Introduction
Renal cell carcinoma (RCC) is regarded as one of the
most common malignancies of the genitourinary system, with an
increasing incidence rate in the United States (1). RCC is generally rare in children and
young adults as it primarily manifested in the elderly (2,3). Some of
the contributing factors of RCC include hypertension, smoking,
familial syndromes, with an unhealthy lifestyle and dietary habits
(4). RCC has multiple subtypes, with
clear cell RCC (ccRCC) as the most prevalent type with 85%
proportion among other subtypes (5).
Chest metastasis is a frequent finding in RCC, however lung
metastasis is the most common form of distant metastasis (6). Additionally, RCC is habitually
asymptomatic until the diagnosis of metastasis by medical
intervention. With expanding alternative treatment options,
surgical intervention persists as the gold standard for the
treatment of RCC (7,8). Therefore, we sought to explore new and
reliable protocols for RCC therapy.
Extracellular vesicles (EVs), fundamentally
recognized as the nanoscale tools composed of exosomes and
microvesicles for intrinsic intercellular communication, are active
contributors in diverse physiological processes, including
regulation of tumor development (9).
A recent study ascertained the vital functionality of EVs in RCC
initiation and progression (10).
Tumor-derived EVs are the focus of increased research due to their
ability to facilitate tumorigenesis and thus are being adopted as
promising markers for tumor treatment, including RCC (11,12).
Furthermore, the role of EVs in mediating
cell-to-cell communication and facilitating tumorigenesis is
achieved via transfer of multiple RNAs, including long noncoding
RNAs (lncRNAs) (13). Accumulating
evidence has validated lncRNAs as crucial in regulating diverse
cancers, including RCC (14,15). Metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) has been validated as a potent
marker for multiple human cancers accounting for its abnormal
upregulation as an indicator of aggravated pathological alterations
in cancerous organs (16). MALAT1
shows an aberrantly high expression and promotes cell malignant
biological behaviors in RCC (17).
Furthermore, as previously evidenced, MALAT1 can be diffused by the
epithelial ovarian cancer cell-secreted exosomes to the target
recipient cells to promote cancer angiogenesis (18). However, the mutual action of
tumor-derived EVs and MALAT1 in RCC has not yet been explored.
Therefore, we hypothesized that tumor-derived EVs
may play a role in RCC with the involvement of lncRNA MALAT1.
Consequently, we performed an array of histological and molecular
experiments to identify the interaction between RCC cell-derived
EVs and MALAT1, and to explore the relative regulatory mechanism,
in an attempt to identify novel therapies against RCC.
Materials and methods
Extraction and identification of
EVs
786-O cells [American Type Culture Collection
(ATCC)] underwent a 72-h culture using Dulbecco's modified Eagle's
medium (DMEM) containing 10% EV-free fetal bovine serum (FBS)
(Thermo Fisher Scientific, Inc.) and 1% penicillin streptomycin.
The supernatant was collected and subjected to 5-min (at 1,100 × g)
and then 30-min (at 40,000 × g) centrifugation regimens to
eliminate the cells and cellular debris, and large vesicles,
respectively. After filtration of the supernatant using a 0.2-µm
filter and centrifugation (at 1,000,000 × g) for 2 h, the EVs
underwent a phosphate-buffered saline (PBS) rinse and a 2-h regimen
of centrifugation (at 100,000 × g), followed by resuspension in
PBS. An EV isolation reagent (Thermo Fisher Scientific, Inc.) was
used for EV isolation. Nanoparticle tracking analysis (NTA) was
applied for EV analysis using the NanoSight NS300 (Malvern
Panalytical Co., Ltd.). The obtained EVs were allocated into the EV
group and the blank group [supplemented with 1 µM GW4869
(MedChemExpress Co., Ltd.)] (19).
The small-interfering (si)-MALAT1-1, si-MALAT1-2 and si-negative
control (NC) (Shanghai GenePharma Co., Ltd.) were transfected into
the 786-O cells, respectively, using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) to attain a final
transfection concentration of 50 nM. The EVs (EVs/si-MALAT1-1,
EVs/si-MALAT1-2 and EVs/si-NC) were extracted respectively by
ultracentrifugation and identified with the preceding protocol. The
morphology of the EVs was observed under transmission electron
microscopy (TEM). The protein determination by the bicinchoninic
acid (BCA) method was performed on all the aforementioned EVs and
the result was 2.2 mg/ml. Subsequent experiments were conducted
using the protein concentration as the concentration of EVs.
Bioinformatics analysis. The lncDisease database
(http://www.cuilab.cn/lncrnadisease)
was applied to search for lncRNAs associated with RCC. The
expression of the candidate lncRNAs in the tumor-EVs was searched
through the exoRBase database (http://www.exorbase.org/). The lncMAP database
(http://bio-bigdata.hrbmu.edu.cn/LncMAP/) was used to
predict the downstream regulatory transcription factors in kidney
renal clear cell carcinoma (KIRC) and kidney renal papillary cell
carcinoma (KIRP). Next, the genes related to RCC were searched
through the DisGeNET database (https://www.disgenet.org/). Gene interaction network
diagram was constructed using the STRING database (https://string-db.org/) and Cytoscape V3.7.1 software
(www.cytoscape.org). RCC microarray
GSE16441 (20) including 17 normal
samples and 17 RCC samples was obtained from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/).
With the normal samples as the control, the R language ‘limma’
package (21) was applied for
differential analysis. TheP-value was corrected using the false
discovery rate (FDR) method, and |logFC|>2 and FDR <0.05 were
used as the screening criteria for the differentially expressed
genes. JASPAR database (http://jaspar.genereg.net/) was used to predict the
E26 transformation specific-1 (ETS1) downstream regulatory genes,
and to obtain the existing binding site between transcription
factor CP2-like 1 (TFCP2L1) promoter and ETS1.
Cell culture and grouping
RCC cell lines A498 and ACHN supplied by ATCC were
cultured in minimal essential medium (MEM; Gibco Co.; Thermo Fisher
Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) in an incubator at 37°C under saturated conditions with 5%
CO2. The A498 and ACHN cells were co-cultured with 10
µg/ml of the above-mentioned 786-O-EVs (22) for 24 h, grouped and named similarly as
the aforementioned EVs.
PKH26 fluorescence labeling
The 786-O-EVs were labeled by a PKH26 red
fluorescence labeling kit (Sigma-Aldrich; Merck KGaA) in compliance
with the provided instructions. Briefly, the EVs underwent a 4-min
incubation regimen with the PKH26 dye, which was terminated by the
addition of EV-free FBS. The EVs were rinsed three times and the
excess PKH26 dye was removed with an Amicon Ultra-4 (100-kDa, Merck
Millipore Corp.), followed by a 4-h incubation regimen with the RCC
cells. The internalization of EVs by RCC cells was observed under
confocal microscopy.
Quantitative real-time polymerase
chain reaction (qPCR)
The total RNA content from the cells in each group
was extracted using the TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and the RNA concentration was determined using a
NanoDrop 2000 (Thermo Fisher Scientific, Inc.). The PrimeScript RT
reagent kit and gDNA Eraser kit (Takara Bio Inc.) were used to
transcribe the total RNA (1 mg) content into cDNA. Genomic DNA
termination was induced by the addition of 5X DNA Eraser buffer and
gDNA Eraser at 42°C for 2 min. Next, the cDNA was synthesized with
the reaction condition as 37°C for 15 min and 85°C for 5 sec. Next,
qPCR was conducted using a SYBR Premix Ex Taq (Tli RNaseH Plus) kit
(Takara) on a ABI 7500 real-time PCR instrument (Thermo Fisher
Scientific, Inc.). Data of each gene were analyzed based on the
2−ΔΔCq method (23). The
primer sequences are listed in Table
I.
 | Table I.Primer sequences for qPCR. |
Table I.
Primer sequences for qPCR.
| Gene | Primer |
|---|
| MALAT1 | F:
5′-GGGGCAGTAGTGTAGAGA-3′ |
|
| R:
5′-CAGTGCGTGTCGTGGAGT-3′ |
| U6 | F:
5′-CTCGCTTCGGCAGCACA-3′ |
|
| R:
5′-GTGTCGTGGAGTCGGCAA-3′ |
| TFCP2L1 | F:
5′-AGGTGCTGACCTCCTGAAGA-3′ |
|
| R:
5′-GTTTTGCTCCAGCTCCTGAC-3′ |
| ETS1 | F:
5′-CATGCTTTTCGTTTGACACCC-3′ |
|
| R:
5′-CTTTGCTTCCACCCGCCCCCC-3′ |
| GAPDH | F:
5′-ACCAGGTATCTGCTGGTTG-3′ |
|
| R:
5′-TAACCATGATGTCAGCGTGGT-3′ |
Western blot analysis (WB)
The total protein content was extracted from cells
in each group, and the protein concentration was measured using a
BCA kit (Thermo Fisher Scientific, Inc.). Total proteins (30 mg)
underwent 35-min (at 80 V) and 45-min (at 120 V) polyacrylamide gel
electrophoresis successively. Next, the proteins were loaded onto
polyvinylidene fluoride membranes (Amersham Pharmacia), blocked for
1 h using 5% skim milk, and probed with the corresponding primary
antibodies [cluster of differentiation (CD)63 (dilution 1:1,000,
ab134045), CD81 (dilution 1:1,000, ab109201) and TFCP2L1 (dilution
1:1,000, ab140197) (all from Abcam Inc.)] at 4°C. Following 3
rinses (10 min each) with PBS and 0.1% Tween-20 (PBST), the
membranes underwent a 1-h incubation regimen with the horseradish
peroxidase-labeled goat anti-rabbit immunoglobulin G (IgG) H&L
(dilution 1:2,000, ab205718; Abcam Inc.), and rinsed 3 times with
PBST (10 min each). An optical illuminator (General Electric) was
utilized for membrane visualization. Image-Pro Plus 6.0 (Media
Cybernetics) was applied for protein band gray value analysis.
Cell Counting Kit-8 (CCK-8) assay
RCC cell proliferation was detected using the CCK-8
kit (Dojindo Molecular Technologies) in strict accordance with the
provided instructions. RCC cells were seeded into 96-well plates
and each well was supplemented with EVs based on cell grouping and
incubation for 24 h. Then approximately 10 µl CCK-8 reagent was
added to each well for a 2-h regimen of incubation at 37°C. The
optical density (OD) at the wavelength of 490 nm was determined
using a microplate reader (Thermo Fisher Scientific, Inc.).
Relative cell viability of each group was determined.
Transwell assays
RCC cell invasion was assessed according to the
passage of the number of transfected cells through Transwell
chambers (8-micron chamber; Corning Inc., Life Sciences). The
Transwell chambers were pre-coated with 100 µl Matrigel (BD
Biosciences) at 37°C for 5 h until gelling was visible. Next,
1×105 A498 and ACHN cells that underwent a 24-h
starvation in 500 µl serum-free MEM medium were seeded in the
apical chamber, respectively (3 technical replicates for each
group). The basolateral chamber was filled with 700 µl MEM with 10%
FBS. Following a 48-h incubation at 37°C, Matrigel and non-invaded
cells were removed from the upper surface of the filters. Cells
that adhered to the lower surface of filters were fixed using 4%
paraformaldehyde and stained with crystal violet. Finally, the
cells were counted and photographed using a Nikon digital camera
(magnification, ×100). Three independent experiments were
repeatedly set. The method for the migration experiment was the
same as that of the invasion experiment, without utilization of
Matrigel.
Immunofluorescence
The cells were seeded on 24-well plates and received
different treatments. Next, the cells were fixed for 15 min in 4%
paraformaldehyde, permeated for 10 min with PBS containing 0.1%
Triton X-100, and sealed for 30 min using the 3% plasma blocking
solution (all performed at room temperature). Subsequently, the
cells were subjected to overnight incubation at 4°C with the
diluted primary antibodies E-cadherin (dilution 1/200, ab1416,
Abcam) and N-cadherin (dilution 1/200, ab76057, Abcam), followed by
a 1-h incubation regimen with the secondary antibody goat
anti-mouse IgG H&L (Alexa Fluor® 488) (dilution
1/200, ab150117, Abcam) in conditions devoid of light at room
temperature. Finally, 4′,6-diamidino-2-phenylindole (DAPI) was
added for nuclear staining. After resting at room temperature for 1
min, the cells were observed under a fluorescence microscope
(Olympus Optical Co., Ltd.).
Fluorescence in situ hybridization
(FISH)
MALAT1 subcellular localization was observed using
FISH assay in strict accordance with the provided instructions of
the Ribo™ lncRNA FISH Probe Mix (Red) (C10920, RiboBio Co., Ltd.).
The cells (6×104 cells/well) were seeded into 24-well
plates. Upon achieving 60–70% confluence, the cells were fixed
using 4% paraformaldehyde, rinsed and permeabilized. The plates
were sealed using the pre-hybridization solution. After elimination
of the pre-hybridization solution, the cells were subjected to
overnight hybridization at 37°C using the probe hybridization
solution supplemented with anti-MALAT1 nucleotide (Wuhan Genecreate
Bioengineering Co., Ltd.) in conditions devoid of light. Next, the
cells were eluted, stained with DAPI, rinsed, and fixed using nail
polish for observation under fluorescence microscopy (Olympus).
Five different fields were selected, and the cells in these fields
were observed and documented.
Dual-luciferase reporter gene
assay
To explore the effect of MALAT1 on TFCP2L1 promoter
activity, overexpressed (oe)-NC, oe-MALAT1, short hairpin (sh)-NC,
and sh-MALAT1 were co-transfected with the TFCP2L1-2 kb luciferase
reporter plasmid respectively into 293T cells. After 48 h of
transfection, the cells were collected and lysed. The
dual-luciferase reporter gene assay was performed using a
luciferase assay kit (K801-200, BioVision Inc.) and a
dual-luciferase reporter gene analysis system (Promega Corp.).
Renilla luciferase was adopted for internal reference. The
activation degree of the target reporter gene was measured using
the ratio of the relative unit of firefly luciferase to that of
Renilla luciferase.
RNA immunoprecipitation (RIP)
A RIP kit (Millipore Corp.) was utilized to assess
the binding of MALAT1 to the ETS1 protein. The cells were lysed in
an ice bath, and then subjected to centrifugation. Next, the
supernatant was collected. The cell extract was incubated with the
corresponding antibody for co-precipitation. Following a rinse and
re-suspension in RIP wash buffer, the magnetic beads were
supplemented with the appropriate antibody for binding. After a
rinse, the magnetic bead-antibody complex was resuspended in RIP
wash buffer, incubated overnight at 4°C with the cell extract, and
harvested. The RNA content was extracted from the sample for
subsequent PCR detection after detachment with proteinase K. The
antibody used above was ETS1 (dilution 1:200, ab225868, Abcam),
with IgG (dilution 1:100, ab172730, Abcam) as the control.
RNA pull-down
Biotinylated RNA sequence probe in the TFCP2L1
promoter region and its negative control (NC) probe were dissolved
in washing/binding buffer at room temperature and incubated with
the streptavidin-coupled magnetic beads for 2 h. Then, the cell
lysate was added and incubated for 2 h. The protein complex
conjugated to magnetic beads was washed. ETS1 content in the
complex was determined by western blot analysis.
Ethics statement
The experimental procedures were approved by the
Ethics Committee of The Second Affiliated Hospital of Harbin
Medical University. Significant efforts were made in order to
minimize both the number of animals used as well as their
respective suffering.
Subcutaneous tumorigenesis in mice and
lung metastasis model establishment
A total of 48 BALB/C nude mice (male, aged 4 weeks,
weighing 15–18 g; 6 each per group) used for animal experiments and
xenograft collection were supplied by Beijing Wantai BioPharm Co.,
Ltd. [Beijing, China; SYXK (Jing) 2017-0041]. Each nude mouse was
housed in a separate cage and provided with free access to food and
water. All mice were fed with clean grade maintenance feed and kept
at a temperature of 26–28°C and humidity at 40–60% with 10/14 h
light/dark cycle. The nude mice were subcutaneously injected with
the 786-O cell suspension (5×106 cells), and treated
with the blank, EVs, EVs/si-NC or EVs/si-MALAT1-1 (20 nM) using
Entranster™-in vivo Transfection Reagent (Engreen Biosystem
Co., Ltd.) after acquiring a tumor volume greater than 100
mm3 (n=6) in each nude mouse. Xenografts were determined
weekly. The nude mice were euthanized via intraperitoneal injection
of 800 mg/kg pentobarbital sodium after 5 weeks and the tumor was
dissected and weighed. The Ki67-positive rate was observed and
counted.
The grouping of the lung metastasis model was the
same as the subcutaneous tumorigenesis. Then, 6 nude mice from each
group were injected with the ACHN cells (2×106 cells)
via tail vein. After 42 days, the mice were euthanized by an
intraperitoneal injection with 800 mg/kg pentobarbital sodium. The
lung tissues were extracted and stained using hematoxylin and eosin
(H&E) (JRDUN Biotechnology Co., Ltd.).
Histological analysis
Immunohistochemical analysis
The removed tumor sample was fixed using 4%
paraformaldehyde and embedded in paraffin, which was then
sectioned, dewaxed, and hydrated independently. The Ki67 antibody
(dilution 1:500, ab15580, Abcam) was used for immunohistochemical
staining. The Ki67-positive rate was observed and counted under a
microscope (BX53M, Olympus; magnification, ×200).
48 H&E staining
The extracted lung tissue was fixed using 4%
paraformaldehyde and embedded in paraffin. Then, the tissue was
sectioned, dewaxed and hydrated independently. The sections were
stained using the hematoxylin solution for 10 min and decolorized
in 70 and 90% ethanol after a rinse with distilled water.
Subsequently, the sections were stained with the eosin solution for
2–3 min, dehydrated with pure ethanol, cleared with xylene, sealed
with neutral gum and then observed under a microscope (BX53M,
Olympus; magnification, ×200).
Statistical analysis
SPSS 21.0 (IBM Corp.) software was used for data
analysis. The experimental data were in normal distribution as
verified by Kolmogorov-Smirnov test, and expressed as mean ±
standard deviation. Comparison between two groups was conducted
with the independent sample t-test. The comparison among groups was
analyzed using one-way analysis of variance (ANOVA), followed by
the Tukey's multiple comparisons test. The P-value was obtained
from a two-sided test and a value of P<0.05 was indicative of
statistical significance.
Results
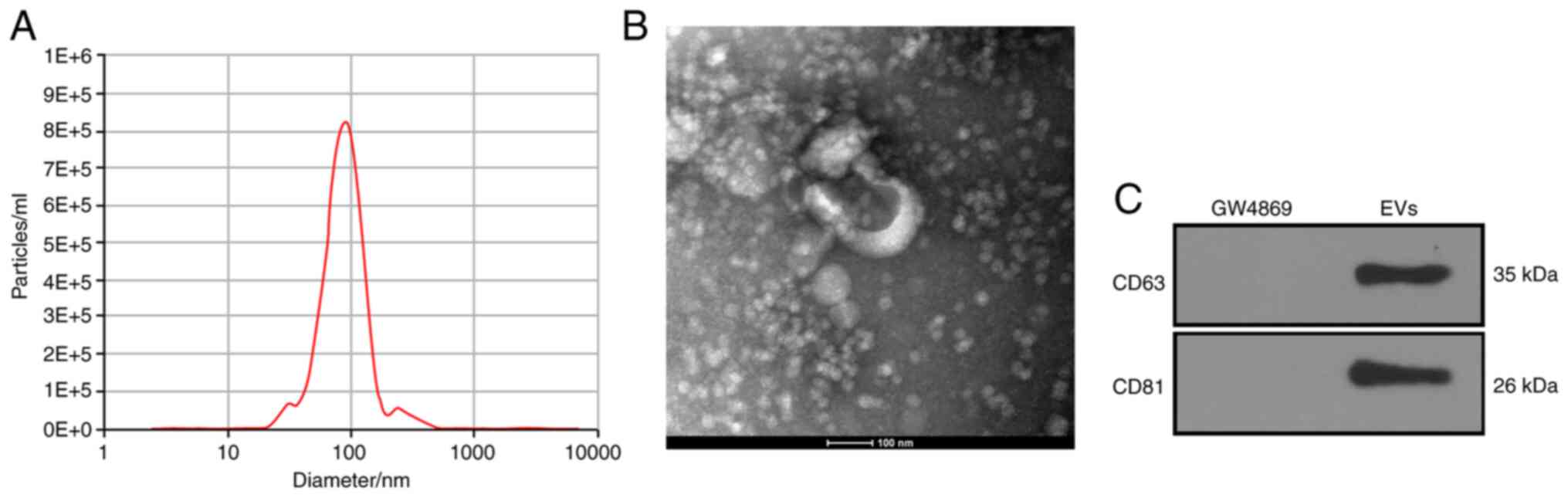
786-O-EVs were successfully
extracted
After a 48-h culture regimen of 786-O cells in
EV-free FBS, the EVs were separated by ultracentrifugation. NTA
confirmed the size of the EVs as ranging between 30–150 nm
(Fig. 1A). Consistently, TEM revealed
that the EVs were oval with a diameter of about 100 nm (Fig. 1B). Next, according to the WB results,
the EV symbolic markers (CD63 and CD81) were positively expressed
in the 786-O-EVs (Fig. 1C). From the
aforementioned findings, the separated granules were identified as
EVs.
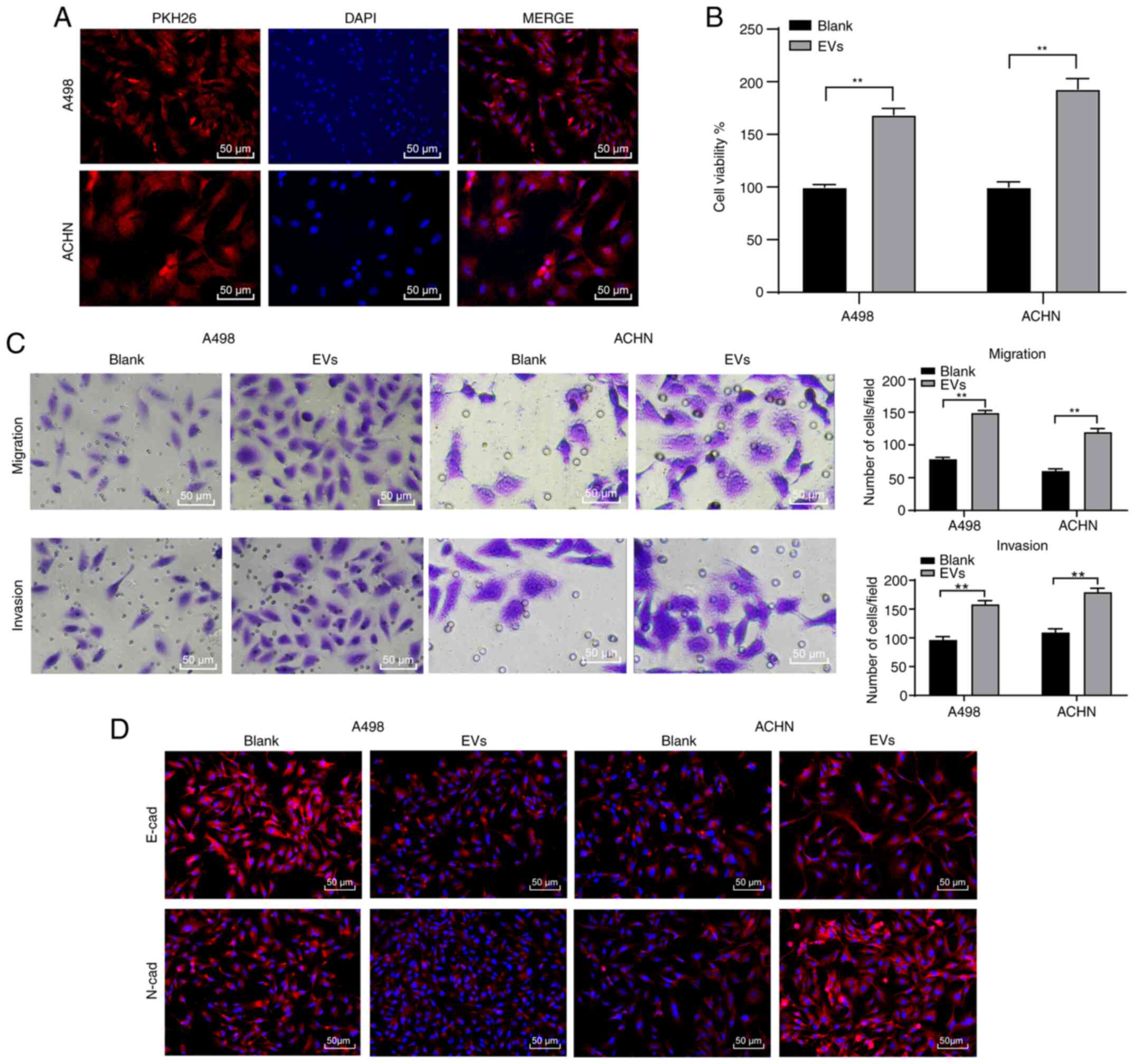
786-O-EVs promote RCC cell invasion
and migration
An existing study reported that the tumor-derived
EVs elicit negative effects on habitual cellular functions
(24). Therefore, we speculated that
RCC cell-derived EVs may have similar effects on the biological
behaviors of RCC cells. To ascertain this speculation, we labeled
the EVs with PKH26 red fluorescence dye after which these EVs were
co-cultured (10 µg/1×105 cells) with the A498 and ACHN
cells for 48 h. Under fluorescence microscopy the A498 and ACHN
cells elicited red fluorescence in their cytoplasm (Fig. 2A), indicating the internalization of
786-O-EVs by the RCC cells. Next, the A498 and ACHN cell viability
was investigated to explore the effect of 786-O-EVs on A498 and
ACHN cell functions. CCK-8 assay verified that the A498 and ACHN
cell viability increased significantly after treatment with the
786-O-EVs (Fig. 2B) (both P<0.05).
Meanwhile, Transwell assays revealed that 786-O-EV treatment
significantly increased A498 and ACHN cell migration and invasion
(Fig. 2C) (all P<0.05). Next, the
A498 and ACHN cell epithelial mesenchymal transition (EMT) was
detected by immunofluorescence, which was evidently promoted after
786-O-EV treatment (Fig. 2D).
Briefly, 786-O-EVs promoted RCC cell invasion and metastasis.
786-O-EVs are the communication
mechanism of MALAT1 in RCC
From the preceding results, it is evident that the
786-O-EVs promoted RCC cell invasion and metastasis. To explore the
underlying mechanism, we referred to previous studies. For
instance, EVs were found to release and carry lncRNAs into the
target cells to manipulate cellular functions (25). MALAT1 was also found to promote RCC
cell malignant biological behaviors (26). Therefore, we searched RCC-related
lncRNAs through the lncDisease database (http://www.cuilab.cn/lncrnadisease), and found the
involvement of lncRNAs such as MALAT1 in the regulation of RCC
(Table SI). Furthermore, the
expression of these candidate lncRNAs in the EVs was searched
through the exoRBase database (http://www.exorbase.org/) (Fig. 3A), which revealed that MALAT1 was
expressed in various tumor-EVs, suggesting that MALAT1 may also
exist in RCC cell-derived EVs. Hence, we speculated that 786-O-EVs
carried MALAT1 and perhaps were internalized by the A498 and ACHN
cells, thus inducing RCC cell malignant biological behaviors.
MALAT1 expression pattern in the A498, ACHN and 786-O cells was
determined by qPCR with no distinctive difference indicated by the
results. Subsequently, the MALAT1 expression pattern in the A498
and ACHN cells before and after EV treatment was initially
detected, which revealed a significant increase after EV treatment
(Fig. 3B) (both P<0.01). RNase
treatment did not affect the MALAT1 expression pattern in the
culture medium of ACHN and A498 cells; while RNase and Triton X-100
treatment showed a significant decreased MALAT1 expression pattern
in the culture medium of ACHN and A498 cells (Fig. 3C) (all P<0.01). These results
indicated that MALAT1 was membrane-encapsulated over direct
release.
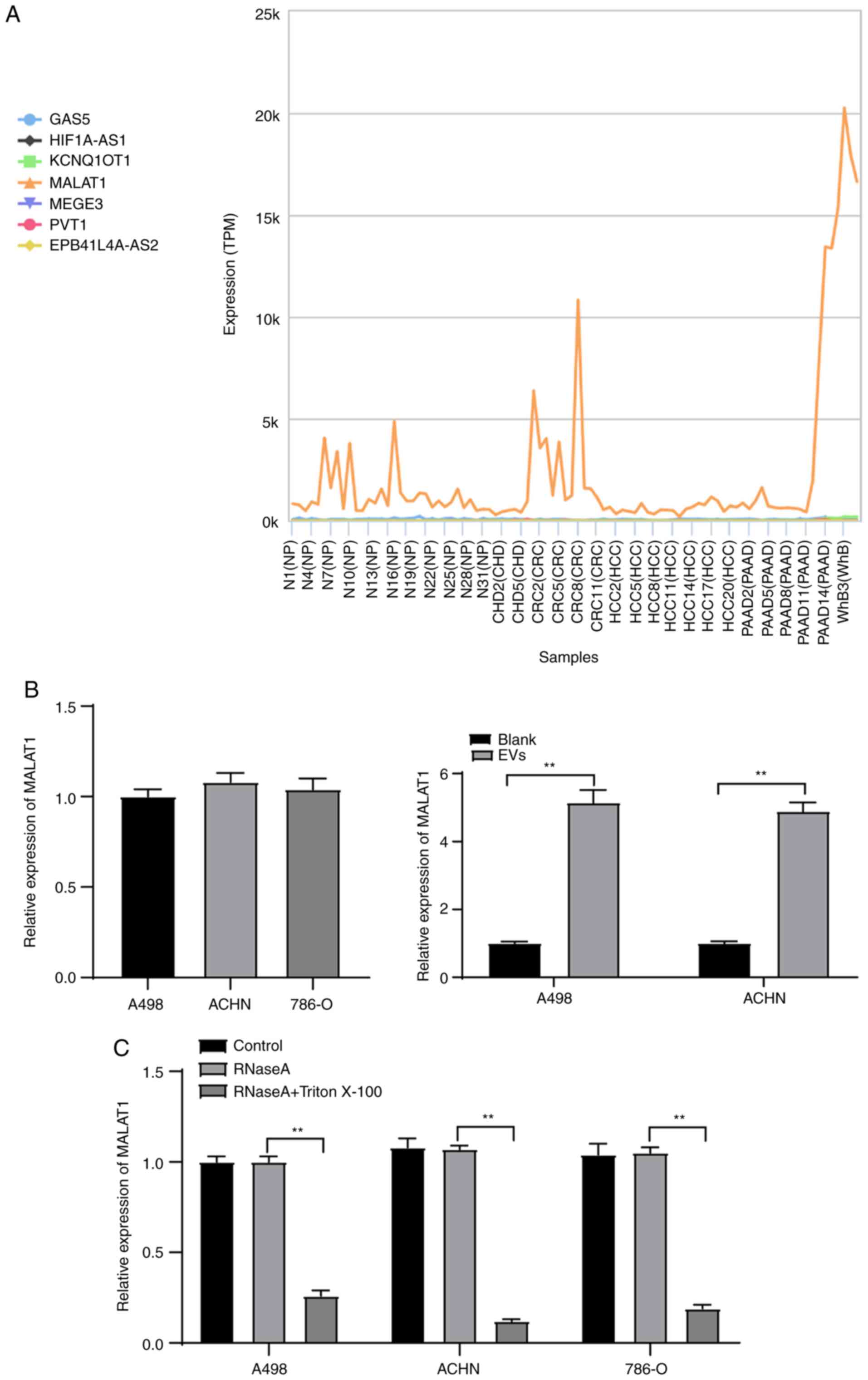 | Figure 3.786-O-EVs are the communication
mechanism of MALAT1 in RCC cells. (A) The expression of candidate
lncRNAs in the EVs evaluated through the exoRBase database
(http://www.exorbase.org/); the
abscissa represents the EVs from different tumors, and the ordinate
represents the expression of different lncRNAs; the left side shows
the diagram. NP, normal person; CHD, coronary heart disease; CRC,
colorectal cancer; HCC, hepatocellular carcinoma; PAAD, pancreatic
adenocarcinoma; WhB, whole blood. (B) Expression of MALAT1 in ACHN
and A498 cells treated with 786-O-EVs was detected using qPCR. (C)
Expression of MALAT1 in 786-O, ACHN and A498 cells after treatment
of RNase and Triton X-100 was detected using qPCR. Each experiment
was repeated three times independently. Data in panels B and C were
analyzed using independent t-test or one-way ANOVA, followed by
Tukey's multiple comparisons test. **P<0.01. EVs, extracellular
vesicles; MALAT1, metastasis-associated lung adenocarcinoma
transcript 1. |
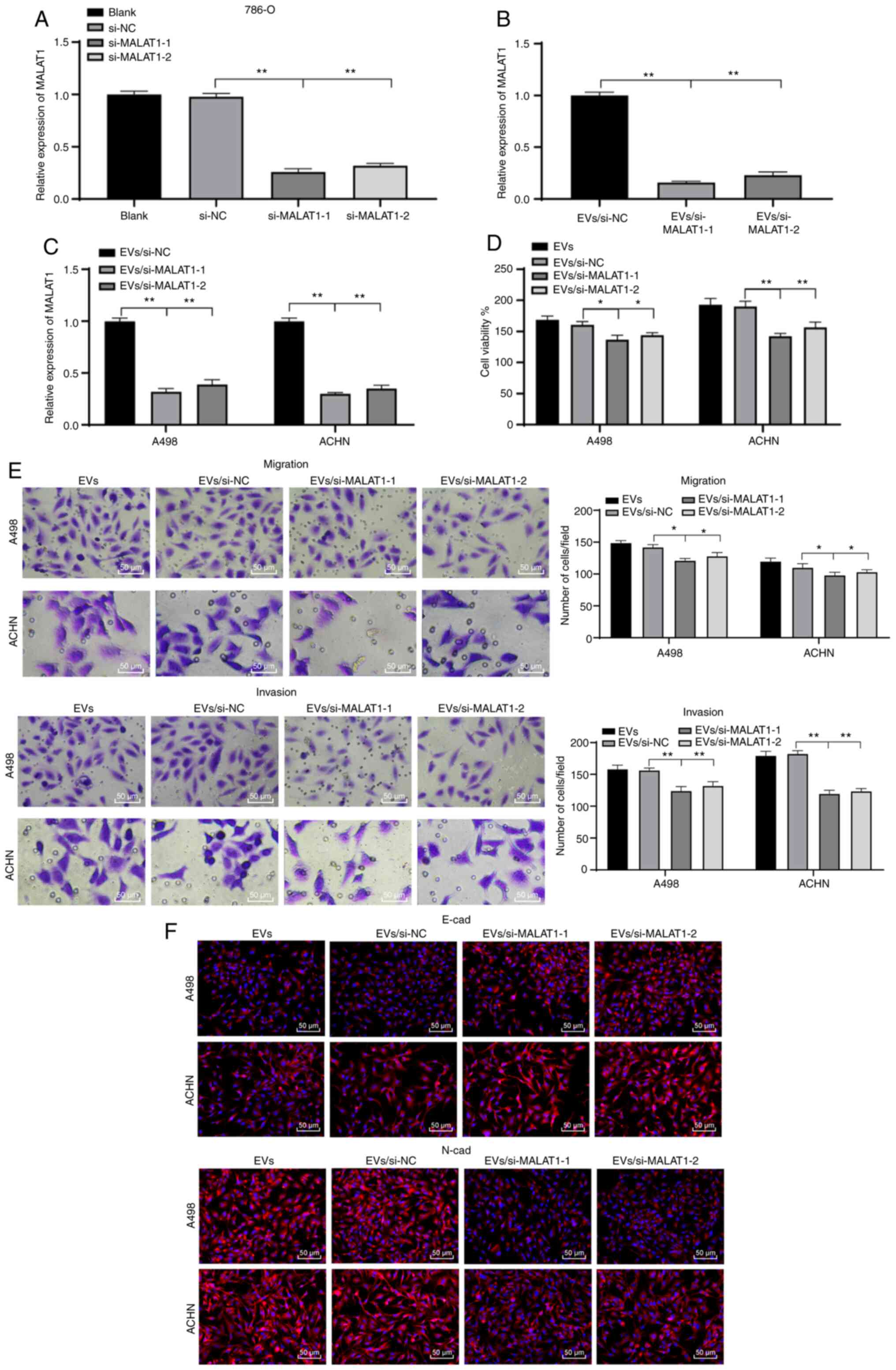
786-O-EVs promote RCC cell invasion
and migration via transferring MALAT1
From the preceding results, we established that
786-O-EVs were the communication medium of MALAT1 between RCC
cells. To further validate the mechanism, we silenced the MALAT1
expression pattern in 786-O cells, and then extracted the EVs
(EVs/si-MALAT1-1/2). We found that the MALAT1 expression pattern in
the 786-O-EVs was significantly decreased after inhibition of
MALAT1 in the 786-O cells (Fig. 4A and
B). Similar reduction in the MALAT1 expression pattern was
evident in RCC cells treated with 786-O-EVs after treatment with
EVs/si-MALAT1-1/2 (Fig. 4C) (all
P<0.01). In addition, the biological behavior changes of cells
in each group were evaluated. EVs/si-MALAT1-1/2 weakened the
explicit effect of simple 786-O-EVs on RCC cell viability, invasion
and migration and EMT capacity (Fig.
4D-F) (all P<0.01). Collectively, 786-O-EVs transferred
MALAT1 to induce RCC cell vitality, invasion and migration and
EMT.
786-O-EVs carry MALAT1 to regulate
transcription factor ETS1 and reduce TFCP2L1 activity
For a comprehensive understanding of the regulatory
mechanism of MALAT1, we predicted the downstream regulatory
transcription factors of MALAT1 in KIRC and KIRP through the lncMAP
database (Table SII). Intersection
of the predicted transcription factors in the two diseases was
evidently intersected (Fig. 5A), from
which four candidate transcription factors were chosen.
Subsequently, the genes relevant to RCC were retrieved through the
DisGeNET database (https://www.disgenet.org/) (Table SIII), where the genes with scores
higher than 0.3 were selected for subsequent analyses. The
interaction between the retrieved known genes and the predicted
four transcription factors was analyzed, after which the gene
interaction network diagram was constructed (Fig. 5B). It was evident that ETS1 had the
most interactions with known genes, suggesting the superior
vitality of ETS1 relative to others. In a previous work, ETS1
served as an oncogene in RCC with intimate relations with a low
survival rate in a large number of RCC samples (27). Meanwhile, a RCC microarray GSE16441
was obtained through the GEO database (https://www.ncbi.nlm.nih.gov/geo/), where 406
differentially downregulated genes were identified by differential
analysis of the GSE16441 chip. Concurrently, the ETS1 downstream
regulatory factors were predicted through the JASPAR database
(http://jaspar.genereg.net/), where the
intersection between the downregulated genes in the chip and JASPAR
prediction results was engaged, and five genes were identified to
be present in the respective intersection (Fig. 5C). Their differential expression
levels in the GSE16441 chip were further studied, where TFCP2L1 was
regarded as the most notably downregulated gene (Table II). As previously highlighted,
TFCP2L1 is a critical developmental transcription factor in normal
kidney development (28). Meanwhile,
JASPAR database predicted a number of binding sites between ETS1
and the TFCP2L1 promoter (Table
SIV). Therefore, we speculated that 786-O-EVs transferred
MALAT1, to subsequently regulate the transcription factor ETS1 to
modulate TFCP2L1 expression, thereby affecting RCC development.
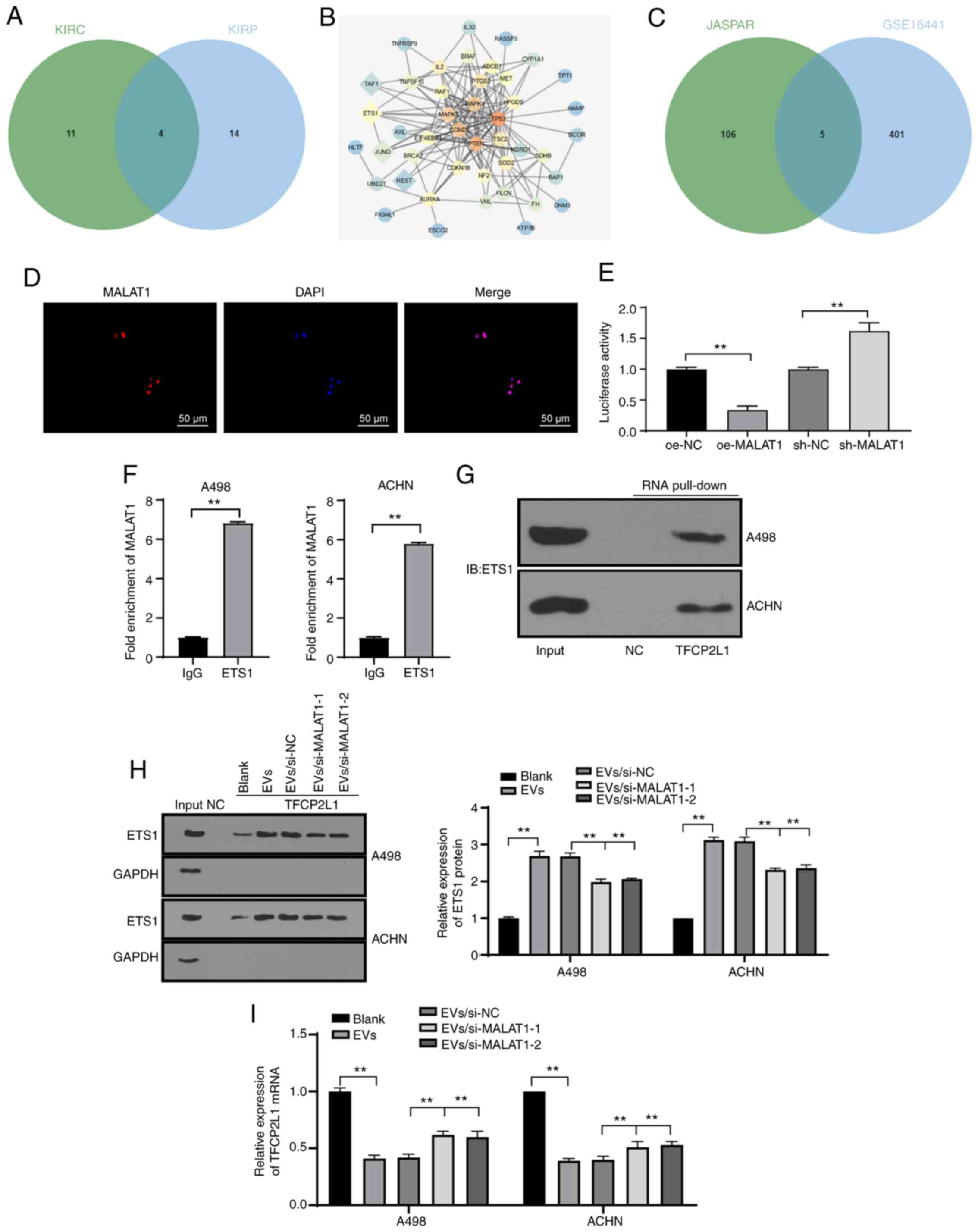 | Figure 5.786-O-EVs carry MALAT1 to regulate
transcription factor ETS1 and reduce TFCP2L1 activity. (A)
Prediction of MALAT1 regulatory transcription factors in KIRC and
KIRP; the two circles in the figure represent the predicted
transcription factors in KIRC and KIRP, and the middle part
represents the intersections of the two sets of data. (B) Network
diagram of interaction analysis between candidate transcription
factors and genes known related to RCC; the diamonds represent the
screened four candidate transcription factors, and the circles
represent RCC-related genes obtained from the DisGeNET database
(https://www.disgenet.org/); the darker
color of the graph represents higher core degree of the gene in the
network diagram and more interaction genes. (C) Prediction of ETS1
candidate target genes; the circle on the right side represents the
downregulated genes in the GSE16441 chip, and the left side shows
the prediction results of JASPAR database (http://jaspar.genereg.net/) on ETS1 target gene, and
the middle part represents the intersection of the two groups of
data. (D) FISH was used to detect subcellular localization of
MALAT1. (E) Dual-luciferase reporter gene assay was used to verify
the effect of MALAT1 on TFCP2L1 promoter activity. (F) RIP was used
to detect the binding of MALAT1 to transcription factor ETS1. (G
and H) RNA pull-down was used to detect the binding relationship
between ETS1 and TFCP2L1 promoter. (I) qPCR was used to detect the
mRNA expression of TFCP2L1. Each experiment was repeated three
times independently. Data in panel E were analyzed using one-way
ANOVA, and data in panel F were analyzed by independent t-test and
data in panels H and I were analyzed using two-way ANOVA, followed
by Tukey's multiple comparisons test. **P<0.01. EVs,
extracellular vesicles; MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; RCC, renal cell carcinoma; TFCP2L1,
transcription factor CP2 like 1; ETS1, ETS proto-oncogene 1,
transcription factor. |
 | Table II.Differential expression of candidate
genes in the chip GSE16441. |
Table II.
Differential expression of candidate
genes in the chip GSE16441.
| Symbol | logFC | P-value | adj.P-value |
|---|
| TFCP2L1 | −5.698466882 | 6.02E-17 | 3.09E-14 |
| ESRRB | −2.343714182 | 4.84E-12 | 3.05E-10 |
| PLAG1 | −2.789086764 | 7.98E-12 | 4.73E-10 |
| KLF5 | −2.069653766 | 8.88E-10 | 2.59E-08 |
| TP63 | −2.593375572 | 1.91E-06 | 1.60E-05 |
Subsequently, FISH revealed nuclear subcellular
localization of MALAT1 in the cell (Fig.
5D). Dual-luciferase reporter gene assay verified that the
MALAT1 overexpression remarkably reduced the TFCP2L1 promoter
activity, while MALAT1 knockdown dramatically increased the TFCP2L1
promoter activity, indicating the negative regulatory effect of
MALAT1 on the TFCP2L1 gene expression pattern (Fig. 5E) (both P<0.01). Next, RIP was
applied to further elucidate the underlying mechanism of MALAT1 to
negatively regulate the TFCP2L1 gene expression pattern, and we
observed that ETS1 would bind to more MALAT1 than IgG (Fig. 5F) (P<0.01), indicating that the
ETS1 protein could specifically bind to MALAT1. RNA pull-down
showed that ETS1 could bind to the TFCP2L1 promoter region
(Fig. 5G). The binding of ETS1 and
the TFCP2L1 promoter in the A498 and ACHN cells treated with EVs
was enhanced whereas the binding of ETS1 and TFCP2L1 promoter was
weakened after silencing of MALAT1 in EVs (Fig. 5H). As shown by qPCR results, 786-O-EV
treatment significantly decreased the TFCP2L1 mRNA expression
pattern. Following knockdown of MALAT1 in the 786-O cells, the
downregulation effect of EVs on TFCP2L1 was alleviated (Fig. 5I). The above results indicated that
lncRNA MALAT1 would bind to the transcription factor ETS1 and
identify the fundamental complex in the sequence of TFCP2L1
promoter to inhibit transcription. Namely, MALAT1 could
specifically bind to ETS1 and inhibit TFCP2L1 expression.
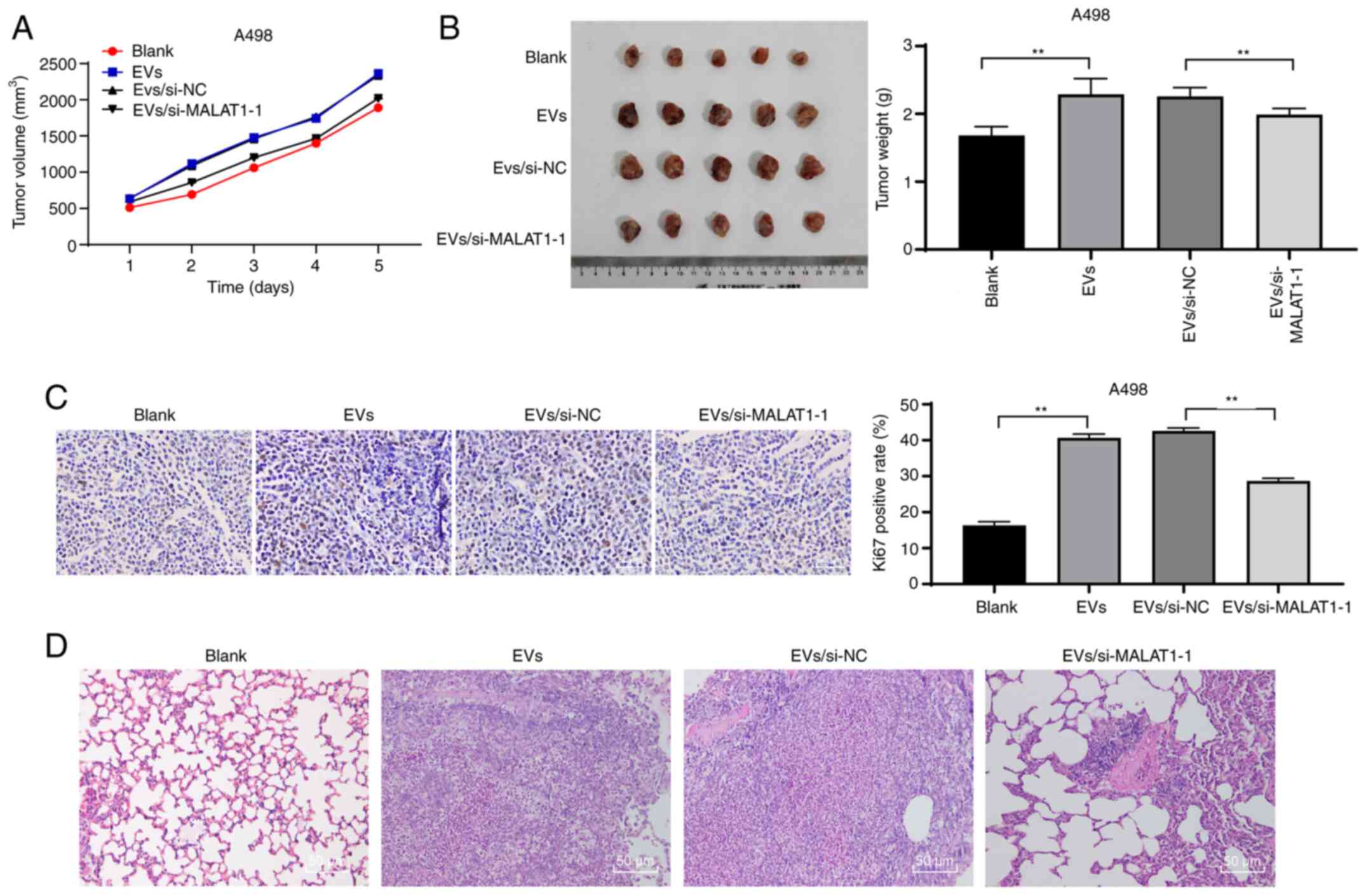
786-O-EVs promote RCC cell growth and
metastasis in vivo through MALAT1
To confirm that 786-O-EVs promote RCC cell invasion
and migration through MALAT1 in vivo, we conducted tumor
xenograft formation assay, and measured the tumor volume weekly for
5 weeks. We observed that 786-O-EVs had notably promoted tumor
growth in vivo (Fig. 6A). Five
weeks after EV injection, the mice were euthanized, and the
xenografts were dissected and weighed. The results revealed that
786-O-EV treatment apparently increased the average tumor weight
(Fig. 6B) (both P<0.01). In
addition, immunohistochemical staining revealed that the 786-O-EVs
resulted in noticeably increased Ki67-positive rates, while the
tumor growth-promoting effect of 786-O-EVs was weakened after
inhibition of MALAT1 in the 786-O cells (Fig. 6C) (both P<0.01). Moreover, the lung
metastasis model was established via tail vein injection. According
to H&E staining results, nude mice exhibited increased lung
tissue lesions after 786-O-EV treatment, while the lung tissue
lesions were reduced after silencing of MALAT1 in the 786-O cells
(Fig. 6D). In conclusion, 786-O-EVs
released MALAT1 to promote RCC cell growth and metastasis in
vivo.
Discussion
Renal cell carcinoma (RCC) ranks as the second most
prevalent cause of fatality associated with urinary tract tumors,
owing to its intricate diagnosis illustrated by several
asymptomatic characteristics (8).
Research has solidified the vital contributions of tumor-derived
extracellular vesicles (EVs) in facilitating tumor progression and
metastasis via promoting pre-metastatic niche preparation, which is
induced by their accommodating capacity of multiple molecules,
including lncRNAs (29). In this
study, our findings confirmed that 786-O-EVs promoted RCC invasion
and metastasis via transport of metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) to facilitate the binding of
transcription factor ETS proto-oncogene 1, transcription factor
(ETS1) and transcription factor CP2 like 1 (TFCP2L1) promoter and
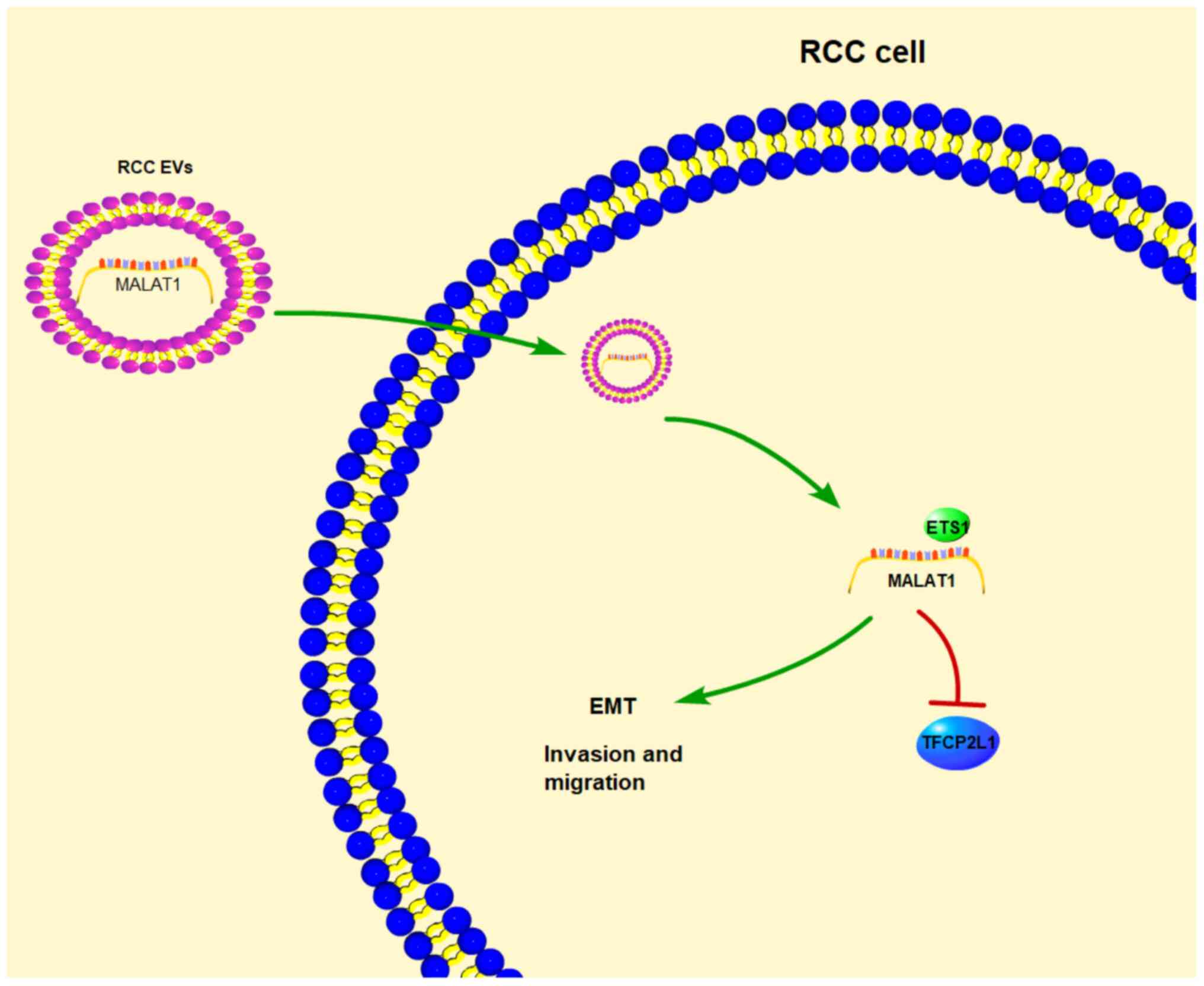
downregulate TFCP2L1 (Fig. 7).
Tumor-derived EVs are vital modulators for
intercellular communication and essentially participate in
promoting primary tumor growth and metastasis (30). As elicited by our results, after
exposure to 786-O-EVs, the A498 and ACHN cells showed evidently
increased cell viability, improved migration and invasion
abilities, and promotion of epithelial mesenchymal transition
(EMT). Consistently, an existing study flagged the significant
contributions of autologous cancer cell-secreted EVs in promotion
of the malignant behaviors of cancer cells (31). Tumor-derived EVs induce cancer cell
invasion and metastasis, and promote EMT (32,33).
Research has extensively established the ability of
lncRNAs to be packaged into EVs and then transferred to the
recipient cells, possessing considerable aptitude for cancer
treatment (34). LncRNA MALAT1 has
been indicated to serve as an oncogenic mediator in RCC with an
aberrant overexpression (35). Our
study documented a notably elevated MALAT1 expression in A498 and
ACHN cells after treatment of 786-O-EVs, which elicited a profound
reduction after silencing of MALAT1 in the 786-O cells, with
membrane-encapsulated release of MALAT1 over direct release.
Consistently, as evidenced by numerous studies, MALAT1 shows a
predominant expression in EVs secreted by multiple cancer cells,
such as thyroid cancer and epithelial ovarian cancer, which is
transported to the recipient cells to manipulate the progression of
cancers (18,36). Altogether, 786-O-EVs can radically
mediate the intercellular communication in RCC via delivery of
MALAT1.
To further validate the communicative ability of
786-O-EVs of MALAT1 in RCC, we silenced the MALAT1 expression in
786-O cells to extract the EVs. Our data validated the weakening
effect of MALAT1 depletion in 786-O cells on the promotive effects
of 786-O-EVs on RCC cell viability, invasion and migration, as well
as EMT. In consistency, numerous studies have confirmed that the
high-level MALAT1 is tightly bound with enhanced viability,
migration and invasion, and EMT of RCC cells (17). MALAT1 knockdown helps suppress the
malignant biological behaviors of RCC cells (37). Similarly, the role of 786-O-EV-carried
MALAT1 was further verified in nude mice in vivo, as
evidenced by inhibited tumor growth and alleviated lung metastasis
after inhibition of MALAT1 in the 786-O cells. The depletion of
MALAT1 exercises inhibitory effects on tumor metastasis to vividly
suppress xenograft growth in nude mice with RCC (38). Briefly, 786-O-EVs transferred MALAT1
as a facilitator of the viability, invasion and migration and EMT
of RCC cells, as well as RCC tumor growth and lung metastasis in
vivo.
For a comprehensive understanding of the regulatory
mechanism of MALAT1, we retrieved several databases, and performed
FISH, dual-luciferase reporter gene assay, RIP and RNA pull-down to
identify the downstream factors. Our findings identified the ETS1
gene as a solemn regulatory transcription factor of MALAT1
alongside TFCP2L1 as the most significantly downregulated gene in
the RCC expression chip GSE16441. JASPAR database supported the
transcriptional regulation of TFCP2L1 by ETS1. Moreover, our data
verified that the TFCP2L1 gene promoter possessed multiple binding
sites with ETS1. The effect of MALAT1 on TFCP2L1 activity was
verified by the dual-luciferase reporter assay, and it could be
speculated that the MALAT1/ETS1 axis exerts a malignant phenotype,
such as proliferation and invasiveness. As previously, ETS1 as an
oncogene is identified in close association with poor survival in
multiple RCC specimens (27).
Elevation of ETS1 via lncRNA-mediated regulation fosters the cell
malignant behaviors and tumor growth in clear cell renal cell
carcinoma (ccRCC) (39). Deregulated
TFCP2L1 is intimately bound with various cancers, including ccRCC
(40). Furthermore, our data
demonstrated that MALAT1 negatively regulated TFCP2L1 via specific
binding to ETS1; 786-O-EV treatment enhanced the inhibition of ETS1
transcription and evidently reduced the TFCP2L1 level in RCC cells,
which were reversed after knockdown of MALAT1 in the 786-O cells.
LncRNAs participate in epigenetic modification via chromosome
remodeling, transcriptional regulation via transcription factor
modulation and post-transcriptional regulation via mRNA alternative
splicing respectively (41). LncRNA
CASC19, as a competitive endogenous RNA, was found to upregulate
ETS1 expression by sponging miR-532 (39), a participant of post-transcriptional
regulation. The current study explored the direct interaction
between RNA and certain transcription factors. LncRNA MALAT1
localized to the nucleus, bound with the ETS1 protein and
participate in transcriptional regulation by complex formation and
localization to specific gene sequences. In consistency with the
preceding finding, TFCP2L1 was proposed to be essential in normal
renal development (28), with
abnormal downregulation in ccRCC (42). However, the interplay between lncRNA
MALAT1 and ETS1, and the relationship between ETS1 and TFCP2L1 have
not been elucidated yet, which, conversely, validates the novelty
of this study. Briefly, we concluded that mechanically 786-O-EVs
shuttled MALAT1 to downregulate TFCP2L1 expression by promoting the
binding of transcription factor ETS1 and TFCP2L1 promoter in
RCC.
Collectively, our study demonstrated that 786-O-EVs
promoted RCC cell invasion and metastasis via transporting MALAT1
and regulating the ETS1/TFCP2L1 axis. These results identified a
novel tumor-derived EV-based therapy for RCC patients, where the
development of a blockade of the MALAT1/ETS1/TFCP2L1 axis might
serve as a promising therapeutic approach for RCC. Although the
present study provided therapeutic value for RCC treatment, the
experimental results and clinical application need further
verification.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data that support the findings of this study are
available from the corresponding author upon reasonable
request.
Authors' contributions
CJ, LS and KL conceived and designed the study. CJ,
WL, YQ, YZ, BZ, ZL, YL and QZ performed the experiments. CJ, LS and
KL wrote the manuscript. CL and LS reviewed and edited the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study received approval from the Ethics
Committee of The Second Affiliated Hospital of Harbin Medical
University (HM-2018-0524). Significant efforts were made to
minimize both the number of animals and their suffering. All
procedures were strictly conducted in accordance with the code of
ethics.
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing
interests.
References
|
1
|
Hanna KS: A review of checkpoint
inhibitors in the management of renal cell carcinoma. J Oncol Pharm
Pract. 26:445–458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdellah A, Selma K, Elamin M, Asmae T,
Lamia R, Abderrahmane M, Sanaa el M, Hanan E, Tayeb K and
Noureddine B: Renal cell carcinoma in children: Case report and
literature review. Pan Afr Med J. 20:842015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Abdulfatah E, Kennedy JM, Hafez K,
Davenport MS, Xiao H, Weizer AZ, Palapattu GS, Morgan TM, Mannan R,
Wang XM, et al: Clinicopathological characterisation of renal cell
carcinoma in young adults: A contemporary update and review of
literature. Histopathology. 76:875–887. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gray RE and Harris GT: Renal cell
carcinoma: Diagnosis and management. Am Fam Physician. 99:179–184.
2019.PubMed/NCBI
|
|
5
|
Makhov P, Joshi S, Ghatalia P, Kutikov A,
Uzzo RG and Kolenko VM: Resistance to systemic therapies in clear
cell renal cell carcinoma: Mechanisms and management strategies.
Mol Cancer Ther. 17:1355–1364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Price M, Wu CC, Genshaft S, Sadow PM, Xie
L, Shepard JO and McDermott S: Imaging and management of
intrathoracic renal cell carcinoma metastases. AJR Am J Roentgenol.
210:1181–1191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adibi M, Thomas AZ, Borregales LD, Matin
SF, Wood CG and Karam JA: Surgical considerations for patients with
metastatic renal cell carcinoma. Urol Oncol. 33:528–537. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Zhang M, Liu X, Sun H, Guo Z, Tang
X, Wang Z, Li J, Li H, Sun W and Zhang Y: Urine metabolomics for
renal cell carcinoma (RCC) prediction: Tryptophan metabolism as an
important pathway in RCC. Front Oncol. 9:6632019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watson DC, Bayik D, Srivatsan A,
Bergamaschi C, Valentin A, Niu G, Bear J, Monninger M, Sun M,
Morales-Kastresana A, et al: Efficient production and enhanced
tumor delivery of engineered extracellular vesicles. Biomaterials.
105:195–205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qin Z, Xu Q, Hu H, Yu L and Zeng S:
Extracellular vesicles in renal cell carcinoma: Multifaceted roles
and potential applications identified by experimental and
computational methods. Front Oncol. 10:7242020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grange C, Brossa A and Bussolati B:
Extracellular vesicles and carried miRNAs in the progression of
renal cell carcinoma. Int J Mol Sci. 20:18322019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sheehan C and D'Souza-Schorey C:
Tumor-derived extracellular vesicles: Molecular parcels that enable
regulation of the immune response in cancer. J Cell Sci.
132:jcs2350852019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma P, Pan Y, Li W, Sun C, Liu J, Xu T and
Shu Y: Extracellular vesicles-mediated noncoding RNAs transfer in
cancer. J Hematol Oncol. 10:572017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu X, Hao Y, Yu W, Yang X, Luo X, Zhao J,
Li J, Hu X and Li L: Long non-coding RNA emergence during renal
cell carcinoma tumorigenesis. Cell Physiol Biochem. 47:735–746.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y and Tang L: The application of
lncRNAs in cancer treatment and diagnosis. Recent Pat Anticancer
Drug Discov. 13:292–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao M, Wang S, Li Q, Ji Q, Guo P and Liu
X: MALAT1: A long non-coding RNA highly associated with human
cancers. Oncol Lett. 16:19–26. 2018.PubMed/NCBI
|
|
17
|
Li Z, Ma Z and Xu X: Long non-coding RNA
MALAT1 correlates with cell viability and mobility by targeting
miR-22-3p in renal cell carcinoma via the PI3K/Akt pathway. Oncol
Rep. 41:1113–1121. 2019.PubMed/NCBI
|
|
18
|
Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY
and Hua KQ: Exosomal metastasis-associated lung adenocarcinoma
transcript 1 promotes angiogenesis and predicts poor prognosis in
epithelial ovarian cancer. Int J Biol Sci. 14:1960–1973. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Essandoh K, Yang L, Wang X, Huang W, Qin
D, Hao J, Wang Y, Zingarelli B, Peng T and Fan GC: Blockade of
exosome generation with GW4869 dampens the sepsis-induced
inflammation and cardiac dysfunction. Biochim Biophys Acta.
1852:2362–2371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu H, Brannon AR, Reddy AR, Alexe G,
Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: With
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei JX, Lv LH, Wan YL, Cao Y, Li GL, Lin
HM, Zhou R, Shang CZ, Cao J, He H, et al: Vps4A functions as a
tumor suppressor by regulating the secretion and uptake of exosomal
microRNAs in human hepatoma cells. Hepatology. 61:1284–1294. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yeon JH, Jeong HE, Seo H, Cho S, Kim K, Na
D, Chung S, Park J, Choi N and Kang JY: Cancer-derived exosomes
trigger endothelial to mesenchymal transition followed by the
induction of cancer-associated fibroblasts. Acta Biomater.
76:146–153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan L, Liang W, Fu M, Huang ZH, Li X,
Zhang W, Zhang P, Qian H, Jiang PC, Xu WR and Zhang X:
Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes
gastric cancer progression. J Cancer Res Clin Oncol. 143:991–1004.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Li W, Gu W, Yan Y, Yao X and
Zheng J: MALAT1 accelerates the development and progression of
renal cell carcinoma by decreasing the expression of miR-203 and
promoting the expression of BIRC5. Cell Prolif. 52:e126402019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhai W, Ma J, Zhu R, Xu C, Zhang J, Chen
Y, Chen Z, Gong D, Zheng J, Chen C, et al: MiR-532-5p suppresses
renal cancer cell proliferation by disrupting the ETS1-mediated
positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br J
Cancer. 119:591–604. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tun HW, Marlow LA, von Roemeling CA,
Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ and
Copland JA: Pathway signature and cellular differentiation in clear
cell renal cell carcinoma. PLoS One. 5:e106962010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu R, Rai A, Chen M, Suwakulsiri W,
Greening DW and Simpson RJ: Extracellular vesicles in
cancer-implications for future improvements in cancer care. Nat Rev
Clin Oncol. 15:617–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Becker A, Thakur BK, Weiss JM, Kim HS,
Peinado H and Lyden D: Extracellular vesicles in cancer:
Cell-to-cell mediators of metastasis. Cancer Cell. 30:836–848.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li YJ, Wu JY, Hu XB, Wang JM and Xiang DX:
Autologous cancer cell-derived extracellular vesicles as
drug-delivery systems: A systematic review of preclinical and
clinical findings and translational implications. Nanomedicine
(Lond). 14:493–509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen J, Fei X, Wang J and Cai Z:
Tumor-derived extracellular vesicles: Regulators of tumor
microenvironment and the enlightenment in tumor therapy. Pharmacol
Res. 159:1050412020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Syn N, Wang L, Sethi G, Thiery JP and Goh
BC: Exosome-mediated metastasis: From epithelial-mesenchymal
transition to escape from immunosurveillance. Trends Pharmacol Sci.
37:606–617. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Zhu X and Huang S: Extracellular
vesicle long non-coding RNAs and circular RNAs: Biology, functions
and applications in cancer. Cancer Lett. 489:111–120. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hardin H, Helein H, Meyer K, Robertson S,
Zhang R, Zhong W and Lloyd RV: Thyroid cancer stem-like cell
exosomes: Regulation of EMT via transfer of lncRNAs. Lab Invest.
98:1133–1142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang LT, Wan CH, Guo QH, Yang SJ, Wu JD
and Cai J: Long noncoding RNA metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) promotes renal cell carcinoma
progression via sponging miRNA-429. Med Sci Monit. 24:1794–1801.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen S, Ma P, Zhao Y, Li B, Jiang S, Xiong
H, Wang Z, Wang H, Jin X and Liu C: Biological function and
mechanism of MALAT-1 in renal cell carcinoma proliferation and
apoptosis: role of the MALAT-1-Livin protein interaction. J Physiol
Sci. 67:577–585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Luo Y, Liu F, Yan C, Qu W, Zhu L, Guo Z,
Zhou F and Zhang W: Long non-coding RNA CASC19 sponges microRNA-532
and promotes oncogenicity of clear cell renal cell carcinoma by
increasing ETS1 expression. Cancer Manag Res. 12:2195–2207. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kotarba G, Krzywinska E, Grabowska AI,
Taracha A and Wilanowski T: TFCP2/TFCP2L1/UBP1 transcription
factors in cancer. Cancer Lett. 420:72–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fanelli GN, Gasparini P, Coati I, Cui R,
Pakula H, Chowdhury B, Valeri N, Loupakis F, Kupcinskas J,
Cappellesso R and Fassan M: long-noncoding RNAs in gastroesophageal
cancers. Noncoding RNA Res. 3:195–212. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zaravinos A, Lambrou GI, Mourmouras N,
Katafygiotis P, Papagregoriou G, Giannikou K, Delakas D and Deltas
C: New miRNA profiles accurately distinguish renal cell carcinomas
and upper tract urothelial carcinomas from the normal kidney. PLoS
One. 9:e916462014. View Article : Google Scholar : PubMed/NCBI
|