Introduction
Cervical cancer is the fourth most frequently
diagnosed type of cancer and the fourth leading cause of
cancer-associated mortality among women, with an estimated 604,000
new cases and 342,000 related mortalities worldwide in 2020
(1). In Brazil, cervical cancer is
the third most common type of cancer among women (1). Despite improved prevention programs,
numerous patients are diagnosed at a locally advanced stage, and
the prognosis and 5-year survival rate of patients with locally
advanced cervical cancer (LACC) remain poor (2). The standard treatment for LACC is
radiotherapy concomitant with platinum-based chemotherapy (3,4).
However, >50% of patients on this treatment regimen may
experience resistance to therapy (5).
It is well understood that, although human
papillomavirus (HPV) infection is necessary to immortalize cervical
cells, this infection is not sufficient to lead to their
transformation (6). For this
reason, studies have indicated that epigenetic mechanisms may be
involved in the process of cervical tumor progression (7). Therefore, it is essential to elucidate
the molecular mechanisms underlying the progression of cervical
cancer.
Recently, the concept of the non-mutational
epigenetic regulation of gene expression was demonstrated as a
novel hallmark of cancer (8). There
is increasing evidence to indicate that these epigenetic
alterations may contribute to tumor development and malignant
progression. MicroRNAs (miRNAs or miRs) can modulate the
pathophysiological mechanisms of cervical cancer, which may provide
novel approaches for the diagnosis and treatment of patients with
cervical cancer in the future (9,10).
miRNAs are short (~22 nucleotides in length) non-coding RNAs that
generally control gene expression at the post-transcriptional level
through mRNA degradation or translational repression (11), and have been reported to function as
oncomiRs or tumor-suppressor miRNAs (12). miRNAs can play essential roles in
cell proliferation, migration and invasion in cervical cancer
(9,13). Therefore, it is necessary to
investigate the functional role of previously identified miRNAs
that are differentially expressed and associated with a higher odds
ratio for the progression of high-grade cervical cancer lesions
and, consequently, invasive cervical cancer.
The canonical delta-like Notch1 ligand (DLL1)
is a transmembrane protein (14).
DLL1 plays a role in mediating cell fate decisions during
hematopoiesis, and may participate in cell-to-cell communication
(15). The uncontrolled activation
of Notch1 signaling has been associated with developmental
disorders and cancer (16,17), suggesting that Notch and its ligand
may also be promising therapeutic targets (18). A previous study demonstrated that
the increased expression of HPV E6 can trigger a decrease in Notch1
receptor activation in basal cells (19). However, the contribution of Notch1
and its δ-like ligands to the progression of cervical cancer, as
well as the possible regulatory targets of this gene remain
unclear.
Recently, the authors reported that seven miRNAs
were highly expressed in cervical intraepithelial neoplasia grade 3
(CIN3), and two miRNAs had a low expression level in these samples
(20). Among the highly expressed
miRNAs, miR-130a-3p conferred a high risk for CIN3 lesions in
liquid-based cytology cervical samples (20). In addition, other studies have
demonstrated that miR-130a-3p plays a critical role in cervical
cancer progression (21,22). Considering these findings, the aim
of the present study was to evaluate the role of miR-130a-3p in
cervical cancer progression in vitro.
Materials and methods
Cells, cell culture and
transfection
A total of five commercial human cervical cancer
cell lines (HeLa, SiHa, CaSki, C-4I and C-33A; American Type
Culture Collection), a primary Brazilian cervical cancer
immortalized cell line (HCB-514), kindly provided by the research
group of R.M.R. [Rosa et al (23)] and a commercial non-tumorigenic
epithelial cell line (HaCaT), kindly provided by the research group
of Dr Laura Sichero and Dr Luisa Lina Villa (Center for
Translational Investigation in Oncology, Instituto do Cancer do
Estado de Sao Paulo, Hospital das Clinicas da Faculdade de Medicina
da Universidade de São Paulo, São Paulo, Brazil) were used in the
present study. All commercial cells were cultured in DMEM (Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS) (Thermo Fisher Scientific, Inc.) in a humidified incubator
with 5% CO2 at 37°C, while the primary cell line,
HCB-514, was cultured as previously described (24). The cell lines were negative for
mycoplasma contamination (MycoAlert Mycoplasma Detection Kit, Lonza
Group, Ltd.; tested monthly) and were authenticated using short
tandem-repeat analysis at the Molecular Oncology Research Center,
Barretos Cancer Hospital facilities (Barretos, São Paulo, Brazil),
as previously reported (24)
(Table SI).
For functional analysis, the cells were seeded in
six-well plates and cultured until reaching 70% confluency.
Transfection was then performed using 20 nM anti-miR-130a-3p locked
nucleic acid (LNA) inhibitor and a negative control A LNA (Qiagen,
Inc.) in combination with 2.5 µl siPORT™ NeoFX™ transfection
reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. According to the manufacturer, miRCURY LNA
miRNA inhibitors are antisense oligonucleotides with perfect
sequence complementary to their target. When introduced into cells,
they sequester their miRNA target in highly stable heteroduplexes,
effectively preventing the miRNA from hybridizing with its normal
cellular interaction partners (25). Briefly, the adherent cells were
trypsinized and resuspended in DMEM (Thermo Fisher Scientific,
Inc.) containing 10% FBS and 1% streptomycin (Sigma-Aldrich; Merck
KGaA) and penicillin (Sigma-Aldrich; Merck KGaA) and incubated at
37°C for ≤1 h. The transfection reagent was diluted in Opti-MEM
(Thermo Fisher Scientific, Inc.), as indicated in Table SII. Subsequently, the miR-130a-3p
inhibitor (anti-miR-130a-3p, catalog number: 339122) and negative
control A (identical to the former Scramble-miR control): No hits
of >70% homology to any sequence in any organism in the NCBI and
miRBase databases (cat. no. 339127) were diluted at a concentration
of 20 µM in Opti-MEM (Thermo Fisher Scientific, Inc.), following
the manufacturer's instructions. The diluted transfection agent and
anti-miR-130a-3p were combined to form a transfection complex.
Finally, the transfection complex and the cervical cancer cells
were distributed in the plate according to the volumes indicated in
Table SIII. Transfection was
confirmed using reverse transcription-quantitative PCR (RT-qPCR).
All subsequent experiments were performed after 48 h of
transfection.
RNA isolation
Total RNA was extracted from the cervical cancer
cells using the RecoverAll Total Nucleic Acid Isolation kit (cat.
no. AM1975; Thermo Fisher Scientific, Inc) following the
manufacturer's instructions. The purity and concentration of the
total RNA were evaluated using a NanoDrop Spectrophotometer v3.7
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.).
RT-qPCR
RT-qPCR was performed to detect the relative
transcript levels of miR-130a-3p. Briefly, 10 ng total RNA were
reverse transcribed into cDNA using MystiCq® microRNA
cDNA Synthesis Mix (Sigma-Aldrich; Merck KGaA). qPCR was performed
using SYBR®-Green I GoTaq® qPCR Master Mix
(Promega Corporation) according to the manufacturer's instructions
using the StepOnePlus™ Real-Time PCR System (Thermo Fisher
Scientific, Inc.). In the reaction, 0.5 µl primers at 10 mg/ml were
used (Table SIV). The cycling
conditions of RT and qPCR are presented in the Table SV. The relative expression levels
of miR-130a-3p were quantified based on the 2−ΔΔCq
method (26,27) and normalized to U6 (28).
Colony formation assay
Adhesion-independent cell proliferation was
evaluated using colony formation assay. The transfected cervical
cancer cells (CaSki and C-4I) were seeded in six-well plates at a
density of 1,000 cells in 2 ml and were incubated for 14 days in a
humidified incubator with 5% CO2 at 37°C. The medium was
replaced every 7 days. Cell colonies were fixed with methanol
(100%) and stained with crystal violet (Sigma-Aldrich; Merck KGaA)
(0.1% in PBS) both for 10 min at room temperature. Colonies
containing >50 cells were photographed under a light microscope
(Eclipse 2200; (Nikon Corporation), and the number of colonies was
analyzed using open-source software (OpenCFU; http://opencfu.sourceforge.net/) (29). The results are presented as the mean
of ≥3 independent experiments and their respective standard
deviations.
Transwell migration and invasion
assays
The cell migratory and invasive abilities were
evaluated using Transwell assay with a 24-well Transwell insert
with an 8-µm pore size membrane (Corning, Inc.). For the invasion
assay, the chambers were pre-coated with Matrigel solution (BD
Biosciences). For the preparation of the inserts, Matrigel (BD
Biosciences) was diluted at 1:10 in 0.01 M Tris buffer (pH 8.0)
with 0.7% NaCl at 4°C on ice. Subsequently, 100 µl diluted Matrigel
were pipetted onto the membranes of each insert and incubated for 1
h in an oven at 37°C. The excess fluid was removed, and the
invasion assay was then carried out. For migration assay, Transwell
assay was used with a 24-well Transwell insert with an 8-µm pore
size membrane (Corning, Inc.) without Matrigel solution. The
transfected cells (2.5×104 CaSki cells, and
4×105 C-4I and HCB-514 cells) were seeded into the upper
chamber in serum-free medium, while in the lower chamber, a
serum-supplemented medium (DMEM culture medium + 10% FBS) (Thermo
Fisher Scientific, Inc.) was added for both assays. Following
incubation for 24 h in a humidified incubator with 5%
CO2 at 37°C, those cells that had migrated or invaded
the lower surface of the membrane were fixed with methanol (100%),
stained with crystal violet (0.1%) both for 10 min at room
temperature.. The Transwell inserts were photographed under a light
microscope (Eclipse 2200; Nikon Corporation), and the number of
cells was analyzed with the open-source software CFU (http://opencfu.sourceforge.net/). The results are
represented as the mean of ≥3 independent experiments.
Wound healing assay
The alteration of cell migration induced by
miR-130a-3p inhibition was also evaluated using a wound healing
assay. The transfected CaSki and C-4I cells were seeded in six-well
plates at a density of 1×106 cells/ml. When the cells
were confluent, a scratch was made on the monolayer using a sterile
plastic 1,000-µl pipette tip. The cells were washed three times
with PBS, and complete medium with 0.5% FBS was then added.
Supplementation with FSB was maintained due to transfection with
the inhibitors or the negative control. Other studies have also
used FSB supplementation in the wound healing assay (30,31).
Images of the scratch were captured at ×10 magnification at 0, 24,
48 and 72 h using an Olympus IX71 inverted microscope (BioTek
Instruments, Inc.). The widths of the scratches were measured using
ImageJ Software version 1.49 (National Institutes of Health). The
relative migration distance was calculated as previously described
(32).
Gene expression profiling
To verify the differentially expressed genes in
cervical cancer cells, the HCB-514 cell line was selected and
compared with the non-tumorigenic epithelial cell line HaCat. For
gene expression profiling, the nCounter® Analysis System
(NanoString Technologies, Inc.) was used with nCounter®
PanCancer Pathways panel, which comprises 730 cancer
pathway-related genes in addition to 40 housekeeping genes curated
from data in The Cancer Genome Atlas (TCGA), as well as six
positive and eight negative controls. All procedures regarding
sample preparation, hybridization, detection and scanning were
performed according to the manufacturer's instructions (NanoString
Technologies, Inc.) (33). Briefly,
100 ng RNA from HCB-514 or HaCaT cells was incubated with reporter
and capture probes for 21 h at 65°C. Subsequently, the hybridized
samples were automatically purified and loaded into a cartridge in
nCounter® PrepStation (NanoString Technologies, Inc.).
Finally, the cartridge containing immobilized and aligned reporter
complexes was transferred to nCounter® Digital Analyzer
(NanoString Technologies, Inc.), with 280 fields of view. Raw data
were collected and pre-processed using nSolver™ Analysis Software
v4.0 (NanoString Technologies, Inc.), and further normalization and
differential expression analysis were performed using R-project
v3.6.3 (The R Foundation). Data were normalized using the
housekeeping method with the NanoStringNorm package (Bioconductor)
(https://www.bioconductor.org/).
Normalized data were log2-transformed and used as input for
differential expression analysis. DLL1 and WNTA10A
expression validation upon transfection of the HCB-514 cells with
anti-miR-130a-3p and the negative control was evaluated from counts
of the expression of these genes using the nCounter®
PanCancer Pathways panel. Data were normalized with the
housekeeping method using the GPATCH3 and TRIM39
genes. The gene expression data are available on the National
Center for Biotechnology Information (NCBI)-Gene Expression Omnibus
(GEO) platform (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) under
the Accession no. GSE224301.
In silico target prediction and
pathways enrichment
The bidirectional analysis between miR-130a-3p and
differentially expressed genes was performed using the online tool
MicroRNA Data Integration Portal (mirDIP) v.5.1.16.1 (34) to predict a suitable target for
miR-130a-3p. The top 5% of target genes were considered as the high
score. By searching for potential miRNAs, the miR-130a-3p targeting
site was identified within the 3′ untranslated region (UTR) of DLL1
and Wnt Family Member 10A (WNT10A), as detected using
TargetScan version 8.0 (http://www.targetscan.org) (35). Subsequently, to determine the
association between miRNA targets and cervical cancer, the plugin
ReactomeFI on Cytoscape version 3.9.1 (https://cytoscape.org/) was employed. Molecular
pathways enrichment was performed with a false discovery rate
(FDR)-corrected P-value of P<0.001. The interaction network was
performed using Cytoscape (36).
External in silico validation of DLL1
and WNT10A genes
To evaluate the expression status of DLL1 and
WNT10A genes in cervical squamous cell carcinoma and
endocervical adenocarcinoma (CESC) samples compared with 13 normal
cervical tissues, TCGA and The Genotype-Tissue Expression datasets
available in the Gene Expression Profiling Interactive Analysis
(GEPIA) 2 database (http://gepia2.cancer-pku.cn/#index) were used
(37). These databases provided 13
normal samples for comparison with 306 CESC tissue samples. For the
validation of DLL1 and WNT10A gene expression in a panel of
cervical cancer cell lines (HeLa, SiHa, CaSki, C-4I and C-33A)
compared with the HaCaT cell line, gene expression data available
on the Gene Expression patterns across Normal and Tumor tissues
(GENT) 2 platform (http://gent2.appex.kr/gent2/) were used (38).
Gene set enrichment analysis
(GSEA)
All expressed genes, regardless of whether they were
differentially expressed in either case, were used for GSEA
analysis. Gene set analysis was analyzed using GSEA software
(http://software.broadinstitute.org/gsea/index.jsp)
(39,40) based on KEGG gene set collections
(MSigDB v7.0, Broad Institute) (41,42).
GSEA first ranked all expressed genes according to the significance
of differential gene expression between the HCB-514 and HaCaT
cells. The enrichment score (ES) for each gene set is then
calculated using the entire ranked list, which reflects how the
genes for each set are distributed in the ranked list. Normalized
enriched score (NES) was determined for each gene set (39). The significant enrichment of gene
set was selected based on the absolute values of NES >1, nominal
P-value of NES ≤0.05 and rate FDR ≤0.05.
Statistical analysis
Single comparisons between the conditions studied
were performed using an unpaired Student's t-test, while
comparisons between multiple groups were performed using ANOVA and
Tukey's post hoc. For the analysis of differential gene expression,
the Student's t-test was used, and P<0.01 corrected by FDR and
FC≥2 was considered. For the correlation analysis, the Spearman's
correlation was used. Significantly enriched pathways terms are
shown in -log10 with Benjamini-Hochberg false discovery
rate-corrected P-values. Statistical analyses were performed using
GraphPad Prism version 9.0.0 (GraphPad Software; Dotmatics).
P<0.05 was considered to indicate a statistically significant
difference. The results are represented as the mean of ≥3
independent experiments.
Results
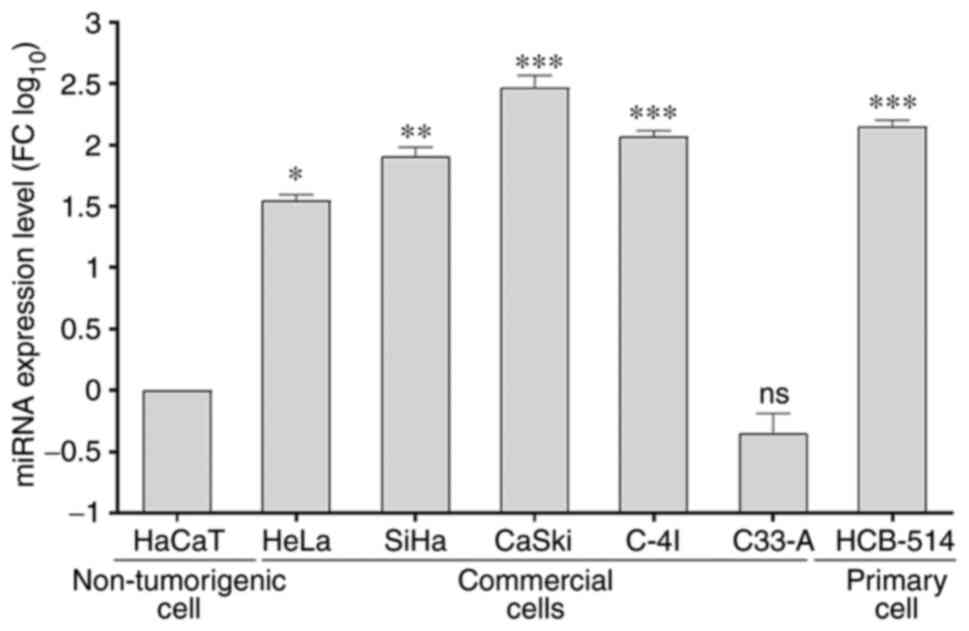
miR-130a-3p is highly expressed in
cervical cancer cell lines
The relative expression levels of miR-130a-3p in
cervical cancer cell lines and a non-tumorigenic epithelial cell
line were determined using RT-qPCR. Following log-scale
normalization, miR-130a-3p expression was found to be significantly
higher in the cervical cancer cell lines, HeLa (P=0.0332), SiHa
(P=0.0021), CaSki (P=0.0001), C-4I (P=0.0002) and HCB-514
(P=0.0002), compared with that in the non-tumorigenic epithelial
cell line, HaCaT (Fig. 1). The
CaSki, C-4I and HCB-514 cell lines were further selected for
subsequent functional analyses.
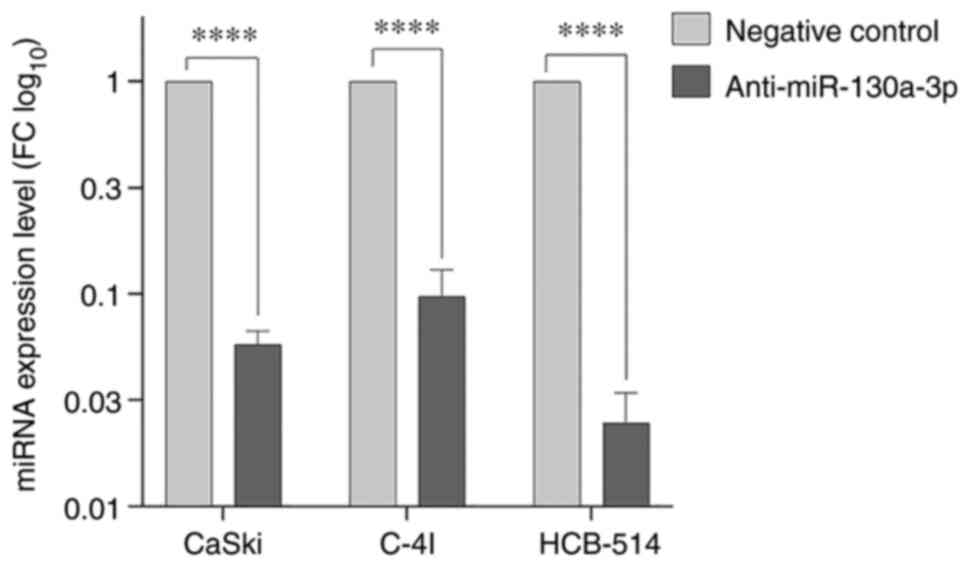
Anti-miR-130a-3p knocks down the
expression of miR-130a-3p in cervical cancer cells
The expression of miR-130a-3p was detected using
RT-qPCR following transfection with anti-miR in the CaSki, C-4I and
HCB-514 cells. As shown in Fig. 2,
in the cervical cancer cell lines, miR-130a-3p expression was lower
in the anti-miR-130a-3p group than in the negative control cells
(P<0.0001).
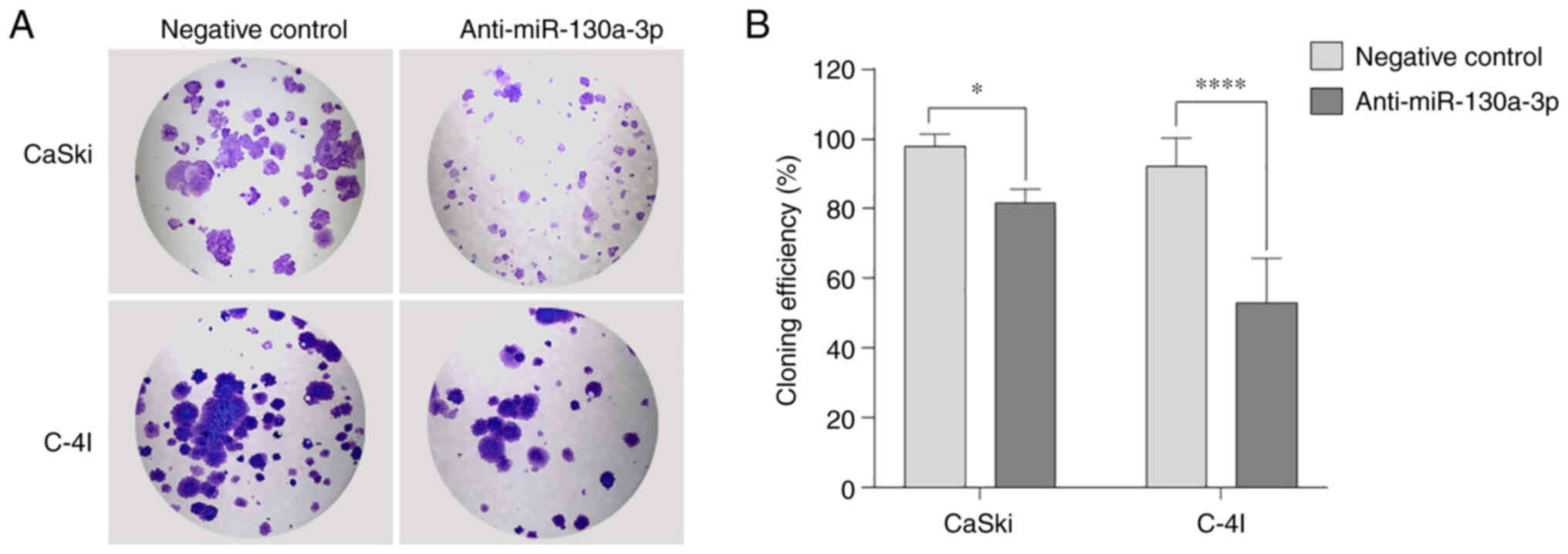
Anti-miR-130a-3p inhibits the
proliferation of cervical cancer cells
The proliferative ability of the CaSki and C-4I
cells was evaluated using colony formation assay. The results
demonstrated that the ability of a single cell to form colonies was
reduced following the inhibition of miR-130a-3p in the CaSki (81%;
P=0.0100) and C-4I (69.5%; P<0.0001) cells compared with that in
the negative control group (Fig.
3).
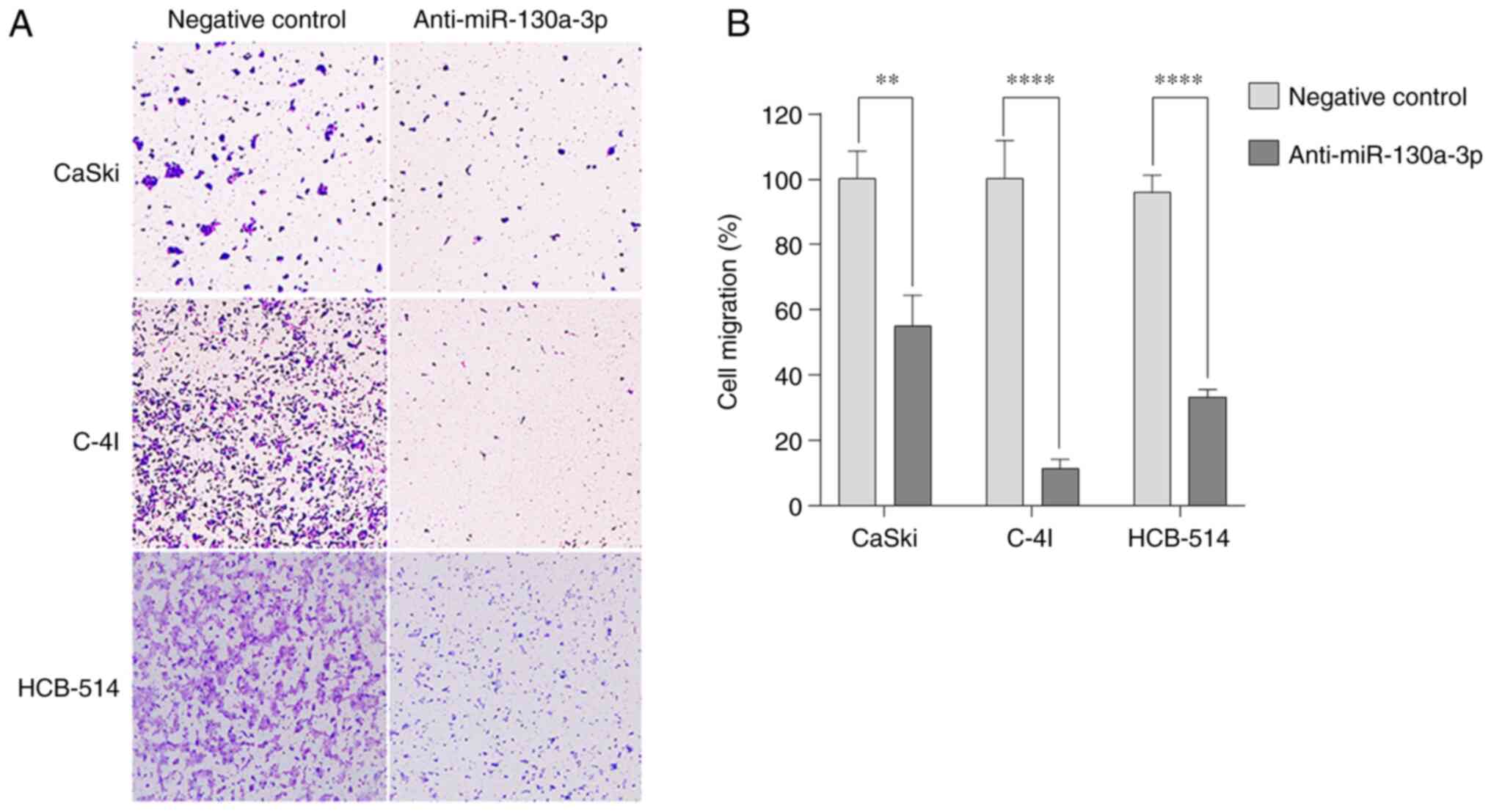
Anti-miR-130a-3p inhibits cervical
cancer cell migration
To evaluate the ability of cervical cancer cells to
migrate, Transwell assays were performed in the absence of
Matrigel. Anti-miR-130a-3p inhibited the migration of the CaSki,
C-4I and HCB-514 cells by 55% (P=0.0012), 11.3% (P<0.0001) and
33.3% (P<0.0001), respectively, compared with that of negative
control cells (Fig. 4).
In the wound healing assay, the open wound in the
control group was narrowed at 48 h in the CasKi cells (Fig. S1A) and at 24 h in the C-4I cells
(Fig. S1B). However, after
knocking down miR-130a-3p in these cell lines, the scar healing
rate was markedly attenuated. It was possible to observe the almost
complete healing of the wound at 72 h in the negative control
group, while in the anti-miR-130a-3p group, the wound was not
completely healed. Moreover, the distance between the wound edges
decreased at a slower rate in the anti-miR-130a-3p group than in
the negative control group. These results indicated that
anti-miR-130a-3p suppressed the migration of cervical cancer
cells.
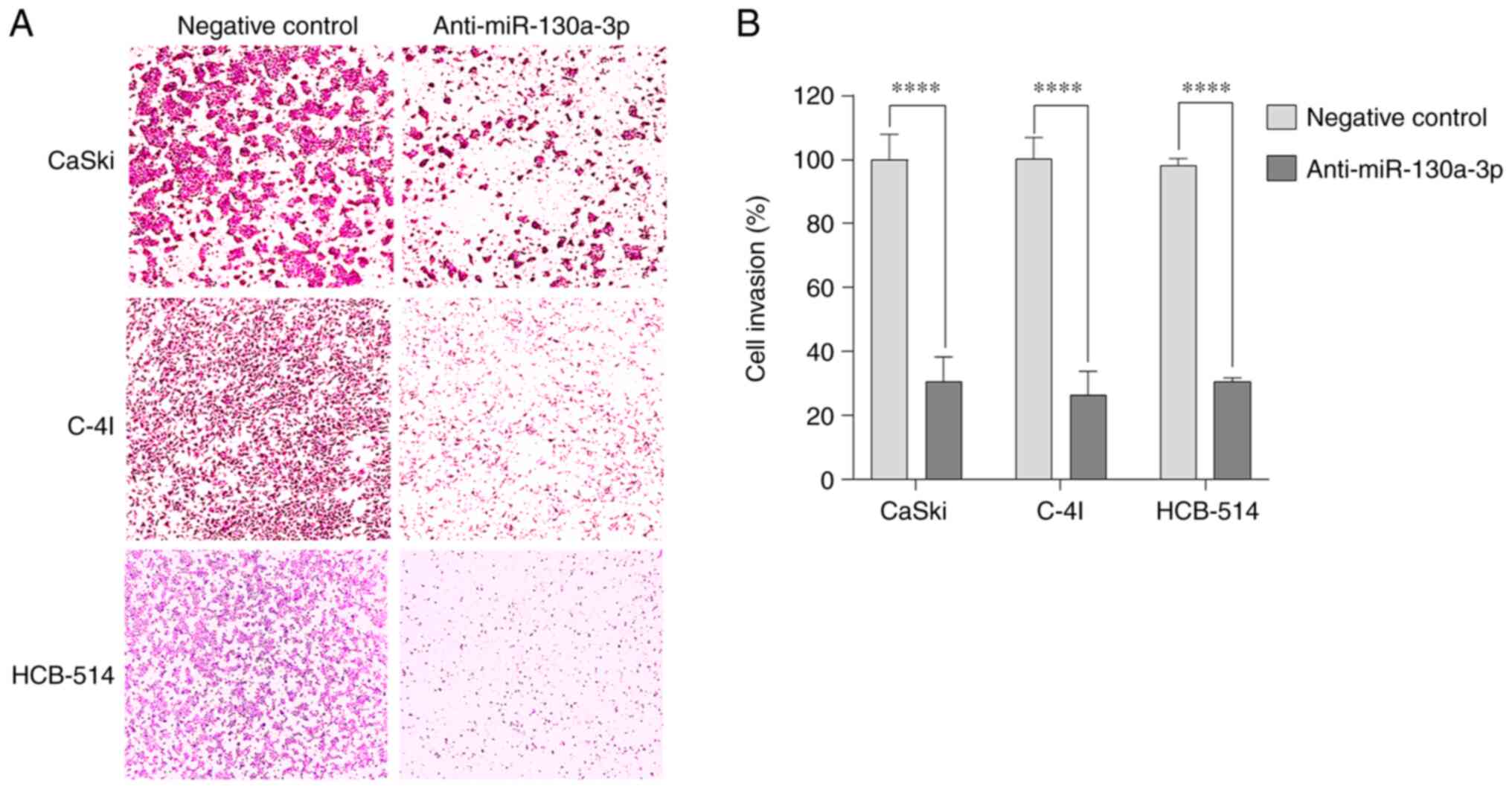
Anti-miR-130a-3p inhibits cervical
cancer cell invasion
The present study also examined the in vitro
invasiveness of CaSki, C-4I and HCB-514 cells transfected with
anti-miR-130a-3p or negative control using Matrigel invasion assay.
Cell invasion was measured by the number of cells that migrated
through the Matrigel insert. Anti-miR-130a-3p inhibited the
invasion of the CaSki, C-4I and HCB-514 cells by 30.7%
(P<0.0001), 26.3% (P<0.0001) and 30.7% (P<0.0001),
respectively, compared with that of the negative control cells
(Fig. 5). These data suggested that
miR-130a-3p contributed to the invasiveness of cervical cancer
cells.
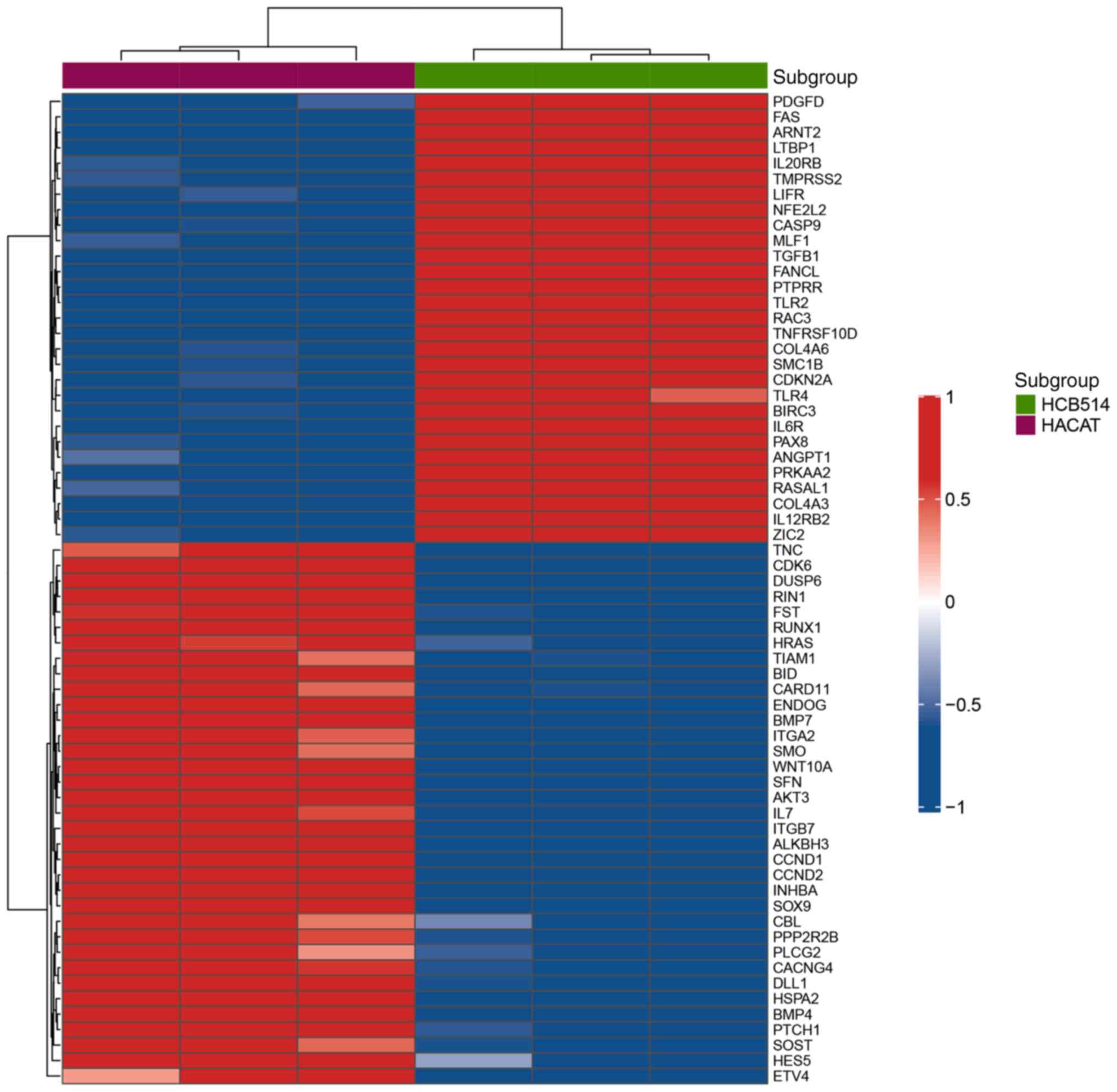
Gene expression profile in HCB-514
cells
Gene expression analysis using nCounter®
PanCancer Pathways revealed a different gene expression profile
between HCB-514 cells and the HaCaT control cell line (FC≥2 and
P<0.01; Fig. 6). The
differential genes expressed were then filtered using a P-value
corrected by FDR <0.01. In total, 22 differentially expressed
genes were identified. Among these 22 genes, 15 genes (CCND2,
FST, INHBA, BMP7, BMP4, CACNG4, SOX9, TNC, DUSP6, WNT10A, DLL1,
ITGB7, CARD11, CCND1 and CDK6) were significantly
downregulated in the HCB-514 cells compared with the HaCaT cell
line, while seven genes (IL12RB2, IL6R, ANGPT1, PTPRR, TLR2,
PAX8 and COL4A3) were upregulated (Table I).
 | Table I.Genes differentially expressed in
HCB-514 cells compared to the HaCaT control cell line using a
NanoString Technologies analysis system. |
Table I.
Genes differentially expressed in
HCB-514 cells compared to the HaCaT control cell line using a
NanoString Technologies analysis system.
| Gene | Fold change | Status | P-value | FDR P-value |
|---|
| CCND2 | −8.0 | Downregulated | 0.0001 | 0.0089 |
| FST | −6.5 | Downregulated | 0.0019 | 0.0350 |
| INHBA | −5.7 | Downregulated | 0.0020 | 0.0350 |
| BMP7 | −5.3 | Downregulated | 0.0004 | 0.0200 |
| BMP4 | −5.0 | Downregulated | 0.0023 | 0.0350 |
| CACNG4 | −4.4 | Downregulated | 0.0022 | 0.0350 |
| SOX9 | −4.4 | Downregulated | <0.0001 | 0.0041 |
| TNC | −4.2 | Downregulated | 0.0038 | 0.0490 |
| DUSP6 | −4.0 | Downregulated | 0.0016 | 0.0330 |
| WNT10A | −4.0 | Downregulated | 0.0022 | 0.0350 |
| DLL1 | −3.2 | Downregulated | 0.0012 | 0.0330 |
| ITGB7 | −2.2 | Downregulated | 0.0006 | 0.0200 |
| CARD11 | −2.1 | Downregulated | 0.0036 | 0.0480 |
| CCND1 | −2.0 | Downregulated | <0.0001 | 0.0041 |
| CDK6 | −2.0 | Downregulated | 0.0002 | 0.0130 |
| IL12RB2 | 2.2 | Upregulated | 0.0006 | 0.0200 |
| IL6R | 2.3 | Upregulated | 0.0006 | 0.0220 |
| ANGPT1 | 3.2 | Upregulated | 0.0036 | 0.0480 |
| PTPRR | 4.1 | Upregulated | <0.0001 | 0.0041 |
| TLR2 | 4.3 | Upregulated | 0.0002 | 0.0120 |
| PAX8 | 4.8 | Upregulated | 0.0014 | 0.0330 |
| COL4A3 | 5.2 | Upregulated | 0.0013 | 0.0330 |
In silico analysis demonstrates that
DLL1 and WNT10A are a decoy of miR-130a-3p to CESC progression
mirDIP v.5.1.16.1 predicted that, among the 22
differentially expressed genes identified usin nCounter®
technology (Table I), DLL1
and WNT10A could be targeted by miR-130a-3p (Table II). To establish whether the
DLL1 and WNT10a genes could affect cervical cancer
progression, TargetScan 7.1 was initially used to confirm an
interaction site of DLL1 or WNT10A with miR-130a-3p.
As illustrated in Fig. 7A, a
potential binding site for miR-130a-3p was found in the 3′UTR
regions of both mRNAs. The GEPIA2 database was then used to examine
the expression of DLL1 and WNT10A genes. It was found
that the DLL1 gene was notably expressed at low levels in
CESC tissues compared with normal tissues (Fig. 7B). However, the WNT10A gene
was upregulated in CESC tissues compared with normal tissues
(Fig. 7C). In addition, using the
GEPIA 2 database, the expression of the DLL1 and
WNT10A genes in patients with different stages of CESC was
evaluated, although no statistically significant differences
between the groups were observed; however, a trend towards a
decrease in expression was observed for the DLL1 and
WNT10A genes in the advanced stages of CESC (Fig. 7D and E).
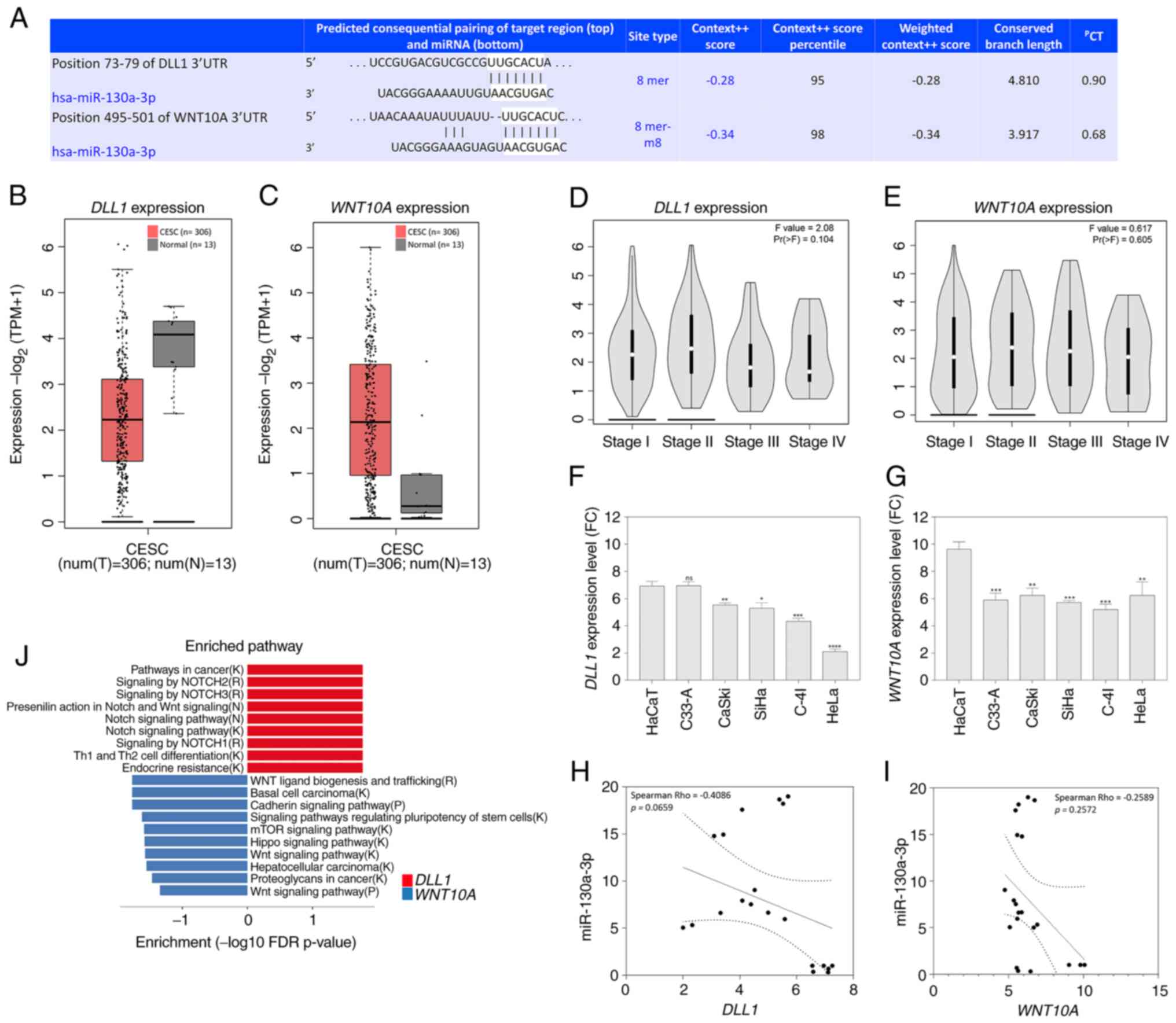 | Figure 7.In silico analysis of
DLL1 and WNT10A, and their association with the
progression of patients with CESC and with cervical cancer cell
lines. (A) Putative miR-130a-3p binding site in the 3′untranslated
regions of DLL1 and WNT10A, as predicted using
TargetScan. (B,C) Expression boxplots of the genes using the GEPIA2
database (P<0.05; CESC tissues, n=306; normal tissues, n=13) (B)
DLL1 expression; (C) WNT10A expression. (D)
Association between DLL1 expression, and pathological stage
in patients with CESC using the GEPIA2 database (P<0.05). (E)
Association between WNT10A expression, and pathological
stage in patients with CESC using the GEPIA2 database (P<0.05).
(F) Data from the GENT2 database regarding DLL1 expression
in a cervical cancer cell line panel in comparison with HaCaT
cells. ns, not significant. *P<0.05, **P<0.01, ***P<0.001
and ****P<0.0001. (G) Data from the GENT2 database regarding
WNT10A expression in a cervical cancer cell line panel in
comparison with HaCaT cells. **P<0.01, ***P<0.001. (H,I)
Spearman's pair-wise correlation analysis of miRNA-mRNA target
pairs in cervical cancer cell lines paired in triplicate (n=5). (H)
Correlation analysis revealed that increased miR-130a-p expression
levels were associated with DLL1 downregulation in cervical
cancer cell lines. In the correlation plots, the solid line
represents the least square estimate, and P-values represent global
significance P<0.05; (I) correlation analysis revealed that
increased miR-130a-p expression levels were associated with
WNT10A downregulation in cervical cancer cell lines. In the
correlation plots, the solid line represents the least square
estimate, and P-values represent global significance P<0.05. (J)
Enrichment pathways using the DLL1 (right panel, red) and
WNT10A (left panel, blue) genes in CESC. Significantly
enriched pathways terms are shown in -log10 with Benjamini-Hochberg
false discovery rate-corrected P-values. The letter in parentheses
after each pathway gene set name corresponds to the source of the
pathway annotations. B, BioCarta; K Kyoto Encyclopedia of Genes and
Genomes; R, Reactome; P, Panther; CESC, cervical squamous cell
carcinoma and endocervical adenocarcinoma; miR, microRNA; GEPIA,
Gene Expression Profiling Interactive Analysis; DLL1, δ-like
Notch1 ligand. |
 | Table II.Bidirectional target prediction for
differentially expressed genes and miR-130a-3p using the mirDIP
tool. |
Table II.
Bidirectional target prediction for
differentially expressed genes and miR-130a-3p using the mirDIP
tool.
| Gene | Uniprot | Rank | Source | Confidence
score |
|---|
| DLL1 | O00548 | 0.6302500 |
miranda_May_2021 | 0.0210342 |
| DLL1 | O00548 | 0.8863533 | mirbase | 0.0417327 |
| DLL1 | O00548 | 0.0976322 | mirzag | 0.0881644 |
| DLL1 | O00548 | 0.2058031 | miRDB_v6 | 0.0930805 |
| DLL1 | O00548 | 0.8913646 | miRTar2GO | 0.1264211 |
| DLL1 | O00548 | 0.3556803 | MBStar | 0.0339947 |
| DLL1 | O00548 | 0.0055481 | MirAncesTar | 0.1070419 |
| DLL1 | O00548 | 0.7009731 | MiRNATIP | 0.0196684 |
| DLL1 | O00548 | 0.4255360 | MultiMiTar | 0.1111873 |
| DLL1 | O00548 | 0.7303563 | RNA22 | 0.0136887 |
| DLL1 | O00548 | 0.2063430 |
TargetScan_v7_2 | 0.3306254 |
| WNT10A | Q9GZT5 | 0.4518441 |
miranda_May_2021 | 0.0254858 |
| WNT10A | Q9GZT5 | 0.6573457 | mirbase | 0.0465710 |
| WNT10A | Q9GZT5 | 0.3362469 | mirzag | 0.0564491 |
| WNT10A | Q9GZT5 | 0.9904704 | miRDB_v6 | 0.0602461 |
| WNT10A | Q9GZT5 | 0.4086939 | miRTar2GO | 0.1266619 |
| WNT10A | Q9GZT5 | 0.0046046 | MirAncesTar | 0.1103192 |
| WNT10A | Q9GZT5 | 0.4074708 | MultiMiTar | 0.1126795 |
| WNT10A | Q9GZT5 | 0.1128482 |
TargetScan_v7_2 | 0.3892957 |
The present study also evaluated the DLL1 and
WNT10A expression levels in cervical cancer cell lines using
data on gene expression available in the GENT2 platform (http://gent2.appex.kr/gent2/). Significantly low
levels of DLL1 expression were observed in the CaSki, SiHa,
C-4I and HeLa cells in comparison with those in the HaCaT cells
(Fig. 7F). Similar results were
found for the WNT10A gene, since all the cervical cell lines
exhibited a decrease in WNT10A expression compared with the
HaCaT cells (Fig. 7G). Although
Spearman's correlation analyses did not reveal any significant
differences, there was a tendency for a negative correlation
between the levels of miR-130a-3p and the DLL1 gene
(Spearman's Rho=−0.4086, P=0.0659; Fig.
7H), and the WNT10A gene (Spearman's Rho=−0.2589, P=
0.0.2572; Fig. 7I) in a cervical
cancer cell line panel.
Subsequently, enrichment analysis was performed
using Cytoscape software with the Reactome plugin of the
DLL1 and WNT10A genes to gain insights into the
pathways that are affected in CESC progression (Fig. 7J and Table SVI). It was found that the
downregulated DLL1 gene in CESC and cervical cancer cell
lines was significantly enriched in the pathway of proteolysis and
signaling pathway of Notch, pathways in cancer, signaling by Notch2
and 3, presenilin action in Notch and Wnt signaling, Notch
signaling pathway, signaling by NOTCH1, T helper (Th)1 and Th2 cell
differentiation, and endocrine resistance. The WNT10A gene
was significantly enriched in pathways in cancer, Wnt ligand
biogenesis and trafficking, basal cell carcinoma, cadherin
signaling pathway, signaling pathways regulating pluripotency of
stem cells, mTOR signaling pathway, Hippo signaling pathway, Wnt
signaling pathway, hepatocellular carcinoma, proteoglycans in
cancer, and Wnt signaling pathway (Fig.
7J and Table SVI).
To validate whether DLL1 and WNT10A
expression was also altered due to miR-130a-3p inhibition, the
expression of DLL1 and WNT10A was evaluated in the HCB-514 cells
following miR-130a-3p inhibition. It was observed that both for the
DLL1 gene (FC=3.03, P=0.0089) and for the WNT10A gene
(FC=4.22, P<0.001), there was a significant increase in
expression when the cells were subjected to the inhibition of
miR-130a-3p in comparison to their respective negative controls
(Fig. S2).
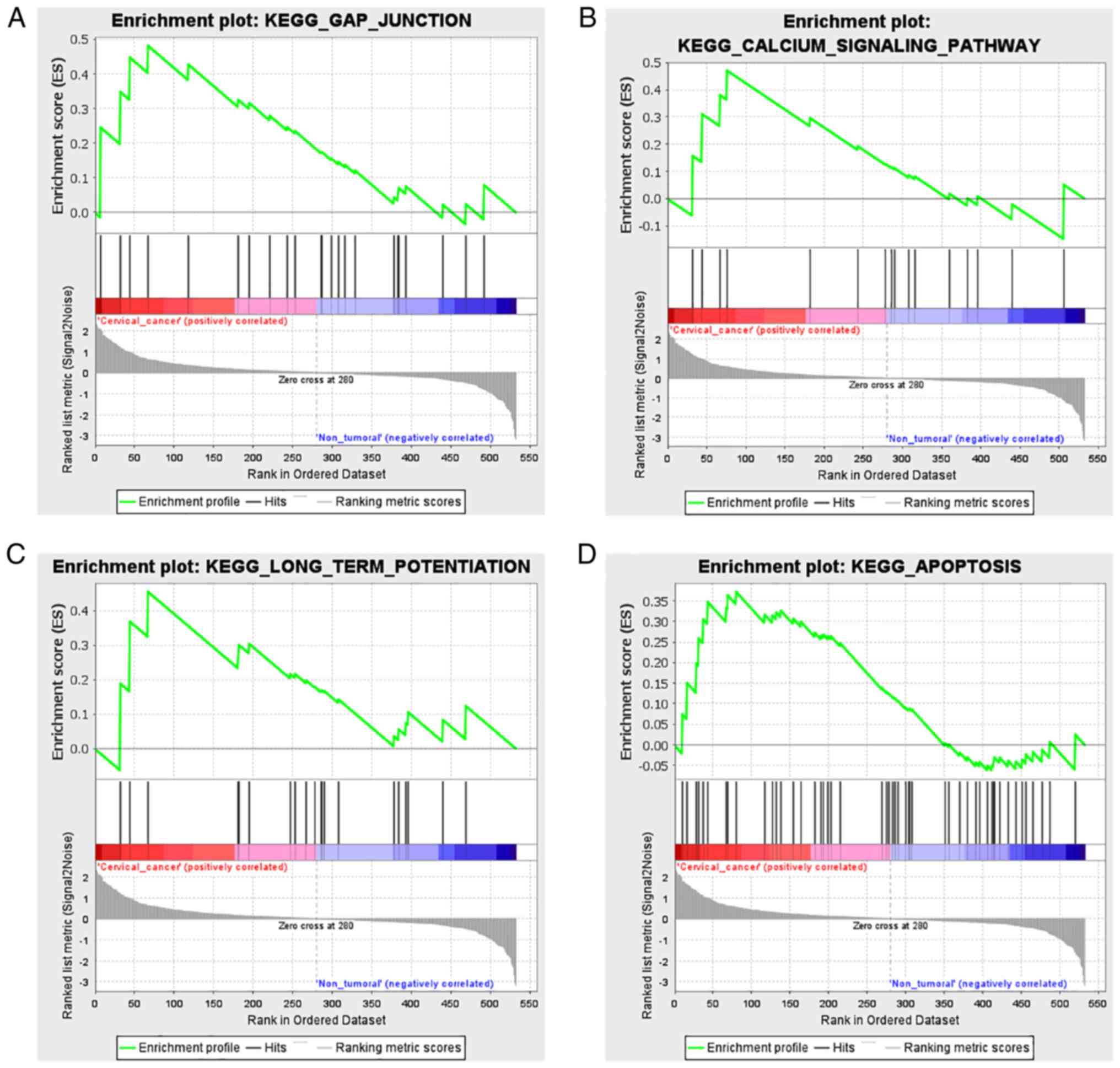
GSEA
The difference in gene expression levels between the
HCB-514 and HaCaT cells was further investigated using GSEA. GSEA
was performed using a KEGG-based list, including 186 gene sets. In
total, 64 KEGG-based gene sets, were identified as significantly
enriched (FDR <0.05; Table
III). NES indicates expression values in HCB-514 vs. HaCaT
cells. Of note, from the KEGG-based list, all gene sets which
demonstrated a high expression in the HCB-514 cells were mainly
related to ‘gap junction’ (Fig.
8A), ‘calcium signaling pathway’ (Fig. 8B), ‘long-term potentiation’
(Fig. 8C) and ‘apoptosis’ (Fig. 8D).
 | Table III.Gene set enrichment analysis between
the HCB-514 and HaCaT cells. |
Table III.
Gene set enrichment analysis between
the HCB-514 and HaCaT cells.
| Gene set | NES | NOM P-value | FDR P-value | Genes |
|---|
| Gap junction | 0.0481 | <0.001 | 0.042 | PDGFD, PLCB1,
PRKCA, PRKX, SOS1, NRAS, MAP2K1, SOS2, GNAS, MAPK3, PRKACB, RAF1,
GRB2, PRKACA, EGFR, PDGFC, MAP2K2, GNA11, KRAS, MAPK1, GNAQ,
HRAS, and PDGFA |
| Calcium signaling
pathway | 0.0470 | <0.001 | 0.036 | PLCB1, PRKCA,
PRKX, PLCE1, PPP3CA, GNAS, PPP3R1, PRKACB, PPP3CC, PRKACA, EGFR,
ERBB2, GNA11, PPP3CB, GNAQ, and PLCG2 |
| Long-term
potentiation | 0.0455 | <0.001 | 0.033 | PLCB1, PRKCA,
PRKX, NRAS, PPP3CA, MAP2K1, EP300, MAPK3, BRAF, PPP3R1, PRKACB,
RAF1, PPP3CC, PRKACA, MAP2K2, KRAS, MAPK1, PPP3CB, GNAQ, and
HRAS |
| Apoptosis | 0.0372 | <0.001 | 0.054 | IRAK3, BIRC3,
IRAK2, CASP9, IL1A, TNFRSF10, PRKX, FAZ, TNFSF10, TNFRSF10, PIK3CA,
IL1R1, AKT2, NFKBIA, PIK3CD, PPP3CA, PIK3R1, BAX, NFKB1, CASP7,
IL1B, CHUK, IL1RAP, PPP3R1, BCL2L1, PRKACB, PPP3CC, CAPN2, PIK3R3,
PIK3CB, PRKACA, IKBKB, CASP3, AKT1, PRKAR2A, RELA, PPP3CB, IKBKG,
TNFRSF10, BAD, TP53, CASP8, PIK3R2, MAP3K14, MYD88, ATM, ENDOG,
PRKAR1B, BID, and AKT3 |
Discussion
The aberrant expression of miR-130a-3p in different
tumor types suggests that this miRNA may play distinct roles in
tumorigenesis, depending on the cell type involved. The role of
miR-130a-3p in tumor development and progression appears to be
contradictory, as it may serve as an oncogene or a tumor suppressor
gene, regulating various canonical signaling pathways or target
genes (43). Previous research has
demonstrated that miR-130a-3p is upregulated in CIN3 (20). In addition, studies performing the
assessment of miR-130a-3p expression in cervical cancer tissue
samples identified an overexpression of this miRNA in comparison
with that in normal samples (22,43,44).
The findings of the present study demonstrated that miR-130a-3p was
highly expressed in HPV-positive cervical cancer cell lines and
played a regulatory role. Thus, the present study evaluated the
effects of miR-130a-3p inhibition on CaSki, C-4I and HCB-514 cells.
These cell lines were selected as they exhibited a significantly
higher expression of miR-130a-3p at the basal level than others.
Furthermore, the C-33A cell line exhibited a low level of
miR-130a-3p expression. Although previous studies reporting the low
expression of miR-130a-3p in C-33A cells are limited, it may be
hypothesized that the high expression of miR-130a-3p may be
associated with the presence of HPV, since C-33A is a negative-HPV
cell line. Therefore, further studies are warranted to confirm this
hypothesis.
The transient transfection of anti-miR-130a-3p
suppressed adhesion-independent cell proliferation, migration and
invasion in cervical cancer cells by decreasing the expression of
miR-130a-3p. In agreement with the findings of a previous study,
decreasing the expression of miR-130a-3p also inhibited cell
proliferation. Furthermore, that study demonstrated that the
inhibition of miR-130a-3p was responsible for the radiosensitivity
of non-small cell lung cancer (45), while another study demonstrated that
miR-130a-3p aggravated gastric cancer cell proliferation, migration
and invasion by reducing GCNT4 expression and activating the
TGF-β1/SMAD3 signaling pathway (46).
The findings of the present study support the
results identified in other studies on the contribution of
miR-130a-3p to cervical tumor progression. Fan et al
(23) observed that miR-130a 3p
expression levels were higher in cervical tumor tissues compared
with those in healthy tissues. Moreover, that study verified that
miR-130a-3p knockdown inhibited cervical cell proliferation and
invasion in vitro and in vivo compared with the
negative control group.
Other studies have demonstrated that miR-130-3p
expression levels can affect the therapeutic response. Huang et
al (47) performed a study on
50 patients with breast cancer before and after epirubicin-based
neoadjuvant chemotherapy. They revealed that the increased
expression of miR-130a enhanced the sensitivity of breast cancer
cells to doxorubicin, and that the inhibition of miR-130a
expression may be associated with resistance to epirubicin-based
chemotherapy in breast cancer tissues. It has been reported that
miR-130a-3p regulates SMAD4 expression in hepatocellular
carcinoma cells, and this regulatory axis plays a critical role in
gemcitabine-mediated sensitivity, in which the overexpression of
miR-130a-3p leads to an increase in the sensitivity to gemcitabine
of these cells (48). Therefore,
the expression of miR-130a-3p is also associated with the treatment
administered.
Furthermore, the present study investigated the role
that miR-130a-3p plays in cervical cancer progression. The results
of colony formation assay revealed that miR-130a-3p inhibition
significantly reduced the proliferation of CaSki and C-4I cells.
Wound healing and Transwell assays were performed to investigate
the mechanisms through which anti-miR-130a-3p affects the migration
of cervical cancer cells, and it was found that anti-miR-130a-3p
inhibited cell migration. The results from the Transwell invasion
assay demonstrated that anti-miR-130a-3p reduced the invasion of
cervical cancer cells. The aforementioned findings are in
accordance with those of previous studies reporting that
miR-130a-3p may function as a tumor promoter (22,45,46).
Bioinformatics analysis predicted that DLL1
and WNT10A may be one of the targets of miR-130a-3p. This
observation was supported by in silico analyses that
demonstrated the reduced expression of DLL1 in patients with
CESC and in a panel of cervical cancer cell lines. However, there
was no reduction in expression in patients with CESC, although
analyses using online databases revealed a significant reduction in
WNT10A expression in cervical cancer cells. Previous
findings have demonstrated that both Notch1 mutation and inhibition
induced by HPV E6 in cervical cancer can drive neoplastic
development in stratified squamous epithelia. However, the
contribution of Notch1 and its d-like ligands to the susceptibility
and progression of cervical cancer remains poorly understood
(19). In comparison with the
results of the present study, Liu et al (48) identified that miR-130b inhibited the
upstream gene, leading to a decrease in the expression of
Notch-DLL1, which enhanced the invasion and metastasis of liver
cancer cells. Taken together, these results suggest that
DLL1 may be the target gene for miR-130a-3p, which is
involved in the proliferation, migration and invasion of cervical
cancer cells in vitro.
By contrast, herein, in silico analysis
revealed that, although WNT10A could be a target of
miR-130a-3p, the WNT10A gene exhibited increased expression
in patients with CESC, and was expressed at low levels in cervical
cancer cells, in which cervical cancer cells demonstrated the
overexpression of miR-130a-3p. In addition, the present study
validated the differential expression of DLL1 and
WNT10A between the HCB-514 cells with miR-130a-3p inhibition
in comparison with negative control A. This result revealed that
the modulation of miR-130a-3p expression was capable of causing a
change in the expression of both evaluated genes.
Thus, these results suggest that the WNT10A
gene may not be the main target of miR-130a-3p. It has been
demonstrated that miRNAs can regulate the expression of the
WNT10A gene. Yu et al (49) reported that the WNT10A gene,
a member of the Wnt/β-catenin signaling pathway, was confirmed to
be a target of miR-378a-3p. It has been observed that the
inhibition of miR-378a-3p expression contributes to the activation
of hepatic stellate cells, which led to increased cell
proliferation, and the expression of α-smooth muscle actin and
collagen (49). Moreover, it has
been demonstrated that miR-361-3p mediates tumor inhibition in
lymphoma, and this regulation can be ascribed to the upregulation
of WNT10A that was identified in lymphoma cell lines and
normal lymphocytes (50).
Despite the novelty of the current findings, the
present study has several limitations. First, cell proliferation
was not evaluated in the HCB-514 cell line. Therefore, it should be
emphasized that this analysis should be performed in the HCB-514
cell line. Although the present study evaluated the expression of
DLL1 and WNT10A using in silico analysis, and also in the
HCB-514 cell line with miR-13a-3p expression modulation, additional
experiments are required to validate the possible regulatory
targets of miR-130a-3p. Therefore, it would be beneficial to
determine whether DLL1 and WNT10A expression are also altered in
the CaSki and C-4I cell lines following miR-13a-3p inhibition. In
this sense, another limitation of this study includes the absence
of western blot analyses in order to validate protein expression
involved in the signaling pathways identified from the pathway
enrichment analysis, and also the GSEA analysis. The current
analyses were performed in vitro; thus, future in
vivo experiments are also warranted to further support the
present conclusions. However, bearing in mind that the present
study was a hypothesis-generating study, it is considered that the
data identified herein are sufficiently robust and can be evaluated
in a more complete manner in a future study.
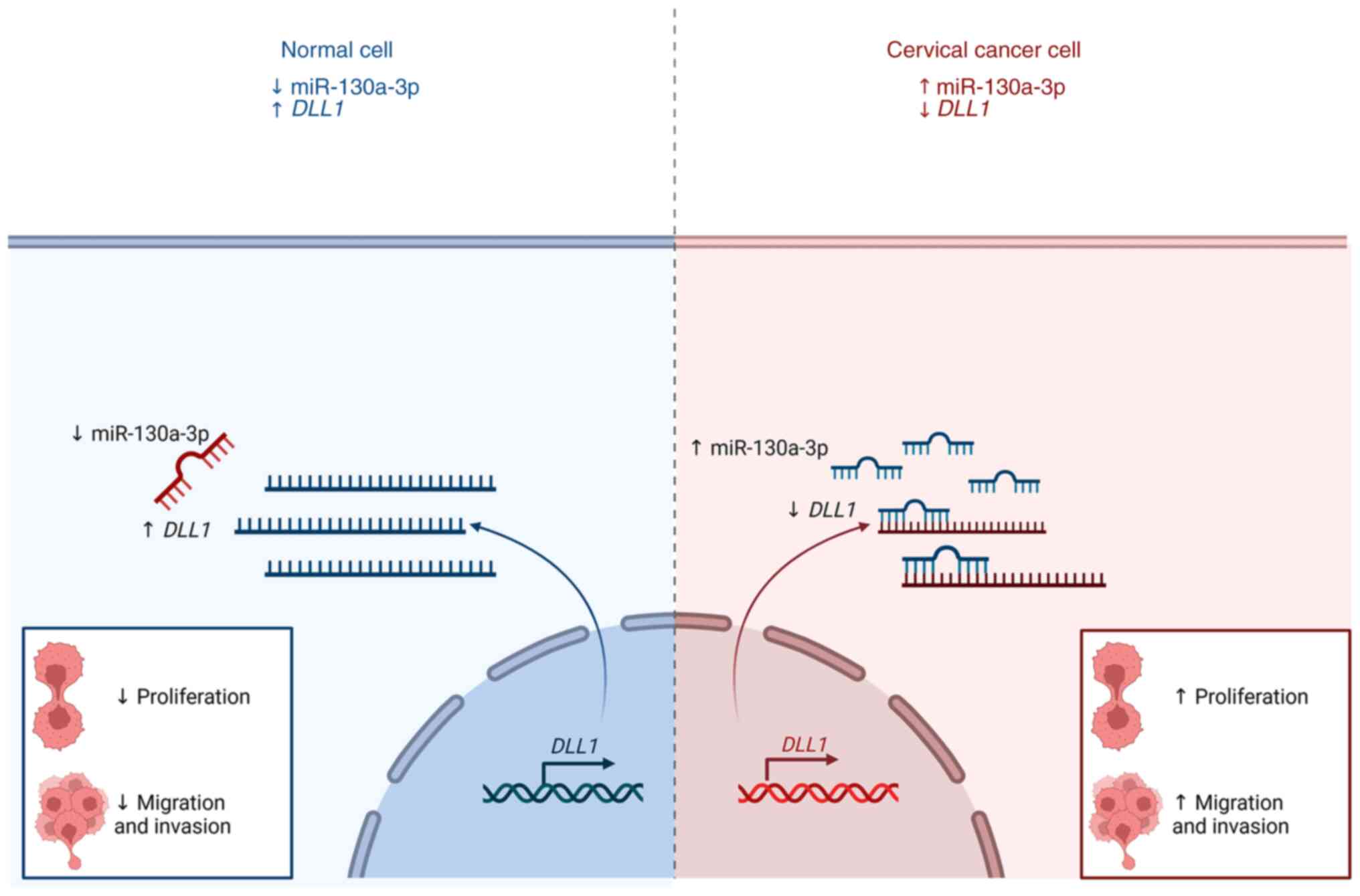
In conclusion, the present study demonstrated that
miR-130a-3p was highly expressed in cervical cancer cells. Cervical
cancer adhesion-independent cell proliferation, migration and
invasion may be suppressed by the inhibition of miR-130a-3p
expression (Fig. 9). Thus, the
results presented herein may provide novel insight into the role of
miR-130a-3p in cervical cancer progression and its potential value
for clinical prognosis.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors are grateful to the researchers, Dr
Laura Sicherond Dr Luisa Lina Villa (Center for Translational
Investigation in Oncology, Instituto do Cancer do Estado de Sao
Paulo, Hospital das Clinicas da Faculdade de Medicina da
Universidade de São Paulo, São Paulo, Brazil), for kindly providing
the HaCaT cell line for the initial testing in the present
study.
Funding
The present study was funded by FAPESP (grant no. 2016/15831-3),
the Research Incentive Program of Barretos Cancer Hospital (PAIP)
and PRONON. The present study was also funded by the São Paulo
Research Foundation (FAPESP; grant no. 2018/19476-9) and this
research was funded by the Brazilian Ministry of Health supported
by PRONON/MS (NUP-25000.023997.2018/34) entitled: ‘Identificação de
biomarcadores para screening e detecção precoce de tumores no
contexto do Sistema Único de Saúde (SUS).’ RMR and MMCM are
recipients of a CNPq Productivity fellowship.
Availability of data and materials
Data regarding gene expression analysis are
available on the National Center for Biotechnology Information
(NCBI)-Gene Expression Omnibus (GEO) platform (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) under
the Accession no. GSE224301. The other datasets used and/or
analyzed during the current study are available from the
corresponding author on reasonable request.
Authors' contributions
RLC developed and led the overall study, conducted
the data reviews and the analysis, and was involved in the
preparation of the draft. AVHL participated in the design of the
study, in functional assays and in developing the study, as well as
in the critical reviewing of the manuscript. INFG participated in
the conception and design of the study, as well as in the critical
reviewing of the manuscript. AJADF participated in the functional
assays, and in the critical reviewing of the manuscript. MNR
participated in the conception and design of the study, as well as
in the critical reviewing of the manuscript. RDR participated in
designing and developing the study, and in the critical reading of
the manuscript. RMR participated in designing and developing the
study, as well as in funding, and the critical reading of the
manuscript. MMCM conceived the study, provided advice during the
study's development, and participated in the preparation the
manuscript. RLC and MMCM confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have not competing
interests.
Glossary
Abbreviations
Abbreviations:
|
UTR
|
untranslated region
|
|
CESC
|
cervical squamous cell carcinoma and
endocervical adenocarcinoma
|
|
CIN3
|
cervical intraepithelial neoplasia
grade 3
|
|
DLL1
|
delta-like notch 1 ligand
|
|
FDR
|
false discovery rate
|
|
HPV
|
human papillomavirus
|
|
LACC
|
locally advanced cervical cancer
|
|
mirDIP
|
microRNA Data Integration Portal
|
|
miRNA/miR
|
microRNA
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
TCGA
|
The Cancer Genome Atlas
|
|
WNT10A
|
Wnt family member 10a
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Espenel S, Garcia MA, Trone JC, Guillaume
E, Harris A, Rehailia-Blanchard A, He MY, Ouni S, Vallard A,
Rancoule C, et al: From IB2 to IIIB locally advanced cervical
cancers: report of a ten-year experience. Radiat Oncol. 13:162018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Robin TP, Amini A, Schefter TE, Behbakht K
and Fisher CM: Disparities in standard of care treatment and
associated survival decrement in patients with locally advanced
cervical cancer. Gynecol Oncol. 143:319–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koh WJ, Abu-Rustum NR, Bean S, Bradley K,
Campos SM, Cho KR, Chon HS, Chu C, Clark R, Cohn D, et al: Cervical
cancer, Version 3.2019, NCCN clinical practice guidelines in
oncology. J Natl Compr Canc Netw. 17:64–84. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He Y, Han SB, Liu Y, Zhang JJ and Wu YM:
Role of APOA1 in the resistance to platinum-based chemotherapy in
squamous cervical cancer. BMC Cancer. 22:4112022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
International Agency for Research on
Cancer I, organizador, . IARC monographs on the evaluation of
carcinogenic risks to humans. 90. Human papillomaviruses: This
publication represents the views and expert opinions of an IARC
Working Group on the Evaluation of Carcinogenic Risks to Humans
which met in Lyon, 15–22 February 2005. IARC; Lyon: pp.
pp6702007
|
|
7
|
Da Silva MLR, De Albuquerque BHDR, Allyrio
TADMF, De Almeida VD, Cobucci RNDO, Bezerra FL, Andrade VS, Lanza
DCF, De Azevedo JCV, De Araújo JMG and Fernandes JV: The role of
HPV-induced epigenetic changes in cervical carcinogenesis (Review).
Biomed Rep. 15:602021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanahan D: Hallmarks of Cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J and Chen L: The role of miRNAs in
the invasion and metastasis of cervical cancer. Biosci Rep.
39:BSR201813772019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao C, Zhou C, Zhuang J, Liu L, Liu C, Li
H, Liu G, Wei J and Sun C: MicroRNA expression in cervical cancer:
Novel diagnostic and prognostic biomarkers. J Cell Biochem.
119:7080–7090. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siomi H and Siomi MC: Posttranscriptional
regulation of microRNA biogenesis in animals. Mol Cell. 38:323–332.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ardekani AM and Naeini MM: The Role of
MicroRNAs in human diseases. Avicenna J Med Biotechnol. 2:161–179.
2010.PubMed/NCBI
|
|
13
|
Causin RL, de Freitas AJA, Trovo Hidalgo
Filho CM, dos Reis R, Reis RM and Marques MMC: A systematic review
of MicroRNAs involved in cervical cancer progression. Cells.
10:6682021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kovall RA, Gebelein B, Sprinzak D and
Kopan R: The canonical notch signaling pathway: Structural and
biochemical insights into shape, sugar, and force. Dev Cell.
41:228–241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bray SJ: Notch signalling in context. Nat
Rev Mol Cell Biol. 17:722–735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Penton AL, Leonard LD and Spinner NB:
Notch signaling in human development and disease. Semin Cell Dev
Biol. 23:450–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Capaccione KM and Pine SR: The Notch
signaling pathway as a mediator of tumor survival. Carcinogenesis.
34:1420–1430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Briot A and Iruela-Arispe ML: Blockade of
specific NOTCH ligands: A new promising approach in cancer therapy.
Cancer Discov. 5:112–114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khelil M, Griffin H, Bleeker MCG,
Steenbergen RDM, Zheng K, Saunders-Wood T, Samuels S, Rotman J, Vos
W, van den Akker BE, et al: Delta-like ligand-notch1 signaling is
selectively modulated by HPV16 E6 to promote squamous cell
proliferation and correlates with cervical cancer prognosis. Cancer
Res. 81:1909–1921. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Causin RL, da Silva LS, Evangelista AF,
Leal LF, Souza KCB, Pessôa-Pereira D, Matsushita GM, Reis RM,
Fregnani JHTG and Marques MMC: MicroRNA biomarkers of high-grade
cervical intraepithelial neoplasia in liquid biopsy. Biomed Res
Int. 2021:66509662021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He L, Wang HY, Zhang L, Huang L, Li JD,
Xiong Y, Zhang MY, Jia WH, Yun JP, Luo RZ and Zheng M: Prognostic
significance of low DICER expression regulated by miR-130a in
cervical cancer. Cell Death Dis. 5:e1205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan Q, Huang T, Sun X, Yang X, Wang J, Liu
Y, Ni T, Gu S, Li Y and Wang Y: miR-130a-3p promotes cell
proliferation and invasion by targeting estrogen receptor α and
androgen receptor in cervical cancer. Exp Ther Med. 21:4142021.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosa MN, Evangelista AF, Leal LF, De
Oliveira CM, Silva VAO, Munari CC, Munari FF, Matsushita GM, Dos
Reis R, Andrade CE, et al: Establishment, molecular and biological
characterization of HCB-514: A novel human cervical cancer cell
line. Sci Rep. 9:19132019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Silva-Oliveira RJ, Silva VAO, Martinho O,
Cruvinel-Carloni A, Melendez ME, Rosa MN, de Paula FE, de Souza
Viana L, Carvalho AL and Reis RM: Cytotoxicity of allitinib, an
irreversible anti-EGFR agent, in a large panel of human
cancer-derived cell lines: KRAS mutation status as a predictive
biomarker. Cell Oncol (Dordr). 39:253–263. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
miRCURY LNA miRNA Inhibitors and Power
Inhibitors. [Citado 9 de fevereiro de 2023]. Disponível em.
https://www.qiagen.com/us/products/discovery-and-translational-research/functional-and-cell-analysis/mirna-functional-anal-sis/mircury-lna-mirna-inhibitors/mircury-lna-mirna-inhibitors?catno=339126
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Causin RL, Pessôa-Pereira D, Souza KCB,
Evangelista AF, Reis RMV, Fregnani JHTG and Marques MMC:
Identification and performance evaluation of housekeeping genes for
microRNA expression normalization by reverse
transcription-quantitative PCR using liquid-based cervical cytology
samples. Oncol Lett. 18:4753–4761. 2019.PubMed/NCBI
|
|
29
|
Geissmann Q: OpenCFU, a new free and
open-source software to count cell colonies and other circular
objects. PLoS One. 8:e540722013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu Y, Sun X, Mao C, Guo G, Ye S, Xu J, Zou
R, Chen J, Wang L, Duan P and Xue X: Upregulation of long noncoding
RNA TUG1 promotes cervical cancer cell proliferation and migration.
Cancer Med. 6:471–482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yew TL, Hung YT, Li HY, Chen HW, Chen LL,
Tsai KS, Chiou SH, Chao KC, Huang TF, Chen HL and Hung SC:
Enhancement of wound healing by human multipotent stromal cell
conditioned medium: The paracrine factors and p38 MAPK activation.
Cell Transplant. 20:693–706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Martinho O, Silva-Oliveira R,
Miranda-Gonçalves V, Clara C, Almeida JR, Carvalho AL, Barata JT
and Reis RM: In vitro and in vivo analysis of RTK inhibitor
efficacy and identification of its novel targets in glioblastomas.
Transl Oncol. 6:187–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Andrade DAP, da Silva LS, Laus AC, de
Lima MA, Berardinelli GN, da Silva VD, Matsushita GM, Bonatelli M,
da Silva ALV, Evangelista AF, et al: A 4-Gene signature associated
with recurrence in low- and intermediate-risk endometrial cancer.
Front Oncol. 11:7292192021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tokar T, Pastrello C, Rossos AEM, Abovsky
M, Hauschild AC, Tsay M, Lu R and Jurisica I: mirDIP
4.1-integrative database of human microRNA target predictions.
Nucleic Acids Res. 46:D360–D370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Park SJ, Yoon BH, Kim SK and Kim SY:
GENT2: An updated gene expression database for normal and tumor
tissues. BMC Medical Genomics. 12 (Suppl 5):S1012019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–1550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liberzon A, Subramanian A, Pinchback R,
Thorvaldsdóttir H, Tamayo P and Mesirov JP: Molecular signatures
database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
da Zhang H, Jiang LH, Sun DW, Li J and Ji
ZL: The role of miR-130a in cancer. Breast Cancer. 24:521–527.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang M, Wang X and Liu W: MicroRNA-130a-3p
promotes the proliferation and inhibits the apoptosis of cervical
cancer cells via negative regulation of RUNX3. Mol Med Rep.
22:2990–3000. 2020.PubMed/NCBI
|
|
45
|
Zhao X, Jin X, Zhang Q, Liu R, Luo H, Yang
Z, Geng Y, Feng S, Li C, Wang L, et al: Silencing of the lncRNA H19
enhances sensitivity to X-ray and carbon-ions through the
miR-130a-3p /WNK3 signaling axis in NSCLC cells. Cancer Cell Int.
21:6442021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hu W, Zheng X, Liu J, Zhang M, Liang Y and
Song M: MicroRNA MiR-130a-3p promotes gastric cancer by targeting
Glucosaminyl N-acetyl transferase 4 (GCNT4) to regulate the
TGF-β1/SMAD3 pathway. Bioengineered. 12:11634–1147. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang J, Zhao M, Hu H, Wang J, Ang L and
Zheng L: MicroRNA-130a reduces drug resistance in breast cancer.
Int J Clin Exp Pathol. 12:2699–2705. 2019.PubMed/NCBI
|
|
48
|
Liu Y, Li Y, Wang R, Qin S, Liu J, Su F,
Yang Y, Zhao F, Wang Z and Wu Q: MiR-130a-3p regulates cell
migration and invasion via inhibition of Smad4 in gemcitabine
resistant hepatoma cells. J Exp Clin Cancer Res. 35:192016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu F, Fan X, Chen B, Dong P and Zheng J:
Activation of hepatic stellate cells is inhibited by
microRNA-378a-3p via Wnt10a. Cell Physiol Biochem. 39:2409–2420.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou H, Tang H, Li N, Chen H, Chen X, Gu
L, Zhang L, Tian G and Tao D: MicroRNA-361-3p inhibit the
progression of lymphoma by the Wnt/β-catenin signaling pathway.
Cancer Manag Res. 12:12375–12384. 2020. View Article : Google Scholar : PubMed/NCBI
|