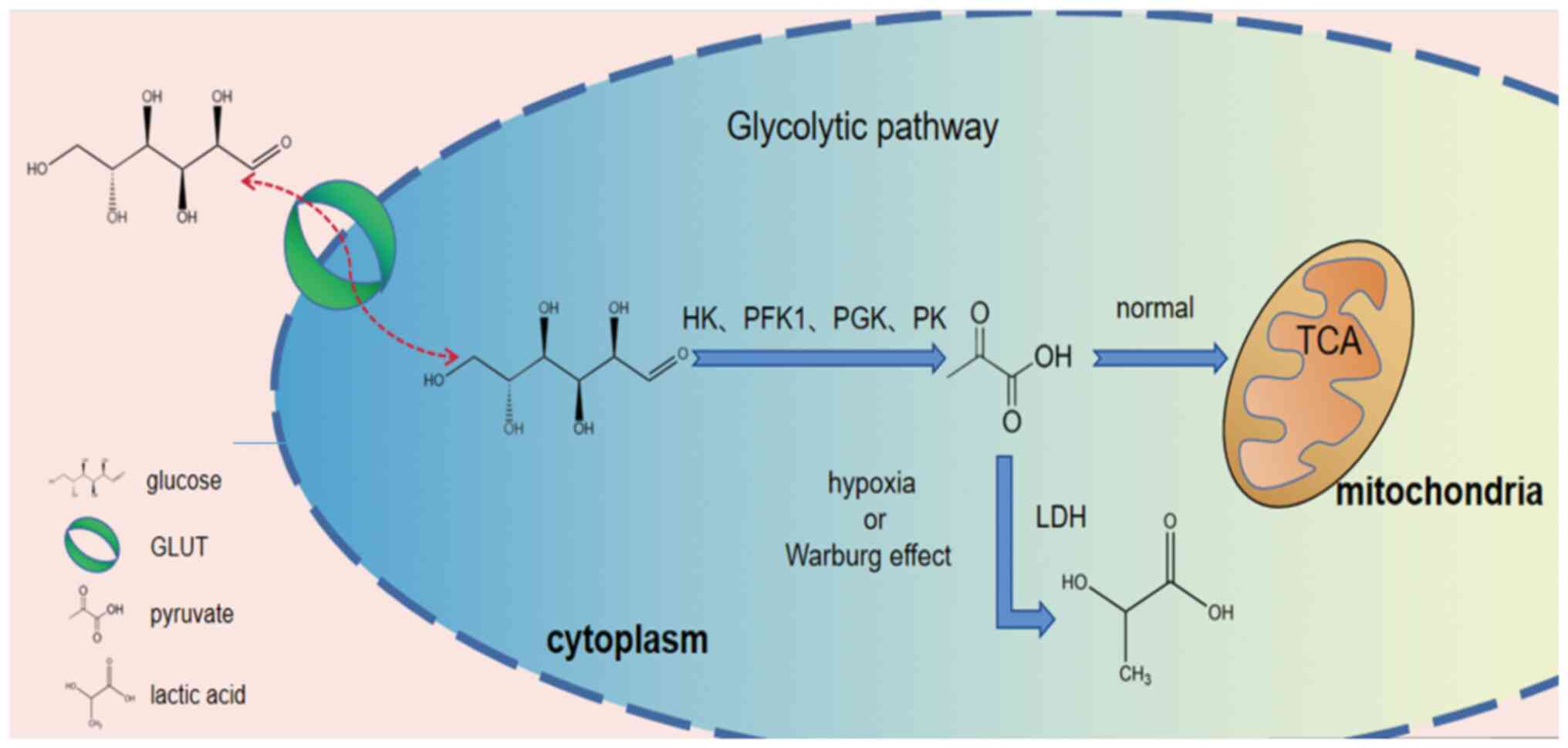
In recent years, due to advancements being made in
research on tumors, the reprogramming of glucose metabolism in
tumor cells has gradually become a focus of research. In normal
cells, glucose-converted pyruvate enters the tricarboxylic acid
cycle (TCA) for oxidative phosphorylation while in an aerobic
environment and undergoes glycolysis to produce lactate only during
hypoxia. However, tumor cells still prefer the glycolytic pathway
for energy even in an aerobic environment (1). This change in the energy pathway
provides certain advantages for the occurrence and development of
tumors. First, aerobic glycolysis produces more adenosine
triphosphate (ATP) than oxidative phosphorylation for consumption
by tumor cells when the substrate is sufficient and the recycling
efficiency is sufficiently high (1,2).
Second, aerobic glycolysis can improve cellular tolerance to
hypoxia and avoid oxidative phosphorylation-induced apoptosis
(3). In addition, glycolytic
intermediates can also provide the raw materials for anabolism, and
the increased production of lactic acid can also decompose and
destroy the cellular matrix around tumor cells, which promotes
tumor cell migration (4). The
scientist, Otto Warburg, discovered the phenomenon of aerobic
glycolysis in tumor cells as early as the 1920s and defined it as
the Warburg effect (5).
For tumor cells to carry out the Warburg effect, two
necessary conditions must be met. On the one hand, due to the
increased consumption of glucose by tumor cells, greater amounts of
glucose need to be taken up from the extracellular environment. On
the other hand, there are numerous enzymatic reactions in the
glycolytic pathway, and these glycolytic enzymes play a crucial
regulatory role. Therefore, the regulation of these two aspects has
become critical for the regulation of the Warburg effect.
Glucose transporters (GLUTs) have been reported to
enable glucose to cross hydrophobic cell membranes into cells,
thereby mediating the use of glucose (6). Previous research has found that the
upregulation of GLUTs promotes the Warburg effect in multiple
cancer types (7). Other studies
have indicated that the transcriptional upregulation of glycolytic
enzymes is closely related to the occurrence of tumors, and that
changes in their activity and stability have a direct impact on the
Warburg effect (8–10). In addition, the abnormal expression
of certain upstream molecules of GLUTs and glycolytic enzymes will
affect them to a certain extent, thereby indirectly regulating the
Warburg effect (11).
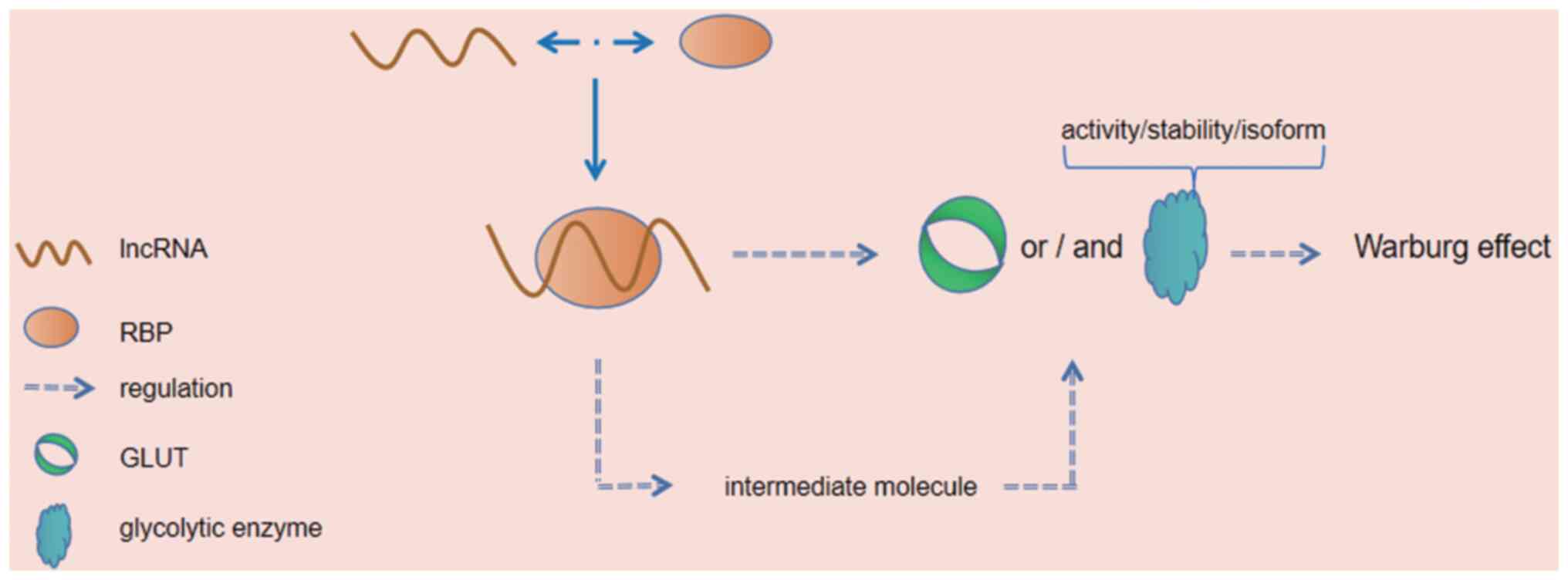
Long non-coding RNAs (lncRNAs) can participate in
tumor metabolism by regulating the metabolic remodeling of various
substances, including the metabolic reprogramming of glucose
(12,13). lncRNAs refer to a class of RNAs
whose transcripts are >200 nucleotides in length and have
limited protein-coding capacity; they are one of the most widely
studied classes of non-coding RNAs (14). It is necessary for lncRNAs to
interact with other cellular macromolecules, such as DNA, proteins
and RNA, to drive the cancer phenotype, and their functional effect
is to interact with proteins. Through lncRNA-protein interactions,
lncRNAs can regulate the functions of proteins directly or regulate
protein interactions between other proteins (15). Such RNA-binding and interacting
proteins are termed RNA-binding proteins (RBPs), which bind to RNA
through one or more globular RNA-binding domains and alter the fate
or function of the bound RNA (16). At present, the intricate
interaction mechanisms of RNA-RBPs have been reported by a large
number of studies (17). The
present review article summarizes and discusses the interaction
between lncRNAs and RBPs in tumor cells, and their direct or
indirect effects on the Warburg effect. Furthermore, the present
review article elaborates on the different mechanisms in the cited
articles. The topics covered in the present review article are
summarized in Figs. 1 and 2, and Table
I.
Tumor cells have a higher demand for glucose as
their metabolic rate is substantially greater than that of normal
cells, which require a constant supply of energy and nutrients. It
has been demonstrated that the first step in energy metabolism is
the uptake of glucose into cells through GLUTs. The gene name of
the GLUT family is solute carrier 2 (SLC2A). The family consists of
14 members, of which GLUT1-4 are the four most well-known subtypes,
and the expression of GLUT family members is increased in various
types of cancer (7). There are
nunerous cases of lncRNAs affecting glucose metabolism by
regulating GLUT, such as lncRNA HOTAIR promoting glycolysis by
upregulating GLUT and activating mammalian target of rapamycin
(mTOR) signaling, whereas the knockdown of HOTAIR suppresses this
effect (18). However, it was
found that when lncRNAs interact with RBPs to modulate the Warburg
effect through the modulation of GLUTs, the predominantly regulated
isoform is GLUT1 (19,20).
It has been found that lncRNA CASC11 interacts with
eukaryotic translation initiation factor 4A3 (EIF4A3) in
hepatocellular carcinoma (HCC) and modulates the Warburg effect and
tumor progression by affecting the expression of GLUT1 (19). EIF4A3 has been reported to be an
important exon junction complex involved in the development of mRNA
secondary structure in the 5′untranslated region (UTR) region and
promotes the initiation of protein translation (21,22).
Mechanistically, the interaction of CASC11 with EIF4A3 leads to an
increased expression of the downstream transcription factor E2F1
due to its enhanced mRNA stability, which in turn enhances GLUT1
expression and aerobic glycolysis. Following the silencing of
CASC11, GLUT1 expression was found to be downregulated, and tumor
progression was also significantly inhibited (19). E2F1 mentioned is an important
promoter for cells to enter S phase, can promote the reprogramming
of glycolysis and the energy metabolism of tumor cells (23), and plays a critical role in the
occurrence and development of various malignant tumors (24,25),
as well as being able to function directly as an RBP. In lung
adenocarcinoma, lncRNA Gas6-AS1 is downregulated as a tumor
suppressor (20). It has been
found that Gas6-AS1 directly interacts with E2F1 and downregulates
GLUT1 transcription by inhibiting the binding of E2F1 to the GLUT1
promoter region, thereby inhibiting tumor progression and the
Warburg effect (20).
The activity and stability of glycolytic enzymes
directly affect the entire glycolytic metabolic pathway, and they
are categorized as rate-limiting enzymes according to whether they
can affect the overall speed of the metabolic pathway. Hexokinase
(HK) is the first rate-limiting enzyme in aerobic glycolysis,
catalyzing the conversion of glucose to glucose-6-phosphate
(G-6-P). It has four subtypes, HK1, HK2, HK3 and HK4 (11,26),
of which HK2 is more dominant than the other subtypes in promoting
aerobic glycolysis (27). The
expression level of HK2 has been found to be significantly
increased in a variety of cancer tissues, and the silencing of HK2
can effectively reduce the level of aerobic glycolysis and promote
the transformation of the metabolic mode of cancer cells to
oxidative phosphorylation (28–30).
Phosphofructokinase-1 (PFK1) is the second rate-limiting enzyme in
glycolysis, which catalyzes fructose-6-phosphate (F-6-P) to
fructose-1,6-diphosphate (F-1,6-BP) through the use of ATP(31). PFK1 exists as three isoforms
(PFK-M, PFK-P and PFK-L) in mammals, and the proportions of these
isoforms may vary in tissues with varying metabolic needs (32). Fully activated PFK1 exists as a
tetramer, and the formation and stabilization of tetramers largely
affects the rate of glycolytic flux (33). The third rate-limiting enzyme in
glycolysis is pyruvate kinase (PK), which catalyzes the
dephosphorylation of phosphoenolpyruvic acid (PEP) to generate ATP
and pyruvate, including the four subtypes of PKM1, PKM2, PKR and
PKL (34). Previous research has
demonstrated that PKM2 isoforms are highly upregulated in tumor
cells and are associated with poor prognosis (35). There are two forms of PKM2: One is
a tetramer, which is located in the cytoplasm, has high catalytic
activity and can rapidly convert PEP to pyruvate, generating more
glycolytic flux and ATP (36),
while the other is a monomer or dimer that can translocate into the
nucleus, has lower catalytic activity, and acts as a coactivator of
several transcription factors (34,37).
In addition, the regulatory effects of enzymes that
are not rate-limiting on glycolysis cannot be ignored.
Phosphoglycerate kinase (PGK) mainly catalyzes the formation of ATP
in the aerobic glycolysis pathway. It has two isoforms: PGK1 and
PGK2. PGK1 is ubiquitously expressed in all cells and catalyzes the
reversible transfer of phosphate groups from
1,3-bisphosphoglycerate (1,3-BPG) to adenosine diphosphate to
generate 3-phosphate glyceric acid (3-PG) and ATP (38). PGK1 is associated with the
occurrence and progression of various cancers, and it mediates the
ATP produced by glycolysis for use by tumor cells (39). PGK1 and PKM2 are the only two
enzymes that control ATP production during aerobic glycolysis in
cancer cells (40). The final
conversion of pyruvate to lactate depends on lactate dehydrogenase
A (LDHA). The abnormal expression and activation of LDHA are
closely related to a variety of tumors, and the upregulation of
LDHA promotes the malignant progression of tumors by increasing
lactate production, accelerating glycolysis, regulating the
production of reactive oxygen species, and regulating numerous
cancer-related proteins (41).
It is well known that phosphorylation is a main
method of protein post-translational modification. By regulating
the degree of protein phosphorylation, the activity of the protein
can be significantly affected, thereby participating in the
regulation of intracellular biological effects. It has been
reported that fibroblast growth factor receptor 1 (FGFR1) can
directly phosphorylate LDHA, increasing the binding of LDHA to the
substrate nicotinamide adenine dinucleotide dehydrogenase to
enhance its enzymatic activity (42), and the intracellular kinase domain
of FGFR1 can also phosphorylate PKM2, preventing F-1,6-BP binding
to PKM2 and inhibiting active tetramer formation (43). In HCC, lncRNA HULC directly binds
and interacts with LDHA and PKM2, and HULC functions as a linker
molecule that enhances the binding of LDHA and PKM2 to FGFR1, and
increases the phosphorylation levels of LDHA and PKM2 (44). The activity of LDHA is enhanced due
to phosphorylation; however, the activity of PKM2 is reduced due to
the reduction of the tetrameric form, which ultimately contributes
to the Warburg effect and cell proliferation (44). FGFR1 is a membrane receptor of the
tyrosine kinase group and can also function as an RBP to interact
with lncRNAs to regulate the Warburg effect (45). lncRNA CASC8 exerts a tumor
suppressive effect by interacting with FGFR1. Mechanistically, when
CASC8 binds to FGFR1, it inhibits the FGFR1-mediated
phosphorylation of LDHA and attenuates the conversion of pyruvate
to lactate, thereby inhibiting glycolysis and bladder cancer cell
growth (45). In addition, it has
been demonstrated that lncRNA CRYBG3 interacts with LDHA in lung
cancer, and CRYBG3 specifically upregulates the activity of LDHA
and promotes aerobic glycolysis and cell proliferation in tumor
cells (46).
In addition to changes in the activity of glycolytic
enzymes, certain lncRNAs interact with RBPs to regulate the
expression of glycolytic enzymes by affecting the mRNAs of the
enzymes. N6-methyladenosine (M6A) is an mRNA
modification that can regulate the metabolism of RNAs (mRNAs,
miRNAs and lncRNAs) (47–49). Insulin-like growth factor 2
mRNA-binding protein (IGF2BP1/2/3, also known as IMP1/2/3) belongs
to the K homology domain family and is one of the M6A
readers involved in the occurrence and development of cancers by
interacting with different RNAs (50,51).
lncRNA CASC9 has a M6A site in glioblastoma multiforme
(GBM), which is recognized by IGF2BP2 and enhances the stability of
CASC9 (52). CASC9 interacts with
IGF2BP2 to form an IGF2BP2/CASC9 complex, which can bind to the
M6A modification site of HK2, promote the expression of
HK2 by improving the stability of HK2 mRNA, and ultimately promote
the Warburg effect (52). The
circular isoform of lncRNA CDKN2B-AS1, circCDKN2B-AS1, interacts
with IMP3 (IGF2BP3) in cervical cancer, and the recruitment of IMP3
(IGF2BP3) to the 3′UTR of HK2 mRNA stabilizes HK2 mRNA and
upregulates HK2 expression, promoting a malignant cell phenotype
and aerobic glycolysis (53). In
addition, lncRNA HCG22 binds and interacts with human antigen R
(HUR) in bladder cancer; HUR can positively regulate the expression
of polypyrimidine tract-binding protein 1 (PTBP1) (54), and studies have demonstrated that
PTBP1 enhances the Warburg effect in certain types of cancer
(55,56). When HCG22 is overexpressed, it
promotes HUR protein degradation, thereby reducing the expression
level of PTBP1 and blocking the Warburg effect mediated by PTBP1,
and has been demonstrated that this is achieved by downregulating
the mRNA and protein levels of PKM2 (54).
In addition to affecting the state of its mRNA, the
stable expression of glycolytic enzymes is also the key to whether
or not they will be degraded. It has been reported that >80% of
protein degradation is related to the ubiquitin-proteasome pathway
(57). For HK, the lncRNA
LINC00470 activates protein kinase B (AKT) after interacting with
fused in sarcoma protein in GBM (58), and the serine/threonine kinase AKT
is a well-established regulator of glucose metabolism that
stimulates aerobic glycolysis in tumor cells (59–61).
These three proteins work together to form a ternary complex. AKT
has an enhanced activity due to its increased phosphorylation, and
high levels of phosphorylated AKT can inhibit HK1 ubiquitination
and attenuate the degradation rate of HK1 protein, thereby
affecting the progress of glycolysis (58). In addition, lncRNA KCNQ1OT1
directly binds and interacts with HK2, inhibits the ubiquitination
and subsequent degradation of HK2 through the proteasome pathway,
and increases the stability of HK2, thereby promoting aerobic
glycolysis and cell proliferation in colorectal cancer (CRC)
(62). For PKM2, lncRNA AC020978
in non-small cell lung cancer and lncRNA FEZF1-AS1 in CRC both
promote tumor progression and aerobic glycolysis, both of which
directly bind to PKM2 in mechanism and maintain the stability of
PKM2 protein by inhibiting ubiquitin-mediated proteasomal
degradation (63,64). The role of PGK1 enzymes has been
investigated in papillary thyroid carcinoma (PTC), where the
expression of the lncRNA PTCSC3, a tumor suppressor, was found to
be significantly downregulated. It was experimentally verified that
PTCSC3 directly binds to PGK1, inhibits aerobic glycolysis, and
inhibits tumor progression by promoting ubiquitin-mediated
degradation of PGK1 (65).
Additionally, the STIP1 homology and U-box containing protein 1
(STUB1) has been reported to be an E3 ubiquitin ligase that
regulates ubiquitination of multiple substrates (66). In breast cancer cells, STUB1 and
PGK1 function together as RBPs to specifically interact with the
lncRNA LINC00926, resulting in an enhanced ability of STUB1 to
mediate PGK1 ubiquitination, thereby reducing PGK1 expression.
LINC00926 inhibits breast tumor growth and metastasis by inhibiting
the PGK1-mediated Warburg effect and induces a switch in glucose
metabolism from glycolysis to mitochondrial respiration (67).
There are four isoforms of PK, of which PKR and PKL
are expressed from the same gene under the control of two different
promoters, while PKM1 and PKM2 are generated by the alternative
splicing of the PKM gene transcript (68). PKM2 is highly upregulated in cancer
cells, and it has been demonstrated that replacing PKM2 with PKM1
can markedly reduce lactate production in tumor cells and the
overall tumor size, suggesting that the selection of PKM1 or PKM2
is directly related to the metabolic phenotype of the tumor
(69).
Heterogeneous nuclear ribonucleoprotein (hnRNP) is
one of the most crucial and classical regulators of alternative
splicing. lncRNA SNHG6 is highly expressed in CRC, and it has ben
experimentally demonstrated that SNHG6 can interact with hnRNPA1,
specifically target the 3′UTR of PKM pre-mRNA, and induce
hnRNPA1-specific splicing of PKM pre-mRNA, leading to an increased
ratio of PKM2/PKM1, which in turn enhances aerobic glycolysis in
CRC cells and promotes tumor development (70). In addition, Ser/Arg-rich splicing
factor 3 (SRSF3), known as an alternative splicing regulator,
functions as an RBP in various cancer types to play oncogenic roles
(71). SRSF3 has been reported to
be a PKM splicer that induces the conversion of PKM from PKM1 to
PKM2; it induces the conversion of PKM to PKM2 after directly
binding to lncRNA LNCAROD in HCC, enhancing aerobic glycolysis,
tumorigenesis and chemoresistance in HCC (72).
In the process of lncRNAs binding to RBPs to
regulate the tumor Warburg effect, certain studies have found that
GLUT and glycolytic enzymes can be affected simultaneously. lncRNA
SNHG14 interacts with RBP Lin28A with an enhanced stability in
glioma cells. Stable SNHG14 promotes the degradation of downstream
interferon regulatory factor 6 (IRF6) mRNA through the
STAU1-mediated degradation pathway and inhibits the expression of
IRF6 in cells and tissues, thereby disabling the effect of IRF6,
which targets the GLUT1 and PKM2 promoters to repress their
expression, and ultimately promoting aerobic glycolysis and cell
proliferation in gliomas (73).
Among these, IRF6, as a tumor suppressor, has the functions of
regulating innate immunity, cell cycle arrest and tumor biological
behavior (74–76). Additionally, hepatocyte nuclear
factor 4 alpha (HNF4A) was identified in neuroblastoma (NB) as a
transcription factor that promotes the expression of the glycolytic
genes SLC2A1 (GLUT1) and HK2 (77). However, it was found that the
expression of HNF4A in NB was regulated by the transcription
factor, CCCTC-binding factor (CTCF). Mechanistically, lncRNA
HNF4A-AS1 interacts with hnRNPU to promote hnRNPU-mediated CTCF
transactivation, and CTCF then increases HNF4A transcription
through epigenetic regulation and promotes the Warburg effect and
tumor progression through the HNF4A-AS1/hnRNPU/CTCF axis (77).
In summary, the abnormal expression of GLUT and
glycolytic enzymes can directly regulate the Warburg effect in
tumor cells. However, there is also the indirect regulation of the
Warburg effect by affecting the upstream molecules targeting GLUT
and glycolytic enzymes. This chapter focuses on hypoxia inducible
factor-1α (HIF-1α) and c-Myc due to their central regulatory roles
in cancer cell glycolysis (78).
Due to the rapid proliferation and expansion of
cancer cells, there is a phenomenon of hypoxia in the core of tumor
tissue, and this abnormal change in the microenvironment will cause
continuous metabolic stress on tumor cells. HIF-1α is a
transcription factor that is active under specific hypoxic
conditions and is a key molecule for cells to adapt to hypoxic
conditions (79). It is
hydroxylated at proline residues by prolyl hydroxylase under
normoxic conditions, and hydroxylated HIF-1α is recognized and
bound by von Hippel-Lindau (VHL) proteins that function as
ubiquitin E3 ligases, followed by rapid degradation via the
ubiquitin-proteasome pathway (80–83).
However, HIF-1α exhibits an increased stability in hypoxia and
binds to the hypoxia response element of target gene promoters,
leading to the transcription of related genes involved in
overcoming hypoxia effects (32).
According to previous research, HIF-1α is mainly involved in the
regulation of the Warburg effect in the following aspects. First,
HIF-1α can promote the transcription of GLUT1, which promotes
glycolysis by enhancing the uptake of glucose by tumor cells
(84). Second, HIF-1α can control
the transcription of several glycolytic enzymes, such as HK2, PFK1,
PKM2, LDHA, etc., by upregulating the expression of these genes,
thus further increasing the level of glycolysis (85,86).
Finally, HIF-1α can promote the expression of pyruvate
dehydrogenase kinase (PDK1) and inhibit pyruvic dehydrogenase (PDH)
activity, leading to the conversion of pyruvate to lactate and
inhibiting the TCA (87,88). According to previous research,
lncRNA CASC9 binds to HIF-1α as an upstream activator of HIF-1α in
nasopharyngeal carcinoma, enhancing the stability of HIF-1α and
reprogramming cellular glucose metabolism (89). lncRNA LINK-A directly interacts
with tyrosine protein kinase 6 (also known as breast tumor kinase)
and leucine-rich repeat kinase 2 in triple-negative breast cancer,
which activates HIF-1α and prevents its degradation under normoxic
conditions, promoting the enhanced Warburg effect (90). In addition, lncRNA lincRNA-p21 and
lncRNA GHET1 both block VHL-mediated HIF-1α degradation by
interacting with VHL protein in tumors, thereby improving the
protein stability and protein level of HIF-1α, and accelerating
aerobic glycolysis (91,92). The expression of VHL is also
regulated by some indirect mechanisms. It has been reported that
lncRNA EPB41L4A-AS1 is a repressor of the Warburg effect and
interacts with histone deacetylase 2 (HDAC2). In a previous study,
following the knockdown of EPB41L4A-AS1, it was found that the
occupancy of HDAC2 on the VHL promoter was increased; histone
modification then reduced the expression of VHL and finally
activated the HIF-1α pathway to promote glycolysis in tumor cells
(93).
c-Myc is the most common oncogene in human
carcinogenesis and is involved in the regulation of the cell cycle,
cell survival, proliferation and metabolic reprogramming (94). It binds to almost all active
promoters and most enhancers, thereby regulating the expression of
key genes in the process of cell growth (95–97).
The regulatory mechanisms of c-Myc in the Warburg effect are
similar to those of HIF-1α. Studies have indicated that c-Myc can
promote the expression of GLUT1, HK2, PKM2 and LDHA, thereby
increasing the glycolytic flux (87,98,99),
increasing the cellular uptake of glucose and the production of
pyruvate and lactate. c-Myc can also synergistically activate PDK1
with HIF-1α, thereby downregulating PDH and inhibiting the aerobic
oxidation of pyruvate, promoting lactate production, and increasing
the acidity of the extracellular environment (100). It has been demonstrated that
lncRNA FILNC1 is specifically expressed in the kidneys and that
FILNC1 interacts with AU-rich elements/poly(U)-binding/degradation
factor 1 (AUF1) during energy stress (101). AUF1 has been reported to bind to
(A+U)-rich elements within the 3′UTR of c-Myc mRNA and promote its
translation without affecting c-Myc mRNA levels (102). FILNC1 reduces c-Myc protein
levels by sequestering AUF1 from binding to the c-Myc 3′-UTR;
however, this regulatory circuit is dysregulated in renal cancer
with a reduced FILNC1 expression (101). Additionally, IGF2BP1/2/3 all
physically prevent the degradation of c-Myc mRNA by binding to a
consensus sequence containing the ‘GGAC’ M6A core motif
(103). lncRNA LINC00261, a
widely reported tumor suppressor (104–106), interacts with IGF2BP1 in
pancreatic cancer and attenuates the c-Myc-mediated Warburg effect
by regulating c-Myc mRNA stability (107). Similarly, the interaction of
lncRNA FGF13-AS1 with IGF2BP1 in breast cancer shortens the
half-life of c-Myc mRNA and ultimately impairs c-Myc-associated
glycolysis to inhibit tumor progression (78). lncRNA LINRIS and IGF2BP2 bind and
interact in CRC to ensure the stabilization of c-Myc mRNA by
preventing the degradation of IGF2BP2, thereby maintaining the
c-Myc-mediated Warburg effect and cell proliferation (108).
Among studies on the regulation of glucose
metabolism reprogramming following the binding of lncRNAs to RBPs,
some did not involve a clear target for regulating the Warburg
effect. Rather, changes in glucose consumption, extracellular
acidification rates, and lactate production were experimentally
demonstrated to explain their effects on the Warburg effect. In HCC
tissues, lncRNA UCA1 binds to the tumor suppressor Up-frameshift
protein 1 (UPF1), and their expression is inversely correlated. The
knockdown of UPF1 has been found to increase the rate of glucose
consumption and lactate production (109). In addition, it has been reported
that the Obg-like ATPase 1 (OLA1) is an ATP hydrolase that binds to
ATP through an amino acid site (110), which plays a crucial role in the
production of lactate in tumor cells (111). The stability of lncRNA ZFAS1 is
improved after binding to IGF2BP2 in CRC cells, and the binding of
stable ZFAS1 to the OBG-type functional domain of OLA1 enhances its
ATP hydrolysis ability, thereby promoting the accumulation of
lactate and the release of ATP synthesis raw materials, activating
the Warburg effect (51).
Glucose metabolic reprogramming is one of the
recognized metabolic features of tumor cells, and enhanced
glycolysis produces an acidic and hypoxic microenvironment that
promotes tumorigenesis, invasion and metastasis. lncRNAs interact
with RBPs in a variety of tumor cells, directly or indirectly
affecting the Warburg effect by regulating GLUTs, glycolytic
enzymes, or their upstream molecules. However, there are still a
number of studies that have not yet clearly defined the targets
that regulate glycolysis in tumor cells, and further research is
still required to elucidate the underlying molecular mechanisms.
Interfering with the Warburg effect of tumor cells by affecting the
expression of lncRNAs and RBPs or inhibiting GLUTs and glycolytic
enzymes can cut off the energy supply, thereby interfering with
tumor progression. This provides a potential novel direction and
strategy for the targeted therapy of tumors. In the tumor glucose
metabolism pathway, there are numerous different molecular targets
to choose from to inhibit tumor growth and progression, which may
contribute to the advancement of clinical treatment of patients and
the improvement of prognosis.
Not applicable.
The present was supported by the National Natural Science
Foundation of China (grant no. 82160575) and the Outstanding Young
Technological and Innovative Talent Cultivation Project of Zunyi
Municipal Science and Technology Bureau, 2021 (no. 10).
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
WW and KW conceived the review. WW was involved in
the collection of references. WW wrote the manuscript and
constructed the figures. WW and KW checked and revised the
manuscript. WW was responsible for the organization, revision and
submission of this manuscript. Both authors have read and approved
the final manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:703–707. 2008. View Article : Google Scholar
|
|
3
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar
|
|
4
|
Gatenby RA, Gawlinski ET, Gmitro AF,
Kaylor B and Gillies RJ: Acid-mediated tumor invasion: A
multidisciplinary study. Cancer Res. 66:5216–5223. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Koppenol WH, Bounds PL and Dang CV: Otto
Warburg's contributions to current concepts of cancer metabolism.
Nat Rev Cancer. 11:325–337. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mueckler M and Thorens B: The SLC2 (GLUT)
family of membrane transporters. Mol Aspects Med. 34:121–138. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ancey PB, Contat C and Meylan E: Glucose
transporters in cancer-from tumor cells to the tumor
microenvironment. FEBS J. 285:2926–2943. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li L, Liang Y, Kang L, Liu Y, Gao S, Chen
S, Li Y, You W, Dong Q, Hong T, et al: Transcriptional regulation
of the warburg effect in cancer by SIX1. Cancer Cell.
33:368–385.e7. 2018. View Article : Google Scholar
|
|
9
|
Akins NS, Nielson TC and Le HV: Inhibition
of glycolysis and glutaminolysis: An emerging drug discovery
approach to combat cancer. Curr Top Med Chem. 18:494–504. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng Y, Liu P, Wang N, Wang S, Yang B, Li
M, Chen J, Situ H, Xie M, Lin Y and Wang Z: Betulinic acid
suppresses breast cancer metastasis by targeting GRP78-mediated
glycolysis and ER stress apoptotic pathway. Oxid Med Cell Longev.
2019:87816902019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng J, Li J, Wu L, Yu Q, Ji J, Wu J, Dai
W and Guo C: Emerging roles and the regulation of aerobic
glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res.
39:1262020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu C, Li H, Chu F, Zhou X, Xie R, Wei Q,
Yang S, Li T, Liang S and Lü M: Long noncoding RNAs: Key regulators
involved in metabolic reprogramming in cancer (Review). Oncol Rep.
45:542021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Z and Sun X: Non-coding RNAs Operate in
the crosstalk between cancer metabolic reprogramming and
metastasis. Front Oncol. 10:8102020. View Article : Google Scholar
|
|
14
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar
|
|
16
|
Hentze MW, Castello A, Schwarzl T and
Preiss T: A brave new world of RNA-binding proteins. Nat Rev Mol
Cell Biol. 19:327–341. 2018. View Article : Google Scholar
|
|
17
|
Ferre F, Colantoni A and Helmer-Citterich
M: Revealing protein-lncRNA interaction. Brief Bioinform.
17:106–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei S, Fan Q, Yang L, Zhang X, Ma Y, Zong
Z, Hua X, Su D, Sun H, Li H and Liu Z: Promotion of glycolysis by
HOTAIR through GLUT1 upregulation via mTOR signaling. Oncol Rep.
38:1902–1908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song H, Liu Y, Li X, Chen S, Xie R, Chen
D, Gao H, Wang G, Cai B and Yang X: Long noncoding RNA CASC11
promotes hepatocarcinogenesis and HCC progression through
EIF4A3-mediated E2F1 activation. Clin Transl Med. 10:e2202020.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo J, Wang H, Wang L, Wang G, Yao Y, Xie
K, Li X, Xu L, Shen Y and Ren B: lncRNA GAS6-AS1 inhibits
progression and glucose metabolism reprogramming in LUAD via
repressing E2F1-mediated transcription of GLUT1. Mol Ther Nucleic
Acids. 25:11–24. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu WT, Wilczynska A, Smith E and Bushell
M: The diverse roles of the eIF4A family: you are the company you
keep. Biochem Soc Trans. 42:166–172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chan CC, Dostie J, Diem MD, Feng W, Mann
M, Rappsilber J and Dreyfuss G: eIF4A3 is a novel component of the
exon junction complex. RNA. 10:200–209. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu M, Seto E and Zhang J: E2F1 enhances
glycolysis through suppressing Sirt6 transcription in cancer cells.
Oncotarget. 6:11252–11263. 2015. View Article : Google Scholar
|
|
24
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Farra R, Grassi G, Tonon F, Abrami M,
Grassi M, Pozzato G, Fiotti N, Forte G and Dapas B: The role of the
transcription factor E2F1 in hepatocellular carcinoma. Curr Drug
Deliv. 14:272–281. 2017.PubMed/NCBI
|
|
26
|
Lis P, Dylag M, Niedzwiecka K, Ko YH,
Pedersen PL, Goffeau A and Ułaszewski S: The HK2 dependent ‘Warburg
Effect’ and mitochondrial oxidative phosphorylation in cancer:
Targets for effective therapy with 3-Bromopyruvate. Molecules.
21:17302016. View Article : Google Scholar
|
|
27
|
Gong L, Cui Z, Chen P, Han H, Peng J and
Leng X: Reduced survival of patients with hepatocellular carcinoma
expressing hexokinase II. Med Oncol. 29:909–914. 2012. View Article : Google Scholar
|
|
28
|
Wolf A, Agnihotri S, Micallef J, Mukherjee
J, Sabha N, Cairns R, Hawkins C and Guha A: Hexokinase 2 is a key
mediator of aerobic glycolysis and promotes tumor growth in human
glioblastoma multiforme. J Exp Med. 208:313–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wolf A, Agnihotri S and Guha A: Targeting
metabolic remodeling in glioblastoma multiforme. Oncotarget.
1:552–562. 2010. View Article : Google Scholar
|
|
30
|
Patra KC, Wang Q, Bhaskar PT, Miller L,
Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen EL, et al:
Hexokinase 2 is required for tumor initiation and maintenance and
its systemic deletion is therapeutic in mouse models of cancer.
Cancer Cell. 24:213–228. 2013. View Article : Google Scholar
|
|
31
|
Kanai S, Shimada T, Narita T and
Okabayashi K: Phosphofructokinase-1 subunit composition and
activity in the skeletal muscle, liver, and brain of dogs. J Vet
Med Sci. 81:712–716. 2019. View Article : Google Scholar
|
|
32
|
Al Hasawi N, Alkandari MF and Luqmani YA:
Phosphofructokinase: A mediator of glycolytic flux in cancer
progression. Crit Rev Oncol Hematol. 92:312–321. 2014. View Article : Google Scholar
|
|
33
|
Bartrons R, Rodríguez-García A,
Simon-Molas H, Castaño E, Manzano A and Navarro-Sabaté À: The
potential utility of PFKFB3 as a therapeutic target. Expert Opin
Ther Targets. 22:659–674. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van Niekerk G and Engelbrecht AM: Role of
PKM2 in directing the metabolic fate of glucose in cancer: A
potential therapeutic target. Cell Oncol (Dordr). 41:343–351. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shang RZ, Qu SB and Wang DS: Reprogramming
of glucose metabolism in hepatocellular carcinoma: Progress and
prospects. World J Gastroenterol. 22:9933–9943. 2016. View Article : Google Scholar
|
|
36
|
Anastasiou D, Yu Y, Israelsen WJ, Jiang
JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, et al:
Pyruvate kinase M2 activators promote tetramer formation and
suppress tumorigenesis. Nat Chem Biol. 8:839–847. 2012. View Article : Google Scholar
|
|
37
|
Azoitei N, Becher A, Steinestel K, Rouhi
A, Diepold K, Genze F, Simmet T and Seufferlein T: PKM2 promotes
tumor angiogenesis by regulating HIF-1α through NF-κB activation.
Mol Cancer. 15:32016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
He Y, Luo Y, Zhang D, Wang X, Zhang P, Li
H, Ejaz S and Liang S: PGK1-mediated cancer progression and drug
resistance. Am J Cancer Res. 9:2280–2302. 2019.PubMed/NCBI
|
|
39
|
Daly EB, Wind T, Jiang XM, Sun L and Hogg
PJ: Secretion of phosphoglycerate kinase from tumour cells is
controlled by oxygen-sensing hydroxylases. Biochim Biophys Acta.
1691:17–22. 2004. View Article : Google Scholar
|
|
40
|
Hu H, Zhu W, Qin J, Chen M, Gong L, Li L,
Liu X, Tao Y, Yin H, Zhou H, et al: Acetylation of PGK1 promotes
liver cancer cell proliferation and tumorigenesis. Hepatology.
65:515–528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Feng Y, Xiong Y, Qiao T, Li X, Jia L and
Han Y: Lactate dehydrogenase A: A key player in carcinogenesis and
potential target in cancer therapy. Cancer Med. 7:6124–6136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fan J, Hitosugi T, Chung TW, Xie J, Ge Q,
Gu TL, Polakiewicz RD, Chen GZ, Boggon TJ, Lonial S, et al:
Tyrosine phosphorylation of lactate dehydrogenase A is important
for NADH/NAD(+) redox homeostasis in cancer cells. Mol Cell Biol.
31:4938–4950. 2011. View Article : Google Scholar
|
|
43
|
Hitosugi T, Kang S, Vander Heiden MG,
Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et
al: Tyrosine phosphorylation inhibits PKM2 to promote the warburg
effect and tumor growth. Sci Signal. 2:ra732009. View Article : Google Scholar
|
|
44
|
Wang C, Li Y, Yan S, Wang H, Shao X, Xiao
M, Yang B, Qin G, Kong R, Chen R and Zhang N: Interactome analysis
reveals that lncRNA HULC promotes aerobic glycolysis through LDHA
and PKM2. Nat Commun. 11:31622020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu R, Zhong P, Xiong L and Duan L: Long
Noncoding RNA cancer susceptibility candidate 8 suppresses the
proliferation of bladder cancer cells via regulating glycolysis.
DNA Cell Biol. 36:767–774. 2017. View Article : Google Scholar
|
|
46
|
Chen H, Pei H, Hu W, Ma J, Zhang J, Mao W,
Nie J, Xu C, Li B, Hei TK, et al: Long non-coding RNA CRYBG3
regulates glycolysis of lung cancer cells by interacting with
lactate dehydrogenase A. J Cancer. 9:2580–2588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dai F, Wu Y, Lu Y, An C, Zheng X, Dai L,
Guo Y, Zhang L, Li H, Xu W and Gao W: Crosstalk between RNA
m6A Modification and Non-coding RNA Contributes to
Cancer Growth and Progression. Mol Ther Nucleic Acids. 22:62–71.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang J, Liu J, Zhao S and Tian F: NN
6-Methyladenosine METTL3 modulates the proliferation and apoptosis
of lens epithelial cells in diabetic cataract. Mol Ther Nucleic
Acids. 20:111–116. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhong L, He X, Song H, Sun Y, Chen G, Si
X, Sun J, Chen X, Liao W, Liao Y and Bin J: METTL3 Induces AAA
development and progression by modulating
N6-methyladenosine-dependent primary miR34a processing. Mol Ther
Nucleic Acids. 21:394–411. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang J, Chen L and Qiang P: The role of
IGF2BP2, an m6A reader gene, in human metabolic diseases and
cancers. Cancer Cell Int. 21:992021. View Article : Google Scholar
|
|
51
|
Lu S, Han L, Hu X, Sun T, Xu D, Li Y, Chen
Q, Yao W, He M, Wang Z, et al: N6-methyladenosine reader IMP2
stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect:
Implication in colorectal cancer. J Hematol Oncol. 14:1882021.
View Article : Google Scholar
|
|
52
|
Liu H, Qin S, Liu C, Jiang L, Li C, Yang
J, Zhang S, Yan Z, Liu X, Yang J and Sun X: m 6 A reader
IGF2BP2-stabilized CASC9 accelerates glioblastoma aerobic
glycolysis by enhancing HK2 mRNA stability. Cell Death Discov.
7:2922021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang Y, Zhao L, Yang S, Cen Y, Zhu T,
Wang L, Xia L, Liu Y, Zou J, Xu J, et al: CircCDKN2B-AS1 interacts
with IMP3 to stabilize hexokinase 2 mRNA and facilitate cervical
squamous cell carcinoma aerobic glycolysis progression. J Exp Clin
Cancer Res. 39:2812020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiang D, Zhang Y, Yang L, Lu W, Mai L, Guo
H and Liu X: Long noncoding RNA HCG22 suppresses proliferation and
metastasis of bladder cancer cells by regulation of PTBP1. J Cell
Physiol. 235:1711–1722. 2020. View Article : Google Scholar
|
|
55
|
Minami K, Taniguchi K, Sugito N, Kuranaga
Y, Inamoto T, Takahara K, Takai T, Yoshikawa Y, Kiyama S, Akao Y
and Azuma H: MiR-145 negatively regulates Warburg effect by
silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget.
8:33064–33077. 2017. View Article : Google Scholar
|
|
56
|
Taniguchi K, Sakai M, Sugito N, Kumazaki
M, Shinohara H, Yamada N, Nakayama T, Ueda H, Nakagawa Y, Ito Y, et
al: PTBP1-associated microRNA-1 and −133b suppress the Warburg
effect in colorectal tumors. Oncotarget. 7:18940–18952. 2016.
View Article : Google Scholar
|
|
57
|
Wang J and Maldonado MA: The
ubiquitin-proteasome system and its role in inflammatory and
autoimmune diseases. Cell Mol Immunol. 3:255–261. 2006.
|
|
58
|
Liu C, Zhang Y, She X, Fan L, Li P, Feng
J, Fu H, Liu Q, Liu Q, Zhao C, et al: A cytoplasmic long noncoding
RNA LINC00470 as a new AKT activator to mediate glioblastoma cell
autophagy. J Hematol Oncol. 11:772018. View Article : Google Scholar
|
|
59
|
Wang RC, Wei Y, An Z, Zou Z, Xiao G,
Bhagat G, White M, Reichelt J and Levine B: Akt-mediated regulation
of autophagy and tumorigenesis through Beclin 1 phosphorylation.
Science. 338:956–959. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Le Grand M, Berges R, Pasquier E, Montero
MP, Borge L, Carrier A, Vasseur S, Bourgarel V, Buric D, André N,
et al: Akt targeting as a strategy to boost chemotherapy efficacy
in non-small cell lung cancer through metabolism suppression. Sci
Rep. 7:451362017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Polivka J Jr and Janku F: Molecular
targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol
Ther. 142:164–175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen C, Wei M, Wang C, Sun D, Liu P, Zhong
X and Yu W: Long noncoding RNA KCNQ1OT1 promotes colorectal
carcinogenesis by enhancing aerobic glycolysis via hexokinase-2.
Aging (Albany NY). 12:11685–11697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hua Q, Mi B, Xu F, Wen J, Zhao L, Liu J
and Huang G: Hypoxia-induced lncRNA-AC020978 promotes proliferation
and glycolytic metabolism of non-small cell lung cancer by
regulating PKM2/HIF-1α axis. Theranostics. 10:4762–4778. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bian Z, Zhang J, Li M, Feng Y, Wang X,
Zhang J, Yao S, Jin G, Du J, Han W, et al: LncRNA-FEZF1-AS1
promotes tumor proliferation and metastasis in colorectal cancer by
regulating PKM2 signaling. Clin Cancer Res. 24:4808–4819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiang B, Chen Y, Xia F and Li X:
PTCSC3-mediated glycolysis suppresses thyroid cancer progression
via interfering with PGK1 degradation. J Cell Mol Med.
25:8454–8463. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ullah K, Chen S, Lu J, Wang X, Liu Q,
Zhang Y, Long Y, Hu Z and Xu G: The E3 ubiquitin ligase STUB1
attenuates cell senescence by promoting the ubiquitination and
degradation of the core circadian regulator BMAL1. J Biol Chem.
295:4696–4708. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chu Z, Huo N, Zhu X, Liu H, Cong R, Ma L,
Kang X, Xue C, Li J, Li Q, et al: FOXO3A-induced LINC00926
suppresses breast tumor growth and metastasis through inhibition of
PGK1-mediated Warburg effect. Mol Ther. 29:2737–2753. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen M, Zhang J and Manley JL: Turning on
a fuel switch of cancer: HnRNP proteins regulate alternative
splicing of pyruvate kinase mRNA. Cancer Res. 70:8977–8980. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Christofk HR, Vander Heiden MG, Harris MH,
Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL and
Cantley LC: The M2 splice isoform of pyruvate kinase is important
for cancer metabolism and tumour growth. Nature. 452:230–233. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lan Z, Yao X, Sun K, Li A, Liu S and Wang
X: The interaction between lncRNA SNHG6 and hnRNPA1 contributes to
the growth of colorectal cancer by enhancing aerobic glycolysis
through the regulation of alternative splicing of PKM. Front Oncol.
10:3632020. View Article : Google Scholar
|
|
71
|
Zhou Z, Gong Q, Lin Z, Wang Y, Li M, Wang
L, Ding H and Li P: Emerging roles of SRSF3 as a therapeutic target
for cancer. Front Oncol. 10:5776362020. View Article : Google Scholar
|
|
72
|
Jia G, Wang Y, Lin C, Lai S, Dai H, Wang
Z, Dai L, Su H, Song Y, Zhang N, et al: LNCAROD enhances
hepatocellular carcinoma malignancy by activating glycolysis
through induction of pyruvate kinase isoform PKM2. J Exp Clin
Cancer Res. 40:2992021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lu J, Liu X, Zheng J, Song J, Liu Y, Ruan
X, Shen S, Shao L, Yang C, Wang D, et al: Lin28A promotes
IRF6-regulated aerobic glycolysis in glioma cells by stabilizing
SNHG14. Cell Death Dis. 11:4472020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ferretti E, Li B, Zewdu R, Wells V, Hebert
JM, Karner C, Anderson MJ, Williams T, Dixon J, Dixon MJ, et al: A
conserved Pbx-Wnt-p63-Irf6 regulatory module controls face
morphogenesis by promoting epithelial apoptosis. Dev Cell.
21:627–641. 2011. View Article : Google Scholar
|
|
75
|
Rotondo JC, Borghi A, Selvatici R, Magri
E, Bianchini E, Montinari E, Corazza M, Virgili A, Tognon M and
Martini F: Hypermethylation-Induced inactivation of the IRF6 gene
as a possible early event in progression of vulvar squamous cell
carcinoma associated with lichen sclerosus. JAMA Dermatol.
152:928–933. 2016. View Article : Google Scholar
|
|
76
|
Bailey CM, Abbott DE, Margaryan NV,
Khalkhali-Ellis Z and Hendrix MJ: Interferon regulatory factor 6
promotes cell cycle arrest and is regulated by the proteasome in a
cell cycle-dependent manner. Mol Cell Biol. 28:2235–2243. 2008.
View Article : Google Scholar
|
|
77
|
Song H, Li D, Wang X, Fang E, Yang F, Hu
A, Wang J, Guo Y, Liu Y, Li H, et al: HNF4A-AS1/hnRNPU/CTCF axis as
a therapeutic target for aerobic glycolysis and neuroblastoma
progression. J Hematol Oncol. 13:242020. View Article : Google Scholar
|
|
78
|
Ma F, Liu X, Zhou S, Li W, Liu C, Chadwick
M and Qian C: Long non-coding RNA FGF13-AS1 inhibits glycolysis and
stemness properties of breast cancer cells through
FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 450:63–75. 2019.
View Article : Google Scholar
|
|
79
|
Ruan K, Song G and Ouyang G: Role of
hypoxia in the hallmarks of human cancer. J Cell Biochem.
107:1053–1062. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kaelin WG Jr and Ratcliffe PJ: Oxygen
sensing by metazoans: The central role of the HIF hydroxylase
pathway. Mol Cell. 30:393–402. 2008. View Article : Google Scholar
|
|
81
|
Ivan M, Kondo K, Yang H, Kim W, Valiando
J, Ohh M, Salic A, Asara JM, Lane WS and Kaelin WG Jr: HIFalpha
targeted for VHL-mediated destruction by proline hydroxylation:
Implications for O2 sensing. Science. 292:464–468. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Jaakkola P, Mole DR, Tian YM, Wilson MI,
Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji
M, Schofield CJ, et al: Targeting of HIF-alpha to the von
Hippel-Lindau ubiquitylation complex by O2-regulated prolyl
hydroxylation. Science. 292:468–472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Maxwell PH, Wiesener MS, Chang GW,
Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER and
Ratcliffe PJ: The tumour suppressor protein VHL targets
hypoxia-inducible factors for oxygen-dependent proteolysis. Nature.
399:271–275. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yao J, Man S, Dong H, Yang L, Ma L and Gao
W: Combinatorial treatment of Rhizoma Paridis saponins and
sorafenib overcomes the intolerance of sorafenib. J Steroid Biochem
Mol Biol. 183:159–166. 2018. View Article : Google Scholar
|
|
85
|
Marín-Hernández A, Gallardo-Pérez JC,
Ralph SJ, Rodríguez-Enríquez S and Moreno-Sánchez R: HIF-1alpha
modulates energy metabolism in cancer cells by inducing
over-expression of specific glycolytic isoforms. Mini Rev Med Chem.
9:1084–1101. 2009. View Article : Google Scholar
|
|
86
|
Zheng F, Chen J, Zhang X, Wang Z, Chen J,
Lin X, Huang H, Fu W, Liang J, Wu W, et al: The HIF-1α antisense
long non-coding RNA drives a positive feedback loop of HIF-1α
mediated transactivation and glycolysis. Nat Commun. 12:13412021.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kim JW, Tchernyshyov I, Semenza GL and
Dang CV: HIF-1-mediated expression of pyruvate dehydrogenase
kinase: A metabolic switch required for cellular adaptation to
hypoxia. Cell Metab. 3:177–185. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yeung SJ, Pan J and Lee MH: Roles of p53,
MYC and HIF-1 in regulating glycolysis-the seventh hallmark of
cancer. Cell Mol Life Sci. 65:3981–3999. 2008. View Article : Google Scholar
|
|
89
|
Su X, Li G and Liu W: The long noncoding
RNA cancer susceptibility candidate 9 promotes nasopharyngeal
carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 36:394–400.
2017. View Article : Google Scholar
|
|
90
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar
|
|
91
|
Yang F, Zhang H, Mei Y and Wu M:
Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the
Warburg effect. Mol Cell. 53:88–100. 2014. View Article : Google Scholar
|
|
92
|
Liu D and Li H: Long non-coding RNA GEHT1
promoted the proliferation of ovarian cancer cells via modulating
the protein stability of HIF1α. Biosci Rep. 39:2019.
|
|
93
|
Liao M, Liao W, Xu N, Li B, Liu F, Zhang
S, Wang Y, Wang S, Zhu Y, Chen D, et al: LncRNA EPB41L4A-AS1
regulates glycolysis and glutaminolysis by mediating nucleolar
translocation of HDAC2. EBioMedicine. 41:200–213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yoshida GJ: Emerging roles of Myc in stem
cell biology and novel tumor therapies. J Exp Clin Cancer Res.
37:1732018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Dang CV: Gene regulation: Fine-tuned
amplification in cells. Nature. 511:417–418. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Sabò A, Kress TR, Pelizzola M, de Pretis
S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et
al: Selective transcriptional regulation by Myc in cellular growth
control and lymphomagenesis. Nature. 511:488–492. 2014. View Article : Google Scholar
|
|
97
|
Walz S, Lorenzin F, Morton J, Wiese KE,
von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M,
et al: Activation and repression by oncogenic MYC shape
tumour-specific gene expression profiles. Nature. 511:483–487.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Dang CV, Kim JW, Gao P and Yustein J: The
interplay between MYC and HIF in cancer. Nat Rev Cancer. 8:51–56.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Grüning NM, Lehrach H and Ralser M:
Regulatory crosstalk of the metabolic network. Trends Biochem Sci.
35:220–227. 2010. View Article : Google Scholar
|
|
100
|
Kim JW, Gao P, Liu YC, Semenza GL and Dang
CV: Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively
induce vascular endothelial growth factor and metabolic switches
hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol.
27:7381–7393. 2007. View Article : Google Scholar
|
|
101
|
Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y,
Baddour J, Nagrath D, Wood CG, Gu J, Wu X, et al: Energy
stress-induced lncRNA FILNC1 represses c-Myc-mediated energy
metabolism and inhibits renal tumor development. Nat Commun.
8:7832017. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liao B, Hu Y and Brewer G: Competitive
binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat
Struct Mol Biol. 14:511–518. 2007. View Article : Google Scholar
|
|
103
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar
|
|
104
|
Guo C, Shi H, Shang Y, Zhang Y, Cui J and
Yu H: LncRNA LINC00261 overexpression suppresses the growth and
metastasis of lung cancer via regulating miR-1269a/FOXO1 axis.
Cancer Cell Int. 20:2752020. View Article : Google Scholar
|
|
105
|
Yu Y, Li L, Zheng Z, Chen S, Chen E and Hu
Y: Long non-coding RNA linc00261 suppresses gastric cancer
progression via promoting Slug degradation. J Cell Mol Med.
21:955–967. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Yan D, Liu W, Liu Y and Luo M: LINC00261
suppresses human colon cancer progression via sponging miR-324-3p
and inactivating the Wnt/β-catenin pathway. J Cell Physiol.
234:22648–22656. 2019. View Article : Google Scholar
|
|
107
|
Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng
C, Li H, Chen H, Shen B and Deng X: Epigenetic silencing of LncRNA
LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating
miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene.
40:277–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen
YX, Liu J, Luo XJ, Meng Q, Pu HY, et al: LncRNA LINRIS stabilizes
IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer.
Mol Cancer. 18:1742019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhou Y, Li Y, Wang N, Li X, Zheng J and Ge
L: UPF1 inhibits the hepatocellular carcinoma progression by
targeting long non-coding RNA UCA1. Sci Rep. 9:66522019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Koller-Eichhorn R, Marquardt T, Gail R,
Wittinghofer A, Kostrewa D, Kutay U and Kambach C: Human OLA1
defines an ATPase subfamily in the Obg family of GTP-binding
proteins. J Biol Chem. 282:19928–19937. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Whitaker-Menezes D, Martinez-Outschoorn
UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S,
Tsirigos A, Ertel A, Pestell RG, et al: Hyperactivation of
oxidative mitochondrial metabolism in epithelial cancer cells in
situ: Visualizing the therapeutic effects of metformin in tumor
tissue. Cell Cycle. 10:4047–4064. 2011. View Article : Google Scholar : PubMed/NCBI
|