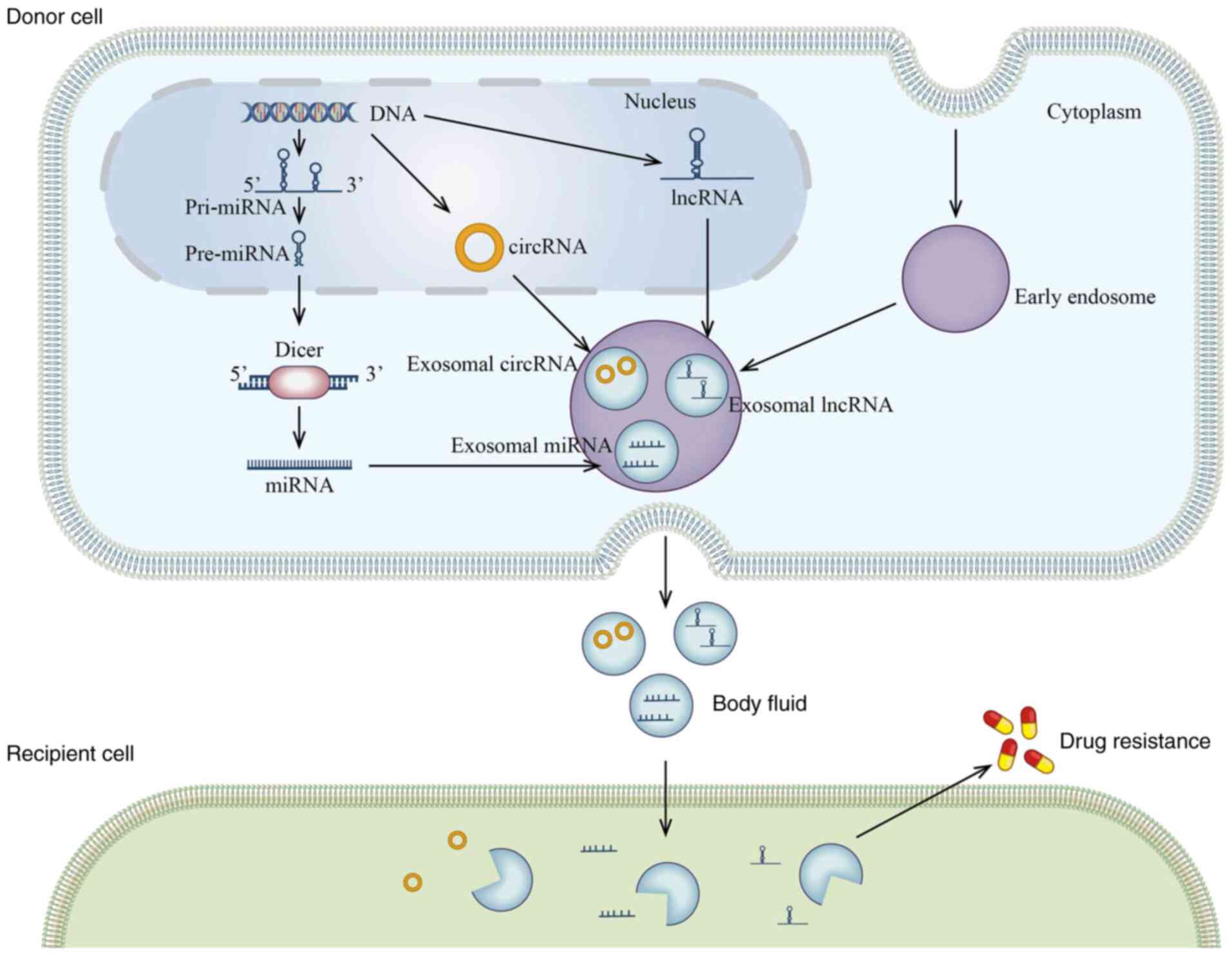
Cancer is a leading cause of death, with increasing
incidence and mortality rates, worldwide. Globally, there were an
estimated 18.1 million new cancer cases and 9.6 million
cancer-associated deaths in 2018. More than 50% of the
cancer-associated deaths occurred in Asia (1,2).
Effective chemotherapy is beneficial against cancer; however, the
development of chemoresistance, whether intrinsic or acquired, is
the primary concern to ensure good outcomes, and contributes to
distant metastasis, tumor recurrence, and disease deterioration,
resulting in cancer progression (3). Chemoresistance may result from various
factors, including increased drug efflux pumps, resistance to
apoptosis, DNA repair defects, and mutations affecting drug targets
(4–7). Thus, it is crucial to identify
mechanisms underlying chemotherapy to improve cancer outcomes.
Advances in whole genome and transcriptome
sequencing technologies have revealed that non-coding (nc) RNAs
modulate disease pathogenesis at the cellular and molecular levels
(8). ncRNAs include microRNAs
(miRNA/miR), long non-coding (lnc) RNAs, circular (circ) RNAs,
small nuclear RNAs, small nucleolar RNAs, small interfering RNAs,
and piwi-interacting RNA. Of these, miRNAs, lncRNAs, and circRNAs
are known to regulate cancer cell proliferation, invasion and
metastases, at the transcriptional and post-transcriptional levels
(9). Recent studies have shown that
ncRNAs was associated with cancer cell chemoresistance. For
example, GBCDRlnc1, a lncRNA, was reported to induce
chemoresistance in gallbladder cancer cells by interacting with
phosphoglycerate kinase 1 and upregulating autophagy-related gene
levels (10). CRIM1, a circRNA, has
been shown to promote nasopharyngeal cancer metastasis and
docetaxel chemoresistance by competitively binding Forkhead box Q1
(11). While these findings show
that ncRNAs may modulate cancer chemotherapy, the underlying
mechanisms are unclear.
Exosomes, which are 30-100 nm in diameter, are
nanoscale membrane vesicles derived from endosomal multivesicular
bodies released by various cell types, including immune cells,
neurons, mesenchymal stem cells, Schwann cells and epithelial cells
(12) and have been associated with
cancer growth, metastasis and chemoresistance (13–15).
Exosomes carry various cargo, including DNAs, RNAs and proteins,
that are involved in the exchange of genetic information between
donor and recipient cells in tumor microenvironments, which may
transmit drug resistance (13,16).
In the present review, research on exosomal ncRNAs in different
types of cancer, will be discussed, with the aim to highlight their
role in cancer treatment.
Non-coding RNAs comprise ~98% of the human genome,
which was once considered to be transcriptional noise (17). Extensive research has revealed that
ncRNAs regulate various molecular processes and human diseases
(18).
miRNAs are small ncRNAs, comprised of 20-22
nucleotides in length, excised from 60-110 nucleotide foldback RNA
precursor structures (19) and are
widely expressed in various types of cancer, including breast,
gastric, and colorectal cancer, and have been associated with
prognosis (20–22).
circRNAs are single-stranded covalently closed RNA
molecules, that were first identified in RNA viruses in 1976 using
electron microscopy (29). circRNAs
fall into four different types: Exon circRNAs, circular intronic
RNAs, exon-intron circRNAs, and intergenic circRNAs or fusion
circRNAs (30–32). circRNAs are protected from RNase
digestion, as they lack free 5′ and 3′ structures, therefore they
are more stable than linear RNAs (33). Functionally, circRNAs modulate gene
expression transcriptionally and post-transcriptionally, in
physiological and pathological contexts, primarily by acting as
miRNA sponges. For example, circ-TCF4.85 has been reported to
promote hepatocellular carcinoma (HCC) development by competitively
binding to miR-486-5p (34).
Hsa_circ_0091570 knockdown significantly enhanced cell
proliferation and migration, and inhibited apoptosis in HCC by
sponging miR-1307 (35).
There is an increasing number of studies which have
shown that exosomes contain specific miRNAs in response to cancer
therapy, resulting in chemoresistance or chemosensitivity in body
fluids (48,49) (Table
I). Exosomes have been found to transfer miR-196a to recipient
cells, promoting cisplatin resistance by sponging CDKN1B and ING5
in plasma and tissues from patients with head and neck cancer
(50). Exosomal miR-1238 derived
from resistant cancer cells could confer drug resistance to
sensitive cells by activating the EGFR-PI3K-Akt-mTOR signaling
pathway (51). In tumors from the
digestive system, miR-374a-5p was found to be upregulated in the
serum from patients with gastric cancer and was associated with
poor prognosis (52). miR-374a-5p
suppressed apoptosis and increased the expression of multi-drug
resistant and topoisomerase II by negatively regulating its target
gene, Neurod1. In addition, inhibition of exosome-mediated
miR-374a-5p activity may re-sensitize gastric cancer cells to
oxaliplatin (52). Reduced miR-744
expression levels were reported in the exosomes from serum and
tissues in patients with HCC (53,54).
PAX2 was found to be upregulated in HCC tissue and was a miR-744
target. Knockdown and overexpression functional assays found that
HepG2-resistant cells treated with miR-744-overexpressing exosomes
were more sensitive to sorafenib. Treating HCC cells with exosomes
from mature adipocytes transferred miR-23a/b to them, conferring
5-fluorouracil resistance and an aggressive phenotype. These
findings indicated that exosomal miR-744 or miR-23a/b are potential
HCC biomarkers (53,54). miR-196b-5p promoted chemoresistance
to 5-fluorouracil by targeting negative regulators of SOCS
(suppressor of cytokine signaling)-1 and −3 in colorectal cancer,
activating the STAT3 signaling pathway. In addition, miR-196b-5p
expression in serum exosomes from patients with colorectal cancer
was markedly higher compared with that in healthy controls
(55).
Gene function and bioinformatics analyses revealed
that miR-23a-3p, miR-27a-3p, miR-30a-5p, and miR-320a were markedly
upregulated in exosomes derived from adriamycin-resistant cells
compared with that in parental breast cancer cells (56). These miRNAs may contribute to
chemoresistance via multiple signaling pathways, including the
MAPK, and Wnt signaling pathways (56). Furthermore, 14 miRNAs were found to
be significantly elevated in oxaliplatin/5-fluorouracil resistant
colorectal cancer cells compared with that in the control parental
cells (49). Of these, miR-21-5p,
miR-1246, miR-1229-5p, miR-135b, miR-425, and miR-96-5p were highly
abundant in exosomes from the resistant cells. In addition,
exosomal miR-21-5p, miR-1246, miR-1229-5p and miR-96-5p levels in
the serum of patients with colorectal cancer (CRC) who were
chemoresistant were markedly higher compared with that in patients
who were chemosensitive. Notably, Gene Ontology and Kyoto
Encyclopedia of Genes and Genomes analysis revealed that these
miRNAs were enriched in the PI3K-AKT, FOXO, and autophagy pathways.
These 4 exosomal miRNAs could be potential biomarkers for
chemoresistance restoration in colorectal cancer (49). Signaling pathways have been found to
be involved in miRNA-mediated chemoresistance via tumor exosomes.
For example, exosome-mediated transfer of miR-21 promoted cisplatin
resistance via the PTEN/PI3K/AKT signaling pathway (57). Exosomal transfer of miR-142-3p from
bone marrow-derived MSCs suppressed Numb expression, activating the
Notch signaling pathway (58).
Intercellular communication was found to be
initiated by surface interactions between circulating exosomes and
transmembrane molecules expressed on target cells. The release and
uptake of exosomes, containing miRNAs, are a crucial form of
cell-cell communication in tumors, as cells acquire malignant
phenotypes by engulfing tumor-derived oncogenic factors in the
exosomes (59). In addition to
cancer cells, cancer-associated fibroblasts (CAFs) are innately
chemoresistant and contribute to cancer cell chemoresistance
(60). CAF-derived miR21 was
reported to be significantly upregulated in ovarian cancer cells.
Exosomes carrying miR21 from CAFs to recipient cells conferred
chemoresistance and aggressive phenotypes (61). Furthermore, as CAFs are
intrinsically resistant to cisplatin, their exosomes accelerated
resistance (50). CAF-derived
exosomes transferred miR-196a to head and neck cancer (HNC) cells,
promoting proliferation and cisplatin resistance, which was
mediated by nuclear ribonucleoprotein A1. Functionally, miR-196a
conferred cisplatin resistance by suppressing the expression level
of CDKN1B and ING5, both in the plasma and tissues from patients
with HNC, indicating that miR-196a might be an independent
predictor for chemoresistance in patients with head and neck cancer
(50). CAFs constitute the bulk of
pancreatic ductal adenocarcinoma (PDAC) tumors and promote
epithelial cancer cell proliferation and drug resistance.
Gemcitabine-treated CAF exosomes enhanced proliferation and
survival of chemoresistant PDAC cells (62). miRNA-sequencing analysis found that
miR-146a was markedly elevated in gemcitabine-treated fibroblasts.
Compared with that in untreated CAFs, gemcitabine-treated CAFs
expressed significantly higher miR-146a expression levels.
Gemcitabine-treated CAFs derived exosomes were also found to
elevate Snail and miR-146a expression levels. These data suggest
that CAF-derived exosomes are key modulators of PDAC
chemoresistance. However, blocking exosome release may suppress
chemoresistance caused by exosome-mediated signaling (62). In recent decades, cancer stem cells
(CSCs) have emerged as a unique cellular subtype capable of
self-renewal and multi-differentiation. CSCs are important drivers
of cancer recurrence, metastases, and chemoresistance (63). Santos et al (64) found that breast cancer cells were
more resistant to doxorubicin (DOX) upon receiving exosomes from
DOX- and paclitaxel-resistant CSCs compared with that in sensitive
cells, suggesting that exosomes transfer drug-resistance. In
addition, miR-155 overexpression increased its content in exosomes,
leading to epithelial-mesenchymal transition (EMT)-associated
chemoresistance. Furthermore, co-culture assays revealed that the
exosome-transferred miR-155 significantly induced DOX and
paclitaxel resistance in breast cancer cells. The study by Santos
et al (64) highlighted the
importance of inhibiting transfer of drug-resistance from resistant
breast cancer cells to sensitive ones. One cut homeobox 2 promoted
breast cancer cell growth and its overexpression may suppress
stemness-associated gene expression via chemoresistant
extracellular vesicles (EVs), aiding the reverse of chemoresistance
(65). This mechanism was
negatively mediated by miR-9-5p, miR-195-5p, and miR-203a-3p in
circulating EVs (65). Thus,
cell-cell communication via exosomal miRNAs may be an important
mechanism for cancer chemotherapy.
EMT contributes to cancer chemoresistance.
miR-128-3p has been reported to suppress EMT and enhance
intracellular oxaliplatin accumulation in colorectal cancer.
Delivery of miR-128-3p to resistant cells via secreted exosomes
improved oxaliplatin response (66). Delivery of miR-155-5p to MGC-803
cells, in tumor-derived exosomes induced EMT and chemoresistance by
suppressing the expression level of GATA binding protein 3
expression and tumor protein p53-inducible nuclear protein 1,
highlighting a prospective strategy for reversing paclitaxel
resistance in gastric cancer (67).
An increasing number of studies have shown that
exosomal miRNAs also trigger tumor drug resistance via autophagy,
angiogenesis, inflammation and hypoxia induction (68–70).
Cisplatin elevated circulating exosomal miR-425-3p levels and
exo-miRNA release in non-small cell lung cancer (68). Furthermore, exosomal miR-425-3p may
target AKT1, activating autophagy by negatively regulating the
AKT/mTOR signaling pathway and causing resistance to
cisplatin-induced apoptosis (68).
miR-126a was released from myeloid-derived suppressor cell
exosomes, promoting lung metastasis induced by DOX (69). Hypoxia has been reported to induce
macrophage M2-polarization and enhance the level of
tumor-associated macrophage (TAM)-derived exosomal miR-223
(70). In addition, TAM-derived
exosomal miR-223 was reported to modulate chemoresistance and
exosmal miR-223 has been shown to inactivate the PI3K/AKT signaling
pathway by targeting PTEN. Furthermore, circulating exosomal
miR-223 predicted the response to chemotherapy in patients with
advanced epithelial ovarian cancer (70). Taken together, exosomal miRNAs are
potential non-invasive biomarkers and therapeutic targets in a new
anti-cancer paradigm. The value of exosomal miRNAs as
anti-chemoresistance targets warrants further Investigation.
lncRNAs, which were previously regarded as
transcriptional junk, due to their lack of protein coding capacity,
are involved in numerous biological processes at the
transcriptional, post-transcriptional, and chromatin levels. An
increasing number of studies have indicated that exosomal lncRNAs
modulate chemoresistance (Table
II).
Trastuzumab, a humanized monoclonal antibody against
the extracellular region of human epidermal growth factor receptor
2 (HER-2) is used to treat HER2-positive breast cancer; however,
its effects are limited by resistance (71). The lncRNA, actin filament associated
protein 1 antisenseRNA 1 (AFAP1-AS1) has been reported to be
elevated in trastuzumab-resistant cells compared with that in
sensitive cells, and was associated with poor prognosis and
survival time in patients with breast cancer (72). In addition, knockdown of AFAP1-AS1
has been shown to reverse chemoresistance. Furthermore, AFAP1-AS1
was transferred from resistant cells to recipient cells by the
incorporation of exosomes, inducing trastuzumab resistance.
Mechanically, AFAP1-AS1 has been reported to promote ERBB2
translation by combining with AU-binding factor 1 (AUF1) protein.
Thus, exosomal AFAP1-AS1 promoted trastuzumab resistance by binding
to AUF1 (72). Similarly, the
lncRNA, AGAP2 antisense RNA 1 (AGAP2-AS1) promoted trastuzumab
resistance in breast cancer cells via packaging into exosomes
(73). Exosomal small nucleolar RNA
host gene 14 (SNHG14) lncRNA induced trastuzumab resistance by
upregulating the Bcl-2/Bax signaling pathway. Serum exosomal SNHG14
has biomarker potential in breast cancer (74).
Breast cancer cell-derived exosomes might enhance
DOX resistance by transferring lncRNA H19. Serum exosomal H19 is
highly expressed in patients who are non-responsive to DOX compared
with that in patients who are responsive (75). Crosstalk between CAFs and cancer
cells has been reported to promote tumor progression and induce
chemoresistance in the tumor microenvironment. CAFs promoted
stemness and chemoresistance by transmitting exosomal H19 in CRC.
Mechanically, H19 mediated oxaliplatin resistance by activating the
WNT/β-catenin signaling pathway by sponging miR-141 (76). CAFs transferred CRC-associated
lncRNA to tumor cells in exosomes, conferring oxaliplatin and
5-fluorouracil resistance and activating the WNT/β-catenin
signaling pathway (59). The
WNT/β-catenin signaling pathway modulates various tumor processes,
including cancer growth, invasion and angiogenesis (77,78)
and its abnormal activation may result in cancer drug resistance
(79,80).
Exosomal lncRNA dysregulation has been observed in
different types of cancer, where they act as competitive endogenous
RNAs against active factors, such as miRNAs (76,81).
In gastric cancer (GC), exosomes have been reported to downregulate
HOXA at the distal tip, a lncRNA, hence conferring cisplatin
resistance by sponging miR-218, highlighting a potential exosome
lncRNA-directed therapy against GC (82). The lncRNA, prostate
androgen-regulated transcript 1 (PART1) was abundantly expressed in
gefitinib-resistant cells compared with that in parental esophageal
squamous cell carcinoma cells (83). By binding to PART1's promoter
region, signal transducer and activator of transcription
participated in chemoresistance. In addition, exosome-mediated
transfer of PART1 induced resistance by targeting miR-129.
Furthermore, serum exosomal PART1 was stably upregulated in
patients with esophageal squamous cell carcinoma who were also
gefitinib-resistant (83). A
binding site is often located in the promoter region of lncRNAs,
and a gene can bind to this site to regulate expression, thus
affecting tumor biology. For example, lncRNA SBF2 antisense RNA 1
(SBF2-AS1) overexpression contributed to temozolomide (TMZ)
resistance in glioblastoma (GBM) cells, while its downregulation
could reverse this effect (84).
The transcription factor, ZEB1 combined with the promoter region of
SBF2-AS1 to alter chemoresistance. SBF2-AS1-containing exosomes
secreted by cancer cells transmitted TMZ to the recipient cells.
SBF2-AS1-containing serum exosomes were associated with poor
response to TMZ treatment in patients with GBM (84). Notably, acetylated histone 3 lysine
27 was enriched at the promoter region of AFAP1-AS1, which might
trigger drug resistance (72). In
non-small cell lung cancer (NSCLC), exosome-mediated transfer of
lncRNA RP11-838N2.4 induced erlotinib resistance. Forkhead box
protein O1 could bind to the promoter region of RP11-838N2.4, and
silenced it by recruiting histone deacetylase. Therefore, lncRNA
RP11-838N2.4 may be a valuable target for reversing NSCLC
chemoresistance (85). Taken
together, exosomal lncRNAs play important roles in transmitting
chemoresistance. However, the regulatory mechanism of
chemoresistance is unclear and warrants further investigation.
Numerous studies have suggested that circRNAs are
novel therapeutic and prognostic biomarkers for various types of
cancer, due to their characteristics of closed loop structure and
insensitivity to RNase. It is now known that exosomes might be
carriers of circRNAs, which are subsequently secreted into body
fluids, enabling them to modulate tumor cell proliferation,
migration, invasion, and apoptosis, and to affect drug resistance.
For example, the circRNA, Cdr1as, has been reported to be
downregulated in serum exosomes from patients with
cisplatin-resistant ovarian cancer (86). In CRC, hsa_circ_0000338 has been
reported to have tumor suppressive function in CRC cells, so that
they are sensitive to fluorouracil plus oxaliplatin. Clinical data
has validated that hsa_circ_000338 was markedly downregulated in
serum exosomes in patients with CRC who are chemo-resistant
(87). Exosomes, from resistant CRC
cells, transmitted ciRS-122 to recipient cells by sponging miR-122,
thereby resulting in oxaliplatin resistance (88). Serum exosomal circNFIX has been
reported to be upregulated in patients with glioma and who are
TMZ-resistant (89). Extracellular
circNFIX secretion in exosomes also conferred TMZ resistance in
sensitive cells, while circNFIX silencing promoted TMZ sensitivity
in resistant cells by negatively interacting with miR-132. This
finding highlighted the role of prognostic biomarkers and promising
therapeutic targets in improving the clinical benefits of TMZ in
patients with glioma (89).
However, further studies are required to improve the understanding
of the roles of exo-circRNAs in overcoming chemoresistance.
Since their discovery, extensive research has shown
that ncRNAs regulate multiple molecular processes. In addition, due
to sequence conservation, they are more stably expressed in various
types of tissues, cells, and body fluids. Recent studies have
indicated that ncRNAs affect tumor chemoresistance; therefore, they
are promising biomarkers for cancer therapy and reversal of
chemoresistance (97–99). Despite the increasing number of
studies on ncRNAs chemoresistance, they have not yet been targeted
clinically.
Exosomes deliver various cargo, such as DNA, RNA and
proteins, from donor cells to recipient cells, thus exerting
biological effects. In recent years, it has emerged that
exosome-mediated transfer of ncRNAs may enhance chemoresistance.
The present review described the importance of exosome-contained
ncRNA in cancer chemotherapy. However, their effects in cancer are
still controversial, and most of the evidence only comes from
cell-based and animal studies. Thus, comprehensive studies into the
roles and mechanisms of exosome ncRNAs in different types of cancer
may uncover novel effective treatment strategies. Thus,
comprehensive clinical studies in this area are warranted.
Not applicable.
This study was supported by grants from the Natural
Science Foundation of Anhui Province (grant no. 2008085QH404) and
the Science Research Project of Bengbu Medical College (grant no.
BYKY2019048ZD).
Not applicable.
YYW and QL designed the study. YYW drafted the
manuscript. YYW and QL designed the tables and figures. FCW
critically discussed and revised the manuscript. All authors read
and approved the final manuscript for publication.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sarmento-Ribeiro AB, Scorilas A, Gonçalves
AC, Efferth T and Trougakos IP: The emergence of drug resistance to
targeted cancer therapies: Clinical evidence. Drug Resist Updat.
47:1006462019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hall MD, Okabe M, Shen DW, Liang XJ and
Gottesman MM: The role of cellular accumulation in determining
sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol
Toxicol. 48:495–535. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mikamori M, Yamada D, Eguchi H, Hasegawa
S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K,
et al: MicroRNA-155 controls exosome synthesis and promotes
gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci
Rep. 7:423392017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu T, Chen G, Sun D, Lei M, Li Y, Zhou C,
Li X, Xue W, Wang H, Liu C and Xu J: Exosomes containing miR-21
transfer the characteristic of cisplatin resistance by targeting
PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim
Biophys Sin (Shanghai). 49:808–816. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu X, Lu Y, Xu Y, Hou S, Huang J, Wang B,
Zhao J, Xia S, Fan S, Yu X, et al: Exosomal transfer of miR-501
confers doxorubicin resistance and tumorigenesis via targeting of
BLID in gastric cancer. Cancer Lett. 459:122–134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Tong X, Zhou Z, Wang S, Lei Z,
Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al: Circular RNA
hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced
epithelial-mesenchymal transition and metastasis by controlling
TIF1γ in non-small cell lung cancer. Mol Cancer. 17:1402018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu J, Li Y, Tong J, Gao J, Guo Q, Zhang
L, Wang B, Zhao H, Wang H, Jiang E, et al: Long non-coding
RNA-dependent mechanism to regulate heme biosynthesis and
erythrocyte development. Nat Commun. 9:43862018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang
J, Tang Z and Quan Z: Long non-coding RNA GBCDRlnc1 induces
chemoresistance of gallbladder cancer cells by activating
autophagy. Mol Cancer. 18:822019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong X, Liu N, Liang Y, He Q, Yang X, Lei
Y, Zhang P, Zhao Y, He S, Wang Y, et al: Circular RNA CRIM1
functions as a ceRNA to promote nasopharyngeal carcinoma metastasis
and docetaxel chemoresistance through upregulating FOXQ1. Mol
Cancer. 19:332020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Raposo G and Stoorvogel W: Extracellular
vesicles: Exosomes, microvesicles, and friends. J Cell Biol.
200:373–383. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Wang Y, Wang Q, Liu Y, Bao W and Wu
S: Exosomes in cancer: Small transporters with big functions.
Cancer Lett. 435:55–65. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bandari SK, Purushothaman A, Ramani VC,
Brinkley GJ, Chandrashekar DS, Varambally S, Mobley JA, Zhang Y,
Brown EE, Vlodavsky I and Sanderson RD: Chemotherapy induces
secretion of exosomes loaded with heparanase that degrades
extracellular matrix and impacts tumor and host cell behavior.
Matrix Biol. 65:104–118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mashouri L, Yousefi H, Aref AR, Ahadi AM,
Molaei F and Alahari SK: Exosomes: Composition, biogenesis, and
mechanisms in cancer metastasis and drug resistance. Mol Cancer.
18:752019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H, Deng T, Liu R, Ning T, Yang H,
Liu D, Zhang Q, Lin D, Ge S, Bai M, et al: CAF secreted miR-522
suppresses ferroptosis and promotes acquired chemo-resistance in
gastric cancer. Mol Cancer. 19:432020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei LH and Guo JU: Coding functions of
‘noncoding’ RNAs. Science. 367:1074–1075. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bandres E, Bitarte N, Arias F, Agorreta J,
Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ,
et al: microRNA-451 regulates macrophage migration inhibitory
factor production and proliferation of gastrointestinal cancer
cells. Clin Cancer Res. 15:2281–2290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baltruskeviciene E, Schveigert D,
Stankevicius V, Mickys U, Zvirblis T, Bublevic J, Suziedelis K and
Aleknavicius E: Down-regulation of miRNA-148a and miRNA-625-3p in
colorectal cancer is associated with tumor budding. BMC Cancer.
17:6072017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-beta promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Niu Y, Ma F, Huang W, Fang S, Li M, Wei T
and Guo L: Long non-coding RNA TUG1 is involved in cell growth and
chemoresistance of small cell lung cancer by regulating LIMK2b via
EZH2. Mol Cancer. 16:52017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 166:1055–1056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao J, Dai C, Yu X, Yin XB and Zhou F:
Circ-TCF4.85 silencing inhibits cancer progression through
microRNA-486-5p-targeted inhibition of ABCF2 in hepatocellular
carcinoma. Mol Oncol. 14:447–461. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang YG, Wang T, Ding M, Xiang SH, Shi M
and Zhai B: hsa_circ_0091570 acts as a ceRNA to suppress
hepatocellular cancer progression by sponging hsa-miR-1307. Cancer
Lett. 460:128–138. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harding C, Heuser J and Stahl P:
Receptor-mediated endocytosis of transferrin and recycling of the
transferrin receptor in rat reticulocytes. J Cell Biol. 97:329–339.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pan BT and Johnstone RM: Fate of the
transferrin receptor during maturation of sheep reticulocytes in
vitro: Selective externalization of the receptor. Cell. 33:967–978.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang M, Zhou L, Yu F, Zhang Y, Li P and
Wang K: The functional roles of exosomal long non-coding RNAs in
cancer. Cell Mol Life Sci. 76:2059–2076. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Colombo M, Moita C, van Niel G, Kowal J,
Vigneron J, Benaroch P, Manel N, Moita LF, Théry C and Raposo G:
Analysis of ESCRT functions in exosome biogenesis, composition and
secretion highlights the heterogeneity of extracellular vesicles. J
Cell Sci. 126:5553–5565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Barile L and Vassalli G: Exosomes: Therapy
delivery tools and biomarkers of diseases. Pharmacol Ther.
174:63–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khare D, Or R, Resnick I, Barkatz C,
Almogi-Hazan O and Avni B: Mesenchymal stromal cell-derived
exosomes affect mRNA expression and function of B-lymphocytes.
Front Immunol. 9:30532018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Burrello J, Monticone S, Gai C, Gomez Y,
Kholia S and Camussi G: Stem cell-derived extracellular vesicles
and immune-modulation. Front Cell Dev Biol. 4:832016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jayaseelan VP: Emerging role of exosomes
as promising diagnostic tool for cancer. Cancer Gene Ther.
27:395–398. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lakkaraju A and Rodriguez-Boulan E:
Itinerant exosomes: Emerging roles in cell and tissue polarity.
Trends Cell Biol. 18:199–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kahroba H, Hejazi MS and Samadi N:
Exosomes: From carcinogenesis and metastasis to diagnosis and
treatment of gastric cancer. Cell Mol Life Sci. 76:1747–1758. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Z, Chen Z, Hu G and Jiang Y: Roles of
circular RNA in breast cancer: Present and future. Am J Transl Res.
11:3945–3954. 2019.PubMed/NCBI
|
|
48
|
Zeng A, Wei Z, Yan W, Yin J, Huang X, Zhou
X, Li R, Shen F, Wu W, Wang X and You Y: Exosomal transfer of
miR-151a enhances chemosensitivity to temozolomide in
drug-resistant glioblastoma. Cancer Lett. 436:10–21. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jin G, Liu Y, Zhang J, Bian Z, Yao S, Fei
B, Zhou L, Yin Y and Huang Z: A panel of serum exosomal microRNAs
as predictive markers for chemoresistance in advanced colorectal
cancer. Cancer Chemother Pharmacol. 84:315–325. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qin X, Guo H, Wang X, Zhu X, Yan M, Wang
X, Xu Q, Shi J, Lu E, Chen W and Zhang J: Exosomal miR-196a derived
from cancer-associated fibroblasts confers cisplatin resistance in
head and neck cancer through targeting CDKN1B and ING5. Genome
Biol. 20:122019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yin J, Zeng A, Zhang Z, Shi Z, Yan W and
You Y: Exosomal transfer of miR-1238 contributes to
temozolomide-resistance in glioblastoma. EBioMedicine. 42:238–251.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J,
Qian H, Xu W, Qian J and Lin J: miR-374a-5p: A new target for
diagnosis and drug resistance therapy in gastric cancer. Mol Ther
Nucleic Acids. 18:320–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang G, Zhao W, Wang H, Qiu G, Jiang Z,
Wei G and Li X: Exosomal MiR-744 inhibits proliferation and
sorafenib chemoresistance in hepatocellular carcinoma by targeting
PAX2. Med Sci Monit. 25:7209–7217. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Y, Tan J, Ou S, Chen J and Chen L:
Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth
in hepatocellular cancer by targeting the VHL/HIF axis. J Physiol
Biochem. 75:391–401. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y,
Liang Y, Cao L, Li X, Li R, et al: Maintenance of cancer stemness
by miR-196b-5p contributes to chemoresistance of colorectal cancer
cells via activating STAT3 signaling pathway. Oncotarget.
8:49807–49823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen WX, Xu LY, Qian Q, He X, Peng WT, Zhu
YL and Cheng L: Analysis of miRNA signature differentially
expressed in exosomes from adriamycin-resistant and parental human
breast cancer cells. Biosci Rep. 38:BSR201810902018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie
G, Ma Y and Shen L: Exosomal transfer of tumor-associated
macrophage-derived miR-21 confers cisplatin resistance in gastric
cancer cells. J Exp Clin Cancer Res. 36:532017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li H and Li F: Exosomes from BM-MSCs
increase the population of CSCs via transfer of miR-142-3p. Br J
Cancer. 119:744–755. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng
H, Zhang X, Kong F and Guan M: Long noncoding RNA CCAL transferred
from fibroblasts by exosomes promotes chemoresistance of colorectal
cancer cells. Int J Cancer. 146:1700–1716. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kulkarni B, Kirave P, Gondaliya P, Jash K,
Jain A, Tekade RK and Kalia K: Exosomal miRNA in chemoresistance,
immune evasion, metastasis and progression of cancer. Drug Discov
Today. 24:2058–2067. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Au Yeung CL, Co NN, Tsuruga T, Yeung TL,
Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, et al: Exosomal
transfer of stroma-derived miR21 confers paclitaxel resistance in
ovarian cancer cells through targeting APAF1. Nat Commun.
7:111502016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Richards KE, Zeleniak AE, Fishel ML, Wu J,
Littlepage LE and Hill R: Cancer-associated fibroblast exosomes
regulate survival and proliferation of pancreatic cancer cells.
Oncogene. 36:1770–1778. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nio K, Yamashita T and Kaneko S: The
evolving concept of liver cancer stem cells. Mol Cancer. 16:42017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Santos JC, Lima NDS, Sarian LO, Matheu A,
Ribeiro ML and Derchain SFM: Exosome-mediated breast cancer
chemoresistance via miR-155 transfer. Sci Rep. 8:8292018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shen M, Dong C, Ruan X, Yan W, Cao M,
Pizzo D, Wu X, Yang L, Liu L, Ren X and Wang SE:
Chemotherapy-induced extracellular vesicle miRNAs promote breast
cancer stemness by targeting ONECUT2. Cancer Res. 79:3608–3621.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu T, Zhang X, Du L, Wang Y, Liu X, Tian
H, Wang L, Li P, Zhao Y, Duan W, et al: Exosome-transmitted
miR-128-3p increase chemosensitivity of oxaliplatin-resistant
colorectal cancer. Mol Cancer. 18:432019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang M, Qiu R, Yu S, Xu X, Li G, Gu R, Tan
C, Zhu W and Shen B: Paclitaxel-resistant gastric cancer MGC-803
cells promote epithelial-to-mesenchymal transition and
chemoresistance in paclitaxel-sensitive cells via exosomal delivery
of miR-155-5p. Int J Oncol. 54:326–338. 2019.PubMed/NCBI
|
|
68
|
Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan
M, Xue W, Wang Y, Li W, Shu Y, et al: Exosomal transfer of
cisplatin-induced miR-425-3p confers cisplatin resistance In NSCLC
through activating autophagy. Int J Nanomedicine. 14:8121–8132.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Deng Z, Rong Y, Teng Y, Zhuang X,
Samykutty A, Mu J, Zhang L, Cao P, Yan J, Miller D and Zhang HG:
Exosomes miR-126a released from MDSC induced by DOX treatment
promotes lung metastasis. Oncogene. 36:639–651. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhu X, Shen H, Yin X, Yang M, Wei H, Chen
Q, Feng F, Liu Y, Xu W and Li Y: Macrophages derived exosomes
deliver miR-223 to epithelial ovarian cancer cells to elicit a
chemoresistant phenotype. J Exp Clin Cancer Res. 38:812019.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Adamczyk A, Kruczak A, Harazin-Lechowska
A, Ambicka A, Grela-Wojewoda A, Domagała-Haduch M, Janecka-Widła A,
Majchrzyk K, Cichocka A, Ryś J and Niemiec J: Relationship between
HER2 gene status and selected potential biological features related
to trastuzumab resistance and its influence on survival of breast
cancer patients undergoing trastuzumab adjuvant treatment. Onco
Targets Ther. 11:4525–4535. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Han M, Gu Y, Lu P, Li J, Cao H, Li X, Qian
X, Yu C, Yang Y, Yang X, et al: Exosome-mediated lncRNA AFAP1-AS1
promotes trastuzumab resistance through binding with AUF1 and
activating ERBB2 translation. Mol Cancer. 19:262020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zheng Z, Chen M, Xing P, Yan X and Xie B:
Increased expression of exosomal AGAP2-AS1 (AGAP2 antisense RNA 1)
in breast cancer cells inhibits trastuzumab-induced cell
cytotoxicity. Med Sci Monit. 25:2211–2220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dong H, Wang W, Chen R, Zhang Y, Zou K, Ye
M, He X, Zhang F and Han J: Exosome-mediated transfer of
lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast
cancer. Int J Oncol. 53:1013–1026. 2018.PubMed/NCBI
|
|
75
|
Wang X, Pei X, Guo G, Qian X, Dou D, Zhang
Z, Xu X and Duan X: Exosome-mediated transfer of long noncoding RNA
H19 induces doxorubicin resistance in breast cancer. J Cell
Physiol. 235:6896–6904. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen
S, Wang Y, Wang T and Hou Y: Carcinoma-associated fibroblasts
promote the stemness and chemoresistance of colorectal cancer by
transferring exosomal lncRNA H19. Theranostics. 8:3932–3948. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ramachandran I, Thavathiru E, Ramalingam
S, Natarajan G, Mills WK, Benbrook DM, Zuna R, Lightfoot S, Reis A,
Anant S and Queimado L: Wnt inhibitory factor 1 induces apoptosis
and inhibits cervical cancer growth, invasion and angiogenesis in
vivo. Oncogene. 31:2725–2737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Tenbaum SP, Ordóñez-Morán P, Puig I,
Chicote I, Arqués O, Landolfi S, Fernández Y, Herance JR, Gispert
JD, Mendizabal L, et al: β-catenin confers resistance to PI3K and
AKT inhibitors and subverts FOXO3a to promote metastasis in colon
cancer. Nat Med. 18:892–901. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Arend RC, Londoño-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wickström M, Dyberg C, Milosevic J, Einvik
C, Calero R, Sveinbjörnsson B, Sandén E, Darabi A, Siesjö P, Kool
M, et al: Wnt/β-catenin pathway regulates MGMT gene expression in
cancer and inhibition of Wnt signalling prevents chemoresistance.
Nat Commun. 6:89042015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang
X, Chen K, Zhang Y, Liu H, Gan L, et al: Tumor-derived exosomal
lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in
pancreatic ductal adenocarcinoma. Oncogene. 37:3822–3838. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang J, Lv B, Su Y, Wang X, Bu J and Yao
L: Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin
resistance in gastric cancer cells by regulating HMGA1/miR-218
axis. Onco Targets Ther. 12:11325–11338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kang M, Ren M, Li Y, Fu Y, Deng M and Li
C: Exosome-mediated transfer of lncRNA PART1 induces gefitinib
resistance in esophageal squamous cell carcinoma via functioning as
a competing endogenous RNA. J Exp Clin Cancer Res. 37:1712018.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang Z, Yin J, Lu C, Wei Y, Zeng A and
You Y: Exosomal transfer of long non-coding RNA SBF2-AS1 enhances
chemoresistance to temozolomide in glioblastoma. J Exp Clin Cancer
Res. 38:1662019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang W, Cai X, Yu J, Lu X, Qian Q and
Qian W: Exosome-mediated transfer of lncRNA RP11-838N2.4 promotes
erlotinib resistance in non-small cell lung cancer. Int J Oncol.
53:527–538. 2018.PubMed/NCBI
|
|
86
|
Zhao Z, Ji M, Wang Q, He N and Li Y:
Circular RNA Cdr1as upregulates SCAI to suppress cisplatin
resistance in ovarian cancer via miR-1270 suppression. Mol Ther
Nucleic Acids. 18:24–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal
R and Abu N: Extracellular Vesicle-derived circular RNAs confers
chemoresistance in colorectal cancer. Sci Rep. 9:164972019.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang X, Zhang H, Yang H, Bai M, Ning T,
Deng T, Liu R, Fan Q, Zhu K, Li J, et al: Exosome-delivered circRNA
promotes glycolysis to induce chemoresistance through the
miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 14:539–555.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z,
Zhang G, Gu J and Kang D: Exosome-mediated transfer of circRNA
CircNFIX enhances temozolomide resistance in glioma. Cancer Lett.
479:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang X, Zhang H, Bai M, Ning T, Ge S, Deng
T, Liu R, Zhang L, Ying G and Ba Y: Exosomes serve as nanoparticles
to deliver anti-miR-214 to reverse chemoresistance to cisplatin in
gastric cancer. Mol Ther. 26:774–783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Jingyue S, Xiao W, Juanmin Z, Wei L,
Daoming L and Hong X: TFAP2E methylation promotes 5-fluorouracil
resistance via exosomal miR-106a-5p and miR-421 in gastric cancer
MGC-803 cells. Mol Med Rep. 20:323–331. 2019.PubMed/NCBI
|
|
92
|
Patel GK, Khan MA, Bhardwaj A, Srivastava
SK, Zubair H, Patton MC, Singh S, Khushman M and Singh AP: Exosomes
confer chemoresistance to pancreatic cancer cells by promoting ROS
detoxification and miR-155-mediated suppression of key
gemcitabine-metabolising enzyme, DCK. Br J Cancer. 116:609–619.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Xu Y and Zhu M: Novel exosomal miR-46146
transfer oxaliplatin chemoresistance in colorectal cancer. Clin
Transl Oncol. 22:1105–1116. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen WX, Xu LY, Qian Q, He X, Peng WT, Fan
WQ, Zhu YL, Tang JH and Cheng L: d Rhamnose β-hederin reverses
chemoresistance of breast cancer cells by regulating
exosome-mediated resistance transmission. Biosci Rep.
38:BSR201801102018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kanlikilicer P, Bayraktar R, Denizli M,
Rashed MH, Ivan C, Aslan B, Mitra R, Karagoz K, Bayraktar E, Zhang
X, et al: Exosomal miRNA confers chemo resistance via targeting
Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine.
38:100–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Weiner-Gorzel K, Dempsey E, Milewska M,
McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A,
Sharpe D, et al: Overexpression of the microRNA miR-433 promotes
resistance to paclitaxel through the induction of cellular
senescence in ovarian cancer cells. Cancer Med. 4:745–758. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai
D, Xu W, Jiang T, Feng L, Shin VY, et al: Impaired autophagic
degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in
gastric cancer. Cell Death Dis. 10:3832019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lee JW, Guan W, Han S, Hong DK, Kim LS and
Kim H: MicroRNA-708-3p mediates metastasis and chemoresistance
through inhibition of epithelial-to-mesenchymal transition in
breast cancer. Cancer Sci. 109:1404–1413. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Shen Z, Zhou L, Zhang C and Xu J:
Reduction of circular RNA Foxo3 promotes prostate cancer
progression and chemoresistance to docetaxel. Cancer Lett.
468:88–101. 2020. View Article : Google Scholar : PubMed/NCBI
|