Introduction
With rapidly growing morbidity and mortality, cancer
remains the leading cause of mortality globally. Worldwide, there
are ~19.3 million new cancer cases and 10 million cancer-related
deaths annually (1). In recent
decades, continuous efforts have been made to improve the outcomes
and prognosis of cancer patients. However, following curative
treatments, patients still face a high risk of disease recurrence,
progression and therapeutic resistance, owing to relapse and the
development of metastases. The ability to monitor disease
recurrence or progression following primary curative treatment is
key to providing immediate effective interventions to eliminate or
control residual tumor cells in patients with a high risk of
relapse. The current standard-of-care mainly relies on traditional
diagnostic methods, such as regular clinical examination, and
radiological imaging to detect disease recurrence or metastasis.
However, a large proportion of patients with successful curative
treatment may harbor clinical micrometastases or minimal residual
disease (MRD) (2,3), a potential source of subsequent early
relapse locally and/or metastatic relapse at distant sites, which
cannot be reliably assessed and monitored by standard radiological
imaging or regular clinical examination due to the lack of
sensitivity. In various types of tumors, there is ample evidence to
support the clinical validity of MRD detection in anticipating
future recurrence in patients treated for early-stage cancer
(4–8). MRD detection is well-established and
widely employed in hematological cancers; however, it remains
challenging in solid tumors, although MRD detection has been
mentioned in the National Comprehensive Cancer Network (NCCN)
guidelines for solid tumors. Performing serial biopsies of solid
tumors for MRD detection is not practical, as it is invasive, and
there is intratumor heterogeneity and difficultly in sampling once
the tumor has metastasized to a distant site.
Liquid biopsy, which detects blood-based
tumor-specific biomarkers, has exhibited good potential in
identifying MRD in cancer patients. Analytes of liquid biopsies
include circulating tumor cells, proteins, extracellular vesicles,
cell-free DNA (cfDNA), circulating tumor DNA (ctDNA), exosomes,
circulating tumor RNA, metabolites, or other biological components
that are shed into the bloodstream by tumor cells (9,10).
Among these, ctDNA is a widely used biomarker as it carries the
comprehensive genetic and epigenetic aberrations of cancers,
including point mutation, deletion, amplification, gene fusion,
hypermethylation and hypomethylation (10), which are representative of the
entire tumor profile, including the intratumor heterogeneity of a
patient. Therefore, it can be utilized as a substitution of tumor
DNA to detect and characterize MRD in a number of cancer types.
There are several potential benefits of detecting MRD using ctDNA
as a biomarker compared with traditional diagnostic methods. First,
ctDNA-based MRD detection can predict disease relapse several
months or even years before current radiological imaging procedures
(11,12). Second, ctDNA may depict a more
comprehensive genetic and epigenetic landscape of tumor
heterogeneity, possibly allowing for the quantitative and
qualitative real-time evaluation of the characteristics of multiple
cancer types in a particular patient. Third, ctDNA-based MRD
detection is safer, easier to obtain, less invasive, and less
dangerous and painful than a tumor biopsy. Finally, ctDNA-based MRD
detection may monitor the molecular evolution of residual tumor
cells in real time during tumor progression, thereby aiding in
precision treatment to delay or eliminate metastatic
recurrence.
Herein, the current approaches and commercial
platforms of plasma-based ctDNA MRD assays are discussed, and the
evidence of using ctDNA as a promising biomarker for the detection
and characterization of MRD in solid tumors is summarized. The
present review also discusses the experimental workflow
considerations and the technical and biological challenges of
ctDNA-based MRD analysis using plasma and its future horizon.
Biology of cfDNA and ctDNA
cfDNA was first discovered by Mandel and Métais
(13) in human blood plasma in
1948. cfDNA are fragments of extracellular nucleic acids, which are
released into biological fluids through multiple mechanisms, such
as cellular apoptosis, necrosis, phagocytosis and possibly active
secretion (14,15) (Fig.
1). cfDNA exists in various bodily fluids, including blood
plasma (16), saliva (17), urine (18), cerebrospinal fluid (19) and pleural fluid (20), but mostly in blood plasma. cfDNA is
typically double-stranded, highly fragmented and ~150–200 base
pairs (bp) in length. The prominent length of cfDNA fragments is
166 bp, corresponding to the length of DNA wrapped in a
chromatosome (21). The half-life
of cfDNA is relatively short, from 16 min to 2.5 h (22). This may be prolonged up to 10-fold
by binding to protein complexes, cell membranes, or extracellular
vesicles (22,23). In healthy individuals, cfDNA mainly
originates from cells of hematopoietic lineage, such as
erythrocytes, leukocytes and endothelial cells (24–26).
The concentration of cfDNA is usually low, ranging from a
negligible amount to 10 ng/ml of plasma, and is rarely >30 ng/ml
(27). Under certain physiological
and pathological conditions, including intense exercise, trauma,
pregnancy, inflammation, infection and cancer, the concentration of
cfDNA significantly increases (28,29).
In a malignant state, the levels of cfDNA may increase 50-fold over
the normal levels to 50–1,000 ng/ml plasma (14), presumably due to a higher cellular
turnover rate.
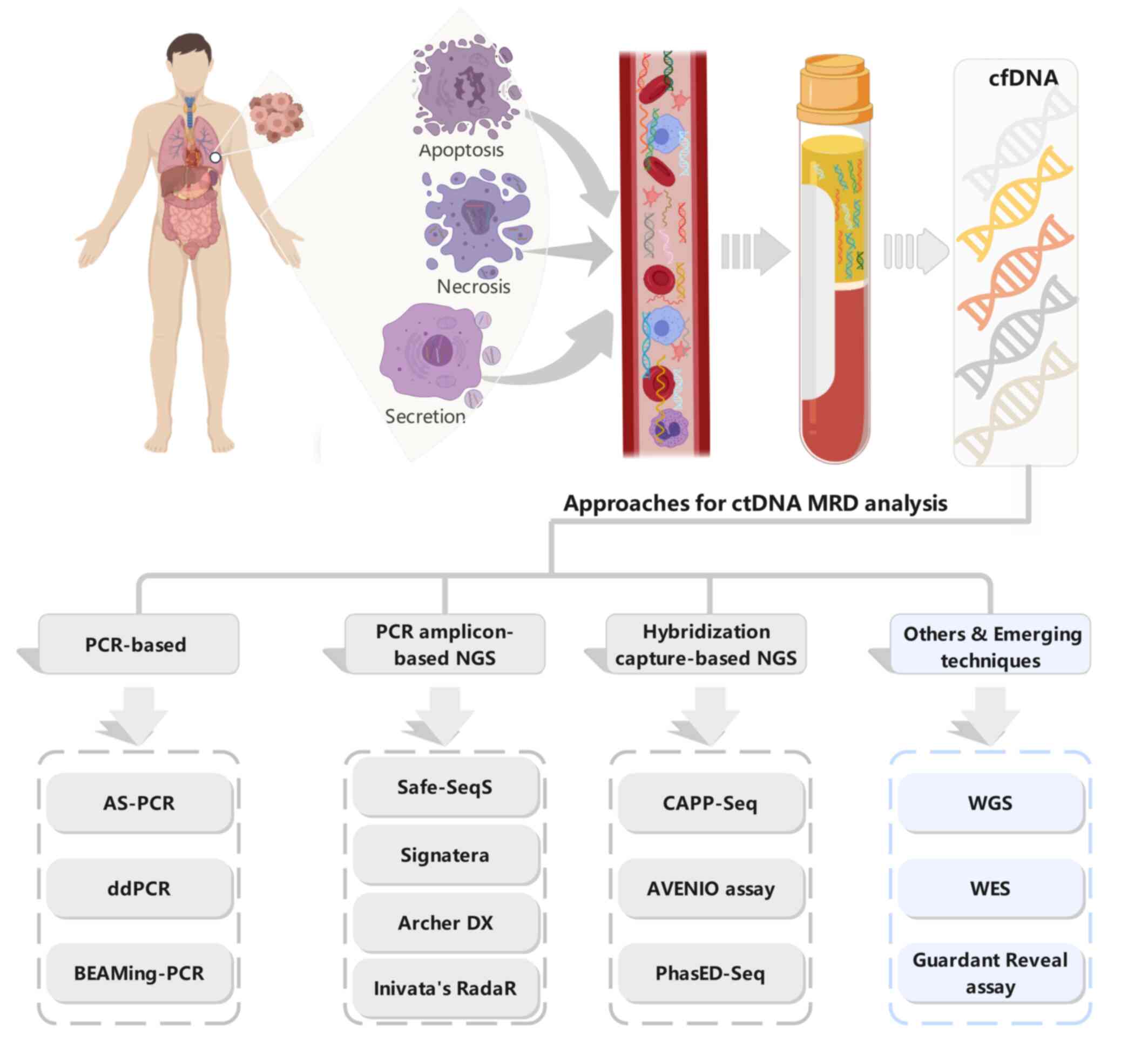 | Figure 1.Origins of ctDNA and technologies for
ctDNA MRD detection. Top panel: ctDNA, released from tumor cells
via apoptosis, necrosis, and active secretion, can be extracted
from the plasma of patients with cancer. Tumor-associated genetic
aberrations can be analyzed in the isolated ctDNA. Bottom panel:
Several different technologies for ctDNA MRD analysis in solid
tumor patients with definitive therapy.cfDNA, cell-free DNA; ctDNA,
circulating tumor DNA; MRD, minimal residual disease; NGS,
next-generation sequencing; AS-PCR, allele-specific PCR; ddPCR,
droplet digital PCR; BEAMing-PCR, Beads, Emulsions, Amplification,
and Magneticsing PCR; Safe-SeqS, Safe-Sequencing; CAPP-Seq, Cancer
Personalized Profiling by Deep Sequencing; PhasED-Seq, Phased
variant Enrichment and detection Sequencing; WGS, whole-genome
sequencing; WES, whole-exome sequencing. |
ctDNA, released by tumor cells, is a fraction of
cfDNA. A higher concentration cfDNA was found in cancer patients by
Leon et al (30) in 1977,
and Stroun et al (31) first
identified tumor-derived cfDNA, referring to it as ctDNA in 1989.
The fraction of ctDNA is highly variable, ranging from <0.05%
(32) to 90% (14). It is influenced by a number of
factors, including the type of tumor, tumor size and staging,
localization and vascularization, tumor microenvironment, antitumor
treatments, and hepatic, splenic and renal clearance (33). ctDNA is more fragmented than healthy
cfDNA, with a shorter fragment length distribution of 132–145 bp,
and the mean length is ~143 bp (34). ctDNA analysis is considered as a
‘real-time’ reflection of tumor burden for the short half-life.
ctDNA can be enriched through some DNA extraction and fragment
enrichment procedures, considering the size difference between
ctDNA and healthy cfDNA. To prevent the degradation, purified ctDNA
should be stored at −20°C, with no more than one freeze-thaw
cycle.
Current technologies/approaches and
commercial platforms for ctDNA-based MRD analysis in solid
tumors
Strategies for ctDNA-based MRD assays are based on
the detection and/or tracking of tumor-specific genomic alterations
in the ctDNA of patients with definitive therapy. The detection of
ctDNA of MRD requires a significantly higher sensitivity to
increase the likelihood of detecting low-variant allele frequency
(VAF). The relatively lower concentration of ctDNA in plasma
following curative-intent treatments, even <0.01% of total cfDNA
(9), poses a key challenge to MRD
detection. With immense efforts being made over the past decades,
multiple approaches based on different technologies have been
developed and utilized for ctDNA-based MRD detection (Fig. 1, bottom panel). These include
PCR-based methods, such as allele-specific PCR, droplet digital PCR
(ddPCR), and beads, emulsions, amplification and magnetics
(BEAM)ing-PCR (35); PCR
amplicon-based next-generation sequencing (NGS) approaches, such as
Safe-Sequencing (Safe-SeqS) (36),
Signatera (Natera Inc), ArcherDX's personalized cancer monitoring
assay and RaDaR assay (Inivata); hybridization capture-based NGS
approaches, such as Cancer Personalized Profiling by Deep
Sequencing (CAPP-Seq), AVENIO assay (Roche Diagnostics), and Phased
variant Enrichment and detection Sequencing (PhasED-Seq) (37); and whole-genome sequencing (WGS),
whole-exome sequencing (WES), and other approaches based on
epigenetic features, such as the Guardant Reveal assay. PCR-based
methods, particularly ddPCR, have a high possibility to reliably
detect known genomic alterations with high sensitivity, down to a
VAF of 0.01% (38). PCR-based
methods to assess MRD have been broadly used in hematological
malignancies with a or several specific mutations, such as BRAF
V600E in hairy cell leukemia (39).
In addition, promising results were observed in one study on solid
tumors with well-established hotspot driver mutations (40). The NGS-based approaches with
high-throughput ability and hypersensitivity are currently the most
broadly used ctDNA detection techniques (16,41).
All those approaches can be classified into two broad categories:
tumor-informed and tumor-agnostic approaches.
Tumor-informed ctDNA-based MRD
approaches
Tumor-informed approaches are based on prior
knowledge of the tumor-specific mutational profile. Thus, the
sensitivity and specificity can be enhanced by tracking
patient-specific genomic alterations expected to be present in
plasma vs. de novo potential false-positive alterations
originated from non-tumor, such as clonal hematopoiesis of
indeterminate potential (CHIP) (42,43).
Typically, tumor biopsy or surgical tissue is sequenced by WGS,
WES, or a large NGS panel to identify the genomic alterations that
were not found in the corresponding germline sample, and the
identified alterations are monitored by tumor-informed fixed or
bespoke panels, or custom-based PCR assays in post-treatment
plasma. The use of WGS, WES, or a large NGS panel to identify
somatic mutations detected as ctDNA in plasma by custom-based panel
broad WGS, WES, or large NGS panel and/or custom-based panel
integrated as a combinatorial approach has demonstrated the
feasibility to detect MRD in previous clinical studies (44–46).
The outstanding advantage of this design is the comprehensive
exploration of genomic alterations of the corresponding tumor to
provide multiple follow-up targets in the subsequent plasma,
thereby improving the sensitivity and reliability of detection.
Moreover, using a small target-sized patient-specific panel allows
for a lower limit of detection (LOD) by deep sequencing at a lower
cost. Some approaches, such as Safe-SeqS (36) and targeted digital sequencing
(TARDIS) (46), which combine
ultra-deep sequencing with unique molecular identifier (UMI)
barcoding, or MRDetect (47) and
the Integration of Variant Reads (INVAR) pipeline (48), which increase the number of
informative targets in an assay, can detect tumor VAF as low as
0.002%. However, the selection of follow-up tumor variants (tumor
variants that should be included in the patient-specific panel) and
the threshold of ctDNA positivity (the limit number of variants per
sample to follow) are subjective or arbitrary, mostly based on
investigator's personal criterion and/or disease-pertinence
bioinformatic algorithms. The number of tracking variants ranges
from 1 to 115 in the currently used tumor-informed approaches
(36,49,50). A
previous study found that tracking at least two variants in plasma
increased the ability to identify MRD by 87.5% (6). The sensitivity of MRD detection by
tracking multiple somatic variants (both driver and passenger) has
been found to be significantly higher than that of MRD detection by
tracking a single mutation (94 vs. 58%, respectively) (51).
The tumor-informed approach offers a cost-effective
solution to examine only tumor-specific genomic alterations present
in the primary tumor in ctDNA, which lowers the risk of
false-positive findings, decreases demands for blood volume, and
increases sensitivity compared with the tumor-agnostic approach.
However, the tumor-informed approach requires tumor biopsy or
surgical tissue for genotyping and may pose several limitations.
First, the turnaround time (TAT) is longer, and more resources are
required for custom personalized assay development compared with
the tumor-agnostic approach, where generally only plasma is
required. Second, in numerous cases, biopsy or surgical tumor
specimens with limited tumor cellularity, particularly in tumors
with neoadjuvant therapy, are insufficient for molecular analysis,
which hinders the application of the tumor-informed approach. In a
previous study, up to 9% of the specimens were reported as
inadequate for tissue sequencing, given their insufficient tumor
content, DNA yield, or DNA quality (52). Third, NGS panels based on a
particular part of the tested tissue cannot capture the whole
genomic alteration view due to tumor heterogeneity (subclones in
different parts of the same tumor or distant metastases). A
patient-specific panel developed according to the genomic profile
of primary tumor may miss new potentially informative targets in
the plasma that occur during the evolution of tumor metastases or
the natural selection of tumor clones during treatment. Despite
these drawbacks, the tumor-informed approach still has immense
potential to become the gold standard of ctDNA MRD detection for
its ability to reliably detect very low quantities of tumor
variants (53).
Tumor-agnostic ctDNA-based MRD
approaches
Tumor-agnostic or tumor-uninformed (tumor-naïve)
approaches detect plasma only and are conducted without a prior
tumor genomic profiling in the tumor of a particular patient.
Currently available liquid biopsy assays designed for identifying
actionable tumor mutations in advanced tumors are not suitable for
ctDNA-based MRD detection due to a low reliability (<0.5% VAF)
(54). Tumor-agnostic approaches
need to be developed for the primary purpose of ctDNA-based MRD
detection, with high specificity and low LOD. NGS combined with
specific enrichment strategies and/or other supplementary methods,
e.g., using UMI allows tumor-agnostic assays to maintain high
specificity and accuracy, while detecting the lower ctDNA allele
fraction. Targeted sequencing approaches examining a larger number
of loci of interest with deep sequencing are optimal for
tumor-agnostic ctDNA MRD detection. Typically, multiple PCR
amplicons [e.g., simple, multiplexed, PCR-based barcoding of DNA
for sensitive mutation detection using sequencing or SiM Sen-seq
(55)] and hybrid capture with
molecular barcoding [e.g., CAPP-Seq (56)] are the two main enrichment
strategies.
Multiplexed PCR-based assays generally demand
multiple separate reaction pools running simultaneously to cover
the large genomic space of designed gene panels, considering the
limited multiplex-ability of PCR in a single reaction pool up to
2,000 bp of genomic breadth (57,58),
which may be challenging when the total cfDNA quantity is limited.
In comparison, hybrid capture-based assays have the ability to
enrich and sequence tens of thousands to millions of bp in a single
reaction. However, these assays may have a higher false-positive
rate (due to the increased opportunity for artifactual mutations to
arise across a large genomic space), and a longer TAT for target
enrichment compared with PCR-based assays (58). Incorporation with single-strand
and/or double-strand UMI or other molecular barcodes and
bioinformatics pipelines, can reduce the artifactual mutations and
technical noise derived from enrichment and sequencing errors,
allowing for the detection of molecular alterations present at very
low allele frequency in high sequencing depth. The CAPP-seq
approach enables the detection of a ctDNA mutant allele fraction
from 0.1% to as low as ~0.0001% and yields very promising results
in specific cancer-type MRD detection (56). Tumor-agnostic assays integrating
both standard genomic alterations and DNA methylation status, such
as the Guardant Health Reveal test (59), exhibit an increased sensitivity
compared with those addressing genomic alterations alone.
Commercial platforms of ctDNA-based
MRD detection
With the advancement and distinct advantages of each
strategy, multiple companies have adopted these technologies to
develop their own ctDNA-based MRD assays. To date, several
commercial platforms are available, including tumor-informed assays
Signatera™ (Natera), PCM™ (ArcherDX), RaDaR™ (Inivata), MRDetect™
(C2i Genomics) and PhasED-Seq™ (Foresight Diagnostic), and
tumor-agnostic assays predicineALERT™ (predicine), AVENIO™ (Roche),
Guardant reveal™ (Guardant Health) and NavDX™ (Naveris) (Table SI).
Tumor-informed commercial
platforms
Signatera™, which has received three US Food and
Drug Administration (FDA) breakthrough device designations for MRD
testing (one in May, 2019 and two in March, 2021), utilizes a
patient's top 16 somatic variants from the primary tumor detected
by WES to personalize a 16-plex multiplex PCR-based NGS approach
for the detection of MRD in plasma. Each target can achieve an
average depth of 100,000× with ultra-deep sequencing, and this
approach has exhibited a sensitivity >98% at 0.01-0.02% ctDNA
concentrations (60). The total TAT
of the Signatera™ assay is between 4–5 weeks, including ~2 weeks
for tumor sequencing results, 1 week for designing the
patient-specific PCR primers and assay, and 1–2 weeks for
post-treatment blood sample testing. Promising results and evidence
of clinical validity were observed using the Signatera™ assay in
several studies for multiple solid tumor types, including
colorectal cancer (CRC), non-small cell lung cancer (NSCLC), breast
cancer and bladder cancer. Moreover, recurrence was detected by the
Signatera™ assay prior to clinical evidence by a median lead time
of 2.3-10.1 months, depending on the tumor type (11,61–63).
ArcherDX received a breakthrough device designation
by the FDA in January, 2019 for its Personalized Cancer Monitoring
(PCM™) assay that uses the company's proprietary anchored-multiplex
PCR technology for ctDNA MRD detection. PCM™ uses patient-specific
anchored-multiplex PCR enrichment panels developed based on somatic
variants via WES of the resected tumor to identify MRD in plasma,
which has served as companion diagnostics in the MERMAID-1 study
(NCT04385368). Inivata's RaDaR™ assay, a multiplex PCR-based assay
based on InVision® platform, was granted breakthrough
device designation by the FDA in March, 2021 as an assay for ctDNA
MRD detection. RaDaR™ tracks a set of up to 48 patient-specific
variants for assessing ctDNA MRD in various types of solid tumor,
with a LOD of 0.0011% VAF reported by Inivata. Similar to
Signatera™, the total TAT of RaDaR™ is 5 weeks, including 4 weeks
for initial bespoke personalized assay design and development and 7
calendar days for plasma cfDNA analysis (64).
MRDetectTM is a WGS-based cfDNA assay for MRD
detection, and all somatic single-nucleotide alterations and copy
number alterations (CNAs) of the tumor tissue via WGS are used to
inform each personalized ctDNA assay. For integrating genome-wide
genomic signature with machine-learning artificial
intelligence-based error suppression models, this assay requires
only 2–3 ml peripheral blood and exhibits a great LOD of 0.001%
tumor fraction at a genome-wide sequencing depth of 35× (47). PhasED-Seq, a hybrid capture-based
sequencing assay, detects the phased variants [two or more SNVs
that occur with 150 bp regions on the same individual DNA molecule
(in cis)] to improve the sensitivity, allowing a 100-fold
improvement over traditional SNV-based ctDNA MRD assays, with a LOD
of below 1 part-per-million (<0.0001%) (ppm) tumor fraction
(37). With high sensitivity,
PhasED-Seq offers the potential for detecting ctDNA during and
immediately after curative intent treatment.
Tumor-agnostic commercial
platforms
Predicine ALERT™ is an integrated MRD assay,
including a targeted panel covering hotspot mutations and important
genes, PredicineCNB™ (a companion LP-WGS assay for copy number
burden), and PredicineEPIC™ (a whole-genome methylation assay). It
detects the genomic variants, aberrant methylation and chromosomal
abnormalities simultaneously for ctDNA MRD detection. With a LOD of
0.005% VAF, Predicine ALERT™ can be a personalized assay based on
the molecular profile of the patient's plasma, urine, or tissue or
a baseline-agnostic assay with a simple blood draw to predict
residual disease (65). The AVENIO
ctDNA Surveillance kit, one of three assays in the AVENIO ctDNA
assay portfolio, is a hybrid capture-based NGS assay designed and
optimized for the longitudinal monitoring of tumor burden in lung
cancer and CRC. The surveillance kit, containing 197 genes with a
panel size of 198 kb, exhibits >99% specificity and >99%
positive predictive value (PPV) for all four classes of alterations
(SNVs, indels, fusions, and CNAs) with a LOD of 0.1% VAF. The
clinical utility and validity of the platform are currently being
investigated. In a previous study on 24 patients with
oligometastatic CRC, the use the tumor-agnostic approach based on
the AVENIO assay to detect plasma and urine samples yielded a
sensitivity and specificity of 95 and 100% for plasma-based ctDNA
analysis, respectively, and a sensitivity and specificity of 64 and
100% for urine-based ctDNA analysis, respectively, due to the lower
ctDNA levels in urine (~11-fold lower than in plasma) (66). Guardant Reveal™, a 500 kb hybrid
capture-based gene panel, is a blood-only MRD assay for MRD
assessment in early-stage CRC, breast cancer and lung cancer.
Utilizing Guardant Health proprietary bioinformatics software,
Guardant Reveal™ can query the genomic and epigenomic signatures
simultaneously and filter out biological noise derived from CHIP
without the paired blood mononuclear cell sequencing data. In
early-stage CRC, Guardant Reveal™ exhibits 91% sensitivity at a LOD
of 0.01%. In addition, favorable sensitivity and specificity for
recurrence were observed in a prospective observational study that
enrolled 103 patients with stage I–IV CRC with curative-intent
therapies (59).
Evidence supporting ctDNA MRD in solid
tumors
The significant development of cfDNA-sequencing
methods expands the potential clinical use of ctDNA profiling for
the detection of MRD and molecular relapse. Multiple studies using
a variety of ctDNA-based MRD approaches in various types of solid
tumors have indicated that ctDNA has the potential as a predictor
of cancer recurrence with high specificity and sensitivity, and the
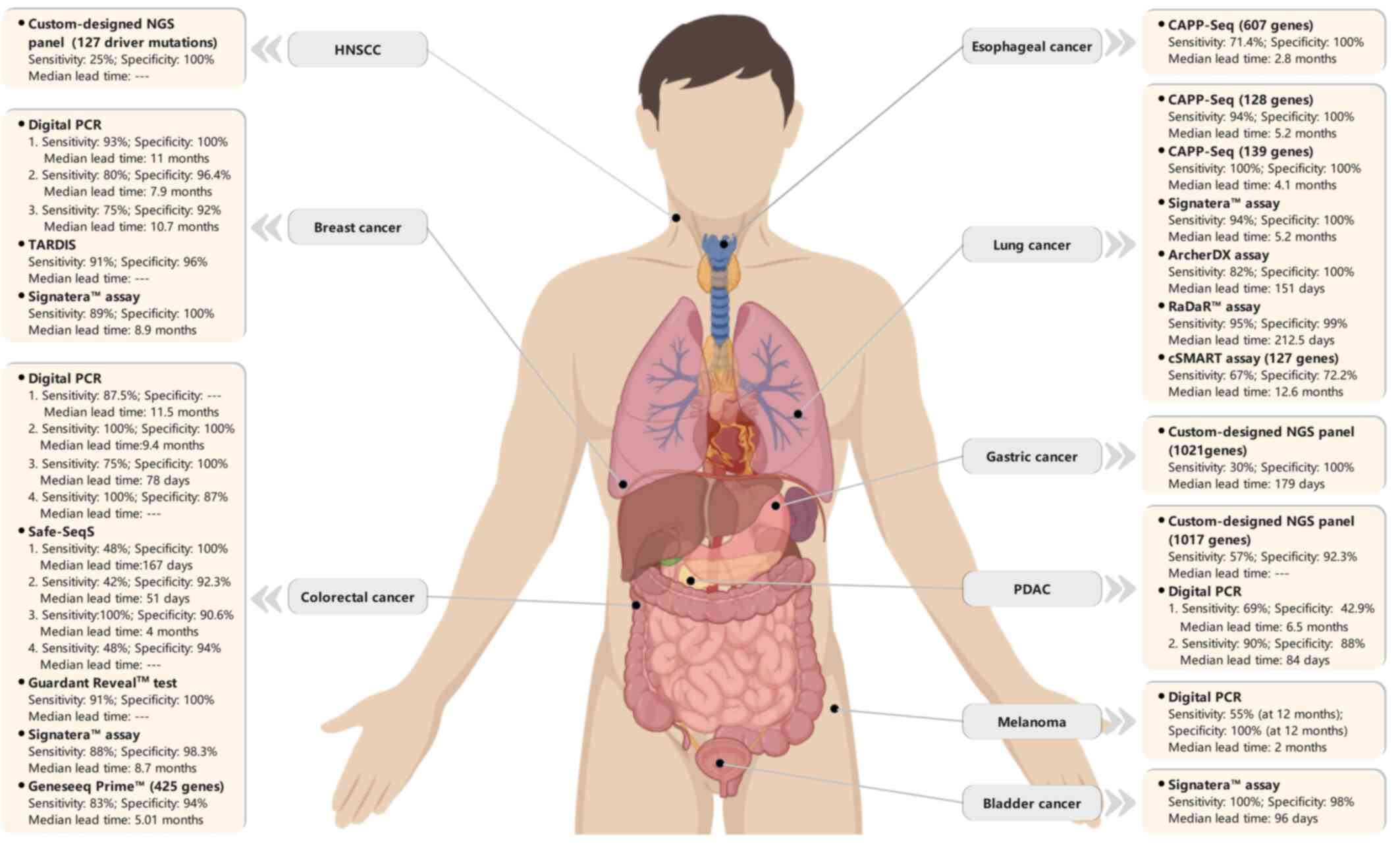
detailed performances are shown in Table SII and Fig. 2.
Lung cancer
The TRACERx study (NCT01888601), a multicenter
prospective trial that aimed to enroll >800 patients with
primary stage I–III NSCLC treated with surgery for tracking tumor
evolutionary dynamics in ctDNA through cancer relapse and
metastases, was one of the first studies demonstrating the clinical
utility of ctDNA MRD in patients with early-stage NSCLC following
curative-intent treatment. In 2017, based on the analysis of a
subgroup of 24 patients using a tumor-informed multiplex-PCR NGS
approach (Signatera™ assay), Abbosh et al (50) reported that 13/14 (93% sensitivity)
patients with confirmed NSCLC relapse had positive ctDNA detection
prior to or at clinical relapse. The median lead time of ctDNA
detection prior to NSCLC relapse confirmed by radiographic evidence
was 70 days (range, 10–346 days); one patient was ctDNA-positive
among 10 patients without clinical evidence of relapse (90%
specificity) (50). The updated
data of that study using Archer DX technology revealed that 37/45
(82.2%) patients who suffered relapse of their primary NSCLC were
ctDNA-positive before or at the clinical relapse, with a median
ctDNA lead time of 151 days (range, 0–984 days). For 23
relapse-free patients at a median of 1,184 days of study follow-up,
ctDNA was detected in 1 of 199 time points analyzed, reflecting the
specificity of the ctDNA MRD assay (67).
In a surveillance study on 40 patients treated with
curative intent for stage I–III localized lung cancer, ctDNA MRD
detection using CAPP-seq ctDNA analysis with a LOD of ~0.002% VAF
revealed that 94% sensitivity and 100% specificity were achieved by
tracking multiple somatic variants each patient at the MRD landmark
(defined as first ctDNA detected within 4 months of treatment
completion). Compared with patients with detectable ctDNA at the
MRD landmark, ctDNA-negative patients had an improved
disease-specific survival and overall survival (OS). The serial
ctDNA analysis of 37 patients with detectable pretreatment ctDNA
revealed that 20 patients (54%) were ctDNA-positive at least at one
post-treatment time point during follow-up, and all of these 20
patients ultimately recurred. Recurrence detected by ctDNA was
earlier than that detected by radiographic imaging in 72% of the
recurred patients by a median of 5.2 months (4). Peng et al (68) found that among 19 patients with
recurrent NSCLC, 17 were ctDNA-positive within 2 weeks after
surgery, and post-operative ctDNA detection preceded the
radiographic findings by a median of 12.6 months. A significantly
improved progression-free survival (PFS) and OS were observed in
patients with undetectable postoperative ctDNA (68). In a recent surveillance study on 88
patients with localized early-stage (IA-IIIB) NSCLC, serial ctDNA
in post-operative plasma samples was analyzed using personalized
RaDaR™ assay to detect residual disease and recurrence. ctDNA MRD
detection yielded 95% PPV and 98.7% specificity for relapse
prediction of primary tumor, with a median lead time of 212.5 days
relative to clinical recurrence (69).
CRC
Although the majority of CRC cases are diagnosed at
early stages (I–III), a higher recurrence rate (30–40%) remains for
patients with stage IIB and higher CRC following surgical resection
plus adjuvant chemotherapy, and in the majority of cases (80%),
recurrence occurs within the first 2 years of surgery (70). Several studies have demonstrated
that ctDNA MRD detection can predict the risk of relapse in
patients with resected, localized CRC with 82–100% sensitivity and
89–100% specificity (Table SII).
In a cohort of 230 patients with stage II CRC treated with surgical
resection, ctDNA in plasma was detected by designed personalized
Safe-SeqS assay. ctDNA was detected in 14/178 (7.9%) patients
without chemotherapy treatment, and 11/14 (78.6%) had clinical
recurrence. In 164 of 178 (92.1%) post-operative ctDNA-negative
patients, disease recurrence occurred in 16 (9.8%) patients. The
post-operative ctDNA status was an independent predictor of
inferior recurrence-free survival (RFS). ctDNA-based MRD analysis
yielded a 48% sensitivity and 100% specificity in predicting
recurrence at 36 months after surgery. The median lead time of
ctDNA over radiological recurrence was 167 days (81–279 days),
longer than that of carcinoembryonic antigen elevation (61 days;
0–207 days) (71). In another
prospective study on 130 patients with stage I–III CRC following
definitive therapy, longitudinal ctDNA analysis using Signatera™
technology identified 14 of 16 (87.5%) patients with clinical
recurrence, and ctDNA-positive patients exhibited 40-fold increased
risk of recurrence. Serial ctDNA analyses with 88% sensitivity and
98% specificity for relapse provided a median lead time of 8.7
months (0.8-16.5 months) relative to the radiographic recurrence
(11).
Recently, Parikh et al (59) provided a plasma-only MRD assay
(Guardant Reveal™) in patients with stage I–IV CRC post-definitive
therapy. At the landmark time point (blood drawn 1 month after
definitive therapy; median, 31.5 days), 15 patients were
ctDNA-positive, and all of them recurred at a median of 162 days
following definitive therapy. The assay demonstrated 55.6%
sensitivity and 100% specificity for landmark recurrence.
Integrating serial longitudinal (blood drawn subsequently after the
‘landmark’ time point) and surveillance (blood drawn within 4
months of clinical recurrence) analyses improved the sensitivity
from 55.6 to 69.0 and 91%, respectively, with the specificity
remaining at 100%. Incorporating epigenomic signatures improved MRD
detection sensitivity by 25–36% compared with the assessment of
genomic variants alone. The results revealed that the sensitivity
of ctDNA-based MRD for recurrence prediction could be improved by
combining the genomic and epigenomic signatures and by integrating
serial longitudinal and surveillance samples. This assay with
acceptable assay performance characteristics is currently
commercially available for clinical use to identify CRC patients at
high risk of recurrence after curative-intent resection and is used
in multiple ongoing prospective studies to collect additional
datasets for further validation of the assay performances and
clinical utility. ctDNA-MRD analysis is also a powerful prognostic
factor in patients with resectable colorectal liver metastasis
(CRLM), as well as in patients with non-metastatic CRC. The serial
analysis of ctDNA using Safe-Sequencing (Safe-SeqS) assay in 54
patients with resectable CRLM revealed that post-operative ctDNA
clearance was significantly associated with an improved PFS. All 8
patients with persistently detectable ctDNA of serial analysis
during adjuvant chemotherapy recurred (72).
Breast cancer
An initial study on ctDNA MRD detection in breast
cancer was performed in 2015 by Garcia-Murillas et al
(73). In this prospective cohort
of 55 patients with high-risk early-stage breast cancer with
neoadjuvant chemotherapy (NAC), personalized tumor-specific dPCR
assays were designed for each somatic mutation identified in the
corresponding primary tumor. Tracking these mutations in the single
or serial blood samples obtained at different post-curative therapy
time points yielded a high early relapse prediction accuracy
(hazard ratio, 25.1). Mutation tracking in serial blood samples
improved the sensitivity of relapse prediction compared with a
single postoperative sample (from 50 to 80%), with a median lead
time of 7.9 months (range, 0.03-13.6 months) relative to clinical
relapse (73). In the same year, a
retrospective study on 20 patients with primary breast cancer
revealed that serial ctDNA analysis using a quantitative
ddPCR-based personalized rearrangement analysis had a 93%
sensitivity and 100% specificity for recurrence prediction, with a
median lead time of 11 months (range, 0–37 months) relative to the
clinical recurrence in 86% (12/14) of patients (51). In another pilot study on 38 patients
with early-stage triple-negative breast cancer who received
multiple-agent NAC, the Oncomine Research Panel consisting of 134
cancer genes was used to detect the mutations in the primary tumor
and track the mutations in the following plasma. A total of 4 of
the 33 patients with mutations identified in their primary tumors
were ctDNA-positive in their plasma, and all four patients had
disease recurrence (100% specificity) within 9 months. However,
sensitivity was limited to detect only 4 of 13 patients who
clinically relapsed (31% sensitivity) (74). Another study revealed that the
detection of ctDNA during follow-up was associated with the risk of
relapse in all early-stage breast cancer subtypes, with a median
lead time of 10.7 months (8.1-19.1 months) prior to clinical
relapse. A total of 22 of 23 patients (96%) with distant
extracranial metastatic relapse were ctDNA-positive compared with 1
of 6 (17%) patients with brain-only metastasis, suggesting that
relapse sites may affect the sensitivity of ctDNA MRD detection
(75).
In a study on 49 patients with high-risk with stage
I–III breast cancer, serial plasma ctDNA analysis by Signatera™
yielded 88.9% sensitivity and 100% specificity for predicting
relapse, with a lead time of up to 2 years (median, 8.9 months;
range, 0.5-24.0 months) ahead of clinical or radiologic relapse
(63). A personalized ctDNA
analysis using TARDIS, developed by McDonald et al (45), demonstrated excellent accuracy for
identifying molecular response and residual disease in patients
with stage I–III breast cancer treated with curative intent. In
addition, a novel, ultrasensitive assay for tracking hundreds of
patient-specific mutations to detect MRD in patients with
early-stage breast cancer revealed that tracking a larger number of
individualized tumor mutations in cfDNA could increase the
reliability and improve the sensitivity of ctDNA MRD detection
(76). This approach demonstrated a
100-fold higher sensitivity than ddPCR when tracking 488 mutations.
The presence of ctDNA MRD soon after curative surgery and at a
post-operative landmark (1 year after surgery) was highly
predictive of distant relapse. The median lead time of ctDNA
detection over clinical relapse was 18.9 months (3.4-39.2 months)
(76). In the multicenter I-SPY 2
trial (NCT01042379), serial ctDNA testing was able to predict
pathological complete response (pCR) and metastatic recurrence risk
in high-risk early breast cancer patients treated with NAC.
Patients who were ctDNA-positive at 3 weeks following the
initiation of paclitaxel treatment were significantly more likely
to have residual disease after NAC compared with patients with
cleared ctDNA (83% non-pCR vs. 52% non-pCR). Among the 43 patients
who failed to achieve pCR, 14% patients who were ctDNA MRD-positive
experienced a significantly higher risk of metastatic recurrence.
Notably, the remaining 86% patients with cleared ctDNA had a
favorable prognosis, similar to those who achieved pCR. The lack of
ctDNA clearance was a strong predictor of poor treatment response
and higher metastatic recurrence (46).
Other solid tumors
ctDNA-based MRD detection has also shown the ability
of reliably predicting recurrence in a number of other solid
tumors, such as pancreatic, bladder, head and neck, and esophageal
cancer. In a study on 68 patients with localized advanced bladder
cancer treated with NAC and surgery, serial ctDNA analysis by
Signatera™ during surveillance following cystectomy demonstrated
100% sensitivity (13/13 patients) and 98% specificity (48/49
patients) in identifying metastatic relapse, with a median lead
time of 96 days relative to radiographic imaging (61). Positive ctDNA at diagnosis before
chemotherapy, after chemotherapy and before cystectomy, and during
disease surveillance after cystectomy was significantly associated
with a poor DFS and inferior OS, and in a multivariate analysis,
positive ctDNA was the strongest predictor of RFS after cystectomy
[hazard ratio (HR), 129.6; P<0.001] (61). In a prospective study that enrolled
45 patients with localized esophageal cancer (ESCA) treated with
esophagectomy or chemo-radio therapy (CRT), ctDNA analysis used a
designed CAPP-Seq ESCA panel targeted 802 regions of 607 genes
(77). The detected ctDNA post-CRT
was significantly associated with an increased risk of disease
progression (HR, 18.7), distant metastasis (HR, 32.1) and
disease-specific mortality (HR, 23.1). The detection of ctDNA
post-CRT was able to detect relapse on an average of 2.8 months
before radiographic evidence, with 71.4% sensitivity and 100%
specificity for recurrence prediction (77).
The value of ctDNA in predicting relapse was
investigated using panel-captured sequencing in patients with
pancreatic ductal adenocarcinoma following surgical treatment
(78). Specifically, 8/9 (88%)
patients with detected post-operative ctDNA ultimately recurred,
and post-operative positive ctDNA was an independent prognostic
factor for DFS (HR, 3.60) and was significantly associated with
disease relapse in univariate analysis and multivariate analyses
(78). For gastric cancer, ctDNA
MRD detection could identify early-stage patients with a high risk
of recurrence and facilitate new adjuvant therapy studies to
improve survival in the adjuvant treatment setting (12). In a prospective cohort study on 46
patients with stage I–III, resectable gastric cancer, positive
ctDNA after surgery was significantly associated with a higher risk
of relapse. All patients with ctDNA positivity in the immediate
postoperative period eventually recurred, with a median time of 179
days prior to radiographic recurrence. Positive ctDNA at any
post-operative subsequent longitudinal time point was associated
with a poor DFS and OS. Post-operative ctDNA positivity yielded
time-dependent sensitivity and specificity in predicting
recurrence, with 30% sensitivity and 100% specificity at 30 months
after surgery (12). In the
chemotherapy vs. chemoradiotherapy after surgery and preoperative
chemotherapy for resectable gastric cancer (CRITICS) study
(NCT00407186), all 11 patients with operable gastric cancer with
ctDNA clearance at the median post-operative 42-month follow-up
were alive and free of recurrence, and 6/9 patients with detected
ctDNA at the post-operative time point experienced disease
recurrence and succumbed due to metastatic disease. Moreover,
patients with detectable tumor-specific mutations after surgery
exhibited a significantly shorter median OS and event-free survival
(28.7 months and 18.7 months vs. median not reached, respectively),
as well as a 21.8-fold higher risk of relapse. ctDNA analysis
determined disease recurrence at 1.3 months, with a median lead
time of 8.9 months relative to clinical recurrence (79).
In another study, serial ctDNA analyses were
conducted in 20 patients with locally advanced head and neck
squamous cell carcinoma (HNSCC) treated with definitive
radio-chemotherapy (80).
Furthermore, 2/8 (25%) patients suffering from a relapse were
ctDNA-positive, and both had a significant number of tumor
fragments with ctDNA MRD scores of 12.16 (recurrence within 101
days) and 2.44 (distant relapse after 833 days). All 8 patients
(100%) who were relapse-free were ctDNA-negative. The same dynamic
properties were observed in circulating HPV DNA (cvDNA) and ctDNA
levels during treatment in that study (80). In another single-center prospective
cohort study of 17 patients with stage III–IVb HNSCC, plasma ctDNA
was detected in all 5 patients with clinical recurrence prior to
disease progression, with the lead time ranging from 108 to 253
days (44). In a study on 133
patients with resected melanoma, positive ctDNA at the
post-operative time point was a stronger predictor of recurrence
than that at the baseline time point and was significantly
associated with distant metastasis-free survival. All patients with
post-operatively detected ctDNA eventually experienced recurrence.
The time-dependent accuracy of post-operative ctDNA in predicting
clinical relapse was observed during 6–30 months of follow-up after
resection, and, 55% sensitivity and 94% specificity at 12 months
after resection were observed (81).
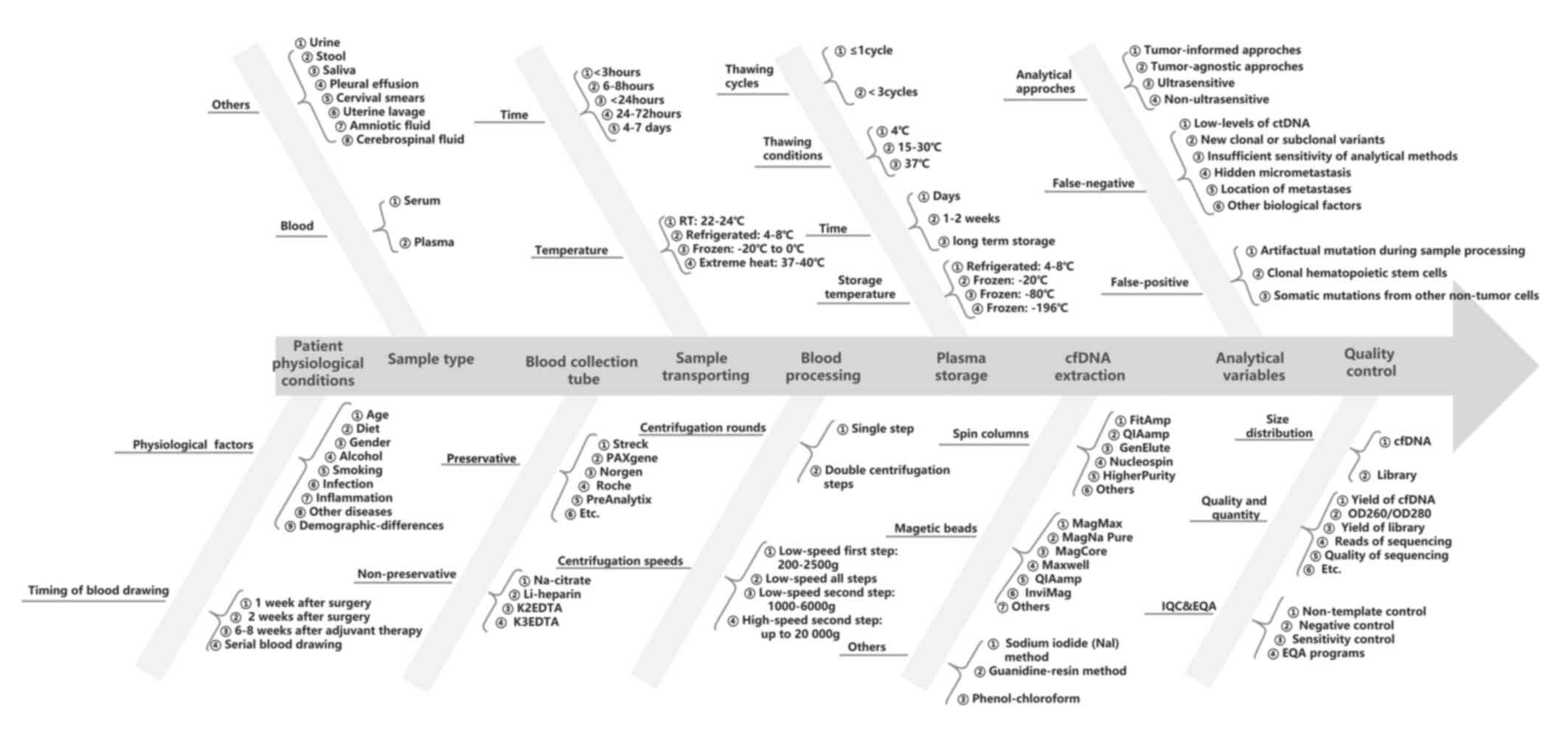
Technical considerations and challenges of
ctDNA analysis
Extensive research has demonstrated the high
potential of ctDNA in determining MRD in solid tumors; however,
ctDNA-based MRD detection is still complex as there are a number of
technical and biological challenges to its widespread clinical
application. An accurate ctDNA analysis remains a critical
technical challenge, particularly in patients with curative
treatments, due to the ultra-low ctDNA level in body fluids.
Pre-analytical workflows are crucial for reliable ctDNA MRD
analysis and analytical approaches. All parameters of the entire
experimental workflow that may impact the accuracy and
reproducibility of the final result, including pre-analytical
factors, should be considered (Fig.
3).
Pre-analytical variables and
considerations
The timing of sample collection, sample collection
tubes, storage and transportation conditions, centrifuge processing
and extraction protocols are the main pre-analytical variables of
plasma ctDNA MRD detection (Fig.
3). The ctDNA level and fraction may be affected by various
factors, such as the physiological condition of a patient and
concurrent inflammatory processes. The timing of post-treatment
sampling is significantly associated with the clinical sensitivity
and specificity of ctDNA MRD assay, particularly in studies
employing an MRD landmark analysis (82). The blood draw timing after the
completion of curative therapy was heterogenous across the reported
ctDNA MRD studies, and there is still no standard for the first and
serial longitudinal blood draw timing. The European Society for
Medical Oncology (EMSO) recommends that for ctDNA MRD detection
after surgery, the ideal timing of blood sampling is at least 1
week after surgery and at least 2 weeks for major surgeries for
longer healing time (83). Plasma
is the preferred source for cfDNA analysis in blood, given that
cfDNA in serum is usually contaminated with larger DNA fragments
originating from the ex vivo rupture of leukocytes and other
cells during coagulation. Storage conditions, extraction options,
sample integrity, and the quality and quantity of cfDNA are highly
dependent on the blood collection tube type. There are various
blood plasma collection tubes available, and the choice of
collection tubes should be compatible with the ctDNA assays to be
deployed. Ethylene diaminetetraacetic acid (EDTA, K2EDTA
and K3EDTA) tubes are widely used non-preservative
collection tubes for blood, which require plasma isolation within 6
h, as recommended in the majority of studies if stored at room
temperature following venipuncture (84). If stored at 4°C, the time can be
extended to 24 or 48 h, as previously reported (85). Cell-preservative blood collection
tubes, such as Cell-Free DNA BCT (Streck) and PAXgene Blood ccfDNA
tubes (PreAnalytix), allow the extension of the time required for
plasma isolation to several days or weeks (7–30 days) (86).
An initial slow centrifuge speed (3,000 × g),
followed by a high-speed centrifuge (>10,000 × g), is a
recommended and broadly used protocol to isolate plasma from blood.
There is also a report demonstrating that a second low-speed
centrifuge at 3,000 × g provides similar cfDNA yields as second
high-speed centrifuge (87).
Temperature variations or exposure to high temperatures may lead to
damage and the degradation of cfDNA; thus, plasma should be stored
at −80°C and freeze-thawing should be avoided. The DNA integrity
index is significantly decreased after three freeze–thaw cycles
compared with only one cycle (88).
Numerous commercial cfDNA extraction methods are available,
including the manual and automated methods. The majority of these
use silicon membrane-based spin columns or magnetic beads. Size
discrimination, reproducibility and recovery efficiency are highly
variable among these methods. Generally, magnetic bead-based
methods appear to have a higher recovery efficiency for small cfDNA
fragments (50–250 bp) than silicon membrane-based spin columns
(86). Selecting the cfDNA
extraction methods or kits needs to be based on the consideration
of their compatibility with the blood collection tubes used and the
analytical approach to be used. The optimization and
standardization of pre-analytical procedures are as critical as the
analytical approach. More importantly, the whole pre-analytical
sample handling and processing should always follow validated
standard operation procedures and be performed in dedicated areas
of laboratories to reduce the risk of contamination.
Analytical variables and
considerations
There are multiple technical and biological factors
that may generate false-negative or false-positive results in ctDNA
MRD analysis. A low level of ctDNA in the limited volume of plasma
is one of the most common causes of false-negative results and a
great technical challenge of ctDNA MRD analysis. NGS panels with
high-depth sequencing, monitoring numerous patient-specific
mutations, and serial testing may improve the assay sensitivity and
enhance the reliability of plasma ctDNA MRD detection. Several
ctDNA analysis methods with ultra-deep sequencing are being
developed, which can detect ctDNA as low as ~0.0001% (37,56).
The emergence of new clonal or subclonal variants is another cause
of false-negative results, particularly in tumor-informed
ctDNA-based MRD approaches. In addition, some biological factors,
such as variable ctDNA levels between tumors or even between
patients with the same tumor, hidden micrometastasis and the
location of metastasis itself, can cause unavoidable false-negative
results. Higher ctDNA levels have been found in patients with CRC
with liver metastases than in those with nodal or lung metastases
(89). Moreover, ctDNA MRD analysis
has revealed a higher sensitivity for distant metastasis than for
local recurrence in some studies (77,90).
The biological mechanism remains unclear, although it is
hypothesized that a higher tumor burden may exist in metastases
than in residual local disease.
The introduction of artifactual mutations during
sample processing, such as unrepaired DNA polymerase errors arising
during PCR amplification and/or oxidative DNA damage in library
preparation and sequencing, is still the main cause of
false-positive results. Several error-suppression strategies, such
as using unique molecular identifier, duplex sequencing, or the
in silico elimination of stereotypical background artifacts
(91,92), have been developed and utilized to
decrease the artifactual alterations arising ex vivo during
the various cfDNA profiling steps. Somatic alterations derived from
CHIP are one of the most common biological sources of
false-positive mutations. The number of variants, allele fractions
and genes involved in CHIP is prone to change over time, and is
associated with smoking, previous cancer therapy and an increasing
age (93,94). These false-positive mutations can be
reduced by the paired sequencing of white blood cell (WBC) DNA or
matched tumor tissues or by utilizing advanced bioinformatics
analysis. The synchronous profiling of plasma ctDNA and paired WBC
DNA to exclude CHIP-associated variants is highly recommended in
ctDNA MRD analysis, particularly in plasma-only approaches.
Nevertheless, accurately separating all non-tumor variants from
tumor variants remains difficult. Somatic mutations from diverse
non-malignant cell types, such as epithelial, endothelial and
stromal cells, can be detected in cfDNA. A previous study found
that ~10% of variants detected in cfDNA were not present in matched
WBCs (95). Tumor-informed
approaches that track specific mutations identified in tumor
tissues can effectively guard against these sources of biological
background.
Conclusions and future perspectives
There is rapidly increasing evidence demonstrating
the capability of ctDNA-based MRD to predict future relapse several
months or even years prior to clinical or radiologic recurrence in
various types of solid tumors, with high sensitivity and
specificity. However, there are still several hurdles to be
overcome before this approach can be integrated into the current
clinical practice workflow. First, although the clinical validity
of ctDNA-based MRD testing has clearly been demonstrated through a
number of studies (11,51,63,68,69,71,72,74,78,81),
the clinical utility remains to be fully established. Although
preliminary data on the clinical benefit of ctDNA-based MRD testing
for personalizing consolidation systemic therapies or adjuvant in
ctDNA-positive patients are promising, the available studies
involved mostly a small proportion of participants, restricted to
limited applications and lacked validation cohorts. In addition,
there is less evidence of the use of ctDNA to guide the
de-escalation or discontinuation of adjuvant, consolidation
systemic therapies in ctDNA-negative patients. Hence, multiple
carefully designed large-scale prospective randomized clinical
trials are critical to firmly establish and validate the clinical
utility of ctDNA-based MRD analysis for treatment personalization
in various types of solid tumors. Second, the lack of
standardization is another major issue for ctDNA-based MRD
detection in clinical practice. As aforementioned, multiple
pre-analytical and analytical factors have an effect on the
sensitivity and specificity of ctDNA MRD. Optimizations of the
standardization of pre-analytical variables, NGS standards and
post-analytical ctDNA interpretation are the key improvement areas
to use ctDNA MRD as robust standalone blood-based biopsy in solid
tumor management. The basic standardization of pre-analytical
conditions has been established in ctDNA MRD commercial assays
(e.g., plasma is the required source and Streck tubes are the
required blood collection tubes) (96). However, multiple aspects, such as
assay timing, number of collection time points, variable ctDNA
shedding between cancer types and patients, and LODs of the
detection approaches, still have to be optimized and considered by
commercial and academic partners before ctDNA MRD is incorporated
into clinical practice workflow.
Multiple studies (12,46,50,59,61,67,73,77,80)
suggest that ctDNA is a stronger predictor of relapse, and ctDNA
MRD following curative-intent treatments has high PPV for the risk
of recurrence in multiple types of solid tumors. With immense
efforts being made over the past decade, several academic and
commercial ctDNA MRD assays/platforms have been developed and
implemented in clinical trials/practice. Although ctDNA MRD assays
are still facing multiple technical difficulties due to the very
low ctDNA concentration at post-treatment time points, no
standardized methods, the lack of validated large-scale clinical
trials and other obstacles, there is no doubt that ctDNA MRD assays
will play an increasing role in the personalized management of
consolidation systemic therapies and/or adjuvant for various types
of solid tumors in the coming years.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was support by Cancer Genome Atlas of China (CGAC)
project (YCZYPT [2018]06) from the National Human Genetic Resources
Sharing Service Platform (2005DKA21300).
Availability of data and materials
Not applicable.
Authors' contributions
Both authors (HC and QZ) were involved in the
collection and interpretation of data to be included in the review,
in reviewing the literature, as well as in drafting and revising
the manuscript. Both authors agree to be accountable for all
aspects of the work. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qiu B, Guo W, Zhang F, Lv F, Ji Y, Peng Y,
Chen X, Bao H, Xu Y, Shao Y, et al: Dynamic recurrence risk and
adjuvant chemotherapy benefit prediction by ctDNA in resected
NSCLC. Nat Commun. 12:6770–6780. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li H, Ma ZL, Li B, Pan YJ, Xiang JQ, Zhang
YW, Sun YH, Hou T, Lizaso A, Chen Y, et al: Potential utility of
longitudinal somatic mutation and methylation profiling for
predicting molecular residual disease in postoperative non-small
cell lung cancer patients. Cancer Med. 10:8377–8386. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaudhuri AA, Chabon JJ, Lovejoy AF,
Newman AM, Stehr H, Azad TD, Khodadoust MS, Esfahani MS, Liu CL,
Zhou L, et al: Early detection of molecular residual disease in
localized lung cancer by circulating tumor DNA profiling. Cancer
Discov. 7:1394–1403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gale D, Heider K, Ruiz-Valdepenas A,
Hackinger S, Perry M, Marsico G, Rundell V, Wulff J, Sharma G,
Knock H, et al: Residual ctDNA after treatment predicts early
relapse in patients with early-stage non-small cell lung cancer.
Ann Oncol. 33:500–510. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tarazona N, Gimeno-Valiente F, Gambardella
V, Zuñiga S, Rentero-Garrido P, Huerta M, Roselló S,
Martinez-Ciarpaglini C, Carbonell-Asins JA, Carrasco F, et al:
Targeted next-generation sequencing of circulating-tumor DNA for
tracking minimal residual disease in localized colon cancer. Ann
Oncol. 30:1804–1812. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gögenur M, Hadi NA, Qvortrup C, Andersen
CL and Gögenur I: ctDNA for risk of recurrence assessment in
patients treated with neoadjuvant treatment: A systematic review
and meta-analysis. Ann Surg Oncol. 29:8666–8674. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cullinane C, Fleming C, O'Leary DP, Hassan
F, Kelly L, O'Sullivan MJ, Corrigan MA and Redmond HP: Association
of circulating tumor DNA with disease-free survival in breast
cancer: A systematic review and meta-analysis. JAMA Netw Open.
3:e20269212020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pantel K and Alix-Panabières C: Liquid
biopsy and minimal residual disease-latest advances and
implications for cure. Nat Rev Clin Oncol. 16:409–424. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heitzer E, Haque IS, Roberts CES and
Speicher MR: Current and future perspectives of liquid biopsies in
genomics-driven oncology. Nat Rev Genet. 20:71–88. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reinert T, Henriksen TV, Christensen E,
Sharma S, Salari R, Sethi H, Knudsen M, Nordentoft I, Wu HT, Tin
AS, et al: Analysis of plasma cell-free DNA by ultradeep sequencing
in patients with stages I to III colorectal cancer. JAMA Oncol.
5:1124–1131. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang J, Gong Y, Lam VK, Shi Y, Guan Y,
Zhang Y, Ji L, Chen Y, Zhao Y, Qian F, et al: Deep sequencing of
circulating tumor DNA detects molecular residual disease and
predicts recurrence in gastric cancer. Cell Death Dis. 11:346–354.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
MandeL P and Metais P: Les acides
nucléiques du plasma sanguin chez l'homme (Nuclear Acids in Human
Blood Plasma). C R Seances Soc Biol Fil. 142:241–243. 1948.(In
French). PubMed/NCBI
|
|
14
|
Jahr S, Hentze H, Englisch S, Hardt D,
Fackelmayer FO, Hesch RD and Knippers R: DNA fragments in the blood
plasma of cancer patients: Quantitations and evidence for their
origin from apoptotic and necrotic cells. Cancer Res. 61:1659–1665.
2001.PubMed/NCBI
|
|
15
|
Thakur BK, Zhang H, Becker A, Matei I,
Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et
al: Double-stranded DNA in exosomes: A novel biomarker in cancer
detection. Cell Res. 24:766–779. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chin RI, Chen K, Usmani A, Chua C, Harris
PK, Binkley MS, Azad TD, Dudley JC and Chaudhuri AA: Detection of
solid tumor molecular residual disease (MRD) using circulating
tumor DNA (ctDNA). Mol Diagn Ther. 23:311–331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei F, Lin CC, Joon A, Feng Z, Troche G,
Lira ME, Chia D, Mao M, Ho CL, Su WC, et al: Noninvasive
saliva-based EGFR gene mutation detection in patients with lung
cancer. Am J Respir Crit Care Med. 190:1117–1126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dudley JC, Schroers-Martin J, Lazzareschi
DV, Shi WY, Chen SB, Esfahani MS, Trivedi D, Chabon JJ, Chaudhuri
AA, Stehr H, et al: Detection and surveillance of bladder cancer
using urine tumor DNA. Cancer Discov. 9:500–509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Mattos-Arruda L, Mayor R, Ng CKY,
Weigelt B, Martínez-Ricarte F, Torrejon D, Oliveira M, Arias A,
Raventos C, Tang J, et al: Cerebrospinal fluid-derived circulating
tumour DNA better represents the genomic alterations of brain
tumours than plasma. Nat Commun. 6:88392015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Husain H, Nykin D, Bui N, Quan D, Gomez G,
Woodward B, Venkatapathy S, Duttagupta R, Fung E, Lippman SM, et
al: Cell-free DNA from ascites and pleural effusions: Molecular
insights into genomic aberrations and disease biology. Mol Cancer
Ther. 16:948–955. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang P and Lo Y MD: The long and short of
circulating cell-free DNA and the ins and outs of molecular
diagnostics. Trends Genet. 32:360–371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao W, Mei C, Nan X and Hui L: Evaluation
and comparison of in vitro degradation kinetics of DNA in serum,
urine and saliva: A qualitative study. Gene. 590:142–148. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lo YM, Zhang J, Leung TN, Lau TK, Chang AM
and Hjelm NM: Rapid clearance of fetal DNA from maternal plasma. Am
J Hum Genet. 64:218–224. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong FC, Sun K, Jiang P, Cheng YK, Chan
KC, Leung TY, Chiu RW and Lo YM: Cell-free DNA in maternal plasma
and serum: A comparison of quantity, quality and tissue origin
using genomic and epigenomic approaches. Clin Biochem.
49:1379–1386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lam WKJ, Gai W, Sun K, Wong RSM, Chan RWY,
Jiang P, Chan NPH, Hui WWI, Chan AWH, Szeto CC, et al: DNA of
erythroid origin is present in human plasma and informs the types
of anemia. Clin Chem. 63:1614–1623. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Razavi P, Li BT, Brown DN, Jung B, Hubbell
E, Shen R, Abida W, Juluru K, De Bruijn I, Hou C, et al:
High-intensity sequencing reveals the sources of plasma circulating
cell-free DNA variants. Nat Med. 25:1928–1937. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Breitbach S, Tug S, Helmig S, Zahn D,
Kubiak T, Michal M, Gori T, Ehlert T, Beiter T and Simon P: Direct
quantification of cell-free, circulating DNA from unpurified
plasma. PLoS One. 9:e878382014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tug S, Helmig S, Deichmann ER,
Schmeier-Jürchott A, Wagner E, Zimmermann T, Radsak M, Giacca M and
Simon P: Exercise-induced increases in cell free DNA in human
plasma originate predominantly from cells of the haematopoietic
lineage. Exerc Immunol Rev. 21:164–173. 2015.PubMed/NCBI
|
|
29
|
Lo YM, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW, Redman CW and Wainscoat JS: Presence of
fetal DNA in maternal plasma and serum. Lancet. 350:485–487. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leon SA, Shapiro B, Sklaroff DM and Yaros
MJ: Free DNA in the serum of cancer patients and the effect of
therapy. Cancer Res. 37:646–450. 1977.PubMed/NCBI
|
|
31
|
Stroun M, Anker P, Maurice P, Lyautey J,
Lederrey C and Beljanski M: Neoplastic characteristics of the DNA
found in the plasma of cancer patients. Oncology. 46:318–322. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo H, Wei W, Ye Z, Zheng J and Xu RH:
Liquid biopsy of methylation biomarkers in cell-free DNA. Trends
Mol Med. 27:482–500. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Underhill HR, Kitzman JO, Hellwig S,
Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP
and Shendure J: Fragment length of circulating tumor DNA. PLoS
Genet. 12:e10061622016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dressman D, Yan H, Traverso G, Kinzler KW
and Vogelstein B: Transforming single DNA molecules into
fluorescent magnetic particles for detection and enumeration of
genetic variations. Proc Natl Acad Sci USA. 100:8817–8822. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tie J, Cohen JD, Wang Y, Christie M,
Simons K, Lee M, Wong R, Kosmider S, Ananda S, McKendrick J, et al:
Circulating tumor DNA analyses as markers of recurrence risk and
benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol.
5:1710–1717. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kurtz DM, Soo J, Co Ting Keh L, Alig S,
Chabon JJ, Sworder BJ, Schultz A, Jin MC, Scherer F, Garofalo A, et
al: Enhanced detection of minimal residual disease by targeted
sequencing of phased variants in circulating tumor DNA. Nat
Biotechnol. 39:1537–1547. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McDuff SGR, Hardiman KM, Ulintz PJ, Parikh
AR, Zheng H, Kim DW, Lennerz JK, Hazar-Rethinam M, Van Seventer EE,
Fetter IJ, et al: Circulating tumor DNA predicts pathologic and
clinical outcomes following neoadjuvant chemoradiation and surgery
for patients with locally advanced rectal cancer. JCO Precis Oncol.
5:PO.20.00220. 2021.PubMed/NCBI
|
|
39
|
Guerrini F, Paolicchi M, Ghio F, Ciabatti
E, Grassi S, Salehzadeh S, Ercolano G, Metelli MR, Del Re M, Iovino
L, et al: The droplet digital PCR: A new valid molecular approach
for the assessment of B-RAF V600E mutation in hairy cell
leukemia. Front Pharmacol. 7:3632016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huerta M, Roselló S, Sabater L, Ferrer A,
Tarazona N, Roda D, Gambardella V, Alfaro-Cervelló C, Garcés-Albir
M, Cervantes A and Ibarrola-Villava M: Circulating tumor DNA
detection by digital-droplet PCR in pancreatic ductal
adenocarcinoma: A systematic review. Cancers (Basel). 13:9942021.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Elazezy M and Joosse SA: Techniques of
using circulating tumor DNA as a liquid biopsy component in cancer
management. Comput Struct Biotechnol J. 16:370–378. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu Y, Ulrich BC, Supplee J, Kuang Y,
Lizotte PH, Feeney NB, Guibert NM, Awad MM, Wong KK, Jänne PA, et
al: False-positive plasma genotyping due to clonal hematopoiesis.
Clin Cancer Res. 24:4437–4443. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abbosh C, Swanton C and Birkbak NJ: Clonal
haematopoiesis: A source of biological noise in cell-free DNA
analyses. Ann Oncol. 30:358–359. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Flach S, Howarth K, Hackinger S, Pipinikas
C, Ellis P, McLay K, Marsico G, Forshew T, Walz C, Reichel CA, et
al: Liquid BIOpsy for MiNimal RESidual DiSease Detection in head
and neck squamous cell carcinoma (LIONESS)-a personalised
circulating tumour DNA analysis in head and neck squamous cell
carcinoma. Br J Cancer. 126:1186–1195. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McDonald BR, Contente-Cuomo T, Sammut SJ,
Odenheimer-Bergman A, Ernst B, Perdigones N, Chin SF, Farooq M,
Mejia R, Cronin PA, et al: Personalized circulating tumor DNA
analysis to detect residual disease after neoadjuvant therapy in
breast cancer. Sci Transl Med. 11:eaax73922019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Magbanua MJM, Swigart LB, Wu HT, Hirst GL,
Yau C, Wolf DM, Tin A, Salari R, Shchegrova S, Pawar H, et al:
Circulating tumor DNA in neoadjuvant-treated breast cancer reflects
response and survival. Ann Oncol. 32:229–239. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Loomis B, Ishii J, Wolchok JD, Boland G,
Robine N, Altorki NK and Landau DA: Genome-wide cell-free DNA
mutational integration enables ultra-sensitive cancer monitoring.
Nat Med. 26:1114–1124. 2020. View Article : Google Scholar
|
|
48
|
Wan JCM, Heider K, Gale D, Murphy S,
Fisher E, Mouliere F, Ruiz-Valdepenas A, Santonja A, Morris J,
Chandrananda D, et al: ctDNA monitoring using patient-specific
sequencing and integration of variant reads. Sci Transl Med.
12:eaaz80842020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kasi PM, Fehringer G, Taniguchi H,
Starling N, Nakamura Y, Kotani D, Powles T, Li BT, Pusztai L,
Aushev VN, et al: Impact of circulating tumor DNA-Based detection
of molecular residual disease on the conduct and design of clinical
trials for solid tumors. JCO Precis Oncol. 6:e21001812022.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abbosh C, Birkbak NJ, Wilson GA,
Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA,
Veeriah S, Rosenthal R, et al: Phylo genetic ctDNA analysis depicts
early-stage lung cancer evolution. Nature. 545:446–451. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Olsson E, Winter C, George A, Chen Y,
Howlin J, Tang MH, Dahlgren M, Schulz R, Grabau D, van Westen D, et
al: Serial monitoring of circulating tumor DNA in patients with
primary breast cancer for detection of occult metastatic disease.
EMBO Mol Med. 7:1034–1047. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zehir A, Benayed R, Shah RH, Syed A,
Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et
al: Mutational landscape of metastatic cancer revealed from
prospective clinical sequencing of 10,000 patients. Nat Med.
23:703–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Honoré N, Galot R, van Marcke C, Limaye N
and Machiels JP: Liquid biopsy to detect minimal residual disease:
Methodology and impact. Cancers (Basel). 13:53642021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Deveson IW, Gong B, Lai K, LoCoco JS,
Richmond TA, Schageman J, Zhang Z, Novoradovskaya N, Willey JC,
Jones W, et al: Evaluating the analytical validity of circulating
tumor DNA sequencing assays for precision oncology. Nat Biotechnol.
39:1115–1128. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Stahlberg A, Krzyzanowski PM, Egyud M,
Filges S, Stein L and Godfrey TE: Simple multiplexed PCR-based
barcoding of DNA for ultrasensitive mutation detection by
next-generation sequencing. Nat Protoc. 12:664–682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Newman AM, Bratman SV, To J, Wynne JF,
Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et
al: An ultrasensitive method for quantitating circulating tumor DNA
with broad patient coverage. Nat Med. 20:548–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tewhey R, Warner JB, Nakano M, Libby B,
Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB,
Larson JW, et al: Microdroplet-based PCR enrichment for large-scale
targeted sequencing. Nat Biotechnol. 27:1025–1031. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dudley JC and Diehn M: Detection and
diagnostic utilization of cellular and cell-free tumor DNA. Annu
Rev Pathol. 16:199–222. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Parikh AR, Van Seventer EE, Siravegna G,
Hartwig AV, Jaimovich A, He Y, Kanter K, Fish MG, Fosbenner KD,
Miao B, et al: Minimal residual disease detection using a
plasma-only circulating tumor DNA assay in patients with colorectal
cancer. Clin Cancer Res. 27:5586–5594. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sethi H, Salari R, Navarro S, Natarajan P,
Srinivasan R, Dashner S, Tin T, Balcioglu M, Swenerton R and
Zimmermann B: Abstract 4542: Analytical validation of the
SignateraTM RUO assay, a highly sensitive patient-specific
multiplex PCR NGS-based noninvasive cancer recurrence detection and
therapy monitoring assay. Cancer Res. 78:45422018. View Article : Google Scholar
|
|
61
|
Christensen E, Birkenkamp-Demtröder K,
Sethi H, Shchegrova S, Salari R, Nordentoft I, Wu HT, Knudsen M,
Lamy P, Lindskrog SV, et al: Early detection of metastatic relapse
and monitoring of therapeutic efficacy by Ultra-Deep sequencing of
plasma cell-Free DNA in patients with urothelial bladder carcinoma.
J Clin Oncol. 37:1547–1557. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Coombes RC, Page K, Salari R, Hastings RK,
Armstrong A, Ahmed S, Ali S, Cleator S, Kenny L, Stebbing J, et al:
Personalized detection of circulating tumor DNA antedates breast
cancer metastatic recurrence. Clin Cancer Res. 25:4255–4263. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lipsyc-Sharf M, de Bruin EC, Santos K,
McEwen R, Stetson D, Patel A, Kirkner GJ, Hughes ME, Tolaney SM,
Partridge AH, et al: Circulating Tumor DNA and Late Recurrence in
High-Risk Hormone Receptor-Positive, Human Epidermal Growth Factor
Receptor 2-Negative Breast Cancer. J Clin Oncol. 40:2408–2419.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li R, Bonora G, Dai C, Xiang B, Zheng T,
Mo W, Wang X, Zhou K, Jia S, Luo S, et al: 911P the development and
application of a baseline-agnostic minimal residual disease assay.
Ann Oncol. 33:S9642022. View Article : Google Scholar
|
|
66
|
Pellini B, Pejovic N, Feng W, Earland N,
Harris PK, Usmani A, Szymanski JJ, Qaium F, Mudd J, Petty M, et al:
ctDNA MRD detection and personalized oncogenomic analysis in
oligometastatic colorectal cancer from plasma and urine. JCO Precis
Oncol. 5:PO.20.00276. 2021.PubMed/NCBI
|
|
67
|
Abbosh C, Frankell A, Garnett A, Harrison
T, Weichert M, Licon A, Veeriah S, Daber B, Moreau M, Chesh A, et
al: Abstract CT023: Phylogenetic tracking and minimal residual
disease detection using ctDNA in early-stage NSCLC: A lung TRACERx
study. Cancer Res. 80:CT0232020. View Article : Google Scholar
|
|
68
|
Peng M, Huang Q, Yin W, Tan S, Chen C, Liu
W, Tang J, Wang X, Zhang B, Zou M, et al: Circulating tumor DNA as
a prognostic biomarker in localized Non-small Cell lung cancer.
Front Oncol. 10:5615982020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Groot VP, Mosier S, Javed AA, Teinor JA,
Gemenetzis G, Ding D, Haley LM, Yu J, Burkhart RA, Hasanain A, et
al: Circulating tumor DNA as a clinical test in resected pancreatic
cancer. Clin Cancer Res. 25:4973–4984. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Duineveld LA, van Asselt KM, Bemelman WA,
Smits AB, Tanis PJ, van Weert HC and Wind J: Symptomatic and
asymptomatic colon cancer recurrence: A multicenter cohort study.
Ann Fam Med. 14:215–220. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tie J, Wang Y, Tomasetti C, Li L, Springer
S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al:
Circulating tumor DNA analysis detects minimal residual disease and
predicts recurrence in patients with stage II colon cancer. Sci
Transl Med. 8:346ra922016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tie J, Wang Y, Cohen J, Li L, Hong W,
Christie M, Wong HL, Kosmider S, Wong R, Thomson B, et al:
Circulating tumor DNA dynamics and recurrence risk in patients
undergoing curative intent resection of colorectal cancer liver
metastases: A prospective cohort study. PLoS Med. 18:e10036202021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Garcia-Murillas I, Schiavon G, Weigelt B,
Ng C, Hrebien S, Cutts RJ, Cheang M, Osin P, Nerurkar A, Kozarewa
I, et al: Mutation tracking in circulating tumor DNA predicts
relapse in early breast cancer. Sci Transl Med. 7:302ra13322015.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen YH, Hancock BA, Solzak JP, Brinza D,
Scafe C, Miller KD and Radovich M: Next-generation sequencing of
circulating tumor DNA to predict recurrence in triple-negative
breast cancer patients with residual disease after neoadjuvant
chemotherapy. NPJ Breast Cancer. 3:242017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Garcia-Murillas I, Chopra N, Comino-Méndez
I, Beaney M, Tovey H, Cutts RJ, Swift C, Kriplani D, Afentakis M,
Hrebien S, et al: Assessment of molecular relapse detection in
early-stage breast cancer. JAMA Oncol. 5:1473–1478. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Parsons HA, Rhoades J, Reed SC, Gydush G,
Ram P, Exman P, Xiong K, Lo CC, Li T, Fleharty M, et al: Sensitive
detection of minimal residual disease in patients treated for
early-stage breast cancer. Clin Cancer Res. 26:2556–2564. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Azad TD, Chaudhuri AA, Fang P, Qiao Y,
Esfahani MS, Chabon JJ, Hamilton EG, Yang YD, Lovejoy A, Newman AM,
et al: Circulating tumor DNA analysis for detection of minimal
residual disease after chemoradiotherapy for localized esophageal
cancer. Gastroenterology. 158:494–505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang J, Ye S, Xu Y, Chang L, Hu X, Ru G,
Guo Y, Yi X, Yang L and Huang D: Circulating tumor DNA as a
potential marker to detect minimal residual disease and predict
recurrence in pancreatic cancer. Front Oncol. 10:12202020.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Leal A, van Grieken NCT, Palsgrove DN,
Phallen J, Medina JE, Hruban C, Broeckaert MAM, Anagnostou V,
Adleff V, Bruhm DC, et al: White blood cell and cell-free DNA
analyses for detection of residual disease in gastric cancer. Nat
Commun. 11:5252020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hilke FJ, Muyas F, Admard J, Kootz B, Nann
D, Welz S, Rieß O, Zips D, Ossowski S, Schroeder C and Clasen K:
Dynamics of cell-free tumour DNA correlate with treatment response
of head and neck cancer patients receiving radiochemo therapy.
Radiother Oncol. 151:182–189. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Tan L, Sandhu S, Lee RJ, Li J, Callahan J,
Ftouni S, Dhomen N, Middlehurst P, Wallace A, Raleigh J, et al:
Prediction and monitoring of relapse in stage III melanoma using
circulating tumor DNA. Ann Oncol. 30:804–814. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Moding EJ, Nabet BY, Alizadeh AA and Diehn
M: Detecting liquid remnants of solid tumors: circulating tumor DNA
minimal residual disease. Cancer Discov. 11:2968–2986. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Pascual J, Attard G, Bidard FC, Curigliano
G, De Mattos-Arruda L, Diehn M, Italiano A, Lindberg J, Merker JD,
Montagut C, et al: ESMO recommendations on the use of circulating
tumour DNA assays for patients with cancer: A report from the ESMO
Precision Medicine Working Group. Ann Oncol. 33:750–768. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Andersson D, Kristiansson H, Kubista M and
Stahlberg A: Ultrasensitive circulating tumor DNA analysis enables
precision medicine: Experimental workflow considerations. Expert
Rev Mol Diagn. 21:299–310. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Nikolaev S, Lemmens L, Koessler T, Blouin
JL and Nouspikel T: Circulating tumoral DNA: Preanalytical
validation and quality control in a diagnostic laboratory. Anal
Biochem. 542:34–39. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ungerer V, Bronkhorst AJ and Holdenrieder
S: Preanalytical variables that affect the outcome of cell-free DNA
measurements. Crit Rev Clin Lab Sci. 57:484–507. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Risberg B, Tsui DWY, Biggs H,
Ruiz-Valdepenas Martin de Almagro A, Dawson SJ, Hodgkin C, Jones L,
Parkinson C, Piskorz A, Marass F, et al: Effects of collection and
processing procedures on plasma circulating cell-free DNA from
cancer patients. J Mol Diagn. 20:883–892. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chiu RW, Lui WB, El-Sheikhah A, Chan AT,
Lau TK, Nicolaides KH and Lo YM: Comparison of protocols for
extracting circulating DNA and RNA from maternal plasma. Clin Chem.
51:2209–2210. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Vidal J, Muinelo L, Dalmases A, Jones F,
Edelstein D, Iglesias M, Orrillo M, Abalo A, Rodríguez C, Brozos E,
et al: Plasma ctDNA RAS mutation analysis for the diagnosis and
treatment monitoring of metastatic colorectal cancer patients. Ann
Oncol. 28:1325–1332. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tie J, Cohen JD, Lo SN, Wang Y, Li L,
Christie M, Lee M, Wong R, Kosmider S, Skinner I, et al: Prognostic
significance of postsurgery circulating tumor DNA in nonmetastatic
colorectal cancer: Individual patient pooled analysis of three
cohort studies. Int J Cancer. 148:1014–1026. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Abbosh C, Birkbak NJ and Swanton C: Early
stage NSCLC-challenges to implementing ctDNA-based screening and
MRD detection. Nat Rev Clin Oncol. 15:577–586. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Mansukhani S, Barber LJ, Kleftogiannis D,
Moorcraft SY, Davidson M, Woolston A, Proszek PZ, Griffiths B,
Fenwick K, Herman B, et al: Ultra-sensitive mutation detection and
genome-wide DNA copy number reconstruction by error-corrected
circulating tumor DNA sequencing. Clin Chem. 64:1626–1635. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Bolton KL, Ptashkin RN, Gao T, Braunstein
L, Devlin SM, Kelly D, Patel M, Berthon A, Syed A, Yabe M, et al:
Cancer therapy shapes the fitness landscape of clonal
hematopoiesis. Nat Genet. 52:1219–1226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Coombs CC, Zehir A, Devlin SM, Kishtagari
A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, et
al: Therapy-related clonal hematopoiesis in patients with
non-hematologic cancers is common and associated with adverse
clinical outcomes. Cell Stem Cell. 21:374–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Chabon JJ, Hamilton EG, Kurtz DM, Esfahani
MS, Moding EJ, Stehr H, Schroers-Martin J, Nabet BY, Chen B,
Chaudhuri AA, et al: Integrating genomic features for non-invasive
early lung cancer detection. Nature. 580:245–251. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen K, Shields MD, Chauhan PS, Ramirez
RJ, Harris PK, Reimers MA, Zevallos JP, Davis AA, Pellini B and
Chaudhuri AA: Commercial ctDNA assays for minimal residual disease
detection of solid tumors. Mol Diagn Ther. 25:757–774. 2021.
View Article : Google Scholar : PubMed/NCBI
|