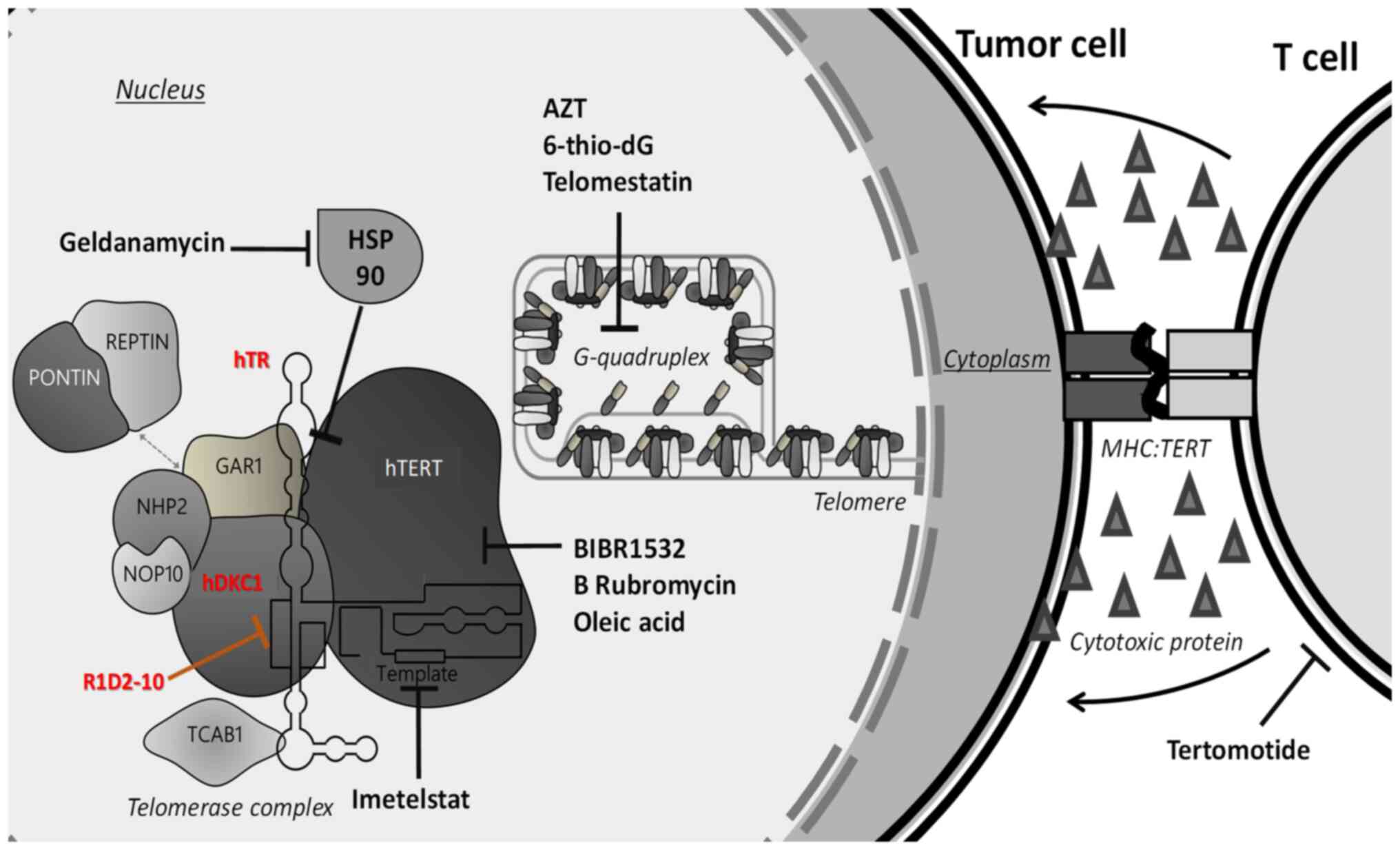
Introduction
Telomerase is one of the primary targets for
developing effective therapies against cancer due to its expression
in most types of tumor, as well as in stem-like tumor cells. As is
shown in Fig. 1, several
strategies were designed to attack positive telomerase cells, by
targeting different sites of the proteins involved in telomere
homeostasis. Additionally, normal cells-including stem cells-have
lower telomerase activity and generally maintain telomeres at
longer lengths in comparison to tumor cells. These properties
confer a benefit that ensures minimum risk for possible telomere
shortening in stem cells, making telomerase a selective target to
cancer cells (1).
Based on experimental evidence that links telomeric
homeostasis with drug resistance and antitumor treatment (2,3), the
development of drugs targeting telomerase represents a promising
tool for antitumor therapy, both alone and in combination with
other treatment. For example, the treatment with
6-thio-2′deoxyguanosine (6-thio-dG), a telomerase substrate
precursor analogue, it is reported as a novel and effective
strategy to treat therapy-resistant melanoma, pediatric brain
tumors and Non-Small Cell Lung Cancer (NSCLC) (4–6).
Furthermore, Wu et al proposed a nanoplatform containing the
specific telomerase inhibitor BIBR1532 as a strategy to reverse
multidrug resistance in breast cancer (7). The telomerase complex is regulated at
multiple levels, by mechanisms such as epigenetic regulation,
transcriptional and post-translational processing, intracellular
compartmentalization, recruitment and substrate accessibility
(2). Considering these processes,
different strategies have been adopted to develop novel telomerase
inhibitor therapies that recognize TERT tumour-associated antigens;
small molecule inhibitors or oligonucleotides that suppress
telomerase activity; disruptors of telomerase regulation or
function such as G-quadruplex stabilizers; targeting TERT gene
expression and utilization of nucleoside analogues to disrupt
elongation of newly extended telomeres (8,9). The
primary aim of antitumor therapy targeting telomerase is to
selectively induce tumor cell death without affecting healthy cells
(10).
Active human telomerase is a ribonucleoprotein
comprising catalytic subunit (human telomerase reverse
transcriptase (hTERT) and template RNA human telomerase RNA (hTR)
(11). However, other proteins are
necessary for in vivo assembly, subcellular trafficking and
telomere recruitment of the functional telomerase holoenzyme. Among
these proteins, dyskerin (hDKC1), which allows correct assembly and
stabilization of mature hTR, is essential for telomerase activity
(11).
Our previous study designed a novel strategy for
rational development of new holoenzyme telomerase assembly
inhibitors based on interruption of the interaction between hDKC1
and hTR. An in silico approach was used to obtain a model of
hDKC1 with accurate model parameters (confidence score, template
modelling score, root mean square deviation and stereochemical
property) to identify candidate compounds with inhibitory effects
on telomerase activity by docking-based virtual screening (DBVS)
(11). The present study aimed to
characterize the effect of two of these candidates: R1D2-10, a
carboxy-phenyl-benzamide, and R1D2-15, an indoquinoline. Both
compounds showed greatest telomerase inhibition at the lowest
concentration. Here, a breast cancer model was selected to validate
these candidates as telomerase is a potential target for diagnosis
and therapy in breast cancer (12). MDA MB 231 breast cancer cell line
was previously used (11) to
determine the effect of these compounds on telomere shortening,
senescence and apoptosis; the aforementioned study determined that
R1D2-10 showed the best performance as a telomerase inhibitor. The
present study aimed to identify novel drugs with potential clinical
use for cancer treatment. Analogs of the parental compound R1D2-10
were designed and interaction with the target, as well as
absorption, distribution, metabolism, excretion (ADME) properties,
toxicity, off-target interaction, and chemical diversity were
assessed.
Materials and methods
Drug preparation
R1D2-10 and R1D2-15 lyophilized compounds (10 mg)
were purchased from Enamine LTD, Ukraine. Each compound was
solubilized in sterile DMSO within a laminar flow to obtain 10 mM
stock solution.
Breast cancer cell lines
Breast cancer is a complex and heterogenous disease
classified into at least five subtypes: Luminal A and B, human
epidermal growth factor receptor 2, basal and normal (13). To represent subtypes three breast
cancer cell lines were selected: Human mammary adenocarcinoma MDA
MB 231, which is a highly aggressive, invasive and poorly
differentiated claudin-low triple-negative breast cancer model; MDA
MB 468, which is less aggressive and belongs to basal type and
MCF-7, a poorly-aggressive and non-invasive Luminal A subtype.
Culture conditions
All cell lines were obtained from the American Type
Culture Collection (cat. nos. HTB-26, HTB-132 and HTB-22). Cells
were grown in DMEM (Thermo Fisher Scientific, Inc.) supplemented
with 10% heat-inactivated FBS (Sigma-Aldrich; Merck KGaA), 2 mM
glutamine and 80 µg/ml gentamicin at 37°C in 5% CO2
atmosphere. Cell cultures were routinely sub-cultured by
trypsinization according to the manufacturer's instructions. For
MDA MB 468 culture, Gibco MEM-Non-essential amino acids 100X
(Thermo Fisher Scientific, Inc.) was added to the medium to a final
concentration of 1X. DMSO was used as a vehicle. Mycoplasma testing
was performed for the cell lines by Indirect HoechstStain (cat. no.
62249, Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions.
Cell proliferation assay
MDA MB 231, MDA MB 468 and MCF-7 cells
(1×104) were plated in 96-well plates. After 24 h, cells
were treated with different concentrations of compound R1D2-10 or
R1D2-15 (0.0,3.1,6.3,12.5,25.0,50.0 µM) for 72 h at 37°C. Cell
survival was measured by colorimetric MTT assay (Sigma-Aldrich;
Merck KGaA). Formazan was dissolved in DMSO and the plate was read
at 570 nm in a microplate reader. The concentration producing 50%
inhibition (IC50), maximmum response (Emax)
and goodness of fit (R2) values were determined by
non-linear regression function of GraphPad Prism 6®
(GraphPad Software, Inc.). A total of of three independent
experiments was performed.
Determination of telomerase
activity
Telomerase activity was determined by real-time
quantitative telomerase repeat amplification protocol (RQ-TRAP)
assay using SYBR-Green (StepOne™ System; Thermo Fisher Scientific,
Inc.). MDA MB 231, MDA MB 468 and MCF-7 cells in logarithmic growth
phase were harvested and washed once with PBS. Cells
(2×106) were transferred to 1.5 ml conical tubes and
centrifuged for 8 min at 450 × g at room temperature. The pellet
was lysed with 200 µl 3-[(3-Cholamidopropyl)
dimethylammonio]-1-propanesulfonate buffer 0.5% p/v with RNaseOUT
(Thermo Fisher Scientific, Inc.) and protease inhibitor
(Sigma-Aldrich; MercK KGaA), quantified by Micro BCA Protein Assay
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions and stored at −20°C until use. RQ-TRAP assay was
performed in a final volume of 10 µl, using 2 µl lysate as a
template, Power SYBR Green Master Mix 1X (Thermo Fisher Scientific,
Inc.), 250 nM alternative complementary primer
(5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′) and 800 nM telomerase
substrate primer (5′-AATCCGTCGAGCAGAGTT-3′). Following 20 min
incubation at 25°C, thermocycling conditions were as follows:
Initial denaturation at 90°C for 10 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 10 sec. The reaction ended with melt
curve analysis in which the temperature was increased from 55 to
95°C at a linear rate of 0.2°C/sec. StepOne Software v2.3 (Thermo
Fisher Scientific, Inc.) was used to analyze results.
Relative telomere length
determination
Telomere length was determined as described by
Cawthon (14). According to the
manufacturer's instructions, high molecular weight DNA from control
(DMSO) and treated MDA MB 231 cells was extracted using Pure gDNA
kit (Productos Bio-Lógicos). Extracted DNA was quantified at 230,
260 and 280 nm absorbance using NanoDrop 1000 (Thermo Fisher
Scientific, Inc.) spectrophotometer. Specific primers for the
repetitive telomere sequence were used to quantify telomere length.
Specific primers for the single copy gen ribosomal protein lateral
stalk subunit P0 (rplp0) were used to determine the genome copies
on the sample. Primer sequences and concentrations were as follows:
Telomere length (500 nM) forward,
5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ and reverse,
5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′ and single copy gen
rplp0 (250 nM) forward, 5′-CAGCAAGTGGGAAGGTGTAATCC-3′ and
reverse, 5′-CCCATTCTATCATCAACGGGTACAA-3′. The PCR thermocycling
conditions were as follows: Initial denaturation at 90°C for 10
min, followed by 40 two-step PCR cycles at 95°C for 15 sec and 60°C
for 10 sec. Results were analyzed using v2.3 StepOne®
software (Thermo Fisher Scientific, Inc.).
RNA extraction and copy DNA
synthesis
Total RNA was extracted from control (DMSO) and
treated MDA MB 231 cells using Bio-Zol (Productos Bio-Lógicos)
according to the manufacturer's instructions. The extracted RNA was
quantified at 230, 260, and 280 nm absorbance using NanoDrop 1000
(Thermo Fisher Scientific, Inc.) spectrophotometer. DNA was
synthesized from 1 µg total RNA in 20 µl reaction mix using
oligodT18 (PB-L) and Superscript III (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions.
Evaluation of senescence
Senescence was evaluated in control (DMSO) and MDA
MB 231 cells treated with R1D2-10 and R1D2-15. Differential
expression of replicative senescence-associated genes, such as
glb1 (galactosidase β1) and p16ink4a
(cyclin-dependent kinase inhibitor 2A), was evaluated by qPCR (ΔΔCq
method) (15) using SYBR-Green
(StepOne™ System; Thermo Fisher Scientific, Inc.). Primer sequences
were as follows: Glb1 (900 nM) foward,
5′-CCACGATCGAGCATATGTTG-3′ and reverse, 5′-CAGAGTGGCTCCAGCTTTC-3′
and p16ink4a (600 nM) forward, 5′-GCGATGTCGCACGGTACCTG-3′
and reverse, 5′-GGCATGGTTACTGCCTCTGG-3′. Thermocycling conditions
were as follows: Initial denaturation at 90°C for 10 min, followed
by 40 two-step PCR cycles at 95°C for 15 sec and 60°C for 10 sec.
Results were analyzed with v2.3 StepOne® software
(Thermo Fisher Scientific, Inc.), using hprt1 (hypoxanthine
phosphoribosyltransferase 1) as a endogenous control (300 nm;
forward, 5′-AACGTCTTGCTCGAGATGTG-3′ and reverse,
5′-GCTTTGATGTAATCCAGCAGG-3′). Quantitative determination of
SA-β-gal (Senescence-associated beta-galactosidase) activity was
performed using Senescence β-Galactosidase Staining kit (Cell
Signaling Technology, Inc.) according to the manufacturer's
instructions. Quantification was made by manually counting
positive-stained cells in four randomly selected fields/well in a
inverted light microscope at a magnification of 100× (Leica
Microsystems). The percentage of positive cells is expressed as the
mean ± SD.
Evaluation of apoptosis
Apoptosis was determined based on differential
expression of apoptosis-associated bcl2 and bax in
treated and control MDA MB 231 cells via qPCR (ΔΔCq method)
(15) using SYBR-Green (StepOne™
System; Thermo Fisher Scientific, Inc.). Primer sequences were as
follows: bcl-2 (125 nM) forward,
5′-GGATGCCTTTGTGGAACTGTAC-3′ and reverse,
5′-TTCACTTGTGGCCCAGATAGG-3′ and bax (500 nM) forward,
5′-GGCCGGGTTGTCGCCCTTTT-3′ and reverse, 5′-CCGCTCCCGGAGGAAGTCCA-3′.
Thermocycling conditions were as follows: Initial denaturation at
90°C for 10 min, followed by 40 two-step PCR cycles at 95°C for 15
sec and 60°C for 10 sec. Results were analyzed with v2.3 StepOne
(Thermo Fisher Scientific, Inc.) software using hprt1 as
endogenous control (300 nm; forward, 5′-AACGTCTTGCTCGAGATGTG-3′ and
reverse, 5′-GCTTTGATGTAATCCAGCAGG-3′). MDA MB 231 apoptosis was
determined via Caspase 3 activity measurement using CaspACE™ Assay
System according to the manufacturer's instructions (Promega
Corporation).
Generation of novel ligands and
screening
With candidate R1D2-10 as the seed compound, a
library of 100 novel derivatives was generated using the LigDream
web server with its default parameters (16). LigDream is a machine learning-based
software that uses convolutional and recurrent neural networks to
generate Simplified Molecular Input Line Entry Specification
(SMILES) sequences of novel compounds based on characteristics of
the seed molecule (playmolecule.com/LigDream/).
SMILES sequences were converted to Protein Data Bank
(PDB) format using RDKit (rdkit.org/), a specific cheminformatics
Python library (17). The 3D
structures of the molecules were optimized using the UFF (universal
force field) (18) and converted
to PDB Partial Charge & Atom Type format using the
AutoDockTools software suite Version 1.5.7 (19).
Docking assay of novel ligands was performed with
the same parameters in DBVS as previously described (11). The interaction between ligands and
protein was analyzed using Protein-Ligand Interaction Profile
(PLIP) software Version 2.2.2 (20) to identify which novel ligands bound
to hDKC1 with a better docking energy than candidate R1D2-10 while
also establishing strong interactions with hDKC1 key residue
(plip-tool.biotec.tu-dresden.de/plip-web/plip/index).
In vitro R1D2-10 cell viability
evaluation in normal primary culture derived from p4 neonatal mouse
brain
Isolation and plating of mixed cortical cells were
carried out following the procedure described by Schildge et
al (21). A total of
2.5×104 cells from primary culture was seeded in DMEM
(Thermo Fisher Scientific, Inc.) supplemented with 10%
heat-inactivated FBS (Sigma-Aldrich; Merck KGaA), 2 mM glutamine
and 80 µg/ml gentamicin at 37°C in 5% CO2 atmosphere.
Following 72 h treatment with compound R1D2-10, viable cells were
determined by trypan blue 0,4% p/v staining (Thermo Fisher
Scientific, Inc.) following supplier's recommendations. Then,
stained cells were load in hemocytometer and manually counting (4
wells/condition) in an inverted light microscope at a magnification
of 200 X (Leica Microsystems GmbH). Data were analyzed by two-way
ANOVA followed by Dunnett's test and are presented as the mean ±
SEM (n=3).
ADME properties, off-target
interaction and toxicity prediction
Compound physicochemical and pharmacokinetic
properties were analyzed using ADME descriptor algorithm
(https://biosig.lab.uq.edu.au/pkcsm/prediction). The
online server pkCSM ADME was used to analyze the ADME descriptors
such as absorption, solubility, cytochrome CYP2D6, plasma protein
binding, distribution volume (VDss), fraction unbound (human) and
central nervous system (CNS) and blood-brain barrier membrane
permeability (22). Off-target
protein interaction was analyzed using Swiss Target Prediction tool
2019 version (23). Toxicity
parameters (hepatotoxicity, skin sensitization, mutagenic and
tumorigenic, reproductive toxicity, irritant) were analyzed by
using DataWarrior software version 5.5.0 (24). The prediction process relies on a
precomputed set of structural fragment that give rise to toxicity
alerts in case they are encountered in the structure drawn.
(openmolecules.org/propertyexplorer/toxicity-assessment.html).
Chemical diversity of compounds
Similarity of candidate analogs was estimated to
identify chemical diversity. Molecular fingerprints encode the
presence or absence of structural features in a molecule as a bit
vector (25,26). Once the molecular fingerprint
describing two molecules is generated, a similarity coefficient is
computed. The present study used Tanimoto coefficient of similarity
(27,28), which ranges from 0 to 1, where 1
implies identity. Thus, low Tanimoto coefficient of similarity
implies dissimilarity.
The present study used four types of fingerprint to
estimate the similarity between molecules using Tanimoto
coefficient of similarity. First, 1,024-bit Morgan 2D fingerprints
with a radius of 2 were calculated using RDKit (17). Furthermore, Murcko scaffold
(29) of each molecule was
extracted using RDKit and 1024-bit Morgan 2D fingerprint was
calculated. Molecular access system (MACCS) 166 keys (30) were calculated using RDKit. MACCS
keys are a coarse-grain fingerprint where each key is associated
with a SMILES arbitrary target specification pattern (31). Lastly, the 1,024-bit Extended 3D
fingerprints (32) were calculated
for the best conformer following UFF energy minimization.
Statistical analysis
All data are presented as the mean ± SEM.
Comparisons between >2 groups, the significance of differences
was determined using one-way ANOVA followed by post hoc Dunnett's
test. In case were analyzed only two groups were determined by
unpaired T-test. The analyses were made using GraphPad Prism
6® (GraphPad Software, Inc.). P<0.05 was considered
to indicate a statistically significant difference.
Results
R1D2-10 and R1D2-15 inhibit MDA MB 231
cell proliferation
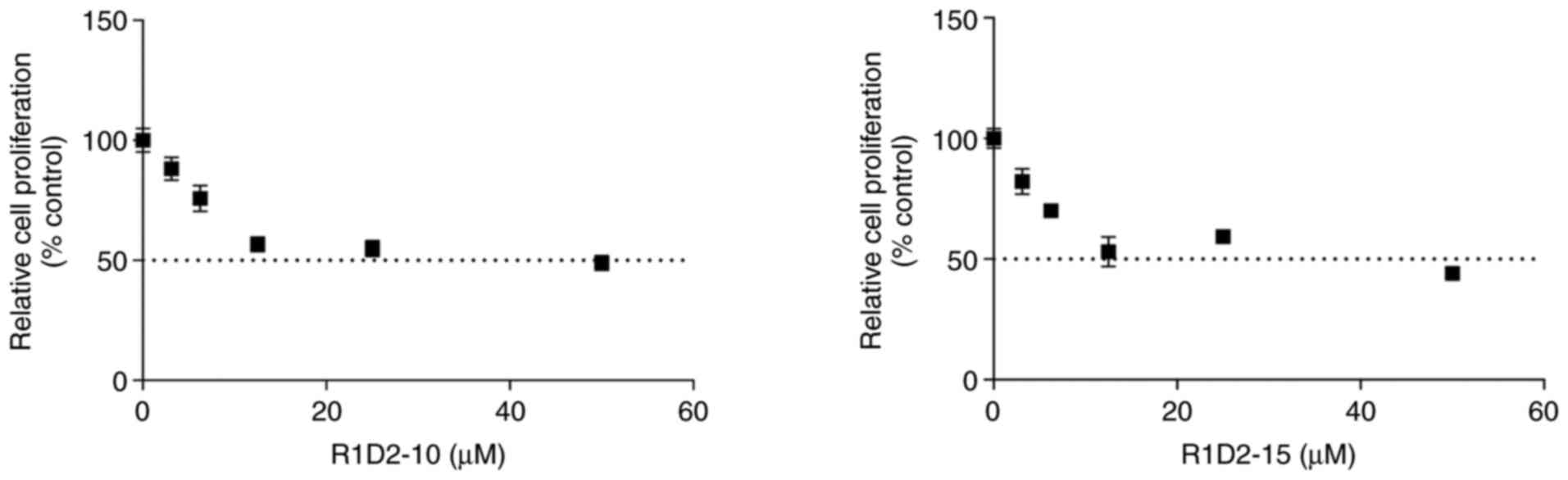
To define concentrations for long-term treatment
assay, proliferation assay was performed on MDA MB 231 cells. At
low concentrations (3–25 µM) there was a cytotoxic effect in a
concentration-dependent manner for both compounds (data not shown).
For compound R1D2-10 and R1D2-15, IC50 value was 9.52
and 12.95 µM, respectively (Fig.
2). Also, maximum response (Emax) and goodness of
fit (R2) were calculted for both curves, obataing a
Emax: 60.15% and R2: 0.76 for R1D2-10 and a
Emax: 62.71%, R2: 0.81 for R1D2-15. Since
chronic treatment requires use of the non-cytotoxic concentrations
to evaluate the long-term effect of the active agents, 2 µM was
selected for both compounds as this concentration was below
IC25 and exhibited minimal cytotoxic effects.
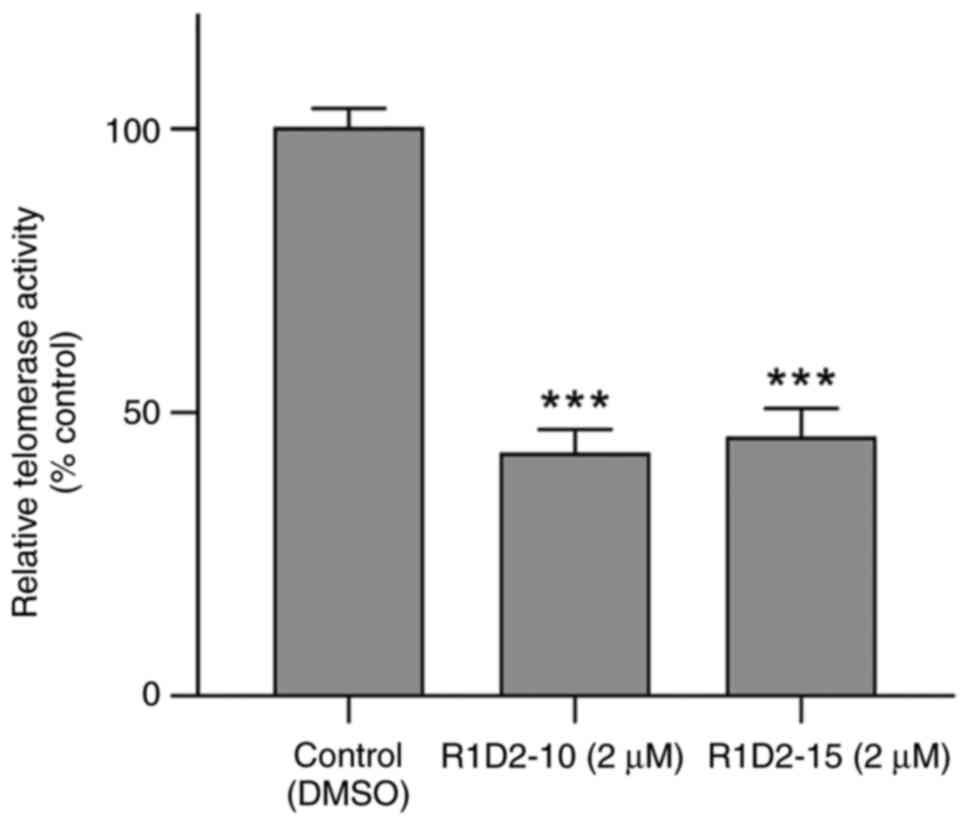
R1D2-10 and R1D2-15 inhibit telomerase
activity of MDA MB 231 cells
Telomerase activity was determined following 48 h
treatment with potential inhibitors. R1D2-10 and R1D2-15 inhibited
telomerase activity of by 57.3 and 54.6%, respectively (P<0.001;
Fig. 3). Therefore 2 µM was used
for chronic exposure of cells to compounds R1D2-10 and R1D2-15 to
evaluate parameters as treatment progresses.
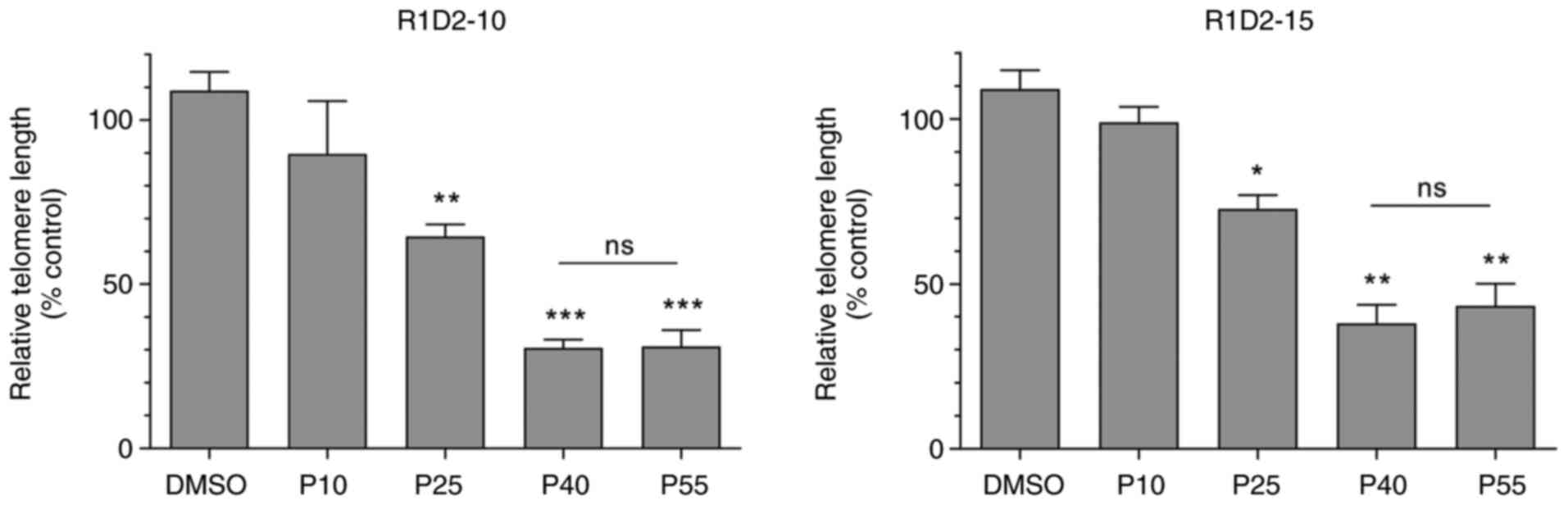
R1D2-10 and R1D2-15 chronic treatment
cause telomere shortening in MDA MB 231 cell line
Telomere length following treatment was determined
at 10, 25, 40 and 55 cell passages. R1D2-10 and R1D2-15 chronic
exposure, exhibit a downward trend in telomere length until passage
40 (Fig. 4). There was no
significant variation between passages 40 and 55 in both to R1D2-10
as R1D2-15 treated cells. Chronic treatment with R1D2-10 or R1D2-15
exhibited a decrease in telomere length of 66 and 43%,
respectively, after 55 passages (P<0.001).
Considering that telomere shortening triggers
senescence and apoptosis (33–35),
entry into these processes was evaluated following cell
treatment.
R1D2-10 treatment induces senescence
in MDA MB 231 cell line
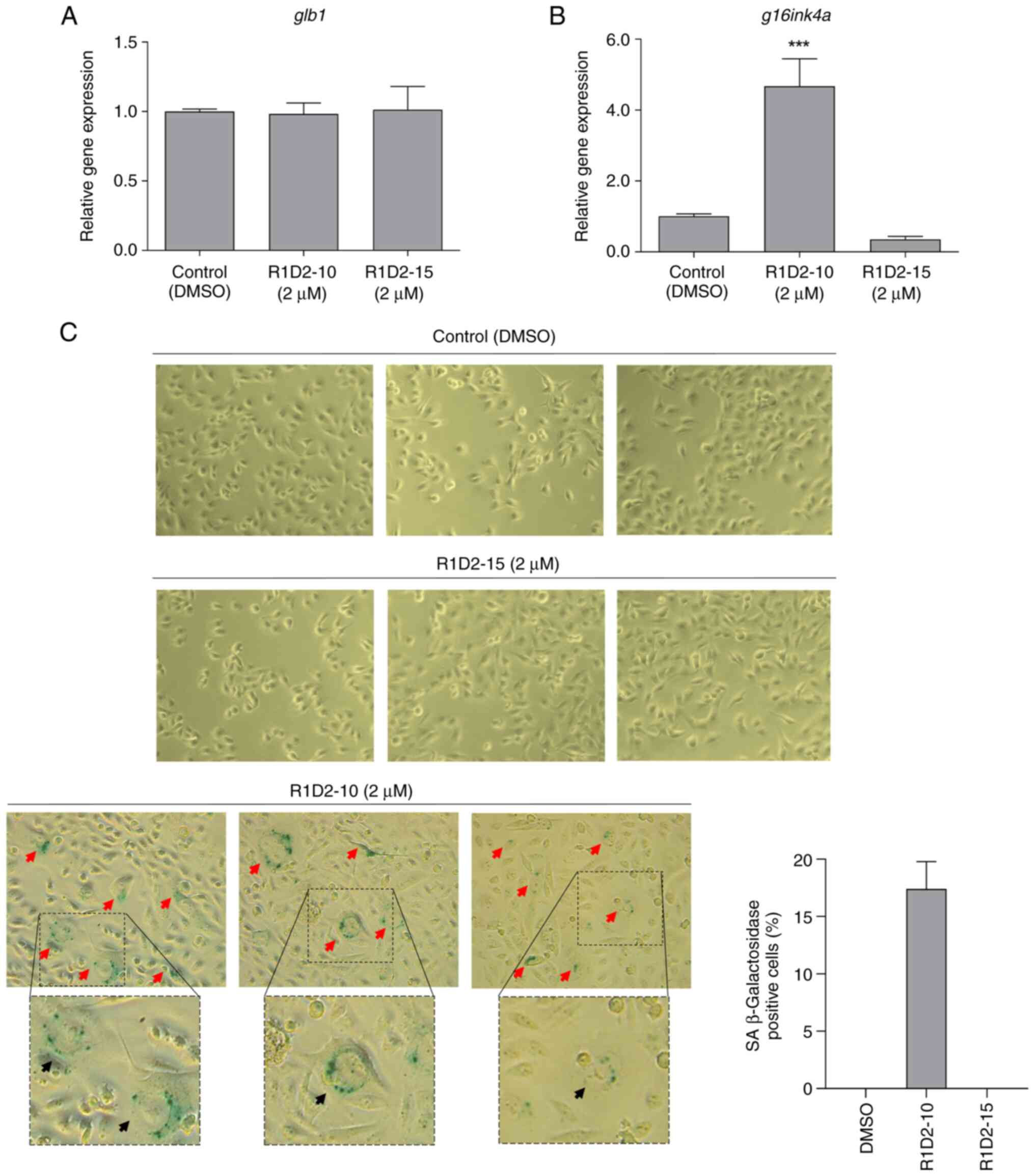
Gene expression of two senescence-associated markers
was analyzed. Expression of glb1 did not change
significantly compared with the control following treatment with
both compounds (Fig. 5A). Same
results were obtained for p16ink4a expression following
treatment with R1D2-15. However, afterchronic treatment with
R1D2-10, expression of this marker increased more than four times
in comparison with untreated cells (Fig. 5B).
Senescence-associated β galactosidase activity of
treated cells was evaluated. Control and cells treated with
compound R1D2-15 did not show positive staining for β galactosidase
activity (Fig. 5C). However,
17.4±4.3% of R1D2-10-treated cells exhibited blue staining
associated with senescence-associated β galactosidase activity (red
arrows), in addition to an enlarged and rounded morphology with
cytoplasmic vacuolization, consistent with senescence (black
arrows; Fig. 5C).
R1D2-10 promotes apoptosis in MDA MB
231 cells
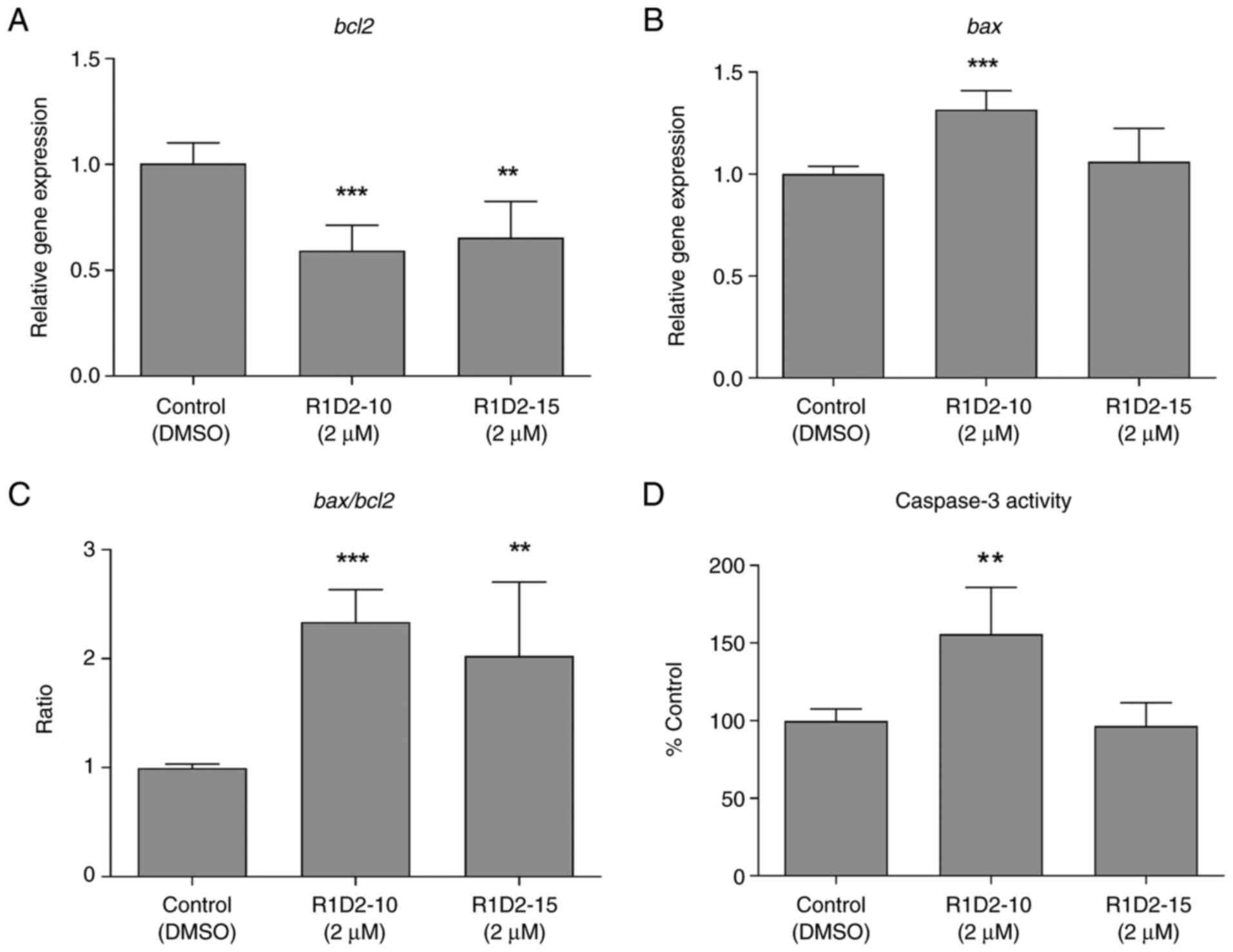
Apoptosis was evaluated by expression of anti- and
pro-apoptotic bcl2/bax, and Caspase 3 activity of
cells following chronic treatment with compounds R1D2-10 and
R1D2-15. R1D2-10 and R1D2-15 decreased Bcl2 expression by 40 and
35%, respectively (P<0.001 and P<0.01, respectively; Fig. 6A). Regarding Bax expression, an
increase of 30% was observed following treatment with R1D2-10,
whereas no difference following R1D2-15 treatment was observed
(Fig. 6B). Bax/Bcl2 ratio was 2.3
and 2.0 following treatment with R1D2-10 and R1D2-15, respectively
(P<0.001 and P<0.01, respectively; Fig. 6C).
Caspase 3 activity was measured in treated cell
lysate. Treatment with compound R1D2-10 led to a 55% increase in
activity compared with control cells (P<0.01; Fig. 6D), whereas R1D2-15 exhibited
similar activity to untreated cells.
Based on in vitro analysis of telomere
length, senescence and apoptosis, R1D2-10 was selected for
subsequent experiments.
R1D2-10 inhibits telomerase in other
breast cancer cell lines
The aforementioned experiments were performed on MDA
MB 231 cells, which correspond to a basal-claudin low breast cancer
subtype. To eliminate the possibility that the inhibitory effect
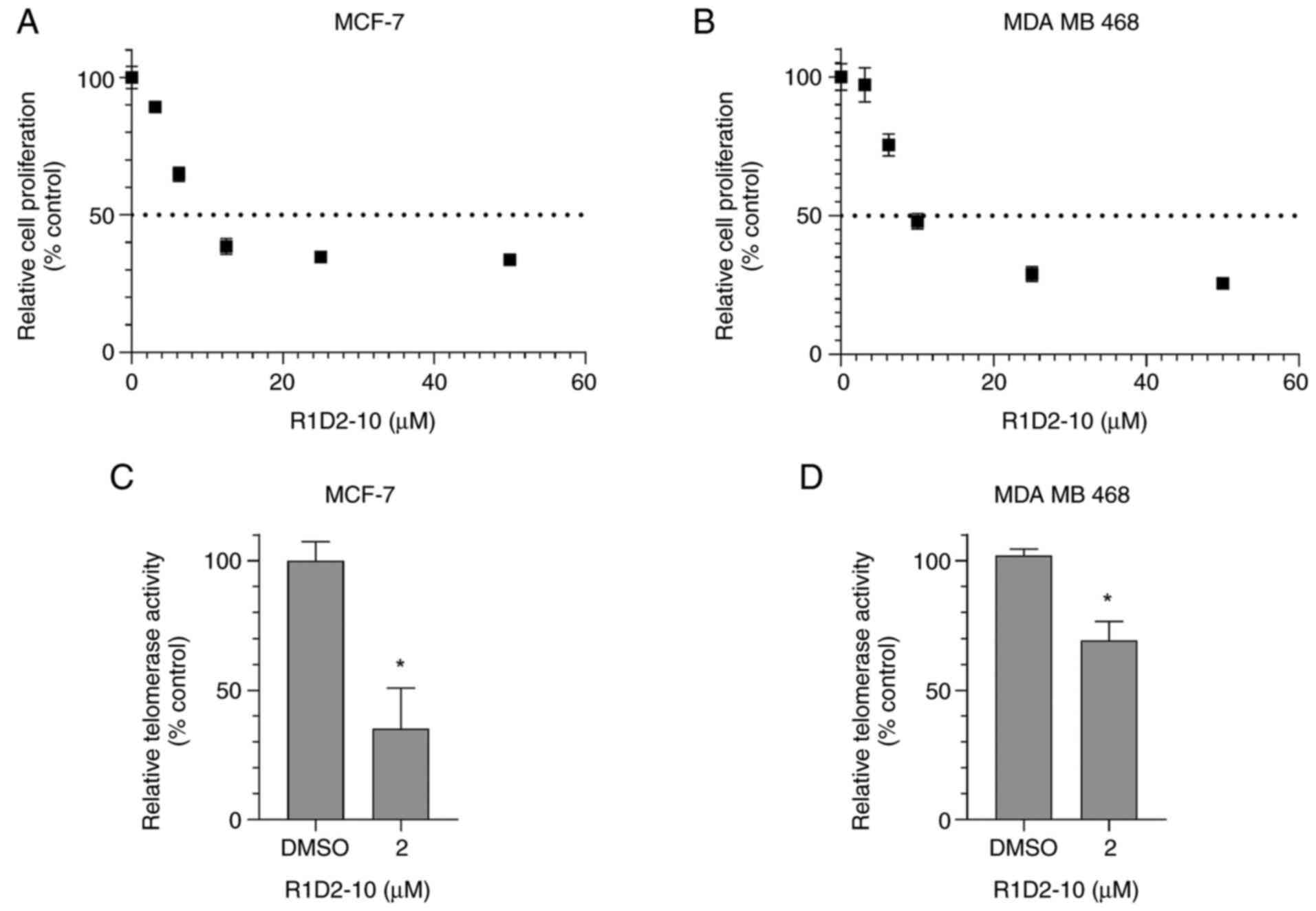
was cell line-dependent, the effect of R1D2-10 on IC50
value and telomerase activity was assessed in luminal A MCF-7 and
basal MDA MB 468 cells. For MCF-7 and MDA MB 468, IC50
values were 5.95 (Emax: 67.56%, R2: 0.90) and
9.79 µM (Emax: 73.36%, R2: 0.91),
respectively (Fig. 7A and B).
Therefore, 2 µM was selected to evaluate telomerase activity.
R1D2-10 treatment for 48 h caused telomerase inhibition of 64.7 in
MCF-7 and 30.8% in MDA MB 468 cells (Fig. 7C and D; P<0.05). Furthermore,
telomerase activity assay was assessed, to validate drug response.
MDA MB 231 cells were treated with increasing concentrations of
R1D2-10 for 48 h and then evaluted by RQ-TRAP. As is shown in
Fig. S1, concentration-dependent
behavior was observed, reaching a significant inhibition of more
than 80% at the higher concentration (P<0.001).
R1D2-10 shows low cytotoxic effect on
nomal culture
A lead compound that exhibits the desired biological
activity and its optimization is a key step in drug discovery.
Absence of toxicity against healthy cells is a sought feature.
Therefore, the present study evaluated in vitro toxicity in
a normal primary culture derived from a p4 neonatal mouse brain.
The results showed no significant cytotoxic effect below 25 µM
(Fig. S2). Furthermore,
preliminary assays in BALB/c mice revealed no toxic effect in
vivo. (data not shown).
R1D2-10 analogs shows improved
affinity to DKC1
Considering the aforementioned results, it was
hypothesized that R1D2-10 serves as a bioactive molecule in
vitro and may be used as a seed molecule for analog design. To
obtain novel candidates with improved activity, R1D2-10 analogs
were generated using LigDream web server. A total of 100 candidates
was obtained and used to perform docking assay using previously
reported parameters (11).
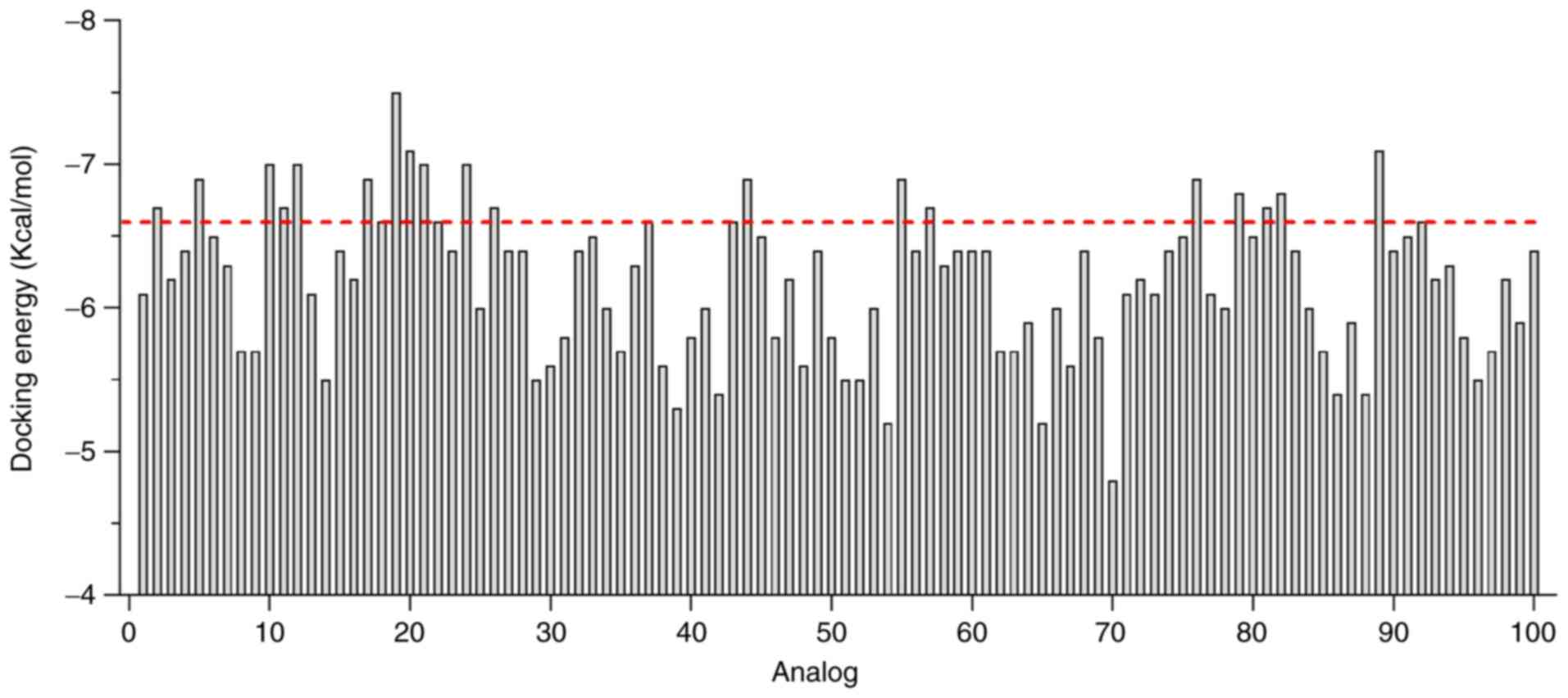
Fig. 8 shows docking energy value
for each analog.
Analogs that exhibited greater affinity than that
predicted for R1D2-10 were selected to analyze whether the novel
compounds exhibited the same interactions with hDKC1 residues as
R1D2-10 (hydrogen bond and π-stacking). Table I summarizes residue interactions of
R1D2-10 and the selected analogs. As a result of this analysis, we
selected analogs that showed improved affinity values for the
target while maintaining the main interactions of R1D2-10.
 | Table I.Analog interaction with specific
residues of hDKC1. |
Table I.
Analog interaction with specific
residues of hDKC1.
|
|
| hDKC1 residue
interaction |
|---|
|
|
|
|
|---|
| Compound | Docking energy,
Kcal/mol | Ala 305 | Ala 308 | Lys 314 | Met 316 | Gly 319 | Tyr 311 |
|---|
| R1D2-10 | −6.6 | H | H | 2HB | HB | HB | π |
| R1D2-10(02) | −6.7 | - | - | - | HB | HB | 2H |
| R1D2-10(05) | −6.9 | - | - | - | H | HB | H |
| R1D2-10(10) | −7.0 | H | - | - | HB | - | π |
| R1D2-10(11)a | −6.7 | H | - | - | - | - | H |
| R1D2-10(12) | −7.0 | H | - | - | H | - | H, π |
| R1D2-10(17) | −6.9 | H | - | - | - | - | H, π |
| R1D2-10(20)a | −7.1 | H | - | - | H | - | 2H |
| R1D2-10(21) | −7.0 | H | - | - | - | - | Π |
| R1D2-10(24) | −7.0 | - | - | H | - | - | - |
| R1D2-10(26) | −6.7 | H | - | - | H | - | H, 2π |
| R1D2-10(44) | −6.9 | - | - | - | H | - | Π |
| R1D2-10(55)a | −6.9 | H | - | - | - | - | 2H |
| R1D2-10(57) | −6.7 | H | - | - | - | - | H, π |
| R1D2-10(76) | −6.9 | H | - | - | - | - | Π |
| R1D2-10(79)a | −6.8 | H | - | - | - | - | 3H |
| R1D2-10(81)a | −6.7 | H | - | - | H | - | H |
| R1D2-10(82) | −6.8 | H | - | - | - | - | H, π |
| R1D2-10(89) | −7.1 | H | - | - | HB | HB | 2H |
R1D2-10 analogs prediction on ADME and
toxicity properties
Pharmacokinetics, drug-likeness and medicinal
chemistry profile of analogs were evaluated using pkCSM ADME
descriptors algorithm protocol (Table
II). All analogs complied with Lipinski's rule of five (data
not shown). All analogs showed suitable profiles of ADME
parameters. Caco-2 permeability, intestinal absorption (human) and
skin permeabilization were used to predict the absorption level of
compounds. When Papp coefficient is >8×10−6, the
predicted value is >0.90; thus, the compound has high Caco-2
permeability and is easy to absorb (36). R1D2-10(10, 12, 44 and 76) were
predicted to have highest Caco-2 permeability, followed by
R1D2-10(2, 26, 82 and 89). Intestinal absorption <30% is
considered to be poorly absorbed (37). All the compounds were predicted to
have absorption >88% (Table
II). Logarithmic skin permeation coefficient (logKp) >-2.5
is considered to indicate relatively low skin permeability. All
analyzed compounds showed logKp<2.5 and were considered to have
suitable skin permeability.
 | Table II.Analog ADME properties and off-target
prediction. Analysis was performed using pkCSM ADMET descriptors
algorithm protocol. All analogs agree with Lipinski's rules. |
Table II.
Analog ADME properties and off-target
prediction. Analysis was performed using pkCSM ADMET descriptors
algorithm protocol. All analogs agree with Lipinski's rules.
|
| Absorption | Distribution | Metabolism | Excretion |
|---|
|
|
|
|
|
|
|---|
| Compound | WS, log mol/l | Caco2 P, log
Papp | HIA, % | SP, log log Kp | PgP S | PgP I | VDss, log log
l/kg | FU | BBB P, log BB | CNS P, log PS | CYP2D6 S | CYP2D6 I | CYP2C9 I | TC, log
ml/min/kg | Renal OCT2 S | Off-target IP |
|---|
| R1D2-10 | −3.828 | 0.764 | 91.456 | −2.735 | Yes | Yes | −0.385 | 0.011 | 0.051 | −1.564 | Yes | No | Yes | 0.554 | No | Low |
| R1D2-10(02) | −4.363 | 0.827 | 88.163 | −2.735 | Yes | Yes | −0.601 | 0.013 | −0.757 | −1.468 | Yes | No | Yes | 0.239 | No | Low |
| R1D2-10(05) | −3.863 | 0.707 | 88.938 | −2.769 | Yes | Yes | 0.044 | 0.117 | −0.553 | −1.922 | Yes | Yes | Yes | 0.301 | No | Low |
| R1D2-10(10) | −3.693 | 1.011 | 92.107 | −2.745 | Yes | Yes | 0.672 | 0.019 | −0.849 | −1.967 | Yes | No | Yes | 0.452 | No | Low |
| R1D2-10(12) | −4.374 | 1.032 | 92.389 | −2.735 | Yes | Yes | −0.494 | 0.015 | −0.518 | −2.181 | No | No | Yes | 0.476 | No | NE |
| R1D2-10(17) | −4.152 | 0.732 | 89.153 | −2.736 | Yes | Yes | −0.400 | 0.000 | −0.569 | −1.749 | Yes | No | Yes | 0.308 | No | NE |
| R1D2-10(21) | −4.391 | 0.556 | 94.679 | −2.759 | Yes | Yes | −0.080 | 0.027 | −0.867 | −1.964 | No | No | Yes | 0.359 | No | Low |
| R1D2-10(24) | −5.371 | 0.609 | 95.679 | −2.735 | Yes | Yes | −1.083 | 0.088 | 0.081 | −1.366 | No | No | Yes | 0.240 | No | Low |
| R1D2-10(26) | −3.587 | 0.893 | 94.051 | −2.735 | Yes | Yes | −0.023 | 0.000 | −0.776 | −1.869 | No | No | Yes | 0.489 | No | NE |
| R1D2-10(44) | −4.059 | 0.917 | 90.807 | −2.870 | Yes | Yes | 0.414 | 0.086 | 0.034 | −1.707 | Yes | No | Yes | 0.358 | No | Low |
| R1D2-10(57) | −5.097 | 0.539 | 93.358 | −2.753 | Yes | Yes | −0.272 | 0.000 | −0.054 | −1.748 | No | No | Yes | 0.284 | No | Low |
| R1D2-10(76) | −4.058 | 1.053 | 91.659 | −2.843 | Yes | No | −0.106 | 0.050 | −1.084 | −1.991 | Yes | No | Yes | 0.294 | No | Low |
| R1D2-10(82) | −4.082 | 0.892 | 92.963 | −2.742 | Yes | Yes | 0.740 | 0.052 | 0.199 | −1.957 | Yes | No | Yes | 1.090 | No | Low |
| R1D2-10(89) | −4.098 | 0.892 | 93.241 | −2.763 | Yes | No | 0.108 | 0.048 | −0.762 | −2.370 | No | No | No | 0.437 | No | Low |
Distribution volume (VDss), fraction unbound (human)
and central nervous system (CNS) and blood-brain barrier membrane
permeability (logBB) were evaluated to characterize the
distribution of compounds. VDss describes the relationship between
the concentration of drug in the plasma and the body (38). VDss<0.71 l/kg (log
VDss<-0.15) is considered to be relatively low; VDss>2.81
l/kg (log VDss >0.45) is considered to be relatively high
(36). VDss of R1D2-10(2, 12, 17,
24 and 79) was low, whereas compounds R1D2-10(10) and (82) showed relatively high values. For
blood-brain barrier membrane permeability, logBB>0.3 indicates
compounds cross the blood-brain barrier easily; logBB<0.3
indicates compounds do not easily cross the blood-brain barrier
(39). All analogs show values of
logBB lower than 0.3, so them were predicted to have low
blood-brain barrier permeability. Regarding CNS crossing,
logarithmic permeability surface-area product (logPS) was analyzed.
All compounds show a logPS value among −1,366 and −2,370.
Suenderhauf et al reported that drugs with logPS<-3 were
predicted to not cross the central nervous system (CNS) (40). All compounds show a lopPS<-3,
therefore, were predicted to cross the CNS.
Cytochrome P450 is a key enzyme system for drug
metabolism in the liver. The primary subtypes of cytochrome P450
are CYP2D6 and CYP3A4 (41).
R1D2-10(12, 21, 24, 26, 57 and 89) were not substrates for CYP2D6,
whereas R1D2-10(5) was predicted
to be a CYP2D6 inhibitor (Table
II). By contrast, all compounds were predicted to be substrates
and inhibitors of CYP3A4 (data not shown).
Drug elimination is associated with molecular weight
and hydrophilicity of compounds. The prediction analysis showed
that total clearance of R1D2-10(82) was highest, followed by R1D2-10(10,
12, 26 and 89). Furthermore, all candidates showed low or
non-existent probability of off-target interaction (Table II).
Toxicity of compounds is shown in Table III. All the compounds were
predicted to generate hepatotoxicity, except R1D2-10(10). Furthermore, R1D2-10(24, 57 and 82)
exhibited high probability of generating tumorigenic effects,
whereas R1D2-10(57 and 82) were also predicted to be mutagenic.
Compound R1D2-10(12) was
associated with reproductive toxicity.
 | Table III.Prediction of analog toxicity.
Analysis was performed using DataWarrior software (24). |
Table III.
Prediction of analog toxicity.
Analysis was performed using DataWarrior software (24).
| Compound | Hepatotoxicity | Skin
sensitization | Mutagenic | Tumorigenic | Reproductive
toxicity | Irritant |
|---|
| R1D2-10 | Yesa | No | None | None | None | None |
| R1D2-10(02) | Yesa | No | None | None | None | None |
| R1D2-10(05) | Yesa | No | None | None | None | None |
| R1D2-10(10) | No | No | None | None | None | None |
| R1D2-10(12) | Yesa | No | None | None | Lowb | None |
| R1D2-10(17) | Yesa | No | None | None | None | None |
| R1D2-10(21) | Yesa | No | None | None | None | None |
| R1D2-10(24) | Yesa | No | None | Higha | None | None |
| R1D2-10(26) | Yesa | No | None | None | None | None |
| R1D2-10(44) | Yesa | No | None | None | None | None |
| R1D2-10(57) | Yesa | No | Higha | Higha | None | None |
| R1D2-10(76) | Yesa | No | None | None | None | None |
| R1D2-10(82) | Yesa | No | Higha | Higha | None | None |
| R1D2-10(89) | Yesa | No | None | None | None | None |
The predicted results indicated that
R1D2-10(10) was the only analog
that did not show any non-desirable side effects. R1D2-10(2, 5, 17,
21, 26, 44, 76 and 89) were predicted to induce only
hepatotoxicity.
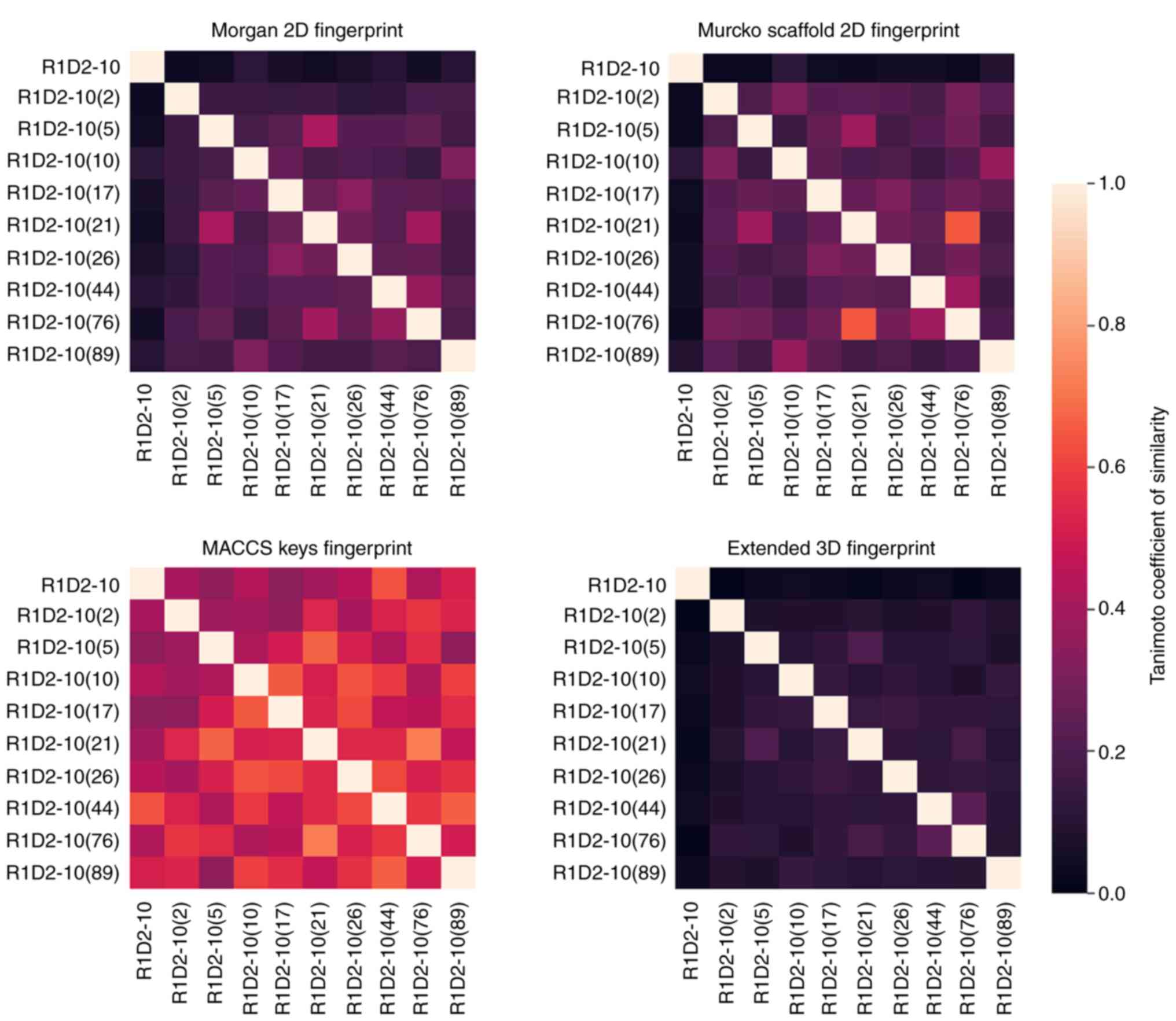
Chemical diversity analysis was performed to
determine whether the selected analogs were different using four
molecular fingerprints and Tanimoto coefficient. MACCS fingerprint
similarity values were notably higher compared with the other
fingerprints (Fig. 9). Morgan 2D
and Murcko Scaffold fingerprints analyze similarity based on
two-dimensional molecular structure and its backbone (32,42),
respectively. These results indicated no similarity among the
selected compounds. Furthermore, 3D extended fingerprint analysis
demonstrated lower levels of similarity than the aforementioned
fingerprints. Given that low similarity values imply dissimilarity
(Tanimoto coefficient <0.8) (43,44),
these results indicated that the selected analogs derived from a
common parent compound represent a diverse analog family and may
exhibit different activity.
Discussion
The use of telomerase as an antitumor target
presents various advantages. It is a key and specific component of
most tumor cells and it is widely expressed in multiple types of
tumor (10). Moreover, telomerase
is the most common mechanism to evade replicative mortality by
tumoral cells. Only one less robust compensatory process exists:
Alternative lengthening of telomeres. This may limit risk of
developing resistance to telomerase inhibition-based therapy
(45). On the other hand, low or
null expression of telomerase in normal tissue promotes specificity
and diminishes risk of toxicity in healthy cells, concluding in a
promising therapeutic window (46). Telomerase inhibitors include
natural compounds, immunotherapies, oligonucleotides, gene therapy,
small molecules, and nucleoside analogs (8).
R1D2-10 and R1D2-15 cytotoxic effect and telomerase
inhibitory capacity were evaluated on MDA MB 231 cells at 0-50 µM.
Following 72 h incubation, IC50 values of 9.52 and 12.95
µM were obtained for R1D2-10 and R1D2-15, respectively. These
results agree with those reported for other telomerase inhibitors
(47–49). Furthermore, both compounds
demonstrated an inhibitory effect on telomerase activity, inducing
inhibition >50% following 48 h treatment. R1D2-10 showed a
concentration-dependent behavior, validating the observed drug
response. This effect has been reported for other telomerase
inhibitors, supporting the present results (50,51).
Furthermore, in comparison with the reported effect
of other telomerase inhibitors, such as Pterostilbene, BIBR1532 and
AZT, R1D2-10 and R1D2-15 exhibited a greater inhibitory effect at
shorter time and lower concentrations (52–54).
Obtaining the desired effect using the lowest possible dose
minimizes undesirable side effects. The present study aimed to
evaluate capacity to inhibit telomerase activity and effect on the
proliferative capacity of cells. To evaluate whether R1D2-10 and
R1D2-15 lead to telomere shortening, senescence and apoptosis,
cells were treated for 55 passages (~22 weeks). Considering that
the aforementioned process depends on cell division, chronic
telomerase inhibitor treatment is usually among this amount of cell
passages (52–56).
To evaluate telomere length, a relative telomere
length determination assay based on qPCR was performed. Telomere
length decreased to 66 and 43% decrease for compounds R1D2-10 and
R1-D2-15, respectively, after 55 passages. Similar results have
been reported for chronic treatment with Imetelstat (57), MST312 (58) and BIBR1532 (59). Cell senescence is primarily caused
by telomere shortening (49).
Senescence involves changes in replicative capacity, cellular
morphology, gene expression and metabolism (60). Senescence is commonly evaluated by
SA-β galactosidase activity due to easy detection at pH 6 with
artificial substrate X-gal (61)
and expression of tumor suppressor gene p16ink4a. This gene
encodes a protein that exhibits tumor suppressor functions by
inhibiting CDK4 and CDK6, and then regulates cell cycle and
senescence (62). The present
study demonstrated a positive staining for SA-β galactosidase
activity and a 4-fold increase in expression for p16ink4a
following treatment with compound R1D2-10. Moreover, positive cells
were bigger and rounded with cytoplasmic vacuolization. These
results indicated that R1D2-10 treatment led to senescence entrance
triggered by telomere shortening; this was comparable with the
effect of other reported telomerase inhibitors (59–65).
Although it has been reported that increased glb1 expression
is associated with the senescent phenotype, treatment with R1D2-10
and R1D2-15 did not affect this marker. However, we consider that
this result does not invalidate our conclusion, since the positive
activity observed by X-gal staining is more relevant to the
senescent phenotype than a direct correlation with glb1 mRNA
levels (66).
Telomere shortening triggers both senescence and
programmed cell death (34). Bcl2
protein family (Bax, Bak, Bcl2, Bcl-xL) and the effector Caspases 3
and 6 are key in regulation of apoptosis (67). Considering telomere shortening was
caused by R1D2-10 and R1D2-15 treatment, the effect on
bcl2/bax expression and Caspase 3 activity was
evaluated. R1D2-10 generated a decrease of 40% in expression of
anti-apoptotic bcl2 and increase of 30% of pro-apoptotic
bax. The ratio of bax/bcl2 was 2.3, indicating
a pro-apoptotic state compared with the control. In addition,
>50% increase in Caspase 3 activity was observed following
treatment with R1D2-10, indicating apoptosis. These results are
consistent with the pro-apoptotic effect of other telomerase
inhibitors in cancer (49,65,68,69),
supporting the potential role of R1D2-10 as a telomerase inhibitor
with an in vitro antitumor effect.
By contrast with R1D2-10, cells treated with R1D2-15
showed no difference in senescence or apoptosis. Although both
compounds were identified by DBVS and used at the same
concentration, they have different chemical structures and
different effectiveness as telomerase inhibitor and on telomere
shortening. Therefore, the effect of R1D2-15 may not be enough to
trigger senescence and apoptosis.
Once R1D2-10 was selected as our lead compound, we
evaluate that inhibitory effect of telomerase is not dependent of
the cell line. R1D2-10 inhibited telomerase activity both in MDA MB
468 and MCF-7 breast cancer cell lines, demonstrating that it could
be used in different subtypes of human breast cancer.
In drug discovery, a primary feature to select a
seed compound is absent or low toxicity against normal cells. As an
in vitro toxicity assay, evaluation in primary cell culture
is a relatively simple method (70). The effect of R1D2-10 was evaluated
in primary cell culture from mouse brain; there was no significant
cytotoxic effect <25 µM. This was considerably higher than the
concentration used define for long-term treatment (2 µM).
Additionally, preliminary data from in vivo toxicity study
how no toxic effect on mice. Therefore, it was hypothesized that
R1D2-10 used in our defined concentration of 2 µM had a low or null
cytotoxic effect on normal cells.
After identifying a bioactive compound, the design
of novel chemical entities based on the hit structure is the
following step in drug discovery. Analogs based on R1D2-10 were
designed using LigDream web server. This strategy is widely
reported in novel drug design (71,72).
A total of 100 novel analogs was evaluated by
docking assay using the same parameters as previously described
(11). Considering the structural
information obtained from the initial DBVS, protein-ligand
interactions were used as criteria for selecting novel ligands with
improved affinity compared with R1D2-10. We selected candidates
that presented, preferably, hydrogen bond and π stacking
interactions with the residues of DKC1, considering that they are
stronger than hydrophobic interactions (73). The selected interaction are
reported to improve the affinity and selectivity of the drug with a
binding site (74).
As ADME and toxicity prediction serve an important
role in facilitating appropriate selection of candidate drugs by
pharmaceutical companies prior to expensive clinical trials, the
ADME and toxicity parameters of novel candidates were predicted
(75). All analogs complied with
Lipinski's rule of five, which indicates a good druggability
profile, and most showed suitable values of ADME properties using
the pkCSM prediction tool, which is broadly reported (22,76–78).
Additionally, target prediction of small bioactive molecules was
analyzed using Swiss Target Prediction (63,79).
Small molecules are designed to bind to proteins or other
macro-molecular targets to modulate their activity, resulting in
phenotypic effects. For this reason, mapping the targets of small
bioactive molecules is a key step for unraveling the molecular
mechanisms underlying bioactivity and predicting potential side
effects or cross-reactivity (80).
All candidates were predicted to exhibit low or null probability of
off-target interaction. Toxicity parameters of the analogs were
assessed by DataWarrior (24,81–83);
analogs that were predicted to exhibit mutagenic, tumorigenic or
reproductive toxicity were discarded. A total of nine analogs with
suitable parameters regarding target affinity, ADME properties,
off-target interaction and undesirable side effects was obtained.
Considering that we want to synthesize these candidates in order to
find one with improved in vitro and in vivo
effectiveness, we decided to analyze their similarity. The
fundamental principle behind similarity-based study is the
‘chemical similarity principle,’ which states that if two molecules
share similar structures, then they will likely have similar
bioactivities (84) In this way,
we would select the analogs that are chemically different in order
to obtain different effectiveness. Several studies proposed that
calculation of molecular similarity could be carried out by using
small-molecule fingerprints, and these fingerprints are calculated
by numerous approaches (85,86).
The present study calculated four types of fingerprints with
Tanimoto coefficient. These strategies are widely reported as
methods of refinement and molecule characterization (87–90).
Particularly, MACCS fingerprint similarity values were notably
higher compared with the other fingerprints. This fact was
expectable considering that MACCS fingerprint bases analysis on
searching common structural patterns in drug molecules. Considering
that we designed analogs from a seed molecule with drug-like
features, this result supports the idea that all the selected
analogs maintain these features. In Summary, analogs exhibited
chemical diversity, so may exhibit different in vitro or
in vivo activity. Based on the aforementioned results, we
propose analogs R1D2-10(2),
(5), (10), (17), (21), (26), (44), (76) and (89) as candidates to be synthesized and
evaluated in further works.
The present study evaluated potential telomerase
inhibitors. The jump from in silico to in vitro assay
constitutes one of the most important steps of the rational design
of novel drugs. Furthermore, the finding of the desired activity
establishes the validation basis of the rational and experimental
design.
The primary goal of antitumor telomerase-based
therapy is to selectively induce cell death in tumor cells
targeting unlimited replicative capacity, which is associated with
apoptosis evasion.
The present in vitro evaluation of drug-like
candidates on MDA MB 231 cells showed that R1D2-10 inhibited
telomerase activity at a dose similar to that of other reported
telomerase inhibitors such as BIBR1532, MST312 and Imetelstat
(47,49,55);
this induced telomere shortening, senescence and apoptosis.
To the best of our knowledge, destabilization of
the interaction of hTR/hDKC1 complex has not been reported as a
strategy for telomerase inhibition. The present results may allow
development of a directed therapy based on telomerase and
contributes to rational design of novel antitumor drugs.
The present study identified the hit R1D2-10, which
showed activity as telomerase inhibitor and induced cell senescence
and apoptosis. From this seed compound, analogs were generated to
identify those with the best profile to be synthesized and
evaluated. A total of nine chemically diverse analogs with suitable
parameters regarding predicted affinity, ADME properties,
off-target interaction and undesirable side effects was identified.
These results provide a basis for preclinical assays to
characterize R1D2-10 and selected analog effectiveness as antitumor
therapy in breast cancer models to demonstrate their potential use
in the clinic.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr. Andrii Buvailo
from Enamine for support in the development and sinthesys of
analogues. Primary mouse culture was were kindly provided by
Chronobiology Laboratory (National Quilmes University, Buenos
Aires, Argentina).
Funding
The present study was supported by grants from Quilmes National
University (PUNQ EXPTE 1297/19), National Research Council (PIP
EXPTE N° 1811/19), the National Agency for the Promotion of Science
and Technology (PICT 2018 N° 2377; PICT START UP 2020 N° 00001) and
Cancer National Institute (EXPTE 827-1533/18) (Argentina).
Availability of data and materials
The data generated in the present study are not
publicly available due to reasons of protection of intellectual
property but are available from the corresponding author on
reasonable request.
Authors' contributions
RA, RV, CP, JM and DMG designed and performed in
vitro experiments and wrote the manuscript. MC, PC and PLM
contributed to R1D2-10 analog generation and in silico
analysis and prediction. DG conceived the study. All authors have
read and approved the final manuscript. RA and DG confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jafri MA, Ansari SA, Alqahtani MH and Shay
JW: Roles of telomeres and telomerase in cancer, and advances in
telomerase-targeted therapies. Genome Med. 8:692016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berardinelli F, Coluzzi E, Sgura A and
Antoccia A: Targeting telomerase and telomeres to enhance ionizing
radiation effects in in vitro and in vivo cancer models. Mutat Res
Rev Mutat Res. 773:204–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lipinska N, Romaniuk A, Paszel-Jaworska A,
Toton E, Kopczynski P and Rubis B: Telomerase and drug resistance
in cancer. Cell Mol Life Sci. 74:4121–4132. 2017. View Article : Google Scholar
|
|
4
|
Mender I, LaRanger R, Luitel K, Peyton M,
Girard L, Lai TP, Batten K, Cornelius C, Dalvi MP, Ramirez M, et
al: Telomerase-Mediated strategy for overcoming non-small cell lung
cancer targeted therapy and chemotherapy resistance. Neoplasia.
20:826–837. 2018. View Article : Google Scholar
|
|
5
|
Sengupta S, Sobo M, Lee K, Senthil Kumar
S, White AR, Mender I, Fuller C, Chow LML, Fouladi M, Shay JW and
Drissi R: Induced telomere damage to treat telomerase expressing
therapy-resistant pediatric brain tumors. Mol Cancer Ther.
17:1504–1514. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang G, Wu LW, Mender I, Barzily-Rokni M,
Hammond MR, Ope O, Cheng C, Vasilopoulos T, Randell S, Sadek N, et
al: Induction of telomere dysfunction prolongs disease control of
therapy-resistant melanoma. Clin Cancer Res. 24:4771–4784. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Y, Zhong D, Li Y, Wu H, Xu X, Yang J
and Gu Z: Tumor-Oriented telomerase-terminated nanoplatform as
versatile strategy for multidrug resistance reversal in cancer
treatment. Adv Healthc Mater. 9:e19017392020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gomez DL, Armando RG, Cerrudo CS,
Ghiringhelli PD and Gomez DE: Telomerase as a cancer target.
Development of new molecules. Curr Top Med Chem. 16:2432–2440.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guterres AN and Villanueva J: Targeting
telomerase for cancer therapy. Oncogene. 39:5811–5824. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jager K and Walter M: Therapeutic
Targeting of Telomerase. Genes. 7:2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Armando RG, Mengual Gomez DL, Juritz EI,
Lorenzano Menna P and Gomez DE: Homology model and docking-based
virtual screening for ligands of human dyskerin as new inhibitors
of telomerase for cancer treatment. Int J Mol Sci. 19:32162018.
View Article : Google Scholar
|
|
12
|
Jaiswal RK and Yadava PK: Assessment of
telomerase as drug target in breast cancer. J Biosci. 45:722020.
View Article : Google Scholar
|
|
13
|
Holliday DL and Speirs V: Choosing the
right cell line for breast cancer research. Breast Cancer Res.
13:2152011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cawthon RM: Telomere measurement by
quantitative PCR. Nucleic Acids Res. 30:e472002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Skalic M, Jimenez J, Sabbadin D and De
Fabritiis G: Shape-Based generative modeling for de novo drug
design. J Chem Inf Model. 59:1205–1214. 2019. View Article : Google Scholar
|
|
17
|
RDKit, . Open-source cheminformatics.
GitHub and SourceForge, 2021. https://www.rdkit.org/
|
|
18
|
Rappé AK, Casewit CJ, Colwell K, Goddard
WA III and Skiff WM: UFF, a full periodic table force field for
molecular mechanics and molecular dynamics simulations. J Am Chem
Soc. 114:10024–10035. 1992. View Article : Google Scholar
|
|
19
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salentin S, Schreiber S, Haupt VJ, Adasme
MF and Schroeder M: PLIP: Fully automated protein-ligand
interaction profiler. Nucleic Acids Res. 43((W1)): W443–447. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schildge S, Bohrer C, Beck K and
Schachtrup C: Isolation and culture of mouse cortical astrocytes. J
Vis Exp. ((71)): 500792013.
|
|
22
|
Pires DE, Blundell TL and Ascher DB:
pkCSM: Predicting small-molecule pharmacokinetic and toxicity
properties using graph-based signatures. J Med Chem. 58:4066–4072.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Daina A, Michielin O and Zoete V:
SwissTargetPrediction: Updated data and new features for efficient
prediction of protein targets of small molecules. Nucleic Acids
Res. 47((W1)): W357–W364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sander T, Freyss J, von Korff M and
Rufener C: DataWarrior: An open-source program for chemistry aware
data visualization and analysis. J Chem Inf Model. 55:460–473.
2015. View Article : Google Scholar
|
|
25
|
Gasteiger J and Engel T: Chemoinformatics:
a textbook. John Wiley & Sons, 2006. Chapter 2.9. Volume.
1:92–110. 2006.
|
|
26
|
Leach AR and Gillet VJ: An introduction to
chemoinformatics. Springer, 2007. Chapter 5 - Similiraty Methods.
Volume. 1:99–117. 2007.
|
|
27
|
Sharma A and Lal SP: Tanimoto based
similarity measure for intrusion detection system. J Inf Sec.
2:195–201. 2011.
|
|
28
|
Willett P, Barnard JM and Downs GM:
Chemical similarity searching. J Chem Inf Comput Sci. 38:983–996.
1998. View Article : Google Scholar
|
|
29
|
Bemis GW and Murcko MA: The properties of
known drugs. 1. Molecular frameworks. J Med Chem. 39:2887–2893.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Durant JL, Leland BA, Henry DR and Nourse
JG: Reoptimization of MDL keys for use in drug discovery. J Chem
Inf Comput Sci. 42:1273–1280. 2002. View Article : Google Scholar
|
|
31
|
BIOVIA, . The keys to understanding MDL
keyset technology. Dassault Systemes; Waltham, MA: 2011, https://docplayer.net/64556108-The-keys-to-understanding-mdl-keyset-technology-white-paper.html
|
|
32
|
Axen SD, Huang XP, Caceres EL, Gendelev L,
Roth BL and Keiser MJ: A Simple Representation of three-dimensional
molecular structure. J Med Chem. 60:7393–7409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deng Y, Chan SS and Chang S: Telomere
dysfunction and tumour suppression: The senescence connection. Nat
Rev Cancer. 8:450–458. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roake CM and Artandi SE: Control of
cellular aging, tissue function, and cancer by p53 downstream of
telomeres. Cold Spring Harb Perspect Med. 7:a0260882017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin J and Epel E: Stress and telomere
shortening: Insights from cellular mechanisms. Ageing Res Rev.
73:1015072022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han Y, Zhang J, Hu CQ, Zhang X, Ma B and
Zhang P: In silico ADME and toxicity prediction of ceftazidime and
its impurities. Front Pharmacol. 10:4342019. View Article : Google Scholar
|
|
37
|
Hou T, Wang J, Zhang W and Xu X: ADME
evaluation in drug discovery. 7. Prediction of oral absorption by
correlation and classification. J Chem Inf Model. 47:208–218. 2007.
View Article : Google Scholar
|
|
38
|
Holt K, Nagar S and Korzekwa K: Methods to
predict volume of distribution. Curr Pharmacol Rep. 5:391–399.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Muehlbacher M, Spitzer GM, Liedl KR and
Kornhuber J: Qualitative prediction of blood-brain barrier
permeability on a large and refined dataset. J Comput Aided Mol
Des. 25:1095–1106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Suenderhauf C, Hammann F and Huwyler J:
Computational prediction of blood-brain barrier permeability using
decision tree induction. Molecules. 17:10429–10445. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McDonnell AM and Dang CH: Basic review of
the cytochrome p450 system. J Adv Pract Oncol. 4:263–268. 2013.
|
|
42
|
Laufkotter O, Sturm N, Bajorath J, Chen H
and Engkvist O: Combining structural and bioactivity-based
fingerprints improves prediction performance and scaffold hopping
capability. J Cheminform. 11:542019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bajusz D, Racz A and Heberger K: Why is
Tanimoto index an appropriate choice for fingerprint-based
similarity calculations? J Cheminform. 7:202015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Snarey M, Terrett NK, Willett P and Wilton
DJ: Comparison of algorithms for dissimilarity-based compound
selection. J Mol Graph Model. 15:372–385. 1997. View Article : Google Scholar
|
|
45
|
Zhang JM and Zou L: Alternative
lengthening of telomeres: From molecular mechanisms to therapeutic
outlooks. Cell Biosci. 10:302020. View Article : Google Scholar
|
|
46
|
Shay JW and Wright WE: Telomeres and
telomerase in normal and cancer stem cells. FEBS Lett.
584:3819–3825. 2010. View Article : Google Scholar
|
|
47
|
Gurung RL, Lim SN, Low GK and Hande MP:
MST-312 alters telomere dynamics, gene expression profiles and
growth in human breast cancer cells. J Nutrigenet Nutrigenomics.
7:283–298. 2014.PubMed/NCBI
|
|
48
|
Kazemi-Lomedasht F, Rami A and Zarghami N:
Comparison of inhibitory effect of curcumin nanoparticles and free
curcumin in human telomerase reverse transcriptase gene expression
in breast cancer. Adv Pharm Bull. 3:127–130. 2013.
|
|
49
|
Wardi L, Alaaeddine N, Raad I, Sarkis R,
Serhal R, Khalil C and Hilal G: Glucose restriction decreases
telomerase activity and enhances its inhibitor response on breast
cancer cells: Possible extra-telomerase role of BIBR 1532. Cancer
Cell Int. 14:602014. View Article : Google Scholar
|
|
50
|
Noureini SK and Wink M: Dose-dependent
cytotoxic effects of boldine in HepG-2 cells-telomerase inhibition
and apoptosis induction. Molecules. 20:3730–3743. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang YL, Huang PH, Chiu HC, Kulp SK and
Chen CS, Kuo CJ, Chen HD and Chen CS: Histone deacetylase inhibitor
AR42 regulates telomerase activity in human glioma cells via an
Akt-dependent mechanism. Biochem Biophys Res Commun. 435:107–112.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bashash D, Ghaffari SH, Mirzaee R,
Alimoghaddam K and Ghavamzadeh A: Telomerase inhibition by
non-nucleosidic compound BIBR1532 causes rapid cell death in pre-B
acute lymphoblastic leukemia cells. Leuk Lymphoma. 54:561–568.
2013. View Article : Google Scholar
|
|
53
|
Chen RJ, Wu PH, Ho CT, Way TD, Pan MH,
Chen HM, Ho YS and Wang YJ: P53-dependent downregulation of hTERT
protein expression and telomerase activity induces senescence in
lung cancer cells as a result of pterostilbene treatment. Cell
Death Dis. 8:e29852017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fang JL and Beland FA: Long-term exposure
to zidovudine delays cell cycle progression, induces apoptosis, and
decreases telomerase activity in human hepatocytes. Toxicol Sci.
111:120–130. 2009. View Article : Google Scholar
|
|
55
|
Hu Y, Bobb D, He J, Hill DA and Dome JS:
The HSP90 inhibitor alvespimycin enhances the potency of telomerase
inhibition by imetelstat in human osteosarcoma. Cancer Biol Ther.
16:949–957. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tejera AM, Alonso DF, Gomez DE and Olivero
OA: Chronic in vitro exposure to 3′-azido-2′, 3′-dideoxythymidine
induces senescence and apoptosis and reduces tumorigenicity of
metastatic mouse mammary tumor cells. Breast Cancer Res Treat.
65:93–99. 2001. View Article : Google Scholar
|
|
57
|
Frink RE, Peyton M, Schiller JH, Gazdar
AF, Shay JW and Minna JD: Telomerase inhibitor imetelstat has
preclinical activity across the spectrum of non-small cell lung
cancer oncogenotypes in a telomere length dependent manner.
Oncotarget. 7:31639–31651. 2016. View Article : Google Scholar
|
|
58
|
Morais KS, Guimaraesb AFR, Ramos DAR,
Silva FP and de Oliveira DM: Long-term exposure to MST-312 leads to
telomerase reverse transcriptase overexpression in MCF-7 breast
cancer cells. Anticancer Drugs. 28:750–756. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mueller S, Hartmann U, Mayer F, Balabanov
S, Hartmann JT, Brummendorf TH and Bokemeyer C: Targeting
telomerase activity by BIBR1532 as a therapeutic approach in germ
cell tumors. Invest New Drugs. 25:519–524. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
van Deursen JM: The role of senescent
cells in ageing. Nature. 509:439–446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sharpless NE and Sherr CJ: Forging a
signature of in vivo senescence. Nat Rev Cancer. 15:397–408. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhao R, Choi BY, Lee MH, Bode AM and Dong
Z: Implications of genetic and epigenetic alterations of CDKN2A
(p16(INK4a)) in cancer. EBioMedicine. 8:30–39. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liang Y, Liang B, Wu XR, Chen W and Zhao
LZ: Network pharmacology-based systematic analysis of molecular
mechanisms of dingji fumai decoction for ventricular arrhythmia.
Evid Based Complement Alternat Med. 2021:55354802021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bernadotte A, Mikhelson VM and Spivak IM:
Markers of cellular senescence. Telomere shortening as a marker of
cellular senescence. Aging (Albany NY). 8:3–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Burchett KM, Yan Y and Ouellette MM:
Telomerase inhibitor imetelstat (GRN163L) limits the lifespan of
human pancreatic cancer cells. PloS One. 9:e851552014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lee BY, Han JA, Im JS, Morrone A, Johung
K, Goodwin EC, Kleijer WJ, DiMaio D and Hwang ES:
Senescence-associated beta-galactosidase is lysosomal
beta-galactosidase. Aging Cell. 5:187–195. 2006. View Article : Google Scholar
|
|
67
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Asghari-Kia L, Bashash D, Safaroghli-Azar
A, Momeny M, Hamidpour M and Ghaffari SH: Targeting human
telomerase RNA component using antisense oligonucleotide induces
rapid cell death and increases ATO-induced apoptosis in APL cells.
Eur J Pharmacol. 809:215–223. 2017. View Article : Google Scholar
|
|
69
|
Bashash D, Ghaffari SH, Zaker F, Kazerani
M, Hezave K, Hassani S, Rostami M, Alimoghaddam K and Ghavamzadeh
A: BIBR 1532 increases arsenic trioxide-mediated apoptosis in acute
promyelocytic leukemia cells: Therapeutic potential for APL.
Anticancer Agents Med Chem. 13:1115–1125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Vairano M, Graziani G, Tentori L, Tringali
G, Navarra P and Dello Russo C: Primary cultures of microglial
cells for testing toxicity of anticancer drugs. Toxicol Lett.
148:91–94. 2004. View Article : Google Scholar
|
|
71
|
Cardama GA, Comin MJ, Hornos L, Gonzalez
N, Defelipe L, Turjanski AG, Alonso DF, Gomez DE and Menna PL:
Preclinical development of novel Rac1-GEF signaling inhibitors
using a rational design approach in highly aggressive breast cancer
cell lines. Anticancer Agents Med Chem. 14:840–851. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ramana M, Lokhande R, Bhar S, Ranade P,
Mehta A and Gadre G: In Silico design, synthesis and bioactivity of
N-(2, 4-Dinitrophenyl)-3-oxo-3-phenyl-N-(aryl) phenyl propanamide
derivatives as breast cancer inhibitors. Curr Comput Aided Drug
Des. 13:112–126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gerbelli BB, Vassiliades SV, Rojas JEU,
Pelin JNBD, Mancini RSN, Pereira WSG, Aguilar AM, Venanzi M,
Cavalieri F, Giuntini F and Alves WA: Hierarchical Self-assembly of
peptides and its applications in bionanotechnology. Bioin Bioba
Mater. 220:19000852019.
|
|
74
|
Smith AJ, Zhang X, Leach AG and Houk KN:
Beyond picomolar affinities: Quantitative aspects of noncovalent
and covalent binding of drugs to proteins. J Med Chem. 52:225–233.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Alqahtani S: In silico ADME-Tox modeling:
Progress and prospects. Expert Opin Drug Metab Toxicol.
13:1147–1158. 2017. View Article : Google Scholar
|
|
76
|
Nandini Asha R, Ravindran Durai Nayagam B
and Bhuvanesh N: Synthesis, molecular docking, and in silico ADMET
studies of 4-benzyl-1-(2,4,6-trimethyl-benzyl)-piperidine:
Potential inhibitor of SARS-CoV2. Bioorg Chem. 112:1049672021.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Almeleebia TM, Shahrani MA, Alshahrani MY,
Ahmad I, Alkahtani AM, Alam MJ, Kausar MA, Saeed A, Saeed M and
Iram S: Identification of new mycobacterium tuberculosis proteasome
inhibitors using a knowledge-based computational screening
approach. Molecules. 26:23262021. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gentile D, Floresta G, Patamia V,
Chiaramonte R, Mauro GL, Rescifina A and Vecchio M: An integrated
pharmacophore/Docking/3D-QSAR approach to screening a large library
of products in search of future botulinum neurotoxin a inhibitors.
Int J Mol Sci. 21:94702020. View Article : Google Scholar
|
|
79
|
Daina A, Michielin O and Zoete V:
SwissADME: A free web tool to evaluate pharmacokinetics,
drug-likeness and medicinal chemistry friendliness of small
molecules. Sci Rep. 7:427172017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gfeller D, Grosdidier A, Wirth M, Daina A,
Michielin O and Zoete V: SwissTargetPrediction: A web server for
target prediction of bioactive small molecules. Nucleic Acids Res.
42:(Web Server Issue). W32–W38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Abdelfatah S, Bockers M, Asensio M,
Kadioglu O, Klinger A, Fleischer E and Efferth T: Isopetasin and
S-isopetasin as novel P-glycoprotein inhibitors against
multidrug-resistant cancer cells. Phytomedicine. 86:1531962021.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Greish KF, Salerno L, Al Zahrani R, Amata
E, Modica MN, Romeo G, Marrazzo A, Prezzavento O, Sorrenti V,
Rescifina A, et al: Novel structural insight into inhibitors of
heme oxygenase-1 (HO-1) by new imidazole-based compounds:
Biochemical and in vitro anticancer activity evaluation. Molecules.
23:12092018. View Article : Google Scholar
|
|
83
|
Verma K, Kannan K, V S, R S, V K and K R:
Exploring β-tubulin inhibitors from plant origin using
computational approach. Phytochem Anal. 28:230–241. 2017.
View Article : Google Scholar
|
|
84
|
Lo YC and Torres JZ: Chemical similarity
networks for drug discovery. Special Topics in Drug Discovery. Chen
T: IntechOpen; Volume 1. pp. 53–70. 2016
|
|
85
|
Duran-Frigola M, Pauls E, Guitart-Pla O,
Bertoni M, Alcalde V, Amat D, Juan-Blanco T and Aloy P: Extending
the small-molecule similarity principle to all levels of biology
with the Chemical Checker. Nat Biotechnol. 38:1087–1096. 2020.
View Article : Google Scholar
|
|
86
|
Jasial S, Hu Y, Vogt M and Bajorath J:
Activity-relevant similarity values for fingerprints and
implications for similarity searching. F1000Res. 5:Chem Inf Sci.
–591. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kropiwnicki E, Evangelista JE, Stein DJ,
Clarke DJB, Lachmann A, Kuleshov MV, Jeon M, Jagodnik KM and
Ma'ayan A: Drugmonizome and Drugmonizome-ML: Integration and
abstraction of small molecule attributes for drug enrichment
analysis and machine learning. Database (Oxford). 2021:baab0172021.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lambert LJ, Grotegut S, Celeridad M,
Gosalia P, Backer LJ, Bobkov AA, Salaniwal S, Chung TD, Zeng FY,
Pass I, et al: Development of a robust high-throughput screening
platform for inhibitors of the striatal-enriched tyrosine
phosphatase (STEP). Int J Mol Sci. 22:44172021. View Article : Google Scholar
|
|
89
|
Lopez-Lopez E, Cerda-Garcia-Rojas CM and
Medina-Franco JL: Tubulin inhibitors: A chemoinformatic analysis
using cell-based data. Molecules. 26:24832021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Thomas M, Smith RT, O'Boyle NM, de Graaf C
and Bender A: Comparison of structure- and ligand-based scoring
functions for deep generative models: A GPCR case study. J
Cheminform. 13:392021. View Article : Google Scholar : PubMed/NCBI
|