Introduction
Clear cell renal cell carcinoma (ccRCC), which
accounts for 70–75% of all diagnosed renal types of cancer, is one
of the most common malignancies in the urinary system. It is an
aggressive cancer derived from the proximal tubular epithelium,
whose metastatic form is associated with high mortality (1–3).
Moreover, the incidence of ccRCC has increased in recent decades,
with 76,080 new cases diagnosed in the United States in 2021
(4). Although targeted therapies
and novel immunotherapeutic agents are gradually being applied, the
efficacy is limited, leading to a low 5-year survival rate of only
10–20% for patients at an advanced stage (5,6).
Therefore, considering the high morbidity and mortality of ccRCC,
it is necessary to determine the potential mechanism of the
occurrence and development of ccRCC and find new biomarkers with
high specificity and sensitivity.
Human yippee-like (YPEL) proteins are members of a
recently discovered clan of putative zinc finger motif coding genes
comprised of YPEL1–5 (7,8). The proteins of the YPEL family are
located in the centrosome, present in a wide range of eukaryotic
species, adjacent to the nucleolus and mitotic apparatus (9,10).
Members of the YPEL gene family are involved in various cell
biological processes, including the cell cycle, senescence,
mammalian development and tumor progression (11,12).
It is worth noting that depending on types of cancer, YPELs may act
as tumor promoters or inhibitors. It has been reported that YPEL1,
a nuclear protein, is involved in the mesenchymal-epithelial
transition in cancer (13).
Compared with normal pancreatic tissues, YPEL1 expression is
significantly reduced in pancreatic cancer tissues (14). Moreover, bioinformatics analysis
showed that YPEL1 is upregulated in epidermal growth factor
receptor (EGFR)-mutant NSCLC samples treated with erlotinib
(15). Tuttle et al
(16) found reduced YPEL3
expression in tumor samples compared with patient-matched normal
tissue. Zhang et al (17)
demonstrated that YPEL3 expression was reduced in nasopharyngeal
carcinoma cell lines and clinical samples, and YPEL3 overexpression
inhibited nasopharyngeal carcinoma cell invasion and metastasis
in vitro and in vivo. YPEL5 was revealed to inhibit
cell proliferation and cell cycle progression (18). However, overexpression of YPEL2 in
breast tumors correlates with breast cancer risk (19). The expression level of YPEL4 in
patients with aldosterone-producing adenomas (APAs) is 2.4-fold
higher than that in nonfunctioning adenomas of the adrenal cortex,
and YPEL4 expression levels in APAs are positively correlated with
tumor diameter (11). These
findings indicated that the YPEL gene plays a vital role in
tumorigenesis. Nevertheless, there is currently no study of YPELs
in ccRCC, which means that the prognostic value of the YPEL family
in ccRCC remains unclear and needs to be further elucidated.
The identification and application of new cancer
biomarkers have become increasingly accurate and valuable with the
development of a great quantity of RNA sequencing technologies and
available databases in the present study. The overall function,
prognosis and distribution of the YPEL genes in humans were
systematically analyzed by pan-cancer analysis. The prognostic
value and potential mechanisms of the YPEL genes in ccRCC were
screened using data from patients with ccRCC in multiple databases.
Furthermore, the effects of YPELs on the proliferation and invasion
of ccRCC were preliminarily verified by observing the cell
phenotype of the ccRCC model in vitro. Paired clinical
samples and multi-group analysis were used to further investigate
factors affecting expression changes.
Materials and methods
Data acquisition and processing
ONCOMINE database (www.oncomine.org) is a comprehensive online cancer
microarray database for DNA or RNA sequence analysis, helping to
find answers from whole gene expression analysis. The
transcriptional expressions of YPELs in tumor tissues and
corresponding adjacent normal samples used as control were obtained
from the ONCOMINE database. The Genotype-Tissue Expression (GTEx)
database (https://www.gtexportal.org) was used
to analyze the distribution of the YPEL genes in human normal organ
tissues. The gene expression RNAseq (HTSeq-FPKM),
clinicopathological data, immune subtype, survival data and
stemness score (RNA based) of 33 types of cancer were downloaded
from The Cancer Genome Atlas (TCGA) (http://portal.gdc.cancer.gov/). Difference analysis
was performed using the Limma package from Bioconductor (version:
3.52.0). Genes with an average count value >1 were excluded.
P<0.05 and |log2 (FC)|>1.0 was taken into consideration.
Patients and sample collection
A total of 20 pairs (13 males and 7 females; age
range 40–70 years) of ccRCC tissues and corresponding non-cancer
tissues were obtained from patients undergoing surgical resection
in the general surgery department of the Second Affiliated Hospital
of Nanchang University (Nanchang, China) from September 2020 to
November 2021. All resected specimens were frozen and stored at
−80°C for further analysis. Written informed consent was provided
from all patients. The present study was approved (approval no.
2020090) by the Ethics and Research Committee of the Second
Affiliated Hospital of Nanchang University (Nanchang, China).
Tumor microenvironment (TME)
analysis
Stromal score and immune score were calculated using
ESTIMATE analysis (20). The
results were visualized using the R package ‘corrplot’.
Stemness indices analysis
Stemness index data were downloaded from UCSC Xena
(http://xena.ucsc.edu/). The Limma and Corplot
packages were used to visualize the results.
Estimation of immune cell type
fractions
CIBERSORT is a method for characterizing the cell
composition from their gene expression profiles and is the most
frequently cited tool for estimating and analysing immune cells
infiltration (21).
TIMER
TIMER (https://cistrome.shinyapps.io/timer/) (22) is a comprehensive database for tumor
immune infiltrating cells (TIIC) analysis of 32 tumors. In the
present study, Spearman's correlation analysis in the gene module
was used to investigate the correlation between YPEL genes
expression and immune infiltration, including tumor purity and six
types of cells of the immune system (B cells, CD8+ T
cells, CD4+ T cells, macrophages, neutrophils and
dendritic cells).
Cell culture and transfection
The renal cell carcinoma (RCC) cell line 786-O was
obtained from the Institute of Biochemistry and Cell Biology,
Chinese Academy of Sciences (Shanghai, China). The cell line was
detected without mycoplasma, and the cell line was verified by STR
detection. The cells were cultured at 37°C in an atmosphere
containing 5% CO2 in DMEM supplemented with 10% fetal
bovine serum (FBS; both from Beijing Solarbio Science &
Technology Co., Ltd.). Lipofectamine™ 3000 Transfection
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection. Briefly, HCC cells were seeded in six-well plates the
day before transfection. The siRNAs and Lipofectamine 3000 were
mixed with Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.)
and incubated for 10 min at room temperature. The siRNA-lipid
complex was diluted in DMEM to achieve a final siRNA concentration
of 10 nM. Cells were incubated for 48 h in a 5% CO2
incubator at 37°C. The YPEL1 siRNA sequences were as follows:
sense, 5′-UGUCUUUGAUCAUAUGAGCAA-3′ and antisense,
5′-GCUCAUAUGAUCAAAGACAAU-3′. The YPEL2 siRNA sequences were as
follows: sense, 5′-AUUAGUUCAUCAUGAUUGGCC-3′ and antisense,
5′-CCAAUCAUGAUGAACUAAUUU-3′. The YPEL5 siRNA sequences were as
follows: sense, 5′-UGAUCAAGGAAAAUUCUGCCC-3′ and antisense,
5′-GCAGAAUUUUCCUUGAUCAUA-3′. The negative control for siRNA
silencing was a non-targeting (scramble) siRNA sequence, with the
sequences were as follows: sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
antisense, 5′-ACGUGACACGUUCGGAGAATT-3′.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from 786-O cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). According to the manufacturer's protocols, total RNA was
reverse transcribed into cDNA using PrimeScript™ RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
protocol, followed by qPCR utilizing the 7500 Real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR
Premix Ex Taq kit (Takara Bio USA, Inc.) according to the
manufacturer's protocol. The following thermocycling conditions
were used: Initial denaturation at 95°C for 30 sec, followed by 40
cycles at 95°C for 5 sec and 60°C for 30 sec. Relative mRNA
expression was normalized to an internal control (β-actin) and
results were expressed as relative expression calculated using the
2−ΔΔCq method (23).
Primer sequences used in the present study are listed in Table SI. Experiments were performed at
least three times independently.
Western blotting
786-O cells were lysed using
radioimmunoprecipitation lysis buffer containing Protease Inhibitor
Single Use Cocktail and Phosphatase Inhibitor Cocktail (Thermo
Fisher Scientific, Inc.) and protein concentration was determined
using a Pierce BCA protein assay kit (Thermo Fisher Scientific,
Inc.). Cell lysates (20 µg/lane) were separated on 8–15% gel by
SDS-PAGE and transferred to polyvinylidene difluoride membranes
(MilliporeSigma). The membranes were blocked with 5% skimmed milk
at room temperature for 1 h. The membranes were incubated with the
following primary antibodies: YPEL1 (1:1,000; cat no. 17743-1-AP),
GADPH (1:1,000; cat no. 60004-1-Ig; both from ProteinTech Group,
Inc.), YPEL2 (1:5,000; cat. no. PA5-34348) and YPEL5 (1:1,000; cat.
no. PA5-34351; both from Invitrogen; Thermo Fisher Scientific,
Inc.). Following the primary incubation, the membranes were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
or mouse polyclonal secondary antibodies (1:10,000; cat. no.
ZB-2301/2305, ZSGB-BIO, Inc.) for 2 h at room temperature.
Immunoreactive bands were detected using the Bio-Rad ChemiDoc MP
Imaging System (Bio-Rad Laboratories, Inc.).
Cell proliferation assay
Proliferation of 786-O cells was detected using the
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.)
according to the manufacturer's protocol. 786-O cells in 100 ml
medium were inoculated in 96-well plates (4×103
cells/well). A total of 100 µl of CCK-8 reagent was added to each
well and plates were incubated for 1.5 h at 37°C. The absorbance of
each well at 0, 24, 48, 72, 96 and 120 h was detected at 450 nm
using an enzyme-linked immunosorbent assay microplate reader
(Thermo Fisher Scientific, Inc.).
5-Ethynyl-2′ deoxyuridine (EdU)
assay
Cell proliferation was determined using EdU assay
kit (Guangzhou RiboBio Co., Ltd.). 786-O cells were seeded in
24-well plates (2×104 cells/well), cultured (5%
CO2; 37°C) in DMEM supplemented with 10% FBS for 24 h
before EdU (50 µmol/l) was added. Cells were fixed with 4%
paraformaldehyde for 30 min and permeabilized with 0.5%
Triton-X-100 in PBS for 20 min at room temperature according to the
manufacturer's protocols. The cell nuclei were stained with Hoechst
dye 33342 and incubated for 30 min in the dark for visualization.
Images of 786-O cells were acquired under a fluorescence microscope
(Leica Microsystems GmbH). Proliferation was analysed using the
average number of cells in three random fields per sample.
Invasion assay
The upper chamber of Transwell system (24 inserts,
8-µm pore size, polycarbonate membrane; Corning, Inc.) was coated
with precooled Matrigel and incubated at 37°C for 30 min. Briefly,
5×104 786-O cells pre-transfected with 50 nm siRNA for
48 h, were suspended in 100 ml serum-free DMEM and seeded in the
upper chamber. A total of 500 ml medium supplemented with 10% FBS
was added to the lower chamber.
After 48 h of incubation at 37°C, the impermeable
cells were wiped off, and the cells on the lower surface of the
filter were fixed with 4% paraformaldehyde for 15 min at room
temperature and then stained with 0.4% crystal violet for 30 min at
room temperature. The numbers of invasive cells were counted in 5
random fields of view in the same chamber (mean ± SE) under a light
microscope for 3 samples.
Migration assay
The migration assay was performed in the same manner
as the aforementioned invasion assay, with the exception that the
membrane was not coated with Matrigel. Briefly, 5×104
786-O cells pre-transfected with 50 nm siRNA for 48 h were
suspended in 100 ml serum-free medium and seeded in the upper
chamber. A total of with 500 ml medium supplemented with 10% FBS
was added to the lower chamber. After 24 h of incubation, cells
were similarly stained and counted for invasion studies.
Statistical analysis
Statistical analysis was performed using the
GraphPad Prism 8.0 (GraphPad Software, Inc.) and the R programming
language version 3.6.3. Student's unpaired t-test and one-way ANOVA
(followed by Tukey's post-hoc test) were used to determine
significance. Survival curves were generated using the Kaplan-Meier
method, and differences between groups were compared with the log
rank test with the cutoff as the median. Each experiment was
performed in triplicate and P<0.05 was considered to indicate a
statistically significant difference.
Results
Pan-cancer analysis of YPEL family
member genes
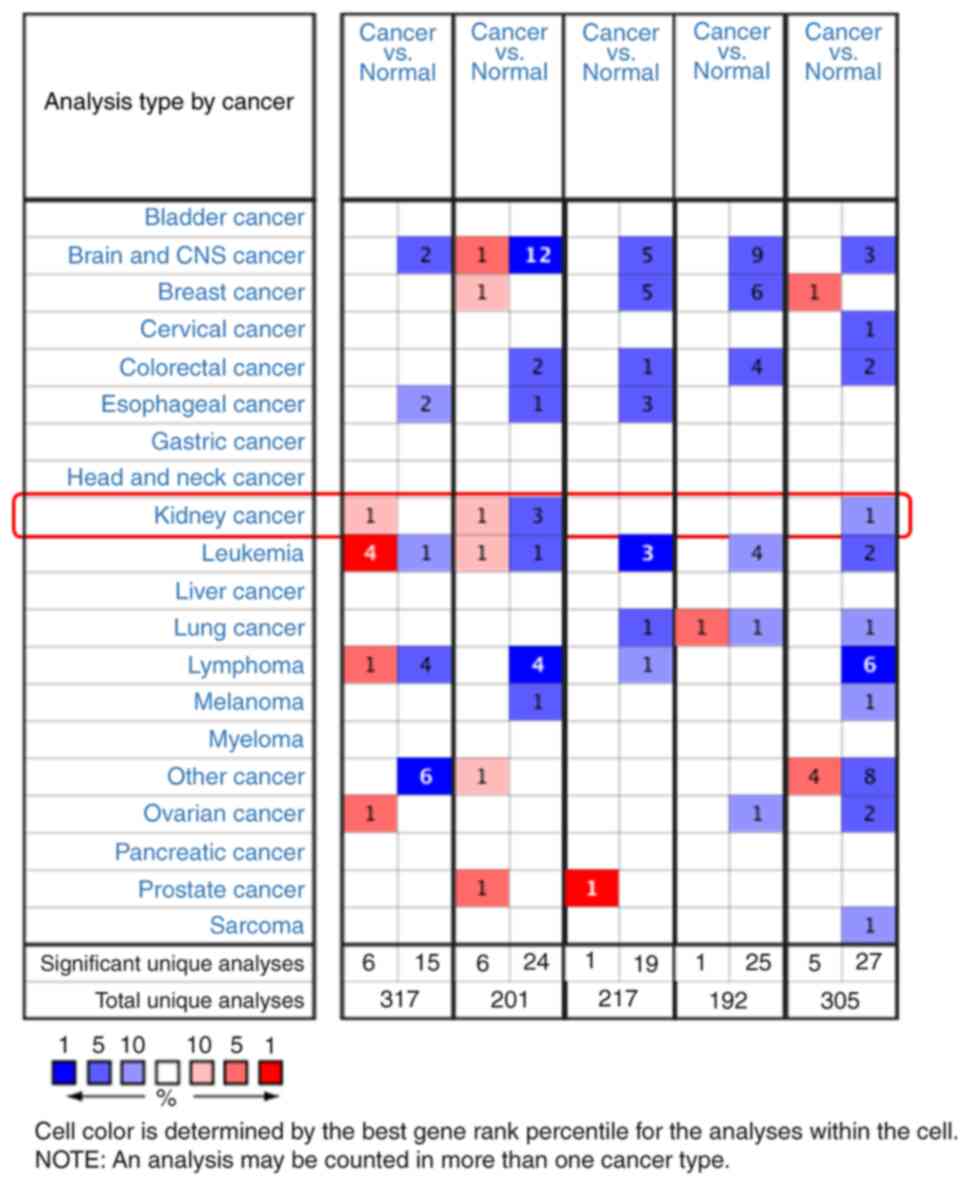
The Oncomine database was used to examine all five
members of the YPEL gene family in 20 cancer samples and compare
them to normal tissue. The apparently different gene expression is
revealed in Fig. 1. The Oncomine
database contains 317, 201, 217, 192 and 305 different studies
involving the genes from YPEL1 to YPEL5. The remarkable unique
analysis between cancer and normal tissue that meets the selection
criteria is revealed in the cell at the bottom of Fig. 1. The case number in the left cell
indicates gene upregulation, and the case number in the right cell
indicates downregulation. The counting results were YPEL1 (6:15),
YPEL2 (6:24), YPEL3 (1:19), YPEL4 (1:25), and YPEL5 (5:27).
Specifically, in renal carcinoma, a significant increase in the
mRNA expression level of YPEL1 was shown in multiple datasets, and
the mRNA expression level of YPEL5 was significantly downregulated,
while the expression level of YPEL2 was significantly downregulated
in 3 independent pancreatic cancer studies and overexpressed in 1
case (P<0.05, fold change >2) (Fig. 1 and Table I).
 | Table I.The significant changes of YPELs in
transcription level (ONCOMINE database). |
Table I.
The significant changes of YPELs in
transcription level (ONCOMINE database).
|
| Types of RCC vs.
kidney | Fold change | P-value | t-test | Reference |
|---|
| YPEL1 | Renal Wilms
tumor | 6.444 | 0.005 | 3.667 | Yusenko renal |
| YPEL2 | Papillary renal
cell carcinoma | 2.042 | 0.003 | 3.138 | Yusenko renal |
|
| Chromophobe renal
cell carcinoma | −2.509 |
6.66×10−4 | −5.240 | Yusenko renal |
|
| Renal Wilms
tumor | −4.393 | 0.002 | −5.363 | Yusenko renal |
|
| Renal
oncocytoma |
| 0.002 | −4.460 | Yusenko renal |
| YPEL5 | Clear cell sarcoma
of the kidney |
| 0.006 | −6.552 | Cutcliffe
renal |
Subsequently, a series of pan-cancer analyses were
performed using the TCGA and GTEx databases to investigate the
distribution, function and prognosis of the YPEL genes in humans.
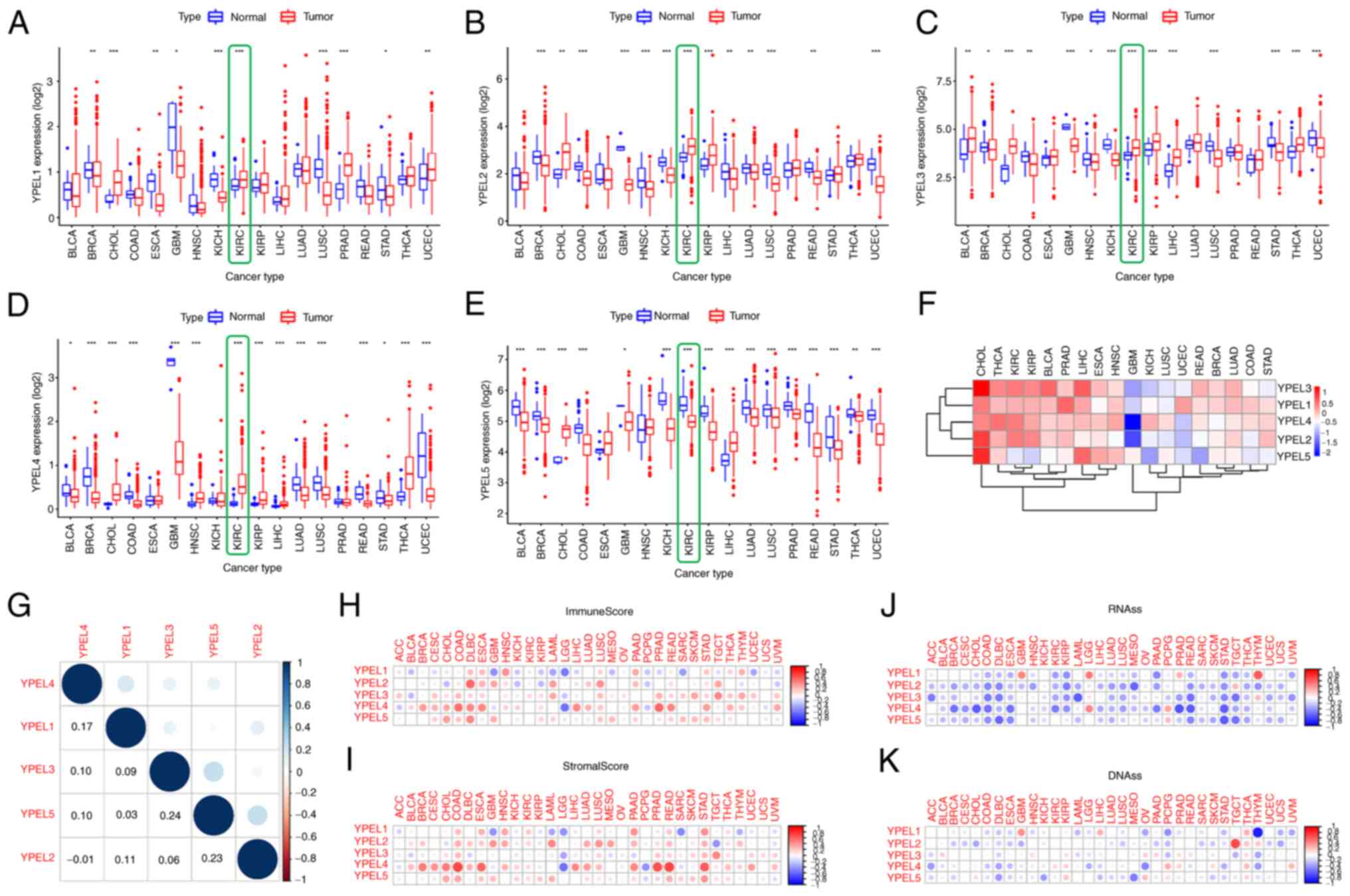
Compared with other organ tissues, the results of gene expression
data analysis of the GTEx database demonstrated that the expression
levels of YPEL1–3 and YPEL5 were above moderate levels in human
kidney tissues, while the expression levels of YPEL4 were lower
(Fig. S1A-E). Furthermore,
combined with the analysis of gene expression data in the TCGA and
GTEx databases, the human tumor tissue was compared with the
corresponding normal organ tissue to detect the expression level of
the YPEL gene family across tumor types. Specifically, it was found
that YPEL1–4 was upregulated and YPEL5 was downregulated in renal
tumor tissue (Fig. 2A-E). In
addition, the mean expression levels of the YPEL gene family were
assessed and a heatmap showing the results of differential analysis
of diffuse cancer data was generated (Fig. 2F). Moreover, Pearson's correlation
coefficients among the YPEL gene family were calculated. As shown
in Fig. 2G, certain genes showed a
certain correlation: YPEL3 and YPEL5 (r=0.24; P<0.05); YPEL2 and
YPEL 5 (r=0.23; P<0.05). Accumulating evidence indicates that
tumor stem cells and the TME play important roles in stimulating
tumor cell heterogeneity, increasing multidrug resistance, and
promoting tumor progression and metastasis. The correlation between
the expression of YPEL genes, tumor stem cells, and the TME was
further verified using pan-cancer analysis. The ESTIMATE algorithm
was used to calculate the stromal and immune scores in pan-cancer
(Fig 2H and I). The results showed
that the RNAss and DNAss of YPEL genes of the YPEL gene family were
calculated by mRNA expression and DNA methylation data. Similarly,
the expression of YPEL genes was also significantly positively or
negatively correlated with RNAss and DNAss using pan-cancer
analysis (Fig. 2J and K).
Differential expression of YPEL family
genes in patients with ccRCC
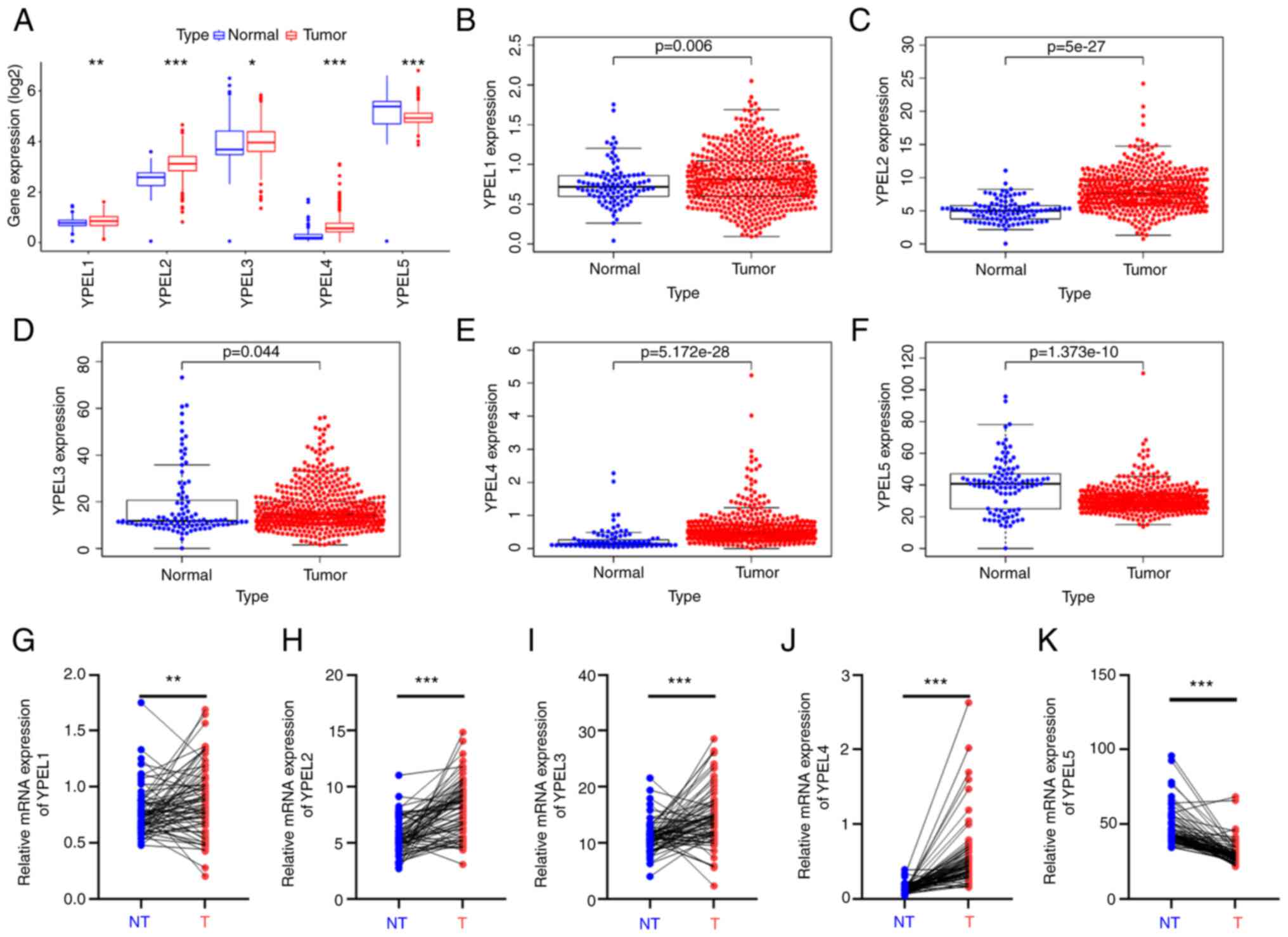
To accurately determine the expression of the YPEL
genes in patients with ccRCC, ccRCC sample data were obtained from
TCGA (TCGA-KIRC: 72 normal and 539 tumor samples). Data were
normalized and subjected to variance analysis using the R package
(version: 3.52.0). It was found that different expression levels of
all YPEL genes were statistically significant (Fig. 3A). Among them, YPEL1–4 were
significantly upregulated in tumor tissues compared with the
control, whereas YPEL5 was significantly downregulated. This result
was consistent with the aforementioned multi-database joint
analysis. In addition, paired expression data were further
extracted and compared from the TCGA-KIRC dataset, and it was
similarly identified that YPEL1–4 expression in tumor tissues was
significantly higher than that in paired non-tumor tissues, while
YPEL5 showed the opposite trend (Fig.
3B-K).
Prognostic analysis of YPEL family
genes in patients with ccRCC
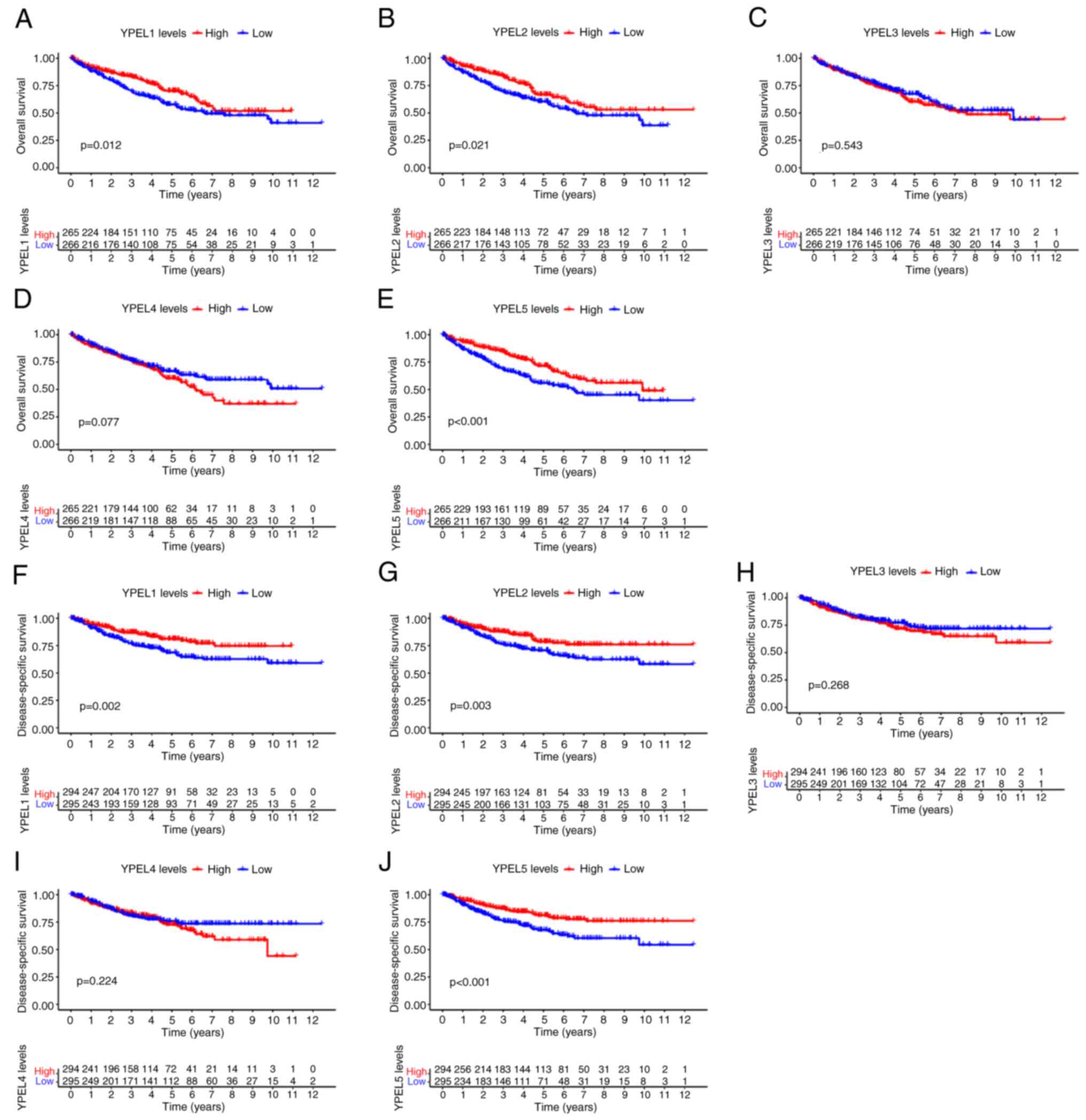
Furthermore, the Kaplan-Meier survival curve and
log-rank test were used to compare the relationship between the
expression of YPEL genes and overall survival (OS) and
disease-specific survival (DSS) and to evaluate the prognostic
significance of YPEL in ccRCC. It was found that the mRNA
expression of YPEL1, YPEL2 and YPEL5 was associated with the
prognosis of patients with ccRCC. Higher expression of YPEL1, YPEL2
and YPEL5 was associated with long-term survival, including OS and
DSS (Fig. 4A-J).
Association between expression of YPEL
family genes and clinicopathological parameters in ccRCC
The relationship between mRNA expression of the YPEL
family genes and clinicopathological parameters was then further
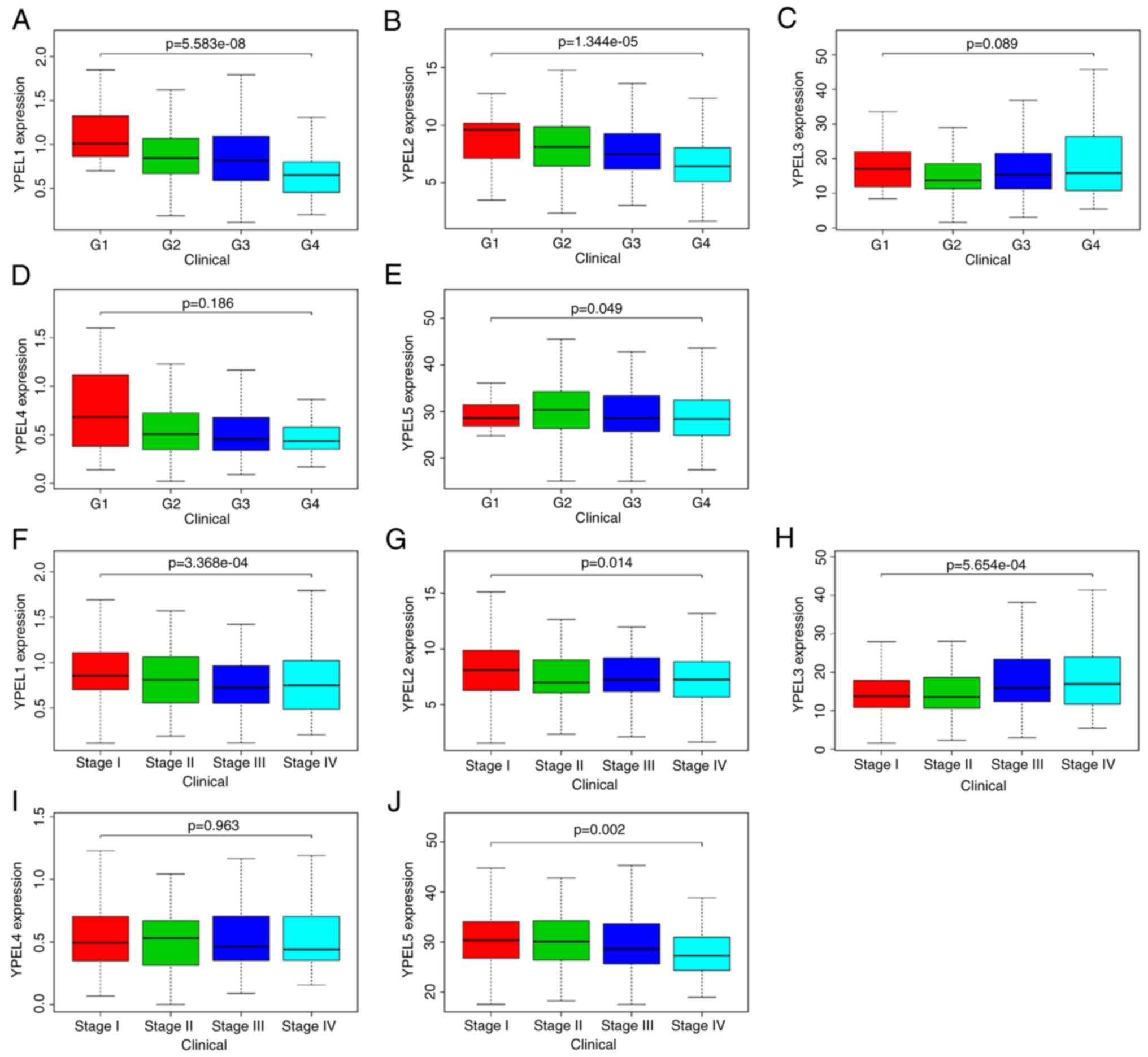
examined, including individual tumor grade and stage, in patients
with ccRCC using the TCGA dataset. As revealed in Fig. 5, the expression of three YPELs with
prognostic value was significantly correlated with individual
clinicopathological parameters and YPEL mRNA expression levels.
Specifically, a decrease in the expression of YPEL1 and YPEL2 mRNA
resulted in increased tumor grade, and YPEL5 expression was
significantly correlated with different tumor grades (Fig. 5A-E). Additionally, the expression
levels of YPEL1, YPEL2 and YPEL5 were significantly different among
ccRCC patients with different tumor stages (Fig. 5F-J). Collectively, these results
suggested that the expression of YPEL1, YPEL2 and YPEL5 may be a
risk factor.
Association between YPEL family genes
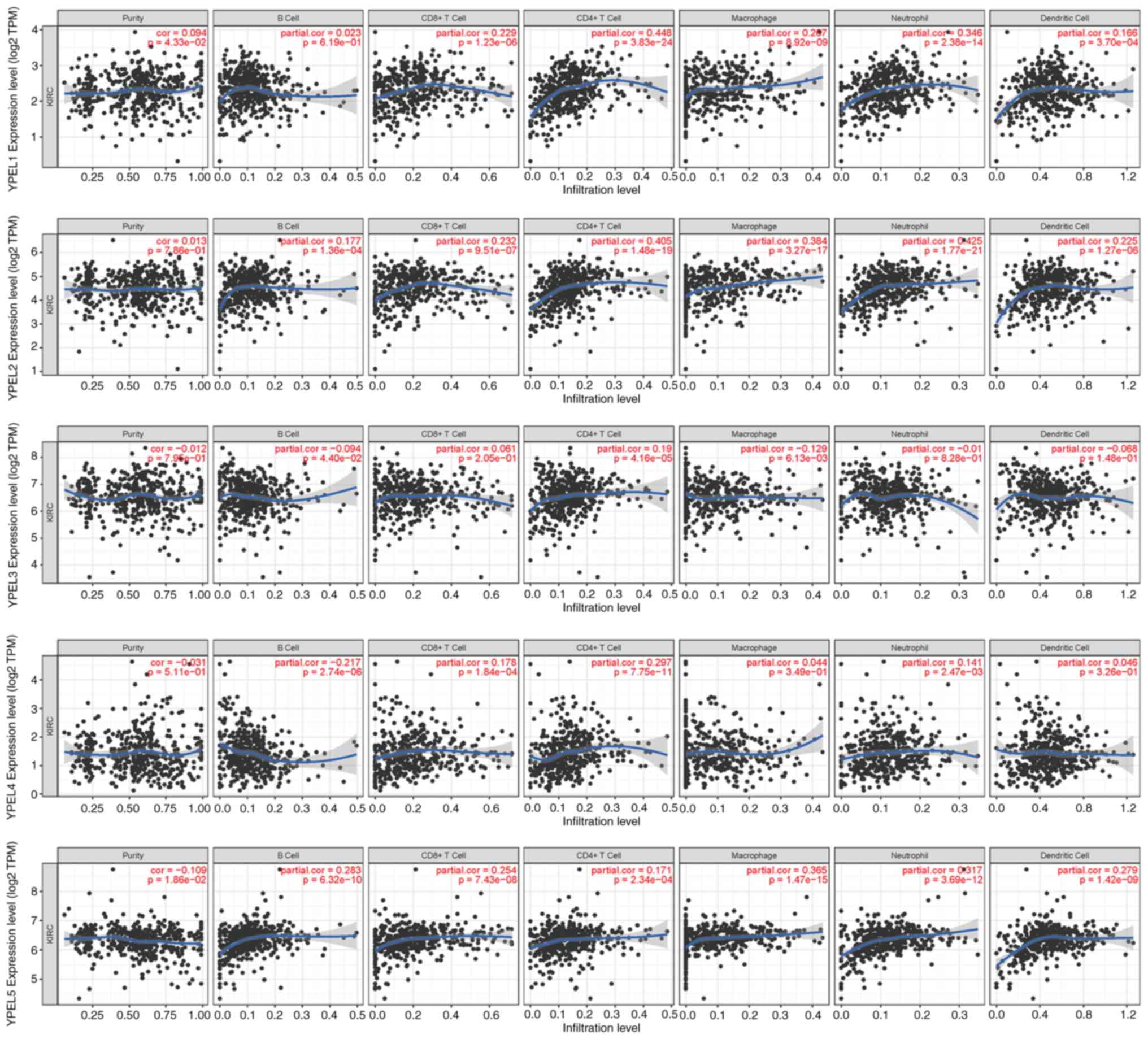
expression and immune infiltration in ccRCC
In recent years, the relationship between the immune
microenvironment and tumor progression has received increasing
attention from researchers. Therefore, TIMER was used to analyze
the association between the YPEL gene family and ccRCC immune
infiltration. It was found that the expression of YPEL1 in ccRCC
was significantly positively correlated with tumor purity and the
degree of infiltration of CD8+ T cells, CD4+
T cells, macrophages, neutrophils and dendritic cells in ccRCC.
YPEL2 was significantly positively correlated with B cells,
CD8+ T cells, CD4+ T cells, macrophages,
neutrophils, and dendritic cells. YPEL3 and YPEL4 were
significantly positively correlated with CD4+ T cells,
and YPEL5 was observably negatively correlated with all TIIC types.
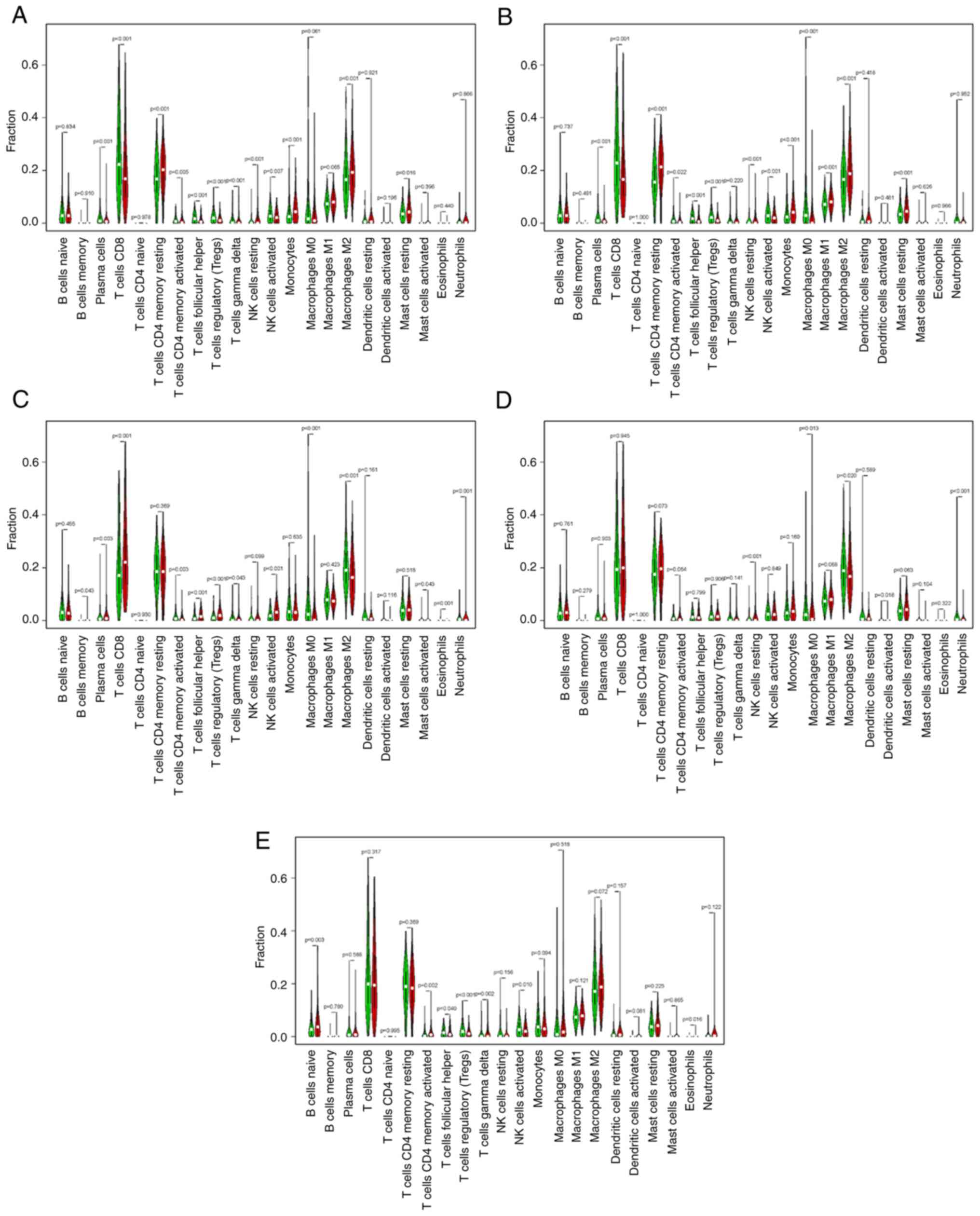
The CIBERSORT results showed a correlation between the YPEL gene
family and 22 immune cell types (Fig.
6). High expression of YPEL1 and resting memory CD4+
T cells, activated memory CD4+ T cells, delta gamma T
cells, resting natural killer cells, M2 macrophages, resting mast
cells and fewer plasma cells, CD8+ T cells, follicular
helper T cells, regulatory T cells (Tregs), activated natural
killer (NK) cells and monocytes were significantly associated. High
YPEL2 expression was associated with more monocytes,
CD8+ T cells, resting memory CD4+ T cells, M1
macrophages, M2 macrophages, M0 macrophages, resting mast cells and
fewer plasma cells, activated memory CD4+ T cells,
follicular helper T cells, Tregs, resting NK cells, and activated
NK cells. High YPEL3 expression was related to more CD8+
T cells, memory B cells, plasma cells, follicular helper T cells,
activated NK cell and Tregs and fewer delta gamma T cells,
activated memory CD4+ T cells, M2 macrophages and M0
macrophages. High YPEL4 expression was associated with more
neutrophils and resting NK cells and fewer M2 macrophages and M0
macrophages. High YPEL5 expression was associated with more naive B
cells, activated memory CD4+ T cells, and fewer
follicular helper T cells, Tregs, delta gamma T cells and activated
NK cells (Fig. 7A-E). The present
results suggested that YPEL genes may be regulators of the ccRCC
immune microenvironment and merit further investigation.
Prognosis-Related YPEL gene functions
as suppressor oncogenes to inhibit proliferation, migration and
invasion of the ccRCC cell line
Given that YPEL1, YPEL2 and YPEL5 are significantly
differentially expressed in ccRCC and strongly associated with
patient outcomes, including both OS and DSS, it was hypothesized
that these genes may play key roles in ccRCC. Therefore, the
expression of YPEL1, YPEL2 and YPEL5 in ccRCC was further studied
and the effect of their expression on cell function was explored.
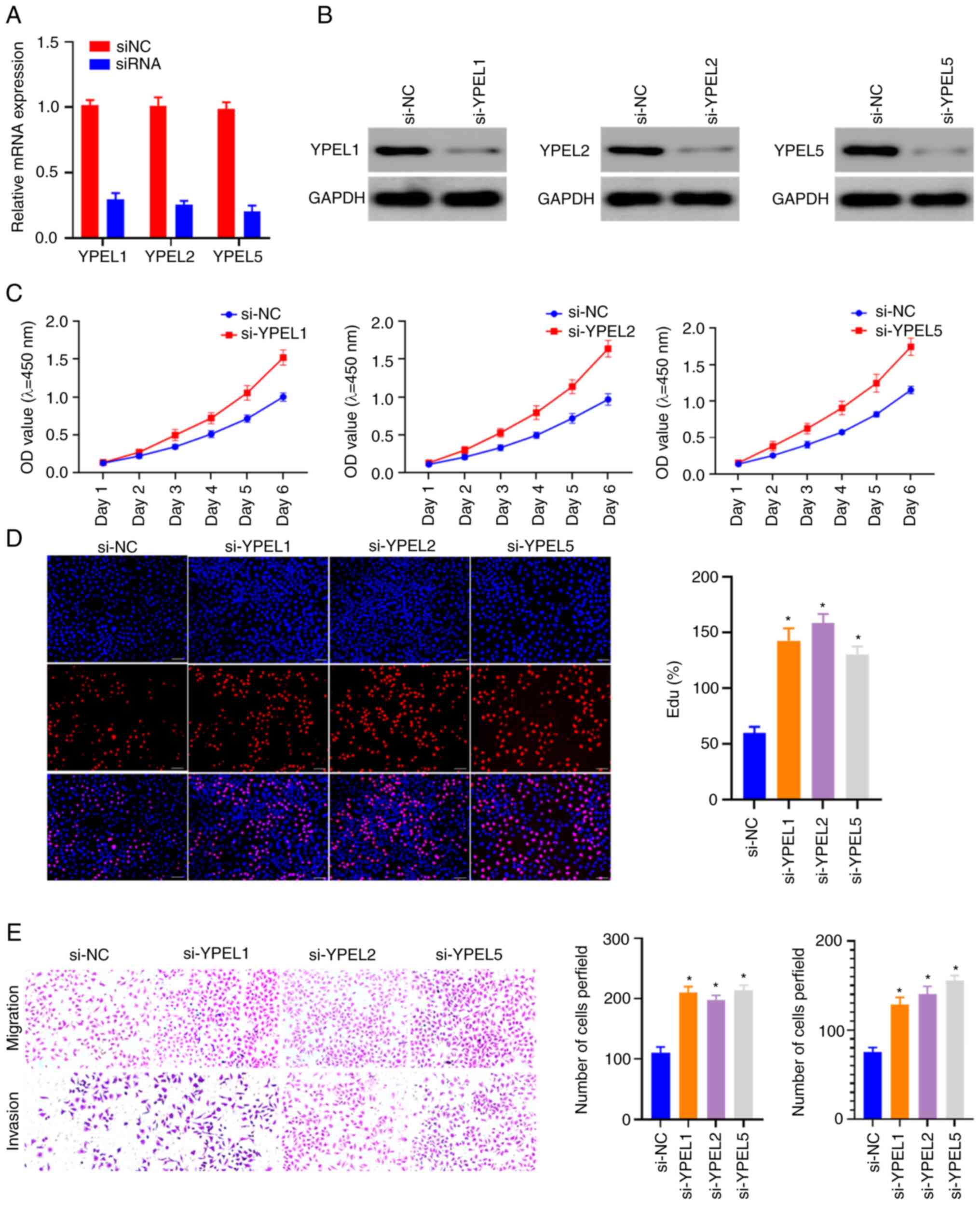
As revealed in Fig. S2, RT-qPCR
revealed that YPEL1 and YPEL2 had significantly elevated expression
in ccRCC tissues compared with adjacent non-tumor tissues, whereas
the expression of YPEL5 was reduced. These results are largely
consistent with the results of the database analysis. In addition,
CCK-8 and EdU assays were performed to analyze cell viability, and
Transwell assays were used to test the migration ability of 786-O
cells. The knockdown efficiency of YPEL1, YPEL2 and YPEL5 was
examined by using RT-qPCR and western blotting (Fig. 8A and B). Knockdown of YPEL1, YPEL2
and YPEL5 significantly promoted cell proliferation, migration and
invasion (Fig. 8C-E). The
experimental results suggested that YPEL1, YPEL2 and YPEL5 may play
an important role in maintaining the characteristics of ccRCC
tumors.
Discussion
The YPEL gene family was discovered in 2001, and
homologs of the Drosophila yippee gene in a variety of eukaryotes
were subsequently identified (7,8).
Experiments on interaction, cloning, and sequence analysis showed
that the YPEL gene family is highly conserved in eukaryotes and
possesses a putative zinc binding RING finger protein with
self-interacting properties (9).
According to previous studies, dysregulation of YPELs can be
observed in a variety of malignancies, suggesting that they may
play a vital role in the genesis and progression of cancer
(13,16–18,24).
However, at present there is no systematic study on the overview of
the whole YPELs in ccRCC. To the best of our knowledge, the present
study was the first to systematically analyze the overall
distribution, function and prognosis of human YPEL genes by mining
public databases, extensively investigating the prognostic value of
YPEL genes, and investigating its potential mechanism in ccRCC,
providing further suggestions for the clinical value of YPEL genes
in ccRCC. The present data indicated that low expression of YPEL1,
YPEL2 and YPEL5 is associated with poor prognosis and may act as an
independent predictor of ccRCC. These results suggested that YPEL1,
YPEL2 and YPEL5 can act as prognostic biomarkers for ccRCC.
Although several studies have investigated the
function and underlying mechanism of YPELs in tumors, their
prognostic value in urological tumors and their function in tumor
cells have not been reported. In the present study, a pan-cancer
analysis of the YPEL gene family was first performed to explore
their overall distribution, function and prognosis in humans. The
differential profiles of YPELs in normal human organs and multiple
tumors and the relationship between the expression of YPELs and
tumor grade, tumor stage and survival status were investigated.
Second, the ccRCC data of the TCGA-KIRC database were analyzed to
confirm the result of the pan-cancer analysis that YPELs have
prognostic significance.
By analyzing and screening YPEL genes associated
with differential expression and prognosis, it was identified that
only YPEL1, YEPL2 and YPEL5, but not YPEL3 and YPLE4, are most
promising for further studies. In addition, based on the cell
phenotype observed in the ccRCC model in vitro, the effect
of YPELs on the proliferation and invasion of ccRCC was
preliminarily verified.
YPEL1 is a nuclear protein considered to be involved
in mesenchymal-epithelial transition during tissue development
(25). As previously reported,
aberrant expression of the YPEL1 gene was observed in invasive
pancreatic cancer cells (14). Li
et al (13) demonstrated
that downregulation of YPEL1 expression inhibited gastric cancer
cell proliferation and invasion. In the present study, it was found
that YPEL1 may be an important inhibitory prognostic factor.
Decreased expression of YPEL1 at the mRNA level is associated with
poor prognosis in patients with ccRCC. Due to the fainted study on
the role of YPEL2 in cancer progression, it is known that
interactions between YPEL2 and other genes expressed from the 17q23
amplicon may be relevant to breast cancer (19). Similarly, the present study found
that aberrant expression of YPEL2 was significantly associated with
the prognosis of patients with ccRCC, and its expression was
markedly inversely correlated with the staging grade of ccRCC.
Studies have shown that YPEL5 protein is expressed at various
subcellular sites in the cell cycle (9,18);
it is localized in the nucleus and centrosome during interphase,
then sequentially translocated during mitosis to spindle poles,
mitotic spindle and spindle midzone, and finally transferred to the
midbody upon cytokinesis. Reduction of YPEL5 expression by siRNA
inhibited the growth of COS-7 cells and the early development of
medaka fish embryos, suggesting that YPEL5 is involved in cell
cycle progression (9). YPEL5 has
been reported to play an important role in tumor development. For
example, YPEL5 was revealed to inhibit cell proliferation and cell
cycle progression in cervical cancer cells (26). Velusamy et al (22) demonstrated that YPEL5 formed a
recurrent reciprocal RNA chimera with PPP1CB and played an
important role in chronic lymphocytic leukemia. Zhou et al
(18) revealed that overexpression
of YPEL5 significantly reduced the expression of CCNB1 and PCNA in
SW620 and HT29 cells, suggesting that YPEL5 was involved in the
regulation of cell proliferation and cell cycle progression in
colorectal carcinoma. These findings may help to understand the
role of YPEL5 in the development of various types of cancer.
Nevertheless, whether it affects the prognosis of patients with
ccRCC needs to be further clarified. Importantly, our data showed
that the expression of YPEL5 at the mRNA level was low in ccRCC,
which was related to poor prognosis of patients. Tumor-infiltrating
immune cells are considered to be a marker of the host antitumor
immune response and prognostic features (27,28).
CD8+ T cells and NK cells play a predominant role in the
antitumoral immune response via immune checkpoints. In the present
study, it was observed that YPEL1, YPEL2, YPEL3 and YPEL4 were
significantly positively correlated with several immune
infiltrating cells, particularly CD4+ T cells, while
YPEL5 was observably negatively correlated with all TIIC types.
These results suggested that YPELs may recruit and regulate
infiltrating immune cells to inhibit or promote the progression of
cancer, which strongly suggests that YPELs serve as a key factor in
cancer immunity. Finally, the present study further investigated
the effects of the expression of YPEL1, YPEL2, and YPEL5 on the
biological function of ccRCC cell lines by siRNA-mediated knockdown
experiments in an in vitro model. CCK-8, EdU and Transwell
assays showed that the proliferation, migration and invasion
abilities of ccRCC cell lines could be promoted by knocking out
YPEL1, YPEL2 and YPEL5.
There are certain limitations to the present study.
First, YPELs that may play an important role in ccRCC were only
selected for experimental verification according to the results of
database analysis and all YPELs in the verification range were not
included, which should be further improved in future research.
Second, the phenomenon that the high expression of YPEL1 and YPEL2
in ccRCC tissues has an improved prognosis only partially explained
by the combination of a previous study by Cao et al
(29) and the present results. The
results of ccRCC immune infiltration analysis showed that there was
a significant positive correlation between YPEL1 and YPEL2 and
CD8+ and CD4+ T cells and macrophages, which
may indicate that YPEL1 and YPEL2 not only play a role in tumor
cells but also play certain key roles in tumor immunity, such as
the recruitment of related immune cells into tumor sites. Indeed,
this requires further research to verify this hypothesis. Finally,
most of the results of the present study are based on
transcriptomics analysis, and more omics data are needed for
validation. Finally, it was only confirmed that that YPEL1, YPEL2
and YPEL5 can affect the phenotype of ccRCC in vitro, and
their potential mechanisms need to be further studied both in
vivo and in vitro.
Collectively, the present study indicated the
abnormal expression and prognostic value of the YPEL gene family in
ccRCC. Furthermore, the relationship between the YPEL gene family
and immunodeficiency may provide another clinical treatment option.
Additionally, in vitro experiments on ccRCC cell lines were
performed to determine the functions and potential mechanisms of
YPELs. The aforementioned results suggested that the value of
YPEL1, YPEL2 and YPEL5 as potential clinical biomarkers and novel
therapeutic targets in patients with ccRCC deserves further
investigation.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Jiangxi Natural Science
Foundation (grant no. 20181BAB215028), and The National Natural
Science Foundation of China (grant no. 81760458).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZH designed the study. LW and ZZ performed the
bioinformatics analyses and wrote the manuscript. XZ and JW
performed the cell experiments. All authors contributed to the
article and read and approved the final version of the manuscript.
ZH and LW confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics and
Research Committee of the Second Affiliated Hospital of Nanchang
University (Nanchang, China). Written informed consent was provided
from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:313–326.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Singh D: Current updates and future
perspectives on the management of renal cell carcinoma. Life Sci.
264:1186322021. View Article : Google Scholar
|
|
4
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:36982018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitchell TJ, Turajlic S, Rowan A, Nicol D,
Farmery JHR, O'Brien T, Martincorena I, Tarpey P, Angelopoulos N,
Yates LR, et al: Timing the landmark events in the evolution of
clear cell renal cell cancer: TRACERx renal. Cell. 173:611–623.e17.
2018. View Article : Google Scholar
|
|
7
|
Hosono K, Sasaki T, Minoshima S and
Shimizu N: Identification and characterization of a novel gene
family YPEL in a wide spectrum of eukaryotic species. Gene.
340:31–43. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roxström-Lindquist K and Faye I: The
Drosophila gene yippee reveals a novel family of putative zinc
binding proteins highly conserved among eukaryotes. Insect Mol
Biol. 10:77–86. 2001. View Article : Google Scholar
|
|
9
|
Hosono K, Noda S, Shimizu A, Nakanishi N,
Ohtsubo M, Shimizu N and Minoshima S: YPEL5 protein of the YPEL
gene family is involved in the cell cycle progression by
interacting with two distinct proteins RanBPM and RanBP10.
Genomics. 96:102–111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Truong L, Zheng YM, Song T, Tang Y and
Wang YX: Potential important roles and signaling mechanisms of
YPEL4 in pulmonary diseases. Clin Transl Med. 7:162018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Oki K, Plonczynski MW, Gomez-Sanchez EP
and Gomez-Sanchez CE: YPEL4 modulates HAC15 adrenal cell
proliferation and is associated with tumor diameter. Mol Cell
Endocrinol. 434:93–98. 2016. View Article : Google Scholar
|
|
12
|
Berberich SJ, Todd A and Tuttle R: Why
YPEL3 represents a novel tumor suppressor. Front Biosci (Landmark
Ed). 16:1746–1751. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li S, Sun MY and Su X: MiR-885-5p promotes
gastric cancer proliferation and invasion through regulating YPEL1.
Eur Rev Med Pharmacol Sci. 23:7913–7919. 2019.
|
|
14
|
Abiatari I, Kiladze M, Kerkadze V, Friess
H and Kleeff J: Expression of YPEL1 in pancreatic cancer cell lines
and tissues. Georgian Med News. 60–62. 2009.
|
|
15
|
Wu X: Up-regulation of YPEL1 and YPEL5 and
down-regulation of ITGA2 in erlotinib-treated EGFR-mutant non-small
cell lung cancer: A bioinformatic analysis. Gene. 643:74–82. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tuttle R, Simon M, Hitch DC, Maiorano JN,
Hellan M, Ouellette J, Termuhlen P and Berberich SJ:
Senescence-associated gene YPEL3 is downregulated in human colon
tumors. Ann Surg Oncol. 18:1791–1796. 2011. View Article : Google Scholar
|
|
17
|
Zhang J, Wen X, Ren XY, Li YQ, Tang XR,
Wang YQ, He QM, Yang XJ, Sun Y, Liu N and Ma J: YPEL3 suppresses
epithelial-mesenchymal transition and metastasis of nasopharyngeal
carcinoma cells through the Wnt/β-catenin signaling pathway. J Exp
Clin Cancer Res. 35:1092016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou D, Tang W, Xu Y, Xu Y, Xu B, Fu S,
Wang Y, Chen F, Chen Y, Han Y and Wang G: METTL3/YTHDF2 m6A axis
accelerates colorectal carcinogenesis through epigenetically
suppressing YPEL5. Mol Oncol. 15:2172–2184. 2021. View Article : Google Scholar
|
|
19
|
Kelemen LE, Wang X, Fredericksen ZS,
Pankratz VS, Pharoah PD, Ahmed S, Dunning AM, Easton DF, Vierkant
RA, Cerhan JR, et al: Genetic variation in the chromosome 17q23
amplicon and breast cancer risk. Cancer Epidemiol Biomarkers Prev.
18:1864–1868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Newman AM, Liu CL, Green MR, Gentles AJ,
Feng W, Xu Y, Hoang CD, Diehn M and Alizadeh AA: Robust enumeration
of cell subsets from tissue expression profiles. Nat Methods.
12:453–457. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Velusamy T, Palanisamy N, Kalyana-Sundaram
S, Sahasrabuddhe AA, Maher CA, Robinson DR, Bahler DW, Cornell TT,
Wilson TE, Lim MS, et al: Recurrent reciprocal RNA chimera
involving YPEL5 and PPP1CB in chronic lymphocytic leukemia. Proc
Natl Acad Sci USA. 110:3035–3040. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kelley KD, Miller KR, Todd A, Kelley AR,
Tuttle R and Berberich SJ: YPEL3, a p53-regulated gene that induces
cellular senescence. Cancer Res. 70:3566–3575. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Farlie P, Reid C, Wilcox S, Peeters J,
Reed G and Newgreen D: Ypel1: A novel nuclear protein that induces
an epithelial-like morphology in fibroblasts. Genes Cells.
6:619–629. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:(Database Issue).
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Braun DA, Hou Y, Bakouny Z, Ficial M,
Sant' Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA,
Elagina L, et al: Interplay of somatic alterations and immune
infiltration modulates response to PD-1 blockade in advanced clear
cell renal cell carcinoma. Nat Med. 26:909–918. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim MC, Jin Z, Kolb R, Borcherding N,
Chatzkel JA, Falzarano SM and Zhang W: Updates on immunotherapy and
immune landscape in renal clear cell carcinoma. Cancers (Basel).
13:58562021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cao Y, Jiao N, Sun T, Ma Y, Zhang X, Chen
H, Hong J and Zhang Y: CXCL11 correlates with antitumor immunity
and an improved prognosis in colon cancer. Front Cell Dev Biol.
9:6462522021. View Article : Google Scholar
|