Introduction
Acute lymphoblastic leukaemia (ALL) is a type of
malignant proliferative disease that originates from B-lineage or
T-lineage lymphoid progenitor cells (1). ALL is mainly composed of primitive and
immature lymphocyte clones with high specificity (2). Patients with ALL, both adults and
paediatrics, face unique clinical challenges. In adults, ALL is a
rare disease with poor prognosis, and the 5-year overall survival
rate is 30–40% for patients <60 years of age and 15% for
patients >60 years of age (3,4). The
prognosis of recurrent disease is very poor, and the median
survival time is only 6 months (5).
Moreover, ALL is the most common malignant tumour in paediatrics,
and thus it is an important cause of cancer-related mortality in
children.
The treatment strategies for leukaemia include
systemic chemotherapy, local radiotherapy and surgical treatment,
among which chemotherapy is the most important clinical method
(6). In recent years, single drug
plus small dose chemotherapy has been improved to which greatly
improves the therapeutic effect in patients with ALL (7). Currently, the chemotherapy drugs for
T-cell Acute lymphoblastic leukaemia (T-ALL) include etoposide and
methotrexate (MTX) (8,9). Although most patients with T-ALL
achieve remission after treatment, the presence of resistant
leukaemia cells means ~20% of patients do not accomplish remission,
which results in pain to patients and their families (10). Therefore, in-depth study of the
mechanism of T-ALL drug resistance has an important social and
medical value.
Early T-cell precursor (ETP) of ALL (ETP-ALL) was
identified to be a subtype of T-ALL in 2009, and this subtype
accounts for up to 15% of T-ALL and has a high risk of becoming
refractory (11,12). To understand and overcome ETP-ALL,
researchers have analysed the differences between ETP-ALL and
classic T-ALL at the genomic level, and with the publication of the
ETP-ALL-related gene expression profile, numerous differentially
expressed genes have been identified; for example, whole-exome
sequencing in adult ETP-ALL revealed a high rate of DNA
methyltransferase 3 α mutations (13,14).
However, the specific functions of a large number of differentially
expressed genes in ETP-ALL, and whether these have potential
effects on treatment requires further research and verification.
Furthermore, to the best of our knowledge, the specific role of
HSH2D in T-ALL has not been previously reported.
Haematopoietic SH2 domain containing (HSH2D) is an
important signalling molecule that affects T-cell activation
(15,16). It has been revealed that HSH2D can
inhibit the transcriptional activation of the IL-2 promoter,
especially at the RE/AP element of IL-2, which is mediated by CD28
(17,18). Considering that the expression of
HSH2D in ETP-ALL is higher compared with that in classical T-ALL,
it was hypothesized that HSH2D may serve a role in IL-2-dependent
human T-lymphoma cells and affect the chemotherapy response.
Materials and methods
Database analysis
The Haferlach leukaemia data set, which included
patients of ALL, from the Oncomine database (https://www.oncomine.org/) was analysed (19). HSH2D expression was assessed in
leukaemia tissue compared with healthy tissue, and differences of
P=1×10−4 were considered to be significant. To further
assess the expression of HSH2D in T-ALL, the results of a
differential microarray based on GEO data were analysed (NCBI GEO
database ETP-ALL; dataset record GDS4299; P=0.0268) (13,20).
Cell culture
HuT-78 cells (BeNa Cell Culture Collection) were
maintained in DMEM (Thermo Fisher Scientific, Inc.) supplementing
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2.
For knockdown of HSH2D, a small interfering (si)RNA
targeting HSH2D (150 nM; siHSH2D, 5′-UGGUUAUUCUGUUCAUCUCUGTT-3′ and
5′-CAGAGAUGAACAGAAUAACCATT-3′; siHSH2D-2,
5′-AGCTGGAGTGGAATGGCACAGTCTATT-3′ and
5′-TAGACTGTGCCATTCCACTCCAGCTTT-3′) and the corresponding negative
control (50 nM; NC; 5′-UUCUCCGAACGUGUCACGUTT-3′ and
5′-ACGUGACACGUUCGGAGAATT-3′) were designed by Shanghai GenePharma
Co. Ltd. For HSH2D overexpression, HSH2D overexpression plasmid
(pcDNA3.1-HSH2D) was also purchased from Shanghai GenePharma Co.,
Ltd. Cells were transfected using Lipofectamine® 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to manufacturer's protocols. Cells were harvested 48 h
after transfection for cell cycle analysis, reverse
transcription-quantitative PCR (RT-qPCR), EdU assay and western
blotting.
According to the manufacturer's instructions, 50
µg/ml recombinant human IL-2 (Beyotime Institute of Biotechnology;
cat. no. P5115) was prepared via dissolution in room temperature.
Briefly, HSH2D overexpressing cells (2×105 cells/well)
were seeded in 12-well plates and incubated at 37°C with 5%
CO2 for 24 h. After carefully removing the medium, 1 µl
IL-2 was added to 999 µl medium [50 ng per ml (100 U)] and the
cells were cultured at 37°C for 48 h.
Cell counting Kit (CCK)-8 assay for
IC50 analyses
The IC50 of HuT-78 cell line to MTX was
evaluated via CCK-8 assay. Briefly, the cells were seeded into
96-well plates at a density of 5×103 cells per well and
left overnight in 37°C. The cells were then treated with different
concentrations (0, 1, 2, 4, 8, 16 and 32 nmol) of MTX (Selleck
Chemicals; cat. no. S1210) at 37°C for 24 h. Then, 10 µl CCK-8
solution (Wuhan Boster Biological Technology, Ltd.) was added into
each well and incubated for 4 h at 37°C. Subsequently, the
absorbance of each well was determined using a microplate reader
(BioTek ELx800; BioTek China) at 450 nm. Each sample was designed
with five repeats and each experiment was performed ≥3 times. The
cytotoxic effect of the treatment was determined as percentage of
viability as compared to untreated cells, with the following
equation (1): Cell viability (%) =
Absorbance of sample cells/Absorbance of untreated cells ×100.
Moreover, the IC50 is 50% inhibition concentration, that
is, the concentration corresponding to Absorbance of sample
cells/Absorbance of untreated cells = 50%.
RT-qPCR
Total RNA was extracted using an RNA Sample Total
RNA kit (Qiagen GmbH) according to the manufacturer's instructions.
GAPDH served as a reference gene. cDNA templates were constructed
via reverse transcription of RNA performed using a BestarTM qPCR RT
kit (Shanghai Xinghan Biological Technology Co., Ltd.) under the
following conditions: 37°C for 15 min and 98°C for 5 min to
synthesize the first strand of cDNA. The final RT-qPCR reaction
mixture had a volume of 20 µl and contained 10 µl
Bestar® SybrGreen qPCR master mix (Shanghai Xinghan
Biological Technology Co., Ltd.), 1 µl each primer (10 µM; IL-2
forward: 5′-TACAAGAACCCGAAACTGACTCG-3′ and reverse,
5′-ACATGAAGGTAGTCTCACTGCC-3′; and CD28 forward,
5′-CTATTTCCCGGACCTTCTAAGCC-3′ and reverse,
5′-GCGGGGAGTCATGTTCATGTA-3′), 1 µl cDNA template and 8 µl
RNase-free H2O. The thermocycling conditions used for
RT-qPCR were as follows: Activation at 50°C for 2 min, Initial
denaturation at 95°C for 2 min, followed by 40 cycles of 94°C for
20 sec and 58°C for 20 sec. Relative expression levels of the
targeted genes were calculated using the 2−ΔΔCq method
(21).
Western blot analysis
Total proteins were extracted using a Total Protein
Extraction kit (Wanleibio Co., Ltd.) according to the
manufacturer's instructions. The protein concentration was
quantified using BCA protein concentration determination kit
(Beyotime Institute of Biotechnology; cat. no. P0012). A 20 µg
sample of total protein was subjected to 10% SDS-PAGE, and the
separated protein bands were transferred onto PVDF membranes, which
were subsequently blocked with 5% skim milk at room temperature for
1 h. After washing, the membranes were incubated with the primary
antibodies against HSH2D (cat. no. ab169172, 1:1,000; Abcam), IL-2
(cat. no. ab92381, 1:1,000; Abcam), CD28 (cat. no. ab243228,
1:1,000; Abcam), β-actin (cat. no. ab8226; 1:5,000; Abcam) or GAPDH
(cat. no. ab8245, 1:2,000; Abcam) at 4°C overnight. Next,
horseradish peroxidase-conjugated secondary antibodies (cat. no.
ab205719, 1:5,000; Abcam) were added and incubated with the
membranes for 45 min at 37°C. The immunostained protein blots were
developed using Beyo ECL Plus reagent (Beyotime Institute of
Biotechnology; cat. no. P0018FS) and detected with a Gel Imaging
system (Thermo Fisher Scientific, Inc.). The relative expression
levels of the targeted proteins were calculated using a
Gel-Pro-Analyzer Plus 4.0 (Media Cybernetics, Inc.).
Flow cytometry analysis
According to the manufacturer's instructions, the
cell cycle was evaluated using Cell Cycle and Apoptosis Analysis
kit (Beyotime Institute of Biotechnology; cat. no. C1052). The
treated HuT-78 cells (2×106 cells/ml) with MTX were
harvested and incubated with 5 µl PI and 10 µg/ml RNase (Beyotime
Institute of Biotechnology; cat. no. ST577) for 15 min at room
temperature in dark.
Cell apoptosis was evaluated using Annexin V FITC/PI
apoptosis detection kit (Beyotime Institute of Biotechnology; cat.
no. C1062). The treated HuT-78 cells (2×106 cells/ml)
with siRNA were resuspended and incubated with 5 µl Annexin V-FITC
and 10 µl PI for 15 min, further incubated at 20–25°C in the dark
for 10–20 min and then place in an ice bath. Cell cycle and
apoptosis were examined using BD FACSCalibur flow cytometer (Becton
Dickinson and Company) and CFLOW plus (Becton Dickinson and
Company).
EdU proliferation assay
The proliferative ability of the siRNA transfected
Hut-78 cells was detected using a Cell-LightTM EdU kit (Guangzhou
RiboBio Co., Ltd.). Transfected HuT-78 cells (1×104
cells/well) were placed into 96-well plates (Costar; Corning, Inc.)
and maintained at 37°C with 5% CO2 for 48 h. Cellular
medium was discarded and HuT-78 cells were then fixed with 4%
paraformaldehyde at room temperature for 30 min (Beyotime Institute
of Biotechnology). The proliferation assay was conducted following
the manufacturer's instructions of the EdU kit. The images were
captured using a digital fluorescent microscope (BX51 Olympus;
Olympus Corporation; magnification, ×200). Each experiment was
repeated for three times, and five images were captured each
time.
Statistical analysis
Data are presented as the mean ± SD, with n=3/group.
Differences between groups were analysed using unpaired Student's
t-test, and differences among ≥3 groups were detected using one-way
ANOVA analysis, followed by a post hoc Tukey's test. Statistical
analysis was conducted using GraphPad Prism 6.0 (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Result
HSH2D is expressed at low levels in
T-ALL
To study the expression of HSH2D in leukaemia, the
Haferlach leukaemia data set from the Oncomine database was
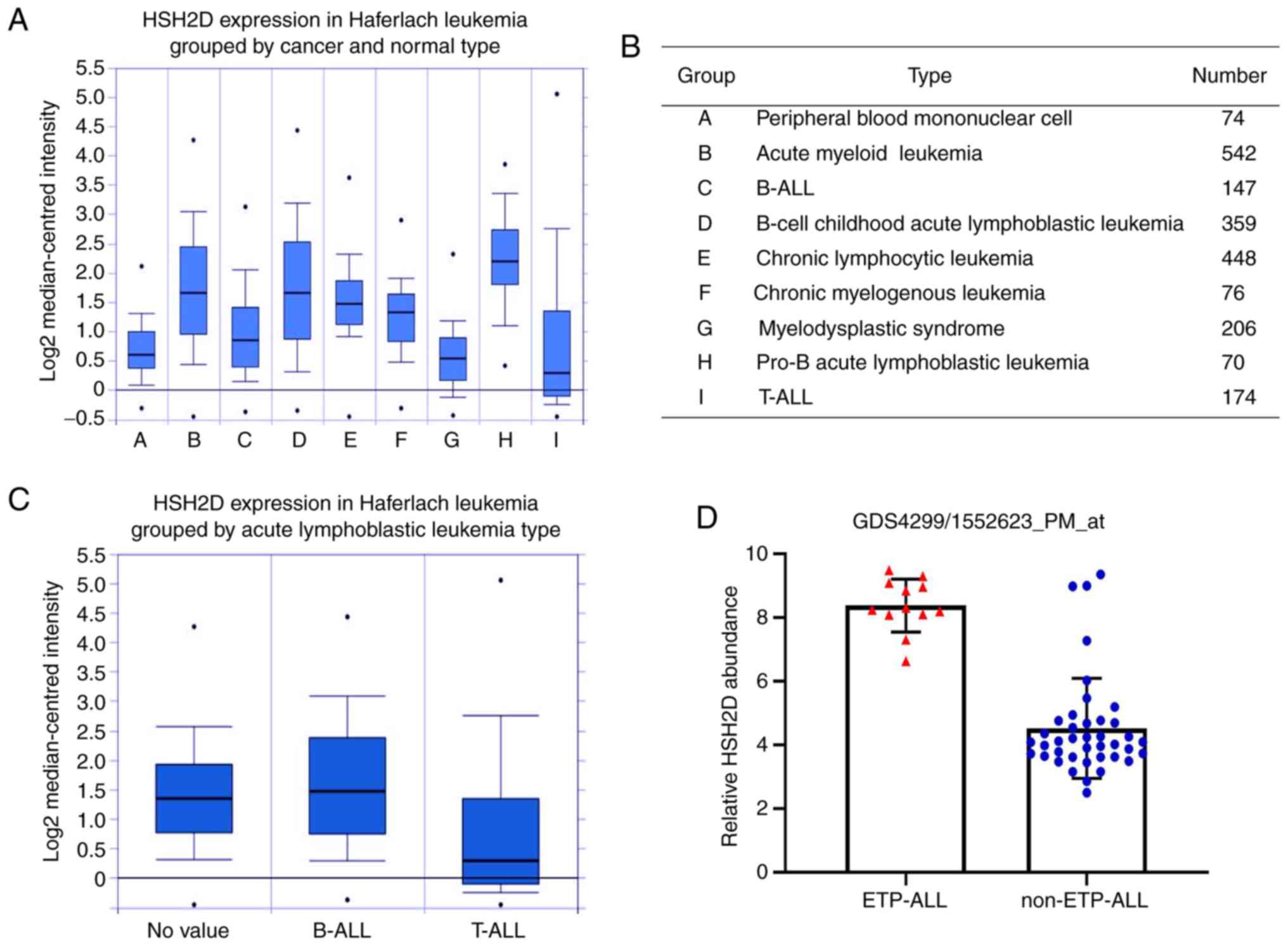
analysed. In the comparison of healthy tissue and cancer tissue,
pro-B ALL had the highest expression of HSH2D (P=1×10−4;
Fig. 1A and B), while the
expression in T-ALL was lower compared with that in peripheral
blood mononuclear cells. Moreover, the expression of HSH2D in
B-cell Acute lymphoblastic leukaemia (B-ALL) was higher compared
with T-ALL, which was also confirmed in a group of ALL cells
(Fig. 1C).
To further assess the expression of HSH2D in T-ALL,
the results of a differential microarray based on GEO data was
evaluated and it was found that the expression of HSH2D in patients
with ETP-ALL was markedly higher compared with patients without ETP
T-ALL (NCBI GEO database ETP-ALL; dataset record GDS4299) (Fig. 1D). The aforementioned results
suggested that the expression of HSH2D was significantly higher in
T-ALL compared with all other patients (NCBI GEO database ETP-ALL;
dataset record GDS4299) (Fig. 1D),
and expression in the T-ALL subtype was lower compared with that in
the B-ALL subtype.
HSH2D inhibits CD28-mediated
activation of IL-2
To investigate the role of HSH2D in T-ALL, an
expression plasmid for HSH2D was constructed and HuT-78 cells were
transfected with this plasmid and with siRNA technology to
construct HSH2D overexpression and knockdown cell lines. Then, the
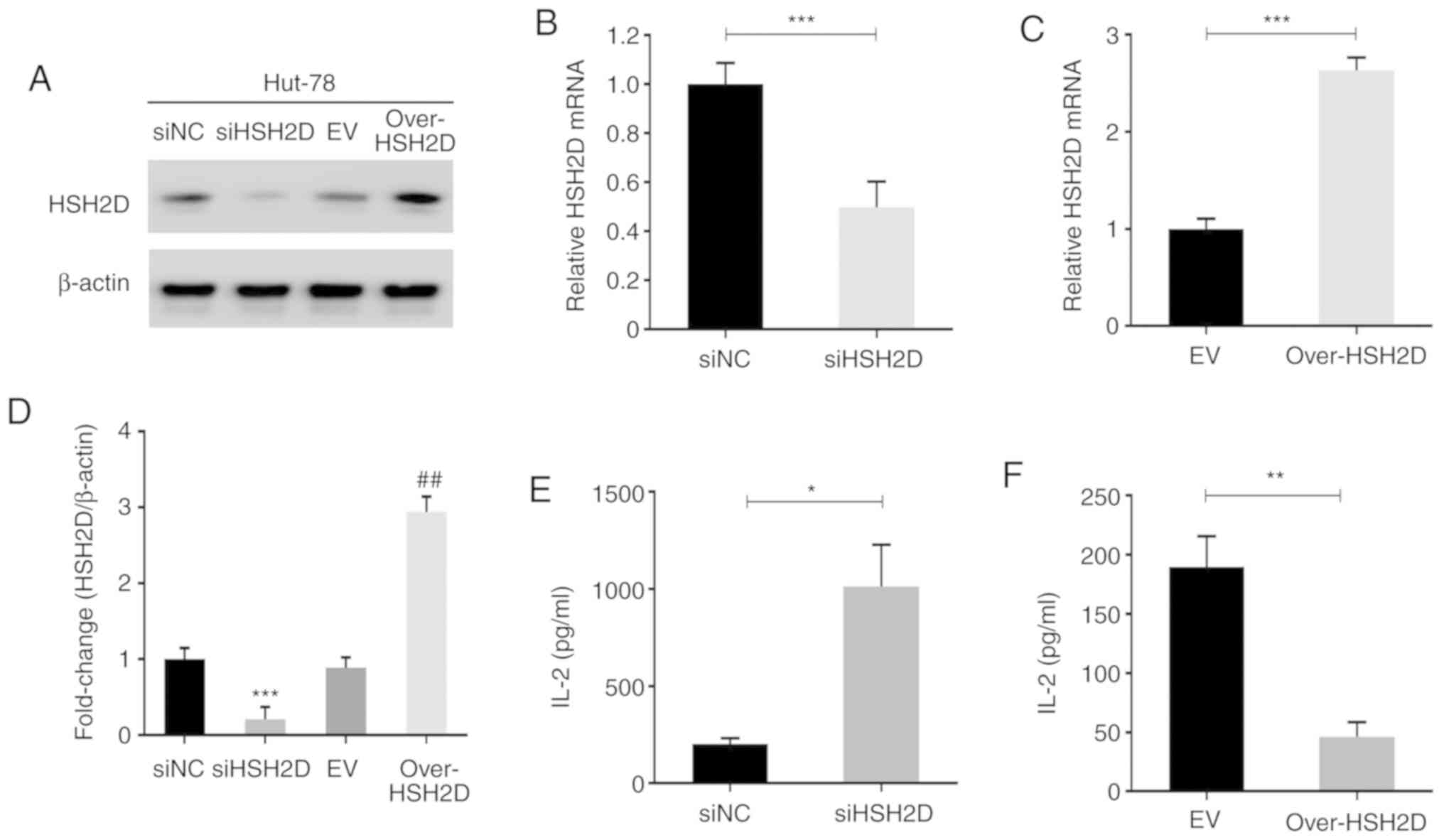
cells were assessed via western blotting, and the effects of
overexpression and knockdown were confirmed (Fig. 2A). Grey scale analysis was performed
on three repeated experiments, and statistical analysis was
performed, which identified that there were significant differences
between the overexpression and knockdown cell lines compared with
the corresponding NC (Fig. 2D).
RT-qPCR was conducted to measure the mRNA expression levels of the
transfected HuT-78 cells, and the results indicated that the
constructed plasmid and interference sequences were effective
(Fig. 2B and C).
As previous studies have reported that HSH2D can
inhibit the transcriptional activation of the IL-2 promoter, the
present study detected the expression and secretion of IL-2 using
RT-qPCR and ELISA. The results demonstrated that overexpression of
HSH2D inhibited IL-2 transcription and that HSH2D knockdown
promoted exocrine IL-2 secretion (Fig.
2E and F). It has been revealed that CD28 serves a key role in
the transcriptional expression of IL-2, and thus in the current
study human recombinant IL-2 [50 ng per ml (100 U)] was added to
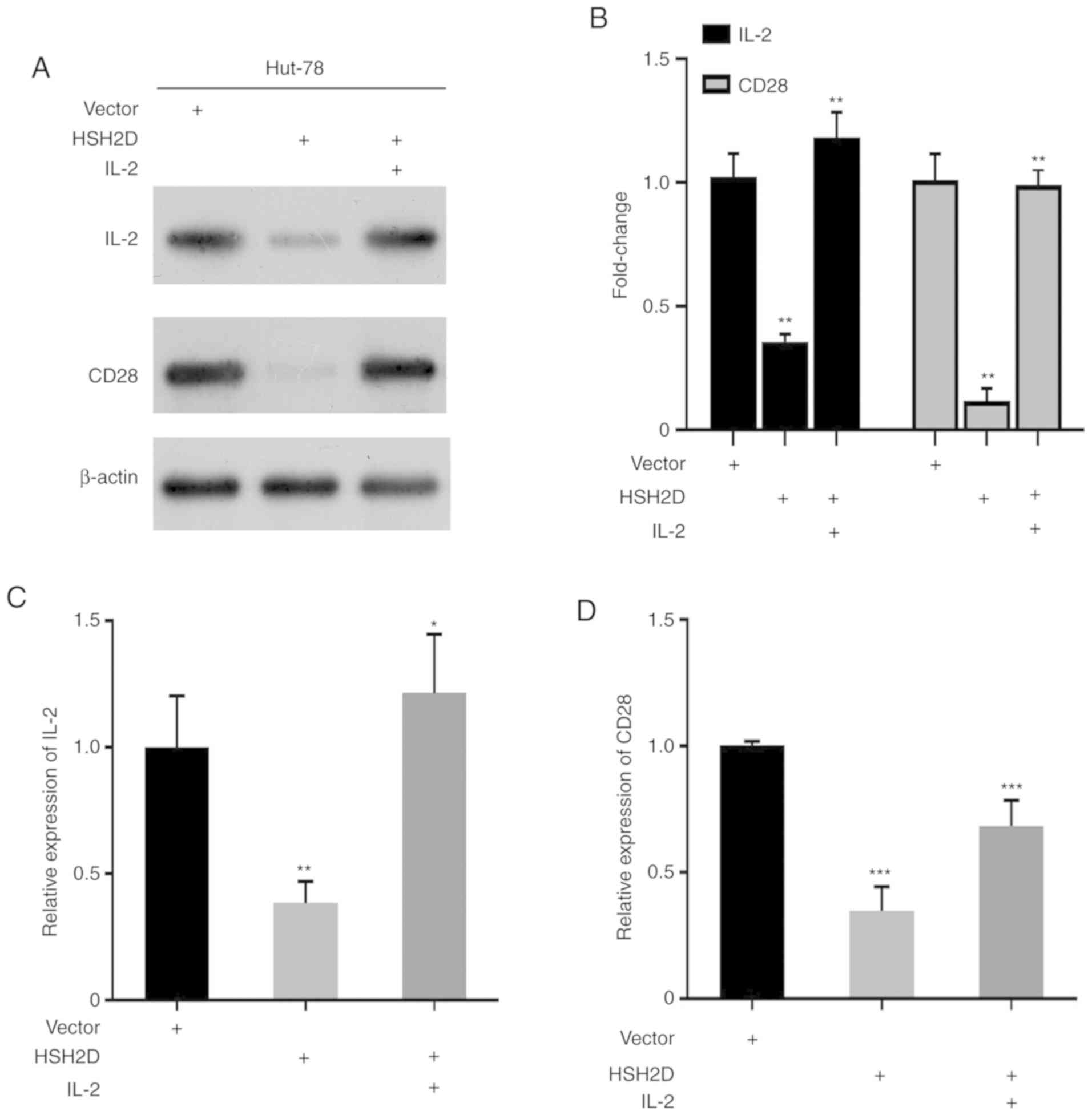
HUT-78 cells transfected with the HSH2D expression vector. Western
blotting results identified that the addition of exogenous IL-2
could restore HSH2D expression (Fig.
3A). Overexpression of HSH2D inhibitsCD28 and IL-2 protein
expression (Fig. 3A and B), and the
RT-qPCR analysis demonstrated the same results (Fig. 3C and D).
Anticancer drug methotrexate inhibits
the proliferation and promotes the apoptosis of HuT-78 cells
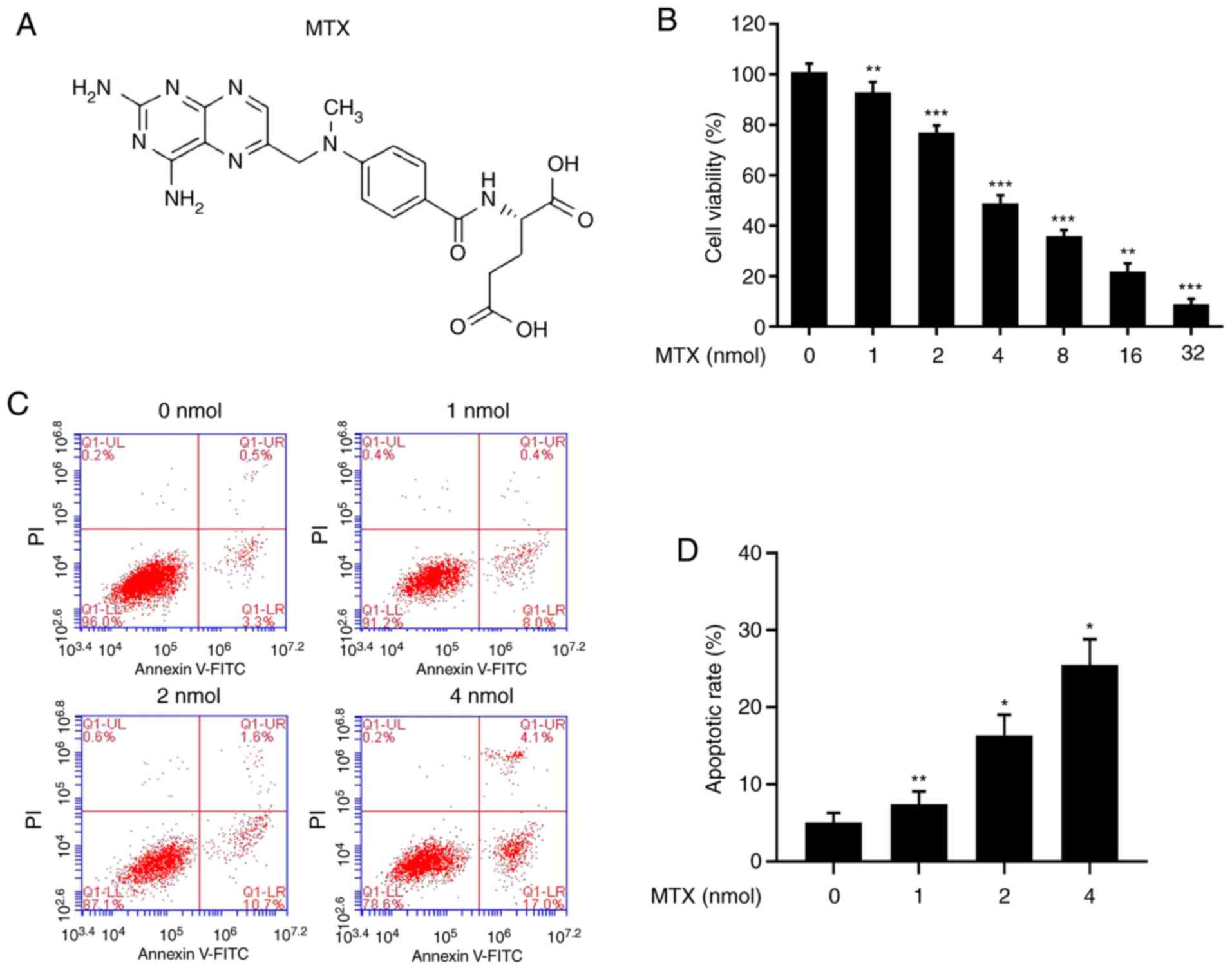
MTX (Fig. 4A) is a
chemical drug commonly used in children with ALL, and it serves a
major role in the treatment of T-ALL. Cells were treated with
different concentrations of MTX (0, 1, 2, 4, 8, 16 and 32 nmol),
and it was identified that the survival rate of the cells was
first-order concentration-dependent: The higher the concentration,
the lower the survival rate (Fig.
4B). The flow cytometry results demonstrated that as the
concentration of MTX increased, the apoptotic rate of the cells
increased (Fig. 4C and D). Thus,
the results indicated that MTX may inhibit the proliferation of
tumour cells by promoting apoptosis.
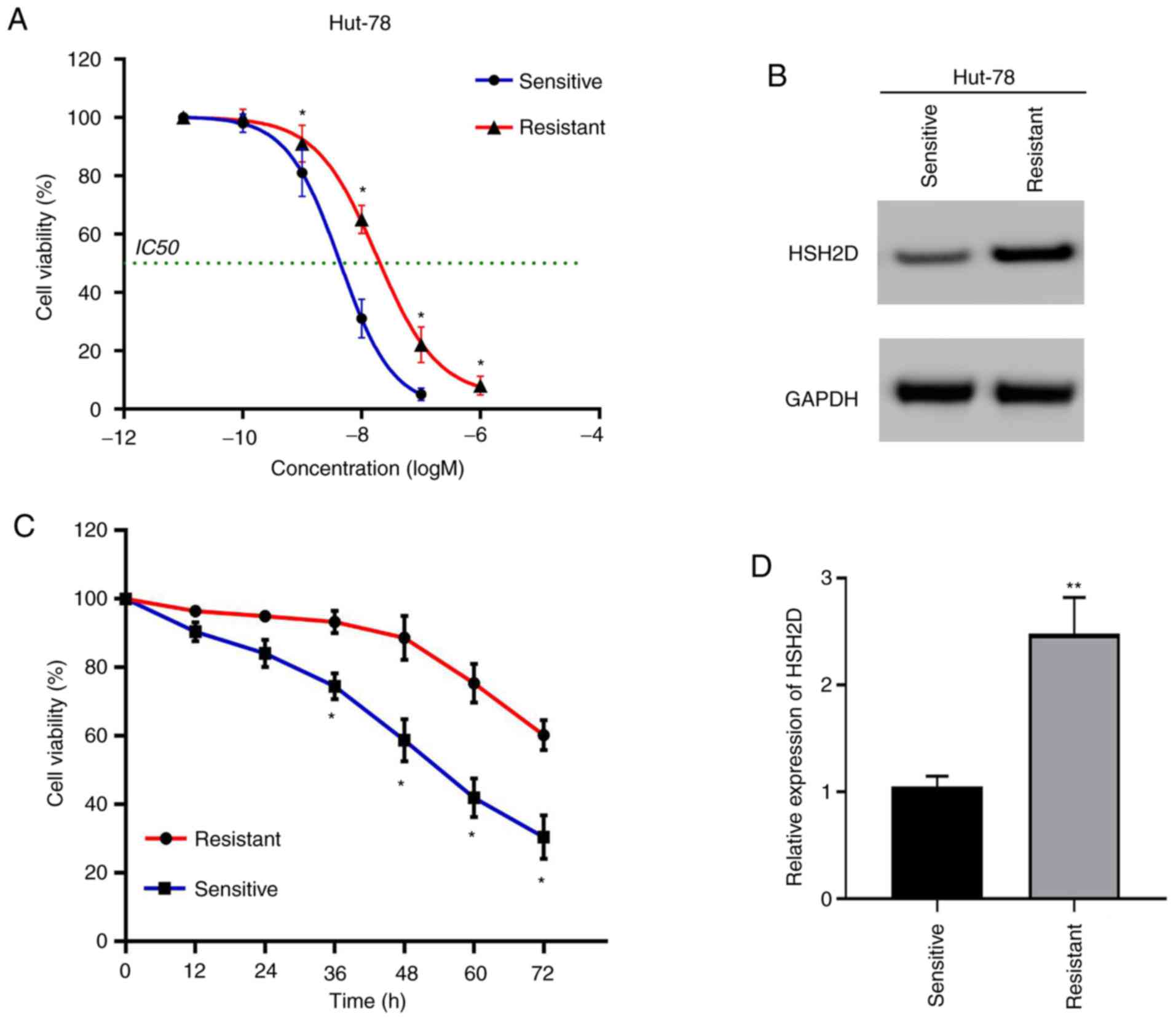
HSH2D is associated with the
resistance of MTX
As drug resistance to MTX is a key factor in the
success of T-ALL treatment, MTX was used to culture resistant cell
lines (22). The drug-resistant
cell lines and drug-sensitive cell lines were compared via the
CCK-8 method, and it was found that the IC50 value of
the resistant cell lines was
1.773×10−8−3.367×10−8 M, and the
IC50 of the sensitive strain was
3.847×10−9−4.802×10−9 M (Fig. 5A). Compared with the experimental
results from the sensitive strain group, the resistant strain
relieved the cytotoxicity induced by MTX (20 nmol) (Fig. 5C).
To determine the association between MTX resistance
and HSH2D, the protein expression of HSH2D was detected via western
blotting. The results demonstrated that the expression of HSH2D was
markedly higher in the resistant cell lines compared with the
sensitive cell lines (Fig. 5B). In
addition, the experimental results of the RT-qPCR analysis
indicated the same conclusion (Fig.
5D).
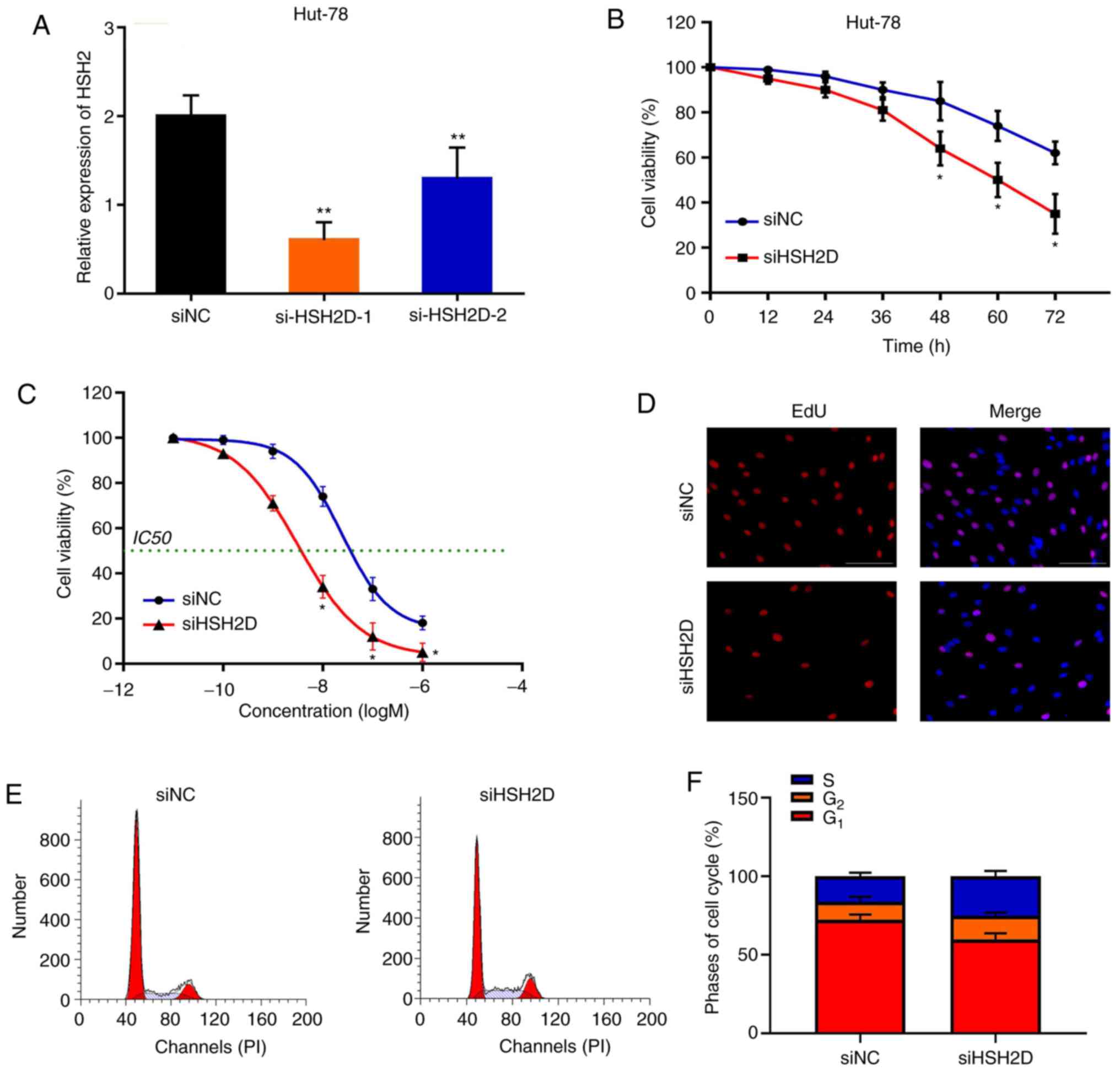
HSH2D is a prerequisite for the
resistance of HuT-78 cells to MTX
To investigate whether HSH2D is important for MTX
resistance in HuT-78 cells, a loss-of-function assay was performed.
Synthetic siRNA was transfected into HuT-78 cells and interfered
with HSH2D expression. It was identified that siHSH2D-1
significantly silenced HSH2D in HuT-78 cells compared with the NC,
and to a greater extent than si-HSH2D-2 (Fig. 6A). Silencing HSH2D promoted MTX
treatment (20 nM)-induced cytotoxicity compared with the control
group (Fig. 6B). Furthermore,
knockdown of HSH2D resulted in a decrease in the IC50
value and in the inhibition of cell proliferation after MTX
treatment (Fig. 6C and D). In
addition, flow cytometry analysis demonstrated that after MTX
treatment and HSH2D silencing in drug-resistant cell lines, the
G1 phase of the cells was shortened and the S phase was
increased (Fig. 6E and F).
Collectively, these results indicated that HSH2D serves a crucial
role in MTX resistance.
Discussion
The treatment strategies for leukaemia include
systemic chemotherapy, local radiotherapy and surgical treatment,
among which chemotherapy is the most important clinical method
(23,24). Single drug low-dose chemotherapy has
been improved to multi-drug high-dose treatment, which greatly
increases the therapeutic effect in paediatric patients with ALL
(25). At present, the chemotherapy
drugs for T-ALL include MTX and etoposide. Although the majority of
paediatric patients with T-ALL go into remission after treatment
due to the presence of drug-resistant leukaemia cells, 20% of
paediatrics do not go into remission, which is a burden on the
patients and their families (26,27).
Therefore, an in-depth study of the mechanism of T-ALL drug
resistance has important social and medical value. In the early
stage, the present results demonstrated that the expression of
HSH2D was significantly increased in lymphoblastic leukaemia cells
and decreased in T-ALL, which suggested that HSH2D may be
associated with the malignancy and drug resistance of T-ALL
tumours.
Immunotherapy has occupied a certain position in the
treatment of leukaemia. IL-2, as a therapeutic agent to improve the
immune function of cells, has been widely used in clinical tumour
treatment (28). Previous studies
have reported that, in ALL, autogenous bone metastasis often leads
to leukaemia recurrence (29). To
produce anti-leukemic immunity, researchers have used recombinant
human IL-2, which is given immediately after transplantation
(30). The clinical toxicity,
haematopoiesis recovery and immune activation of patients treated
with IL-2 have been compared with a group of patients without IL-2
(31). Compared with IL-2 controls,
patients receiving IL-2 tended to recover neutrophils, platelets
and red blood cells earlier and were discharge earlier (28). Moreover, the expression of IL-2Rα
(CD25) on the cell surface is considered a powerful predictor of
poor prognosis in patients with acute myeloid leukaemia (AML)
(32,33). After overexpression and knockdown of
HSH2D in HuT-78 cells, the present study identified that HSH2D
affected the expression of IL-2 and may inhibit the CD28-mediated
activation of the RE/AP element of IL-2. Du et al (34) revealed that serum concentrations of
IL-2 in paediatric patients with ALL decreased significantly, and
along with the analysis of the current findings, it was suggested
that the change of HSH2D expression in patients with ALL results in
the decrease of IL-2. IL-2 receptors, CD25 and CD28, are important
genes for regulatory T-cells (35).
The changes in IL-2 caused by HSH2D may be an important factor in
the changes in the content of regulatory T-cells in ALL, and
previous studies are using the effects of anti-CD3/CD28 coated
beads and IL-2 on expanded T-cells for immunotherapy (36). Thus, HSH2D may be a potential
therapeutic target for ALL.
MTX is an important component in the treatment of
ALL. Currently, 80% of patients diagnosed with early pre-B-ALL are
treated with combined chemotherapy, but this treatment is
ineffective for 20% of patients (37). The failure of the treatment is
partly due to the resistance of the cells to the therapeutic drugs
(38), and drug resistance is more
common in the T-lineage phenotype. Intensive treatment including
high-dose MTX can improve the survival rate of patients with T-ALL
to that of patients with B-ALL (39). Thus, cell resistance to drugs can be
partially overcome by increased doses. The study of MTX resistance
is ongoing (40). It has been
revealed that the increase of dihydrofolate reductase and the
damage of MTX transport are key factors in MTX resistance in
childhood ALL (41). Previous
studies have measured the difference in MTX resistance-related mRNA
expression levels in childhood leukaemia by standardized RT-qPCR
based on a competitive template, and observed that in T-ALL,
compared with ordinary/pre-B-ALL, the difference in MTX resistance,
dihydrofolate reductase (DHFR), thymidylate synthetase (TS) and
folylpolyglutamate synthase mRNA expression levels are increased
(22,42,43).
The present results suggested that HSH2D is one of the important
factors affecting MTX resistance, and identified via gene
manipulation that HSH2D expression is necessary for HuT-78 cells
resistance to MTX. The intracellular accumulation of MTX
polyglutamic acid in leukaemia cells is an important determinant of
the anti-leukaemia activity of MTX in paediatric patients with ALL
(44). The traditional explanation
for MTX resistance is due to inactivating mutations or
downregulation affecting the replication factor C gene, as well as
increased levels of DHFR and TS enzymes and mutations with reduced
affinity for antifolates (45).
Wojtuszkiewicz et al (22)
reported that the low cellular level of long-chain polyglutamates
of MTX is an important predictor of MTX resistance and is
associated with dismal therapeutic outcome. This study demonstrated
the mechanism of MTX resistance to HuT-78 cells from the
perspective of signal regulation. It also suggested that the
regulation of HSH2D on ALL may participate in the expression of RFC
and TS (22). However, this
requires further in-depth investigation.
In conclusion, drug resistance is an obstacle in the
use of chemotherapy for the treatment of lymphoblastic leukaemia.
The present results demonstrated that in HuT-78 cells, HSH2D
inhibited CD28-mediated activation of IL-2, and via the cultivation
of the MTX-resistant cell line, it was identified that HSH2D
expression was necessary for HuT-78 cells to be resistant to MTC.
Therefore, the current results provide a potential target for the
reversal of MTX resistance.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX conceived and designed the experiments. JW
performed the experiments and analyzed the data. JW and YX wrote
the manuscript. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singh N, Frey NV, Grupp SA and Maude SL:
CAR T cell therapy in acute lymphoblastic leukemia and potential
for chronic lymphocytic leukemia. Curr Treat Options Oncol.
17:282016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Swaminathan S, Klemm L, Park E,
Papaemmanuil E, Ford A, Kweon SM, Trageser D, Hasselfeld B, Henke
N, Mooster J, et al: Mechanisms of clonal evolution in childhood
acute lymphoblastic leukemia. Nat Immunol. 16:766–774. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Belver L and Ferrando A: The genetics and
mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer.
16:494–507. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roberts KG and Mullighan CG: Genomics in
acute lymphoblastic leukaemia: Insights and treatment implications.
Nat Rev Clin Oncol. 12:344–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Conter V, Valsecchi MG, Buldini B,
Parasole R, Locatelli F, Colombini A, Rizzari C, Putti MC, Barisone
E, Lo Nigro L, et al: Early T-cell precursor acute lymphoblastic
leukaemia in children treated in AIEOP centres with AIEOP-BFM
protocols: A retrospective analysis. Lancet Haematol. 3:e80–e86.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cooper SL and Brown PA: Treatment of
pediatric acute lymphoblastic leukemia. Pediatr Clin North Am.
62:61–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kantarjian H, Stein A, Gökbuget N,
Fielding AK, Schuh AC, Ribera JM, Wei A, Dombret H, Foà R, Bassan
R, et al: Blinatumomab versus chemotherapy for advanced acute
lymphoblastic leukemia. N Engl J Med. 376:836–847. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pui CH, Yang JJ, Hunger SP, Pieters R,
Schrappe M, Biondi A, Vora A, Baruchel A, Silverman LB, Schmiegelow
K, et al: Childhood acute lymphoblastic leukemia: Progress through
collaboration. J Clin Oncol. 33:2938–2948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grohmann T, Penke M, Petzold-Quinque S,
Schuster S, Richter S, Kiess W and Garten A: Inhibition of NAMPT
sensitizes MOLT4 leukemia cells for etoposide treatment through the
SIRT2-p53 pathway. Leuk Res. 69:39–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartram I, Erben U, Ortiz-Tanchez J,
Blunert K, Schlee C, Neumann M, Heesch S and Baldus CD: Inhibition
of IGF1-R overcomes IGFBP7-induced chemotherapy resistance in
T-ALL. BMC Cancer. 15:6632015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Porter DL, Hwang WT, Frey NV, Lacey SF,
Shaw PA, Loren AW, Bagg A, Marcucci KT, Shen A, Gonzalez V, et al:
Chimeric antigen receptor T cells persist and induce sustained
remissions in relapsed refractory chronic lymphocytic leukemia. Sci
Transl Med. 7:303ra1392015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jain N, Lamb AV, O'Brien S, Ravandi F,
Konopleva M, Jabbour E, Zuo Z, Jorgensen J, Lin P, Pierce S, et al:
Early T-cell precursor acute lymphoblastic leukemia/lymphoma
(ETP-ALL/LBL) in adolescents and adults: A high-risk subtype.
Blood. 127:1863–1869. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Ding L, Holmfeldt L, Wu G,
Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et
al: The genetic basis of early T-cell precursor acute lymphoblastic
leukaemia. Nature. 481:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Neumann M, Heesch S, Schlee C, Schwartz S,
Gökbuget N, Hoelzer D, Konstandin NP, Ksienzyk B, Vosberg S, Graf
A, et al: Whole-exome sequencing in adult ETP-ALL reveals a high
rate of DNMT3A mutations. Blood. 121:4749–4752. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oda T, Muramatsu MA, Isogai T, Masuho Y,
Asano S and Yamashita T: HSH2: A novel SH2 domain-containing
adapter protein involved in tyrosine kinase signaling in
hematopoietic cells. Biochem Biophys Res Commun. 288:1078–1086.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lapinski PE, Oliver JA, Bodie JN, Marti F
and King PD: The T-cell-specific adapter protein family: TSAd, ALX,
and SH2D4A/SH2D4B. Immunol Rev. 232:240–254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pegram HJ, Purdon TJ, van Leeuwen DG,
Curran KJ, Giralt SA, Barker JN and Brentjens RJ: IL-12-secreting
CD19-targeted cord blood-derived T cells for the immunotherapy of
B-cell acute lymphoblastic leukemia. Leukemia. 29:415–422. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shapiro MJ, Powell P, Ndubuizu A, Nzerem C
and Shapiro VS: The ALX Src homology 2 domain is both necessary and
sufficient to inhibit T cell receptor/CD28-mediated up-regulation
of RE/AP. J Biol Chem. 279:40647–40652. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Haferlach T, Kohlmann A, Wieczorek L,
Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK,
Mills KI, et al: Clinical utility of microarray-based gene
expression profiling in the diagnosis and subclassification of
leukemia: Report from the International microarray innovations in
leukemia study group. J Clin Oncol. 28:2529–2537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gutierrez A, Kentsis A, Sanda T, Holmfeldt
L, Chen SC, Zhang J, Protopopov A, Chin L, Dahlberg SE, Neuberg DS,
et al: The BCL11B tumor suppressor is mutated across the major
molecular subtypes of T-cell acute lymphoblastic leukemia. Blood.
118:4169–4173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wojtuszkiewicz A, Peters GJ, Van Woerden
NL, Dubbelman B, Escherich G, Schmiegelow K, Sonneveld E, Pieters
R, van de Ven PM, Jansen G, et al: Methotrexate resistance in
relation to treatment outcome in childhood acute lymphoblastic
leukemia. J Hematol Oncol. 8:612015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ishida T, Jo T, Takemoto S, Suzushima H,
Uozumi K, Yamamoto K, Uike N, Saburi Y, Nosaka K, Utsunomiya A, et
al: Dose-intensified chemotherapy alone or in combination with
mogamulizumab in newly diagnosed aggressive adult T-cell
leukaemia-lymphoma: A randomized phase II study. Br J Haematol.
169:672–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iyer NS, Balsamo LM, Bracken MB and
Kadan-Lottick NS: Chemotherapy-only treatment effects on long-term
neurocognitive functioning in childhood ALL survivors: A review and
meta-analysis. Blood. 126:346–353. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eiser C, Stride CB, Vora A, Goulden N,
Mitchell C, Buck G, Adams M and Jenney MEM; National Cancer
Research Institute Childhood Leukaemia Sub-Group and UK Childhood
Leukaemia Clinicians Network, : Prospective evaluation of quality
of life in children treated in UKALL 2003 for acute lymphoblastic
leukaemia: A cohort study. Pediatr Blood Cancer. 64:2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rodríguez-Lirio A, Pérez-Yarza G,
Fernández-Suárez MR, Alonso-Tejerina E, Boyano MD and Asumendi A:
Metformin Induces cell cycle arrest and apoptosis in drug-resistant
leukemia cells. Leuk Res Treatment. 2015:5164602015.PubMed/NCBI
|
|
27
|
Frismantas V, Dobay MP, Rinaldi A, Tchinda
J, Dunn SH, Kunz J, Richter-Pechanska P, Marovca B, Pail O, Jenni
S, et al: Ex vivo drug response profiling detects recurrent
sensitivity patterns in drug-resistant acute lymphoblastic
leukemia. Blood. 129:e26–e37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee DA: Cellular therapy: Adoptive
immunotherapy with expanded natural killer cells. Immunol Rev.
290:85–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Idris SZ, Hassan N, Lee LJ, Md Noor S,
Osman R, Abdul-Jalil M, Nordin AJ and Abdullah M: Increased
regulatory T cells in acute lymphoblastic leukaemia patients.
Hematology. 21:206–212. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Locatelli F, Moretta F, Brescia L and
Merli P: Natural killer cells in the treatment of high-risk acute
leukaemia. Semin Immunol. 26:173–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Atkins MB: Interleukin-2: Clinical
applications. Semin Oncol. 29 (3 Suppl 7):S12–S17. 2002. View Article : Google Scholar
|
|
32
|
Terwijn M, Feller N, van Rhenen A, Kelder
A, Westra G, Zweegman S, Ossenkoppele G and Schuurhuis GJ:
Interleukin-2 receptor alpha-chain (CD25) expression on leukaemic
blasts is predictive for outcome and level of residual disease in
AML. Eur J Cancer. 45:1692–1699. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cerny J, Yu H, Ramanathan M, Raffel GD,
Walsh WV, Fortier N, Shanahan L, O'Rourke E, Bednarik J, Barton B,
et al: Expression of CD25 independently predicts early treatment
failure of acute myeloid leukaemia (AML). Br J Haematol.
160:262–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Du S, Jia Y, Tang H, Sun Y, Wu W, Sun L,
Du J, Geng B, Tang C and Jin H: Immune regulation of hydrogen
sulfide in children with acute lymphoblastic leukemia. Chin Med J
(Engl). 127:3695–3699. 2014.PubMed/NCBI
|
|
35
|
Shen XH, Xu P, Yu X, Song HF, Chen H,
Zhang XG, Wu MY and Wang XF: Discrepant clinical significance of
CD28+CD8− and CD4+CD25high
regulatory T cells during the progression of hepatitis B virus
infection. Viral Immunol. 31:548–558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Martkamchan S, Onlamoon N, Wang S,
Pattanapanyasat K and Ammaranond P: The effects of anti-CD3/CD28
coated beads and IL-2 on expanded T cell for immunotherapy. Adv
Clin Exp Med. 25:821–828. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Faganel Kotnik B, Grabnar I, Bohanec
Grabar P, Dolzan V and Jazbec J: Association of genetic
polymorphism in the folate metabolic pathway with methotrexate
pharmacokinetics and toxicity in childhood acute lymphoblastic
leukaemia and malignant lymphoma. Eur J Clin Pharmacol.
67:993–1006. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kodidela S, Suresh Chandra P and Dubashi
B: Pharmacogenetics of methotrexate in acute lymphoblastic
leukaemia: Why still at the bench level? Eur J Clin Pharmacol.
70:253–260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Park E, Gang EJ, Hsieh YT, Schaefer P,
Chae S, Klemm L, Huantes S, Loh M, Conway EM, Kang ES, et al:
Targeting survivin overcomes drug resistance in acute lymphoblastic
leukemia. Blood. 118:2191–2199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lopez-Lopez E, Gutierrez-Camino A,
Bilbao-Aldaiturriaga N, Pombar-Gomez M, Martin-Guerrero I and
Garcia-Orad A: Pharmacogenetics of childhood acute lymphoblastic
leukemia. Pharmacogenomics. 15:1383–1398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ansari M, Sauty G, Labuda M, Gagné V,
Laverdière C, Moghrabi A, Sinnett D and Krajinovic M: Polymorphisms
in multidrug resistance-associated protein gene 4 is associated
with outcome in childhood acute lymphoblastic leukemia. Blood.
114:1383–1386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xin N, Fen Z, Li C, Yan X and Runming J:
Intracranial hemorrhage following oral low-dose methotrexate after
multiple toxicities caused by high-dose methotrexate in childhood
acute lymphoblastic leukemia. Front Pharmacol. 10:10722019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gervasini G and Mota-Zamorano S: Clinical
implications of methotrexate pharmacogenetics in childhood acute
lymphoblastic leukaemia. Curr Drug Metab. 20:313–330. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gonen N and Assaraf YG: Antifolates in
cancer therapy: Structure, activity and mechanisms of drug
resistance. Drug Resist Updat. 15:183–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kager L, Cheok M, Yang W, Zaza G, Cheng Q,
Panetta JC, Pui CH, Downing JR, Relling MV and Evans WE: Folate
pathway gene expression differs in subtypes of acute lymphoblastic
leukemia and influences methotrexate pharmacodynamics. J Clin
Invest. 115:110–117. 2005. View Article : Google Scholar : PubMed/NCBI
|