Introduction
Laryngeal squamous cell carcinoma (LSCC) accounts
for ~90% of all laryngeal malignancies, and has the second highest
incidence among all types of head and neck squamous cell carcinoma
(HNSCC) (1,2). The early symptoms of LSCC, such as
hoarseness, dysphagia and cervical lymph node metastasis, are
common and may be easily ignored (3). Over the past 10 years, the treatment
modalities for LSCC have changed, including surgery and
chemotherapy, but the prognosis still remains poor due to its
uncontrolled invasion and metastasis (4,5).
Therefore, it is crucial to understand the underlying molecular
mechanisms of LSCC invasion and metastasis in order to improve the
therapy and overall prognosis of this disease.
Cancer cell invasion and metastasis are complex
processes that involve several factors. One of the most critical
molecular mechanisms that mediate the metastatic cascade in cancer
cells is the epithelial-mesenchymal transition (EMT) (6). It is characterized by decreased
E-cadherin expression and increased expression levels of
nonepithelial cadherins, such as N-cadherin, and these are
considered to be important hallmark changes. Notably, loss of
E-cadherin expression is a rate-limiting step in the progression of
a well-differentiated cancer to invasive carcinoma (7,8).
According to accumulating evidence, long non-coding
RNAs (lncRNAs) are a large class of transcripts that are longer
than 200 nucleotides without protein-coding potential, and exhibit
close association with the occurrence, development and metastasis
of human cancers, including LSCC (9–13).
Hepatic nuclear factor 1α (HNF1A) antisense RNA 1 (HNF1A-AS1) is a
natural antisense transcript of the HNF1A gene, and its start site
is ~5 kb downstream of HNF1A. It is a newly identified lncRNA
located at chromosomal band 12q24.31 with a length of 2,455
nucleotides (14,15). HNF1A-AS1 was initially identified in
the regulation of proliferation and migration of esophageal
adenocarcinoma cells (14).
Furthermore, HNF1A-AS1 is downregulated in both gastric cancer
tissues and cell lines (15). These
findings indicate that HNF1A-AS1 may be involved in the suppression
of gastric cancer occurrence and development (15). Furthermore, in a recent study,
HNF1A-AS1 was demonstrated to be transcriptionally activated by
HNF1α, inhibited the malignant properties of hepatocellular
carcinoma cells both in vitro and in vivo, and
contributed to the antitumor effects of HNF1α by directly
regulating the enzyme activity of Src homology region 2
domain-containing phosphatase 1 (16). These results suggest that HNF1A-AS1
dysregulation serves an important role in carcinogenesis.
Additionally, previous studies have revealed that the promoter
regions of lncRNAs are subjected to DNA methylation-mediated
epigenetic alterations during tumorigenesis in a number of types of
cancer (17,18). However, to the best of our
knowledge, the expression and function of HNF1-AS1, and its
methylation condition, have not been reported in the development
and progression of LSCC.
In the present study, the epigenetic downregulation
of HNF1A-AS1 due to promoter hypermethylation in LSCC was
demonstrated and its biological function was evaluated. The
negative association of HNF1A-AS1 with EMT and inhibition of tumor
invasion and metastasis in LSCC were observed.
Materials and methods
Clinical samples
A total of 30 patients (age range, 47–75 years; mean
age, 61.31±7.0 years; female, 6.67%; male, 93.33%) diagnosed with
LSCC and cervical lymph node metastasis at the Second Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China) between
January 2015 and December 2016 were enrolled in the present study.
All patients provided written informed consent, and the present
study was approved by the Ethics Committee of the Second Affiliated
Hospital of Xi'an Jiaotong University. Samples were collected from
the patients who underwent resection (partial laryngectomy or total
laryngectomy), apart from those whose pathology types were not
squamous cell carcinoma. Patients who only underwent biopsy without
resection were excluded from the present study. Patients who
received radiation or chemotherapy before surgery were also
excluded. Pairs of laryngeal carcinoma tissues, adjacent non-tumor
tissues (normal tissue was obtained 1.0 cm from tumor edge) and
metastatic cervical lymph nodes were either fixed in 10% formalin
at room temperature for 12–24 h or frozen in liquid nitrogen at
−196°C after surgical resection till use.
Cell culture and treatment
The human LSCC TU-686 (Procell Life Science &
Technology Co., Ltd.), AMC-HN-8 (BeNa Culture Collection; Beijing
Beina Chunglian Biotechnology Research Institute), TU-177 (Jennio
Biotech Co., Ltd.) and 293T (Procell Life Science & Technology
Co., Ltd.) cell lines were cultured in DMEM (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (HyClone; Cytiva) and
50 U/ml penicillin-streptomycin at 37°C in a humidified atmosphere
of 5% CO2. Furthermore, the aforementioned cell lines
were treated with different concentrations (0, 1, 2.5, 5 and 10 µM)
of 5-Aza-2′-deoxycytidine (5-Aza-dC; cat. no. S3196; Selleck
Chemicals) for 24, 48 and 72 h at 37°C. Different concentrations of
5-Aza-dC were added to DMEM for treatments.
Transfection of cell lines
To generate lentiviruses that overexpress and target
short hairpin RNA (shRNA) expression of HNF1A-AS1, a full-length
cDNA of HNF1A-AS1 and oligonucleotides that encode HNF1A-AS1 shRNAs
[shRNA-non-specific control (NC), shRNA-853, shRNA-1525 and
shRNA-2048; Table SI] which were
designed by Shanghai GenePharma Co., Ltd. and named according to
the catalogue number were cloned into the
pLVX-mCMV-ZsGreen-PGK-Puro vector or the pLVX-shRNA2-Puro vector
(Biowit Technologies, Ltd.), respectively. All vectors were
verified by Sanger sequencing (19). The lentiviral vectors were
transfected into sub-confluent 293T cells together with packaging
plasmid and envelope plasmid (Biowit Technologies, Ltd.) using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) to produce lentiviral particles. The lentiviruses in the
medium were collected 48 h later and were concentrated by
ultracentrifugation at 448 × g for 10 min at room temperature. The
target cells (cultured in 6-cm plate) were infected by removing the
old culture medium and replacing it with 0.5 ml diluted viral
supernatant (MOI, 20). A total of 0.5 ml complete medium with
Polybrene was added to each well. The plates were placed in a 37°C
incubator with 5% CO2. Fresh media were added at 24 h
after infection. Following another incubation for 48 h, puromycin
(1 µg/ml) selection of stably transduced cells was started. Then,
GFP-positive cells were sorted by flow cytometry. Cells were
suspended in PBS at 1×107 cells/ml and filtered prior to
sorting by FACSAria II (BD Biosciences). FACSDiva software (version
6.1.3; BD Biosciences) was installed on the FACSAria II to operate
the instrument and analyze the measurements.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to detect the expression levels of
HNF1A-AS1 (Homo sapiens; Table
SI) in the clinical specimens and cultured cell lines (cat. no.
9108/9109, RNAiso Plus, total RNA extraction reagent for RNA
extraction; cat. no. RR036A, PrimeScript™ RT Master Mix, Perfect
Real Time for reverse transcription; cat. no. RR820A, TB Green™
Premix Ex Taq™ II, Tli RNaseH Plus for RT-PCR; Takara Bio, Inc.).
Reverse transcription was performed at 37°C for 15 min followed by
85°C for 5 sec and 4°C hold. The RT-qPCR thermocycling conditions
were as follows: Initial denaturation at 95°C for 30 sec; 40 cycles
of denaturation at 95°C for 5 sec, and annealing and elongation at
60°C for 30 sec; final extension 72°C for 15 min (20,21).
β-actin was used as the reference gene (forward,
5′-AGCGAGCATCCCCCAAAGTT-3′; reverse, 5′-GGGCACGAAGGCTCATCATT-3′).
The relative expression levels of HNF1A-AS1 in each group (fold
change compared with the control group) was calculated using the
following formula: RQ=2−ΔΔCq (22). Each reaction was performed in
triplicate.
Immunohistochemical (IHC) staining and
immunofluorescence staining
The expression levels of E-cadherin (cat. no.
ab1416; mouse; dilution, 1:500; Abcam), N-cadherin (cat. no.
ab76057; rabbit; dilution, 1:500; Abcam), Vimentin (cat. no.
ab92547; rabbit; dilution, 1:300; Abcam), Snail (cat. no. ab53519;
goat; dilution, 1:100; Abcam) and Slug (Snail2; cat. no.
12129-1-AP; rabbit; dilution, 1:100; ProteinTech Group, Inc.) in
clinical specimens and cultured cell lines were detected by IHC.
Tissues were fixed in 10% neutral-buffered formalin at room
temperature for 24 h and embedded in paraffin. Sections (4-µm
thick) were deparaffinized in four changes of xylene for 5 min
each. Sections were hydrated in 100, 90 and 80% ethanol for 5 min
each. Subsequently, sections were rinsed in water. Antigens were
retrieved with citrate buffer (cat. no. C9999; Sigma-Aldrich; Merck
KGaA) and sections were microwaved for 15 min at 95°C. Sections
were blocked with 5% BSA (cat. no. A3059; Sigma-Aldrich; Merck
KGaA) in PBS containing 0.1% Tween-20 (cat. no. 85114; Thermo
Fisher Scientific, Inc.) for 1 h at room temperature. Sections were
incubated with the aforementioned primary antibodies at 4°C
overnight. Sections were incubated with the secondary HRP antibody
(dilution, 1:1,000; cat. nos. P0449, K4001 and K4003; Dako; Agilent
Technologies, Inc.) for 1 h at room temperature. Images were
captured under a light microscope (IX51; Olympus Corporation).
Immunoreactivity was scored based on the number of positive cells
and staining intensity, using Image-Pro Plus 6.0 software (Media
Cybernetics, Inc.). Immunohistochemical scores (IHS) were
determined by multiplying the quantity and staining intensity
scores as follows: i) The quantity was rated on a scale of 0–4: 0,
no staining; 1, <10% of cells stained; 2, 10–50% of cells
stained; 3, 51–80% of cells stained; and 4, 81–100% of cells
stained; and ii) staining intensity was rated on a scale of 0–3: 0,
negative; 1, weak; 2, moderate; and 3, strong. Theoretically, the
scores could range between 0 and 12. An IHS of 9–12 was considered
strong immunoreactivity, 5–8 was considered moderate, 1–4 was
considered weak, and 0 was considered negative (23). Immunofluorescence staining was
performed as previously described (24).
Cultured cells were grown on cover slips or chamber
slides. Culture media were removed and cells were rinsed with PBS.
The cells were fixed with 4% formaldehyde for 15 min at room
temperature. Sections were blocked with 5% BSA (cat. no. A3059;
Sigma-Aldrich; Merck KGaA) in PBS containing 0.1% Triton™ X-100
(cat. no. 85111; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. Cells were incubated with the primary antibodies at
the aforementioned concentrations overnight at 4°C in the dark.
Sections were incubated with the secondary Cy5-conjugated antibody
(dilution, 1:500; cat. nos. BA1031 and BA1032; Boster Biological
Technology) for 1 h at room temperature in the dark. Sections were
mounted with Prolong Gold medium-DAPI (cat. no. P36931; Invitrogen;
Thermo Fisher Scientific, Inc.). Images were captured under a
fluorescence microscope (IX51; Olympus Corporation).
Western blotting
The proteins were extracted using a RIPA buffer kit
(cat. no. P0013B; Beyotime Institute of Biotechnology). Protein
concentration was determined using a BCA protein concentration
determination kit (cat. no. P0010; Beyotime Institute of
Biotechnology). Protein (20 µg/lane) was loaded onto an SDS-PAGE
precast gel (8–16%; cat. no. P0058B; Beyotime Institute of
Biotechnology). The proteins were transferred to a nitrocellulose
membrane (cat. no. 88018; Thermo Fisher Scientific, Inc.).
Membranes were blocked with 5% BSA (cat. no. A3059; Sigma-Aldrich;
Merck KGaA) in TBS with 0.1% Tween-20 (cat. no. 85114; Thermo
Fisher Scientific, Inc.) for 1 h at room temperature. The proteins
were assessed by incubation with the specific primary antibodies,
including E-cadherin, Snail1, Vimentin and N-cadherin (all diluted
1:1,000), as well as Slug (cat. no. 9585; rabbit; dilution, 1:800;
Cell Signaling Technology, Inc.), CyclinD1 (cat. no. 55506; rabbit;
dilution, 1:1,000; Cell Signaling Technology, Inc.), proliferating
cell nuclear antigen (PCNA; cat. no. 13110; rabbit; dilution,
1:1,000; Cell Signaling Technology, Inc.) and GADPH (cat. no.
10494-1-AP; rabbit; dilution, 1:1,000; ProteinTech Group, Inc.)
overnight at 4°C. Sections were incubated with the secondary HRP
antibodies (dilution, 1:5,000; cat. no. BA1051, BA1060 and BA1054;
Boster Biological Technology) for 1 h at room temperature. The
membranes were visualized using a ChemiDoc MP system (Bio-Rad
Laboratories, Inc.) with ECL substrate (EMD Millipore).
Densitometric semi-quantification of the bands was performed using
ImageJ software (v. 1.52n; National Institutes of Health).
Cell proliferation assays
Cell proliferation capacities were evaluated using a
Cell Counting Kit-8 assay (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. The results were plotted
as mean ± SE of three separate experiments for each experimental
condition.
Flow cytometry analysis of the cell
cycle
The cell cycle analysis was performed using a Cell
Cycle Staining kit [cat. no. CCS012; PI staining; Hangzhou Multi
Sciences (Lianke) Biotech Co., Ltd.], and analyzed by flow
cytometry (FACScan®; BD Biosciences) with CellQuest
software (version 5.2.1; BD Biosciences). The ratios of cells in
the G0/G1, S and G2/M phases were
calculated and compared with those in the control groups.
Cell migration and invasion
assays
The cell migration ability was measured by scratch
assay analysis with 100% confluence after starving cells in media
containing 0.1% FBS overnight. A 20-µl pipette tip was used to
create the scratch. Then, the cells were cultured in the medium
with a reduced concentration (1%) of serum to control for the
influence of proliferation and avoid apoptosis for 0 (baseline), 24
and 48 h, and then the images of the same view were captured (light
microscope; IX51; Olympus Corporation). ImageJ software was used to
measure the wound area at different time points. The width of the
wound divided by the width of the background field was used to
calculate changes over time. The cell invasion ability was assessed
using 6.5-mm Transwell chambers (Corning Inc.) Matrigel was thawed
for 24 h in ice at 4°C in a refrigerator. A total of 100 µl
Matrigel (0.4 mg/ml) was loaded into the upper chamber and
incubated for 24 h at 4°C in a CO2 incubator. After 24
h, 750 µl DMEM complete medium with 10% FBS was added into the
lower chamber. In the upper chamber, 200 µl cell suspension
(2×105 cells/ml in only DMEM) was added onto the
Matrigel-coated cell culture insert and incubated for 24 h. After
24 h, the medium in the upper chamber was removed, followed by
washing with PBS. Cells were fixed with 4% formaldehyde for 30 min
at room temperature and stained with 0.4% crystal violet for 10 min
at room temperature. Non-invaded cells were removed with cotton
swabs. Invasive cells were counted in four random microscopic
fields (light microscope; IX51; Olympus Corporation). Each assay
was performed in triplicate.
Analysis of CpG islands in the
promoter of HNF1A-AS1
The UCSC genome browser public database (http://genome.ucsc.edu) and Methprimer 1.0 (http://www.urogene.org/methprimer/) tools were
used to identify the CpG islands of HNF1A-AS1 (2 kb upstream of the
transcriptional start site) (25).
The prediction of CpG islands was performed using the following
parameters: >100 bp window, percentage of C plus G >50%, and
observed/expected CpG ratio >0.6. Since the number of patients
with LSCC in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) database was not
sufficient, 530 patients with head and neck squamous cell carcinoma
included in the TCGA HNSC cohort were analyzed retrospectively
(26).
Methylation-specific polymerase chain
reaction (MSP) and bisulfite genomic sequencing (BSP)
Total DNA was extracted from cell lines and tissue
specimens using the MiniBEST Genomic DNA Extraction kit (cat. no.
9765; Takara Bio, Inc.). The EZ DNA Methylation-Gold kit (Zymo
Research Corp.) was used for converting bisulfate for subsequent
methylation analysis. The primers used for MSP and BSP were
complementary to the promoter regions of HNF1A-AS1 (Table SI). The bisulphate-modified DNA was
used as template for amplification in MSP detection (cat. no.
R110A; Takara Bio, Inc.; methylated: 98°C for 10 sec, 61.3°C for 5
sec and 72°C for 30 sec, 40 cycles; unmethylated: 98°C for 10 sec,
58.1°C for 5 sec and 72°C for 30 sec, 40 cycles), and the PCR
products were subjected to 1.5% agarose (cat. no. AG006; Canvax
Biotech) gel electrophoresis (100 V; 40 min). Tris-Acetate-EDTA
buffer was used in the preparation and running of the gel, and 10
µl GelRed (cat. no. 41003-T; Biotium, Inc.) was added to 100 ml
agarose gel. Subsequently, 6 µl sample (5 µl and 1 µl 6X DNA
loading buffer; cat. no. R0611; Thermo Fisher Scientific, Inc.) per
lane was loaded on an agarose gel. The Gel Doc EZ imaging system
(Bio-Rad Laboratories, Inc.) came equipped with Image Lab™ software
version 3.0, with auto image capture and auto analysis, and was
used for semi-quantification. Additionally, the modified DNA was
amplified, purified, subcloned into pMD19-T vector (Takara Bio,
Inc.) and transformed into E. coli (DH5-alpha; Biowit
Technologies, Ltd.), cultured in Luria broth medium (LB; cat. no.
10855001; Thermo Fisher Scientific, Inc.) with 50 µg/ml ampicillin
at 37°C with 0.04% CO2, according to the manufacturer's
protocol. Finally, five isolated colonies that were grown on LB
agar plates (Thermo Fisher Scientific, Inc.) containing ampicillin
with X-gal (0.2 mg/ml)/IPTG (1 mM) were picked, and underwent
sequencing and analysis using an ABI 3730 DNA Sequencer (Applied
Biosystems; Thermo Fisher Scientific, Inc.) for BSP detection.
In vivo xenograft tumorigenicity,
tumor invasion and cervical lymph node metastasis assays
Briefly, 8-week-old male Bal/Bc nude mice (n=44;
weight, 22.95±0.41 g) were supplied by the Institute of Zoology,
Xi'an Jiaotong University Health Science Center (Xi'an, China). The
animals were fed and raised in an ultraclean specific-pathogen-free
laminar flow rack with a constant temperature (20–26°C), humidity
(40–50%) and 12/12-h light/dark cycle. Food and fresh water were
accessible at all times. All animal experiments were performed
according to the Guide for the Care and Use of Laboratory Animals
of the National Institutes of Health (27) and were authorized by the Medical
Ethics Committee of Xi'an Jiaotong University. Animal experiments
were performed in May 2018.
For in vivo tumorigenicity experiments, DMEM
without FBS and Matrigel (cat. no. 356234; BD Biosciences) were
mixed 1:1 as resuspension solution. The cells (TU-686/shRNA-2048,
TU-177/HNF1A-AS1 and control cells;100 µl; 1×106) were
subcutaneously injected into the right flank of each mouse (five
mice per group). The mice were monitored for weight, respiration,
ability to ambulate, taking food, drinking, tumor size, ulceration,
infection, and necrosis by a specialized technician at the animal
facility. The tumor volume of each mouse was determined by
measuring two of its dimensions and calculated using the following
formula: Tumor volume=length × width2/2. The humane
endpoints included a rapid weight loss of 15% within a few days and
a tumor diameter >1.5 cm (subcutaneous xenografts) in any single
dimension. Body weights of mice are shown in Fig. S1A and B. Tumor-bearing mice were
euthanized using CO2 (20% of the volume of the chamber
per min). The time interval between injection and euthanasia was 43
days.
For in vivo tumor cervical lymph node
metastasis, the present study used an orthotopic xenograft model of
head and neck cancers as described previously (28,29).
The indicated cells (TU-686/shRNA-2048, TU-177/HNF1A-AS1 and
control cells; 30 µl; 2×105) (28,29)
were injected submucosally into the tongue of nude mice (6 mice per
group) (30). The resuspension
solution was the same as for flank injection. Tumor-bearing mice
were euthanized by CO2 (20% of the volume of the chamber
per min). The time interval between injection and euthanasia was 21
days. Finally, submucosal tongue tumors and cervical lymph nodes
were surgically excised, weighed and imaged. Pathological
examinations of cervical lymph nodes were performed to confirm
metastasis.
Statistical analysis
Statistical analysis was performed using SPSS v19.0
(IBM Corp.) and GraphPad Prism 5.0 (GraphPad Software, Inc.). All
data are presented as the mean ± SD. In vitro experiments
were performed in triplicate. An unpaired two-tailed Student's
t-test, one-way ANOVA with Tukey's post hoc test and the
χ2 test were used to analyze the data. P<0.05 was
considered to indicate a statistically significant difference.
Results
HNF1A-AS1 expression is downregulated
in LSCC tissues and metastatic cervical lymph nodes
The results of RT-qPCR demonstrated that the
expression levels of HNF1A-AS1 were significantly downregulated in
LSCC tissues and metastatic lymph node samples compared with in the
adjacent non-tumor tissues (P<0.05; Fig. 1A). The results of HE staining are
shown in Fig. 1B. It was
demonstrated that the downregulation of HNF1A-AS1 may serve an
important role in the development and progression of LSCC.
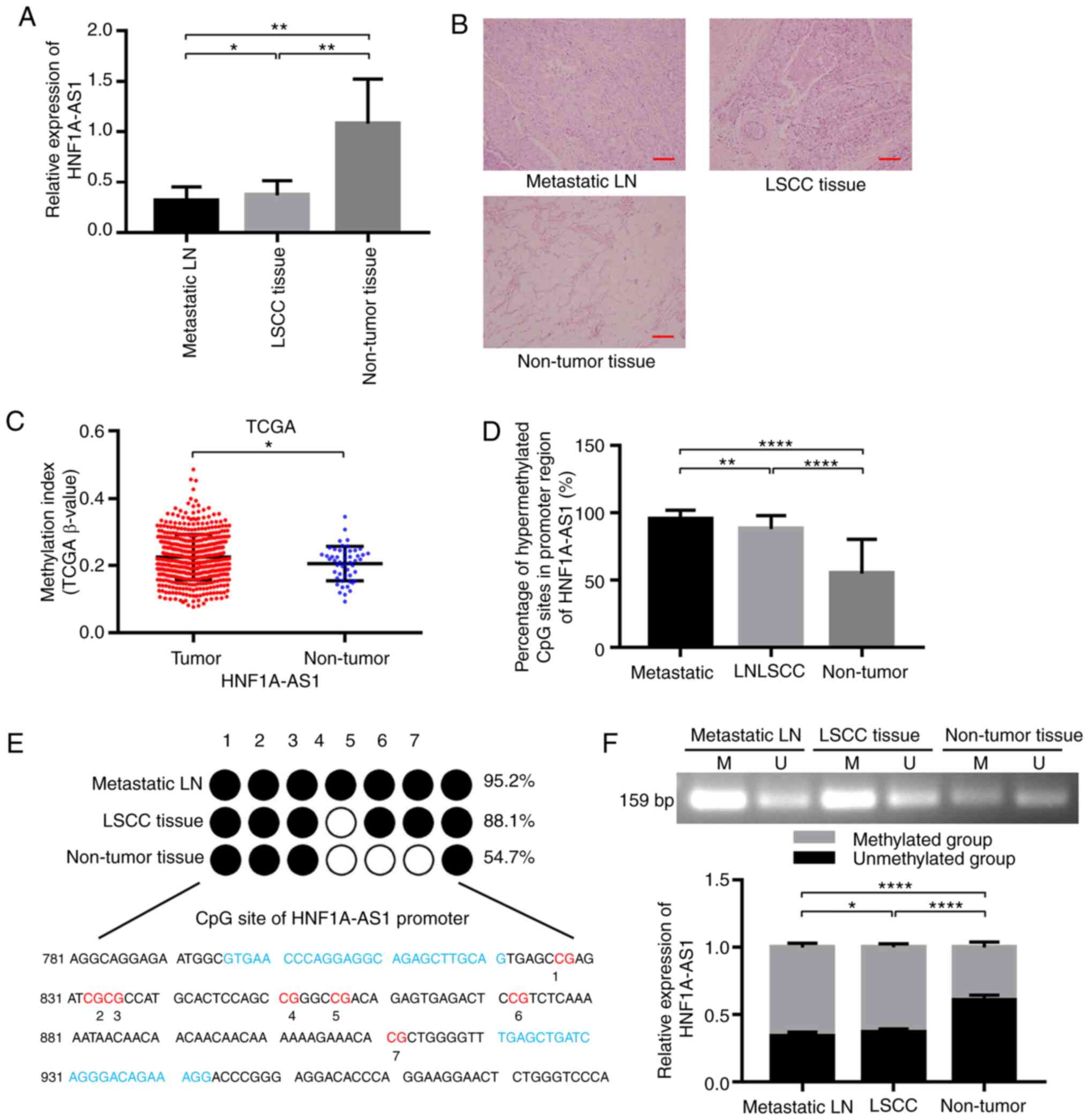 | Figure 1.HNF1A-AS1 is downregulated and
hypermethylated in LSCC tissues and metastatic cervical lymph
nodes. (A) Expression levels of HNF1A-AS1 in primary LSCC tissues,
adjacent metastatic lymph nodes samples and non-tumor tissues were
examined by reverse transcription-quantitative PCR. GAPDH was used
as an internal control (n=30). (B) Representative HE staining in
primary LSCC tissues, adjacent non-tumor tissues and metastatic
cervical lymph nodes of patients with LSCC. Scale bar, 50 µm. (C)
Percentage methylation of HNF1A-AS1 was analyzed in tissues from
530 patients with head and neck squamous cell carcinoma and 50
normal adjacent tissues included in TCGA. (E) Bisulfite genomic
sequencing was performed to examine the methylation status of the
HNF1A-AS1 promoter region in primary LSCC tissues, metastatic
cervical lymph nodes and adjacent non-tumor tissues from 30
patients, and (D) methylation status was analyzed statistically.
(F) The methylation status of the CpG island in the HNF1A-AS1
promoter was determined by methylation-specific polymerase chain
reaction assays in primary LSCC tissues, metastatic cervical lymph
nodes and adjacent non-tumor tissues from 30 patients. For
statistical analysis, one-way ANOVA with Tukey's post hoc test was
used for data in (A, D and F) and an unpaired t-test was used for
data in (C) Data are presented as the mean ± SD. All assays were
performed in triplicate, and the values represent the mean of three
independent experiments. *P<0.05, **P<0.01 and
****P<0.0001. HNF1A-AS1, hepatic nuclear factor 1 α antisense
RNA 1; LN, lymph node; LSCC, laryngeal squamous cell carcinoma;
TCGA, The Cancer Genome Atlas. |
Methylation analysis of the HNF1A-AS1
promoter in vivo
To further ascertain whether the downregulation of
HNF1A-AS1 expression was caused by promoter hypermethylation in
primary LSCC, the methylation frequency in the promoter region of
HNF1A-AS1 in HNSCC tissues and metastatic cervical lymph nodes was
examined and was significantly higher than that in the
corresponding normal tissues according to the TCGA database
(Fig. 1C).
Furthermore, the methylation status of HNF1A-AS1 in
30 paired tissues in patients was analyzed using BSP, CpG
methylation of HNF1A-AS1 promoter was detected in all primary LSCC
tissues (88.1%) and metastatic cervical lymph nodes (95.2%), and
the results revealed significantly lower methylation in the matched
normal adjacent tissues (54.7%; Fig. 1D
and E). Similar to the BSP results, apparent methylation of
HNF1A-AS1 in the promoter region was also detected to be higher in
primary LSCC tissues and metastatic cervical lymph nodes than
normal adjacent tissues by MSP (Fig.
1F). These results implied that hypermethylation of CpG islands
in the promoter region was the major regulatory mechanism of
HNF1A-AS1 downregulation in LSCC.
HNF1A-AS1 inhibits cell proliferation
and cell cycle in laryngeal cancer cells
Compared with HNF1A-AS1 expression in AMC-HN-8
cells, its expression was high in the TU-686 cell line, but was
decreased in the TU-177 cell line as demonstrated by RT-qPCR
(P<0.01; Fig. S2A). Therefore,
TU-686 and TU-177 cell lines were selected as research targets for
further functional studies. To downregulate the expression levels
of HNF1A-AS1 endogenously in laryngeal cells, small shRNA-853,
shRNA-1525 and shRNA-2048 were separately transfected into TU-686
cells and the interference effects were confirmed by RT-qPCR
analysis. At 48 h post-transfection, the expression levels of
HNF1A-AS1 were significantly decreased by shRNA-1525 (50%;
P<0.01) and shRNA-2048 (75%; P<0.0001) compared with
scrambled shRNA-NC (Fig. S2B).
Furthermore, HNF1A-AS1 expression in TU-177 cells was upregulated
10 times following lentiviral vector transfection (pLVX-HNF1A-AS1
vs. pLVX-control; Fig. S2C).
Considering the remarkable interference and overexpression effects
of HNF1A-AS1, three stable cell lines (TU-686/shRNA-1525,
TU-686/shRNA-2048 and TU-177/HNF1A-AS1) were used for subsequent
experiments.
When compared with the negative control group,
knockdown of HNF1A-AS1 resulted in a significant increase in cell
proliferation in TU-686 cells at 48 and 72 h. Conversely,
overexpression of HNF1A-AS1 decreased the cell proliferation in
TU-177 cells at 24, 48 and 72 h (Fig.
2A). Additionally, the downregulation of HNF1A-AS1 promoted
TU-686 cell transition from G0/G1 to S phase
(Fig. 2B and C), but HNF1A-AS1
overexpression arrested cell cycle at G0/G1
phase (Fig. 2B and C). Furthermore,
knockdown of HNF1A-AS1 in TU-686 cells significantly promoted the
expression of cell cycle-related protein cyclin D1 and PCNA;
however, these were inhibited by overexpression of HNF1A-AS1 in
TU-177 cells (Fig. 2D and E).
Overall, overexpression of HNF1A-AS1 could inhibit the
proliferation of LSCC cells and this was partially accompanied by
G0/G1 arrest.
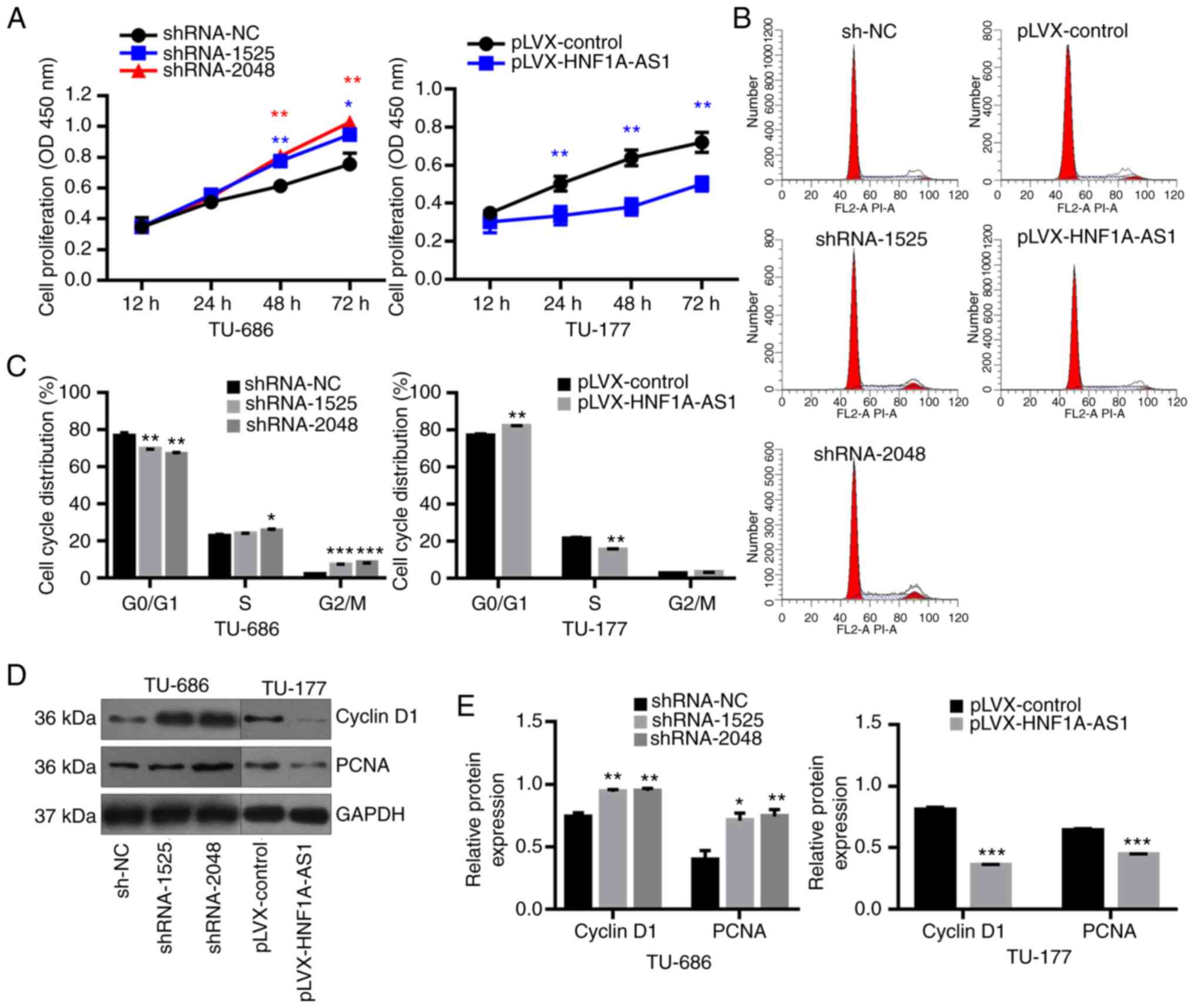 | Figure 2.HNF1A-AS1 inhibits cell proliferation
and cell cycle progression in laryngeal cancer cells. (A) Cell
proliferation was assessed at 12, 24, 48 and 72 h using a Cell
Counting Kit-8 assay. (B) Cell cycle distribution was analyzed by
flow cytometry and (C) results of cell cycle analysis were analyzed
statistically. (D) Western blotting was used to evaluate the
protein expression levels of Cyclin D1 and PCNA following
transfection of sh-NC, shRNA-1525 or shRNA-2048 into TU-686 cells
and pLVX-control or pLVX-HNF1A-AS1 into TU-177 cells and (E)
results of western blotting were analyzed statistically. Data are
presented as the mean ± SD. All assays were performed in
triplicate, and the values represent the mean of three independent
experiments. For statistical analysis, ANOVA with Tukey's post hoc
test was used to compare the sh-NC, shRNA-1525 and shRNA-2048
groups, and an unpaired t-test was used to compare the pLVX-control
and pLVX-HNF1A-AS1 groups. *P<0.05, **P<0.01, ***P<0.001.
HNF1A-AS1, hepatic nuclear factor 1 α antisense RNA 1; NC, negative
control; PCNA, proliferating cell nuclear antigen; shRNA, short
hairpin RNA. |
HNF1A-AS1 inhibits cell migration and
invasion, and reverses epithelial mesenchymal transition in
vitro
The cell migratory abilities were investigated by
scratch assay analysis and the results revealed that HNF1A-AS1
downregulation significantly enhanced the migratory abilities of
LSCC cells (Fig. 3A and B). In
addition, a Transwell invasion assay demonstrated that decreased
expression of HNF1A-AS1 following transfection with
TU-686/shRNA-2048 significantly enhanced the invasive capacity of
TU-686 cells (105.1±0.4 vs. 94.8±1.2; P<0.01; Fig. 3C and D), whereas HNF1A-AS1
overexpression inhibited the invasive abilities of TU-177 cells
(44.9±0.7 vs. 63.4±0.3; P<0.001; Fig. 3C and D). The expression levels of
EMT-related markers were assessed as EMT serves a key role in tumor
invasion and metastasis. The results of western blotting and
immunofluorescence assays demonstrated that knockdown of HNF1A-AS1
notably downregulated E-cadherin expression and upregulated Snail1,
Slug, Vimentin and N-cadherin expression, and vice versa when
HNF1A-AS1 was overexpressed (Fig.
4). These data suggested that HNF1A-AS1 contributed to the
invasion and metastasis of laryngeal cancer cells partly by
affecting the EMT process.
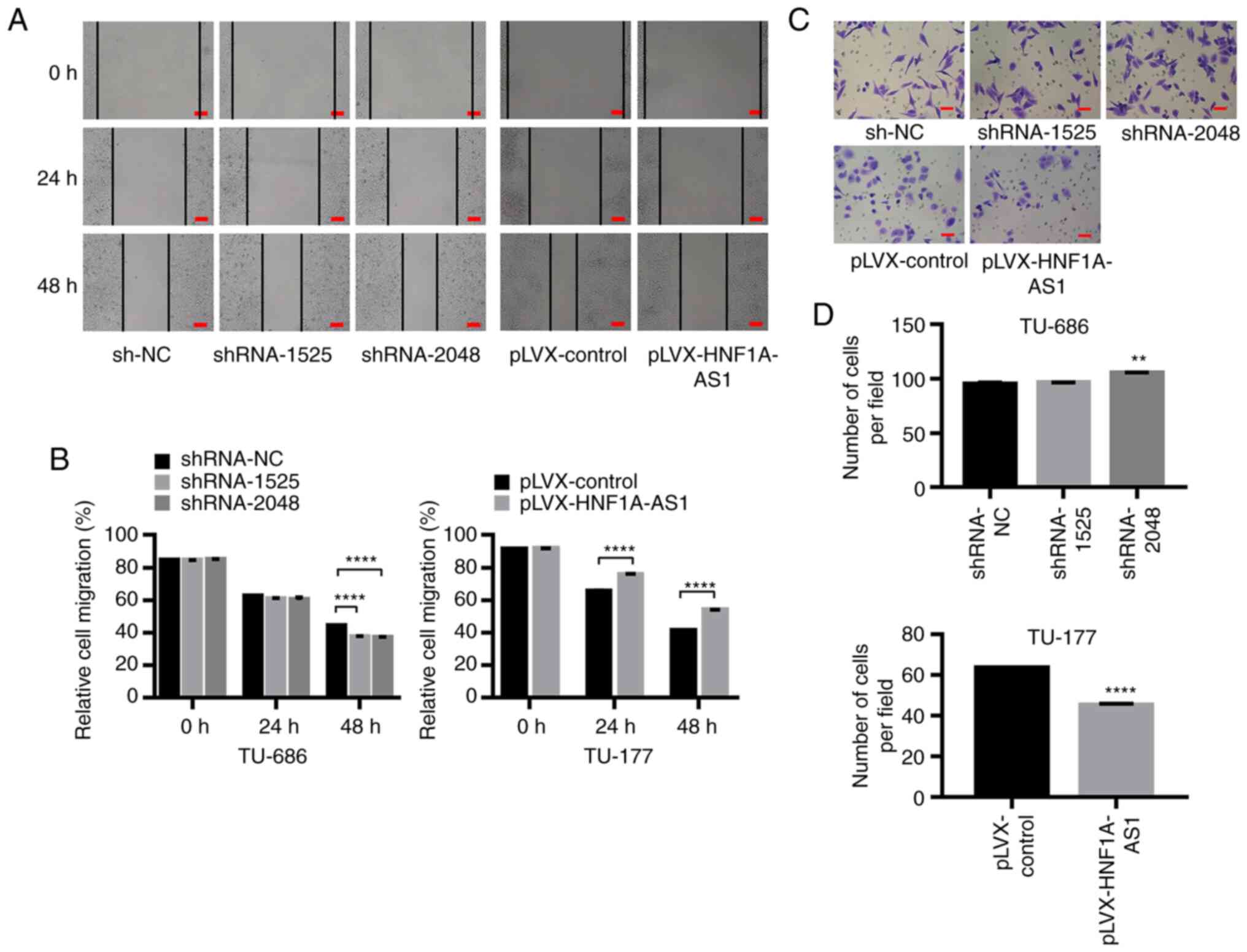 | Figure 3.HNF1A-AS1 suppresses cell migration
and invasion in vitro. (A) Migration of TU-686/shRNA-1525,
TU-686/shRNA-2048, TU-177/HNF1A-AS1 and control cells was assessed
using a scratch assay, and (B) results of migration were analyzed
statistically. Scale bar, 100 µm. (C) Transwell invasion assays
were performed to determine the invasive abilities of
TU-686/shRNA-1525, TU-686/shRNA-2048, TU-177/HNF1A-AS1 and control
cells, and (D) results of the Transwell invasion assay were
analyzed statistically. Scale bar, 50 µm. For statistical analysis,
ANOVA with Tukey's post hoc test was used to compare the sh-NC,
shRNA-1525 and shRNA-2048 groups, and an unpaired t-test was used
to compare the pLVX-control and pLVX-HNF1A-AS1 groups. Each
experiment was performed in triplicates. **P<0.01,
****P<0.0001. HNF1A-AS1, hepatic nuclear factor 1 α antisense
RNA 1; NC, negative control; shRNA, short hairpin RNA. |
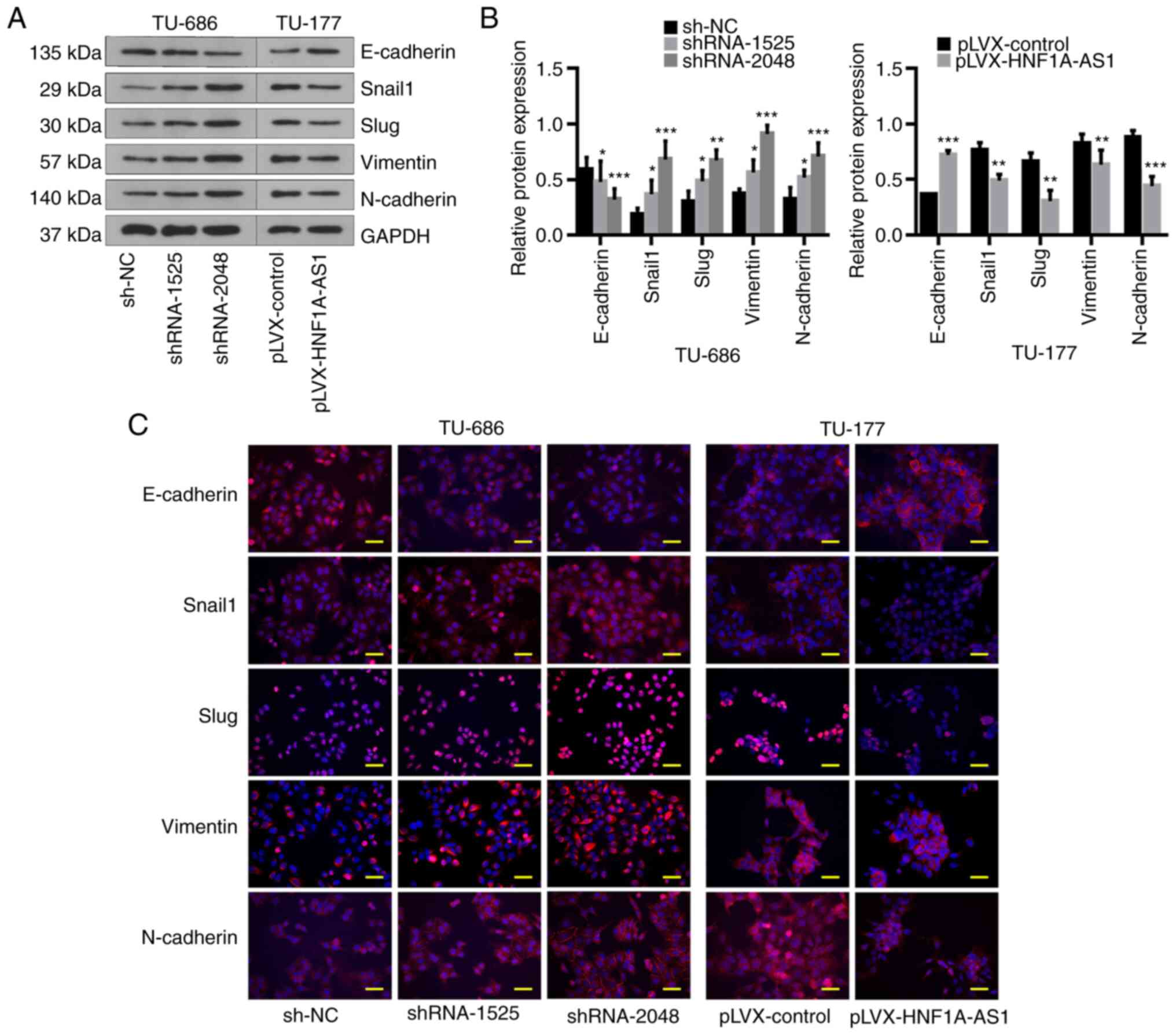 | Figure 4.HNF1A-AS1 suppresses
epithelial-mesenchymal transition in vitro. (A) Relative
protein expression levels of E-cadherin, Snail1, Slug, Vimentin and
N-cadherin were evaluated by western blot analysis in
TU-686/shRNA-1525, TU-686/shRNA-2048, TU-177/HNF1A-AS1 and control
cells, and (B) results of western blotting were analyzed
statistically. (C) Analysis of E-cadherin, Snail1, Slug, Vimentin
and N-cadherin expression (red) in TU-686/sh-NC, TU-686/shRNA-1525,
TU-686/shRNA-2048, TU-177/HNF1A-AS1 and TU-177/HNF1A-AS1-NC cells
by immunofluorescence. Blue DAPI staining shows the nuclei (DNA).
Scale bar, 25 µm. Data are presented as the mean ± SD. All assays
were performed in triplicate, and the values represent the mean of
three independent experiments. For statistical analysis, ANOVA with
Tukey's post hoc test was used to compare the sh-NC, shRNA-1525 and
shRNA-2048 groups in (B), and an unpaired t-test was used to
compare pLVX-control and pLVX-HNF1A-AS1 groups in (B). *P<0.05,
**P<0.01, ***P<0.001. HNF1A-AS1, hepatic nuclear factor 1 α
antisense RNA 1; NC, negative control; shRNA, short hairpin
RNA. |
HNF1A-AS1 suppresses tumor growth, EMT
and lymph node metastasis in vivo
Xenograft tumor (subcutaneous) growth models
revealed that tumor sizes were markedly increased following
injection with TU-686/shRNA-2048 cells, while injection with
TU-177/HNF1A-AS1 cells was associated with a reduction compared
with the control group (Fig. 5A-C).
The cervical lymph node metastasis model (submucosal injection in
tongue) gave similar results to the xenograft subcutaneous model.
Tumor weights were increased for injection with TU-686/shRNA-2048
cells, while they were decreased for injection with
TU-177/HNF1A-AS1 cells (Fig. 5D-F).
This indicated that HNF1A-AS1 could promote laryngeal tumor growth
either by subcutaneous injection or by submucosal injection into
the tongue of nude mice. Additionally, the nude mice injected with
TU-686/shRNA-2048 cells submucosally into the tongue had more
metastatic cervical lymph nodes compared with those injected with
TU-686/sh-NC cells, while pathological examination confirmed the
opposite effects for injection of TU-177/HNF1A-AS1 cells (Fig. 5G and H), demonstrating the
inhibitory effect of HNF1A-AS1 on tumor lymph node metastasis.
Furthermore, compared with that in the control group, E-cadherin
expression was significantly downregulated, while Snail1, Slug,
Vimentin and N-cadherin expression was significantly upregulated in
tumors injected subcutaneously using TU-686/shRNA-2048 cells in
nude mice according to western blotting and IHC. By contrast,
upregulation of HNF1A-AS1 was associated with the opposite effects
in vivo (Fig. 6).
Collectively, these findings demonstrated that HNF1A-AS1 expression
inhibited tumorigenic, as well as metastatic, abilities in a
xenograft tumor model of nude mice via EMT.
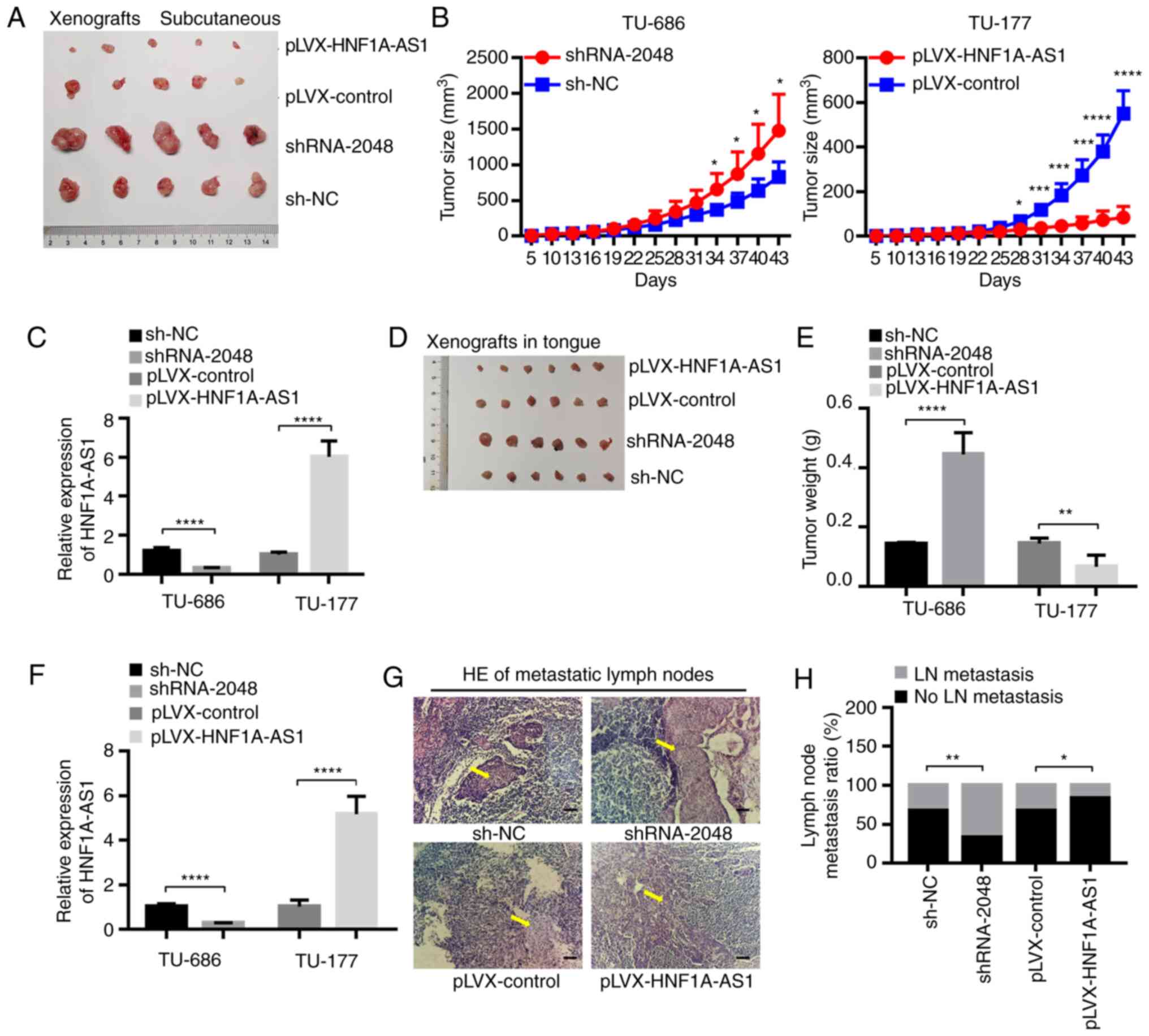 | Figure 5.HNF1A-AS1 suppresses tumor growth and
lymph node metastasis in vivo. (A) Typical specimens in the
subcutaneous xenotransplant tumor model of human laryngeal cancer
in nude mice. (B) Tumor growth curve and (C) RT-qPCR analysis of
HNF1A-AS1 expression in vivo after the TU-686/shRNA-2048,
TU-177/HNF1A-AS1 and control cells were injected subcutaneously
into the right flank of nude mice (n=20). Tumor growth was
calculated from day 5 and tumors were harvested from mice at 43
days after injection. (D) Typical specimens of tumors injected in
tongues submucosally in the cervical lymph node metastasis model in
nude mice. (E) Total weight of tumors submucosally injected into
tongue of nude mice (n=24). (F) RT-qPCR analysis of HNF1A-AS1
expression. Tumors and cervical lymph nodes were harvested from
mice at 15 days after injection. (G) Representative HE staining of
cervical lymph node metastasis in HNF1A-AS1 knockdown or
overexpression cells. Scale bar, 50 µm. Arrows indicate the
metastasis in cervical lymph nodes. (H) Ratios of cervical lymph
node metastasis in nude mice inoculated submucosally with the
indicated cells into the tongue. Data are presented as the mean ±
SD. All assays were performed in triplicate, and the values
represent the mean of three independent experiments. For
statistical analysis, an unpaired t-test was used. *P<0.05,
**P<0.01, ***P<0.001, ****P<0.0001. The xenograft
tumorigenicity experiment in vivo was performed one time.
HNF1A-AS1, hepatic nuclear factor 1 α antisense RNA 1; NC, negative
control; RT-qPCR, reverse transcription-quantitative PCR; shRNA,
short hairpin RNA. |
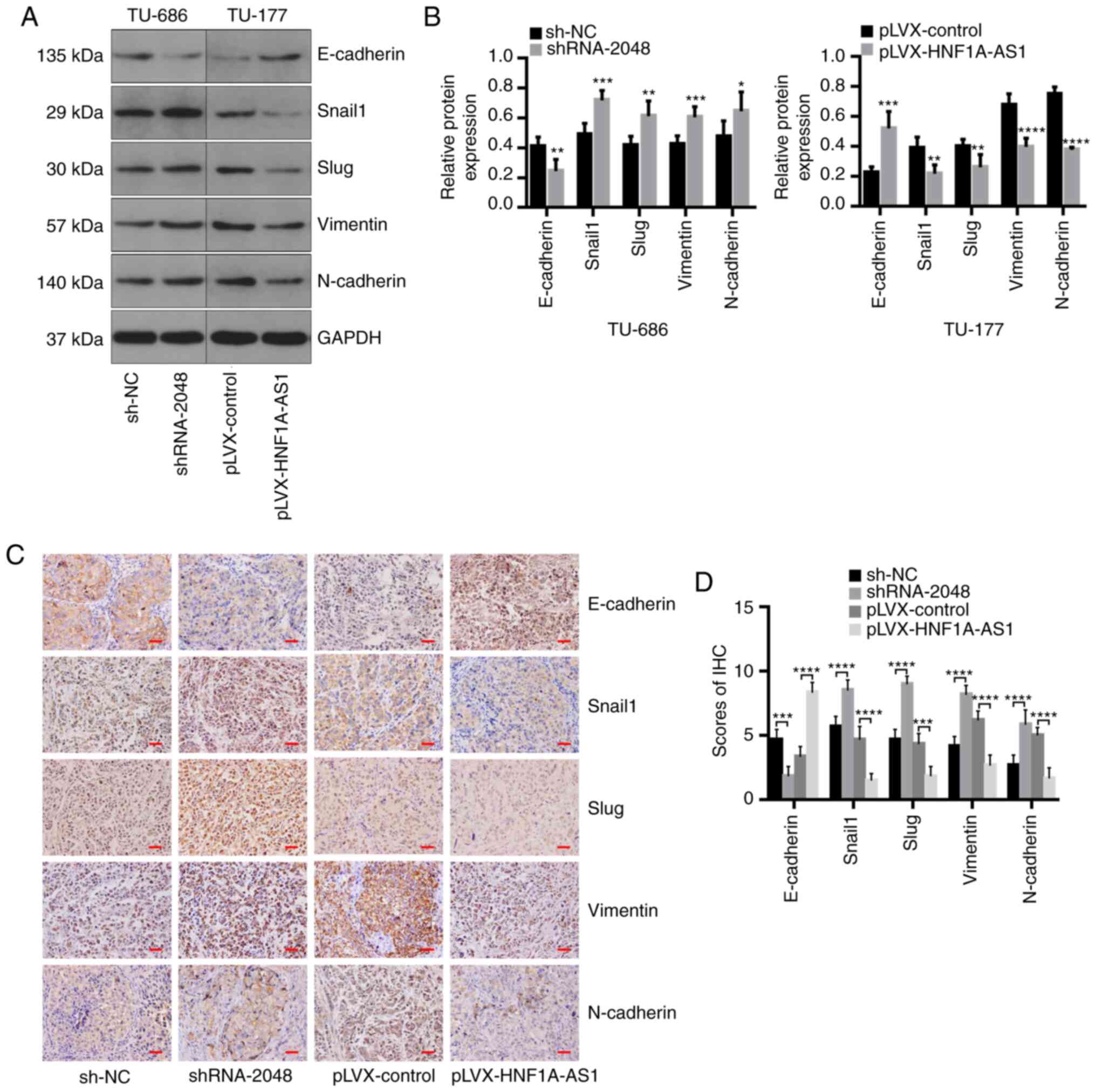 | Figure 6.HNF1A-AS1 suppresses
epithelial-mesenchymal transition in vivo. (A) Relative
protein expression levels of E-cadherin, Snail1, Slug, Vimentin and
N-cadherin in tumors in nude mice injected subcutaneously with
TU-686/shRNA-2048, TU-177/HNF1A-AS1 and control cells were
evaluated by western blot analysis, and (B) results of western
blotting were analyzed statistically. (C) E-cadherin, Snail1, Slug,
Vimentin and N-cadherin immunohistochemical staining in tumors in
nude mice injected subcutaneously with the indicated cells, and (D)
scores of IHC were a analyzed statistically. Scale bar, 25 µm. Data
are presented as the mean ± SD. All assays were performed in
triplicate, and the values represent the mean of three independent
experiments. For statistical analysis, an unpaired t test was used
for data in (B), and a χ2 test was used for data in (D).
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Western
blotting and immunohistochemistry were performed in triplicate.
HNF1A-AS1, hepatic nuclear factor 1 α antisense RNA 1; NC, negative
control; shRNA, short hairpin RNA. |
Downregulation of HNF1A-AS1 is
associated with hypermethylation of CpG island in promoter in
vitro
To explore the inactivation mechanisms of HNF1A-AS1
in LSCC, the sequence of HNF1A-AS1 was analyzed and it was revealed
that the promoter region of HNF1A-AS1 was rich in CpG
dinucleotides. The MethPrimer program was used to predict the
distribution of CpG islands of HNF1A-AS1, and one CpG island (114
bp; −792 to −905 bp) was predicted within the promoter region of
HNF1A-AS1 (Fig. 7A).
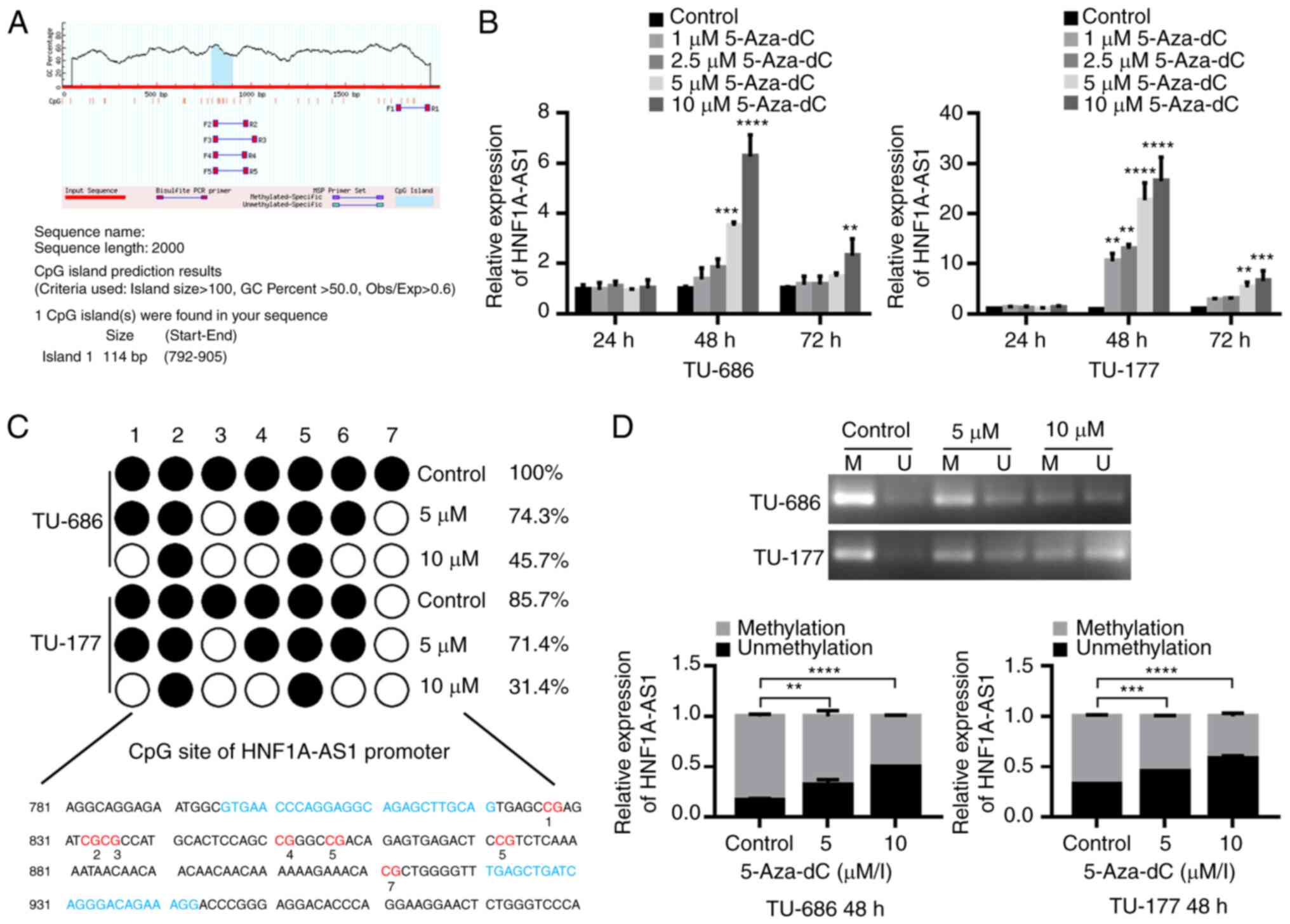 | Figure 7.HNF1A-AS1 is downregulated due to
promoter hypermethylation. (A) Bioinformatics analysis of CpG
island in promoter region of HNF1A-AS1. (B) HNF1A-AS1 expression
was increased following 5-Aza-dC treatment at different
concentrations for 24, 48 and 72 h in TU-686 and TU-177 cells. (C)
Methylation status of BSP for the HNF1A-AS1 CpG island in TU-686
and TU-177 cells treated with or without 5- Aza-dC after 48 h.
White circle, unmethylated CpG dinucleotide; black circle,
methylated CpG dinucleotide. (D) Methylation status of CpG island
in HNF1A-AS1 promoter determined by MSP assay in TU-686 and TU-177
cells treated with or without 5-Aza-dC after 48 h. Data are
presented as the mean ± SD. All assays were performed in
triplicate, and the values represent the mean of three independent
experiments. For statistical analysis, ANOVA with Tukey's post hoc
test was used for data in (B and D) **P<0.01, ***P<0.001,
****P<0.0001. M, amplification with methylated primers; U,
amplification with unmethylated primers; 5-Aza-dC,
5-Aza-2′-deoxycytidine; BSP, bisulfite genomic sequencing;
HNF1A-AS1, hepatic nuclear factor 1 α antisense RNA 1; MSP,
methylation-specific polymerase chain reaction. |
To clarify whether downregulation of HNF1A-AS1 was
regulated by methylation in the CpG islands, TU-686 and TU-177
cells were treated with DNA methyltransferase inhibitor 5-Aza-dC,
and the results demonstrated that the expression levels of
HNF1A-AS1 were significantly increased in TU-686 and TU-177 cells
following treatment with 5-Aza-dC at a dose of 5 and 10 µM for 48
h, revealing the crucial role of abnormal methylation in the
inactivation of HNF1A-AS1 in LSCC cell lines (Fig. 7B).
Subsequently, a 148 bp segment in this region (−796
to −943 bp) that includes 7 CpG sites and spans the core promoter
was analyzed using BSP (Fig. 7C).
The hypermethylated CpG sites of TU-686 and TU-177 cells were 100
and 85.7%. Following treatment with 5 and 10 µM 5-Aza-dC, the
hypermethylated CpG sites of TU-686 cells were decreased to 74.3
and 45.7%, respectively, and the ones of TU-177 cells were
decreased to 71.4 and 31.4%, respectively (Fig. 7C). Similar to the BSP results,
apparent methylation of HNF1A-AS1 in the promoter region was also
detected in TU-686 and TU-177 cells by MSP, in which the
methylation status was partly reversed following 5-Aza-dC
treatment. This revealed the importance of promoter methylation in
the downregulation of HNF1A-AS1 expression (Fig. 7D).
Demethylation of CpG island in the
promoter region of HNF1A-AS1 inhibits cell migration, invasion and
EMT in vitro
The results demonstrated that the migration and
invasion abilities of TU-686 and TU-177 cells were significantly
decreased after demethylation of HNF1A-AS1 by 5-Aza-dC treatment
(Fig. 8A-D). In addition, western
blotting indicated that demethylation of the CpG island in the
promoter of HNF1A-AS1 significantly upregulated E-cadherin
expression and downregulated Snail1, Slug, Vimentin and N-cadherin
expression (Fig. 8E and F). These
data suggested that low HNF1A-AS1 expression which contributed to
the invasion and metastasis of laryngeal cancer cells by affecting
the EMT process may be the result of hypermethylation of HNF1A-AS1
in its promoter region.
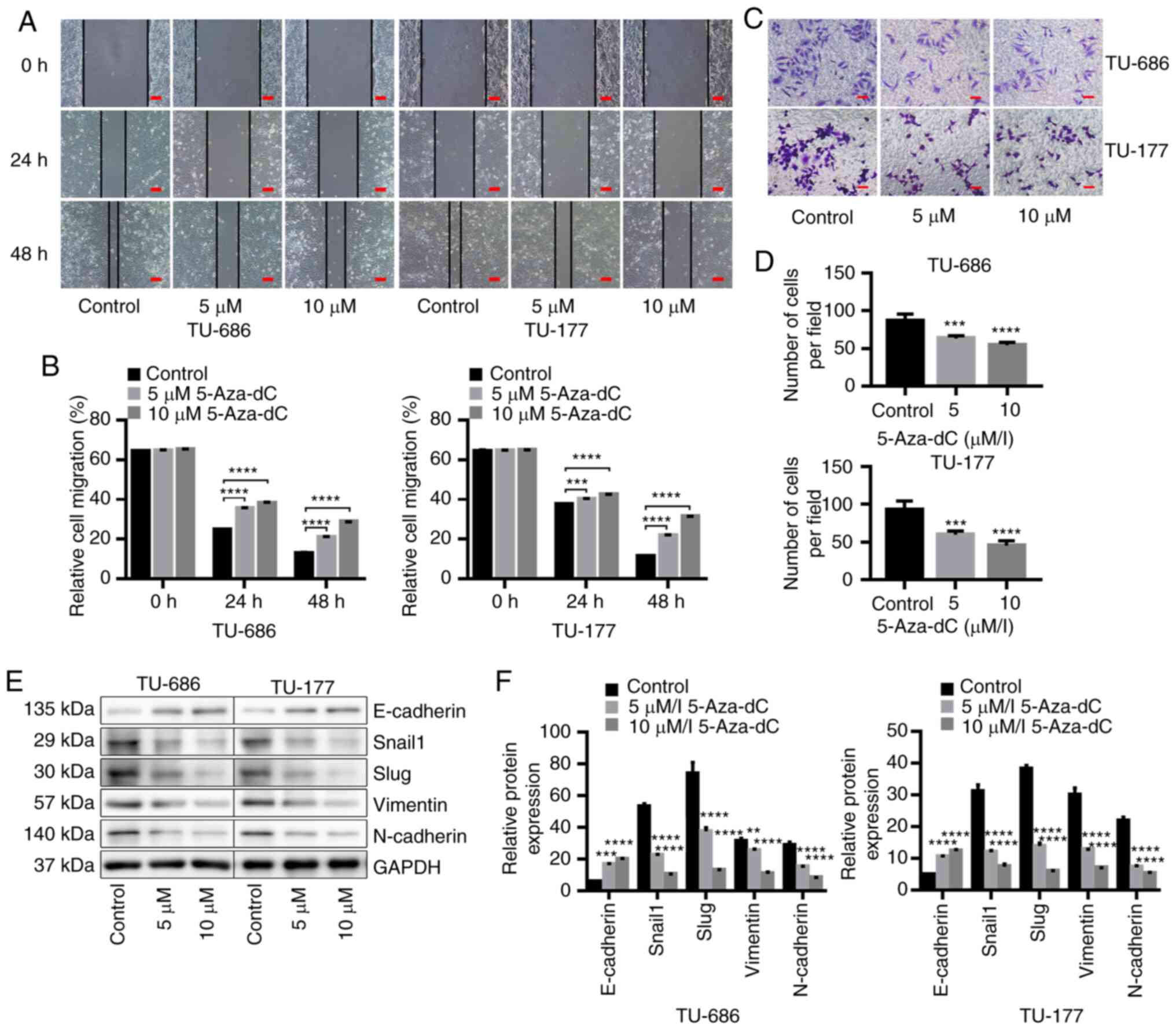 | Figure 8.Demethylation of HNF1A-AS1 inhibits
cell migration, invasion and epithelial-mesenchymal transition
in vitro. (A) Cell migration abilities were tested using a
scratch assay and (B) results of migration assays were analyzed
statistically. Scale bar, 100 µm. (C) Invasive abilities were
determined using a Transwell invasion assay in TU-686 and TU-177
cells treated with or without 5-Aza-dC after 48 h, and (D) the
results of Transwell invasion assays were analyzed statistically.
Scale bar, 50 µm. (E) Relative protein expression levels of
E-cadherin, Snail1, Slug, Vimentin and N-cadherin were evaluated by
western blot analysis in TU-686 and TU-177 cells treated with or
without 5-Aza-dC after 48 h, and (F) results of western blotting
were analyzed statistically. Data are presented as the mean ± SD.
All assays were performed in triplicate, and the values represent
the mean of three independent experiments. For statistical
analysis, an ANOVA with Tukey's post hoc test was used.
**P<0.01, ***P<0.001, ****P<0.0001. 5-Aza-dC,
5-Aza-2-deoxycytidine; HNF1A-AS1, hepatic nuclear factor 1 α
antisense RNA 1. |
Discussion
It is well recognized that enhanced proliferation,
migration and invasion of LSCC cells serve key roles in the
progression of LSCC (31,32). Therefore, the present study first
determined that HNF1A-AS1 expression was low in human primary LSCC
tissues and metastatic cervical lymph nodes compared with that in
the adjacent non-tumor tissues. Due to the limitations of the
difficulty in identifying targets that have low DNA and RNA copies,
poor reproducibility and high expenses of in situ
hybridization assays (33),
RT-qPCR, which has been considered to be a reliable method for
quantification of gene expression due to its accuracy, sensitivity,
specificity and reproducibility (34), was used to assess the expression
levels of HNF1A-AS1.
The present data demonstrated that HNF1A-AS1
decreased cell migration and invasive abilities in vitro,
and reduced tumorigenesis and metastasis of xenograft LSCC tumors
in vivo. The present study used a reduced concentration (1%)
of serum to control for the influence of proliferation and avoid
apoptosis instead of FBS-free medium in the cell migration assay.
Serum starvation can elicit complex, unpredictable time-dependent
and cell-type-dependent effects. Low serum concentrations in cell
medium is the most common method to suppress cell proliferation in
wound healing assays (35). The
duration of serum starvation and the required serum concentrations
need to be rigorously determined for each studied cell line. It is
commonly assumed that serum-starved cells have reduced basal
cellular activity (36), but serum
starvation has also been referred to as an ‘environmental stress’
(37) and ‘apoptotic trigger’
(38), implying dynamism that does
not entirely fit the idea of a passive entry into a dormant
hypoactive state. It remains an open issue whether diversity
observed in the presence of serum also exists when cells are
serum-starved in serum-free medium. The discrepancy may be due to
differences in media composition, since cells were grown in 10% FBS
prior to serum starvation. Furthermore, according to the in
vitro experiments, upregulation of HNF1A-AS1 led to
proliferative inhibition by inducing G0/G1
phase cell cycle arrest in LSCC cells, while overexpression of
HNF1A-AS1 significantly inhibited the protein expression of cyclin
D1 and PCNA. Therefore, these findings suggested that HNF1A-AS1 may
be a suppressor and serves an important role in the development and
progression of LSCC.
EMT increases the invasive and metastatic
capabilities of malignant tumor cells (7). Various epithelial-related proteins are
involved in the EMT process, in which E-cadherin is implicated in
transcriptional suppression, and vimentin is implicated in
activation, leading to tumor invasion and metastasis (39). To investigate the probable
underlying mechanisms, it was revealed that the expression levels
of EMT-associated molecular markers (Snail1, Slug, Vimentin and
N-cadherin) were decreased and E-cadherin remained high when
HNF1A-AS1 was overexpressed in the laryngeal cells and xenografts,
and vice versa when HNF1A-AS1 was downregulated. Overall, these
findings indicated that LSCC tissues and laryngeal cancer cells
with downregulated HNF1A-AS1 expression might undergo the EMT
process, becoming more mobile and invasive, which promotes the
malignant progression of LSCC.
As described in several studies (40,41),
cancer cells often have a gain of methylation in the promoter
regions of selected CpG islands, resulting in silencing of hundreds
of genes, including tumor suppressor genes (42). In the present study, a CpG island
was found in the promoter region of HNF1A-AS1, and the analysis
using TCGA demonstrated that the methylation frequency in the
promoter region of HNF1A-AS1 in HNSCC tissues was significantly
higher than that in the corresponding normal tissues. According to
the BSP and MSP results, CpG methylation of the HNF1A-AS1 promoter
was detected in all primary LSCC tissues. Furthermore, a
significantly different methylation status was identified among the
metastatic cervical lymph nodes, cancer tissues and corresponding
normal control tissues. This demonstrated that HNF1A-AS1 was
downregulated in metastatic cervical lymph nodes and LSCC by
hypermethylation On the other hand, the relationship between loss
of HNF1A-AS1 expression and methylation of HNF1A-AS1 in LSCC cell
lines was confirmed by excellent uniformity among mRNA expression
by RT-qPCR, and DNA methylation in HNF1A-AS1 promoter by BSP and
MSP, which largely accounted for the decreased expression levels of
HNF1A-AS1. Additionally, following treatment with DNA
methyltransferase inhibitor 5-Aza-dC, methylation of HNF1A-AS1 in
TU-686 and TU-177 cells was reduced significantly, and the
expression levels of HNF1A-AS1 were negatively associated with the
methylation status. The migration, invasive abilities and
EMT-associated marker expression in LSCC cell lines were reversed
by treatment with 5-Aza-dC, which indicated that HNF1A-AS1 may act
as a tumor suppressor lncRNA by regulating the EMT process via
hypermethylation in LSCC.
However, the present study had some limitations,
such as that the number of specimens was not sufficient to analyze
the differences of prognosis and staging between HNF1A-AS1 high
expression/hypermethylation and low expression/hypomethylation in
patients with LSCC. Further studies on the overall survival
information and deep functional investigations are required.
It is known that the expression of lncRNAs is
strikingly cell type- and tissue-specific (10). Dang et al (15) reported that HNF1A-AS1 is
downregulated in both gastric cancer tissues and cell lines.
However, in a recent study, HNF1A-AS1 was demonstrated to inhibit
the malignant properties of hepatocellular carcinoma cells both
in vitro and in vivo (16). It is difficult to explain such
phenomena; one possible explanation could be the heterogeneity of
primary tumors. It was hypothesized that HNF1A-AS1 has a two-way
regulatory role in different organs and carcinomas. More
investigations are required to investigate this. Additionally, to
the best of our knowledge, the expression and function of HNF1-AS1
and its methylation condition have not been reported in the
development and progression of LSCC. In the present study,
HNF1A-AS1 was downregulated by hypermethylation in LSCC and
laryngeal cancer cells, and it served as a tumor suppressor lncRNA
in LSCC by inducing the EMT process. These findings imply that
manipulation of HNF1A-AS1 expression may have therapeutic effects
against LSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81402246),
Innovation Capability Support Program of Shaanxi (grant no.
2017KJXX-45), Natural Science Basic Research Program of Shaanxi
(grant no. 2018JM7023), Natural Science Foundation Group-style
Aided Tibet Medical Project of Tibet Autonomous Region [grant no.
XZ2019ZR-ZY72 (Z)], Key Research and Development Program in Social
Development Field of Shaanxi (grant no. 2019SF-084), and Special
Research Fund for Talents Training in the Second Affiliated
Hospital of Xi'an Jiaotong University [grant no. RC (GG)
201707].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YS performed the majority of the experiments, wrote
the original draft, and was responsible for conceptualization and
data curation. QZ, as the co-first author, performed some
experiments and performed data visualization and analysis. MX and
YF assembled the figures and resources, and collected samples. SM
and CY performed formal analysis and visualization. ZW and YL
performed the investigation. Acquisition of data, statistical
analysis of the data and revision of the manuscript were performed
by XL, HLi and HY. Parts of the cell and animal experiments were
performed by HLi, YY and YZ, and they were also involved in
drafting of the manuscript. The corresponding authors, XR and HLu,
were major contributors in funding acquisition, designed
experiments, and were involved in manuscript review and editing.
All authors agreed to be accountable for all aspects of the work in
ensuring that questions related to the accuracy or integrity of any
part of the work are appropriately investigated and resolved. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent. The
study was performed in accordance with the Declaration of Helsinki
and was approved by the Ethics Committee of the Second Affiliated
Hospital of Xi'an Jiaotong University (approval no. 2014001). All
animal experiments were performed according to the Guide for the
Care and Use of Laboratory Animals of the National Institutes of
Health, and were subjected and approved by the Ethics Committee of
Xi'an Jiaotong University (approval no. AE201812-01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LSCC
|
laryngeal squamous cell carcinoma
|
|
EMT
|
epithelial-mesenchymal transition
|
|
lncRNA
|
long non-coding RNA
|
|
HNF1A
|
hepatic nuclear factor 1α
|
|
HNF1A-AS1
|
HNF1A antisense RNA 1
|
|
IHC
|
immunohistochemical
|
|
IHS
|
immunohistochemical scores
|
|
5-Aza-dC
|
5-Aza-2′-deoxycytidine
|
|
MSP
|
methylation-specific polymerase chain
reaction
|
|
BSP
|
bisulfite genomic sequencing
|
References
|
1
|
Lee KW, Kuo WR, Tsai SM, Wu DC, Wang WM,
Fang FM, Chiang FY, Ho KY, Wang LF, Tai CF, et al: Different impact
from betel quid, alcohol and cigarette: risk factors for pharyngeal
and laryngeal cancer. Int J Cancer. 117:831–836. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muscat JE and Wynder EL: Tobacco, alcohol,
asbestos, and occupational risk factors for laryngeal cancer.
Cancer. 69:2244–2251. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shen Z, Hao W, Zhou C, Deng H, Ye D, Li Q,
Lin L, Cao B and Guo J: Long non-coding RNA AC026166.2–001 inhibits
cell proliferation and migration in laryngeal squamous cell
carcinoma by regulating the miR-24-3p/p27 axis. Sci Rep.
8:33752018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rothenberg SM and Ellisen LW: The
molecular pathogenesis of head and neck squamous cell carcinoma. J
Clin Invest. 122:1951–1957. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lefebvre JL, Coche-Dequeant B, Degardin M,
Kara A, Mallet Y and Ton Van J: Treatment of laryngeal cancer: The
permanent challenge. Expert Rev Anticancer Ther. 4:913–920. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perl AK, Wilgenbus P, Dahl U, Semb H and
Christofori G: A causal role for E-cadherin in the transition from
adenoma to carcinoma. Nature. 392:190–193. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou AE, Zheng H, Saad MA, Rahimy M, Ku J,
Kuo SZ, Honda TK, Wang-Rodriguez J, Xuan Y, Korrapati A, et al: The
non-coding landscape of head and neck squamous cell carcinoma.
Oncotarget. 7:51211–51222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang X, Song JH, Cheng Y, Wu W, Bhagat T,
Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al: Long
non-coding RNA HNF1A-AS1 regulates proliferation and migration in
oesophageal adenocarcinoma cells. Gut. 63:881–890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu
Y, Wang L, Wang Y and Huang Q: Expression and clinical significance
of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J
Surg Oncol. 13:3022015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding CH, Yin C, Chen SJ, Wen LZ, Ding K,
Lei SJ, Liu JP, Wang J, Chen KX, Jiang HL, et al: The
HNF1α-regulated lncRNA HNF1A-AS1 reverses the malignancy of
hepatocellular carcinoma by enhancing the phosphatase activity of
SHP-1. Mol Cancer. 17:632018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Subhash S, Andersson PO, Kosalai ST,
Kanduri C and Kanduri M: Global DNA methylation profiling reveals
new insights into epigenetically deregulated protein coding and
long noncoding RNAs in CLL. Clin Epigenetics. 8:1062016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sanger F, Nicklen S and Coulson AR: DNA
sequencing with chain-terminating inhibitors. Proc Natl Acad Sci
USA. 74:5463–5467. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang
TX, Shi WH, Xie HW, Lv J, Wu QQ and Cao XF: Identification of the
long non-coding RNA POU3F3 in plasma as a novel biomarker for
diagnosis of esophageal squamous cell carcinoma. Mol Cancer.
14:32015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou H, Wang F, Chen H, Tan Q, Qiu S, Chen
S, Jing W, Yu M, Liang C, Ye S and Tu J: Increased expression of
long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal
transition of gastric cancer. Aging (Albany NY). 8:2023–2038. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aloysius MM, Zaitoun AM, Bates TE, Ilyas
M, Constantin-Teodosiu D, Rowlands BJ and Lobo DN:
Immunohistochemical expression of mitochondrial membrane complexes
(MMCs) I, III, IV and V in malignant and benign periampullary
epithelium: A potential target for drug therapy of periampullary
cancer? BMC Cancer. 10:802010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Soslow RA, Dannenberg AJ, Rush D, Woerner
BM, Khan KN, Masferrer J and Koki AT: COX-2 is expressed in human
pulmonary, colonic, and mammary tumors. Cancer. 89:2637–2645. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du P, Zhang X, Huang CC, Jafari N, Kibbe
WA, Hou L and Lin SM: Comparison of Beta-value and M-value methods
for quantifying methylation levels by microarray analysis. BMC
Bioinformatics. 11:5872010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Council NR: Guide for the Care and Use of
Laboratory Animals: Eighth edition. National Academies Press;
Washington, DC: 2011, simplehttps://www.ncbi.nlm.nih.gov/books/NBK54050/April.
2018
|
|
28
|
Myers JN, Holsinger FC, Jasser SA, Bekele
BN and Fidler IJ: An orthotopic nude mouse model of oral tongue
squamous cell carcinoma. Clin Cancer Res. 8:293–298.
2002.PubMed/NCBI
|
|
29
|
Sano D and Myers JN: Xenograft models of
head and neck cancers. Head Neck Oncol. 1:322009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen LW, Wang JL, Zhang LY, Yang SM, Li
CS, Yu N, Zhao WJD, Zhao LD, Li K, Liu MB and Zhai SQ:
Establishment of an animal model of spontaneous cervical lymph node
metastasis of laryngeal squamous cell carcinoma and obtaining
laryngocarcinoma cells with high metastatic potential. Neoplasma.
60:504–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li P, Yang Y, Liu H, Yang AK, Di JM, Tan
GM, Wang HF, Qiu JG, Zhang WJ, Jiang QW, et al: MiR-194 functions
as a tumor suppressor in laryngeal squamous cell carcinoma by
targeting Wee1. J Hematol Oncol. 10:322017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng W, Cui W, Zhao L, Chi W, Cao H and
Wang B: Aberrant methylation and downregulation of ZNF667-AS1 and
ZNF667 promote the malignant progression of laryngeal squamous cell
carcinoma. J Biomed Sci. 26:132019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jensen E: Technical review: In situ
hybridization. Anat Rec (Hoboken). 297:1349–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sanders R, Mason DJ, Foy CA and Huggett
JF: Considerations for accurate gene expression measurement by
reverse transcription quantitative PCR when analysing clinical
samples. Anal Bioanal Chem. 406:6471–6483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grada A, Otero-Vinas M, Prieto-Castrillo
F, Obagi Z and Falanga V: Research techniques made simple: Analysis
of collective cell migration using the wound healing assay. J
Invest Dermatol. 137:e11–e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Codeluppi S, Gregory EN, Kjell J,
Wigerblad G, Olson L and Svensson CI: Influence of rat substrain
and growth conditions on the characteristics of primary cultures of
adult rat spinal cord astrocytes. J Neurosci Methods. 197:118–127.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu HS, Hsu PY, Lai MD, Chang HY, Ho CL,
Cheng HL, Chen HT, Lin YJ, Wu TJ, Tzai TS and Chow NH: An unusual
function of RON receptor tyrosine kinase as a transcriptional
regulator in cooperation with EGFR in human cancer cells.
Carcinogenesis. 31:1456–1464. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bousette N, Chugh S, Fong V, Isserlin R,
Kim KH, Volchuk A, Backx PH, Liu P, Kislinger T, MacLennan DH, et
al: Constitutively active calcineurin induces cardiac endoplasmic
reticulum stress and protects against apoptosis that is mediated by
alpha-crystallin-B. Proc Natl Acad Sci USA. 107:18481–18486. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ehrlich M: DNA hypomethylation in cancer
cells. Epigenomics. 1:239–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tycko B: Epigenetic gene silencing in
cancer. J Clin Invest. 105:401–407. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|






















