent promoter activity or response to Rsv. Collectively, these
results indicated that the MCM4 promoter was co-operatively
regulated by the c-ETS element and GC-box to respond positively to
Rsv in both the HeLa S3 and HL-60 cell lines.
Detection of proteins that bind to the
Rsv response element in the MCM4 promoter
To identify proteins that interact with the Rsv
response element, competition and supershift EMSAs were performed
with HeLa S3 cell nuclear extracts. Incubation of the
double-stranded DNA fragment, containing −44 to +2 of the
MCM4 promoter, with Rsv-non-treated cell nuclear extracts
(Fig. 4A, lane 3) gave rise to
retarded bands, which were increased by the Rsv treatment (lane 2).
The d7WT-protein complexes that were generated by incubation with
Rsv-non-treated cell nuclear extract were reduced by the addition
of non-labeled d7WT but not by d7M1, d7M2, and d7MM probes
(Fig. 4A). This result indicated
that formation of the d7WT-protein complex was dependent on the
c-ETS binding sequence GGAA and Sp1-binding sequence GC-box
(31). The addition of the
anti-PU.1 antibody markedly decreased the formation of the
d7WT-protein complexes −1 and −2, whereas anti-ELK and anti-ETS1
antibodies did not (Fig. 4B, lanes
2–4). This result indicated that PU.1 is contained in the complexes
−1 and −2 that bind to the Rsv response element of the MCM4
promoter. The JASPAR program also predicted that the Rsv-responsive
sequence (−44 to +2) contained the GC-box, indicating that Sp1 was
essentially required for the Rsv response. The d7WT-protein complex
was markedly reduced by the addition of anti-Sp1 or anti-STAT1
antibodies, indicating interactions of Sp1 and STAT1 with the d7WT
probe (Fig. 4B, lane 6).
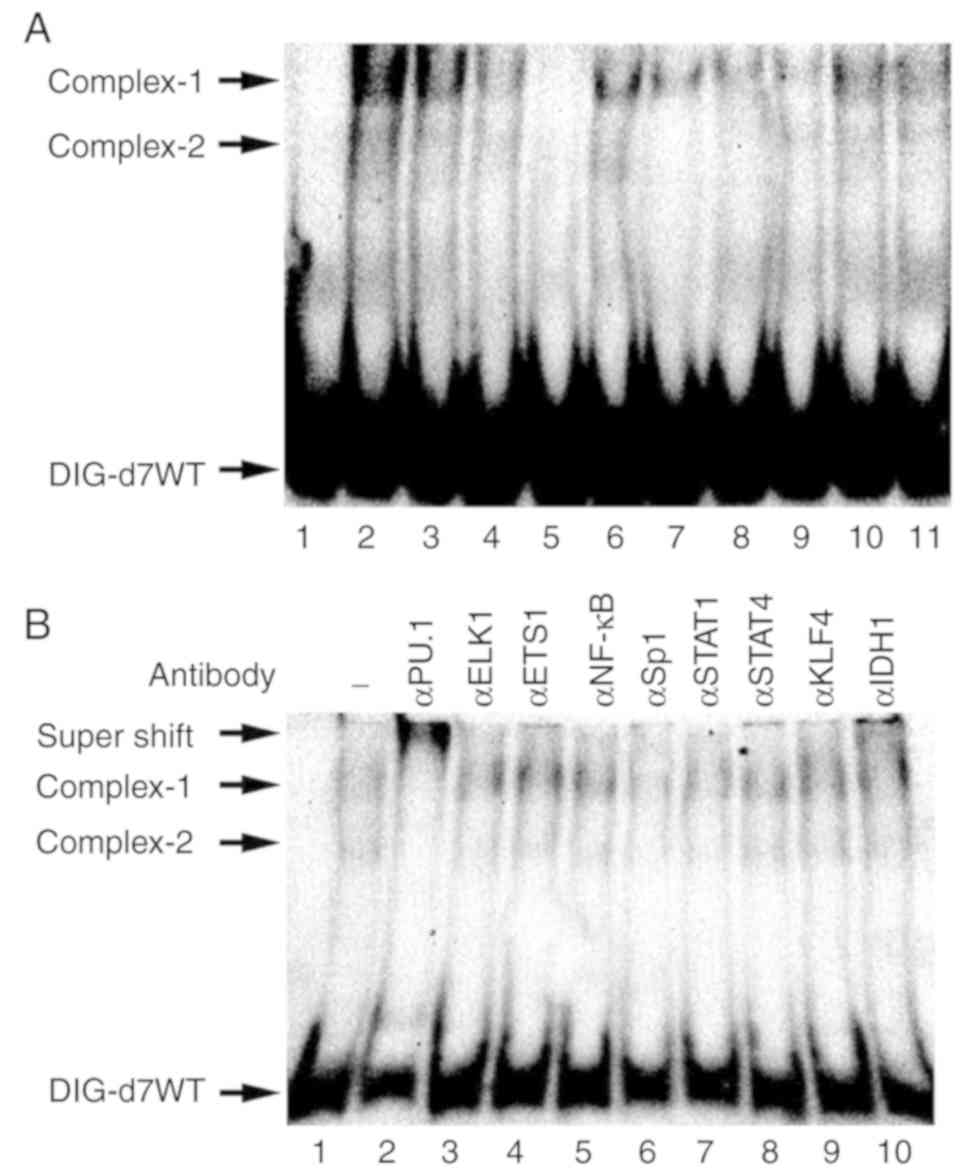 | Figure 4.Sequence-specific DNA-protein complex
formation at the Rsv-responding region of the MCM4 promoter.
(A) Identification of protein-DNA complexes that specifically bind
to the d7WT probe. The sequences of the double-stranded
oligonucleotide probes for EMSA are presented in Table II. Nuclear extracts derived from
HeLa S3 cells, which were either cultured with Rsv (20 µM)
containing DMEM for 24 h (lane 2) or mock stimulated (lanes 3 to
11), were subjected to EMSA with the 3′ end DIG-labeled probe d7WT.
The sequence-specific formations of the complexes were examined by
competition assays with unlabeled specific d7WT (lanes 4 and 5),
and d7M1 (lanes 6 and 7), d7M2 (lanes 8 and 9), and d7MM (lanes 10
and 11) double-stranded probes. The molar excess of unlabeled
competitor was either 5-fold (lanes 4, 6, 8, and 10) or 10-fold
(lanes 5, 7, 9, and 11). (B) Supershift EMSA analysis was performed
with Rsv non-treated HeLa S3 extract and antibodies (1.0 µl)
targeting PU.1, Elk1, ETS1, NF-κB (p50), Sp1, STAT1, STAT4, KLF4,
and IDH1 (lanes 2 to 10, respectively), which were included in the
binding reaction. (A and B) Lane 1 represents a binding reaction
without an antibody. Arrows indicate DIG-labeled d7WT probe,
DNA-protein complexes, and a supershifted complex. Rsv,
trans-resveratrol; EMSA, electrophoretic mobility shift
assay; MCM4, minichromosome maintenance 4. |
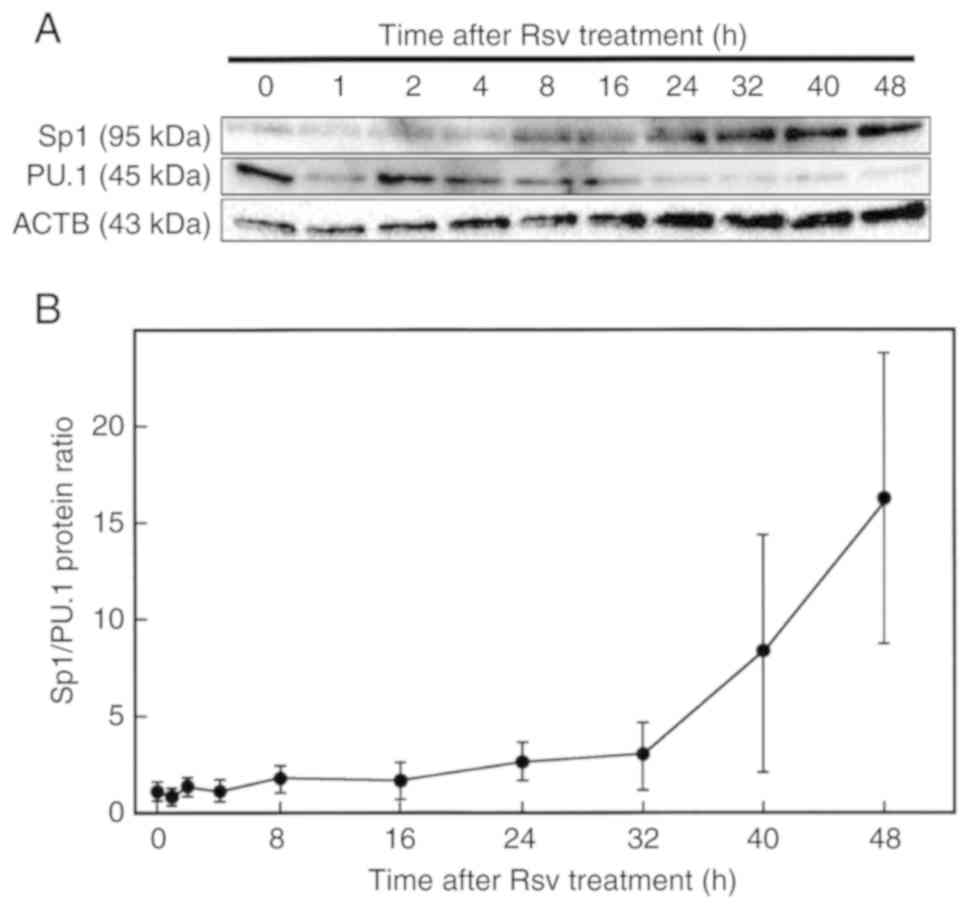
Next, protein amounts after Rsv treatment were
analyzed by western blotting. As revealed in Fig. 5A, an increase of Sp1 and a decrease
of PU.1 were observed. The Sp1/PU.1 ratio was markedly induced 24 h
after the addition of Rsv to the culture medium (Fig. 5B).
Discussion
The present study revealed that treatment with Rsv
(20 µM) induced MCM4 gene and protein expression in HeLa S3
cells. Deletion and mutation analyses revealed that c-Ets and
GC-box elements co-operatively responded to Rsv.
Previously, ChIP (chromatin immunoprecipitation)
analysis of the chicken Mcm4-Prkdc bi-directional
promoter revealed that the c-Myb protein binds to that region
(30). Mutated p53 affects the
amount of MCM4 protein in breast cancer cell lines (32). At present, however, it has not been
elucidated how human MCM4 gene expression is controlled. The
duplicated GGAA (TTCC) motifs are frequently found in the promoter
regions of genes encoding DNA repair and genome maintenance factors
(15,33). The duplicated GGAA motif in the
human TP53 promoter is an essential element that confers
positive response to Rsv in HeLa S3 cells (20). Although the duplicated GGAA (TTCC)
in the 309-bp fragment of the human MCM4 promoter is not
essential for a positive response to Rsv, it was completely
abolished by introduction of mutations on the c-ETS recognition
sequence (−31 to −26) and the GC-box (−10 to −3). Similar results
were observed in the human HELB promoter region (19). We have reported that the GC-box,
which is a target binding sequence motif for Sp1, is commonly
contained in the WRN and TERT promoter regions
(21,23,25).
The Rsv-responsive nucleotide sequence from −35 to −22 in the
MCM4 promoter is 5′-GACCCGGAAATGCC-3′ (Fig. 3B), which can be recognized by Ets
family class IIa proteins, including EHF and ELF1-5 (34). In human cells, co-operative
functioning of the ETS family and Sp1 has been reported in the
PTGIR (35) and PARG
(36) gene promoters. The
duplicated GGAA (TTCC) motif and multiple GC-boxes are present in
the 5′ upstream end of the human TERT gene (21,33).
Notably, mutations on the GGAA (TTCC) motifs or the creation of Ets
binding elements in the TERT promoter are frequently found
in human melanoma (37,38). These observations suggest that
cis-acting functions of the GGAA motifs and GC-boxes
co-operatively regulate promoter activities of DNA
replication/repair factor-encoding genes, including HELB, MCM4,
PRKDC, and TERT, in response to biological stresses.
Moreover, the present study indicated that both Sp1 and PU.1 are
contained in the d7WT-protein complex, which was strengthened by
Rsv treatment. PU.1 can regulate the differentiation and
development of lymphoid cells (39,40),
and it controls fibroblast polarization (41). The induction of PU.1 enforces
differentiation of fibroblasts into a fibrotic phenotype. In the
Rsv-treated HeLa S3 cells, the amount of PU.1 protein was gradually
decreased. PU.1 has both stimulatory and suppressive functions on
gene transcription (42). In the
experimental settings of this study, PU.1 may have acted as a
suppressor for MCM4 gene transcription.
The natural compound Rsv upregulated the expression
of the TP53 and HELB genes and its encoded proteins
in HeLa S3 cells (19,20). The tumor suppressor p53 is a
‘guardian of the genome’ that induces cell cycle regulatory
factor-encoding genes, which regulate cellular senescence,
apoptosis, and autophagy, in response to DNA damage stresses
(17). The human HELB
(HDHB) gene encodes a DNA replication-associated helicase
(18). The dominant negative mutant
HDHB protein, lacking ATPase/helicase activities, inhibited DNA
synthesis when it was micro-injected into the nucleus of cells at
the early G1 phase (18). A recent study indicated that the
recruitment of HELB to sites of DNA double-strand breaks plays a
role in the inhibition of DNA end resection (43). Moreover, the HELB protein has been
revealed to interact with the DNA replication protein factor CDC45
(44). It should be noted that the
MCM complex, whose structure has been recently revealed by
cryo-electron microscopy (45), is
associated with CDC45 and GINS (12,46).
The timing of the CMG complex formation at the origin of
replication should be faithfully limited (47). The 5′ upstream regions of the
RB1 and CDKN1A (p21) genes (22,48),
carrying duplicated GGAA motifs, respond to Rsv in HeLa S3 cells.
These results indicated that the expression of genes encoding p53,
HELB, CDC45, MCM4, RB1, and CDKN1A need to be accurately regulated
before entering the S phase. Additionally, the MCM4 gene has
been revealed to be overexpressed in human cervical (49) and lung (50) cancer cells, suggesting that its
expression should be appropriately controlled. In mice embryo,
genomic instability, which was caused by a deficiency in MCM
complex, triggered an inflammatory response (51). Given that interferon-stimulated
genes are regulated by GGAA motifs (28), the transcription factors, including
PU.1 and Sp1, that regulate MCM4 gene expression may
simultaneously modulate immune responses.
The TP53 gene is inactivated by the human
papillomavirus (HPV) E6 protein (52), and HL-60 cells have large homozygous
deletion of the TP53 gene (53). The p53-deficient HL-60 cells were
selected as well, to examine the effect of Rsv on the MCM4
promoter activity. The results revealed that the MCM4
promoter activation was evident in both cell lines, indicating that
it basically was not dependent on the TP53 gene, whose
mutations are very frequently found in various cancers.
Rsv has an effect on lengthening the life span of
organisms (54,55). Numerous clinical trials suggest that
health-promoting responses, including reduction in the generation
of reactive oxygen species and induction of insulin sensitivity,
are induced by Rsv treatment (56).
Further investigations are required to elucidate the mechanisms by
which Rsv-induced signals regulate DNA
replication/repair-associated gene expression.
Acknowledgements
The authors are grateful to Jun Arakawa, Yutaka
Takihara, and Sakiko Kawahara for their excellent technical
assistance. This study was performed under the permission of
recombinant DNA experimental committee admission no. 1462 of Tokyo
University of Science.
Funding
The present study was supported in part by JSPS
KAKENHI grant no. 24510270 and a Research Fellowship from the
Research Center for RNA Science, RIST, Tokyo University of
Science.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MoA and MK constructed the Luc reporter plasmids.
CK, MoA, MK, and MT performed the experiments and analyzed the data
(transfection assay, RT-PCR, western blotting, and EMSA). FU
interpreted the data and wrote the manuscript. SIT collected and
analyzed/interpreted the data. MaA interpreted the data and edited
the manuscript. All authors have read and approved the final
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CMG complex
|
CDC45-MCM-GINS complex
|
|
CT
|
threshold cycle
|
|
DIG
|
digoxigenin
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
EMSA
|
electric mobility shift assay
|
|
FBS
|
fetal bovine serum
|
|
Luc
|
luciferase
|
|
MCM
|
minichromosome maintenance
|
|
PCR
|
polymerase chain reaction
|
|
Rsv
|
trans-resveratrol
|
|
RT-PCR
|
reverse transcription-polymerase
chain reaction
|
|
RT-qPCR
|
quantitative reverse
transcription-polymerase chain reaction
|
|
TSS
|
transcription start site
|
References
|
1
|
Mcintosh D and Blow JJ: Dormant
replication origins. In: Cold Spring Harbor Perspectives in Biology
‘DNA Replication’. Bell SD, Méchali M and DePamphilis M: Cold
Spring Harbor Laboratory Press; Woodbury, NY: pp. 33–42. 2013
|
|
2
|
Tanaka S and Araki H: Helicase activation
and establishment of replication forks at chromosomal origins of
replication. In: Cold Spring Harbor Perspectives in Biology ‘DNA
Replication’. Bell SD, Méchali M and DePamphilis M: Cold Spring
Harbor Laboratory Press; Woodbury, NY: pp. 81–94. 2013
|
|
3
|
Bell S and Kagni JM: Helicase loading at
chromosomal origins of replication. In: Cold Spring Harbor
Perspectives in Biology ‘DNA Replication’. Bell SD, Méchali M and
DePamphilis M: Cold Spring Harbor Laboratory Press; Woodbury, NY:
pp. 61–80. 2013, PubMed/NCBI
|
|
4
|
Coster G and Diffley JFX: Bidirectional
eukaryotic DNA replication is established by quasi-symmetrical
helicase loading. Science. 357:314–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
You Z, De Falco M, Kamada K, Pisani FM and
Masai H: The mini-chromosome maintenance (Mcm) complexes interact
with DNA polymerase α-primase and stimulate its ability to
synthesize RNA primers. PLoS One. 8:e724082013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sheu Y, Kinney JB and Stillman B:
Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin
activation and DNA replication fork progression. Genome Res.
26:315–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shima N, Alcaraz A, Liachko I, Buske TR,
Andrews CA, Munroe RJ, Hartford SA, Tye BK and Schimenti JC: A
viable allele of Mcm4 causes chromosome instability and mammary
adenocarcinomas in mice. Nat Genet. 39:93–98. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tatsumi R and Ishimi Y: An MCM4 mutation
detected in cancer cells affects MCM4/6/7 complex formation. J
Biochem. 161:259–268. 2017.PubMed/NCBI
|
|
9
|
Lee Y, Park T, Lee S and Lee H:
Characterization of genetic aberrations in a single case of
metastatic thymic adenocarcinoma. BMC Cancer. 17:3302017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhai Y, Cheng E, Wu H, Li N, Yung PY, Gao
N and Tye BK: Open-ringed structure of the Cdt1-Mcm2-7 complex as a
precursor of the MCM double hexamer. Nat Struct Mol Biol.
24:300–310. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan Z, Riera A, Bai L, Sun J, Nandi S,
Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al:
Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6
and Cdt1. Nat Struct Mol Biol. 24:316–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei L and Zhao X: A new MCM modification
cycle regulates DNA replication initiation. Nat Struct Mol Biol.
23:209–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moritani M and Ishimi Y: Inhibition of DNA
binding of MCM2-7 complex by phosphorylation with cyclin-dependent
kinases. J Biochem. 154:363–372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sheu YJ, Kinney JB, Lengronne A, Pasero P
and Stillman B: Domain within the helicase subunit Mcm4 integrates
multiple kinase signals to control DNA replication initiation and
fork progression. Proc Natl Acad Sci USA. 111:E1899–E1908. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Uchiumi F, Larsen S and Tanuma S:
Biological systems that control transcription of DNA repair and
telomere maintenance-associated genes. New Research in Directions
in DNA Repair. Chen C: InTech Open; London: pp. 309–325. 2013
|
|
16
|
Uchiumi F, Arakawa J, Takihara Y, Akui M,
Ishibashi S and Tanuma S: The effect of trans-resveratrol on the
expression of the human DNA-repair associated genes. Int Mol Med.
3:783–792. 2016.
|
|
17
|
Zilfou JT and Lowe SW: Tumor suppressive
functions of p53. Cold Spring Harbor Perspectives in Biology.
Levine AJ and Lane D: Cold Spring Harbor Laboratory Press;
Woodbury, NY: pp. 1–12. 2009
|
|
18
|
Taneja P, Gu J, Peng R, Carrick R, Uchiumi
F, Ott RD, Gustafson E, Podust VN and Fanning E: A
dominant-negative mutant of human DNA helicase B blocks the onset
of chromosomal DNA replication. J Biol Chem. 277:40853–40861. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uchiumi F, Arakawa J, Iwakoshi K,
Ishibashi S and Tanuma S: Characterization of the 5′-flanking
region of the human DNA helicase B (HELB) gene and its response to
trans-Resveratrol. Sci Rep. 6:245102016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uchiumi F, Shoji K, Sasaki Y, Sasaki M,
Sasaki Y, Oyama T, Sugisawa S and Tanuma S: Characterization of the
5′-flanking region of the human TP53 gene and its response to the
natural compound, Resveratrol. J Biochem. 159:437–447. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uchiumi F, Higami Y and Tanuma S:
Regulations of telomerase activity and WRN gene expression.
Telomerase: Composition, Functions and Clinical Implications.
Gagnon AN: Nova Science Publishers Inc; Hauppauge, NY: pp. 95–103.
2010
|
|
22
|
Uchiumi F, Watanabe T and Tanuma S:
Characterization of various promoter regions of the human DNA
helicase-encoding genes and identification of duplicated ets (GGAA)
motifs as an essential transcription regulatory element. Exp Cell
Res. 316:1523–1534. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou B, Ikejima T, Watanabe T, Iwakoshi K,
Idei Y, Tanuma S and Uchiumi F: The effect of 2-deoxy-D-glucose on
Werner syndrome RecQ helicase gene. FEBS Lett. 583:1331–1336. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uchiumi F, Oyama T, Ozaki K and Tanuma S:
Characterization of 5′-flanking regions of various human telomere
maintenance factor-encoding genes. DNA Repair. Kruman I: InTech
Open; London: pp. 585–596. 2011
|
|
25
|
Uchiumi F, Watanabe T, Hasegawa S, Hoshi
T, Higami Y and Tanuma S: The effect of resveratrol on the Werner
Syndrome RecQ helicase gene and telomerase activity. Curr Aging
Sci. 4:1–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uchiumi F, Larsen S and Tanuma S:
Application of DEAE-dextran to an efficient gene transfer system.
Dextran: Chemical Structure, Application and Potential Side
Effects. Figgs GP: Nova Science Publishers Inc.; Hauppauge, NY: pp.
143–156. 2014
|
|
28
|
Larsen S, Kawamoto S, Tanuma S and Uchiumi
F: The hematopoietic regulator, ELF-1, enhances the transcriptional
response to Interferon-β of the OAS1 anti-viral gene. Sci Rep.
5:174972015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takihara Y, Sudo D, Arakawa J, Takahashi
M, Sato A, Tanuma S and Uchiumi F: Nicotinamide adenine
dinucleotide (NAD+) and cell aging. New Research on Cell
Aging and Death. Strakoš R and Lorens B: Nova Science Publishers
Inc.; Hauppauge, NY: pp. 131–158. 2018
|
|
30
|
Gundelach H, Braas D and Klempnauer KH:
The promoter regions of the Myb-regulated Adora2B and Mcm4 genes
co-localize with origins of DNA replication. BMC Mol Biol.
8:752007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wieratra I: Sp1: Emerging roles-Beyond
constitutive activation of TATA-less housekeeping genes. Biochem
Biophys Res Commun. 372:1–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Polotskaia A, Xiao G, Reynoso K, Martin C,
Qiu WG, Hendrickson RC and Bargonetti J: Proteome-wide analysis of
mutant p53 targets in breast cancer identifies new levels of
gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl
Acad Sci USA. 112:E1220–E1229. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Uchiumi F, Larsen S and Tanuma S:
Transcriptional regulation of the human genes that encode DNA
repair- and mitochondrial function-associated proteins. Advances in
DNA Repair. Chen C: InTech Open; London: pp. 129–167. 2015
|
|
34
|
Wei GH, Badis G, Berger MF, Kivioja T,
Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al:
Genome-wide analysis of ETS-family DNA-binding in vitro and in
vivo. EMBO J. 29:2147–2160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Turner EC and Kinsella BT: Transcriptional
regulation of the human prostacyclin receptor gene is dependent on
Sp1, PU.1 and Oct-1 in megakaryocytes and endothelial cells. J Mol
Biol. 386:579–597. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Molloy-Simard V, St-Laurent JF, Vigneault
F, Gaudreault M, Dargis N, Guérin MC, Leclerc S, Morcos M, Black D,
Molgat Y, et al: Altered expression of the poly(ADP-ribosyl)ation
enzymes in uveal melanoma and regulation of PARG gene expression by
the transcription factor ERM. Invest Ophthalmol Vis Sci.
53:6219–6231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin
L and Garraway LA: Highly recurrent TERT promoter mutations in
human melanoma. Science. 339:957–959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Horn S, Figl A, Rachakonda PS, Fischer C,
Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al:
TERT promoter mutations in familial and sporadic melanoma. Science.
339:959–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Carotta S, Wu L and Nutt SL: Surprising
new roles for PU.1 in the adaptive immune response. Immunol Rev.
238:63–75. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rothenberg EV, Hosokawa H and Ungerbäck J:
Mechanisms of action of hematopoietic transcription factor PU.1 in
initiation of T-cell development. Front Immunol. 10:2282019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wohlfahrt T, Rauber S, Uebe S, Luber M,
Soare A, Ekici A, Weber S, Matei AE, Chen CW, Maier C, et al: PU.1
controls fibroblast polarization and tissue fibrosis. Nature.
566:344–349. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hosokawa H, Ungerbäck J, Wang X, Matsumoto
M, Nakayama KI, Cohen SM, Tanaka T and Rothenberg EV: Transcription
factor PU.1 represses and activates gene expression in early T
cells by redirecting partner transcription factor binding.
Immunity. 48:1119–1134 e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tkáč J, Xu G, Adhikary H, Young JTF, Gallo
D, Escribano-Díaz C, Krietsch J, Orthwein A, Munro M, Sol W, et al:
HELB is a feedback inhibitor of DNA end resection. Mol Cell.
61:405–418. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gerhardt J, Guler GD and Fanning E: Human
DNA helicase B interacts with the replication initiation protein
Cdc45 and facilitates Cdc45 binding onto chromatin. Exp Cell Res.
334:283–293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miller TCR, Locke J, Greiwe JF, Diffley
JFX and Costa A: Mechanism of head-to-head MCM double-hexamer
formation revealed by cryo-EM. Nature. 575:704–710. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Araki H: Molecular mechanisms of DNA
replication. DNA Replication, Recombination, and Repair. Hanaoka F
and Sugasawa K: Springer; Japan, Tokyo: pp. 3–22. 2016, View Article : Google Scholar
|
|
47
|
Hayano M, Matsumoto S and Masai H: DNA
replication timing: Temporal and spatial regulation of eukaryotic
DNA replication. DNA Replication, Recombination, and Repair.
Hanaoka F and Sugasawa K: Springer; Japan, Tokyo: pp. 53–69. 2016,
View Article : Google Scholar
|
|
48
|
Deléhouzée S, Yoshioka T, Sawa C, Sawada
J, Ito T, Omori M, Wada T, Yamaguchi Y, Kabe Y and Handa H: GABP,
HCF-1 and YY1 are involved in Rb gene expression during myogenesis.
Genes Cells. 10:717–731. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cheng J, Lu X, Wang J, Zhang H, Duan P and
Li C: Interactome analysis of gene expression profiles of cervical
cancer reveals dysregulated mitotic gene clusters. Am J Transl Res.
9:3048–3059. 2017.PubMed/NCBI
|
|
50
|
Yi J, Wei X, Li X, Wan L, Dong J and Wang
R: A genome-wide comprehensive analysis of alterations in driver
genes in non-small-cell lung cancer. Anticancer Drugs. 29:10–18.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
McNairn AJ, Chuang CH, Bloom JC, Wallace
MD and Schimenti JC: Female-biased embryonic death from
inflammation induced by genomic instability. Nature. 567:105–108.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kajitani K, Honda K, Terada H, Yasui T,
Sumi T, Koyama M and Ishiko O: Human papillomavirus E6 knockdown
restores adenovirus mediated-estrogen response element linked p53
gene transfer in HeLa cells. Asian Pac J Cancer Prev. 16:8239–8245.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wolf D and Rotter V: Major deletions in
the gene encoding the p53 tumor antigen cause lack of p53
expression in HL-60 cells. Proc Natl Acad Sci USA. 82:790–794.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Stefani M, Markus MA, Lin RC, Pinese M,
Dawes IW and Morris BJ: The effect of resveratrol on a cell model
of human aging. Ann NY Acad Sci. 1114:407–418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kaeberlein M: Resveratrol and rapamycin:
Are they anti-aging drugs? Bioessays. 32:96–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vang O: What is new for resveratrol? Is a
new set of recommendations necessary? Ann NY Acad Sci. 1290:1–11.
2013. View Article : Google Scholar : PubMed/NCBI
|