Introduction
Oral squamous cell carcinoma (OSCC), which includes
cancers of the head and neck, is a highly malignant disease with
high morbidity and mortality rates worldwide (1). Of the patients diagnosed with OSCC,
50% exhibit lymph node metastases, which is correlated with poor
prognosis; the OSCC survival rates have not changed during the last
30 years (2). The disease
frequently occurs on the tongue, upper and lower gingiva, palate,
oral floor, and buccal mucosa. Previously, Sasahira and Kirita
(3) reviewed prognostic factors for
OSCC and described the ‘10 hallmarks of cancer’, (sustained
proliferative signaling, evasion of growth suppressors,
circumvention of immune destruction, activation of invasion and
metastasis, tumor-promoting inflammation, enabling of replicative
immortality, induction of angiogenesis, genome instability and
mutation, resistance to cell death, and dysregulation of
energetics). However, treatment to curtail the progression of
cancer has not yet been established, despite an increase in related
research.
Oxygen is essential for energy metabolism in the
body, but rapid tumor growth affects the surrounding vascular
system, which results in reduced oxygen levels and the creation of
hypoxic regions (4). The tumor
microenvironment is composed of various cellular and non-cellular
components, including the normal and immune cells surrounding the
tumor tissue. Most tumors cause hypoxia due to increased oxygen
consumption and inadequate supply (5). Persistent hypoxia increases the
irregular distribution of tumor vasculature and the distance from
capillaries (4). Tumor cells adapt
to hypoxia and have an important effect on various cells and their
physiological functions. The increased expression of
hypoxia-inducible factor 1α (HIF-1α) is an important marker of
hypoxia around tumor cells (4–7); HIF
has a central role in the cellular mechanism that responds to
hypoxia. HIF-1α activation is involved in cellular infiltration,
metastases, metabolic reprogramming and resistance to therapy
(7–10). To adapt to hypoxia, tumor cells
develop into blood vessels. Therefore, an important feature of the
hypoxic response is angiogenesis. Angiogenesis and
lymph-angiogenesis are required for the growth, invasion and
metastasis of tumor cells (3).
Metastasis is the main cause of cancer-related death (6). Hypoxia and HIF are involved in several
different steps of metastasis and induce tumor cell invasion and
degradation of the extracellular matrix (ECM) (6).
Suprabasin (SBSN) is a secreted protein that has
been isolated as a novel gene expressed in differentiated
keratinocytes in mice and humans. It has been detected in the basal
layers of the epithelia of the tongue, esophagus, stomach,
epidermis, and lung, as well as in invasive glioblastoma cells
(11–14). The SBSN isoform is presumed to be a
signaling molecule, similar to AKT (15) and WNT/β-catenin (16), and it induces cellular
proliferation, invasion, metastasis, migration, angiogenesis,
apoptosis, therapy (15–17) and immune resistance. Shao et
al (17) revealed that SBSN is
important for maintaining the ability of invasion and metastasis
and anchorage-independent growth in salivary gland adenoid cystic
carcinoma (ACC). In addition, SBSN expression was shown to be
upregulated by the demethylation of CpG islands, and SBSN was found
to be significantly hypomethylated in primary ACC compared with
that in normal salivary gland tissue (17). Furthermore, it has been reported
that overexpression of SBSN in esophageal squamous cell carcinoma
(ESCC) increases proliferation and tumorigenicity and induces
migration and angiogenesis in human tumor endothelial cells (TEC)
isolated from two carcinomas, renal and colon, and the AKT pathway
is a downstream factor of SBSN (15,16,18).
Liu et al (19) reported
that SBSN mRNA levels increased in OSCC treated with inactive
Prophyromonas gingivalis. Besides, SBSN has been reported to
be involved in the development and progression of cancer and has a
role in treatment resistance (20).
Therefore, SBSN may serve as a promising biomarker for these
cancers. However, there are few studies that show the relationship
between SBSN and cancer under hypoxic conditions, where tumor cells
are known to respond differently during adaptation. Thus, it was
hypothesized that SBSN would affect tumor cells under hypoxic
conditions and its role in OSCC under such conditions was
investigated. The present results revealed that SBSN is important
in cell proliferation, cell migration and vascular induction into
OSCC cells under hypoxia.
Materials and methods
Cell culture
Human OSCC-derived cell lines, SAS (21,22),
HSC-3 (22,23) and HSC-4 (22,23),
were cultured in high-glucose Dulbecco's modified Eagle's medium
(HDMEM) with L-glutamine and phenol red (FUJIFILM Wako Pure
Chemical Corporation) supplemented with 10% fetal bovine serum
(FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C,
with 5% CO2 and 100% humidity. Normal human epidermal
keratinocytes (NHEKs) were purchased from PromoCell GmbH and
cultured in endothelial cell growth medium (PromoCell GmbH),
according to the manufacturer's protocol. Human umbilical vein
endothelial cells (HUVEC) were purchased from Lonza Group, Ltd. and
cultured in an EGM-2 Endothelial Cell Growth Medium-2 BulletKit
(CC-3162) (Lonza Group, Ltd.) according to the manufacturer's
protocol. Hypoxic conditions were set at 37°C, 2% O2,
and 5% CO2 in a BIOLABO mini-multi-gas incubator
(BL-43MD; TOSC Japan Ltd.).
Antibodies
Rabbit monoclonal anti-HIF1-α (cat. no. ab179483)
and anti-HIF-2α (cat. no. ab199) and polyclonal anti-SBSN (cat. no.
ab232771) antibodies were purchased from Abcam. Rabbit polyclonal
anti-E-cadherin (cat. no. 20874-1-AP), anti-N-cadherin (cat. no.
22018-1-AP) and anti-β-actin (cat. no. 20536-1-AP) antibodies were
purchased from Proteintech Group, Inc. Rabbit monoclonal
anti-LC3A/B (D3U4C; cat. no. 12741) and anti-SQSTM1/p62 (D1Q5S;
cat. no. 39749) were purchased from Cell Signaling Technology, Inc.
Rabbit polyclonal anti-HaloTag antibody (cat. no. G9281) was
purchased from Promega Corporation. Mouse monoclonal anti-endocrine
gland-derived vascular endothelial growth factor (EG-VEGF) antibody
was purchased from Santa Cruz Biotechnology, Inc. (E-12: cat. no.
sc-390741). The anti-rabbit IgG and horseradish peroxidase-linked
whole secondary antibody (from donkey) was purchased from Merck
Millipore (cat. no. NA934V). Otherwise, primary and secondary
antibodies for cyclin proteins were used from the Cyclin Antibody
Sampler Kit (cat. no. 9869; Cell Signaling Technology, Inc.).
RNA purification and reverse
transcriptase polymerase chain reaction (RT-qPCR)
Total cellular RNA was purified using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol and stored at −80°C until
use. Total cellular RNA (100 ng) was reverse-transcribed using the
ReverTra Ace qPCR RT Kit (Toyobo Life Science) according to the
manufacturer's protocol. The generated cDNA was subjected to
RT-qPCR using the THUNDERBIRD Probe qPCR Mix (Toyobo Life Science)
according to the manufacturer's protocol. Final concentrations of
the primers were 20 µl each. The PCR cycles were 95°C for 1 min,
95°C for 15 sec, 60°C for 30 sec (for 49 cycles). TaqMan primers
were purchased from Thermo Fisher Scientific, Inc. (SBSN, cat. no.
Hs01078781; VEGF, cat. no. Hs00900055; and β-actin, cat. no.
Hs01060665). After qPCR, statistical analysis was performed using
the CFX Connect Real-Time PCR Detection System (Bio-Rad
Laboratories, Inc.). Gene expression was analyzed using the
2−ΔΔCq method (24) and
normalized to that of β-actin.
Protein preparation and western blot
analysis
Total cellular protein was prepared as previously
described (21). For western blot
analysis, 20 µg of total cellular protein was analyzed using sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with
a 4–20% gradient gel (Bio-Rad Laboratories, Inc.) and blotted onto
a polyvinylidene difluoride membrane with iBlot2 (Thermo Fisher
Scientific, Inc.). After transcription, the membrane was shaken for
1 h in Tris-buffered saline (Takara Bio, Inc.) containing 0.2%
non-fat dry milk (Cell Signaling Technology, Inc.). Primary and
secondary antibody reactions were performed as previously described
(25). Protein bands were
visualized using Amersham ECL Prime Western Blotting Detection
Reagent (Cytiva) and a ChemiDoc XRS Plus Image Lab System (Bio-Rad
Laboratories, Inc.), which was also used to measure the density of
each band.
Gene transfection
Small interfering RNA (siRNA) for human SBSN
(EHU016151) and negative control siRNA (for firefly luciferase,
EHUFLUC) were purchased from Sigma-Aldrich; Merck KGaA. The
expression vectors HaloTag-SBSN (pFN21AE2798) and HaloTag control
vector (G659) were purchased from Promega Corporation. The siRNAs
and expression vectors were transfected into the cells using
Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol and a recent study
(22).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
caspase 3/7, and 5-bromo-2′-deoxyuridine (BrdU) assays
Cells were seeded in six-well tissue culture plates
at a density of 6×105 cells/well for MTT and caspase 3/7
assays and in 48-well tissue culture plates at a density of
1.2×105 cells/well for the BrdU assay. After incubation
under normoxic conditions for 24 h, the cells were transfected with
siRNAs or expression vectors as aforementioned. The day after
transfection, the cells were reseeded in 96-well tissue culture
plates at a density of 5×102 cells/well for MTT and
apoptotic assays and in 96-well tissue culture plates at
5×102 cells/well for the BrdU assay. The cells were
exposed to normoxic or hypoxic conditions. After 0, 1, and 3 days,
MTT and caspase 3/7 assays were performed, as previously described
(21). The BrdU assay was performed
after 3 days using a commercially available kit (CytoSelect BrdU
Cell Proliferation ELISA Kit; Cell Biolabs Inc.).
Crystal violet cell staining
Transfected cells were reseeded in 96-well tissue
culture plates as aforementioned. The cells were exposed to
normoxic or hypoxic conditions, and after 1 and 3 days, they were
fixed with 4% paraformaldehyde phosphate buffer solution (FUJIFILM
Wako Pure Chemical Corporation) at 4°C for 30 min and stained with
0.5% crystal violet (FUJIFILM Wako Pure Chemical Corporation) in
20% methanol at room temperature for 5 min. Images were captured
using an BX51 microscope (Olympus Corporation) and a microscopic
CCD camera (Olympus DP71), and microscopic images were analyzed
using commercial software (Olympus cellSens standard).
Cell cycle assay
Cell seeding and transfection were performed as in
the MTT and caspase 3/7 assays. The day after transfection, the
cells were reseeded in six-well tissue culture plates at a density
of 6×105 cells/well and exposed to either normoxic or
hypoxic conditions for 3 days. The cell cycle assay was performed
by using a commercially available kit (Tali Cell Cycle Kit; cat.
no. A10798; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and a recent study (26), and they were analyzed using a Tali
Image-Based Cytometer (Thermo Fisher Scientific, Inc.).
Cell invasion assay
Cell invasion assay was performed using a
commercially available kit (CytoSelect 24-Well Cell Invasion Assay;
Cell Biolabs Inc.). After transfection, the medium was replaced
with fresh medium and the cells were exposed to either normoxic or
hypoxic conditions for another 48 h. A 1-ml aliquot of condition
medium from each well was collected and centrifuged at 10,000 × g
for 3 min at 4°C, and the supernatant was stored on ice before use.
Next, 1.0×106 SAS cells were suspended in serum-free
high glucose DMEM, and the cell suspension was placed in the upper
chamber (polycarbonate membrane inserts with 8-µm pores, with an
upper surface coated with a uniform layer of dried basement
membrane matrix solution). The chamber was incubated with 500 µl of
conditioned medium (as aforementioned) in a 24-well plate for 48 h
under normoxic or hypoxic conditions at 37°C. After the removal of
non-invasive cells from the basement membrane, invasive cells were
stained and quantified according to the manufacturer's protocol.
Images were captured, as aforementioned.
Wound healing assay
This assay was performed according to previous
studies (21,25,27),
with slight modifications. The transfected cells were reseeded in a
24-well tissue culture plate at a density of 2.4×105
cells/well, were exposed to normoxic conditions for 24 h. Scratches
were made using 1-ml pipette chips, and the wells were washed twice
with phosphate-buffered saline (PBS). The cells were allowed to
proliferate for another 24 h under normoxic or hypoxic conditions.
Then, the cells were fixed at 4°C for 30 min and stained at room
temperature for 30 min with 0.5% crystal violet in 20% methanol,
and images were captured, as aforementioned.
Gelatin zymography
This assay was performed using a commercially
available kit (Gelatin-zymography kit, Code No. AK47; Cosmo Bio
Co., Ltd.). After a 24-h incubation post transfection, the medium
was replaced with fresh medium. The cells were exposed to normoxic
or hypoxic conditions for 48 h. The conditioned medium (500 µl) was
collected, centrifuged 10,000 × g at 4°C for 3 min, and
concentrated to 20 µl by Amicon Ultra-0.5 10k centrifugal filter
unit (Merck Millipore). Next, 10 µl of concentrated aliquot was
subjected to gelatin zymography according to the manufacturer's
protocol.
Tube formation assay
The tube formation assay was performed using a
commercially available kit (Endothelial Tube Formation assay; Cell
Biolabs Inc.) according to the manufacturer's protocol, as
previously described (15). The SAS
cells were seeded into 24-well tissue culture plates at a density
of 1.0×105 cells/well and transfected as aforementioned.
After 24 h, the medium was changed to EGM-2 and the cells were
incubated for an additional 48 h under normoxia. A 1-ml aliquot of
conditioned medium was collected, centrifuged at 10,000 × g for 3
min, and stored on ice until needed. Next, 2.0×105 HUVEC
cells were suspended in each conditioned medium and overlaid onto
an ECM gel set in a 96-well plate, according to the manufacturer's
protocol. The cells were incubated for 24 h under normoxic or
hypoxic conditions, examined, and images were captured using a
phase-contrast microscope system (Nikon Eclipse TS-100 and DS-Ri1;
Nikon Corporation).
Mice
The present study complied with the ARRIVE
guidelines and the AVMA euthanasia guidelines 2020, was approved
(approval nos. 14033 and 14034) by the Institutional Animal Care
and Use Committee of Showa University (Tokyo, Japan) and conducted
in accordance with the Showa University Guidelines for Animal
Experiments. Four-week-old female BALB/cAJcl-nu/nu mice (~20 g)
were purchased from Claire Japan (Tokyo) and maintained under
pathogen-free conditions as previously described (21). Each group included three mice (nine
mice per an experiment) and thereby, thirty-six mice in total were
used. SAS, HSC-3 and HSC-4 cells (~1.0×106) in 100 µl
saline were injected subcutaneously into a unilateral flank, and
the cancer-bearing mice were maintained for 40 days to develop
tumors, according to a previous study by the authors (22). Movement disorders, anorexia, nausea,
and abnormal behavior were to be expected due to cancer growth and
metastasis, causing distress and stress to the mice. If the mice
exhibited body weight loss of more than 10% in 7 days, abnormal
behavior by excessive stress such as self-injury or other injury or
damage to the breeding gauge, or if the size of the primary tumor
exceeded 4 cm3, the experiment was to be terminated
immediately by CO2 asphyxiation, even before the set end
point (day 40). However, in the present study, since no mice showed
these abnormal behaviors, all mice were sacrificed at 40 days
without interrupting the experiment. At the end point, the mice
were sacrificed by CO2 asphyxiation. After confirmation
of death by palpation, the tumor was resected, as described in our
previous study (22). The day when
the cells were injected into the mice was set as day 0. Tumor
volumes were measured every 20 days. Tumor volume was determined by
direct measurement and calculated using the formula: π/6 × (large
diameter) × (small diameter)2 (22).
Histology
Resected specimens were fixed with 10% formalin at
4°C for 2 days, embedded in paraffin, and stained with hematoxylin
and eosin (H&E Stain Kit; Sakura Finetek) for 5 min each as
previously described (21,22).
Statistical analysis
All experiments were performed at least four times,
and data was presented as the mean ± standard deviation.
Statistical analyses were performed using the Tukey's test as post
hoc for two-way analysis of variance (ANOVA). Statistically
significant difference was determined at P<0.05. All analyses
were carried out using the KaleidaGraph version 4.5 software
(Hulinks, Inc.) as previously described (22,28).
Results
Hypoxia increases SBSN expression
Previous studies have shown that SBSN is highly
expressed in tumor cells and affects several cancer cells (13–17).
OSCC cells were transplanted into the right unilateral flanks of
mice (Fig. S1A). The transplanted
SAS cells showed the highest ability to form tumors (Fig. S1B and C). The tumor formed by the
HSC-4 cells was too small to be histopathologically sectioned.
Since H&E staining (Fig. S1D)
showed differences in cell differentiation between the SAS and
HSC-3 cells, an in vitro study was conducted to further
investigate whether hypoxia induced the expression of SBSN genes
and proteins in the OSCC and NHEK cells (Fig. 1). The SBSN mRNA was upregulated in
each cell type under hypoxic conditions (P<0.05). In OSCC cells
(SAS, HSC-3 and HSC-4), SBSN mRNA was strongly increased by hypoxia
in a time-dependent manner, but this effect was weaker in the NHEK
cells (Fig. 1A). Western blot
analysis (Fig. 1B and C) revealed
similar results as observed in RT-qPCR, although expression of
cellular SBSN was faint. In addition, VEGF, which is located at the
downstream of HIF, a marker of hypoxia, was increased under hypoxia
conditions, which was in consistency with previous studies
(29,30). However, the signal strength of VEGF
in OSCC was weak, and in NHEK it was barely detected. These results
indicated that SBSN is more crucial in OSCC cells under hypoxia
than in NHEK cells. Notably, since SBSN mRNA and protein levels
were highest in SAS cells, these cells were used as representatives
of OSCC cells in the following experiments.
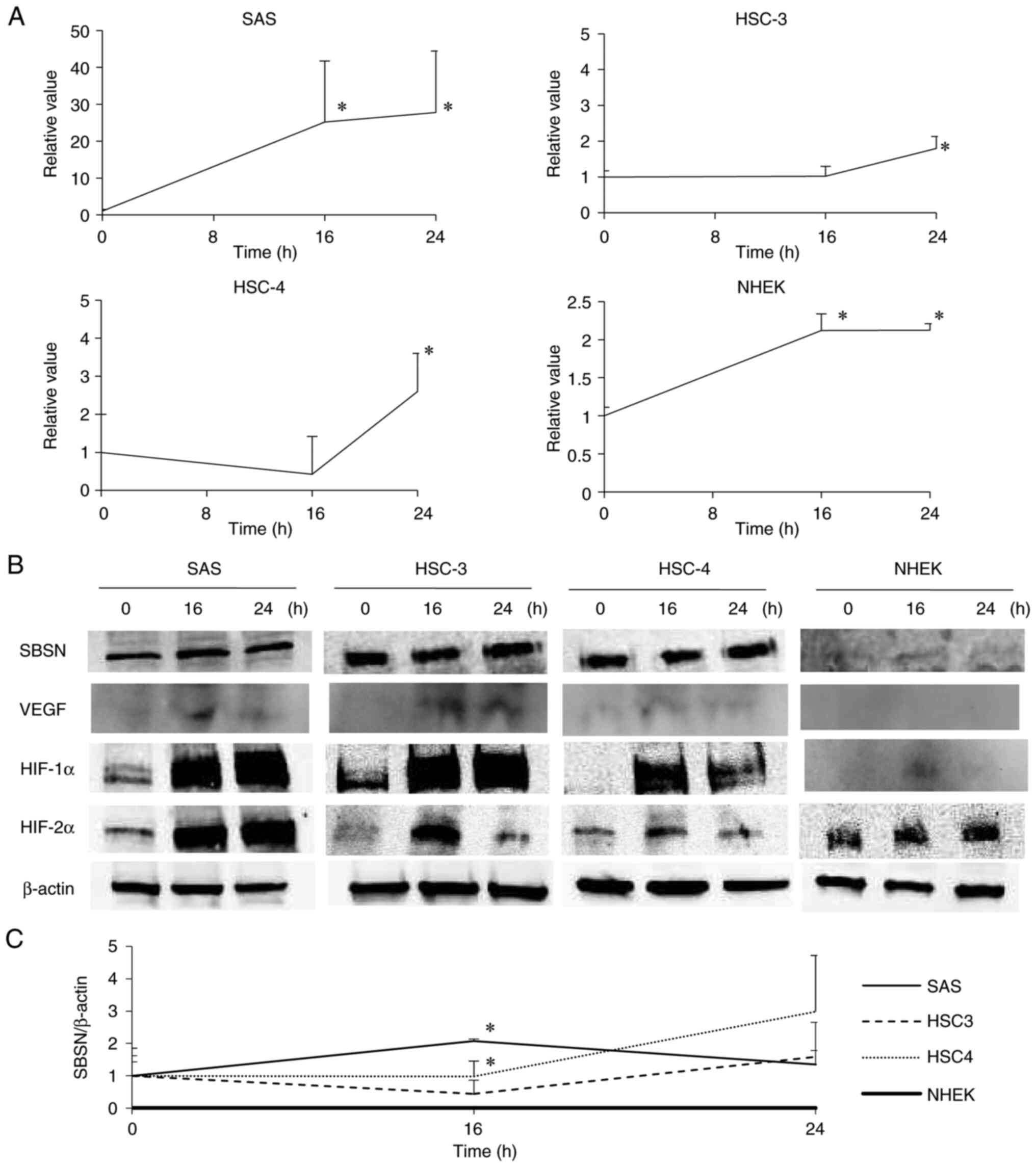 | Figure 1.Induction of SBSN in OSCC and NHEK
cells under hypoxia. SAS, HSC-3, HSC-4 and NHEK cells were exposed
to hypoxia for 0, 16 and 24 h. (A) The SBSN mRNA levels in SAS,
HSC-3, HSC-4 and NHEK cells were analyzed using reverse
transcription-quantitative PCR. (B) The SBSN, VEGF, HIF-1α, HIF-2α
and β-actin protein levels were analyzed using western blotting and
band densities divided by that of β-actin of each band in four
individual experiments are shown with standard deviations (C). The
value at time 0 is designated as ‘1’, and relative values are
shown. *P<0.05 vs. time 0. All experiments were performed four
times at least, and a representative result is shown. SBSN,
suprabasin; OSCC, oral squamous cell carcinoma; HIF,
hypoxia-inducible factor. |
SBSN increases cell proliferation and
decreases apoptosis and autophagy
The effects of SBSN on cell proliferation under
hypoxia were investigated using the MTT (Fig. 2A), BrdU (Fig. 2B), and cell cycle (Fig. 2C and D) assays. Knockdown of SBSN
increased MTT activity (P<0.05) and overexpression of SBSN
decreased MTT activity (P<0.05). However, in the BrdU assay,
which directly examined actively proliferating cells, SBSN
knockdown decreased BrdU incorporation (P<0.05). Additionally,
overexpression of SBSN increased under normoxic and hypoxic
conditions (P<0.05), and this increase was more pronounced under
hypoxia. In addition, the cell cycle assay showed similar results
to the BrdU assay. Western blotting for cyclin-related proteins
(Fig. S2A) indicated involvement
of the cyclin pathways. Crystal violet cell staining (Fig. S2B) showed that the cells under all
conditions were under exponential proliferating phase and that even
on day 3, they did not reach confluency, indicating little
consideration for contact inhibition. These results indicated that
SBSN increased cell proliferation under normoxia and hypoxia. To
investigate this discrepancy between the assays, the effects of
SBSN on representative cell death, apoptosis and autophagy were
examined by using a caspase 3/7 assay (Fig. 3A) and western blotting for p62 and
LC3 (Fig. 3B). Knockdown of SBSN
decreased caspase 3/7 activity (P<0.05); however, SBSN
overexpression had little decreasing effect (P<0.05) under
normoxia. Both SBSN knockdown and overexpression had a slightly
increasing effect (P<0.05) on caspase 3/7 activity under
hypoxia, which was weaker than that under normoxia. In addition,
SBSN knockdown increased both the degradation of p62 and the
conversion of LC3-I to LC3-II, and overexpression of SBSN showed
the opposite effect under normoxia. Similar results were obtained
under hypoxic conditions. These results indicated that although the
effects were weak, SBSN suppresses autophagy in SAS cells,
regardless of the oxygen partial pressure. Western blotting also
showed the successful transfection of siRNAs and expression
vectors.
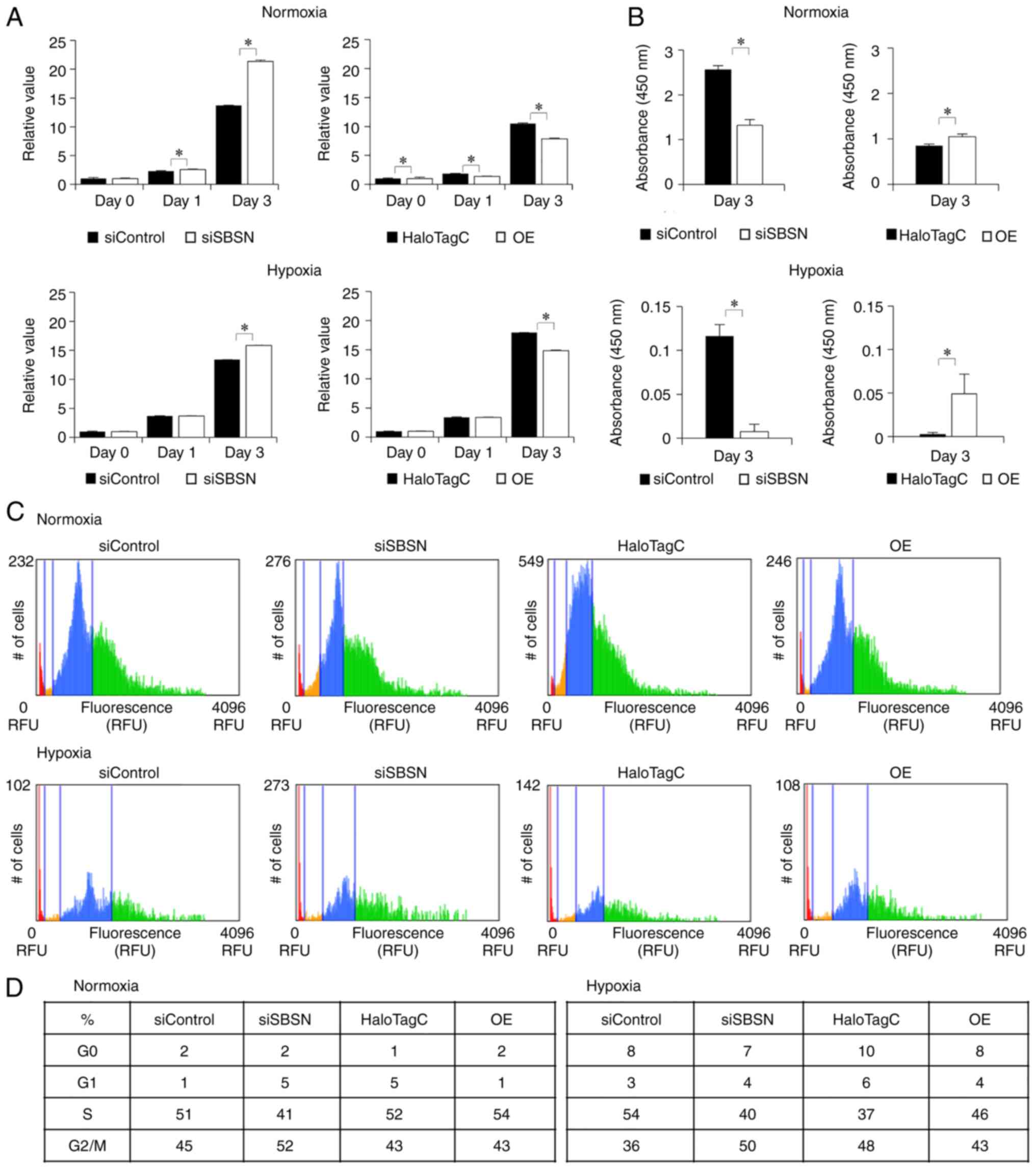 | Figure 2.Effects of knockdown and
overexpression of SBSN on growth of SAS cells. In this experiment,
siRNA for SBSN or control siRNA, and HaloTag-SBSN (OE) or HaloTag
control vector, were transfected into SAS cells and incubated under
normoxic conditions for 24 h. Next, the cells were exposed to
normoxic or hypoxic conditions. After 0, 1 and 3 days, cells were
subjected to the (A) MTT assay and (B) BrdU assay, and the (C and
D) cell cycle assay. For the MTT assay, the value at day 0 is
designated as ‘1’, and relative values are shown. The value of
results was subjected to ANOVA. *P<0.05 vs. control. In panel D,
the percentage of cell-cycle phase (G0 and G1, S, and G2/M) in each
sample is shown in a table. All experiments were performed four
times at least, and a representative result is shown. SBSN,
suprabasin; siRNA, small interfering RNA; OE, overexpression; BrdU,
5-bromo-2′-deoxyuridine; OE, overexpression. |
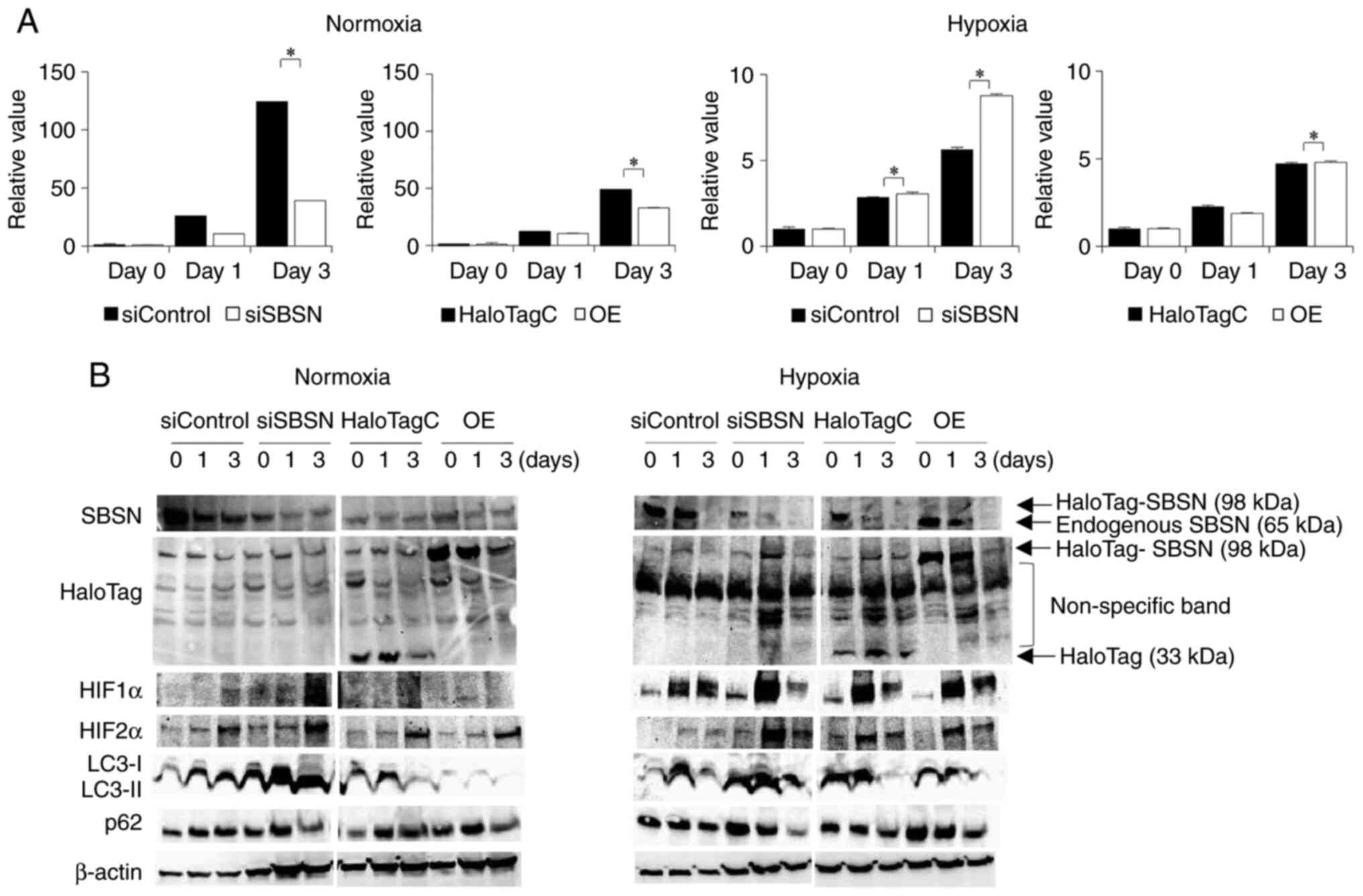 | Figure 3.Effects of knockdown and
overexpression of SBSN on apoptosis and autophagy of SAS cells. (A)
First, siRNA for SBSN or control siRNA, and HaloTag-SBSN (OE) or
HaloTag control vector, were transfected into SAS cells and
incubated under normoxic conditions for 24 h. Next, the cells were
exposed to normoxic or hypoxic conditions. After 0, 1 and 3 days,
either (A) the cells were subjected to caspase 3/7 assays or (B)
total cellular proteins were subjected to western blot analysis to
detect SBSN and HaloTag (overexpressed and endogenous proteins are
indicated by arrows along with each molecular weight on the right
side of right panel), as well as HIF-1α, HIF-2α, LC3-I, LC3-II, p62
and β-actin. For caspase 3/7 assays, the value at day 0 is
designated as ‘1’, and relative values are shown. The value of
results was subjected to ANOVA. *P<0.05 vs. control. All
experiments were performed four times at least, and a
representative result is shown. SBSN, suprabasin; siRNA, small
interfering RNA; OE, overexpression; HIF, hypoxia-inducible
factor. |
SBSN increases cell invasion and
migration
The effects of SBSN on cell invasion were
investigated using a cell invasion assay (Fig. 4A and B). Knockdown of SBSN decreased
cell invasion, and SBSN overexpression increased it under normoxia
or hypoxia (P<0.05). Notably, the increase in cell invasion was
more prominent under hypoxic conditions than under normoxic
conditions. As this effect was considered to be due to increased
cell migration (15), ECM protein
degradation (31–33), and promotion of
epithelial-mesenchymal transition (EMT) (32,34–36),
the effect of SBSN on cell migration, matrix metalloprotease (MMP)
activity, and EMT was investigated using wound healing assay
(Fig. 4C and D), gelatin zymography
(Fig. 4E), and western blotting for
E-cadherin and N-cadherin (Fig.
4F), respectively. The cell migration assay (Fig. 4C and D) revealed that SBSN knockdown
suppressed cell migration and that overexpression of SBSN promoted
migration under both normoxic and hypoxic conditions (P<0.05).
The increasing effect of SBSN was stronger under hypoxia than under
normoxia, as observed in the cell invasion assay. However, gelatin
zymography (Fig. 4E) showed that
knockdown or overexpression of SBSN had no significant or little
effect on the alteration of secreted MMP activity (ProMMP-9,
ProMMP-2, and MMP-2), regardless of normoxia or hypoxia.
Furthermore, western blotting for E-cadherin and N-cadherin, which
are representative EMT markers (Fig.
4F), was performed. E-cadherin expression was increased by both
SBSN knockdown and SBSN overexpression. However, the expression of
N-cadherin was increased by SBSN knockdown and decreased by SBSN
overexpression. These effects were similar under both normoxia and
hypoxia. Importantly, these results showed that SBSN knockdown or
overexpression had no significant involvement in EMT. According to
these results, SBSN increased cell invasion while MMP activity and
EMT did not, and these effects resulted from increased cell
migration.
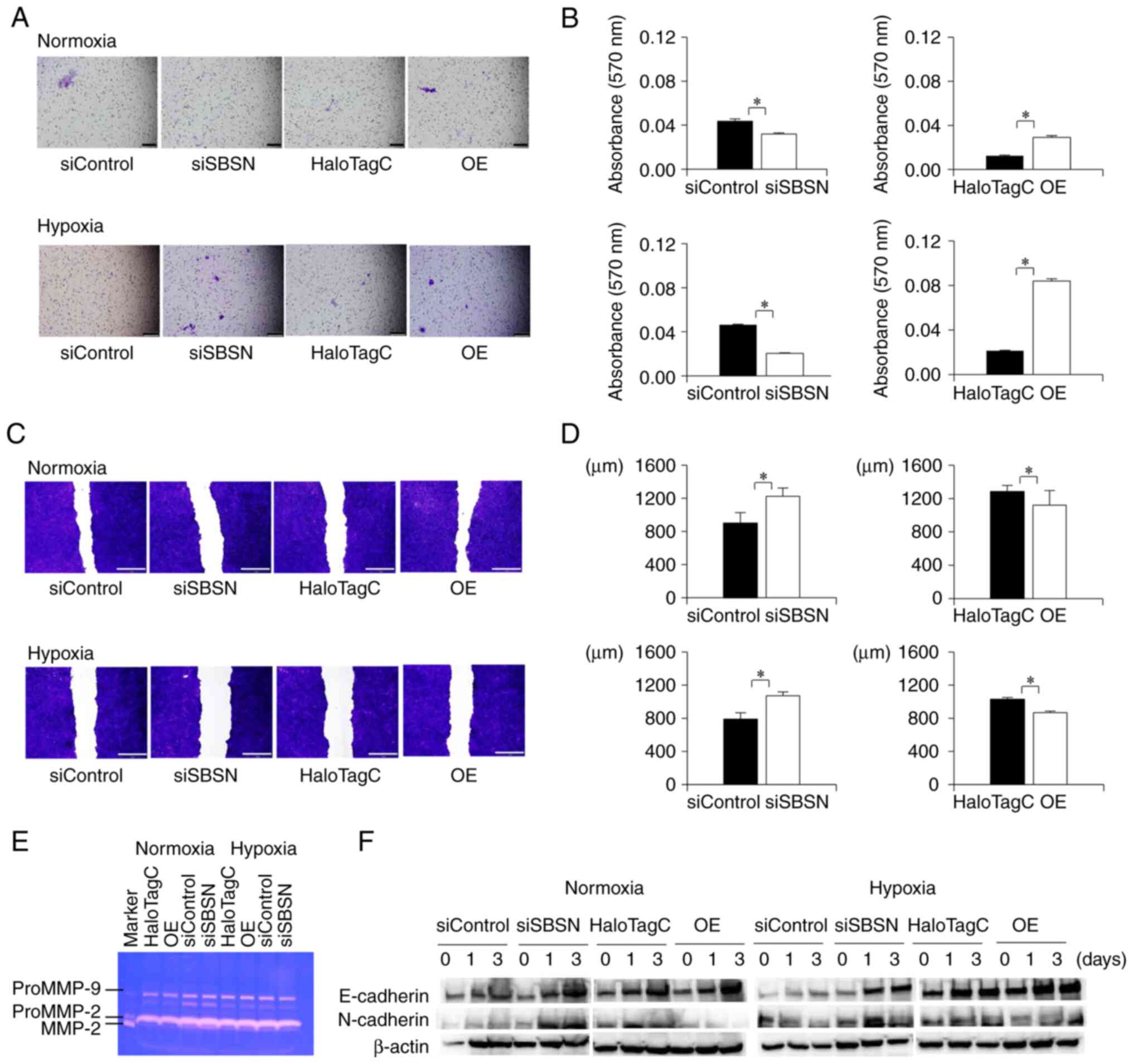 | Figure 4.Effects of knockdown and
overexpression of SBSN on cell invasion, migration, MMP activities,
and EMT. First, siRNA for SBSN or control siRNA, and HaloTag-SBSN
(OE) or HaloTag control vector, were transfected into SAS cells. (A
and B) Cell invasion assay in which the transfected cells were
incubated under normoxic or hypoxic conditions for 48 h. From
these, 1-ml aliquots of the condition medium were collected and
used as chemoattractants. Freshly suspended SAS cells were
subjected to cell invasion assay for 48 h. Then, the invasive cells
were stained by crystal violet. (A) Images of the invasive cells
were captured and (B) the optical absorbance of destained solution
from the membrane was measured. (C and D) Wound healing assay. The
transfected cells were reseeded to a 24-well tissue culture plate,
exposed to normoxic or hypoxic conditions for 24 h, and subjected
to a cell migration assay. (E) Gelatin zymography, in which the
transfected cells were exposed to normoxic or hypoxic conditions
for 48 h, the condition medium was collected and concentrated, and
10-ml aliquots of each concentrated condition medium were subjected
to gelatin-zymography. (F) Western blotting analysis for
epithelial-mesenchymal transition, in which the transfected cells
were exposed to normoxic or hypoxic conditions for 0, 1 and 3 days.
Next, total cellular proteins were subjected to western blot
analysis to detect E-cadherin, N-cadherin and β-actin. The values
in panels (B) and (D) were subjected to ANOVA. *P<0.05 vs.
control. Scale bars: (A) 200 µm (×100) and (C) 1 mm (×40). All
experiments were performed four times at least, and a
representative result is shown. SBSN, suprabasin; MMP, matrix
metalloprotease; siRNA, small interfering RNA; OE,
overexpression. |
SBSN induces angiogenic ability of
OSCC cells
As a previous study has reported that SBSN affects
tube formation in tumor endothelial cells (15), the relationship between SBSN and
angiogenesis was investigated using a tube formation assay
(Fig. 5). In this assay, angiogenic
ability of SBSN in SAS cells was assessed by counting the number of
junctions formed in endothelial tubes. Overexpression of SBSN under
normoxia and hypoxia induced angiogenesis. Knockdown of SBSN
suppressed this effect while overexpression increased this effect.
These effects were more prominent under hypoxic conditions than
under normoxia. This result indicated that SBSN induced
angiogenesis. It has been reported that expression of HIF-1α is
upregulated, which activates transcription of pro-angiogenic
factors such as VEGF (37).
Finally, the relationship between SBSN and VEGF under hypoxia was
investigated (Fig. S3). Knockdown
of SBSN slightly increased, and SBSN overexpression slightly
decreased, VEGF mRNA (P<0.05). These results indicated
that VEGF gene expression is not located downstream of SBSN.
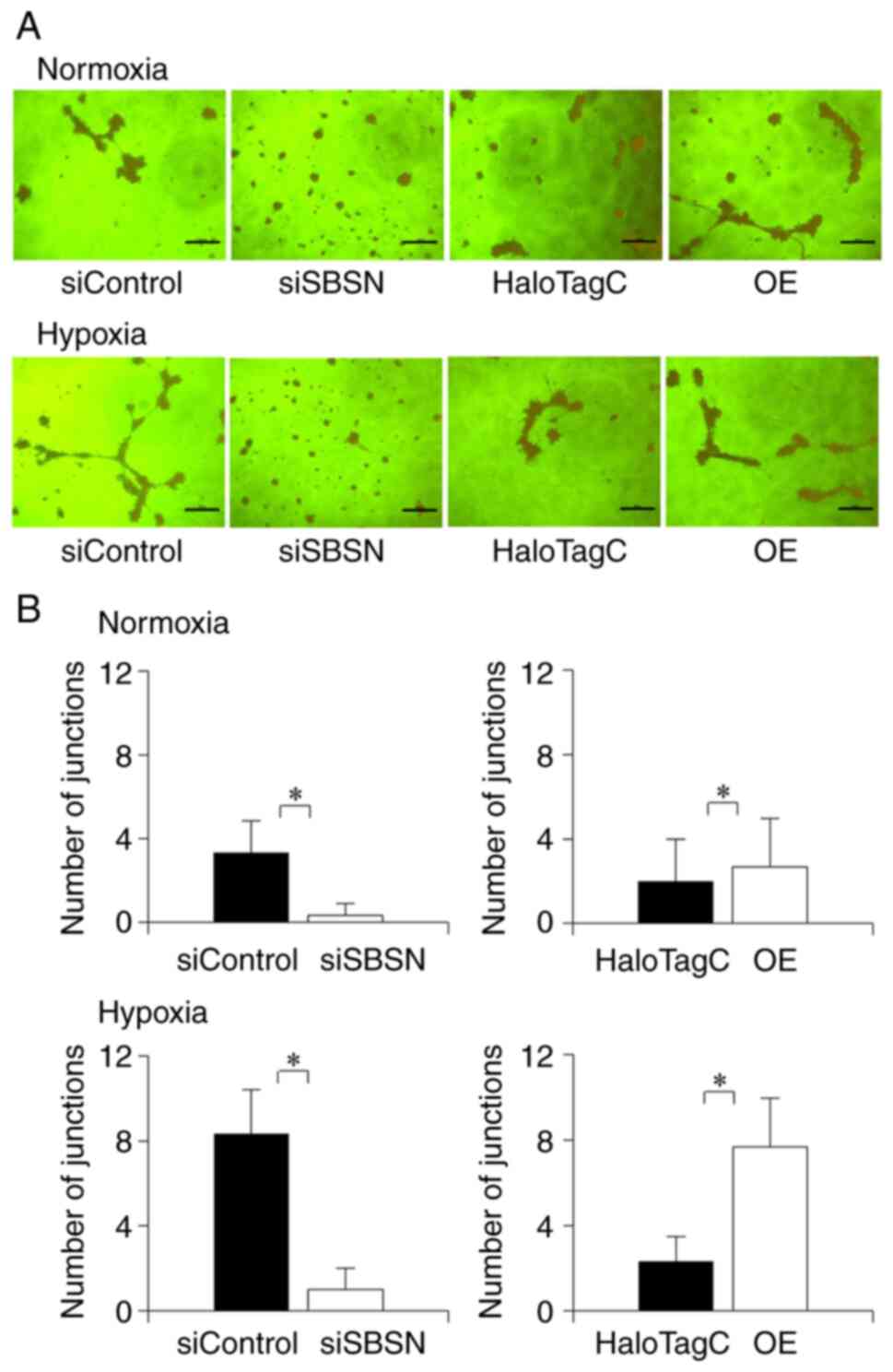 | Figure 5.Effects of knockdown and
overexpression of SBSN on in vitro angiogenesis. (A) First,
siRNA for SBSN or control siRNA, and HaloTag-SBSN (OE) or HaloTag
control vector were transfected into SAS cells and incubated under
normoxic conditions for 24 h. Next, the culture medium was changed
to serum-free EBM-2 medium, and the cells were cultured for another
48 h under normoxic conditions. The conditioned medium of SAS cells
was collected for tube formation assay. The experimental HUVEC
cells were suspended by the conditioned medium of SAS cells and
were subjected to tube formation assay. After 24 h, (Α) images of
the cells were captured, and (B) formed junctions of the
endothelial tubes were counted under a microscope. The results were
subjected to ANOVA. *P<0.05. Scale bars: 200 µm (×200). All
experiments were performed four times at least, and a
representative result is shown. SBSN, suprabasin; siRNA, small
interfering RNA; OE, overexpression. |
Discussion
The center of a tumor is subjected to hypoxia, and
several studies have shown that hypoxia-induced HIF-1α expression
(38,39) is involved in the malignant
progression of OSCC. Tumor cells have been shown to promote
adaptation to hypoxia and contribute to cell invasion, cell
survival and angiogenesis (6,7,39,40).
Accordingly, resistance to treatment and the prognosis of patients
have been revealed to be related to tumor hypoxia (6–10).
Pribyl et al (41) reviewed
the general functions of SBSN, and Tan et al (42) also reviewed the specific roles of
SBSN in cancer and other diseases. However, the role of SBSN under
hypoxic conditions in OSCC cells has not yet been reported.
Therefore, in the present study, focus was addressed on the role of
SBSN under hypoxia and SAS, HSC-3 and HSC-4 cells was used as
representative OSCC cells. It was demonstrated that SAS cells
formed the largest tumor in the tumor xenograft experiment. In
previous studies, SBSN was found to have a poor prognosis in ESCC,
and it was considered that the secretion of SBSN may be positively
correlated with the malignancy of the cancer. The SAS cells are
known to have the highest malignancy in OSCC, and in preliminary
experiments, the different tumor sizes were identified to affect
the malignancy of the cancer. It was hypothesized that the
secretion of SBSN in SAS with the highest malignancy would
correlate with the malignancy of the cancer and the expression of
SBSN in different cell lines. The present in vitro
experiment showed that SAS cells expressed the highest levels of
SBSN. These results are consistent with a previous study revealing
that the expression of SBSN mRNA was upregulated in ESCC cells
(16). Therefore, it was
hypothesized that SBSN is involved in the proliferation and
tumorigenesis of OSCC cells under hypoxic conditions. Among OSCC,
SAS is known to be one of the most malignant (22), thus it was considered that SBSN may
be a prognostic biomarker in cancer cells.
Next, the relationship between SBSN and cell
proliferation was investigated using the MTT, BrdU and cell cycle
assays (Fig. 2). The SBSN decreased
MTT activity but increased BrdU incorporation and accelerated the
cell cycle under hypoxic and normoxic conditions. In addition,
western blotting for cyclin-related proteins was performed and
showed involvement of the cyclin pathways, although further
detailed experiments concerning relationship between cyclin pathway
and cell cycle should be performed. Zhu et al (16) reported that MTT and BrdU assays
showed that SBSN overexpression promoted the growth and
proliferation of ESCC. To address the discrepancy between those
assays in the present study, it was hypothesized that since the MTT
assay examines cell proliferation and viability by measuring
mitochondrial enzyme activity, the results of this assay may not
always reflect cell proliferation alone. It was previously
identified that SAS cells have high proliferation ability and high
malignancy even under hypoxic conditions (21,22,25).
Wigerup et al (6)
demonstrated that hypoxic tumor cells attempt to survive by
decreasing cell proliferation and increasing apoptosis and
autophagy functions. It has been suggested that SBSN function may
be involved in cell survival by increasing apoptosis under hypoxia.
To address this discrepancy, more detailed experiments should be
conducted. It was concluded that the results of the MTT assay may
be related to SBSN secretion rather than to only cell
proliferation. These results suggested that SBSN is slightly
involved in cell proliferation via several signaling pathways such
as the cyclin-related signal.
The autophagosome LC3 is localized to autophagosomes
(28,43), while p62 is selectively integrated
into autophagosomes and degraded by interactions with LC3 (28,44).
Induction of autophagy converts LC3-I to LC3-II, and autophagy
degrades p62. Therefore, LC3 and p62 as autophagy markers were
used. Western blotting for LC3 and p62 (Fig. 3B) showed that SBSN has an inhibitory
role on autophagy in SAS cells under both normoxia and hypoxia. In
addition to normal and cancer cells, autophagy is known to be
involved in the adaptation of normal cells to metabolic stress,
such as hypoxia. This phenomenon may have suppressive and/or
enhancing effects on cancer cells (e.g., suppression of
metastasis and limitation of necrosis and inflammatory cell
infiltration in early-stage cancers and promotion of metastasis in
advanced cancers) (45–49). Furthermore, it was revealed that
SBSN acted in an autophagy suppressive manner. As Su et al
(46) demonstrated that
over-activated autophagy under stress conditions results in cell
death, the results of the present study suggested that SBSN may
suppress over-activated autophagy to protect SAS cells from
excessive cell death under hypoxia.
Alam et al (15) reported that SBSN knockdown inhibited
migration and tube formation in mouse tumor endothelial cells, thus
cell invasion of SBSN under normoxic and hypoxic conditions was
examined using a cell invasion assay. The results demonstrated that
SBSN enhanced cell invasion, and the effects were stronger under
hypoxia than under normoxia. Cell invasion has been reported to
result from cell migration (32,50),
degradation of ECM proteins by MMPs (3,31), and
increased EMT (3,32,34,35).
The effects of SBSN on cell migration, MMP activities, and EMT were
also examined using wound healing assays, gelatin zymography, and
western blotting for E-cadherin and N-cadherin, respectively. It
was found that SBSN enhanced cell migration ability, and the
effects were stronger under hypoxia than under normoxia. However,
SBSN barely altered MMP activity or EMT and had repressive effects
on EMT. Alam et al (15)
reported that SBSN affects cell migration. Importantly, the present
study showed that SBSN enhanced cell invasion and migration, and
its effects were more prominent in hypoxia. Previous studies
(3,6,7,50) have
demonstrated that tumor cells induce degradation of the ECM by MMPs
and increased EMT, as well as cell invasion and migration. The
effects of SBSN on cell invasion and migration may be related to
several other signaling pathways. To clarify this, more detailed
investigations should be performed.
Based on the studies of Alam et al (15) and Takahashi et al (18), wherein SBSN expression induced cell
migration and angiogenesis, the relationship between angiogenesis
and SBSN under normoxic and hypoxic conditions was examined. It was
identified that SBSN induced angiogenesis, which is consistent with
the results of previous studies. Notably, this function is
prominently enhanced under hypoxic conditions. Several previous
studies (3,5,6,37,40,51)
have found that tumor cells induce various transcription factors,
including HIF-1α, to adapt to hypoxia, resulting in induced
angiogenesis (i.e., formation of new blood vessels to
prevent hypoxic conditions). Therefore, angiogenesis is essential
for tumor cell growth, invasion and metastasis, and it is enhanced
under hypoxia (51). Furthermore,
Zhang et al (30)
demonstrated that HIF-1α regulates the transcriptional activation
of the VEGF gene and that hypoxia increases HIF-1α expression,
whereby angiogenesis is promoted, followed by increased VEGF
expression. Melincovici et al (37) also reported that hypoxia is the most
important trigger for the induction of angiogenesis, and Alam et
al (15) reported that SBSN
knockdown had no significant effect on VEGF receptor mRNA
expression in mouse TEC (mTEC). Based on these results, the
relationship between SBSN knockdown or overexpression under hypoxia
and VEGF mRNA (Fig. S3). It was
found that VEGF mRNA was slightly upregulated and downregulated by
SBSN knockdown and overexpression, respectively. These results
suggest that VEGF is not located downstream of SBSN. Alam et
al (15) demonstrated that the
AKT pathway is a downstream factor of SBSN, and Melincovici et
al (37) showed that VEGF
binding to VEGFR-2 leads to downstream PI3K signaling, resulting in
the activation of the AKT pathway for angiogenesis as a cell
survival function. Based on the results of the present study and
those of previous studies, VEGF is not located downstream of SBSN,
and there may be several other signaling pathways between SBSN and
VEGF.
In the present study, focus was addressed on SBSN in
OSCC cells under hypoxia using in vitro experiments and it
was found that SBSN slightly affects cell proliferation and cell
death. More importantly, it was revealed that SBSN was a factor in
cell invasion and angiogenesis, which is in the process of
investigation by the authors using in vivo experiments. This
finding indicated the potency of SBSN as a molecular target for
cancer therapy. However, the functions of SBSN under hypoxic
conditions, such as the relationship between SBSN and HIF and the
signaling pathways, remain to be investigated. Aoshima et al
(52) reported that SBSN expression
in the epidermis is decreased in atopic dermatitis lesional skin
compared with healthy skin, and that decreased expression of SBSN
suggested abnormal differentiation of epidermal keratinocytes and
induction of apoptosis. Pribyl et al (53) reported that SBSN is expressed in the
bone marrow by myeloid cell subpopulations, including
myeloid-derived suppressor cells, and is secreted into bone marrow
plasma and peripheral blood of myelodysplastic syndromes patients.
Furthermore, the aforementioned study revealed that the highest
expression of SBSN is present in a patient group with poor
prognosis, and that SBSN is a candidate biomarker of high-risk
myelodysplastic syndromes with a possible role in disease prognosis
and therapy resistance. Therefore, SBSN may be useful in diagnosing
several diseases, and other roles of SBSN as a secreted protein in
serum should be investigated.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank the staff at the
Department of Oral and Maxillofacial Surgery for their helpful
suggestions, Ms Miho Yoshihara for secretarial assistance, Dr
Shintaro Ohnuma and Dr Kenji Mishima of the Division of Pathology
in the Department of Oral Diagnostic Sciences for their
pathological diagnostic advice, and Dr Kiyohito Sasa and Dr Ryutaro
Kamijyo of Oral Biochemistry for their assistance with several
experiments.
Funding
The present study was supported by Grants-in-Aid for Scientific
Research (KAKENHI) from the Japan Society for the Promotion of
Science (JSPS) (Grant-in-Aid for Scientific Research C) (grant no.
22K10201) and (Grant in-Aid for Early-Career Scientists) (grant no.
20K18738).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AH significantly contributed to and performed the
present study, prepared the figures, and wrote the manuscript. YM
conceived the idea for the study. YA, MW, MN, SM and MK performed
experiments. ToS and TaS helped draft the figures and manuscript.
All the authors have read and approved the final version of the
manuscript. YM and TaS confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved (approval nos. 14033
and 14034) by the Animal Care and Use Committee of of Showa
University (Tokyo, Japan) and was conducted in accordance with the
Showa University Animal Guidelines for Animal Experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Singh P, Rai A, Verma AK, Alsahli MA,
Rahmani AH, Almatroodi SA, Alrumaihi F, Dev K, Sinha A, Sankhwar S
and Dohare R: Survival-based biomarker module identification
associated with oral squamous cell carcinoma (OSCC). Biology
(Basel). 10:7602021.PubMed/NCBI
|
|
2
|
Kim JW, Park Y, Roh JL, Cho KJ, Choi SH,
Nam SY and Kim SY: Prognostic value of glucosylceramide synthase
and P-glycoprotein expression in oral cavity cancer. Int J Clin
Oncol. 21:883–889. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sasahira T and Kirita T: Hallmarks of
cancer-related newly prognostic factors of oral squamous cell
carcinoma. Int J Mol Sci. 19:24132018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jing X, Yang F, Shao C, Wei K, Xie M, Shen
H and Shu Y: Role of hypoxia in cancer therapy by regulating the
tumor microenvironment. Mol Cancer. 18:1572019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arneth B: Tumor microenvironment. Medicina
(Kaunas). 56:152019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wigerup C, Påhlman S and Bexell D:
Therapeutic targeting of hypoxia and hypoxia-inducible factors in
cancer. Pharmacol Ther. 164:152–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilson WR and Hay MP: Targeting hypoxia in
cancer therapy. Nat Rev Cancer. 11:393–410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lagory EL and Giaccia AJ: The
ever-expanding role of HIF in tumour and stromal biology. Nat Cell
Biol. 18:356–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhong H, De Marzo AM, Laughner E, Lim M,
Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL and Simons
JW: Overexpression of hypoxia-inducible factor 1alpha in common
human cancers and their metastases. Cancer Res. 59:5830–5835.
1999.PubMed/NCBI
|
|
10
|
Semenza GL: Hypoxia-inducible factors:
Mediators of cancer progression and targets for cancer therapy.
Trends Pharmacol Sci. 33:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park GT, Lim SE, Jang SI and Morasso MI:
Suprabasin, a novel epidermal differentiation marker and potential
cornified envelope precursor. J Biol Chem. 277:45195–45202. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakazawa S, Shimauchi T, Funakoshi A,
Aoshima M, Phadungsaksawasdi P, Sakabe JI, Asakawa S, Hirasawa N,
Ito T and Tokura Y: Suprabasin-null mice retain skin barrier
function and show high contact hypersensitivity to nickel upon oral
nickel loading. Sci Rep. 10:145592020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Glazer CA, Smith IM, Ochs MF, Begum S,
Westra W, Chang SS, Sun W, Bhan S, Khan Z, Ahrendt S and Califano
JA: Integrative discovery of epigenetically derepressed cancer
testis antigens in NSCLC. PLoS One. 4:e81892009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Formolo CA, Williams R, Gordish-Dressman
H, MacDonald TJ, Lee NH and Hathout Y: Secretome signature of
invasive glioblastoma multiforme. J Proteome Res. 10:3149–3159.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alam MT, Nagao-Kitamoto H, Ohga N, Akiyama
K, Maishi N, Kawamoto T, Shinohara N, Taketomi A, Shindoh M, Hida Y
and Hida K: Suprabasin as a novel tumor endothelial cell marker.
Cancer Sci. 105:1533–1540. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu J, Wu G, Li Q, Gong H, Song J, Cao L,
Wu S, Song L and Jiang L: Overexpression of suprabasin is
associated with proliferation and tumorigenicity of esophageal
squamous cell carcinoma. Sci Rep. 6:215492016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shao C, Tan M, Bishop JA, Liu J, Bai W,
Gaykalova DA, Ogawa T, Vikani AR, Agrawal Y, Li RJ, et al:
Suprabasin is hypomethylated and associated with metastasis in
salivary adenoid cystic carcinoma. PLoS One. 7:e485822012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takahashi K, Asano N, Imatani A, Kondo Y,
Saito M, Takeuchi A, Jin X, Saito M, Hatta W, Asanuma K, et al:
Sox2 induces tumorigenesis and angiogenesis of early-stage
esophageal squamous cell carcinoma through secretion of suprabasin.
Carcinogenesis. 41:1543–1552. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu S, Zhou X, Peng X, Li M, Ren B, Cheng
G and Cheng L: Porphyromonas gingivalis promotes immunoevasion of
oral cancer by protecting cancer from macrophage attack. J Immunol.
205:282–289. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hubackova S, Pribyl M, Kyjacova L, Moudra
A, Dzijak R, Salovska B, Strnad H, Tambor V, Imrichova T, Svec J,
et al: Interferon-regulated suprabasin is essential for
stress-induced stem-like cell conversion and therapy resistance of
human malignancies. Mol Oncol. 13:1467–1489. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kato K, Mukudai Y, Motohashi H, Ito C,
Kamoshida S, Shimane T, Kondo S and Shirota T: Opposite effects of
tumor protein D (TPD) 52 and TPD54 on oral squamous cell carcinoma
cells. Int J Oncol. 50:1634–1646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abe Y, Mukudai Y, Kurihara M, Houri A,
Chikuda J, Yaso A, Kato K, Shimane T and Shirota T: Tumor protein
D52 is upregulated in oral squamous carcinoma cells under hypoxia
in a hypoxia-inducible-factor-independent manner and is involved in
cell death resistance. Cell Biosci. 11:1222021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Momose F, Araida T, Negishi A, Ichijo H,
Shioda S and Sasaki S: Variant sublines with different metastatic
potentials selected in nude mice from human oral squamous cell
carcinomas. J Oral Pathol Med. 18:391–395. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mukudai Y, Kondo S, Fujita A, Yoshihama Y,
Shirota T and Shintani S: Tumor protein D54 is a negative regulator
of extracellular matrix-dependent migration and attachment in oral
squamous cell carcinoma-derived cell lines. Cell Oncol (Dordr).
36:233–245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakamura S, Mukudai Y, Chikuda J, Zhang M,
Shigemori H, Yazawa K, Kondo S, Shimane T and Shirota T:
Combinational anti-tumor effects of chemicals from Paeonia lutea
leaf extract in oral squamous cell carcinoma cells. Anticancer Res.
41:6077–6086. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Lu B, Yang Q, Fearns C, Yates JR
III and Lee JD: Combined integrin phosphoproteomic analyses and
small interfering RNA-based functional screening identify key
regulators for cancer cell adhesion and migration. Cancer Res.
69:3713–3720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kurihara M, Mukudai Y, Watanabe H, Asakura
M, Abe Y, Houri A, Chikuda J, Shimane T and Shirota T: Autophagy
prevents osteocyte cell death under hypoxic conditions. Cells
Tissues Organs. 210:326–338. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eckert AW, Kappler M, Schubert J and
Taubert H: Correlation of expression of hypoxia-related proteins
with prognosis in oral squamous cell carcinoma patients. Oral
Maxillofac Surg. 16:189–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang D, Lv FL and Wang GH: Effects of
HIF-1α on diabetic retinopathy angiogenesis and VEGF expression.
Eur Rev Med Pharmacol Sci. 22:5071–5076. 2018.PubMed/NCBI
|
|
31
|
Di Nezza LA, Jobling T and Salamonsen LA:
Progestin suppresses matrix metalloproteinase production in
endometrial cancer. Gynecol Oncol. 89:325–333. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scheau C, Badarau IA, Costache R, Caruntu
C, Mihai GL, Didilescu AC, Constantin C and Neagu M: The role of
matrix metalloproteinases in the epithelial-mesenchymal transition
of hepatocellular carcinoma. Anal Cell Pathol (Amst).
2019:94239072019.PubMed/NCBI
|
|
33
|
Kim SH, Cho NH, Kim K, Lee JS, Koo BS, Kim
JH, Chang JH and Choi EC: Correlations of oral tongue cancer
invasion with matrix metalloproteinases (MMPs) and vascular
endothelial growth factor (VEGF) expression. J Surg Oncol.
93:330–337. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
González-González R, Ortiz-Sarabia G,
Molina-Frechero N, Salas-Pacheco JM, Salas-Pacheco SM,
Lavalle-Carrasco J, López-Verdín S, Tremillo-Maldonado O and
Bologna-Molina R: Epithelial-mesenchymal transition associated with
head and neck squamous cell carcinomas: A review. Cancers (Basel).
13:30272021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aseervatham J and Ogbureke KUE: Effects of
DSPP and MMP20 silencing on adhesion, metastasis, angiogenesis, and
epithelial-mesenchymal transition proteins in oral squamous cell
carcinoma cells. Int J Mol Sci. 21:47342020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
De Craene BD and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Melincovici CS, Boşca AB, Şuşman S,
Mărginean M, Mihu C, Istrate M, Moldovan IM, Roman AL and Mihu CM:
Vascular endothelial growth factor (VEGF)-key factor in normal and
pathological angiogenesis. Rom J Morphol Embryol. 59:455–467.
2018.PubMed/NCBI
|
|
38
|
Höckel M and Vaupel P: Tumor hypoxia:
Definitions and current clinical, biologic, and molecular aspects.
J Natl Cancer Inst. 93:266–276. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ribeiro M, Teixeira SR, Azevedo MN, Fraga
AC Jr, Gontijo APM and Vêncio EF: Expression of hypoxia-induced
factor-1 alpha in early-stage and in metastatic oral squamous cell
carcinoma. Tumour Biol. 39:10104283176955272017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zimna A and Kurpisz M: Hypoxia-inducible
factor-1 in physiological and pathophysiological angiogenesis:
Applications and therapies. Biomed Res Int. 2015:5494122015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pribyl M, Hodny Z and Kubikova I:
Suprabasin-a review. Genes (Basel). 12:1082021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tan H, Wang L and Liu Z: Suprabasin: Role
in human cancers and other diseases. Mol Biol Rep. 49:1453–1461.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bjørkøy G, Lamark T, Brech A, Outzen H,
Perander M, Overvatn A, Stenmark H and Johansen T: p62/SQSTM1 forms
protein aggregates degraded by autophagy and has a protective
effect on huntingtin-induced cell death. J Cell Biol. 171:603–614.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang X, Yu DD, Yan F, Jing YY, Han ZP, Sun
K, Liang L, Hou J and Wei LX: The role of autophagy induced by
tumor microenvironment in different cells and stages of cancer.
Cell Biosci. 5:142015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Towers CG, Wodetzki D and Thorburn A:
Autophagy and cancer: Modulation of cell death pathways and cancer
cell adaptations. J Cell Biol. 219:e2019090332020.PubMed/NCBI
|
|
49
|
Amaravadi R, Kimmelman AC and White E:
Recent insights into the function of autophagy in cancer. Genes
Dev. 30:1913–1930. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zanotelli MR, Zhang J and Reinhart-King
CA: Mechanoresponsive metabolism in cancer cell migration and
metastasis. Cell Metab. 33:1307–1321. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shih CH, Ozawa S, Ando N, Ueda M and
Kitajima M: Vascular endothelial growth factor expression predicts
outcome and lymph node metastasis in squamous cell carcinoma of the
esophagus. Clin Cancer Res. 6:1161–1168. 2000.PubMed/NCBI
|
|
52
|
Aoshima M, Phadungsaksawasdi P, Nakazawa
S, Iwasaki M, Sakabe JI, Umayahara T, Yatagai T, Ikeya S, Shimauchi
T and Tokura Y: Decreased expression of suprabasin induces aberrant
differentiation and apoptosis of epidermal keratinocytes: Possible
role for atopic dermatitis. J Dermatol Sci. 95:107–112. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pribyl M, Hubackova S, Moudra A, Vancurova
M, Polackova H, Stopka T, Jonasova A, Bokorova R, Fuchs O,
Stritesky J, et al: Aberrantly elevated suprabasin in the bone
marrow as a candidate biomarker of advanced disease state in
myelodysplastic syndromes. Mol Oncol. 14:2403–2419. 2020.
View Article : Google Scholar : PubMed/NCBI
|



















