Introduction
Uterine corpus endometrial carcinoma (UCEC) is a
malignancy that originates from the endometrium and is frequently
symptomatic at advanced stage (1,2). It is
one of the most common gynecological malignancies worldwide with
increasing incidence (1,2). Patients with advanced disease present
with abnormal bleeding in the vagina or pelvic organs (3). For women with localized disease on
diagnosis, the 5-year survival rate is >90%, but the 5-year
overall survival rate decreases to <20% in patients with distal
metastases (4). Therefore,
identifying novel therapeutic targets for UCEC is particularly
important for treating patients with UCEC.
Long non-coding RNAs (lncRNAs) belong to a class of
non-coding RNAs that are typically >200 nucleotides in length
and have little or no protein-coding potential (5). LncRNAs reverse the inhibitory effects
of microRNAs (miRNAs or miRs) on mRNAs by functioning as miRNA
sponges (6). To date, numerous
lncRNAs have been reported to be aberrantly expressed in various
cancers, where lncRNAs serve pivotal regulatory roles in
tumorigenesis and metastasis (7,8).
Bladder cancer-associated transcript 2 (BLACAT2) was first
identified to be an oncogene in bladder cancer, which increases
tumor lymphangiogenesis and lymphatic metastasis (9,10). It
has been previously reported that the expression of BLACAT2 is
upregulated in several malignancies, including hepatocellular
carcinoma, cervical cancer, and nasopharyngeal carcinoma (11–19),
which suggests that BLACAT2 is closely associated with cancer
progression. In fact, BLACAT2 has been demonstrated to aggravate
hepatocellular carcinoma via sponging miR-3619-5p, and plays a
functional role in modulating radiotherapy resistance in cervical
cancer (11,12). Previous RNA-sequencing analysis
indicated that BLACAT2 expression is highly upregulated in the
endometrium of patients with UCEC compared to adjacent normal
controls (20). In addition,
another previous study revealed that BLACAT2 is a possible
oncogenic lncRNA in endometrial cancer (21). However, the regulatory mechanism
underlying the role of BLACAT2 remains unclear.
Transcription factor Yin Yang-1 (YY1) is a zinc
finger protein belonging to the GLI-Kruppel family (22). As a DNA-binding protein, it serves
dual functions and has been documented to regulate cell
differentiation, proliferation, apoptosis and cell division
(23). YY1 can either function as
an activator or repressor of gene transcription, depending on the
context of YY1-binding proteins in proximity (24). Specifically, YY1 can bind to the
promoter region of LINC00673, thereby increasing its transcription
in cis (25). Numerous
studies have demonstrated the upregulation of YY1 in ovarian,
prostate, esophageal, and melanoma cancer (26–29).
In addition, YY1 expression has also been demonstrated to be
upregulated in human endometrioid endometrial adenocarcinoma cell
lines and tumor tissues. YY1 knockdown can inhibit cell
proliferation and migration in endometrioid endometrial carcinoma
(EEC), whereas YY1 overexpression was found to promote EEC cell
proliferation (30).
MicroRNAs (miRNAs or miRs) form another family of
highly conserved non-coding RNAs, which are 20–25 nucleotides in
length (31,32). They typically mediate translational
repression or mRNA transcript degradation by targeting the
3′-untranslated region (UTR) of downstream mRNAs (31,32).
An increasing number of miRNAs have been observed to regulate the
progression of UCEC (33,34). In particular, miR-378a-3p has been
recognized as a potential miRNA that can bind to BLACAT2 according
to bioinformatic analysis. However, its function in UCEC remains
unreported. Recently, BLACAT2 was revealed to bind to miR-378a-3p
in bladder cancer cells, where it regulated cell viability,
migration and invasion (35).
Based on previous studies and findings, it was
hypothesized that BLACAT2 may serve an important biological
function in UCEC by regulating miR-378a-3p and/or YY1. Therefore,
the present study explored the possible functions of BLACAT2, YY1
and miR-378a-3p, in addition to their associations with UCEC
progression.
Materials and methods
Clinical specimens
A total of 40 paired cancer-adjacent and UCEC tissue
samples were obtained from January 2018 to December 2019 and frozen
for further analysis. All volunteers (mean age, 57.78) provided
written informed consent and the present study was approved
(approval no. 2021PS723K) by the Ethics Committee of Shengjing
Hospital of China Medical University (Shengjing, China) and in
accordance with the Declaration of Helsinki. The inclusion and
exclusion criteria were as follows: The patients with UCEC included
in this study were diagnosed by histopathological detection. The
tumor tissues and their adjacent normal tissues were collected by
surgical resection. None of these patients received radiotherapy or
chemotherapy before the surgery. The patients that had undergone
non-curative resection, cancer recurrence, severe injury of vital
organs or had a history of autoimmune diseases were excluded. The
clinicopathological characterisitics of the patients with UCEC
included in the present study are provided in Table I.
 | Table I.Clinicopathological characteristics
of patients with uterine corpus endometrial carcinoma. |
Table I.
Clinicopathological characteristics
of patients with uterine corpus endometrial carcinoma.
| Patient No. | Age | Tumor stage
(T) | Lymph node
metastasis (N) | Distant metastasis
(M) | TNM | Histopathological
grade | Maximum tumor
diameter (cm) | Tumor number |
|---|
| 1 | 65 | T1a | N1a | M0 | IIIC1 | G3 | 9 | Multifocal |
| 2 | 66 | T1b | N0 | M0 | IB | G1 | 5.5 | Unifocal |
| 3 | 63 | T1a | N0 | M0 | IA | G1-G2 | 3 | Unifocal |
| 4 | 60 | T3b | N0 | M0 | IIIB | G3 | 4.5 | Multifocal |
| 5 | 45 | T1a | N0 | M0 | IA | G1 | 1.1 | Unifocal |
| 6 | 77 | T1a | N0 | M0 | IA | G1 | 6 | Unifocal |
| 7 | 53 | T1a | N0 | M0 | IA | G1-G2 | 4.5 | Unifocal |
| 8 | 57 | T1a | N0 | M0 | IA | G1 | 3.5 | Unifocal |
| 9 | 58 | T1a | N0 | M0 | IA | G1-G2 | 3.1 | Multifocal |
| 10 | 48 | T2 | N0 | M0 | II | G1-G2 | 4 | Multifocal |
| 11 | 58 | T1a | N0 | M0 | IA | G2 | 3.5 | Unifocal |
| 12 | 69 | T1a | N0 | M0 | IA | G3 | 6 | Unifocal |
| 13 | 55 | T1b | N0 | M0 | IB | G2-G3 | 2.0 | Unifocal |
| 14 | 68 | T1b | N0 | M0 | IB | G1-G2 | 2.5 | Unifocal |
| 15 | 60 | T1a | N0 | M0 | IA | G3 | 3.5 | Unifocal |
| 16 | 40 | T1a | N0 | M0 | IA | G1 | 3.5 | Unifocal |
| 17 | 52 | T2 | N0 | M0 | II | G1 | 2.5 | Unifocal |
| 18 | 49 | T1a | N0 | M0 | IA | G2 | 5.5 | Unifocal |
| 19 | 73 | T1b | N0 | M0 | IB | G1-G2 | 3.5 | Unifocal |
| 20 | 46 | T3b | N1a | M0 | IIIC1 | G1 | 7 | Multifocal |
| 21 | 67 | T1a | N0 | M0 | IA | G2 | 6 | Unifocal |
| 22 | 65 | T1a | N0 | M0 | IA | G1-G2 | 4 | Unifocal |
| 23 | 54 | T1a | N0 | M0 | IA | G3 | 4.9 | Multifocal |
| 24 | 66 | T1b | N0 | M0 | IB | G2-G3 | 4.5 | Multifocal |
| 25 | 54 | T1a | N0 | M0 | IA | G2-G3 | 5 | Unifocal |
| 26 | 49 | T2 | N2a | M0 | IIIC2 | G1-G2 | 3.5 | Multifocal |
| 27 | 53 | T1b | N0 | M0 | IB | G2-G3 | 4.7 | Unifocal |
| 28 | 54 | T2 | N0 | M0 | II | G1 | 5 | Unifocal |
| 29 | 70 | T2 | N0 | M0 | II | G2 | 7.5 | Multifocal |
| 30 | 65 | T1a | N0 | M0 | IA | G2 | 4 | Unifocal |
| 31 | 54 | T1a | N0 | M0 | IA | G2 | 1.6 | Unifocal |
| 32 | 67 | T1b | N0 | M0 | IB | G1-G2 | 5.9 | Unifocal |
| 33 | 42 | T1a | N0 | M0 | IA | G2 | 5.4 | Unifocal |
| 34 | 58 | T2 | N0 | M0 | II | G1-G2 | 9 | Unifocal |
| 35 | 60 | T1a | N2a | M0 | IIIC2 | G2-G3 | 4.8 | Multifocal |
| 36 | 53 | T1b | N0 | M0 | IB | G2 | 6.9 | Unifocal |
| 37 | 58 | T1a | N0 | M0 | IA | G1 | 4.8 | Unifocal |
| 38 | 44 | T1a | N0 | M0 | IA | G1 | 3.5 | Unifocal |
| 39 | 63 | T1a | N0 | M0 | IA | G1 | 1.3 | Multifocal |
| 40 | 53 | T1b | N0 | M0 | IB | G2 | 6 | Unifocal |
Cell lines and culture conditions
Human endometrial cancer cell lines Ishikawa (cat.
no. iCell-h113), RL95-2 (cat. no. iCell-h182), HEC-1-B (cat. no.
iCell-h084), AN3CA (cat. no. icell-h018), and HEC-1-A (cat. no.
iCell-h083) were purchased from iCell Bioscience, Inc. Ishikawa,
AN3CA, and HEC-1-B cells were grown in MEM medium (Beijing Solarbio
Science & Technology Co., Ltd.). HEC-1-A cells were cultured in
McCoy's 5A medium (Procell Life Science & Technology Co.,
Ltd.). RL95-2 cells were cultured in DMEM/F12 medium (Procell Life
Science & Technology Co., Ltd.). All the culture media were
supplemented with 10% FBS (Sangon Biotech Co., Ltd.) and cells were
cultured in a CO2 incubator at 37°C.
Database analysis
Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/) was used
to analyze the different expression of genes in various human
cancers. There was a binding relationship between BLACAT2 and
hsa-miR-378a-3p, as predicted by starBase (https://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA).
Furthermore, 49 target genes common to miRDB (http://www.mirdb.org/), TargetScan (https://www.targetscan.org/vert_72/) and starBase
were identified. Jasper database (https://jaspar.genereg.net/) was utilized to show the
association between YY1 and BLACAT2 promotor region.
Lentivirus packaging
The helper plasmids pSPAX2 and pMD2.G were obtained
from Hunan Fenghui Biotechnology Co., Ltd. For lentivirus
packaging, 293T cells (cat. no. ZQ0033; https://www.zqxzbio.com/Index/p_more/pid/486.html;
Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd.) were cultured
to a confluence of ~70% and then transfected with pSPAX2 (3 µg),
pMD2.G (2.2 µg), and the plasmids with overexpressed
BLACAT2/Negative control (5 µg) or shRNA against
BLACAT2/YY1/Negative control (5 µg) using Lipofectamine®
3000 (Thermo Fisher Scientific, Inc.) at 37°C according to the
manufacturer's protocols. At 6 h post transfection, the culture
medium of the cells was changed into fresh DMEM medium (Wuhan
Servicebio Technology Co. Ltd.) with 10% FBS (37°C and 5%
CO2). At 48 h post trasfection, the cell supernatant was
harvested as packaged lentivirus and filtered through a 0.45-µm
pore size filter.
Cell treatments
To establish the stable UCEC cell line with
overexpressed BLACAT2 or knocked down BLACAT2, the AN3CA and
HEC-1-A cells were selected and infected with the corresponsing
lentiviruses at a multiplicity of infection (MOI) of 10. The
puromycin (0.9 µg/ml) was used for stable cell line selection,
while 0.3 µg/ml of puromycin was for maintenance.
To clarify the function of miR-378a-3p in UCEC,
HEC-1-A cells were transfected with miR-378a-3p mimics (100 pmol)
or negative control (NC; 100 pmol) mimics using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.).
To investigate the role of YY1 in the regulation of
BLACAT2 underlying UCEC development, AN3CA cells with stably
overexpressed BLACAT2 were infected with lentivirus encoding shRNA
against YY1 (Lv-shYY1) at a MOI of 10.
The shRNAs against BLACAT2, YY1 and NC were
synthesized by General Biological System and inserted into the
lentiviral vector. The sequence information used in the present
study was as follows: shBLACAT2#1, 5′-GGTTAAGACATTTCTACAAAT-3′;
shBLACAT2#2, 5′-GAGTGGAGAGACTCAGCTACC-3′; shYY1,
5′-CGACGACTACATTGAACAAAC-3′; shNC, 5′-TTCTCCGAACGTGTCACGT-3′;
miR-378a-3p mimics sense, 5′-ACUGGACUUGGAGUCAGAAGGC-3′; NC mimics
sense, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′.
All cells after transfection were cultured at 37°C
and 5% CO2. Overexpression/knockdown efficiency was
assessed using reverse transcription-quantitative PCR
(RT-qPCR).
RT-qPCR
UCEC cells (6×105/well; 6-well plate) or
tumor tissues were lysed in TRIpure (BioTeke Corporation) to
isolate the total RNAs. The miRNAs were reverse transcripted into
cDNA using the miRNA First Strand cDNA Synthesis Kit according to
the manufacturer's instructions (Sangon Biotech Co., Ltd.). LncRNA
and mRNA were synthesized into cDNA via reverse transcription PCR.
The thermocycling conditions for reverse transcription PCR were as
follows: Step 1, a total of 12.5 µl reaction was prepared by adding
1 µg RNA, 1 µl oligo (dT)15, 1 µl random primers, and
ddH2O. The reaction was incubated at 70°C for 5 min and
then placed on ice for 2 min; step 2, 2 µl dNTP, 4 µl 5X buffer,
0.5 µl Rnase inhibitor, and 1 µl Super M-MLV reverse transcriptase
(BioTeke Corporation) were added into the reaction. The total
reaction samples were then incubated at 25°C for 10 min, 40°C for
50 min, and 80°C for 10 min.
The cDNAs were amplified and detected using 2X Power
Taq PCR MasterMix (BioTeke Corporation) and SYBR Green fluorescence
(SYBR Green qPCR kit; Sigma-Aldrich; Merck KGaA) on an
Exicycler™ 96 Real-Time System (Bioneer Corporation).
The thermocycling conditions for qPCR were as the following: Step
1, 94°C for 5 min; step 2, 94°C for 15 sec; step 3, 60°C for 25
sec; step 4, 72°C for 30 sec; scan and step 2, 40 cycles; step 5,
72°C for 5 min 30 sec; step 6, 40°C for 2 min 30 sec; step 7,
melting 60–94°C every 1.0°C for 1 sec; and step 8, 25°C for 1–2
min. The mRNA levels of the target genes were quantified using the
2−ΔΔCq method (36).
β-actin was the reference for the quantification of BLACAT2 and
YY1. The reference gene U6 was used for miR-378a-3p quantification.
Primers for the genes assessed were as follows: BLACAT2 forward,
5′-GAGGAGGAGAAGCAATCA-3′ and reverse, 5′-GGAGCCCATCCATTAGAG-3′; YY1
forward, 5′-ACCCACGGTCCCAGAGT-3′ and reverse,
5′-AAAGCGTTTCCCACAGC-3′; miR-378a-3p forward,
5′-ACTGGACTTGGAGTCAGAAGGC-3′. All primers were synthesized by
Genscript.
Western blot analysis
Proteins samples in cells or tumor tissues were
extracted using cell lysis buffer (cat. no. P0013; Beyotime
Institute of Biotechnology) supplemented with 1 mM
phenylmethanesulfonyl fluoride (PMSF; Beyotime Institute of
Biotechnology). Protein concentration was quantified using a BCA
kit (Beyotime Institute of Biotechnology). Subsequently, 20–40 µg
proteins in each lane were separated on the SDS-PAGE gel (11% or
8%). The 11 or 8% gel was respectively selected for protein
separation according the molecular weight of the target proteins.
The proteins were then transferred onto PVDF membranes and blocked
with a 5% skimmed milk solution at room temperature for 1 h. The
membranes were incubated with the primary antibodies against MMP2
(1:500; cat. no. 10373-2-AP; ProteinTech Group, Inc.), MMP9
(1:1,000; cat. no. 10375-2-AP; ProteinTech Group, Inc.), cyclin D1
(1:500; cat. no. A19038; ABclonal Biotech Co., Ltd.), cyclin E
(1:1,000; cat. no. A14225; ABclonal Biotech Co., Ltd.),
phosphorylated (p)-MEK1/2 (1:500; cat. no. AP0209; ABclonal Biotech
Co., Ltd.), MEK1/2 (1:500; cat. no. AF6385; Affinity Biosciences,
Ltd.), p-ERK1/2 (1:500; cat. no. AF1015; Affinity Biosciences,
Ltd.) or ERK1/2 (1:500; cat. no. AF0155; Affinity Biosciences,
Ltd.) overnight at 4°C. This was followed by a 40-min incubation
with the HRP-conjugated anti-rabbit and anti-mouse secondary
antibodies (1:10,000; cat. no. A0208 and A0216, respectively;
Beyotime Institute of Biotechnology) at 37°C. Enhanced
chemiluminescence (ECL)-detecting reagent (Beyotime Institute of
Biotechnology) was then added onto the membrane, before the protein
bands were visualized in WD-9413B gel-imaging system (Beijing LIUYI
Biotechnology Co., Ltd.) and analyzed using Gel-Pro-Analyzer 4.0
software (Media Cybernetics, USA).
Dual-luciferase reporter assay
293T cells (2×105/well; 12-well plate)
were used for dual-luciferase reporter assay. To assess the
potential association between miR-378a-3p and BLACAT2 or YY1, the
wild-type BLACAT2 or 3′UTR of YY1 mRNA containing a putative
binding site for miR-378a-3p and their corresponding mutant
sequences were amplified and cloned into the pmirGLO plasmid
(General Biological System). Vectors containing the wild-type or
mutant sequence were co-transfected with miR-378a-3p mimics or NC
mimics into 293T cells using Lipofectamine® 3000 at 37°C
using the established protocol. At 6 h post transfection, the
culture medium was replaced by fresh DMEM (Wuhan Servicebio
Technology Co., Ltd.) containing 10% FBS (Hangzhou Sijiqing
Biological Engineering Materials Co., Ltd.). In total, 48 h after
transfection, the activity of firefly luciferase in cells was
detected using a Dual-Luciferase Reporter Gene Assay Kit (Nanjing
KeyGen Biotech Co., Ltd.), which was normalized to that of
Renilla luciferase.
Similarly, to assess the binding of YY1 to BLACAT2
promoter, three regions (−500+5, −1,000+5 and −1,500+5) in the
BLACAT2 promoter or the YY1 coding sequence were inserted into the
pmirGLO vector. Vectors encoding each of the BLACAT2 promoters were
co-transfected with the vector encoding YY1 into 293T cells at 37°C
using the established protocol. The luciferase activity was
detected at 48 h post transfection.
Cell counting kit-8 (CCK-8) assay
Cells (3×103/well)were seeded into a
96-well plate and cultured under standard conditions. Cell
viability was measured at 0, 24, 48 and 72 h using the CCK-8
(Sigma-Aldrich; Merck KGaA) solution. In total, 800 µl CCK-8 was
added into each well, before the cells were cultured for 1 h. The
optical density of each well was then detected at 450 nm using a
microplate reader (800Ts; BioTek Instruments, Inc.).
Flow cytometric analysis
Cell cycle distribution was revealed using a Cell
Cycle and Apoptosis Detection kit (cat. no. C1052; Beyotime
Institute of Biotechnology). After the fixing with cold 70% ethanol
for 12 h at 4°C, the cells were harvested by centrifugation at 4°C
for 3 min with a speed of 420 × g, and 500 µl dye buffer was added.
Next, the cells were incubated with 25 µl of propidium iodide
staining solution and 10 µl RNase A for 30 min at 37°C. The
percentage of the cells in G1, S, and G2 phases were
analyzed using a flow cytometer (NovoCyte; ACEA Bioscience, Inc.)
and quantified using the software NovoExpress (version 1.5.0;
Agilent Technologies, Inc.).
Wound healing assay
Cells were seeded into a six-well plate at a density
of 5×105/well and grown to a confluence of >95% at
the start of the assay. The cells were then cultured in serum-free
medium containing 1 µg/ml mitomycin C (Sigma-Aldrich; Merck KGaA)
for 1 h. Subsequently, a 200-µl pipette tip was used to make a
straight vertical scratch down through the cell monolayers. Cell
debris were then removed and an image was recorded using an
inverted phase-contrast microscope (IX53; Olympus Corporation). The
cells were thereafter cultured in fresh serum-free medium at 37°C
and 5% CO2 for 24 h. The images of wounds at 0 and 24 h
were recorded, before the migration rate was calculated as: (Wound
distance at 0 h-wound distance at 24 h)/wound distance at 0 h
×100%.
Transwell assay
Cell invasion in AN3CA and HEC-1-A cells was
assessed using Transwell assays with a 24-well Transwell chamber
(Corning, Inc.). The chamber was pre-coated with Matrigel at 37°C
for 2 h (Corning, Inc.), which wasdiluted with serum-free mediumat
a ratio of 1:3. The cells were then seeded into the pre-coated
upper chamber in their serum-free culture medium at a density of
1.5×104 cells/well. In total, 800 µl culture medium (MEM
for AN3CA cells and McCoy's 5A for HEC-1-A) containing 10% FBS
(Biological Iundustries) was added into the lower chamber. After 24
h of incubation at 37°C, the cells were fixed with 4%
paraformaldehyde for 20 min at room temperature and then stained
with 0.1% crystal violet solution at room temperature for a further
5 min. A total of five fields of view were randomly selected for
each chamber and the number of cells were counted under an inverted
phase-contrast microscope (IX53; Olympus Corporation).
Gelatin zymography assay
MMP-2 and MMP-9 activity in AN3CA and HEC-1-A cells
were determined using gelatin zymography as previously described
(37). Total protein concentration
in the culture supernatants was quantified with a BCA kit (Beyotime
Insitute of Biotechnology). A total of 10 mg/ml gelatin
(Sigma-Aldrich; Merck KGaA) was added to a 10% acrylamide
separating gel. The proteins in the supernatants were separated
using SDS-PAGE. The SDS-PAGE gel was washed twice for 40 min in an
elution solution (2.5% Triton X-100, 50 mmol/l Tris-HCl, 5 mmol/l
CaCl2 and 1 µmol/l ZnCl2; pH 7.6). The gel
was then incubated in the developing buffer (50 mmol/l Tris-HCl, 5
mmol/l CaCl2, 1 µmol/l ZnCl2, 0.02% Brij and
0.2 mol/l NaCl) at 37°C for 40 h. Following incubation, the gel was
stained with a staining solution (0.5% Coomassie blue R250, 30%
methanol and 10% glacial acetic acid; Beijing Solarbio Science
& Technology Co., Ltd.) at room temperature for 3 h. Digestion
bands were visualized using a Gel imaging system (WD-9413B; Beijing
LIUYI Biotechnology Co., Ltd.) and analyzed with a microplate
reader (ELX-800; BioTeke Corporation).
Chromatin immunoprecipitation
(ChIP)
The ChIP assay was conducted using a ChIP assay kit
(cat. no. WLA106a; Wanleibio, Co., Ltd.) according to the
manufacturer's protocol. Briefly, HEC-1-A cells were
formaldehyde-crosslinked and then lysed using lysis buffer from
Wanleibio, Co., Ltd. (cat. no. WLA106a). The chromatin of HEC-1-A
cells was fragmented by sonication and collected, followed by the
incubation with PBS-washed ProteinA+G beads (60 µl) on a rotator at
4°C for 1–2 h. Following centrifugation with a speed of 625 × g at
room temperature for 1 min, the cell supernatants were collected
and 20 µl was used as the Input. The chromatin fragments (100
µl/group) were then incubated with the anti-YY1 antibody (2 µg,
cat. no. 66281-1-Ig; ProteinTech Group, Inc.) on a rotator at 4°C
overnight. The PBS-washed ProteinA+G beads (60 µl) were next added
and maintained on a rotator at 4°C for 1–2 h. IgG (1 µg; ChIP assay
kit; Wanleibio, Co., Ltd.) was used as a negative control antibody.
The protein-DNA complexes were then washed using the wash buffer
from Wanleibio, Co., Ltd. (cat. no. WLA106a) on a rotator at 4°C
for 10 min/time. The supernatant was removed using centrifugation
at a speed of 625 × g at room temperature for 1 min. A total of 100
µl of elution buffer was added and incubated overnight at 65°C to
reverse cross-links. The DNA specimens released from the
protein-DNA complexes were purified and analyzed using ChIP
followed by PCR (ChIP-PCR; BioTeke Corporation) to amplify the
promoter region of BLACAT2. The primer pair for the BLACAT2
promoter was as follows: Forward, 5′-AGTCTCATTCTGTCGCCT-3′ and
reverse, 5′-TGTAATCCCAGCACTGTG-3′. PCR products were detected using
electrophoresis on 2% agarose gel (Biowest).
In vivo tumorigenesis model
For the present study, 4-week-old female BALB/c nude
mice weighing 16±1 g (n=6 per group, with a total of 24 mice) were
purchased from Huafukang Biosciences and adaptively housed for a
week under a controlled environment (humidity, 45–55%; temperature,
22±1°C; and a 12-h day/night cycle), with free access to water and
food. The mice were then randomly divided into four groups: Lv-NC
(AN3CA), Lv-BLACAT2 (AN3CA), Lv-shNC (HEC-1-A), and Lv-shBLACAT2
(HEC-1-A) groups. AN3CA cells with stably overexpressed BLACAT2/NC
and HEC-1-A cells with stably knocked down BLACAT2/NC
(1×107 cells/100 µl) were subcutaneously injected into
the right flank of the nude mice. The tumor size was measured every
4 days. A total of 4 weeks later, all mice were sacrificed by
CO2 asphyxiation. The flow displacement rate of
CO2 was 40% of the chamber volume per min. Death was
confirmed by the cessation of movement, breathing and heartbeat.
The tumor tissues were then separated and weighed, before they were
fixed in 4% paraformaldehyde for further analysis. The tumor
volumes were calculated according to the following formula:
Volume=length × width2 ×0.5. The animal experiments were
approved by the Ethics Committee of Shengjing Hospital of China
Medical University (approval no. 2021PS741K). A tumor size of 20 mm
at the largest diameter was permitted by the Ethics Committee.
After 4 weeks, the maximum tumor diameter observed was not
exceeded.
Immunohistochemistry
The tumor tissues fixed in 4% paraformaldehyde (at
room temperature for 24 h) were embedded in paraffin and then
sectioned into 5-µm slices. After dewaxing, rehydration, and
antigen retrieval, the tumor slices were incubated with 3%
H2O2 at room temperature for 15 min.
Following 15 min of blocking with goat serum (Beijing Solarbio
Science & Technology Co., Ltd.) at room temperature, tumor
slices were incubated with a anti-Ki67 antibody (1:100 dilution;
cat. no. A2094; ABclonal Biotech Co., Ltd.) overnight at 4°C.
Subsequently, the sections were incubated with an HRP-conjugated
secondary antibody (cat. no. 31460; Thermo Fisher Scientific, Inc.)
at 37°C for 1 h. The sections were then stained with
diaminobenzidine (DAB; Beijing Solarbio Science & Technology
Co., Ltd.) at room temperature for 5 min. Following DAB
visualization, the sections were immersed into water and then
re-stained using hematoxylin at room temperature for 3 min. The
Ki67-positive cells were finally observed under a light microscope
(BX53; Olympus Corporation).
Statistical analysis
Data were analyzed using the GraphPad Prism 8.0
software (GraphPad Software, Inc.) and the results are presented as
the mean ± standard deviation. Statistical analyses were performed
using one-way ANOVA followed by Tukey's multiple comparisons test,
two-way ANOVA followed by Sidak's multiple comparisons test or
unpaired Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of BLACAT2 in UCEC
BLACAT2 was found to be upregulated in cervical
cancer, ovarian cancer and UCEC, whereas its expression was not
found to be significantly different in breast cancer (Fig. 1A). Although carcinogenic functions
have been previously identified in cervical cancer (38) and ovarian cancer (39), the role of BLACAT2 in UCEC remains
unclear. BLACAT2 expression was therefore detected in clinical
samples using RT-qPCR. BLACAT2 expression was found to be
significantly increased in UCEC tumors compared with that in the
adjacent normal tissues (P<0.01; Fig. 1B). Furthermore, RT-qPCR confirmed
that BLACAT2 is expressed in human endometrial cancer cell lines
Ishikawa, RL95-2, HEC-1-B, AN3CA and HEC-1-A (Fig. 1C).
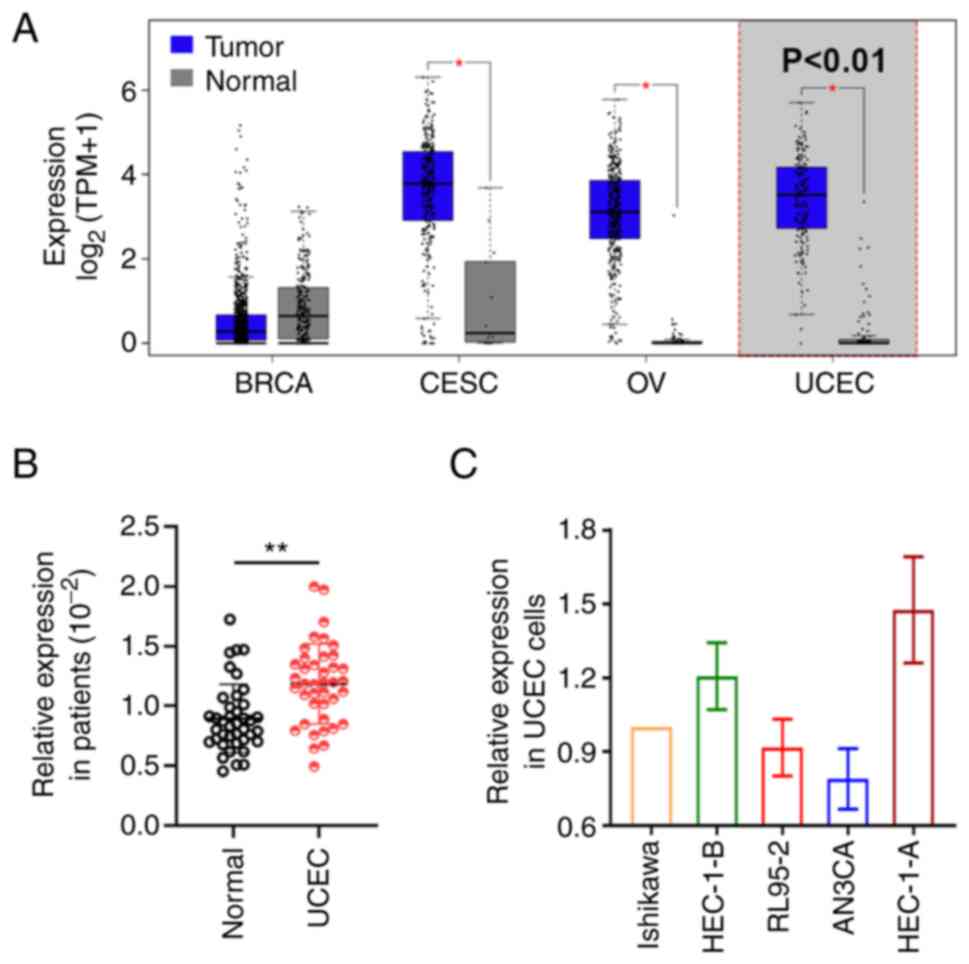 | Figure 1.Expression of BLACAT2 in UCEC tumors
and cell lines. (A) BLACAT2 expression level in BRCA, CESC, OV and
UCEC using the GEPIA database. The expression of BLACAT2 in (B) 40
paired clinical specimens and in (C) the UCEC cell lines Ishikawa,
HEC-1-B, RL95-2, AN3CA and HEC-1-A. *P<0.05 and **P<0.01 vs.
normal. BLACAT2, bladder cancer-associated transcript 2; UCEC,
uterine corpus endometrial carcinoma; BRCA, breast cancer; CESC,
cervical cancer; OV, ovarian cancer. |
BLACAT2 overexpression promotes the
proliferation of UCEC cells
As revealed in Fig.
1C, AN3CA cells exhibited the lowest expression of BLACAT2 than
that in the other UCEC cell lines, while HEC-1-A cells exhibited
the highest expression of BLACAT2. Thus, AN3CA cells were selected
for BLACAT2 overexpression, whereas HEC-1-A cells were selected for
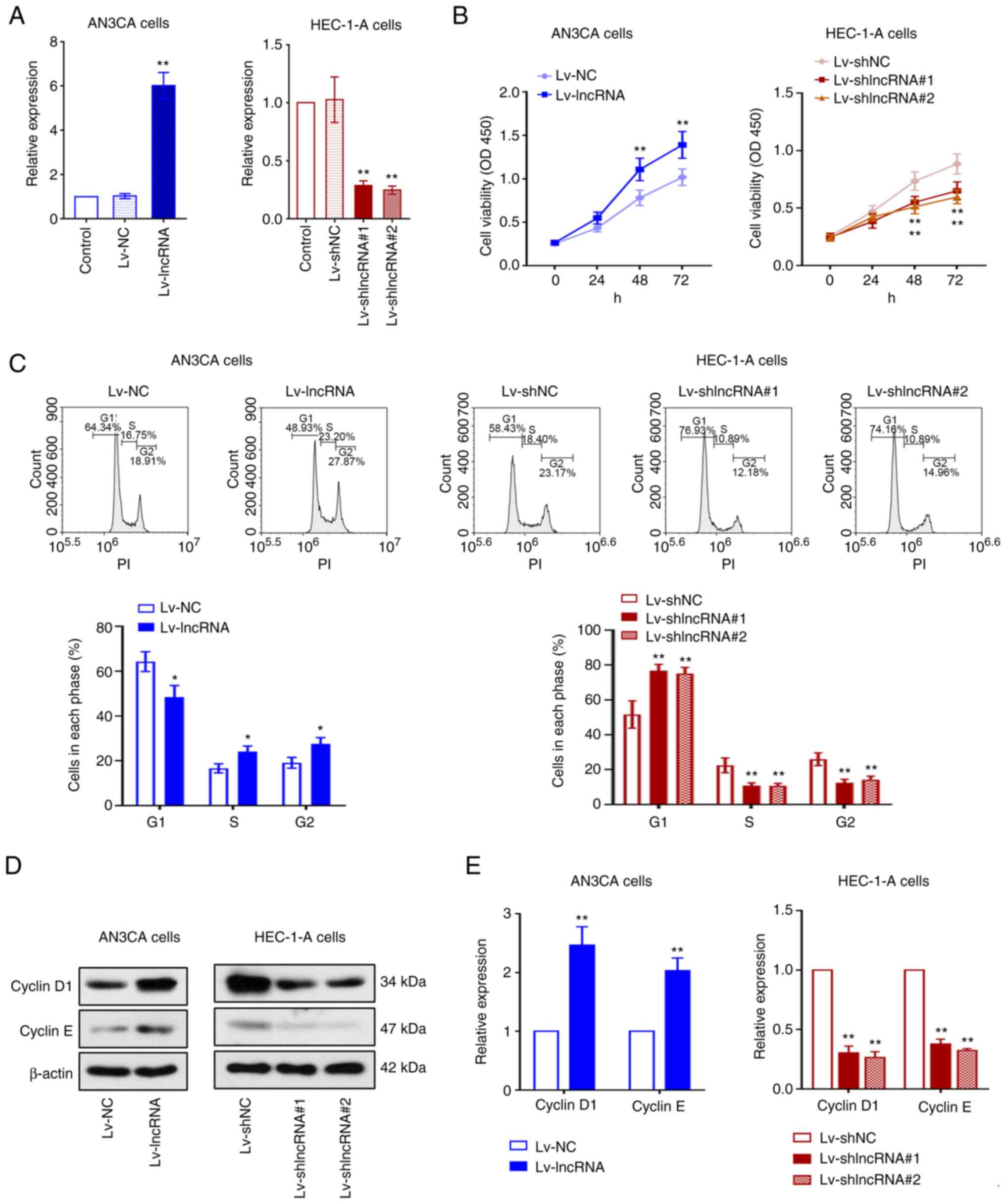
BLACAT2 knockdown. As the results of RT-qPCR revealed, the
expression of BLACAT2 in AN3CA cells was higher when compared to
the Lv-NC group, whereas that in HEC-1-A cells was significantly
lower when compared to the Lv-shNC group (P<0.01; Fig. 2A). Cell viability was then detected
using CCK-8 assay. Notably, BLACAT2 overexpression enhanced UCEC
cell viability, whereas BLACAT2 knockdown reduced cell viability
(P<0.01; Fig. 2B). Cell cycle
progression was next investigated in these cells. BLACAT2
overexpression decreased cell accumulation in the G1
phase and increased the number of cells in the S and G2
phases. By contrast, there were more HEC-1-A cells in the
G1 phase and fewer cells in the S and G2
phases following BLACAT2 knockdown (P<0.05; Fig. 2C). The expression of cyclin D1 and
cyclin E, which are essential regulators of G1/S
transition during cell cycle progression (40), were found to be upregulated in UCEC
cells following BLACAT2 overexpression. However, their protein
expression levels were significantly reduced by BLACAT2 knockdown
(P<0.01; Fig. 2D and E).
Therefore, these data indicated that BLACAT2 overexpression
promotes the proliferation of UCEC cells.
BLACAT2 overexpression facilitates
UCEC cell migration and invasion
The migration and invasion of AN3CA and HEC-1-A
cells were subsequently assessed by wound healing and Transwell
assays. BLACAT2 overexpression was observed to improve the
migration rate of UCEC cells, whilst knockdown of BLACAT2
expression significantly inhibited the migration of these tumor
cells (P<0.05; Fig. 3A). BLACAT2
appeared to mediate similar effects on UCEC cell invasion
(P<0.01; Fig. 3B). MMP9 and MMP2
are proteins that can potentiate cancer cell invasion (41). Therefore, the present study next
examined the protein expression levels of MMP9 and MMP2 in cells,
and determined that BLACAT2 overexpression increased the expression
of MMP9 and MMP2. By contrast, BLACAT2 knockdown inhibited their
protein expression levels in UCEC cells (P<0.01; Fig. 3C). In addition, gelatin zymography
assay revealed that BLACAT2 overexpression enhanced the activity of
MMP9 and MMP2, whilst BLACAT2 knockdown weakened their activities
(P<0.01; Fig. 3D). These
findings indicated that BLACAT2 serves an important role in the
migration and invasion of UCEC cells.
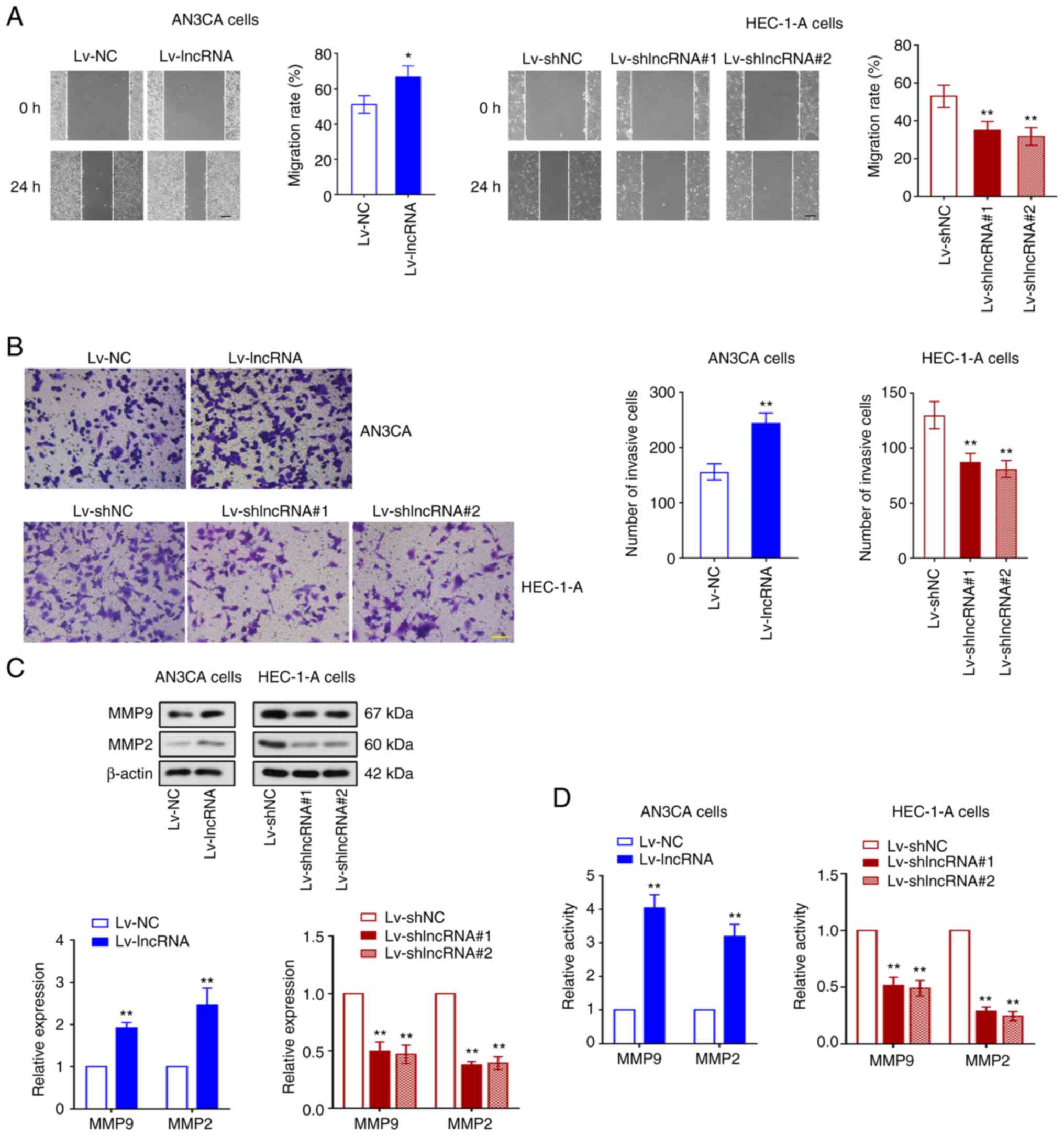 | Figure 3.BLACAT2 facilitates UCEC cell
migration and invasion. The AN3CA stable cell line with
overexpressed BLACAT2 and the HEC-1-A stable cell line with knocked
down BLACAT2 were established, respectively, before the migration
and invasion of UCEC cells were detected. (A) Migration of AN3CA
and HEC-1-A cells was detected by wound healing assay, before the
migration rate was calculated. Magnification, ×100. Scale bar, 200
µm (B) Invasion of AN3CA and HEC-1-A cells was measured using
Transwell assay, before the number of invasive cells was recorded.
Magnification, ×200. Scale bar, 100 µm. (C) Relative protein
expression levels of MMP9 and MMP2 in AN3CA and HEC-1-A cells were
measured by western blot analysis. (D) The activities of MMP9 and
MMP2 in AN3CA and HEC-1-A cells were determined using gelatin
zymography assay. Values are presented as the means ± SD (n=3).
*P<0.05 and **P<0.01 vs. Lv-NC or Lv-shNC. BLACAT2, bladder
cancer-associated transcript 2; UCEC, uterine corpus endometrial
carcinoma; Lv, lentivirus; sh, short-interfering; NC, negative
control. |
BLACAT2 overexpression enhances
MER/ERK signaling in UCEC cells
The ERK signaling pathway has been reported to be
aberrantly activated in cancer, which can promote cell
proliferation and invasion (42).
Therefore, the activation status of ERK and MEK in UCEC cells was
assessed using western blotting (Fig.
4A and C). BLACAT2 overexpression was found to increase the
phosphorylation of MEK1/2 and ERK1/2 without altering the
expression of their corresponding total protein in UCEC cells. The
increased ratio of p-MEK1/2 or p-ERK1/2 to total MEK1/2 or total
ERK1/2, respectively, suggested that the MEK/ERK pathway was
activated by BLACAT2 overexpression (P<0.05; Fig. 4B). By contrast, BLACAT2 knockdown
significantly suppressed the activation of this pathway in UCEC
cells (P<0.01; Fig. 4C and D).
These results indicated that BLACAT2 can regulate MEK/ERK signaling
in UCEC cells.
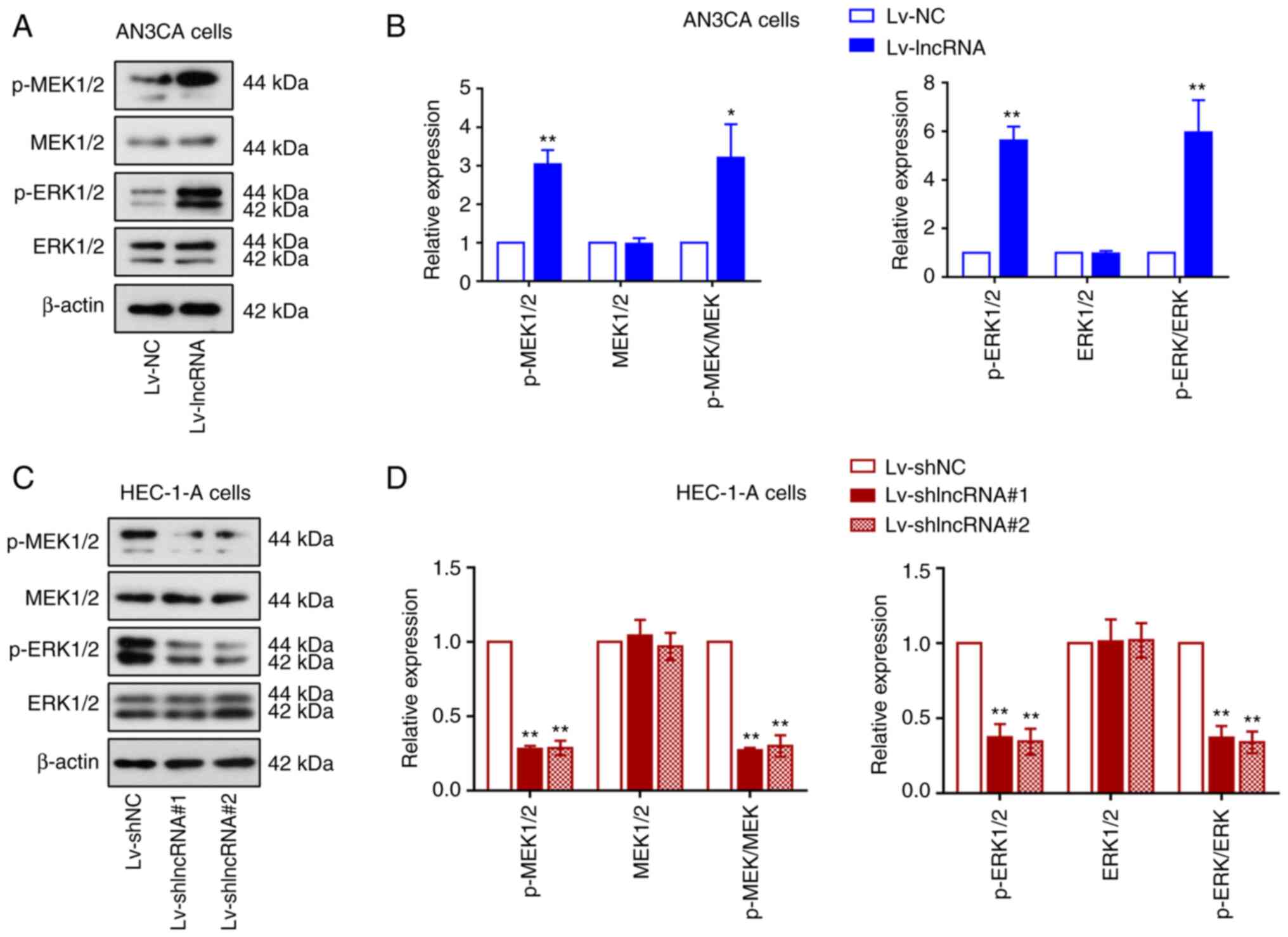 | Figure 4.BLACAT2 overexpression enhances
MER/ERK signaling in uterine corpus endometrial carcinoma cells.
Expression of the protein markers in the MEK/ERK signaling pathway
were next analyzed in AN3CA cells with stably overexpressed BLACAT2
and in HEC-1-A cells with stably knocked down BLACAT2. (A)
Representative images of western blot analysis of p-MEK1/2, total
MEK1/2, p-ERK1/2, and total ERK1/2 both in AN3CA cells, (B) which
were semi-quantified. (C) Representative images of western blot
analysis of p-MEK1/2, total MEK1/2, p-ERK1/2, and total ERK1/2 both
in HEC-1-A cells, (D) which were semi-quantified. Values are
presented as the means ± SD (n=3). *P<0.05 and **P<0.01 vs.
Lv-NC or Lv-shNC. BLACAT2, bladder cancer-associated transcript 2;
p-, phosphorylated; Lv, lentivirus; sh, short-interfering; NC,
negative control. |
BLACAT2 promotes UCEC tumor growth in
vivo
The effect of BLACAT2 on UCEC progression was next
studied in vivo. AN3CA cells with stably overexpressed
BLACAT2/NC and HEC-1-A cells with stably knocked down BLACAT2/NC,
were injected into nude mice. The tumor volumes and weights of the
nude mice injected with cells overexpressing BLACAT2 were found to
be increased compared with those of nude mice injected with cells
expressing Lv-NC. However, BLACAT2 knockdown suppressed tumor
growth in nude mice (P<0.05; Fig.
5A-C). The expression of Ki-67 using immunohitochemical (IHC)
staining indicated that BLACAT2 overexpression promoted, whereas
BLACAT2 knockdown suppressed UCEC tumor cell proliferation
(Fig. 5D). Furthermore, the protein
expression levels of cyclin D1, MMP2 and phosphorylated ERK1/2 were
also increased by BLACAT2 overexpression but inhibited by BLACAT2
knockdown (P<0.01; Fig. 5E and
F). These observations indicated that BLACAT2 promoted UCEC
tumor growth and activation of MEK/ERK signaling in
vivo.
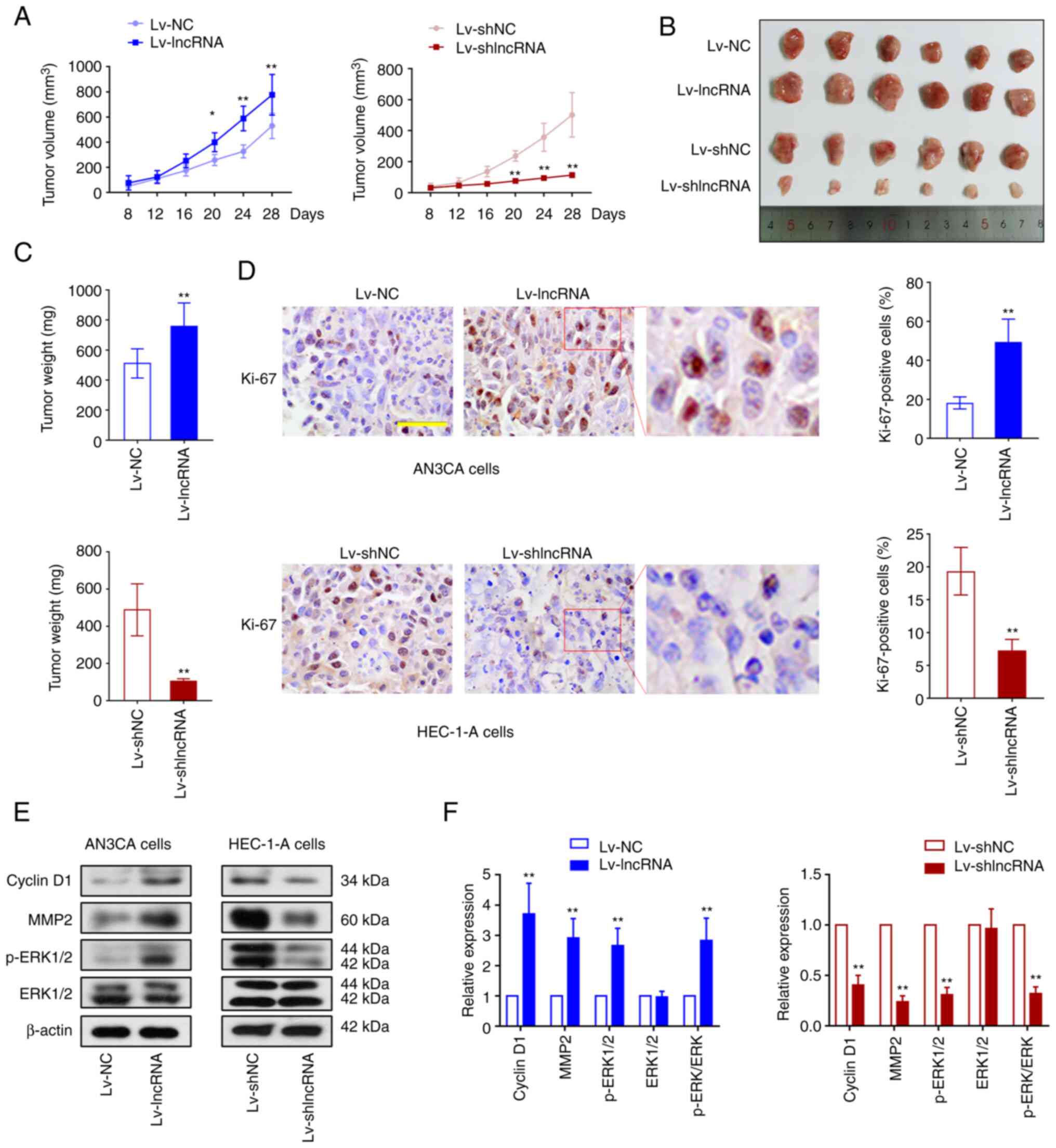 | Figure 5.BLACAT2 promotes UCEC tumor growth
in vivo. AN3CA cells with stably overexpressed BLACAT2 and
HEC-1-A cells with stably knocked down BLACAT2 were injected into
the right flank of nude mice. (A) The volume of UCEC tumors in nude
mice was measured every 4 days. (B) The images of UCEC tumors
removed from the nude mice 4 weeks after injection. (C) The weight
of the tumors in nude mice was measured after sacrifice. (D)
Immunohistochemical staining showed the expression of Ki-67 in the
UCEC tumors in nude mice. Magnification, ×400. Scale bar, 50 µm.
(E) Representative western blotting images and (F) protein
expression semi-quantification of cyclin D1, MMP2, phosphorylated
ERK1/2 and ERK1/2 in the tumors. Values are presented as the means
± SD (n=6). *P<0.05 and **P<0.01 vs. Lv-NC or Lv-shNC.
BLACAT2, bladder cancer-associated transcript 2; UCEC, uterine
corpus endometrial carcinoma; Lv, lentivirus; sh,
short-interfering; NC, negative control; p-, phosphorylated. |
Association among BLACAT2,
miR-378a-3p, and YY1
The predictive binding among BLACAT2, miR-378a-3p,
and YY1 is presented in Fig. 6A.
The putative binding sites of BLACAT2 to miR-378a-3p and
miR-378a-3p to YY1 3′UTR are shown in Fig. 6B. BLACAT2 overexpression was
demonstrated to reduce miR-378a-3p expression, but to increase that
of YY1 in UCEC cells, whilst BLACAT2 knockdown exerted the opposite
effects (P<0.01; Fig. 6C).
BLACAT2 overexpression also led to reduced miR-378a-3p expression
and increased YY1 expression in UCEC tumors, whilst BLACAT2
knockdown enhanced miR-378a-3p expression and reduced YY1
expression (P<0.01; Fig. S1).
The potential targeted association between miR-378a-3p and BLACAT2
or YY1 was next assessed using dual-luciferase reporter assays. The
relative luciferase activity in 293T cells co-transfected with
vectors containing wild-type BLACAT2 3′UTR and miR-378a-3p was
reduced, suggesting that miR-378a-3p can bind to BLACAT2.
miR-378a-3p was also found to bind to the 3′UTR of YY1 (P<0.01;
Fig. 6D). In addition, Fig. 6E revealed the possible binding sites
of YY1 in the BLACAT2 promoter region. The increased relative
luciferase activity in 293T cells co-transfected with the YY1 and
BLACAT2 promoter suggested that YY1, as a transcription factor, may
bind to the BLACAT2 promoter (Fig.
6E). This was further verified using ChIP assay (Fig. 6F).
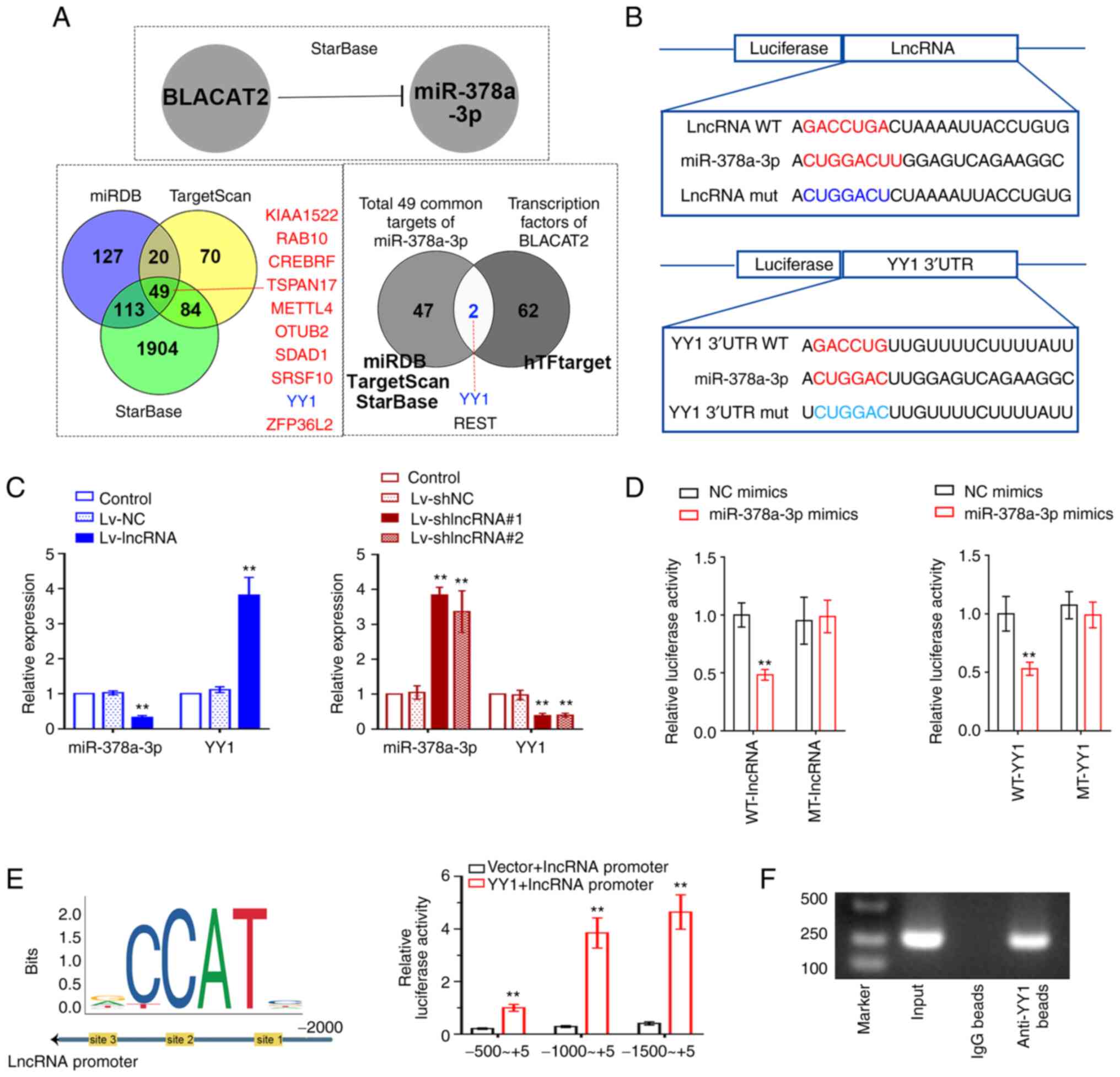 | Figure 6.Association among BLACAT2,
miR-378a-3p and YY1. (A) starBase predicted the binding of BLACAT2
and miR-378a-3p. Venn diagram of the potential targets of
miR-378a-3p was produced after prediction using miRDB, TargetScan
and starBase. Venn diagram showing a total of 49 common possible
targets of miR-378a-3p and the potential transcription factors of
BLACAT2 were predicted by Jasper. (B) Wild-type putative sites of
BLACAT2 or the YY1 3′UTR or their corresponding mutant sequences
were cloned into the pmirGLO luciferase reporter vector. (C)
Relative expression of miR-378-3p and YY1 in AN3CA cells with
stably overexpressed BLACAT2 and in HEC-1-A cells with stably
knocked down BLACAT2. (D) Relative luciferase activity of 293T
cells was detected to assess the association between miR-378a-3p
and BLACAT2 or the YY1 3′UTR. (E) In total, three different regions
in the BLACAT2 promoter were selected and cloned into the pmirGLO
vector, before luciferase reporter assays were performed to
investigate the possible binding between the YY1 and BLACAT2
promoter. (F) ChIP analysis of YY1 binding to the BLACAT2 promoter.
After ChIP, the PCR products were analyzed by gel electrophoresis.
Values are presented as the means ± SD (n=3). **P<0.01 vs.
Lv-NC, Lv-shNC, NC mimics or Vector + BLACAT2 promoter. BLACAT2,
bladder cancer-associated transcript 2; miR, microRNA; YY1, Yin
Yang-1; UTR, untranslated region; ChIP, chromatin
immunoprecipitation; Lv, lentivirus; sh, short-interfering; NC,
negative control; WT-, wild-type; MT-mutant. |
miR-378a-3p overexpression inhibits
UCEC cell proliferation, invasion and the ERK pathway
To study the function of miR-378a-3p in UCEC cells,
HEC-1-A cells were transfected with either miR-378a-3p mimics or NC
mimics. The transfection efficiency was validated and is presented
in Fig. S2. The mRNA expression
level of YY1 in HEC-1-A cells was significantly decreased following
miR-378a-3p overexpressin (P<0.01; Fig. 7A). miR-378a-3p overexpression also
significantly suppressed the viability and invasion of UCEC cells
(P<0.01; Fig. 7B and C). The
expression of cyclin D1, MMP2, and ERK1/2 phosphorylation was found
to be inhibited by miR-378a-3p overexpression (P<0.01; Fig. 7D), suggesting that miR-378a-3p can
suppress cell proliferation, invasion, and ERK signaling in UCEC
cells.
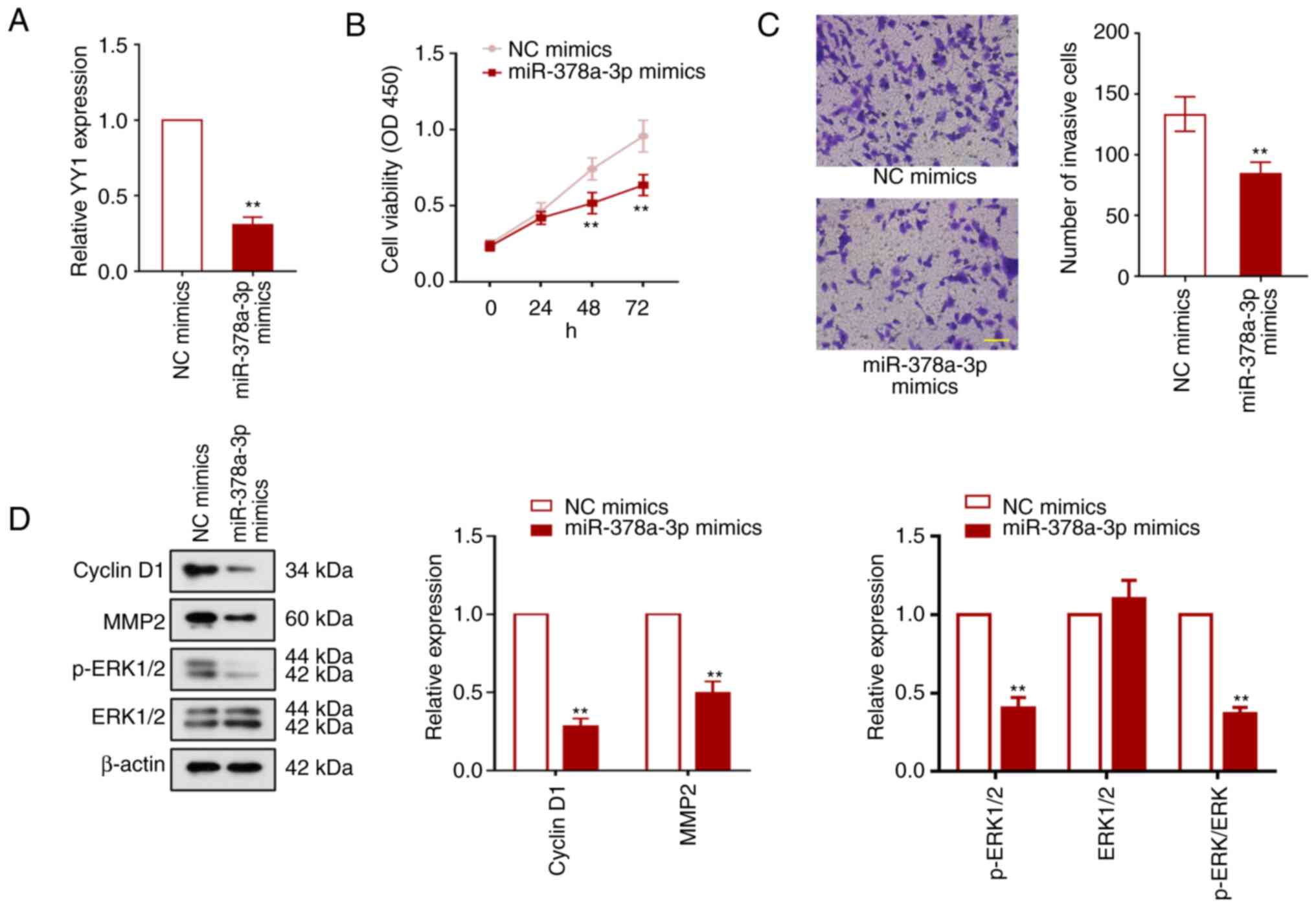 | Figure 7.miR-378a-3p overexpression inhibits
UCEC cell proliferation, invasion and the ERK1/2 pathway. HEC-1-A
cells were transfected with the miR-378a-3p mimics to explore the
function of miR-378a-3p in UCEC progression. (A) Relative
expression of Yin Yang-1 in HEC-1-A cells was detected using
reverse transcription-quantitative PCR 48 h after transfection. (B)
Cell viability was measured using the Cell Counting Kit-8 assay.
(C) The effect of miR-378a-3p overexpression on HEC-1-A cell
invasion. Magnification, ×200. Scale bar, 100 µm (D) Relative
expression levels of cyclin D1, mature MMP2, ERK1/2 and
phosphorylated ERK1/2 in HEC-1-A cells. Values are presented as the
means ± SD (n=3). **P<0.01 vs. NC mimics. miR, microRNA; UCEC,
uterine corpus endometrial carcinoma; YY1, Yin Yang-1; NC, megative
control; p-, phosphorylated. |
YY1 knockdown reverses BLACAT2
overexpression-mediated UCEC development
To investigate whether YY1 is involved in the
regulation of BLACAT2 and therefore UCEC development, Lv-shYY1 was
used to infect AN3CA cells, and YY1 expression was knocked down
effectively in AN3CA cells after the infection (P<0.01; Fig. 8A). AN3CA cells with stably
overexpressed BLACAT2 were then infected with Lv-shYY1 or Lv-shNC.
CCK-8 assay revealed that in BLACAT2-overexpressed UCEC cells, YY1
downregulation significantly reduced cell viability (P<0.01;
Fig. 8B). In addition, YY1
knockdown abrogated the positive effects of BLACAT2 on UCEC cell
migration and invasion (P<0.05; Fig.
8C and D). The protein expression levels of cyclin D1, MMP2 and
ERK1/2 phosphorylation were also significantly supressed by YY1
inhibition (P<0.01; Fig. 8E and
F). These results indicated that YY1 is involved in the
regulation of BLACAT2 expression upstream of UCEC cell
proliferation, migration, invasion, and MER/ERK signaling.
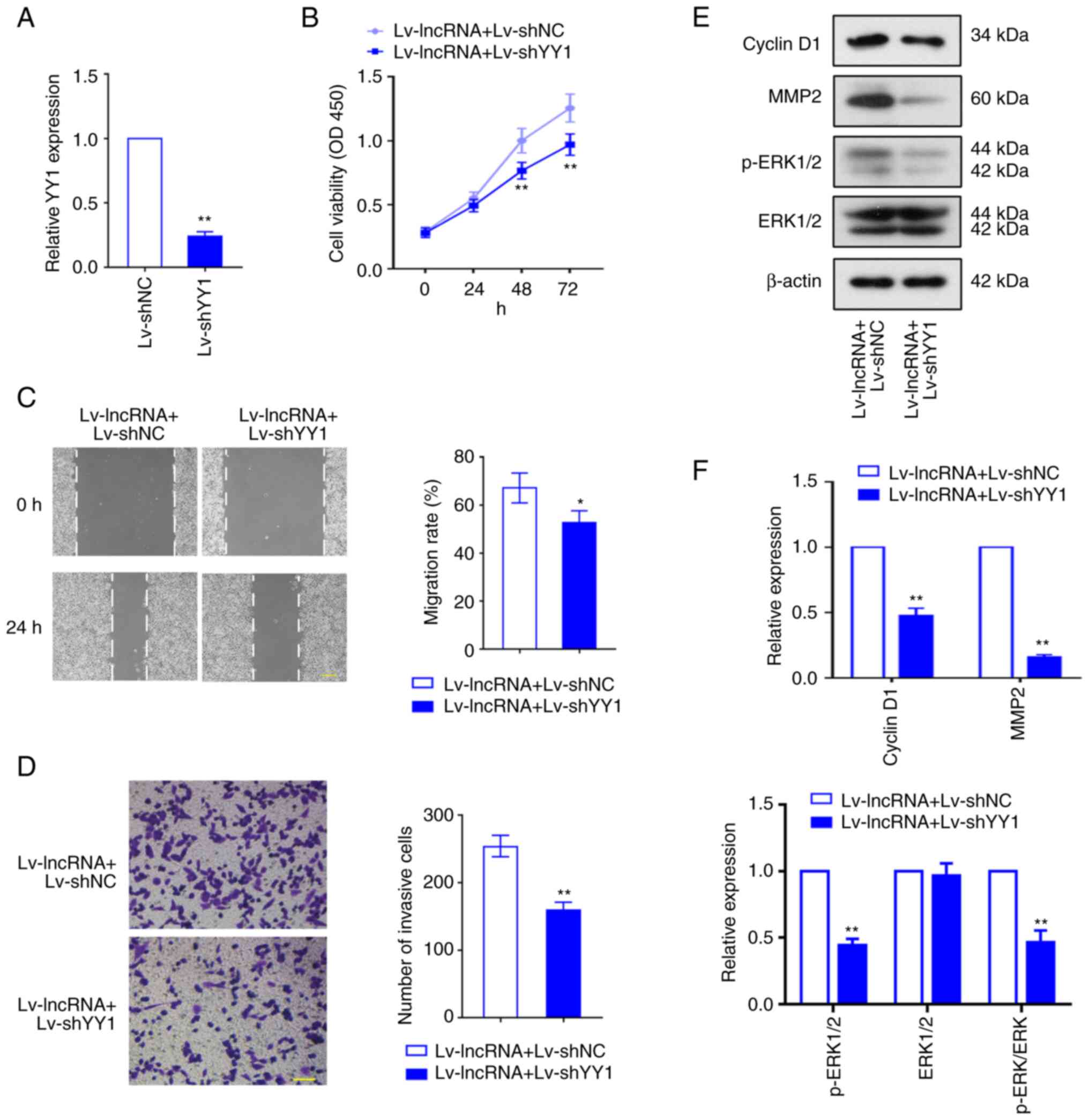 | Figure 8.YY1 knockdown inhibits BLACAT2
overexpression-mediated uterine corpus endometrial carcinoma
development. Lentiviruses encoding shYY1 (Lv-shYY1) were used to
infect AN3CA cells with stably overexpressed BLACAT2. (A) The
infectivity of the shYY1 lentivirus (Lv-shYY1) was verified in
AN3CA cells. (B) The viability of AN3CA cells was determined using
Cell Counting Kit-8 assay. (C) Migration of AN3CA cells was
detected using a wound healing assay, before the migration rate was
calculated. Magnification, ×100. Scale bar, 200 µm. (D) The effect
of YY1 knockdown on AN3CA cell invasion following BLACAT2
overexpression. Magnification, ×200. Scale bar, 100 µm. (E)
Representative images and (F) semi-quantification of protein
expression of cyclin D1, MMP2, phosporylated ERK1/2 and total
ERK1/2 in AN3CA cells after transfection. Values are presented as
the means ± SD (n=3). *P<0.05 and **P<0.01 vs. Lv-BLACAT2 +
Lv-shNC. YY1, Yin Yang-1; BLACAT2, bladder cancer-associated
transcript 2; sh, short-interfering; Lv, lentivirus; NC, negative
control; p-, phosphorylated. |
Discussion
In the present study, BLACAT2 overexpression was
found to promote UCEC cell proliferation, migration, and invasion.
The tumor-promoting effects of BLACAT2 in UCEC are likely to be
mediated by a miR-378a-3p/YY1 axis. This BLACAT2/miR-378a-3p/YY1
axis in UCEC development is also likely to involve the MEK/ERK
signaling pathway. In addition, YY1 may bind to the promoter of
BLACAT2 to regulate its expression.
LncRNAs are expressed in a tissue- and
cell-type-specific manner, where they have been demonstrated to
serve vital roles in multiple cancers (43,44). A
previous study identified seven lncRNAs to be potential prognostic
factors in UCEC according to RNA-sequencing and expression
profiling coupled with analysis of corresponding clinical data of
patients with UCEC from The Cancer Genome Atlas database (45). Illumina paired-end RNA sequencing
demonstrated that BLACAT2 expression is upregulated in endometrial,
ovarian and cervical cancer (20).
BLACAT2 has been found to be differently expressed in UCEC tumor
and normal tissue, but its function and mechanism in UCEC remain
poorly understood. In the present study, BLACAT2 overexpression was
found to suppress miR-378a-3p expression, whilst promoting YY1
expression, UCEC cell viability, cell cycle progression, cell
migration and invasion. By contrast, BLACAT2 knockdown exerted the
opposite effects on all of the aforementioned physiological
processes. As part of the cyclin family of proteins, cyclin D1 and
cyclin E are important regulators of cancer cell proliferation, the
downregulation of which was associated with cell cycle arrest
(46,47). By contrast, the migration and
invasion of cancer cells are mediated by metalloproteinases, MMP2
and MMP9 (48,49). The present study revealed increased
cyclin D1, cyclin E, MMP2, and MMP9 in UCEC cells following BLACAT2
overexpression, whereas they were all reduced by BLACAT2 knockdown.
This suggests that BLACAT2 overexpression can promote UCEC cell
proliferation, migration, and invasion. In addition, it was found
that the BLACAT2-mediated UCEC cell proliferation was associated
with increases in p-MEK/MEK and p-ERK/ERK ratios, suggesting that
BLACAT2 overexpression can also activate MEK/ERK signaling in UCEC
cells. The tumor-promoting effects of the activated MEK/ERK pathway
have been demonstrated in previous studies (50,51).
Therefore, the results from the present study revealed a potential
novel signaling pathway regulated by BLACAT2 in UCEC.
LncRNAs typically function as competing endogenous
RNAs with other RNA transcripts for the same miRNAs (52). LncRNAs can function as miRNA sponges
and block the regulatory effects of miRNAs on mRNAs (53). The regulation of transcription is a
complex process that involves interactions among different types of
signaling components, including mRNA, lncRNA and miRNA. Liu et
al (54) previously proposed a
novel method for constructing genome-wide interactive networks and
found the cross-regulation of lncRNA, miRNA and mRNA, called the
‘mRNA-lncRNA-miRNA triplet’. miR-378a-3p was predicted to bind to
BLACAT2 using bioinformatic analysis, which was then confirmed
using a dual-luciferase report assay. The interaction of BLACAT2
with miR-378a-3p was also found in a previous study by Cui et
al (35), which demonstrated
that BLACAT2 can regulate cancer development by sponging
miR-378a-3p. Previous studies demonstrated that tumor promotion may
involve miR-378a-3p inhibition (55,56).
In the present study, miR-378a-3p overexpression resulted in the
suppression of YY1 expression, UCEC cell proliferation and
invasion, suggesting the antitumor role of miR-378a-3p in UCEC. Hu
et al (16) previously
showed that BLACAT2 can serve as an oncogene by modulating the
miR-193b-5p/methyltransferase 3, N6-adenosine-methyltransferase
complex catalytic subunit axis (16). In the present study, the function of
BLACAT2 in UCEC progression may be achieved by the regulation of
miR-378a-3p and YY1 downstream, which further affected the MEK/ERK
pathway. Therefore, the differential roles of BLACAT2 in human
cancers may depend on its different downstream miRNAs and target
mRNAs.
YY1 was shown to be a potential target of
miR-378a-3p. YY1 was previously found to be overexpressed in tumors
and identified to be an oncogenic factor. This was potentially
mediated by the recruitment of enhancer of zeste homolog 2 and the
activation of histone H3K27me3 in the adenomatous polyposis coli
gene promoter region in EEC, which silences the expression of this
gene. In addition, YY1 depletion was found to suppress EEC cell
proliferation and migration, whilst YY1 overexpression could
promote these processes (30). YY1
can also function as a positive regulator of AKT1, a member of the
PI3K/AKT/mTOR signaling pathway activated in endometrial tumors
(57). These previous studies
suggest that YY1 is an important regulator of endometrial carcinoma
progression. Consistently, the present study showed that YY1
knockdown abolished the BLACAT2 overexpression-induced UCEC cell
proliferation, invasion and activation of MEK/ERK signaling,
suggesting that YY1 was involved in the regulation of BLACAT2 and
therefore UCEC progression. In gastric cancer cells, YY1 knockdown
resulted in the inhibition of ERK/MAPK signaling, a major oncogenic
pathway (58). Therefore, the
present study revealed that the induction of YY1 expression may
activate the ERK pathway to promote UCEC tumor growth.
The direct binding of YY1 to the gene promoter of
BLACAT2 was identified using dual-luciferase reporter and ChIP
assays in the present study, suggesting that YY1 can regulate
BLACAT2 transcription. The regulatory relationship among BLACAT2,
miR-378a-3p, and YY1 appears to form a feedback loop instead of an
axis. Therefore, BLACAT2 not only controls UCEC development by
regulating the miR-378a-3p/YY1 axis, but is also under the control
of the transcription factor YY1. Based on these findings, BLACAT2
may bind to miR-378a-3p and function as a sponge of this miRNA,
resulting in the abolishment of the inhibitory effects of
miR-378a-3p on YY1 in UCEC. However, the regulatory mechanism of
YY1 in BLACAT2 expression requires further investigation. In
addition, the association of the BLACAT2/miR-378a-3p/YY1 loop with
the ERK signaling pathway in UCEC cells warrants further
exploration in the future.
Findings from the present study suggest the
potential of BLACAT2 as a biomarker in UCEC. However, the present
study also has various limitations. It is essential to enroll an
increased number of cancer cases in future studies. In addition,
the majority of lncRNAs can regulate >1 target gene, but only
the preliminary regulatory mechanism of the effect of BLACAT2 was
revealed in the present study. The specific mechanisms involved in
the effect of BLACAT2 in UCEC require further investigation.
Furthering the understanding of the function and potential
molecular mechanisms of BLACAT2 will facilitate the development of
lncRNA-based therapeutics in the clinic.
In conclusion, in the present study it was revealed
that BLACAT2 regulated the progression of UCEC through the
miR-378a-3p/YY1 axis. However, downstream YY1 can bind to BLACAT2
promoter to promote its expression as a transcription factor. In
addition, MEK/ERK is involved downsteam of the
BLACAT2/miR-378a-3p/YY1 loop in UCEC progression. The present study
therefore provides novel therapeutic targets for UCEC and insights
into UCEC tumorigenesis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Public Health Research
and Development Project of Shenyang (Medical Technology Improvement
Project of Common Multiple Diseases), grant no. 22-321-33-15.
Availability of data and materials
The data used to support the findings of this study
are included in the manuscript.
Authors' contributions
CZ and QY conceived and designed the experiments. CZ
and RW performed the experiments. CZ, RW and ML analyzed the data,
and performed the bioinformatics analysis. CZ wrote the manuscript.
QY helped edit the manuscript. CZ and QY confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The research protocol was approved (approval no.
2021PS723K) by the Ethics Committee of Shengjing Hospital of China
Medical University (Shenyang, China) and was in accordance with the
Declaration of Helsinki. The informed consent was obtained from all
volunteers. Animal experimental procedures were approved (approval
no. 2021PS741K) by the Animal Ethics Committee of Shengjing
Hospital of China Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
miRNAs
|
microRNAs
|
|
YY1
|
transcription factor Yin Yang-1
|
|
UCEC
|
uterine corpus endometrial
carcinoma
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
ChIP
|
chromatin immunoprecipitation
|
|
ERK
|
extracellular signal-regulated
kinase
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aoki Y, Kanao H, Wang X, Yunokawa M,
Omatsu K, Fusegi A and Takeshima N: Adjuvant treatment of
endometrial cancer today. Jpn J Clin Oncol. 50:753–765. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Slavoff SA, Mitchell AJ, Schwaid AG,
Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL and
Saghatelian A: Peptidomic discovery of short open reading
frame-encoded peptides in human cells. Nat Chem Biol. 9:59–64.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seitz AK, Christensen LL, Christensen E,
Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM,
Palmfeldt J, Thomson M, et al: Profiling of long non-coding RNAs
identifies LINC00958 and LINC01296 as candidate oncogenes in
bladder cancer. Sci Rep. 7:3952017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He W, Zhong G, Jiang N, Wang B, Fan X,
Chen C, Chen X, Huang J and Lin T: Long noncoding RNA BLACAT2
promotes bladder cancer-associated lymphangiogenesis and lymphatic
metastasis. J Clin Invest. 128:861–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zuo X, Chen Z, Gao W, Zhang Y, Wang J,
Wang J, Cao M, Cai J, Wu J and Wang X: M6A-mediated upregulation of
LINC00958 increases lipogenesis and acts as a nanotherapeutic
target in hepatocellular carcinoma. J Hematol Oncol. 13:52020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao H, Zheng GH, Li GC, Xin L, Wang YS,
Chen Y and Zheng XM: Long noncoding RNA LINC00958 regulates cell
sensitivity to radiotherapy through RRM2 by binding to
microRNA-5095 in cervical cancer. J Cell Physiol. 234:23349–23359.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen M, Xu Z, Zhang Y and Zhang X:
LINC00958 promotes the malignancy of nasopharyngeal carcinoma by
sponging microRNA-625 and thus upregulating NUAK1. Onco Targets
Ther. 12:9277–9290. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu
FN, Yan ML, Tian YF, Peng CH, Shen BY, Chen YL and Wang YD:
Silencing of long noncoding RNA LINC00958 prevents tumor initiation
of pancreatic cancer by acting as a sponge of microRNA-330-5p to
down-regulate PAX8. Cancer Lett. 446:49–61. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo Z, Han Z, Shou F, Li Y and Chen Y:
LINC00958 accelerates cell proliferation and migration in non-small
cell lung cancer through JNK/c-JUN signaling. Hum Gene Ther
Methods. 30:226–234. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu H, Kong Q, Huang XX, Zhang HR, Hu KF,
Jing Y, Jiang YF, Peng Y, Wu LC, Fu QS, et al: Longnon-coding RNA
BLACAT2 promotes gastric cancer progression via the
miR-193b-5p/METTL3 pathway. J Cancer. 12:3209–3221. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ren Y, Zhao C, He Y, Xu H and Min X: Long
non-coding RNA bladder cancer-associated transcript 2 contributes
to disease progression, chemoresistance and poor survival of
patients with colorectal cancer. Oncol Lett. 18:2050–2058.
2019.PubMed/NCBI
|
|
18
|
Zhu J, Zhu Z, Cai P, Gu Z and Wang J:
Bladder cancer-associated transcript 2 contributes to
nephroblastoma progression. J Gene Med. 24:e32922022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Binang HB, Wang YS, Tewara MA, Du L, Shi
S, Li N, Nsenga AGA and Wang C: Expression levels and associations
of five long non-coding RNAs in gastric cancer and their clinical
significance. Oncol Lett. 19:2431–2445. 2020.PubMed/NCBI
|
|
20
|
Chen BJ, Byrne FL, Takenaka K, Modesitt
SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL and Janitz M:
Transcriptome landscape of long intergenic non-coding RNAs in
endometrial cancer. Gynecol Oncol. 147:654–662. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang Y, Qiao Z, Jiang J and Zhang J:
LINC00958 promotes endometrial cancer cell proliferation and
metastasis by regulating the miR-145-3p/TCF4 axis. J Gene Med.
23:e33452021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Khachigian LM: The Yin and Yang of YY1 in
tumor growth and suppression. Int J Cancer. 143:460–465. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sarvagalla S, Kolapalli SP and
Vallabhapurapu S: The two sides of YY1 in cancer: A friend and a
foe. Front Oncol. 9:12302019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thomas MJ and Seto E: Unlocking the
mechanisms of transcription factor YY1: Are chromatin modifying
enzymes the key? Gene. 236:197–208. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qiao K, Ning S, Wan L, Wu H, Wang Q, Zhang
X, Xu S and Pang D: LINC00673 is activated by YY1 and promotes the
proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo
signaling pathway. J Exp Clin Cancer Res. 38:4182019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berchuck A, Iversen ES, Lancaster JM,
Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M,
et al: Patterns of gene expression that characterize long-term
survival in advanced stage serous ovarian cancers. Clin Cancer Res.
11:3686–3696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seligson D, Horvath S, Huerta-Yepez S,
Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L and
Bonavida B: Expression of transcription factor Yin Yang 1 in
prostate cancer. Int J Oncol. 27:131–141. 2005.PubMed/NCBI
|
|
28
|
Luo J, Zhou X, Ge X, Liu P, Cao J, Lu X,
Ling Y and Zhang S: Upregulation of Ying Yang 1 (YY1) suppresses
esophageal squamous cell carcinoma development through heme
oxygenase-1. Cancer Sci. 104:1544–1551. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao G, Li Q, Wang A and Jiao J: YY1
regulates melanoma tumorigenesis through a miR-9~RYBP axis. J Exp
Clin Cancer Res. 34:662015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang
P, Chan LK, Zhang T, Yu J, Kwong J, et al: A novel
miR-193a-5p-YY1-APC regulatory axis in human endometrioid
endometrial adenocarcinoma. Oncogene. 32:3432–3442. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Masood N, Basharat Z, Khan T and Yasmin A:
Entangling relation of micro RNA-let7, miRNA-200 and miRNA-125 with
various cancers. Pathol Oncol Res. 23:707–715. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao M, Iglinski-Benjamin KC, Sharpe O,
Robinson WH and Abrams GD: Exogenous micro-RNA and antagomir
modulate osteogenic gene expression in tenocytes. Exp Cell Res.
378:119–123. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang T, Sui D, You D, Yao S, Zhang L,
Wang Y, Zhao J and Zhang Y: MiR-29a-5p inhibits proliferation and
invasion and induces apoptosis in endometrial carcinoma via
targeting TPX2. Cell Cycle. 17:1268–1278. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang P, Zeng Z, Shen X, Tian X and Ye Q:
Identification of a multi-RNA-type-based signature for
recurrence-free survival prediction in patients with uterine corpus
endometrial carcinoma. DNA Cell Biol. 39:615–630. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cui Y, Xie M and Zhang Z: LINC00958
involves in bladder cancer through sponging miR-378a-3p to elevate
IGF1R. Cancer Biother Radiopharm. 35:776–788. 2020.PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thomas GJ, Lewis MP, Hart IR, Marshall JF
and Speight PM: AlphaVbeta6 integrin promotes invasion of squamous
carcinoma cells through up-regulation of matrix
metalloproteinase-9. Int J Cancer. 92:641–650. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Zhong Y, Yang B, Zhu Y, Zhu X, Xia
Z, Xu J and Xu L: LINC00958 facilitates cervical cancer cell
proliferation and metastasis by sponging miR-625-5p to upregulate
LRRC8E expression. J Cell Biochem. 121:2500–2509. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xie M, Fu Q, Wang PP and Cui YL:
STAT1-induced upregulation lncRNA LINC00958 accelerates the
epithelial ovarian cancer tumorigenesis by regulating Wnt/β-catenin
signaling. Dis Markers. 2021:14050452021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang D, Ma Q, Wang Z, Zhang M, Guo K,
Wang F and Wu E: β2-adrenoceptor blockage induces G1/S phase arrest
and apoptosis in pancreatic cancer cells via Ras/Akt/NFκB pathway.
Mol Cancer. 10:1462011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sato H, Takino T, Okada Y, Cao J,
Shinagawa A, Yamamoto E and Seiki M: A matrix metalloproteinase
expressed on the surface of invasive tumour cells. Nature.
370:61–65. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Roberts PJ and Der CJ: Targeting the
Raf-MEK-ERK mitogen-activated protein kinase cascade for the
treatment of cancer. Oncogene. 26:3291–3310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3:52015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ouyang D, Li R, Li Y and Zhu X: A 7-lncRNA
signature predict prognosis of uterine corpus endometrial
carcinoma. J Cell Biochem. 120:18465–18477. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S,
Zhang M, Yu F, Peng Z and Zhou C: Elevated HOXA1 expression
correlates with accelerated tumor cell proliferation and poor
prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer
Res. 35:152016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Park GH, Song HM and Jeong JB: The coffee
diterpene kahweol suppresses the cell proliferation by inducing
cyclin D1 proteasomal degradation via ERK1/2, JNK and
GKS3β-dependent threonine-286 phosphorylation in human colorectal
cancer cells. Food Chem Toxicol. 95:142–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu H, Zeng Z, Wang S, Li T, Mastriani E,
Li QH, Bao HX, Zhou YJ, Wang X, Liu Y, et al: Main components of
pomegranate, ellagic acid and luteolin, inhibit metastasis of
ovarian cancer by down-regulating MMP2 and MMP9. Cancer Biol Ther.
18:990–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang X, Yang B, She Y and Ye Y: The lncRNA
TP73-AS1 promotes ovarian cancer cell proliferation and metastasis
via modulation of MMP2 and MMP9. J Cell Biochem. 119:7790–7799.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Y, Zhu Y, Zhang L, Tian W, Hua S,
Zhao J, Zhang H and Xue F: Insulin promotes proliferation,
survival, and invasion in endometrial carcinoma by activating the
MEK/ERK pathway. Cancer Lett. 322:223–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang F, Chen Q, Huang G, Guo X, Li N, Li Y
and Li B: BKCa participates in E2 inducing endometrial
adenocarcinoma by activating MEK/ERK pathway. BMC Cancer.
18:11282018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao Z, Sun W, Guo Z, Zhang J, Yu H and
Liu B: Mechanisms of lncRNA/microRNA interactions in angiogenesis.
Life Sci. 254:1169002020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA-LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu C, Zhang YH, Deng Q, Li Y, Huang T,
Zhou S and Cai YD: Cancer-related triplets of mRNA-lncRNA-miRNA
revealed by integrative network in uterine corpus endometrial
carcinoma. Biomed Res Int. 2017:38595822017.PubMed/NCBI
|
|
55
|
Kong D, Hou Y, Li W, Ma X and Jiang J:
LncRNA-ZXF1 regulates P21 expression in endometrioid endometrial
carcinoma by managing ubiquitination-mediated degradation and
miR-378a-3p/PCDHA3 axis. Mol Oncol. 16:813–829. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rong D, Dong Q, Qu H, Deng X, Gao F, Li Q
and Sun P: m6A-induced LINC00958 promotes breast cancer
tumorigenesis via the miR-378a-3p/YY1 axis. Cell Death Discov.
7:272021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Painter JN, Kaufmann S, O'Mara TA, Hillman
KM, Sivakumaran H, Darabi H, Cheng THT, Pearson J, Kazakoff S,
Waddell N, et al: A common variant at the 14q32 endometrial cancer
risk locus activates AKT1 through YY1 binding. Am J Hum Genet.
98:1159–1169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bhaskar Rao D, Panneerpandian P,
Balakrishnan K and Ganesan K: YY1 regulated transcription-based
stratification of gastric tumors and identification of potential
therapeutic candidates. J Cell Commun Signal. 15:251–267. 2021.
View Article : Google Scholar : PubMed/NCBI
|