Introduction
Colorectal cancer (CRC) has a high global incidence
and mortality rate (1). Tumor
metastasis is often associated with poor prognosis and is the
primary cause of death in patients with cancer (2). Existing research shows that the tumor
matrix has profound influences on tumor growth and metastasis
(3). As the core constituent of the
tumor matrix, collagen provides a mechanical or signaling support
for tumor growth and metastasis and is related to prognosis
(4,5). Several studies have shown that
collagen is the migratory channel of cancer cells, controlling the
metastasis of various tumor cells (6–8).
Bonnans et al (9) revealed
that dense deposition of COL I may increase the risk of tumor
metastasis and worse prognosis.
Tumor progression is accompanied by an abnormal
remodeling of the matrix collagen. Histologically, abnormal
remodeling of collagen mainly results in excessive deposition,
altered-proportions and changed-arrangement of collagen (10–12).
In normal tissues, the collagen fibers of the tumor matrix are
curly with an irregular arrangement, while in tumor tissues,
especially those with metastasis, the fibers are linearized and
dense with a directional arrangement (13). Tanjore and Kalluri (14) identified numerous hidden
carcinogen-containing domains within collagen that were exposed
following collagen remodeling, and which subsequently facilitated
tumor metastasis.
Collagen remodeling is induced by collagenase. The
most common collagenases are the matrix metalloproteinase (MMP) and
lysine oxidase (LOX) families, which facilitate the degradation or
cross-link of collagen, respectively (15,16).
In various types of cancers, such as breast cancer and
hepatocellular carcinoma, the overproduction of these collagenases
has been found to promote abnormal collagen remodeling (17–19).
The changes in the deposition and the ratio of subtypes of
collagens are also regulated by their associated coding genes. The
mRNA level of these coding genes can affect tumor occurrence and
prognosis (20,21), but their prognostic significance in
CRC is not totally clear.
The tumor invasion front is the area at the edge of
the tumor which represents a critical interface at tumor
progression and tumor cell dissemination (22). Changes in the environment of the
tumor invasion front also affect the behavior of tumor cells and
patient prognosis (23). However,
there are few studies focused on the association between the matrix
collagen at the tumor invasion front and CRC development and
prognosis. Based on these perspectives, the changes in collagen
arrangement and expression at the tumor invasion front were
analyzed in CRC patients with and without metastasis, as well as
the corresponding differences in collagenase expression. In
addition, bioinformatics analysis from multiple biometric databases
was performed to study the expression and prognostic significance
of matrix collagen and collagenase genes in CRC.
Materials and methods
Patients and specimens
A total of 56 patients (age, 26–86 years) undergoing
colonoscopic polypectomy at the First Affiliated Hospital of
Guangzhou University of Chinese Medicine (Guangzhou, China) between
July 2018 and January 2020 were enrolled in the present study.
Patients who had received neoadjuvant radiotherapy, chemotherapy or
chemoradiotherapy were excluded. Clinical and pathological data
including pathology reports, sex, age at surgical intervention,
macroscopic classification, tumor location, tumor size, tumor
differentiation, lymphovascular infiltration and depth of invasion,
were collected from medical records. The American Joint Commission
on Cancer TNM staging system was used to clinical stage the tumors
(24). Tissues from the tumor
invasion front were obtained from the edge of each tumor, and
normal-adjacent tissues were obtained 5.0–10.0 cm away from the
primary tumor. The 112 samples were then divided into metastatic,
non-metastatic and normal groups (from metastatic patients) and
normal groups (from non-metastatic patients). This study was
approved by the Ethics Committee of First Affiliated Hospital of
Guangzhou University of Chinese Medicine [No. Y (2019)172] and
written informed consent was obtained from all patients. The study
was performed in accordance with the Declaration of Helsinki.
Immunohistochemistry (IHC)
Tissues were fixed with 4% paraformaldehyde at room
temperature for 24 h and serially cut into 4-µm-thick sections for
IHC. The primary antibodies are displayed in Table SI. The paraffin-embedded sections
were dewaxed, rehydrated in a descending alcohol series, and heated
in citrate buffer (pH 6.0) for 20 min at 95°C. The sections were
quenched to block endogenous peroxidase activity (3% endogenous
peroxidase blocker at room temperature for 10 min), and then
blocked with normal goat serum for 20 min at 37°C. The sections
were incubated with primary antibodies for 16 h at 4°C, rinsed with
wash buffer, and then incubated with secondary antibodies at 37°C
for 15 min, horseradish peroxidase-labeled streptomycin was
subsequently added, and the slides were visualized using a
3,3-diaminobenzidine tetrahydrochloride substrate kit (DAB;
ZLI-9018; ZSGB-BIO; OriGene Technologies, Inc.). The sections were
lightly counterstained with hematoxylin, dehydrated, cleared and
mounted with resin. Negative controls were prepared by omitting the
primary antibodies, while keeping all other procedures the
same.
For immunohistochemical quantification, images of
three randomly selected microscopic fields per slide were captured
using a an Olympus BX 51 fluorescence microscope (Olympus
Corporation; magnification, ×200); and evaluated by independent
pathologists. Image Pro Plus 6.0 (Media Cybernetics, Inc.) was used
for digital image analysis. The scores for staining intensity and
the percentage of positive cells were multiplied; the yellow
density reflects the expression level of the target protein. The
expression levels of COL I, III, and IV, MMP-1, MMP-2, MMP-7, and
MMP-9, as well as LOXL2 were quantified via the average optical
density (AOD).
Sirius red staining and
quantification
The tissue sections were deparaffinized and
rehydrated in a graded ethanol series, and then incubated for 1 h
in Sirius red stain (G1018; Servicebio). The stained sections were
analyzed using an Olympus IX73 inverted microscope (Olympus
Corporation; magnification, ×200 or ×400) with a linear polarizer.
To avoid missing any details, the filter was tilted to an angle
between 0 and 90 until the birefringence became evident and the
background became completely black; the focus was then corrected
once more (25). The halogen lamp
intensity and exposure time were constant within each image. Under
polarized light, COL I appears red or yellow with strong
birefringence, while COL III is green with weak birefringence. The
areas of COL I and III staining were analyzed using Image Pro Plus
6.0 (Media Cybernetics, Inc.).
Protein functional annotation
The Search Tool for Recurring Instances of
Neighbouring Genes (STRING) database (https://string-db.org) is designed to evaluate the
integration of protein-protein interactions, including direct
(physical) and indirect (functional) associations (26). The STRING database was used to
detect the potential associations among MMP-1, MMP-2, MMP-7 and
MMP-9 and LOXL2.
Differential mRNA expression between
tumor and normal tissues
The Oncomine database (https://www.oncomine.org/) was used to analyze the
mRNA expression levels of coding genes (COL1A1-2, COL3A1,
COL4A1-6, LOXL2, and MMP1, MMP2, MMP7 and MMP9)
in different types of cancer. The search was conducted using the
following criteria: i) type of analysis: Cancer vs. normal tissues;
ii) type of data: mRNA; iii) thresholds: Fold change=2 and P=0.01.
Then UCSC Xena Browser (27)
(https://xenabrowser.net/datapages/)
was then used to obtain the TCGA-COADREAD gene expression dataset
and corresponding clinical data, which includes 383 tumor samples
and 51 normal samples. The data were used to verify the
differential mRNA expression levels of the target genes in CRC and
control normal tissues. P<0.01 was considered to indicate a
statistically significant difference.
Functional and pathway enrichment
analysis
The Database for Annotation, Visualization and
Integrated Discovery (DAVID) (https://david.ncifcrf.gov) integrates biological data
and analysis tools to generate systematic and comprehensive
functional annotations (28). In
the presents study, DAVID was used to conduct Gene Ontology (GO)
and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of
related coding genes. GO analyses classified the coding genes into
three categories, biological process (BP), cellular component (CC)
and molecular function (MF). KEGG analyses were conducted to
identify the pathways in which the coding genes were significantly
enriched; P<0.05 and an enrichment score (ES) >1.0 were used
as the cutoff criteria for significance.
Cox proportional hazards regression
analysis
The gene expression profiles and corresponding
clinical data of the GSE17536 dataset (29) were downloaded using the Gene
Expression overview database (GEO, http://www.ncbi.nlm.nih.gov/geo/). Samples without
overall survival (OS) events (time from surgery to death) and OS
times were removed, and the remaining 177 CRC samples were included
in the present study. Next, univariate and multivariate Cox
proportional risk regression analyses were used to analyze the
coding genes. Based on the expression and coefficients of these
coding genes, the ‘coxph’ function in the R ‘survival’ package
(http://cran.r-project.org/package=survival) (30) was used to calculate the risk score
for each patient and to establish an optimal prognostic model.
Survival analysis
Survival analysis for the coding genes in CRC
(COL1A1-2, COL3A1, COL4A1-6, LOXL2, and MMP1, MMP2,
MMP7 and MMP9) was performed using the PROGgeneV2
prognostic database (http://genomics.jefferson.edu/proggene/index.php)
(31). Briefly, the following
parameters were selected in the first interface: ‘COL1A1,
COL1A2, COL3A1, COL4A1, COL4A2, COL4A3, COL4A4, COL4A5, COL4A6,
MMP1, MMP2, MMP7, MMP9 and LOXL2’ in multiple user input
genes; ‘combined signature graphs only’; ‘colorectal cancer’ in
cancer type; ‘death’ in survival measure; and ‘median’ in bifurcate
gene expression. Then, in the second interface, filter ‘GSE17536’
was selected, and the plot was created. The log-rank P-value and
hazard ratio (HR) with 95% confidence interval were calculated and
displayed on the webpage. Only data with P<0.05 were selected
for analysis.
Data from the GSE39582 dataset (32) in the Gene Expression overview
database (GEO, http://www.ncbi.nlm.nih.gov/geo/) were used to
externally validate the survival effects of six genes in the
prognostic model. Analysis was performed using the R ‘survminer’
package (https://CRAN.R-project.org/package=survminer).
Statistical analysis
Statistical differences between COL I, COL III and
COL IV, and COL I/COL III, COL I area/COL III area, MMP-1, MMP-2,
MMP-7, MMP-9 and LOXL2 between metastasis and non-metastasis were
evaluated by unpaired t-tests; while between metastasis and normal
sample (metastasis), or non-metastasis and normal sample
(non-metastasis), paired t-tests were utilized. In addition, COL IV
(Fig. 2I) was further separated
into six groups: Distant-metastasis, lymphatic-metastasis,
non-metastasis and normal sample (distant-metastasis), normal
sample (lymphatic-metastasis), normal sample (non-metastasis). The
comparison among distant-metastasis, lymphatic-metastasis,
non-metastasis was evaluated by Welch's ANOVA, and Dunnett's T3
test was used to post hoc comparison; while the comparison between
distant-metastasis and normal sample (distant-metastasis),
lymphatic-metastasis and normal sample (lymphatic-metastasis) or
non-metastasis and normal sample (non-metastasis) was analyzed by
paired t-tests. P-values from Welch's ANOVA were adjusted for
multiple comparisons using Bonferroni corrections. Pearson's
correlation analysis was conducted to determine the associations
between the following: The expression of MMP-7 and MMP-1, MMP-2 or
MMP-9; and the expression of LOXL2 and MMP-1, MMP-2, MMP-7 or
MMP-9. The associations between the clinical parameters and
immunohistochemical results were analyzed using with the
Chi-squared test or Fisher's exact test (one or more cell contains
a count of 5 or fewer; such as the age of MMP-2, MMP-7, MMP-9 and
LOXL2; the differentiation of all index; the clinical stage of COL
I, COL I area, COL III area, COL I/COL III, COL I area/COL III
area, MMP-1, MMP-9 and LOXL2; the T-stage of COL III; the condition
of metastasis of all index). Statistical analyses were conducted
using SPSS version 25.0 for Windows (IBM, Corp.), and P<0.05 was
considered to indicate a statistically significant difference.
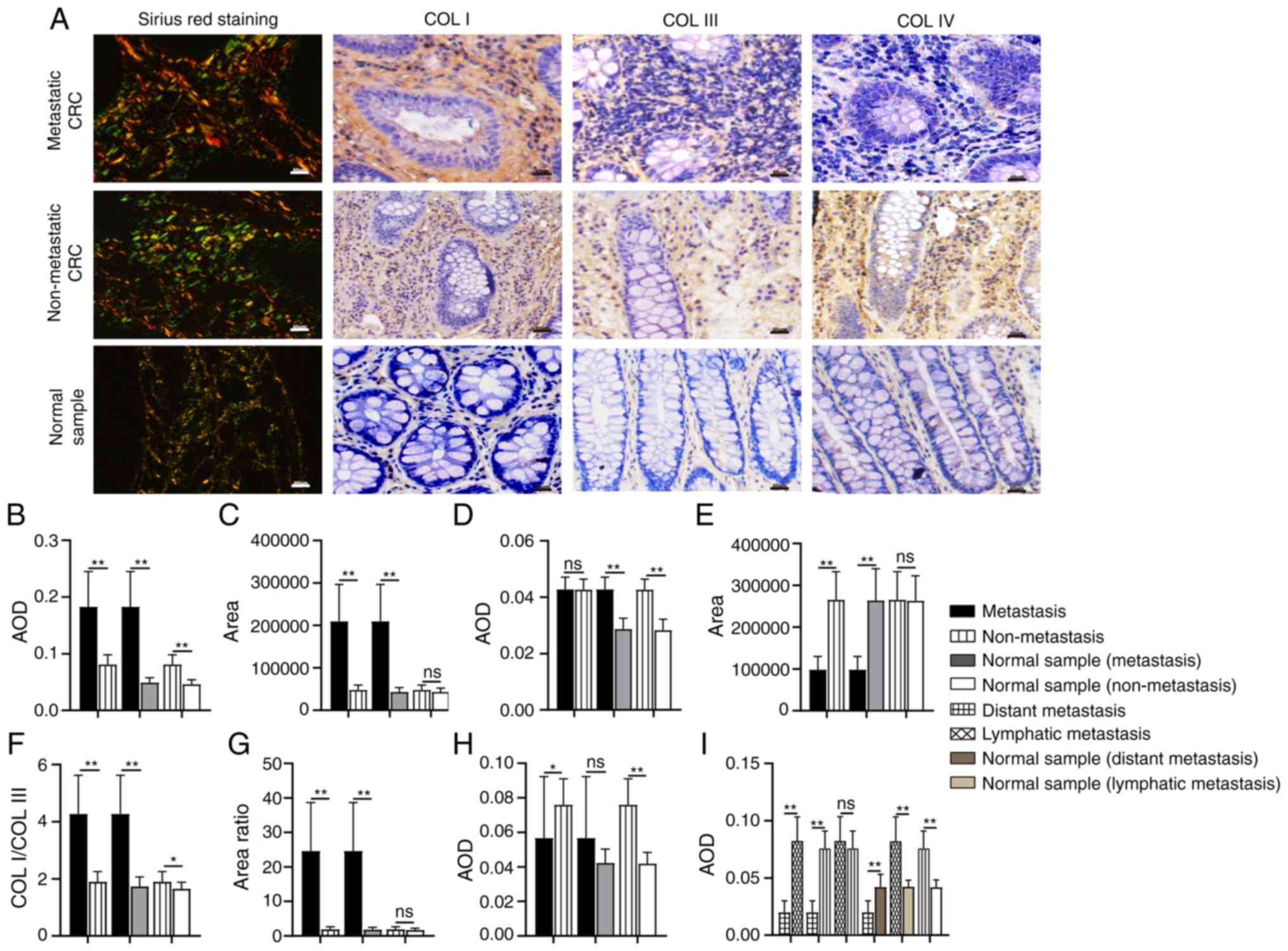 | Figure 2.Comparison of collagen expression in
different tissue samples. (A) Images and expression levels of COL
I, COL III, and COL IV in different tissue samples. (B-I)
Differences in the expression of (B) COL I, (C) COL I area, (D) COL
III, (E) COL III area, (F) the ratio of COL I/COL III, (G) the
ratio of COL I area/COL III area, and (H and I) COL IV.
Magnification, ×200. Scale bar, 50 µm. COL, collagen; AOD, average
optical density; CRC, colorectal cancer. *P<0.05; **P<0.01;
nsP>0.05. |
Differentially expressed coding genes between CRC
and normal tissue samples were assessed using the R ‘limma’ package
(version 3.32.10) (33). Volcano
plots were constructed using the volcano plotting tool (https://shengxin.ren), and the Bioinformatics and
Evolutionary Genomes website (http://bioinformatics.psb.ugent.be/webtools/Venn/) was
used to create the Venn diagram. OS was analyzed by Kaplan-Meier
survival curve analysis and significance was determined using the
log-rank test. Time-dependent receiver operating curve (ROC)
analysis was performed to assess the sensitivity and specificity of
the signature prognosis prediction. Statistical analysis was
performed: **P<0.01; *P<0.05; nsP>0.05 (as
indicated in the images with the relevant symbols).
Results
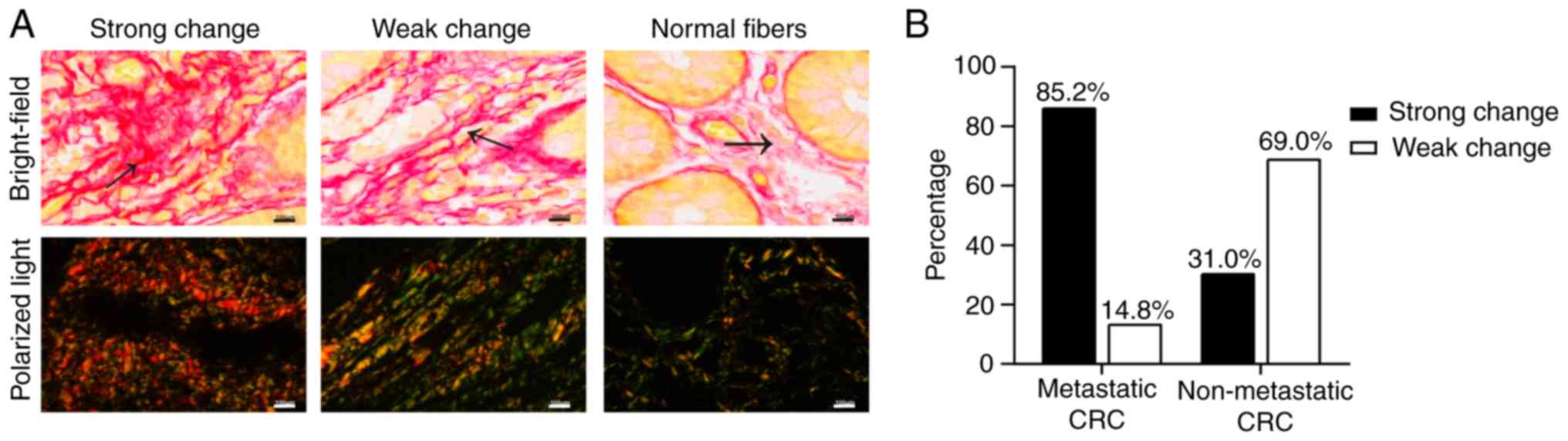
Alterations in collagen arrangement at
the CRC tumor invasion front
As one of its essential characteristics, collagen
arrangement is subsequently influenced by collagen remodeling. In
the present study, the collagen fibers in the normal intestinal
mucosa were thin, wavy and dispersed (Fig. 1A). However, at the CRC tumor
invasion front, the collagens fibers were arranged differently; the
fibers with weak changes were more linearized and denser than
normal collagen fibers (Fig. 1A).
The collagen fibers with strong changes exhibited an evident
increase in density and were crosslinked into bundles with a more
uniform arrangement (Fig. 1A).
Furthermore, among the 27 metastatic CRC samples, 23 exhibited
strong changes in collagen fibers, while only 4 samples exhibited
weak changes (Fig. 1B). Of the 29
non-metastatic samples, 20 exhibited weak changes, and 9 possessed
strong changes (Fig. 1B). These
results indicate that collagen arrangement affects the development
of CRC.
Expression of COL I, III and IV in
CRC
Collagen expression patterns are another essential
aspect of collagen characteristics. During tumor development, the
expression level, distribution and the ratio of collagens are
altered. COL I, III and IV are the most common matrix collagens. In
the present study, the expression level and distribution of COL I,
III, and IV was detected by IHC and Sirius red staining,
respectively (Fig. 2A). COL I is
the most abundant collagen type in the tumor matrix and is mainly
distributed in the interstitial matrix (IM), while COL III is
mostly distributed along with COL I. In addition, a changed COL
I/III ratio influences the hardness of the tumor matrix, regulating
tumor growth and migration (10,11).
The results of IHC showed that the expression of COL I in
metastatic CRC was significantly higher than that in non-metastatic
CRC (P<0.01; Fig. 2B) and normal
tissues (P<0.01; Fig. 2B); COL I
expression in non-metastatic CRC was also higher than that in the
normal samples (P<0.01). There was no significant difference in
the expression of COL III between metastatic and non-metastatic CRC
samples (P=0.928; Fig. 2D),
although its expression in metastatic and non-metastatic CRC was
higher than that in corresponding normal tissues, respectively
(P<0.01). Moreover, the COL I/COL III ratio was significantly
increased in the metastatic CRC group (P<0.01; Fig. 2F), although there were significant
differences in expression between the non-metastatic CRC and
corresponding normal tissue samples (P<0.05).
Sirius red staining reveals thick, strongly
birefringent COL I fibers (red), while COL III appears as thin,
weakly birefringent green fibers (34). In the present study, collagens in
metastatic CRC were mostly reddish or yellowish-orange, while
collagens in the normal samples were a yellowish-green color. In
non-metastatic CRC, the proportions of the two colors were similar
(Fig. 2A). Moreover, compared with
non-metastasis CRC, in metastatic CRC, the area of COL I was
significantly expanded (P<0.01; Fig.
2C), the area of COL III was significantly decreased
(P<0.01; Fig. 2E), and the ratio
of COL I area/COL III area was significantly increased (P<0.01;
Fig. 2G). In the non-metastatic CRC
and corresponding normal samples, there was no significant
difference in COL I area (P=0.051; Fig.
2C) and COL III area (P=0.901; Fig.
2E), or in the ratio between the two (P=0.197; Fig. 2G).
COL IV is mainly distributed in the basal membrane
(BM). The remodeling of COL IV would destroy the continuity and
integrity of the BM and contribute to tumor metastasis (35). The expression of COL IV in
non-metastatic CRC was significantly higher than that in metastatic
CRC (P<0.05; Fig. 2H) and
corresponding normal tissue samples (P<0.01, Fig. 2H), while the difference between the
metastatic CRC and corresponding normal samples was not
statistically significant (P=0.053). To verify the significance of
COL IV in distant metastasis, the metastatic CRC group was divided
into ‘distant metastasis’ and ‘lymph node metastasis only’
according to the degree of metastasis. The expression of COL IV in
subjects with distant metastases was significantly decreased
(P<0.01; Fig. 2I), while the
expression of COL IV in those with lymph node metastasis only was
marginally (but not significantly) higher than that in the
non-metastatic group (P=0.282).
High expression levels of MMP-1,
MMP-2, MMP-7, MMP-9 and LOXL2 in metastatic CRC
MMPs and LOX family are the members of collagenases
which are responsible for the collagen remodeling. MMP-1 is an
interstitial collagenase which degrades COL I and III (15), while MMP-2 and MMP-9 largely degrade
COL IV (36). As a stromelysin,
MMP-7 not only degrades COL IV, promoting tumor invasion, but also
regulates the activities of MMP-1, MMP-2 and MMP-9 (37–39).
Lysyl oxidase-like 2 (LOXL2) is a member of the LOX family, which
influences COL I, III and IV cross-linking and expression (40,41).
IHC was used to detect the expression of different collagenases in
metastatic CRC, non-metastatic CRC and non-tumor tissues (Fig. 3A). The results illustrate that the
expression level of MMP-1, MMP-2, MMP-7, MMP-9 and LOXL2 in
metastatic CRC was higher than that in non-metastatic CRC and
corresponding normal tissues (P<0.01; Fig. 3B-F). Furthermore, the expression of
these proteins in non-metastatic CRC was also higher than that in
corresponding normal tissues (P<0.01; Fig. 3B-F). The results of STRING database
analysis revealed that there were interactions between MMP-1,
MMP-2, MMP-7, and MMP 9, or LOXL2 and MMP-2 (Fig. 3G). Pearson's correlation analysis
was used to evaluate the association between MMP-7 and MMP-1, MMP-2
and MMP-9 expression. A strong correlation was found between MMP-7
and MMP-1 (r=0.887; P<0.01; Fig.
3H). There was also a positive correlation between MMP-7 and
MMP-2 (r=0.855; P<0.01; Fig.
3I), and MMP-7 and MMP-9 (r=0.855; P<0.01; Fig. 3J). According to the correlation
analysis between LOXL2 and MMP-1, MMP-2, MMP-7 and MMP-9, the
expression changes in these collagenases were consistent (Fig. 3K-N), and a strong association was
identified between LOXL2 and MMP-1 (r=0.935; P<0.01; Fig. 3K). There were also positive
correlations between LOXL2 and MMP-7 expression (r=0.894;
P<0.01; Fig. 3M), LOXL2 and
MMP-9 expression (r=0.840; P<0.01; Fig. 3N) and LOXL2 and MMP-2 (r=0.826;
P<0.01; Fig. 3L).
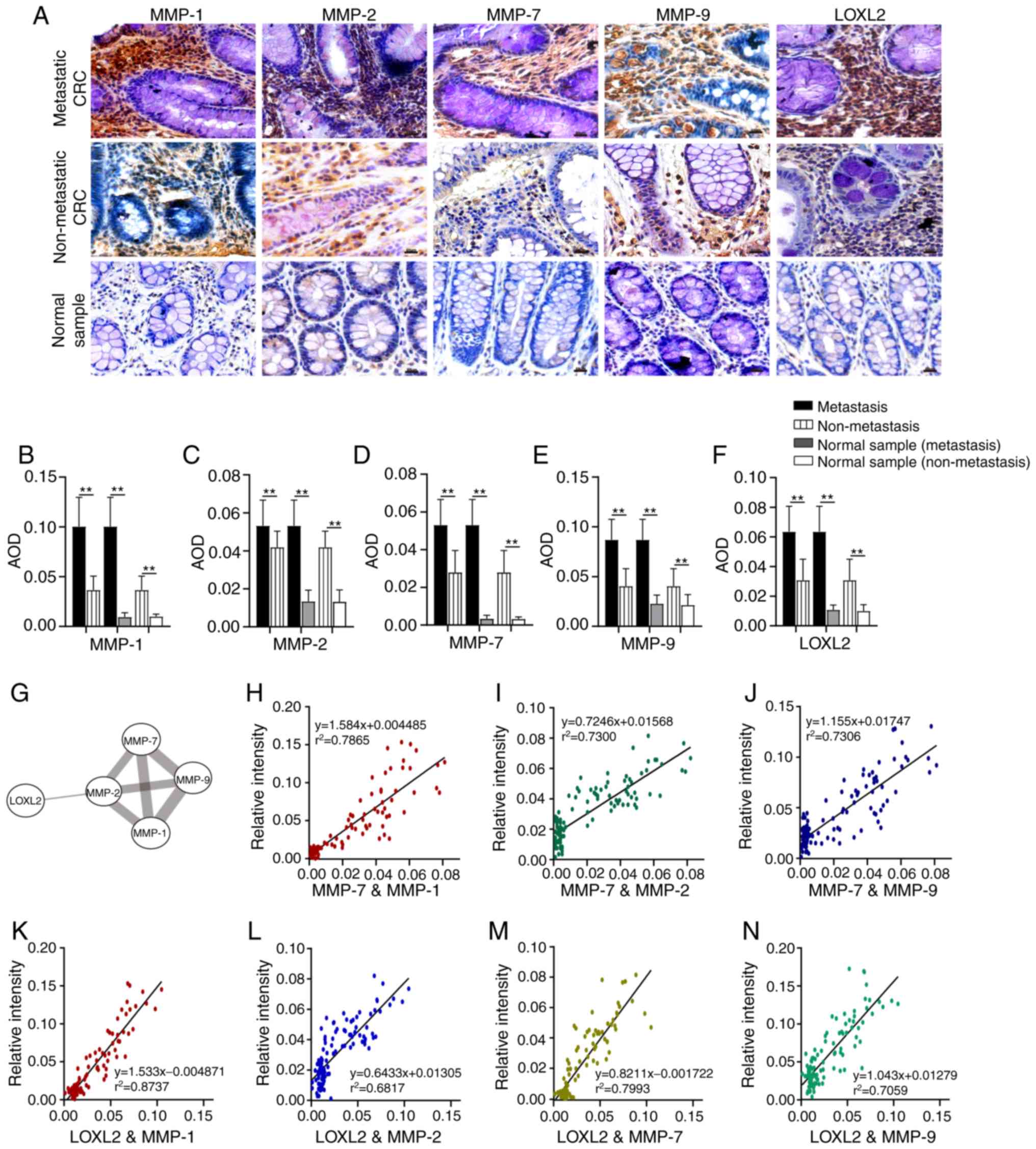 | Figure 3.Comparison of collagenase expression
levels in different tissue samples. (A) Images of MMP-1, MMP-2,
MMP-7, MMP-9 and LOXL2 in different tissue samples. (B-F) The
expression levels of (B) MMP-1, (C) MMP-2, (D) MMP-7, (E) MMP-9 and
(F) LOXL2 in different tissue samples. (G) Interactions between
LOXL2 and MMP-1, MMP-2, MMP-7 and MMP-9. (H-J) Correlation analysis
between the expression of (H) MMP-7 and MMP-1, (I) MMP-7 and MMP-2
and (J) MMP-7 and MMP-9. (K-N) Correlation analysis between the
expression of (K) LOXL2 and MMP-1, (L) LOXL2 and MMP-2, (M) LOXL2
and MMP-7 and (N) LOXL2 and MMP-9. Magnification, ×200; Scale bar,
50 µm. MMP, matrix metalloproteinase; LOXL2, lysyl oxidase-like 2;
AOD, average optical density; CRC, colorectal cancer.
**P<0.01. |
Association between the expression of
collagens and associated collagenases and the clinicopathological
characteristics of CRC
Patient clinicopathological characteristics are
summarized in Table I. The cutoffs
for low and high expression were set according to the median value.
As shown in Table IIA and B, the
expression and area of COL I and COL III, the ratio of COL I/COL
III and COL I area/COL III area, and the expression of COL IV,
MMP-1, MMP-2, MMP-7, MMP-9 and LOXL2 in 56 specimens were not
significantly correlated with patient sex, age or tumor
differentiation. The expression of COL I, COL I area, the ratio of
COL I/COL III, the ratio of COL I area/COL III area, MMP-1, MMP-7,
MMP-9, and LOXL2 was increased in the middle and late stages of CRC
(P<0.01), while COL III area was decreased (P<0.01).
Moreover, high expression of COL III and MMP-7 was associated with
increased infiltration depth (P<0.05). The expression of COL I,
COL I area, COL III area, the ratio of COL I/COL III, the ratio of
COL I area/COL III area, COL IV, MMP-1, MMP-2, MMP-7, MMP-9, and
LOXL2 differed between cases with distant metastasis, lymph node
metastasis and non-metastasis, indicating that their expression is
associated with tumor metastasis (P<0.05).
 | Table I.Clinicopathological characteristics
of the CRC patients (N=56). |
Table I.
Clinicopathological characteristics
of the CRC patients (N=56).
|
Characteristics | n | % |
|---|
| Sex |
|
Male | 31 | 55.4 |
|
Female | 25 | 44.6 |
| Age (years) |
| <50
(mean age, 45) | 11 | 19.6 |
| ≥50
(mean age, 63) | 45 | 71.4 |
| Condition of
metastasis |
|
Metastasis | 27 | 48.2 |
|
Non-metastasis | 29 | 51.8 |
|
Differentiation |
| Well
differentiation | 2 | 3.6 |
|
Moderate | 46 | 82.1 |
|
Poor | 8 | 14.3 |
| Clinical stage |
| I | 12 | 21.4 |
| II | 17 | 30.4 |
|
III | 16 | 28.6 |
| IV | 11 | 19.6 |
| T-stage |
| T1 | 5 | 8.9 |
| T2 | 12 | 21.4 |
| T3 | 38 | 67.8 |
| T4 | 1 | 1.9 |
 | Table II.Relationship between the expression
of collagens or correlated enzymes and clinicopathological
parameters in 56 patients with colorectal cancer. |
Table II.
Relationship between the expression
of collagens or correlated enzymes and clinicopathological
parameters in 56 patients with colorectal cancer.
| A, Collagens and
clinicopathological parameters |
|---|
|
|---|
|
| COLI | COLI area | COLIII | COLIII area | COLIV | COLI/COLIII | COLI area/COLIII
area |
|---|
|
|
|
|
|
|
|
|
|
|---|
|
| Expression |
| Expression |
| Expression |
| Expression |
| Expression |
| Expression |
| Expression |
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Variable | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value |
|---|
| Sex |
|
Male | 18 | 13 | 0.179 | 17 | 14 | 0.42 | 17 | 14 | 0.42 | 14 | 17 | 0.42 | 16 | 15 | 0.788 | 18 | 13 | 0.179 | 17 | 14 | 0.42 |
|
Female | 10 | 15 |
| 11 | 14 |
| 11 | 14 |
| 14 | 11 |
| 12 | 13 |
| 10 | 15 |
| 11 | 14 |
|
| Age (years) |
|
≥50 | 6 | 5 | 0.737 | 6 | 5 | 0.737 | 6 | 5 | 0.737 | 5 | 6 | 0.737 | 6 | 5 | 0.737 | 6 | 5 | 0.737 | 6 | 5 | 0.737 |
|
<50 | 22 | 23 |
| 22 | 23 |
| 22 | 23 |
| 23 | 22 |
| 22 | 23 |
| 22 | 23 |
| 22 | 23 |
|
|
Differentiation |
|
Poor | 25 | 23 | 0.705 | 25 | 23 | 0.705 | 23 | 25 | 0.705 | 23 | 25 | 0.705 | 23 | 25 | 0.705 | 25 | 23 | 0.705 | 25 | 23 | 0.705 |
|
Advanced/Medium | 3 | 5 |
| 3 | 5 |
| 5 | 3 |
| 5 | 3 |
| 5 | 3 |
| 3 | 5 |
| 3 | 5 |
|
| Clinical stage |
|
III+IV | 27 | 2 |
<0.001b | 28 | 1 |
<0.001b | 15 | 14 | 0.789 | 1 | 28 |
<0.001b | 11 | 18 | 0.061 | 27 | 2 |
<0.001b | 28 | 1 |
<0.001b |
|
I+II | 1 | 26 |
| 0 | 27 |
| 13 | 14 |
| 27 | 0 |
| 17 | 10 |
| 1 | 26 |
| 0 | 27 |
|
| T-stage |
|
T3+T4 | 10 | 7 | 0.383 | 10 | 7 | 0.383 | 14 | 3 | 0.003b | 7 | 10 | 0.383 | 8 | 9 | 0.771 | 10 | 7 | 0.383 | 10 | 7 | 0.383 |
|
T1+T2 | 18 | 21 |
| 18 | 21 |
| 14 | 25 |
| 21 | 18 |
| 20 | 19 |
| 18 | 21 |
| 18 | 21 |
|
| Condition of
metastasis |
|
Lymphatic metastasis | 0 | 11 |
<0.001b | 0 | 11 |
<0.001b | 3 | 8 | 0.216 | 11 | 0 |
<0.001b | 11 | 0 |
<0.001b | 0 | 11 |
<0.001b | 0 | 11 |
<0.001b |
|
Non-metastasis | 1 | 15 |
| 0 | 16 |
| 10 | 6 |
| 16 | 0 |
| 6 | 10 |
| 1 | 15 |
| 0 | 16 |
|
| Distant
metastasis | 27 | 2 |
| 28 | 1 |
| 15 | 14 |
| 1 | 28 |
| 11 | 18 |
| 27 | 2 |
| 28 | 1 |
|
| B, Correlated
enzymes and clinicopathological parameters |
|
|
|
| MMP-1 | MMP-2 | MMP-7 | MMP-9 | LOXL2 |
|
|
|
|
|
|
|
|
|
|
|
Expression |
|
Expression |
|
Expression |
|
Expression |
|
Expression |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Variable | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value | Low | High | P-value |
|
| Sex |
|
Male | 17 | 14 | 0.42 | 14 | 17 | 0.42 | 15 | 16 | 0.788 | 16 | 15 | 0.788 | 17 | 14 | 0.42 |
|
Female | 11 | 14 |
| 14 | 11 |
| 13 | 12 |
| 12 | 13 |
| 11 | 14 |
|
| Age (years) |
|
<50 | 7 | 4 | 0.503 | 7 | 4 | 0.503 | 7 | 4 | 0.503 | 4 | 7 | 0.503 | 4 | 7 | 0.503 |
|
≥50 | 21 | 24 |
| 21 | 24 |
| 21 | 24 |
| 24 | 21 |
| 24 | 21 |
|
|
Differentiation |
|
Advanced/ Medium | 25 | 23 | 0.705 | 23 | 25 | 0.705 | 24 | 24 | >0.999 | 25 | 23 | 0.705 | 25 | 23 | 0.705 |
|
Poor | 3 | 5 |
| 5 | 3 |
| 4 | 4 |
| 3 | 5 |
| 3 | 5 |
|
| Clinical stage |
|
I+II | 27 | 2 |
<0.001b | 18 | 11 | 0.061 | 23 | 6 |
<0.001b | 25 | 4 |
<0.001b | 25 | 4 |
<0.001b |
|
III+IV | 1 | 26 |
| 10 | 17 |
| 5 | 22 |
| 3 | 24 |
| 3 | 24 |
|
| T-stage |
|
T1+T2 | 10 | 7 | 0.383 | 9 | 8 | 0.771 | 12 | 5 | 0.042a | 11 | 6 | 0.146 | 10 | 7 | 0.383 |
|
T3+T4 | 18 | 21 |
| 19 | 20 |
| 16 | 23 |
| 17 | 22 |
| 18 | 21 |
|
| Condition of
metastasis |
| Distant
metastasis | 0 | 11 |
<0.001b | 1 | 10 | 0.007b | 0 | 11 |
<0.001b | 0 | 11 |
<0.001b | 0 | 11 |
<0.001b |
|
Lymphatic metastasis | 1 | 15 |
| 9 | 7 |
| 5 | 11 |
| 3 | 13 |
| 3 | 13 |
|
|
Non-metastasis | 27 | 2 |
| 18 | 11 |
| 23 | 6 |
| 25 | 4 |
| 25 | 4 |
|
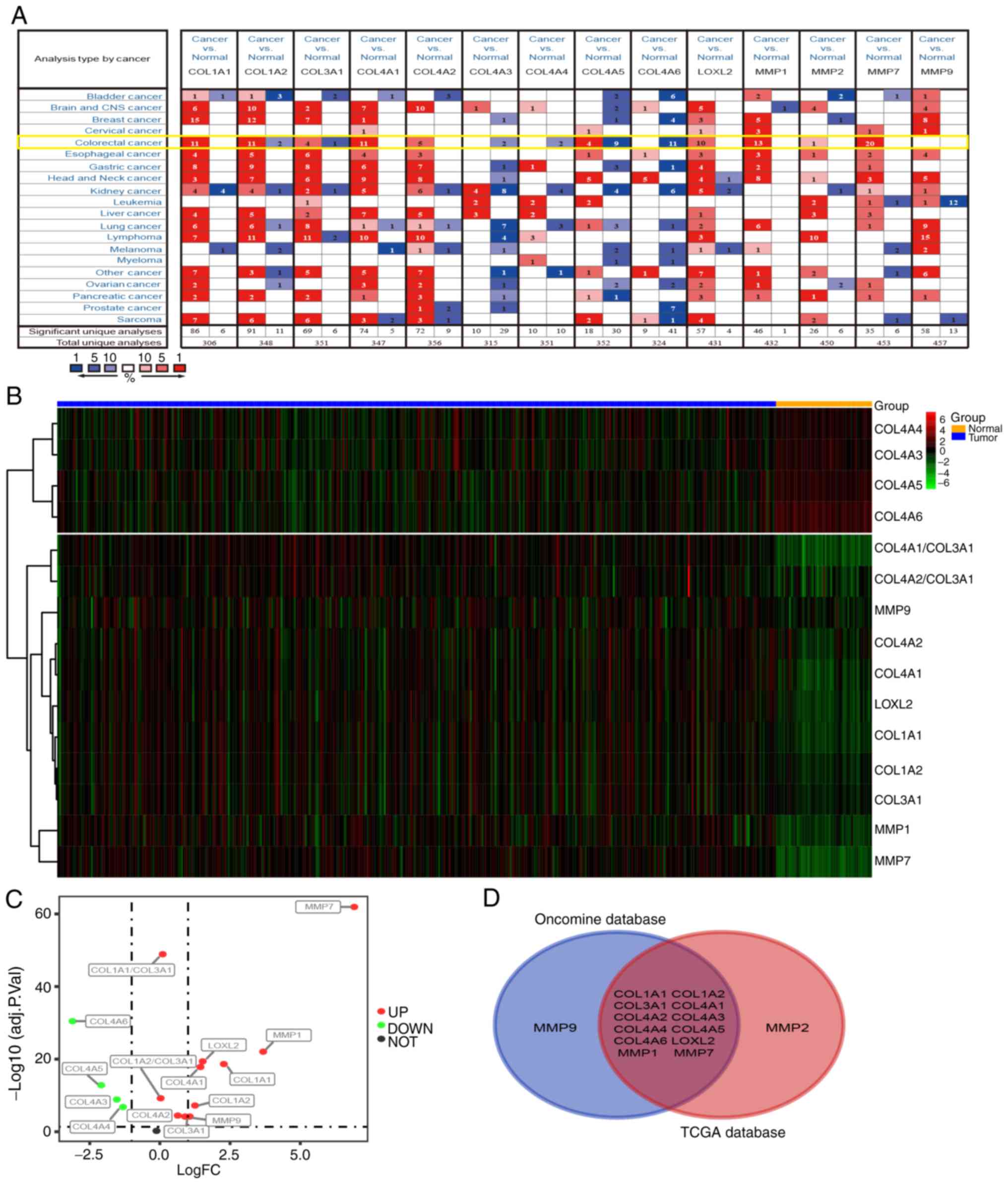
Differential expression of
collagen-related coding genes in tumor and normal tissues
In order to verify whether the expression changes in
COL I, III and IV, and related collagenases influence the
occurrence and development of tumors at the mRNA level, the
Oncomine database (https://www.oncomine.org) was used to analyze the mRNA
levels of related coding genes in different types of tumor and
normal tissues. COL I has two coding genes (COL1A1 and
COL1A2), and the gene for COL III is COL3A1.
COL4A1-A6 are all coding genes for COL IV. The coding genes
for MMP-1, MMP-2, MMP-7, MMP-9 and LOXL2 are MMP1, MMP2, MMP7,
MMP9 and LOXL2, respectively. As shown in Fig. 4A, the database contains 20 cancer
types, (including CRC). COL1A1, COL1A2, COL3A1, COL4A1, COL4A2,
LOXL2, MMP1, MMP2, MMP7 and MMP9 were upregulated in the
majority of tumor types. On the contrary, the expression of
COL4A3-6 was decreased in tumor tissues. The data from CRC
samples revealed that the expression levels of COL1A1-2, COL3A1,
COL4A1-6, LOXL2, and MMP1, MMP2, and MMP7 were
different in tumor tissues and normal intestinal tissues, while the
expression of MMP9 was not significantly altered.
Next, CRC RNA-seq data were downloaded from The
Cancer Genome Atlas (TCGA) database and subsequently analyzed. The
results show that the expression of MMP7, MMP1, MMP9, LOXL2,
COL1A1, COL1A2, COL3A1, COL4A1-6, COL1A1/COL3A1 and
COL1A2/COL3A1 differed between tumor and normal tissues
(logFC>0; P<0.01; Fig. 4B).
The levels of COL4A3, COL4A4, COL4A5 and COL4A6 were
downregulated in tumor tissues, while the expression of COL1A1,
COL1A2, COL3A1, COL4A1, COL4A2, LOXL2, MMP1, MMP7, MMP9,
COL1A1/COL3A1 and COL1A2/COL3A1 were upregulated
(Fig. 4C). The Venn diagram
demonstrates the differential expression of COL1A1-2, COL3A1,
COL4A1-6, LOXL2, MMP1 and MMP7 in both the Oncomine and TCGA
datasets (Fig. 4D), indicating that
these genes may have a greater influence on CRC than MMP2
and MMP9.
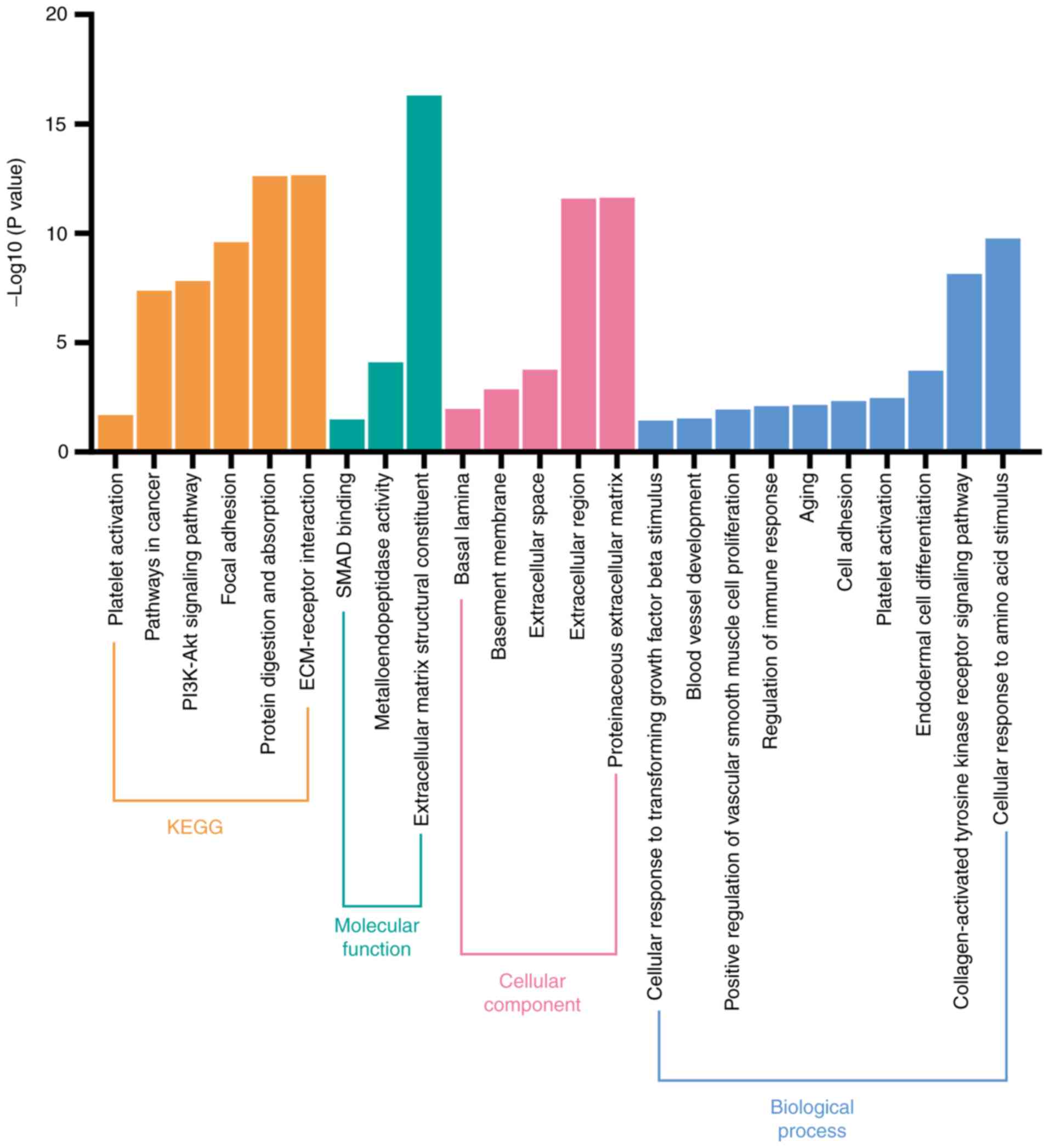
Biological pathways involving coding
genes associated with collagens and collagenases
The potential biological signaling pathways of 14
collagen-associated coding genes (COL1A1-2, COL3A1, COL4A1-6,
LOXL2, MMP1, MMP2, MMP7 and MMP9) were investigated
using DAVID. As shown in Fig. 5,
KEGG analysis revealed that these coding genes were significantly
associated with various pathways, such as those involved in
‘platelet activation’, ‘ECM-receptor interaction’, ‘PI3K-Akt
signaling pathway’, and ‘focal adhesion’. Furthermore, GO analysis
revealed that these genes were significantly enriched in ‘platelet
activation’, ‘blood vessel development’, and ‘collagen-activated
tyrosine kinase receptor signaling pathway’ to name but a few.
Hence, we hypothesized that these genes may affect the progression
of patients with CRC primarily via platelet activation and by
promoting angiogenesis.
Identification of prognostic genes and
establishment of a prognostic model
The PROGgeneV2 database (http://genomics.jefferson.edu/proggene/index.php)
was used to determine the impact of collagen and collagenase genes
on patient prognosis and survival. Combination analysis of these
coding genes suggested that the OS times (Fig. 6A) of the high-expression group were
shorter than those in the low-expression group. Moreover,
univariate Cox regression analysis of 177 CRC samples from the
GSE17536 dataset revealed that seven mRNAs (COL1A1, COL1A2,
COL3A1, COL4A1, COL4A2, LOXL2, MMP2) were not conducive to the
survival of patients with CRC, while the other three mRNAs
(COL4A5, COL4A6 and COL4A3) were beneficial to survival
(P<0.05, Table III). In order
to improve the accuracy of the prognostic effect of these mRNA,
multivariate Cox analysis was conducted using the 10 candidate
mRNAs. Finally, a total of six mRNAs were screened as candidate
factors significantly associated with CRC prognosis (P=6.335e-05,
Table IV). Concurrently, a new
risk scoring formula was established based on the mRNA expression
levels and the coefficients evaluated by multivariate Cox analysis.
The mRNA risk score signature=((2.409e+01)*COL3A1) +
((−1.099e+01)*COL4A3) + ((−1.144e+01)*COL4A6) +
((−1.046e+01)*COL1A1) + ((−4.291e+00)*MMP2) + ((2.163e+00)*COL1A2).
According to the prognostic model formula, patient risk scores were
calculated and ranked, and the risk score distribution is displayed
in Fig. 6B. The expression levels
of four high-risk mRNAs were higher in the patients with high-risk
scores, while the expression levels of two protective mRNAs were
higher in the patients with low-risk scores (Fig. 6E).
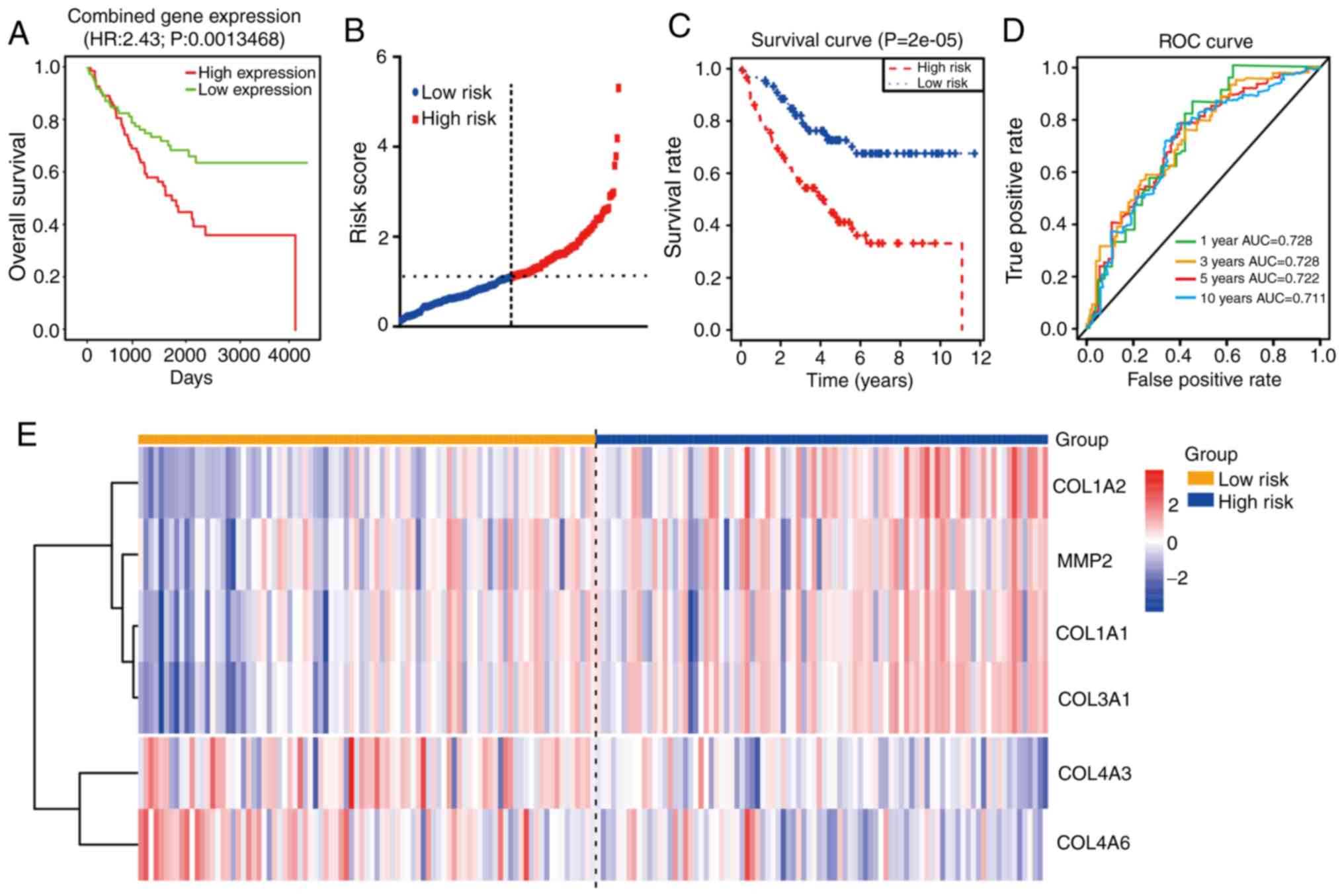 | Figure 6.Prognostic analysis of
collagen-related coding genes in CRC. (A) OS analysis of combined
expression of genes including COL1A1-2, COL3A1, COL4A1-6, LOXL2,
MMP1, MMP2, MMP7 and MMP9 using PROGgenev2. (B)
Time-dependent ROC analysis of the sensitivity and specificity of
the six-mRNA signature (COL1A1-2, COL3A1, COL4A3, COL4A6 and
MMP2). (C) Kaplan-Meier analysis of the six-mRNA signature.
(D) ROC curve of the prognostic model. (E) Heatmap of the
expression of the six-mRNA signature. CRC, colorectal cancer; OS,
overall survival; ROC, receiver operating characteristic; AUC, area
under the curve. |
 | Table III.Univariable Cox regression analysis
to assess the prognostic value of each mRNA. |
Table III.
Univariable Cox regression analysis
to assess the prognostic value of each mRNA.
| Gene | HRa | z | P-value |
|---|
| COL1A2 | 16.48555 | 3.440846 | 0.00058 |
| COL3A1 | 1954.201 | 3.115704 | 0.001835 |
| LOXL2 | 36.23624 | 2.900018 | 0.003731 |
| COL4A5 | 0.000416 | −2.68185 | 0.007322 |
| COL1A1 | 48.30721 | 2.586627 | 0.009692 |
| COL4A6 | 2.82E-05 | −2.39273 | 0.016723 |
| COL4A3 | 0.00033 | −2.22433 | 0.026126 |
| MMP2 | 12.9414 | 2.038733 | 0.041477 |
| COL4A2 | 21.90964 | 2.018873 | 0.0435 |
| COL4A1 | 23.51048 | 1.998489 | 0.045664 |
| COL4A4 | 3.191615 | 1.31461 | 0.188641 |
| MMP1 | 1.728113 | 1.10983 | 0.267072 |
| MMP9 | 0.398133 | −0.91069 | 0.362457 |
| MMP7 | 1.382114 | 0.539802 | 0.589333 |
 | Table IV.The coefficient of each gene. |
Table IV.
The coefficient of each gene.
| Gene | Coefficient | exp(coef) | se(coef) | z | P-value |
|---|
| COL3A1 | 2.409e+01 | 2.908e+10 | 8.180e+00 | 2.945 | 0.00323 |
| COL4A3 | −1.099e+01 | 1.688e-05 | 4.124e+00 | −2.665 | 0.00771 |
| COL4A6 | −1.144e+01 | 1.072e-05 | 4.663e+00 | −2.454 | 0.01412 |
| COL1A1 | −1.046e+01 | 2.855e-05 | 4.450e+00 | −2.351 | 0.01870 |
| MMP2 | −4.291e+00 | 1.369e-02 | 2.374e+00 | −1.808 | 0.07066 |
| COL1A2 | 2.163e+00 | 8.701e+00 | 1.414e+00 | 1.530 | 0.12612 |
In order to determine the value of this prognostic
model, a total of 177 patients were then separated into high (n=88)
and low (n=89) risk score groups. The results showed that the
patients in the high-risk group possessed shorter OS times and
poorer prognoses than those in the low-risk group (P<0.0001;
Fig. 6C). Moreover, time-dependent
ROC curve analysis revealed that the prognostic accuracy of this
model was 0.728 at 1 or 3 years, 0.722 at 5 years and 0.711 at 10
years (Fig. 6D). These results
suggest that this prognostic model (using COL1A1, COL1A2,
COL3A1, COL4A3, COL4A6 and MMP2 mRNA expression) has
high specificity and sensitivity, and may be used effectively
predict the prognosis of patients with CRC.
Verification of survival analysis
To further verify the effects of these six signature
mRNAs (COL1A1, COL1A2, COL3A1, COL4A3, COL4A6 and
MMP2) on the prognosis of patients with CRC, the effects of
these genes on OS were assessed using 562 CRC patient samples from
the GSE39582 dataset. As shown in Fig.
7, high expression of COL1A1, COL1A2, COL3A1 and
MMP2 had adverse effects on the OS of patients with CRC,
while high expression of COL4A3 and COL4A6 was beneficial to OS.
Collectively, the analyses indicate that COL1A1, COL1A2, COL3A1,
COL4A3, COL4A6 and MMP2 may be used as prognostic
biomarkers for CRC.
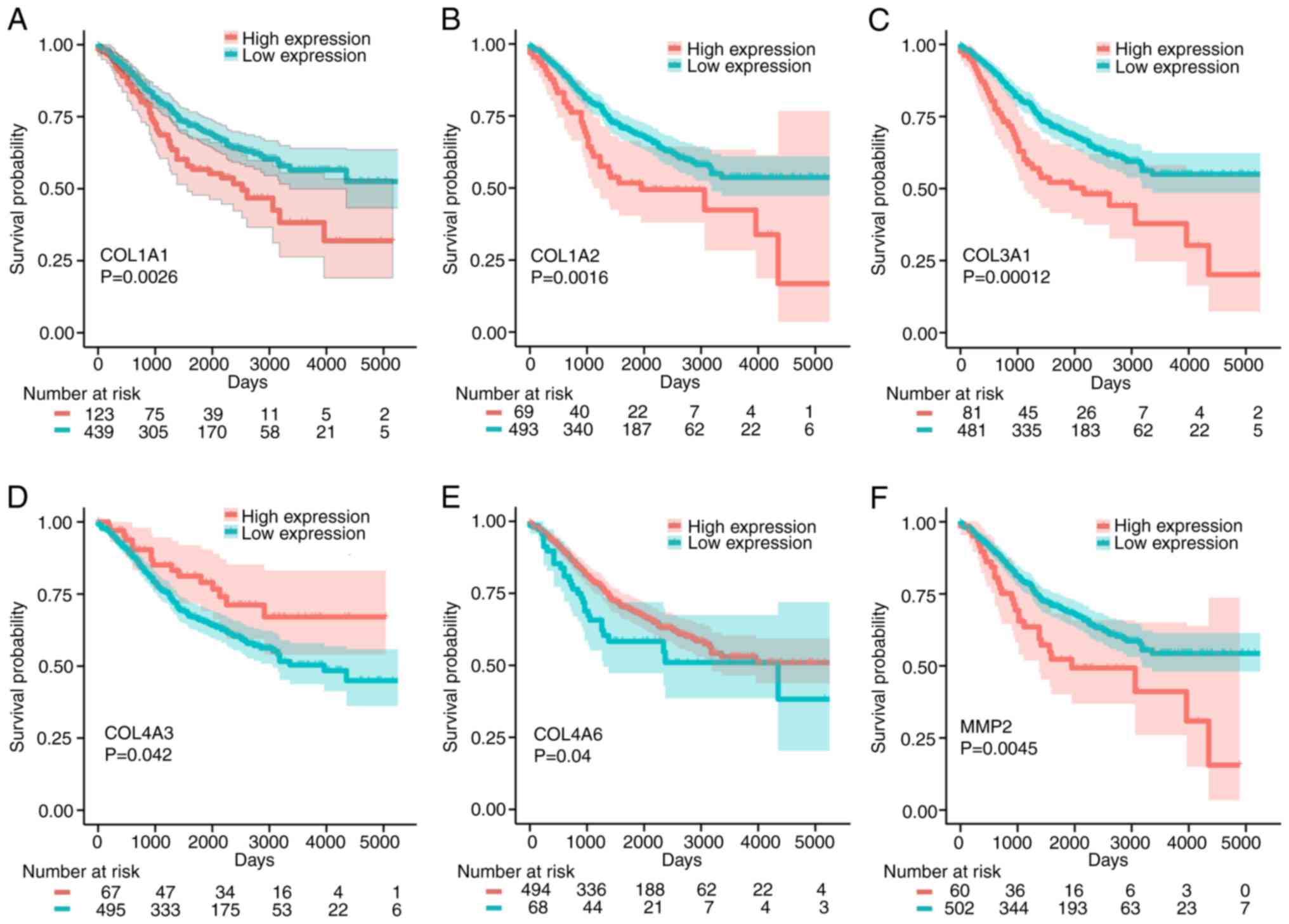 | Figure 7.Kaplan-Meier analysis of the
association between COL1A1, COL1A2, COL3A1, COL4A3, COL4A6
and MMP2 and the OS times of patients with CRC. (A) COL1A1,
(B) COL1A2, (C) COL3A1, (D) COL4A3, (E) COL4A6 and (F) MMP2. OS,
overall survival; CRC, colorectal cancer. |
Discussion
Remodeling of the matrix environment at the tumor
invasion front is considered to be a significant predictor of tumor
development (23,42). In the present study, tissues were
serially cut into 4-µm-thick sections for immunohistochemistry
(IHC) and Sirius red staining. By integrating histological and
bioinformatics analyses, the characteristics and coding genes of
matrix collagen (and related collagenases) at the tumor invasion
front were found to be associated with colorectal cancer (CRC)
progression and metastasis.
Collagen characteristics in tumors differ greatly
from those in healthy tissues, and are characterized by increased
deposition and linearization, as well as considerable cross-linking
(43). In CRC, tumors with stronger
invasion ability exhibited significantly increased collagen density
at the tumor invasion front, and collagen fiber arrangement was
linearized and bunched with reduced curvature. Moreover, the
expression and distribution of COL I was increased significantly in
the tumors, compared with normal tissues. Highly cross-linked and
linearized collagen fibers act as channels for tumor cells, and
restriction to these narrow channels, facilitates more precise
migration of tumor cells to the preferred destination (44,45).
Collagen also promotes tumor invasion and metastasis by expanding
the distribution range of tumor cells (46). In addition, changes in the ratio of
different matrix collagens also affect tumor progression. The
results of the present study demonstrate that the ratio of COL
I/COL III increased significantly in the metastatic group. Current
studies have also shown that an increased COL I/COL III ratio
promotes the migration and distribution of tumor cells by
increasing the stiffness of the matrix (10,34).
It is also worth noting that the expression of COL III in the
matrix also affects the density and arrangement of COL I (47). When studying the properties of
matrix collagen in tumors, the effects of more than one type of
collagen should be considered.
In addition, changes in matrix collagen fluctuate
during the progression of CRC. Fan et al (48) indicated that a decrease in COL IV
facilitates the invasion of tumor cells, while Öhlund et al
(49) showed that the excessive
accumulation of COL IV was one of the necessary conditions for
cancer cell survival. In the present study, the expression of COL
IV at the CRC tumor invasion front was significantly decreased in
patients with distant metastasis. The expression of COL IV in
patients with lymph node metastasis was not significantly different
from that in patients without metastasis, and expression was higher
than that in distal normal tissues. Based on the aforementioned
findings, we hypothesize that unlike the consistent increase in COL
I and COL III expression, both the upregulation and absence of COL
IV expression can promote CRC development, which is detrimental to
patient prognosis. Therefore, in patients with CRC, changes in the
remodeling and expression of different collagens should be
comprehensively analyzed at different stages of tumor
progression.
Matrix collagen remodeling is largely regulated by
the LOX and MMP family (50). The
present study revealed that the expression of LOXL2, MMP-1, MMP-2,
MMP-7 and MMP-9 at the tumor invasion front was significantly
higher in the metastatic group than in the non-metastatic and
normal groups. Previous studies have also shown that these proteins
may enhance tumor cell invasiveness by remodeling the tumor
microenvironment, and may also be associated with poor prognosis
(51,52). Although the cross-linking effect of
LOXL2 opposes the degradation abilities of MMP-1, MMP-2, MMP-7 and
MMP-9, the expression of one does not restrict the abilities of the
other. Liu et al (53) also
found that LOXL2 regulates the activity of MMPs, and when LOXL2 was
highly expressed, MMP-9 activity was also increased. In the current
study, correlation analysis of LOXL2, MMP-1, MMP-2, MMP-7 and MMP-9
indicated that LOXL2 expression was positively correlated with that
of MMP-1, MMP-2, MMP-7 and MMP-9. STRING database analysis also
revealed an interaction between LOXL2 and MMP-2. Although the
mechanism of LOXL2 and MMPs remains unclear, these data show that
LOXL2 and the MMPs jointly regulate collagen remodeling. Collagen
remodeling is more active during metastasis (4), and the activities of LOXL2, MMP-1,
MMP-2, and MMP-9 are also significantly increased.
The synthesis of matrix collagen and associated
collagenases is regulated by their coding genes. The results of the
present study indicate that abnormal changes in the expression of
these genes (COL1A1-A2, COL3A1, COL4A1-A6, MMP1, MMP2, MMP7,
MMP9 and LOXL2) are involved in CRC development, and
also affect the prognosis of patients with CRC. This result further
verified that abnormal expression of collagen and related collagens
is related to the occurrence and prognosis of CRC. The effects of
these coding genes on CRC prognosis were comprehensively analyzed,
and a prognostic model of CRC was constructed with six mRNA
signatures (COL1A1, COL1A2, COL3A1, COL4A3, COL4A6 and
MMP2). Among them, COL1A1, COL1A2, COL3A1 and
MMP2 have been reported to serve vital roles in tumor
invasion, and are thus associated with poor prognosis (54–57).
COL4A3 and COL4A6, encoding COL IV alpha chains 3 and
6, respectively, are required for correct basement membrane (BM)
formation, and mutations within these genes affect BM stability
(58,59). Moreover, GO and KEGG analyses showed
that these coding genes are primarily associated with platelet
activation pathways. It has also been reported that the collagen or
collagenases on the surface of tumor cells binds to integrins on
the surface of platelets, inducing platelet activation (60,61).
Following activation, platelets induces tumor angiogenesis by
releasing associated growth factors, which subsequently promotes
tumor growth (62). The binding of
platelets to cancer cells protects the cells from shear-induced
damage and facilitates cancer cell colonization (63). In addition, these collagens and
collagenases are also involved in multiple tumor-associated
pathways, such as the PI3K-Akt and tyrosine kinase receptor
signaling pathways. Therefore, matrix collagens and their
associated collagenases can affect the development and progression
of CRC in multiple ways.
The present study provides multi-level evidence for
the importance of abnormal collagen remodeling in the development
and prognostic evaluation of CRC from the aspects of collagen
arrangement, related proteins and their coding genes. Dense matrix
collagen at the tumor invasion front can promote tumor invasion,
which provides a novel pathological direction of analysis for
clinicians. Because different types of collagen and collagenases
interact with each other in the matrix, combination analysis of
multiple biomarkers may improve the reliability of clinical
evaluation, suggesting that the evaluation of matrix collagen in
CRC should not focus on only a single type collagen. Finally,
matrix collagens and collagenases can influence the development of
CRC by activating platelets and other tumor-associated pathways,
which may provide the reference direction for subsequent
treatment.
The main limitation of the present study is the
relatively small sample size. Larger-scale functional studies are
required to improve our understanding of the effects of matrix
collagens and their associated collagenases in CRC. However, the
results provide some interesting suggestions for future studies
with an increased sample size.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The study was supported by National Natural Science
Foundation of China, No. 81673944 (Sponsor: Bin Wen).
Availability of data and materials
The STRING, Oncomine, TCGA, GEO and PROGgeneV2
material is publicly available. The datasets used and analyzed
during the current study are available from the corresponding
author on reasonable request.
Authors' contributions
YL conceived and designed the experiments. YL and ZL
performed the experiments and analyzed the data. GH, HL and JQ
contributed to the acquisition of the data. YL, ZL and FN have made
substantial contribution to the collection of tissue samples. YL
and GH wrote the manuscript. BW, as the corresponding author, was
responsible for project funding, design, writing and checking. All
authors have examined the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The experiment was approved by the First Affiliated
Hospital of Guangzhou University of Chinese Medicine. This study
was approved by the Ethics Committee of the First Affiliated
Hospital of Guangzhou University of Chinese Medicine [No. Y
(2019)172] and informed consent was obtained from all patients.
This study was performed in accordance with the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors have declared that no competing interest
exists.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Turajlic S and Swanton C: Metastasis as an
evolutionary process. Science. 352:169–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crotti S, Piccoli M, Rizzolio F, Giordano
A, Nitti D and Agostini M: Extracellular matrix and colorectal
cancer: How surrounding microenvironment affects cancer cell
behavior? J Cell Physiol. 232:967–975. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Provenzano PP, Eliceiri KW, Campbell JM,
Inman DR, White JG and Keely PJ: Collagen reorganization at the
tumor-stromal interface facilitates local invasion. BMC Med.
4:382006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen D, Chen G, Jiang W, Fu M, Liu W, Sui
J, Xu S, Liu Z, Zheng X, Chi L, et al: Association of the collagen
signature in the tumor microenvironment with lymph node metastasis
in early gastric cancer. JAMA Surg. 154:e1852492019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huynh RN, Yousof M, Ly KL, Gombedza FC,
Luo X, Bandyopadhyay BC and Raub CB: Microstructural densification
and alignment by aspiration-ejection influence cancer cell
interactions with three-dimensional collagen networks. Biotechnol
Bioeng. 117:1826–1838. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Conklin MW and Keely PJ: Why the stroma
matters in breast cancer: Insights into breast cancer patient
outcomes through the examination of stromal biomarkers. Cell Adh
Migr. 6:249–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whatcott CJ, Diep CH, Jiang P, Watanabe A,
LoBello J, Sima C, Hostetter G, Shepard HM, Von Hoff DD and Han H:
Desmoplasia in primary tumors and metastatic lesions of pancreatic
cancer. Clin Cancer Res. 21:3561–3568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonnans C, Chou J and Werb Z: Remodelling
the extracellular matrix in development and disease. Nat Rev Mol
Cell Biol. 15:786–801. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Beam J, Botta A, Ye J, Soliman H, Matier
BJ, Forrest M, MacLeod KM and Ghosh S: Excess linoleic acid
increases collagen I/III ratio and ‘stiffens’ the heart muscle
following high fat diets. J Biol Chem. 290:23371–23384. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Najafi M, Farhood B and Mortezaee K:
Extracellular matrix (ECM) stiffness and degradation as cancer
drivers. J Cell Biochem. 120:2782–2790. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brauchle E, Kasper J, Daum R, Schierbaum
N, Falch C, Kirschniak A, Schäffer TE and Schenke-Layland K:
Biomechanical and biomolecular characterization of extracellular
matrix structures in human colon carcinomas. Matrix Biol.
68-69:180–193. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paul CD, Hruska A, Staunton JR, Burr HA,
Daly KM, Kim J, Jiang N and Tanner K: Probing cellular response to
topography in three dimensions. Biomaterials. 197:101–118. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tanjore H and Kalluri R: The role of type
IV collagen and basement membranes in cancer progression and
metastasis. Am J Pathol. 168:715–717. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hua H, Li M, Luo T, Yin Y and Jiang Y:
Matrix metalloproteinases in tumorigenesis: An evolving paradigm.
Cell Mol Life Sci. 68:3853–3868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tjin G, White ES, Faiz A, Sicard D,
Tschumperlin DJ, Mahar A, Kable EPW and Burgess JK: Lysyl oxidases
regulate fibrillar collagen remodelling in idiopathic pulmonary
fibrosis. Dis Model Mech. 10:1301–1312. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wong CC, Tse AP, Huang YP, Zhu YT, Chiu
DK, Lai RK, Au SL, Kai AK, Lee JM, Wei LL, et al: Lysyl
oxidase-like 2 is critical to tumor microenvironment and metastatic
niche formation in hepatocellular carcinoma. Hepatology.
60:1645–1658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shiozawa J, Ito M, Nakayama T, Nakashima
M, Kohno S and Sekine I: Expression of matrix metalloproteinase-1
in human colorectal carcinoma. Mod Pathol. 13:925–933. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Moreno-Bueno G, Salvador F, Martín A,
Floristán A, Cuevas EP, Santos V, Montes A, Morales S, Castilla MA,
Rojo-Sebastián A, et al: Lysyl oxidase-like 2 (LOXL2), a new
regulator of cell polarity required for metastatic dissemination of
basal-like breast carcinomas. EMBO Mol Med. 3:528–544. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Zhang J, Chen Y, Sohel H, Ke X,
Chen J and Li YX: The correlation and role analysis of COL4A1 and
COL4A2 in hepatocarcinogenesis. Aging (Albany NY). 12:204–223.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao S and Yu M: Identification of MMP1 as
a potential prognostic biomarker and correlating with immune
infiltrates in cervical squamous cell carcinoma. DNA Cell Biol.
39:255–272. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugai T, Yamada N, Eizuka M, Sugimoto R,
Uesugi N, Osakabe M, Ishida K, Otsuka K, Sasaki A and Matsumoto T:
Vascular invasion and stromal S100A4 expression at the invasive
front of colorectal cancer are novel determinants and tumor
prognostic markers. J Cancer. 8:1552–1561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiaopei H, Kunfu D, Lianyuan T, Zhen L,
Mei X and Haibo Y: Tumor invasion front morphology: A novel
prognostic factor for intrahepatic cholangiocarcinoma. Eur Rev Med
Pharmacol Sci. 23:9821–9828. 2019.PubMed/NCBI
|
|
24
|
Brierley J, Gospodarowicz M and Wittekind
C: International Union Against Cancer: TNM Classification of
Malignant Tumours. 8th. Wiley-Blackwell; West Sussex, United
Kingdom: 2017
|
|
25
|
Coelho PGB, Souza MV, Conceição LG,
Viloria MIV and Bedoya SAO: Evaluation of dermal collagen stained
with picrosirius red and examined under polarized light microscopy.
An Bras Dermatol. 93:415–418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cline MS, Craft B, Swatloski T, Goldman M,
Ma S, Haussler D and Zhu J: Exploring TCGA Pan-Cancer data at the
UCSC cancer genomics browser. Sci Rep. 3:26522013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smith JJ, Deane NG, Wu F, Merchant NB,
Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al:
Experimentally derived metastasis gene expression profile predicts
recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mizukami A, Matsue Y, Naruse Y, Kowase S,
Kurosaki K, Suzuki M, Matsumura A, Nogami A, Aonuma K and Hashimoto
Y: Kaplan-Meier survival analysis and Cox regression analyses
regarding right ventricular septal pacing: Data from Japanese
pacemaker cohort. Data Brief. 8:1303–1307. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goswami CP and Nakshatri H: PROGgeneV2:
Enhancements on the existing database. BMC Cancer. 14:9702014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Marisa L, de Reyniès A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Junqueira LC, Cossermelli W and Brentani
R: Differential staining of collagens type I, II and III by Sirius
Red and polarization microscopy. Arch Histol Jpn. 41:267–274. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kalluri R: Basement membranes: Structure,
assembly and role in tumour angiogenesis. Nat Rev Cancer.
3:422–433. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hsu TI, Lin SC, Lu PS, Chang WC, Hung CY,
Yeh YM, Su WC, Liao PC and Hung JJ: MMP7-mediated cleavage of
nucleolin at Asp255 induces MMP9 expression to promote tumor
malignancy. Oncogene. 36:875–876. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou D, Tian Y, Sun L, Zhou L, Xiao L, Tan
RJ, Tian J, Fu H, Hou FF and Liu Y: Matrix metalloproteinase-7 is a
urinary biomarker and pathogenic mediator of kidney fibrosis. J Am
Soc Nephrol. 28:598–611. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Imai K, Yokohama Y, Nakanishi I, Ohuchi E,
Fujii Y, Nakai N and Okada Y: Matrix metalloproteinase 7
(matrilysin) from human rectal carcinoma cells activation of the
precursor, interaction with other matrix metalloproteinases and
enzymic properties. J Biol Chem. 270:6691–6697. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim YM, Kim EC and Kim Y: The human lysyl
oxidase-like 2 protein functions as an amine oxidase toward
collagen and elastin. Mol Biol Rep. 38:145–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
López-Jiménez AJ, Basak T and Vanacore RM:
Proteolytic processing of lysyl oxidase-like-2 in the extracellular
matrix is required for crosslinking of basement membrane collagen
IV. J Biol Chem. 292:16970–16982. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chiang WF, Liu SY, Fang LY, Lin CN, Wu MH,
Chen YC, Chen YL and Jin YT: Overexpression of galectin-1 at the
tumor invasion front is associated with poor prognosis in
early-stage oral squamous cell carcinoma. Oral Oncol. 44:325–334.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou ZH, Ji CD, Xiao HL, Zhao HB, Cui YH
and Bian XW: Reorganized collagen in the tumor microenvironment of
gastric cancer and its association with prognosis. J Cancer.
8:1466–1476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pang MF, Siedlik MJ, Han S, Stallings-Mann
M, Radisky DC and Nelson CM: Tissue stiffness and hypoxia modulate
the integrin-linked kinase ILK to control breast cancer stem-like
cells. Cancer Res. 76:5277–5287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cackowski FC, Wang Y, Decker JT, Sifuentes
C, Weindorf S, Jung Y, Wang Y, Decker AM, Yumoto K, Szerlip N, et
al: Detection and isolation of disseminated tumor cells in bone
marrow of patients with clinically localized prostate cancer.
Prostate. 79:1715–1727. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ray A, Slama ZM, Morford RK, Madden SA and
Provenzano PP: Enhanced directional migration of cancer stem cells
in 3D aligned collagen matrices. Biophys J. 112:1023–1036. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tilbury K, Lien CH, Chen SJ and Campagnola
PJ: Differentiation of Col I and Col III isoforms in stromal models
of ovarian cancer by analysis of second harmonic generation
polarization and emission directionality. Biophys J. 106:354–365.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan HX, Li HX, Chen D, Gao ZX and Zheng
JH: Changes in the expression of MMP2, MMP9, and ColIV in stromal
cells in oral squamous tongue cell carcinoma: Relationships and
prognostic implications. J Exp Clin Cancer Res. 31:902012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Öhlund D, Franklin O, Lundberg E, Lundin C
and Sund M: Type IV collagen stimulates pancreatic cancer cell
proliferation, migration, and inhibits apoptosis through an
autocrine loop. BMC Cancer. 13:1542013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang J, Boddupalli A, Koelbl J, Nam DH, Ge
X, Bratlie KM and Schneider IC: Degradation and remodeling of
epitaxially grown collagen fibrils. Cell Mol Bioeng. 12:69–84.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Barker HE, Chang J, Cox TR, Lang G, Bird
D, Nicolau M, Evans HR, Gartland A and Erler JT: LOXL2-mediated
matrix remodeling in metastasis and mammary gland involution.
Cancer Res. 71:1561–1572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou CX, Gao Y, Johnson NW and Gao J:
Immunoexpression of matrix metalloproteinase-2 and matrix
metalloproteinase-9 in the metastasis of squamous cell carcinoma of
the human tongue. Aust Dent J. 55:385–389. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu M, Hu Y, Zhang MF, Luo KJ, Xie XY, Wen
J, Fu JH and Yang H: MMP1 promotes tumor growth and metastasis in
esophageal squamous cell carcinoma. Cancer Lett. 377:97–104. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma HP, Chang HL, Bamodu OA, Yadav VK,
Huang TY, Wu ATH, Yeh CT, Tsai SH and Lee WH: Collagen 1A1 (COL1A1)
is a reliable biomarker and putative therapeutic target for
hepatocellular carcinogenesis and metastasis. Cancers (Basel).
11:7862019. View Article : Google Scholar
|
|
55
|
Misawa K, Mochizuki D, Imai A, Mima M,
Misawa Y and Mineta H: Analysis of site-specific methylation of
tumor-related genes in head and neck cancer: Potential utility as
biomarkers for prognosis. Cancers (Basel). 10:272018. View Article : Google Scholar
|
|
56
|
Su B, Zhao W, Shi B, Zhang Z, Yu X, Xie F,
Guo Z, Zhang X, Liu J, Shen Q, et al: Let-7d suppresses growth,
metastasis, and tumor macrophage infiltration in renal cell
carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 13:2062014.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang T, Hou J, Jian S, Luo Q, Wei J, Li Z,
Wang X, Bai P, Duan B, Xing J and Cai J: miR-29b negatively
regulates MMP2 to impact gastric cancer development by suppress
gastric cancer cell migration and tumor growth. J Cancer.
9:3776–3786. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nabais Sá MJ, Storey H, Flinter F, Nagel
M, Sampaio S, Castro R, Araújo JA, Gaspar MA, Soares C, Oliveira A,
et al: Collagen type IV-related nephropathies in Portugal:
Pathogenic COL4A3 and COL4A4 mutations and clinical
characterization of 25 families. Clin Genet. 88:456–461. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Takeuchi M, Yamaguchi S, Yonemura S,
Kakiguchi K, Sato Y, Higashiyama T, Shimizu T and Hibi M: Type IV
collagen controls the axogenesis of cerebellar granule cells by
regulating basement membrane integrity in Zebrafish. PLoS Genet.
11:e10055872015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mammadova-Bach E, Zigrino P, Brucker C,
Bourdon C, Freund M, De Arcangelis A, Abrams SI, Orend G, Gachet C
and Mangin PH: Platelet integrin α6β1 controls lung metastasis
through direct binding to cancer cell-derived ADAM9. JCI Insight.
1:e882452016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lavergne M, Janus-Bell E, Schaff M, Gachet
C and Mangin PH: Platelet integrins in tumor metastasis: Do they
represent a therapeutic target? Cancers (Basel). 9:1332017.
View Article : Google Scholar
|
|
62
|
Repsold L, Pool R, Karodia M, Tintinger G
and Joubert AM: An overview of the role of platelets in
angiogenesis, apoptosis and autophagy in chronic myeloid leukaemia.
Cancer Cell Int. 17:892017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Egan K, Cooke N and Kenny D: Living in
shear: Platelets protect cancer cells from shear induced damage.
Clin Exp Metastasis. 31:697–704. 2014. View Article : Google Scholar : PubMed/NCBI
|