Introduction
Gastric cancer remains a serious threat to human
health, with its incidence and mortality ranking fifth and fourth
globally, respectively (1). Cancer
sustains its proliferative and metastatic capacities depending on
its surrounding microenvironment, which renders tumor stromal cells
attractive therapeutic targets for cancer (2). Previously, our research group
successfully obtained and identified mesenchymal stem cell-like
cells (MSCs) from primary gastric cancer tissues (GC-MSCs)
(3), and demonstrated that these
stromal cells directly promote gastric cancer cell proliferation,
migration, invasion and cancer stem cell properties (4–7). These
cells also create an immunosuppressive microenvironment for immune
escape by disrupting the Treg/Th17 balance, inducing M2-like
macrophage polarization, and impairing CD8+ T-cell
antitumor activity (8–10). GC-MSCs represent a promising
intervention target for the treatment of gastric cancer. The
exploration of GC-MSC-derived abnormal molecules may be crucial for
uncovering the underlying mechanisms of gastric cancer
progression.
Emerging evidence indicates that circular RNAs
(circRNAs) and metabolic reprogramming are two critical research
hotspots in the field of cancer. CircRNAs are non-coding RNAs that
are characterized by their covalently closed circular structure. To
date, a large number of circRNAs have been screened and identified
in gastric cancer cells, tissues and plasma through high-throughput
sequencing technologies and bioinformatics analyses. It is clear
that circRNAs play either oncogenic or tumor-suppressive roles, and
hold promise as diagnostic, prognostic biomarkers and therapeutic
targets for gastric cancer (11,12).
However, circRNA expression profiles, function and regulatory
mechanisms in gastric cancer-associated stromal cells remain
elusive. Metabolic reprogramming is one of the hallmarks of cancer.
Previous studies have suggested that metabolic alteration is not
unique to cancer cells, and immune cells undergo metabolic changes
to impair their antitumor immunity, thus forming an
immunosuppressive microenvironment to support gastric cancer
progression (13,14). Notably, a recent bioinformatics
analysis demonstrated that the metabolic reprogramming signature of
stromal cells could be explored to calculate prognosis risk scores,
which had good a performance in predicting the overall survival
(OS) of patients with gastric cancer (15). This finding highlights the critical
role of the metabolic reprogramming of stromal cells in gastric
cancer. However, the metabolic reprogramming of stromal cells and
the mechanisms responsible for the promotion of gastric cancer
metastasis remain unclear.
The present study investigated aberrant circRNAs and
metabolic disorders in GC-MSCs, explored the role and mechanisms of
circRNAs in modulating the metabolic reprogramming of GC-MSCs and
determined the clinical significance of the circRNA-regulatory axis
in gastric cancer. It is hoped that the findings presented herein
may help to uncover a novel mechanism and provide a potential
therapeutic strategy for gastric cancer from the perspective of the
circRNA mediated-metabolic reprogramming of GC-MSCs.
Materials and methods
Cell culture, conditioned medium (CM)
preparation and etomoxir treatment
Bone marrow-derived MSCs (BM-MSCs), GC-MSCs, and
gastric cancer cell lines AGS (cat. no. TCHu232, The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences) and
HGC-27 (cat. no. TCHu22, The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences) were obtained and cultured as
previously described (3,16). Briefly, fresh gastric cancer tissues
were washed with phosphate-buffered saline (PBS), cut into
1-mm3-sized pieces and floated in DMEM with low glucose
(LG-DMEM) (Invitrogen; Thermo Fisher Scientific, Inc.) containing
10% FBS, penicillin and streptomycin. The tissue pieces were
subsequently incubated at 37°C in humid air with 5% CO2,
and the medium was replaced every 3 days after the initial plating.
When adherent fibroblast-like cells appeared after 10 days of
culture, the cells were trypsinized and passaged into a new flask
for further expansion. When these cells reached confluence at the
third passage, a homogeneous cell population was obtained and used
for the subsequent experiments. The collection of tissues from
gastric cancer patients was approved by the Affiliated Tumor
Hospital of Nantong University (approval no. 2021-017). Informed
consent was obtained from all patients prior to surgery. MSC-CM was
prepared as previously specified (8). When the MSC confluency reached ~60%,
the cells were washed with PBS (Gibco; Thermo Fisher Scientific,
Inc.) and incubated with DMEM (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS (Biological Industries Israel Beit Haemek
Ltd.). After 48 h, the MSC-CM was harvested separately, centrifuged
at 300 × g for 5 min, at 4°C, and filtered through a 0.45-µm
membrane (cat. no. SLFH050, Merck KGaA). The final CM was stored at
−20°C. The MSCs and gastric cancer cells were attached overnight
and treated with etomoxir (cat. no. HY-50202, MedChemExpress) at
final concentrations of 100 and 40 µM for 24 h, respectively. The
MSCs were then subjected to transfection (as described below),
while the gastric cancer cells were further treated with MSC-CM for
subsequent analyses.
Clinical tissues
A total of 40 gastric cancer tissues and paired
adjacent non-cancerous tissues were provided by the Affiliated
Tumor Hospital of Nantong University from March, 2021 to October,
2022 (Nantong, China). All the obtained tissues were independently
diagnosed by two pathologists and stored at −80°C. None of the
patients had received any therapy prior to surgery. The collection
of tissues from gastric cancer patients was approved by the
Affiliated Tumor Hospital of Nantong University (approval no.
2021-017). Informed consent was obtained from all patients prior to
surgery.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from clinical tissues and
cell lines using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). Reverse transcription reactions were
performed using the PrimeScript™ RT reagent kit (Takara Bio, Inc.)
using random primers and oligo dT primers for circRNA and mRNA
detection, respectively, as well as specific stem-loop primers
(GenePharma Co., Ltd.) for miRNA detection. qPCR was carried out
using TB Green® Premix Ex Taq™ II (Takara Bio, Inc.) on
the Bio-Rad fluorescence thermal cycler CFX-96 (Bio-Rad
Laboratories, lnc.) with β-actin or U6 as internal controls. The
PCR cycle conditions were an initial denaturation at 95°C for 10
min; 40 cycles of 94°C 30 sec, 60°C 30 sec and 72°C 30 sec; and a
final extension at 72°C for 1 min. The relative RNA expression
levels were determined using the 2−ΔΔCq method (17). The primers for circRNA and mRNA
detection are presented in Table
SI. miRNA primers were purchased from GenePharma Co., Ltd.
RNase R treatment and subcellular
fractionation assay
For RNA digestion assay, 2 µg RNA isolated from
GC-MSCs were mixed with RNase R (Epicentre; Illumina, Inc.) and
incubated at 37°C for 20 min. Cytoplasmic and nuclear RNA were
separately obtained from the GC-MSCs using the PARIS™ kit
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. The relative expression of RNA in the two
fractions was detected using RT-qPCR. U6 and β-actin were used as
controls to evaluate the efficiency of subcellular fractionation
isolation.
Microarray analysis
Microarray analysis was conducted by Shanghai
Biotechnology Corporation using SBC Human (4×180K) ceRNA array
(Agilent Technologies, Inc.). In brief, total RNA was extracted,
purified and examined for a RIN number. The RNA was then amplified
and labeled. Each slide was hybridized with Cy3-labeled cRNA,
washed in staining dishes and then scanned. Raw data were
normalized using the Quantile algorithm, limma packages in R. The
threshold value for significance that was used to define
upregulation or downregulation was a fold change >2 and
P<0.05.
circRNA overexpression plasmid
construction, oligonucleotides and cell transfection
Human circ_0024107 cDNA was amplified and cloned
into the GV486 vector by Genechem (Shanghai Genechem Co., Ltd.).
The corresponding empty vector was used as the control. Two small
interfering RNA (siRNA) duplexes targeting the back-splice junction
of circ_0024107, miRNA mimics, miRNA inhibitor and corresponding
negative controls were designed and purchased from GenePharma Co.,
Ltd. and were transiently transfected into the MSCs using
Lipofectamine 2000® (Invitrogen, Thermo Fisher
Scientific, Inc.). siRNAs and inhibitor were transfected at a final
concentration of 100 nM, while mimics were transfected at a final
concentration of 5 nM. A total of 1 µg of the plasmid was
transfected into MSCs per well in a six-well plate. After 4 to 6 h,
the cells were washed with PBS and refreshed with DMEM containing
10% FBS. After 48 h, the cell supernatant was collected,
centrifuged at 1,000 × g at 4°C for 20 min, filtered and finally
stored at −20°C until use. The transfection efficiency was
determined using RT-qPCR and western blot analysis. The sequences
of these oligonucleotides are presented in Table SII.
Transwell assay, carnitine
palmitoyltransferase 1 (CPT1) activity and β-oxidation rate
detection
Migration and invasion were examined using Transwell
assays, and CPT1 activity and β-oxidation rate detection were
conducted as previously described (16,18).
Briefly, 8×104 gastric cancer cells suspended in
serum-free culture media (Gibco; Thermo Fisher Scientific, Inc.)
were plated into the upper chambers of Transwell inserts (Corning,
USA) with or without pre-coating with Matrigel (BD Biosciences),
and separately incubated with the bottom chambers containing fresh
cell culture media (Gibco; Thermo Fisher Scientific, Inc.) for 10
and 24 h at 37°C for migration and invasion assays. The cells were
fixed with 4% paraformaldehyde at room temperature for 30 min, and
stained with 4% crystal violet (Beyotime Institute of
Biotechnology) for 15 min. A light microscope (Nikon Corporation)
was used to observe the cells. For CPT1 activity and β-oxidation
rate assays, cell mitochondria were isolated and separately
subjected to detection using a CPT1 Spectrophotometric Detection
kit (Zikerbio) and the Fatty Acid β-oxidation Rate Colorimetric
Assay kit (Genmed Scientifics, Inc.) as per the manufacturers'
instructions.
Animal tumor model
Male BALB/c nude mice (4 weeks old; 5 mice per
group) were housed in a pathogen-free facility (26°C; 50% humidity;
with food and water provided ad libitum) at the Animal
Center of Jiangsu University and randomly divided into two groups,
including the si-circ_0024107 group injected with HGC-27 cells
subjected to circ_0024107 silencing-GC-MSC-CM and the NC group
inoculated with HGC-27 cells pre-treated with negative control
oligonucleotide-infected GC-MSC-CM. A total of 2×106
HGC-27 cells were suspended in 200 µl PBS and injected into the
left footpads of the nude mice to establish animal models of lymph
node metastasis. After 3 to 4 weeks, the draining popliteal lymph
nodes became swollen and there were palpable lumps, and the mice
were then sacrificed by cervical dislocation, and the draining
popliteal and inguinal lymph nodes were harvested for imaging,
weighing and immunohistochemical staining. The animal experiment
was approved by the Committee on Use and Care of Animals of Jiangsu
University [approval no. SYXK(Su) 2018-0053].
Western blot analysis
Cellular proteins were extracted from the cells
using RIPA lysis buffer pre-mixed with protease and phosphatase
inhibitors (Beyotime Institute of Biotechnology). The concentration
of proteins were determined using a BCA Protein Assay kit (cat. no.
CW0014S, Beijing Kangwei Century Biotechnology Co., Ltd.) and the
mass of protein loaded per lane was 8 µg. The PVDF membranes
bearing proteins were blocked using 5% BSA at room temperature for
1 h and then immersed in 5% BSA containing primary antibody against
CPT1A (cat. no. ab128568, dilution: 1:1,000, Abcam) and β-actin
(cat. no. AC038, dilution: 1:50,000, ABclonal Biotech Co., Ltd.)
overnight at 4°C followed by incubation with the corresponding
secondary antibody at 37°C. The secondary antibodies
HRP-conjugated-goat anti-mouse IgG (H+L) (cat. no. AS003, ABclonal
Biotech Co., Ltd.) and HRP-conjugated-goat anti-rabbit IgG (H+L)
(cat. no. AS014, ABclonal Biotech Co., Ltd.) were used at a
dilution of 1:5,000. β-actin was used as the loading control.
Signaling was detected using ECL Substrate (Bio-Rad Laboratories,
Inc.).
Immunohistochemical staining
The tumor tissues fixed in 4% paraformaldehyde (at
room temperature for 24 h) were embedded in paraffin and then
sectioned into 5-µm-thick slices. Immunohistochemistry was
performed using an Instant SABC-POD kit (cat. no. SA1020, Boster
Biological Technology) according to the manufacturer's
instructions. Briefly, after dewaxing, rehydration and antigen
retrieval, the tumor sections were blocked and were then separately
incubated with primary antibodies against pan-cytokeratin (AE1/AE3;
cat. no. ab27988, Abcam, dilution: 1:200) and CPT1A (cat. no.
ab128568, Abcam, dilution: 1:200) overnight at 4°C. After washing,
the sections were incubated with biotin-conjugated secondary
antibody (cat. no. SA1020, Boster Biological Technology) at 37°C
for 30 min. After washing, the sections were incubated with SAB at
37°C for 30 min and stained with DAB (cat. no. AR1027, Boster
Biological Technology) at room temperature for 5 min. Following DAB
visualization, the sections were immersed into water and then
re-stained using hematoxylin at room temperature for 3 min. The
AE1/AE3-positive cells or the CPT1A-positive cells were finally
observed under a light microscope (Nikon Corporation).
RNA immunoprecipitation (RIP)
assay
Anti-Argonaute 2 (AGO2) (cat. no. C34C6, Cell
Signaling Technology, Inc.) and anti-IgG antibodies (cat. no. 8726S
Cell Signaling Technology, Inc.) were incubated with protein A+G
beads at 4°C for 1 h following the instructions provided with the
RNA Immunoprecipitation kit (cat. no. P0101, Geneseed Biotech Co.,
Ltd.). Briefly, 1 ml Buffer A working solution contained 1% volume
protease inhibitor and 1% volume RNase inhibitor before use. A
total of 1×107 GC-MSCs cells were used for each IP
reaction and added 1 ml of the configured RIP lysis buffer. The
lysate was centrifuged at 14,000 × g for 10 min, at 4°C. The
resulting supernatant and antibody-attached magnetic beads were
incubated for 1 h at 4°C. The product was obtained by
centrifugation at 12,000 × g for 1 min at 4°C. The captured RNAs
and target protein were finally eluted and purified for RT-qPCR and
western blot analyses.
Luciferase reporter assay
Wild-type or mutant circ_0024107 fragments were
amplified by PCR and inserted into the luciferase reporter vector,
GV272, provided by GeneChem. Wild-type or mutant 3′-untranslated
region (3′UTR) CPT1A fragments were synthesized, annealed and
inserted into the pmirGLO Dual-Luciferase miRNA Target Expression
Vector (Promega Corporation). A total of 2 µg of the
dual-luciferase gene vectors were co-transfected with 50 nM miRNA
mimics into 293T cells (National Collection of Authenticated Cells
Cultures) using Lipofectamine 2000®. After 5 h, cell
culture media was refreshed. Cells were harvested for luciferase
activity assay using a Dual-Luciferase Reporter Assay System
(Promega Corporation) after transfection for 24 h. Luciferase
activity was normalized to that of Renilla luciferase
activity.
gDNA extraction and amplification
using PCR and agarose gel electrophoresis
Genomic DNA small volume extraction kit (cat. no.
D0063, Shanghai Biyuntian Biotechnology Co., Ltd.) was used to
extract gDNA. The gDNA was then amplified by PCR. The 2% gel was
prepared with 0.5X TAE. A total of 8 µl sample was added and 6 µl
of 2,000 bp marker (Takara Bio, Inc.) at each end, and the voltage
was set at 110 V for 30 min. Following electrophoresis, the gel was
stained with 0.5 µg/ml ethidium bromide for 5 min, and imaged using
an automatic gel imager (Biobase).
Bioinformatics analysis
Circ_0024107 annotation information was obtained
using the circBase database (http://www.circbase.org). The Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway database
(https://www.kegg.jp/kegg/pathway.html) was used to
analyze signaling pathway enrichment analysis. miRanda (http://www.microrna.org) and TargetScan (http://www.targetscan.org/) were used to predict
potential binding miRNAs connecting circ_0024107 to CPT1A.
Kaplan-Meier Plotter database (http://kmplot.com/) was used to assess the association
between CPT1A, miR-5572 and miR-6855-5p and the survival of
patients based on MSC abundance in gastric cancer tissues
Statistical analysis
SPSS software 22.0 and GraphPad Prism software 8.0
were used for statistical analyses. The data are expressed as the
mean, mean ± SD, mean ± SEM, or median ± QR, as indicated in the
figure legends. Statistically significant differences between
parametric variables were calculated using an unpaired Student's
t-test or one-way ANOVA followed by Tukey's post-hoc test, and
non-parametric variables were analyzed using the Wilcoxon
matched-pairs signed rank test was conducted to compare the
expression of circ_0024107, miR-5572, miR-6855 and CPT1A between
paired cancer tissues and adjacent non-cancerous tissues. The
Chi-squared test was applied to analyze clinical significance
between ordinal variables. The correlation of gene expression was
examined using Spearman's correlation analysis. The Kaplan-Meier
method and log rank test were used to evaluate the survival. A
value of P<0.05 was considered to indicate a statistically
significant difference.
Results
Screening and identification of
GC-MSC-derived circ_0024107
Previous studies by the authors have demonstrated
that BM-MSCs are the critical cellular origin of GC-MSCs (6,16). In
the present study, to identify deregulated circRNAs in GC-MSCs, a
circRNA array was used to screen for differentially expressed
circRNAs between GC-MSCs and BM-MSCs. A total of 5,497 circRNAs
with a fold change ≥2 and P<0.05 were identified, including
2,545 upregulated and 2,952 downregulated circRNAs in the GC-MSCs
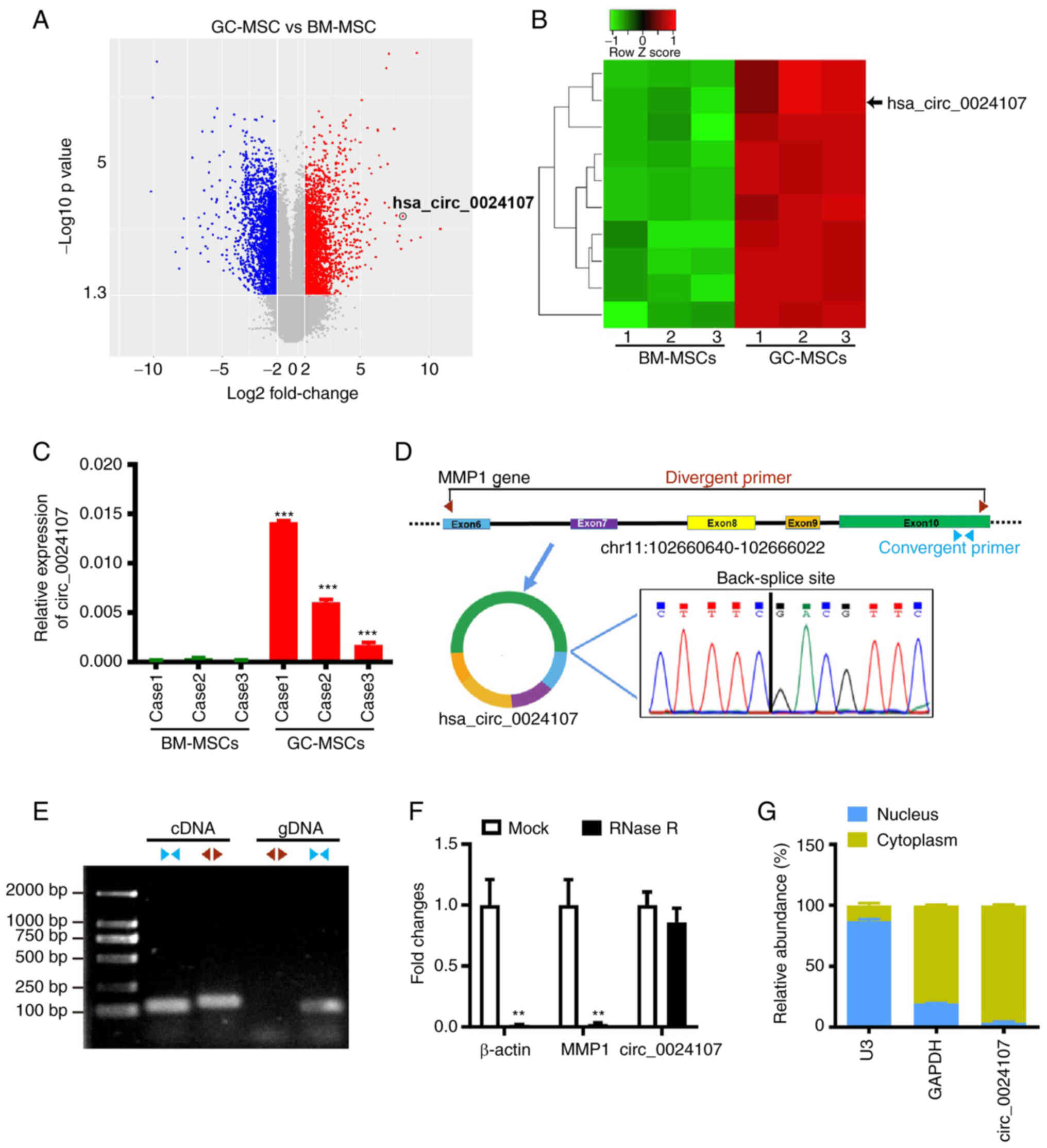
(Fig. 1A and Table SIII). The top 10 upregulated
circRNAs were selected for further verification using RT-qPCR
(Fig. 1B); a consistently increased
expression of hsa_circ_0024107 was observed in GC-MSCs (Fig. 1C). According to the annotation from
the circBase database hsa_circ_002417 is generated from the host
gene MMP1, being composed of exons 6-10 and having a length of
1,131 nucleotides. The amplified PCR products sequencing results
verified the existence of back-splicing site (Fig. 1D). Furthermore, gDNA and cDNA were
obtained from GC-MSCs and convergent primers and divergent primers
were used to perform PCR. The electrophoresis results revealed that
circ_0024107 was only amplified by divergent primers from the cDNA,
not from gDNA (Fig. 1E). RNase R
treatment confirmed that circ_0024107 was more stable than linear
transcript MMP1 and β-actin (Fig.
1F). Subcellular fractionation assay verified that circ_0024107
was mainly located in the cytoplasm of the GC-MSCs (Fig. 1G). The aforementioned data indicate
the successful identification and characterization of
GC-MSC-derived circ_0024107.
circ_0024107 is pivotal for GC-MSCs to
promote gastric cancer cell metastasis in vitro and in vivo
To determine the role of circ_0024107 in GC-MSCs,
two siRNAs targeting back-splicing sites were synthesized and
transfected into the GC-MSCs to knock down circ_0024107 (Fig. 2A and B). CM from the
siRNA-transfected GC-MSCs was prepared and used for the culture of
the AGS and HGC-27gastric cancer cells. In vitro, Transwell
assays revealed that the knockdown of circ_0024017 markedly
suppressed the GC-MSC-mediated stimulation of gastric cancer cell
migration and invasion (Fig. 2C-E).
In addition, circ_0024107-overexpressing plasmid was transfected
into BM-MSCs to overexpress circ_0024107 (Fig. 2F and G). Notably, circ_0024107
conferred migration- and invasion-promoting roles to the BM-MSCs
(Fig. 2H and I). In vivo,
models of lymph node metastasis were established by inoculating
HGC-27 cells treated with different GC-MSC-CM into the left
footpads of nude mice. The reduced volume and weight of popliteal
lymph nodes and inguinal lymph nodes were observed, as well as a
smaller positive expression area of pan-cytokeratin AE1/AE3 in the
lymph node tissues in the si-circ_0024107-transfected groups
compared with those in the negative control-transfected groups
(Fig. 2K-M). These findings suggest
that circ_002417 is crucial for the promoting effects of GC-MSCs on
gastric cancer cell metastasis.
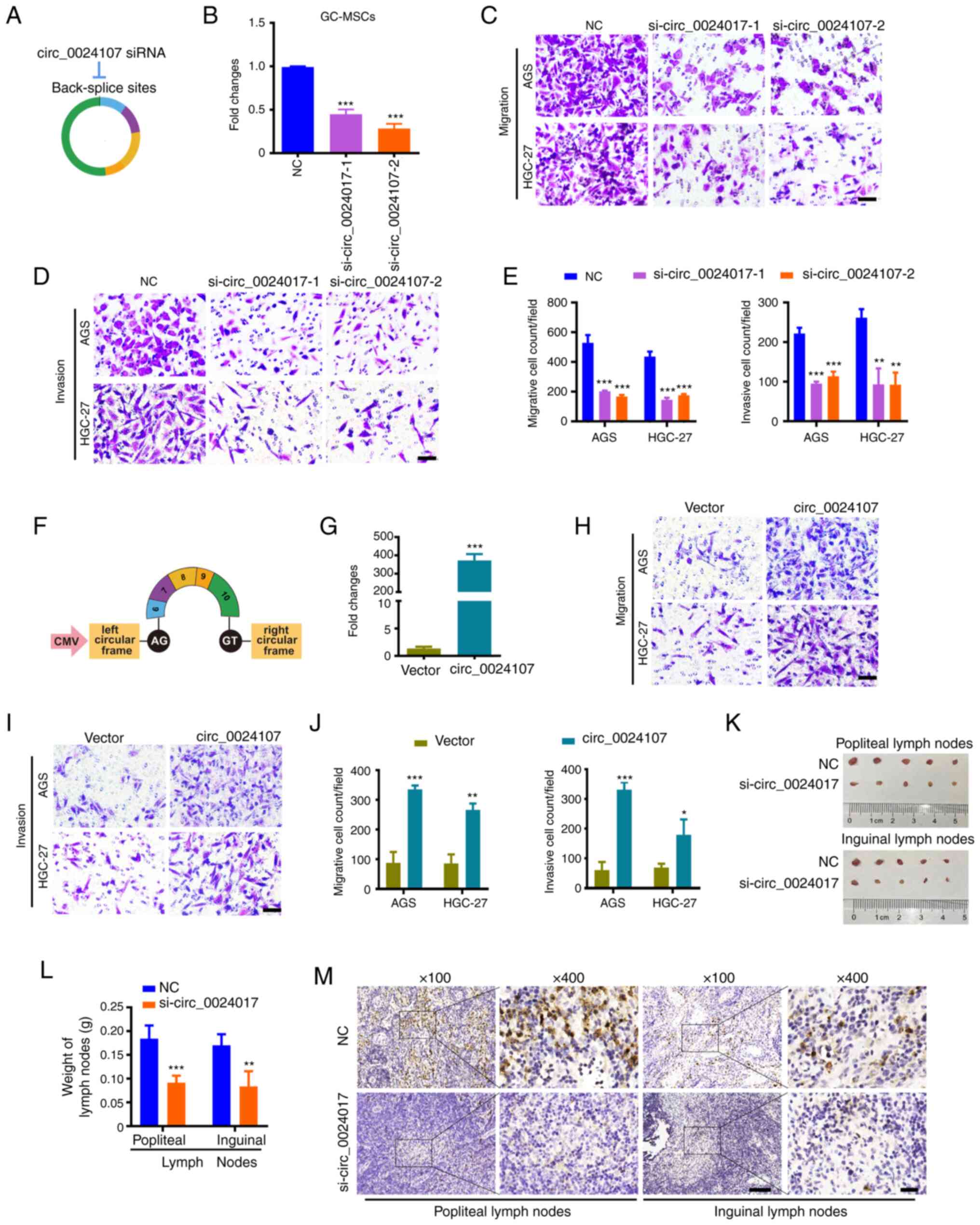 | Figure 2.circ_0024107 mediates the promotion
of gastric cancer cell metastasis by GC-MSCs in vitro and
in vivo. (A) Schematic diagram of the specific siRNAs
targeting the back-splicing site of circ_0024107. (B) RT-qPCR of
circ_0024107 in GC-MSCs following transfection with two different
siRNAs against circ_0024107. NC was the negative control of the two
siRNAs. (C-E) Transwell assays of the migration and invasion of
gastric cancer cell lines, AGS and HGC-27, following treatment with
CM from circ_0024107-silencing GC-MSCs. Scale bars, 100 µM;
magnification, ×200. (F) Schematic diagram of circ_0024107
overexpression vector. (G) RT-qPCR of circ_0024107 in BM-MSCs
following transfection with the overexpression vector. (H-J)
Migration and invasion analysis of gastric cancer cells following
treatment with CM from circ_0024107-overexpressing BM-MSCs. Scale
bars, 100 µM; magnification, ×200. (K-M) Lymphatic metastasis of
HGC-27 treated with CM from circ_0024107-silenced GC-MSCs in
vivo. (K) Lymph nodes; (L) weight of lymph nodes; (M)
representative images of pan-cytokeratin AE1/AE3 staining in lymph
nodes. Magnification, ×100 and scale bars, 100 µM; or
magnification, ×400 and scale bars, 20 µM. Values are presented as
the mean ± SD (n=3) *P<0.05, **P<0.01 and ***P<0.001, vs.
respective control. circRNA, circular RNA; GC-MSCs, gastric
cancer-derived mesenchymal stem cells; BM-MSCs, bone marrow-derived
mesenchymal stem cells; RT-qPCR, reverse transcription-quantitative
PCR; CM, conditioned medium. |
GC-MSCs undergo fatty acid oxidation
(FAO) metabolic reprogramming to exert tumor-promoting regulatory
effects
To investigate whether GC-MSCs undergo metabolic
reprograming, a cDNA array was applied to screen differentially
expressed mRNAs between the GC-MSCs and BM-MSCs. Compared to the
BM-MSCs, a total of 2,276 differentially expressed mRNAs (1,162
upregulated and 1,114 downregulated) were identified in the GC-MSCs
based on the same criteria for circRNA screening (Fig. 3A and Table SIV). KEGG pathway enrichment
analysis revealed that lipid metabolism was dominant in the
metabolism module (Fig. 3B). Recent
research has suggested that FAO emerges as a critical aspect of the
metabolic landscape of cancer (18). Heatmap analysis revealed that the
majority of genes driving FAO metabolism were upregulated in the
GC-MSCs, including the limiting enzyme, CPT1A (Fig. 3C). CPT1A mRNA and protein levels
were verified to be increased in the GC-MSCs relative to those in
the BM-MSCs (Fig. 3D and E). CPT1
activity and β-oxidation rate assays confirmed that FAO was highly
activated in the GC-MSCs compared to the BM-MSCs (Fig. 3F and G). In order to clarify whether
FAO mediates the oncogenic function of GC-MSCs, etomoxir (CPT1A
inhibitor) was used to block FAO in GC-MSCs. As demonstrated by the
results, the stimulated invasion of the gastric cancer cells by
GC-MSCs was significantly attenuated by treatment with etomoxir
(Figs. 3H and S1A). These findings indicate that FAO
metabolic reprogramming is crucial for the oncogenic function of
GC-MSCs.
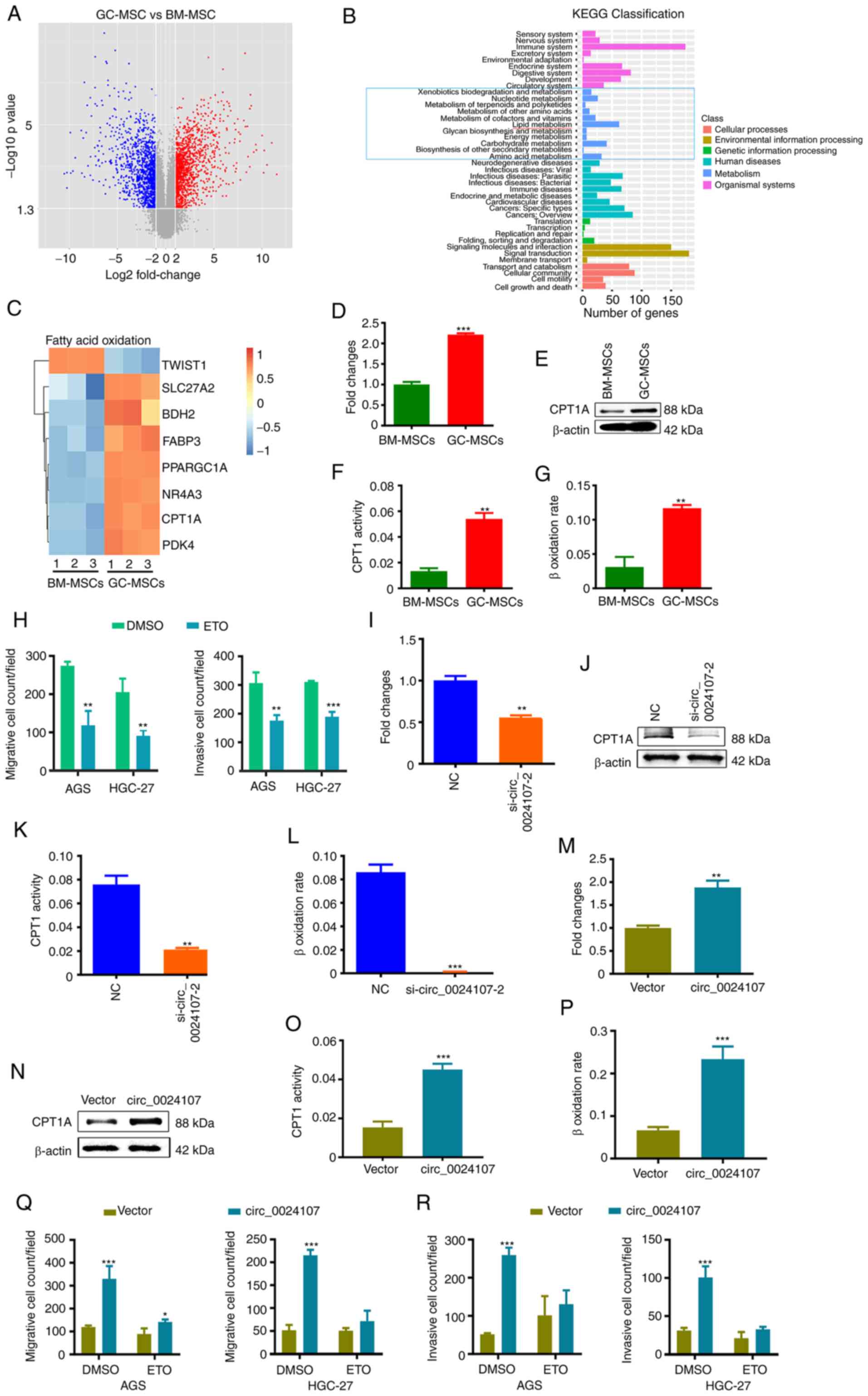 | Figure 3.FAO reprogramming is crucial for the
oncogenic role of GC-MSCs and circ_0024107 modulates the FAO
metabolic reprogramming of GC-MSCs by upregulating CPT1A
expression. (A) Volcano plot of differentially expressed mRNAs with
fold changes ≥2 and P-values <0.05 between GC-MSCs and BM-MSCs.
(B) KEGG classification analysis of the deregulated mRNAs; the blue
bars in the light blue box are related to metabolism; (C) Heatmap
of different genes involved in FAO metabolism. (D) RT-qPCR
verification of CPT1A mRNA levels in GC-MSCs and BM-MSCs. (E)
Western blot analysis of CPT1A protein levels in the two types of
MSCs. (F,G) FAO activity was compared between GC-MSCs and BM-MSCs
by (F) CPT1 activity assay and (G) β-oxidation rate detection. (H)
Count of the migrated and invaded gastric cancer cells following
incubation with CM from etomoxir-treated GC-MSCs. (I-L) The effects
of circ_0024107 silencing on (I) CPT1A mRNA levels, (J) protein
levels, (K) CPT1 activity and (L) β-oxidation rate in GC-MSCs.
(M-P) The effects of circ_0024107 overexpression on (M) CPT1A mRNA
levels, (N) protein levels, (O) CPT1 activity and (P) β-oxidation
rate in BM-MSCs. (Q,R) Count of (Q) migrated and (R) invaded
gastric cancer cells following culture with CM from
circ_0024107-overexpressing BM-MSCs treated with etomoxir. Values
are presented as the mean ± SD (n=3). *P<0.05, **P<0.01 and
***P<0.001, vs. respective control. circRNA, circular RNA;
GC-MSCs, gastric cancer-derived mesenchymal stem cells; BM-MSCs,
bone marrow-derived mesenchymal stem cells; RT-qPCR, reverse
transcription-quantitative PCR; CM, conditioned medium; CPT1A,
carnitine palmitoyltransferase 1A; FAO, fatty acid oxidation; ETO,
etomoxir. |
circ_0024107 increases FAO activity in
GC-MSCs by upregulating CPT1A expression
The aforementioned findings suggested that
circ_0024107 and FAO are critical for GC-MSC function. The present
study then wished to determine whether there is a regulatory
association between them. Cells in which circ_0024107 was knocked
down or overexpressed were separately subjected to CPT1A expression
and FAO activity analyses. The CPT1A mRNA and protein expression
levels were consistently altered along with the knockdown or
overexpression of circ_0024107 in MSCs (Fig. 3I, J, M and N). CPT1 activity and
β-oxidation rate were suppressed in the circ_0024107-silenced
GC-MSCs, whereas they were increased in the
circ_0024107-overexpressing BM-MSCs (Fig. 3K, L, O and P). Furthermore,
pre-treatment with etomoxir notably impaired the effects of
circ_0024107 overexpression in BM-MSCs on gastric cancer cell
invasion (Figs. 3Q and R, and
S1B). In summary,
circ_0024107-induced FAO metabolic reprogramming is crucial for the
oncogenic function of GC-MSCs.
circ_0024107 acts as a sponge of
miR-5572 and miR-6855-5p to upregulate CPT1A expression
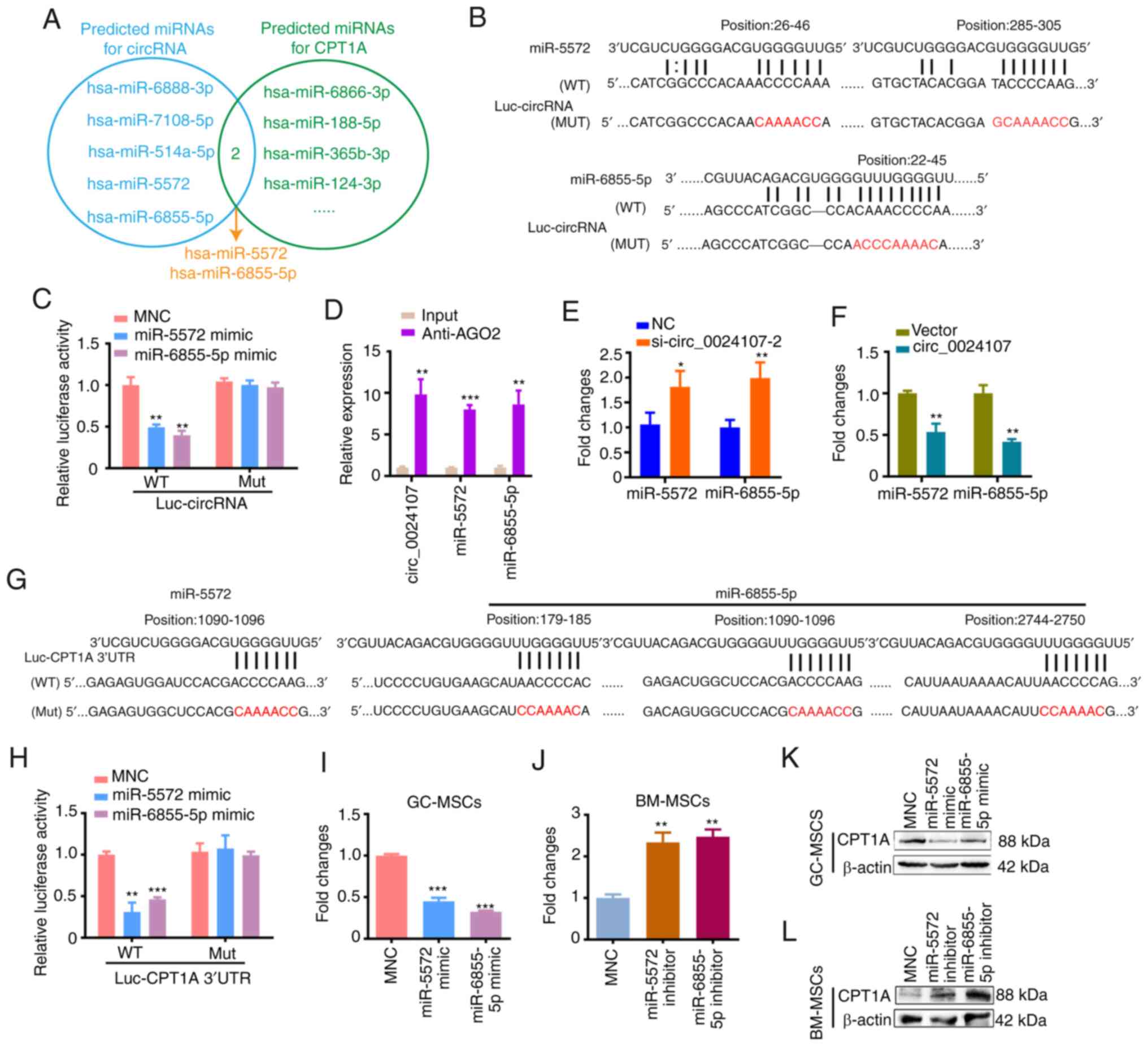
To evaluate whether circ_0024107 induced FAO by
acting as a miRNA sponge to upregulate CPT1A expression, miRanda
and TargetScan were used to predict potential binding miRNAs
connecting circ_0024107 to CPT1A. The overlapped hsa_miR-5572 and
hsa_miR-6855-5p were eventually selected as candidate miRNAs for
subsequent analyses (Fig. 4A).
circ_0024107-luciferase reporter vectors containing the wild-type
or mutant binding sequences were constructed (Fig. 4B) and co-transfected with miRNA
mimics. The luciferase activity of the wild-type vectors was
evidently reduced by miRNA mimics compared to that of miRNA mimics
negative control (MNC). No marked differences were observed for the
mutant-type vectors (Fig. 4C).
Moreover, the results of RIP assay revealed that circ_0024107 and
the two miRNAs were pulled down by anti-AGO2 compared with control
IgG (Fig. 4D). The miR-5572 and
miR-6855-5p levels were markedly increased in the
circ_0024107-silenced GC-MSCs, whereas they were reduced in the
circ_0024107-overexpressing BM-MSCs (Fig. 4E and F). These data illustrate that
circ_0024107 physically interacts with and negatively regulates
miR-5572 and miR-6855-5p.
To examine whether CPT1A is the target of miR-5572
and miR-6855-5p, dual-luciferase reporter plasmids containing the
wild-type or mutant CPT1A 3′UTR fragments were separately
constructed according to the prediction sites for the two miRNAs
(Fig. 4G). Both miR-5572 and
miR-6855-5p mimics notably suppressed the luciferase activities of
the wild-type plasmids, but not those of the mutant types (Fig. 4H). The overexpression of miR-5572
and miR-6855-5p in GC-MSCs following mimic transfection resulted in
reduced CPT1A mRNA and protein levels (Fig. 4I and K). Conversely, the reduced
expression of the two miRNAs in the BM-MSCs following inhibitor
transfection increased CPT1A expression (Fig. 4J and L). These results indicate that
miR-5572 and miR-6855-5p negatively regulate CPT1A mRNA by binding
to its 3′UTR.
miR-5572 and miR-6855-5p reverse the
circ_0024107-induced FAO metabolic reprogramming of GC-MSCs
Based on the aforementioned findings, it was
hypothesized that the two miRNAs may suppress FAO and attenuate the
oncogenic role of GC-MSCs. The corresponding miRNA mimics and
inhibitors were utilized to overexpress and suppress the two miRNAs
in the GC-MSCs and BM-MSCs, respectively. FAO activity was
negatively associated with the levels of both miRNAs (Fig. 5A-D). The pro-invasive role of the
GC-MSCs was attenuated by miRNA mimics, whereas the tumor-promoting
role was conferred to the BM-MSCs by miRNA inhibitors (Figs. 5E and F, and S2A and B). Furthermore, treatment with
etomoxir significantly reversed the effects induced by the BM-MSCs
transfected with miRNA inhibitors (Figs. 5G and S2C). To further elucidate whether
circ_0024107 functions as a competitive endogenous RNA (ceRNA) to
upregulate CPT1A, the circ_0024107 overexpression vector was
separately co-transfected with the two miRNA mimics into BM-MSCs.
The CPT1A mRNA and protein levels induced by circ_0024107
overexpression were significantly reversed by transfection with
miRNA mimics (Fig. 5H and I). A
similar tendency was observed for CPT1 activity and β-oxidation
rate (Fig. 5J and K). In addition,
the promotion of gastric cancer cell migration and invasion induced
by circ_0024107-overexpressing BM-MSCs was eliminated by the two
miRNA mimics (Fig. 5L and M). These
data thus indicate that circ_0024107 elicits the FAO reprograming
of GC-MSCs by sponging miR-5572 and miR-6855-5p.
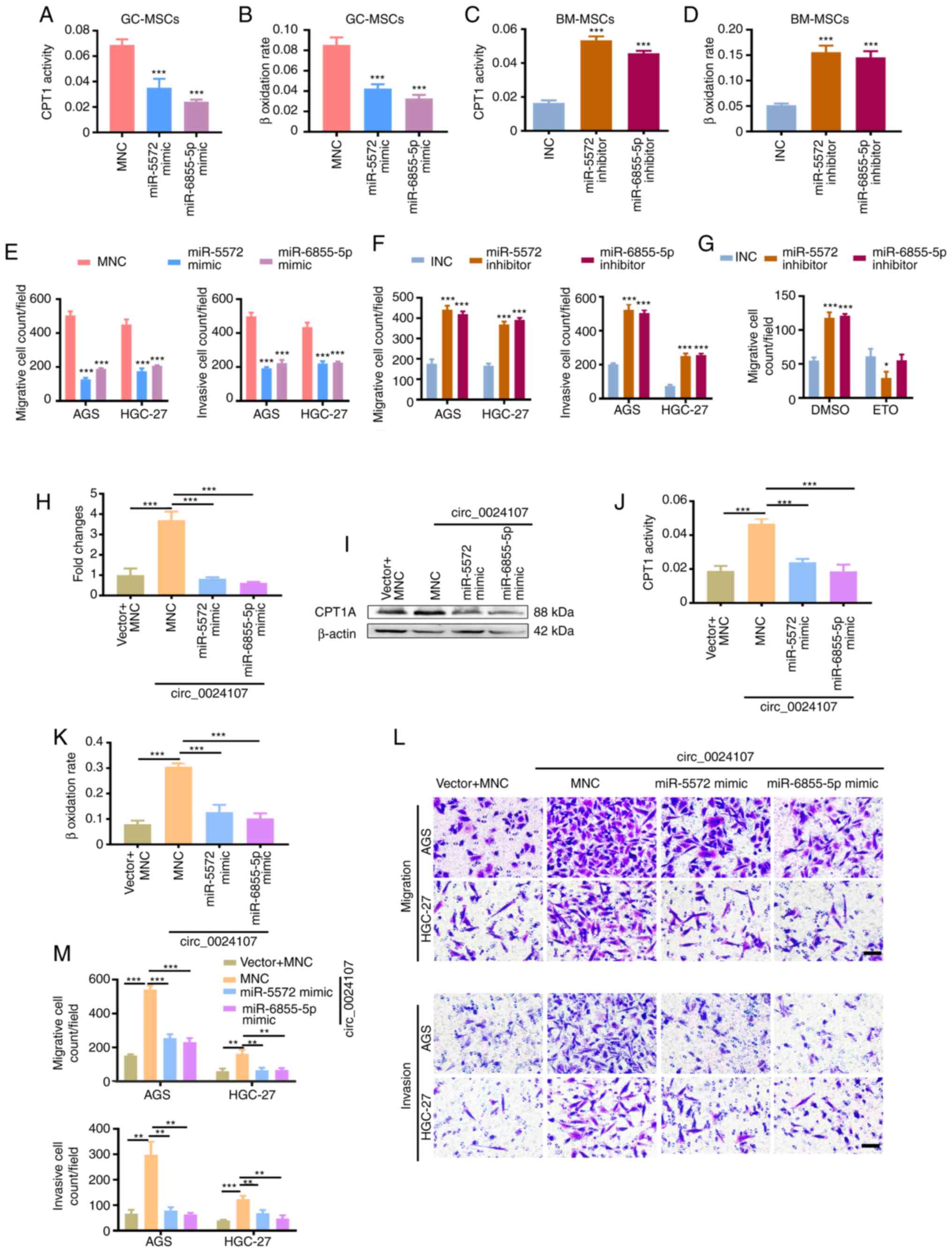 | Figure 5.miR-5572 and miR-6855-5p reverse the
circ_0024107-induced FAO reprograming of GC-MSCs. (A-D) CPT1
activity and β-oxidation rate assay in (A and B) miRNA
mimic-transfected GC-MSCs and (C and D) inhibitor-transfected
BM-MSCs. (E and F) Count of the migrated and invaded gastric cancer
cells following incubation with CM from (E) miRNA mimic-transfected
GC-MSCs and (F) inhibitor-transfected BM-MSCs. (G) Count of the
migrated gastric cancer cells following incubation with CM from
BM-MSCs in which the two miRNAs were silenced and which were
treated with etomoxir. Detection of (H) CPT1A mRNA and (I) protein
levels in BM-MSCs co-transfected with circ_0024107 overexpression
vector and miRNA mimics. Measurement of FAO activity assay by
determining (J) CPT1 activity and (K) β-oxidation rate in the
co-transfected BM-MSCs. (L and M) Transwell assay of the migration
and invasion of gastric cancer cells following incubation with CM
from the co-transfected BM-MSCs. Scale bars, 100 µM; magnification,
×200. Values are presented as the mean ± SD (n=3). *P<0.05,
**P<0.01 and ***P<0.001, vs. respective control. circRNA,
circular RNA; GC-MSCs, gastric cancer-derived mesenchymal stem
cells; BM-MSCs, bone marrow-derived mesenchymal stem cells; CPT1A,
carnitine palmitoyltransferase 1A; CM, conditioned medium; FAO,
fatty acid oxidation; ETO, etomoxir. |
GC-MSC-derived circ_0024107 modulates
FAO in gastric cancer cells
A previous study by the authors demonstrated that
increased FAO levels are indispensable for lymphatic metastatic
gastric cancer cells (19). GC-MSCs
possibly promote lymphatic metastasis by inducing FAO of gastric
cancer cells. In the present study, as was expected, the HGC-27
cells cultured with GC-MSC-CM exhibited higher levels of CPT1
activity and β-oxidation rate than those incubated with BM-MSC-CM
(Fig. 6A and B). Pre-treatment with
etomoxir eliminated the promoting effects of GC-MSCs on gastric
cancer cell migration and invasion (Fig. 6C and D). The results of RT-qPCR
verified that the circ_0024017 and CPT1A levels were notably
increased, while those of miR-5572 and miR-6855-5p were reduced in
the HGC-27 cells treated with GC-MSC-CM (Fig. 6E and F). circ_0024107 silencing in
HGC-27 cells not only suppressed the migrative and invasive
capacities and FAO activity, but also blocked the effects of
GC-MSCs on cell migration, invasion and FAO (Figs. 6G-J and S3A-G). The miR-5572 and miR-6855-5p
expression levels were negatively associated with the circ_0024107
and CPT1A levels in circ_0024107-silenced HGC-27 cells (Figs. 6K and L and S3H). The overexpression of the two miRNAs
not only significantly suppressed CPT1A expression at the mRNA and
protein level, but also inhibited HGC-27 cell migration and
invasion (Fig. S4). Moreover, the
knockdown of circ_0024017 in the GC-MSCs markedly weakened their
effects on inducing circRNA and CPT1A expression, enhancing FAO
activity and suppressing miRNAs in the HGC-27 cells (Fig. 6M-P). On the whole, these data
suggest that circ_0024107 mediates the crosstalk between GC-MSCs
and gastric cancer cells, and may synergistically promote gastric
cancer lymphatic metastasis.
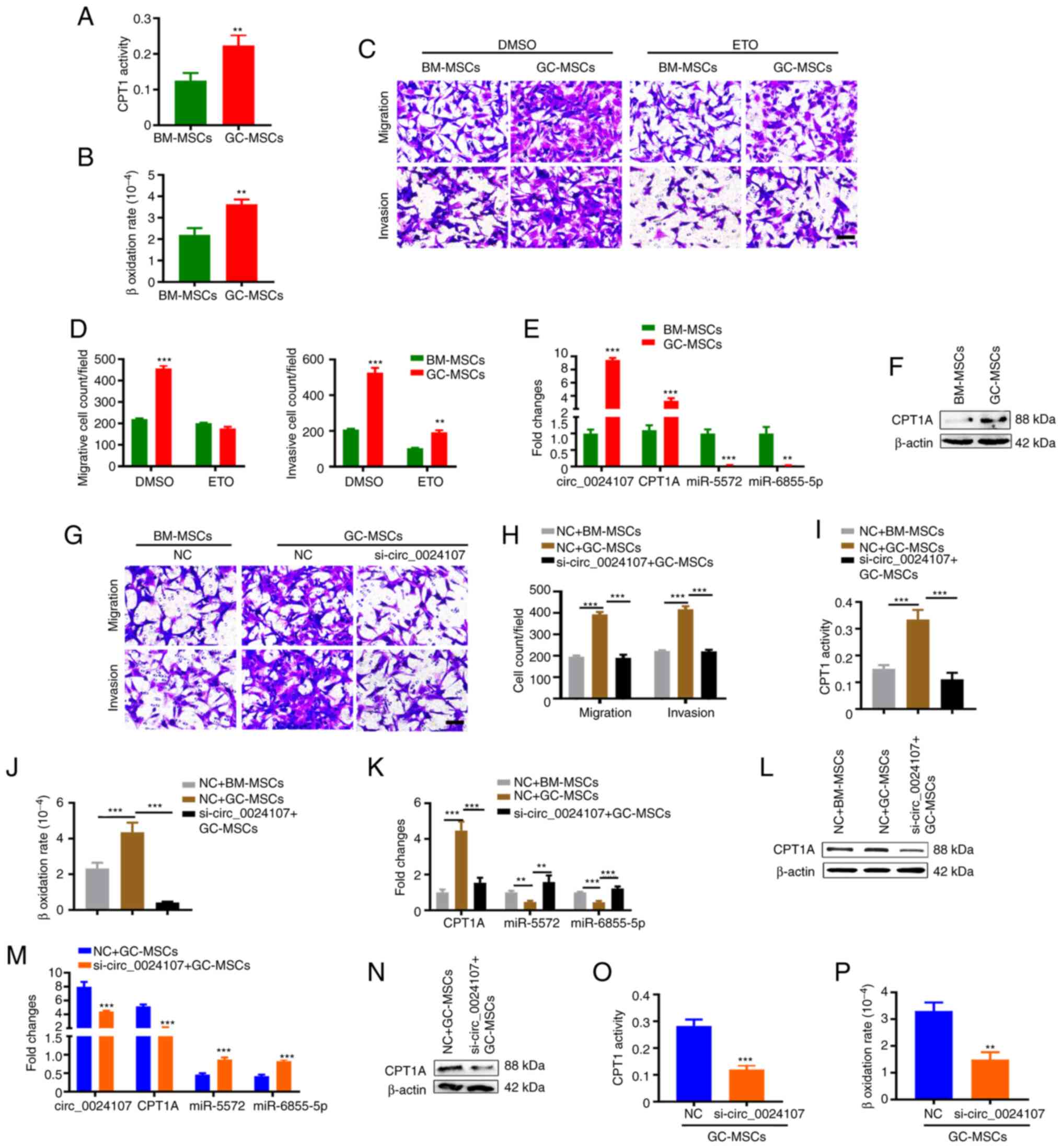 | Figure 6.GC-MSCs promote metastasis by
modulating the circ_0024107/miR-5572/6855-5p/CPT1A axis in gastric
cancer cells. (A) CPT1 activity and (B) β-oxidation rate detection
in HGC-27 cells following treatment with GC-MSC-CM and BM-MSC-CM.
(C and D) Migration and invasion of HGC-27 cells pre-treated with
etomoxir and incubated with MSC-CM. (E and F) Comparison of
circ_0024107, CPT1A, miR-5572 and miR-6855-5p levels in HGC-27
cells following treatment with GC-MSC-CM and BM-MSC-CM using (E)
reverse transcription-quantitative PCR and (F) western blot
analysis. (G and H) Migration and invasion of circ_0024107-silenced
HGC-27 cells following incubation with GC-MSC-CM. (I and J) FAO
activity of circ_0024107-silenced HGC-27 cells following incubation
with GC-MSC-CM. (K) Detection of CPT1A, miR-5572 and miR-6855-5p
levels, and (L) CPT1A protein content in circ_0024107-silenced
HGC-27 cells treated with GC-MSC-CM. (M) Detection of circ_0024107,
CPT1A, miR-5572 and miR-6855-5p levels, and (N) CPT1A protein
content in HGC-27 cells following incubation with the CM from
circ_0024107-silenced GC-MSCs. (O and P) FAO activity of HGC-27
cells following incubation with the CM from circ_0024107-silenced
GC-MSCs. Scale bars, 100 µM; magnification, ×200. Values are
presented as the mean ± SD (n=3). **P<0.01 and ***P<0.001.
circRNA, circular RNA; GC-MSCs, gastric cancer-derived mesenchymal
stem cells; BM-MSCs, bone marrow-derived mesenchymal stem cells;
CPT1A, carnitine palmitoyltransferase 1A; CM, conditioned medium;
FAO, fatty acid oxidation; ETO, etomoxir. |
Expression profiles and clinical
implications of the circ_0024107/miR-5572/6855-5p/CPT1A axis in
gastric cancer
RT-qPCR analysis revealed that the circ_0024107 and
CPT1A levels were aberrantly increased, whereas the levels of the
two miRNAs were significantly decreased in cancer tissues compared
to paired adjacent normal tissues (Fig.
7A). The circ_0024107 expression levels positively correlated
with the CPT1A levels, whereas the miR-5572 and miR-6855-5p levels
negatively correlated with the CPT1A levels (Fig. 7B-D). A significant negative
correlation was detected between circ_0024107 and miR-5572;
however, the correlation between circ_0024107 and miR-6855-5p was
not significant (Fig. 7E and F).
The circ_0024107 level was also found to be associated with lymph
node metastasis status and TNM stage, whereas CPT1A was only
associated with lymph node metastasis status. However, no
association was observed between the two miRNAs and the clinical
features of patients with gastric cancer (Table SV). The Kaplan-Meier Plotter
database was used to assess the association between CPT1A, miR-5572
and miR-6855-5p and the survival of patients based on MSC abundance
in gastric cancer tissues. Of note, CPT1A was a favorable factor,
not only associated with 3-year OS of patients with stage II
gastric cancer, whose cancer tissues contained decreased numbers of
MSCs, but also with the 3-year recurrence-free survival of patients
at the same stage, irrespective of the MSC enrichment status
(Fig. 7G and H). The two miRNAs
only served as favorable prognostic factors for patients with a MSC
enrichment status (Fig. 7I and J).
Furthermore, GC-MSCs another 5 patients and BM-MSCs from another 7
patients were randomly selected to evaluate the expression profile
of the axis. Compared to the BM-MSCs, the circ_0024107 and CPT1A
levels were markedly increased in the GC-MSCs, whereas the
miR-6855-5p level was significantly decreased. By contrast, the
miR-5572 levels were decreased in the GC-MSCs, although without a
significant difference (Fig. 7K-N).
In addition, a positive correlation was detected between
circ_0024107 and CPT1A expression, while a negative correlation was
only observed between miR-6855-5p and CPT1A, as well as between the
miRNAs and circ_0024107 (Fig. S5).
These results suggest that the regulatory axis is deregulated in
gastric cancer tissues and GC-MSCs, and is associated with lymph
node metastasis and prognosis of gastric cancer.
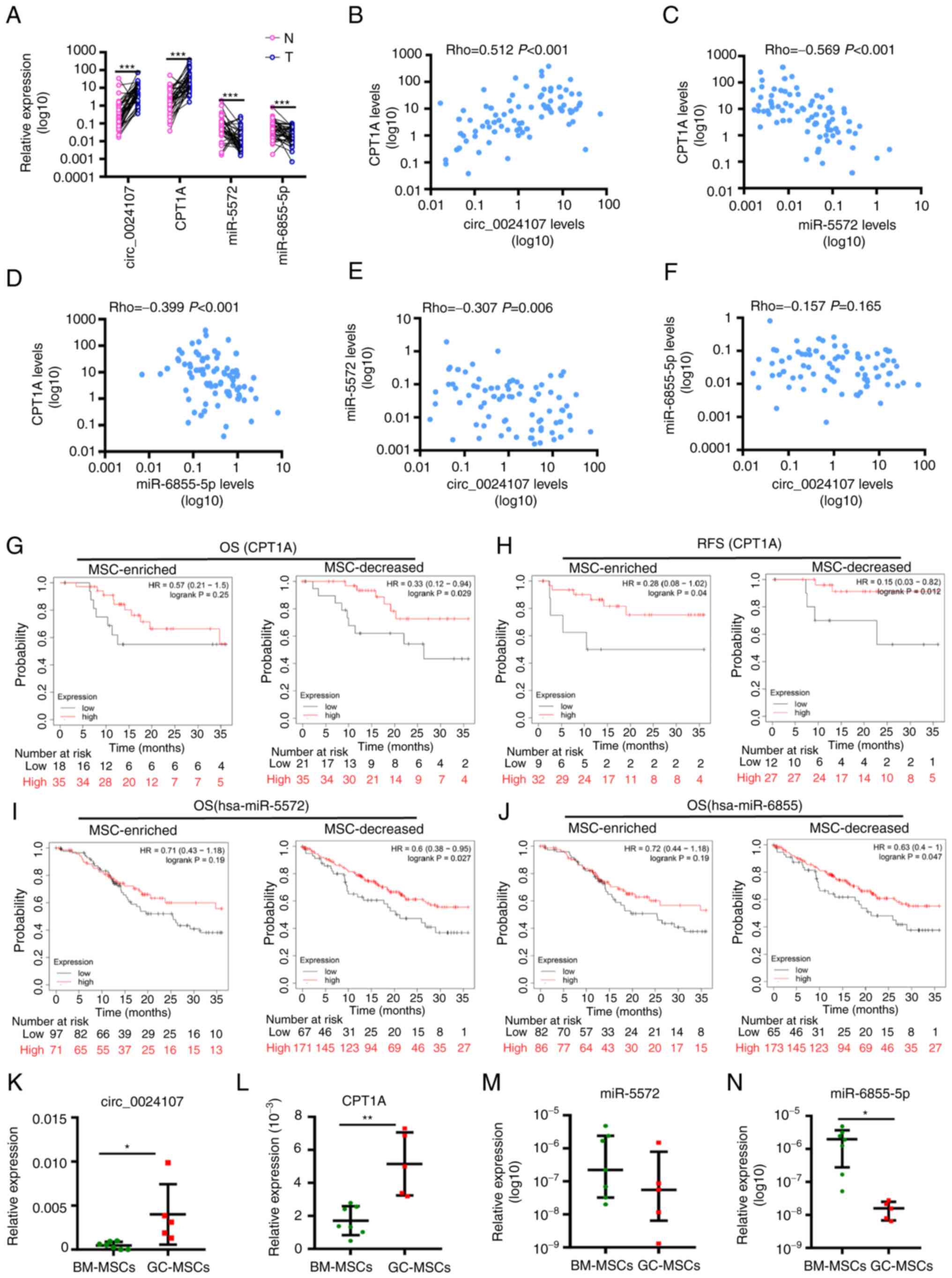 | Figure 7.Expression profiles and clinical
implications of the circ_0024107/miR-5572/6855-5p/CPT1A axis in
gastric cancer. (A) Comparison of circ_0024107, CPT1A, miR-5572 and
miR-6855-5p levels between 40 gastric cancer tissues (T) and paired
adjacent gastric tissues (N). (B-F) Correlation analysis of
molecules involved in this axis. (G-J) Survival analysis of (G and
H) CPT1A, (I) miR-5572 and (J) miR-6855-5p in gastric cancer
tissues with an MSC-enriched status or and with decreased numbers
of MSCs using the Kaplan-Meier Plotter database. (K-N) Comparison
of circ_0024107-miR-5572/6855-5p/CPT1A axis expression levels
between GC-MSCs and BM-MSCs. (A-F) Values are presented as the mean
(n=3); (K, L and N) Values are presented as mean ± SEM (n=3); (M)
Values are presented as the median ± QR (n=3). *P<0.05,
**P<0.01 and ***P<0.001, vs. respective control. circRNA,
circular RNA; GC-MSCs, gastric cancer-derived mesenchymal stem
cells; BM-MSCs, bone marrow-derived mesenchymal stem cells; CPT1A,
carnitine palmitoyltransferase 1A; OS, overall survival; RFS,
recurrence-free survival. |
Discussion
During the metastasis process and during
chemoradiotherapy, cancer cells strive for survival, depending on
interactions with surrounding stromal cells to overcome stressful
conditions, such as nutrient deficiency (20). Cancer cells alter metabolic
procedures to adapt to different environments, while stromal cells
need to undergo metabolic reprogramming to release supportive
signaling or provide necessary metabolites for cancer cell
biosynthesis (21,22). Therefore, tumor stromal cells are a
critical for the understanding of the mechanisms through which
cancer evolves and for the development of an effective therapeutic
strategy. The present study focused on gastric cancer-associated
MSCs and revealed that GC-MSC-derived circ_0024107 promoted gastric
cancer cell lymphatic metastasis via FAO metabolic reprogramming
mediated by the miR-5572/6855-5p-CPT1A axis.
Lymph node metastasis is an independent poor
prognostic predictor of gastric cancer. Over the past decades, an
increasing number of tissue- and plasma-derived circRNAs have been
demonstrated to be associated with the lymphatic metastasis of
gastric cancer (23,24). CircRNAs are sometimes used to
clarify the regulatory mechanisms of specific molecules involved in
the lymph node metastasis of gastric cancer (25). A recent study determined circRNA
profiles in cancer-associated fibroblasts (CAFs) and identified
that CAF-specific circCUL2 conferred the CAF phenotype on normal
fibroblasts and mediated the CAF oncogenic effects by modulating
the miR-203a-3p/MyD88/NF-κB/IL6 axis (26). Likewise, herein, it was proved that
a large number of circRNAs were aberrantly expressed in GC-MSCs,
and GC-MSC-derived circ_0024107 was found to be was pivotal for
GC-MSCs to promote gastric cancer cell lymphatic metastasis. Ba
et al (27) revealed that
gastric cancer cell-derived exosomes probably promoted the
migration and homing of adipose-derived MSCs by inducing their
circRNA deregulation. These findings support the results of the
present study, in that circRNAs are actually deregulated in gastric
cancer-associated MSCs and may play multiple roles in gastric
cancer progression. Furthermore, the alteration of circ_0024017
expression in gastric cancer cells upon GC-MSC-CM treatment and the
knockdown of circ_0024107 in gastric cancer cells eliminated the
tumor-promoting effects of GC-MSCs, which suggested that
circ_0024107 mediates the crosstalk between GC-MSCs and gastric
cancer cells to synergistically promote gastric cancer
metastasis.
A previous study indicated that immune cells, such
as regulatory CD4+ T-cells (Tregs), M2 macrophages,
myeloid-derived suppressor cells and dendritic cells relied on FAO
to adopt immunosuppressive phenotypes to help cancer cell immune
evasion (18). GC-MSCs are
essentially immune cells. Consistently, the data from the present
study verified that GC-MSCs acquired oncogenic functions, dependent
on FAO metabolic reprogramming. A previous study by the authors
confirmed that FAO was indispensable for gastric cancer cells to
sustain a lymphatic metastatic capacity and its activity was
increased along with the enhanced lymphatic metastatic capacity of
gastric cancer cells (19). Recent
studies have reported that MSCs induced FAO in gastric cancer cells
to acquire stemness and chemoresistance (28,29),
which supports the current findings that GC-MSCs promoted cancer
cell migration and invasion by inducing FAO in gastric cancer
cells. Compared to the findings of these two previous studies
(28,29), the present study revealed a
different mechanism regulated by GC-MSC-derived circRNA. These data
further highlight the critical role of FAO metabolic reprogramming
in stromal cells and cancer cells for malignant progression. The
targeted blocking of FAO thus represents a promising alternative
approach for cancer therapy.
It has been demonstrated that a large number of
circRNAs are aberrantly expressed in almost all cancer types and
have been shown to play indispensable roles in almost all aspects
of cancer cell biological behavior, including metabolic
reprogramming (30,31). However, only a limited number of
studies have examined the mechanisms of fatty acid metabolism in
cancer by the exploration of circRNAs (32,33).
Herein, it was demonstrated that the FAO mediated by the
circ_0024107-miR-5572/6855-5p-CPT1A axis not only conferred an
oncogenic function on GC-MSCs, but was also involved in gastric
cancer cells treated with GC-MSC-CM. Relative to the gastric cancer
cells treated with BM-MSC-CM, higher levels of circ_0024107 in
gastric cancer cells were induced after GC-MSC-CM treatment.
Therefore, the knockdown of circ_0024107 hampered the oncogenic
role of GC-MSCs attributed to the direct and indirect blocking of
FAO reprogramming in GC-MSCs and gastric cancer cells, which at
least provides an explanation for the unilateral intervention of
circ_0024107 in GC-MSCs being sufficient to inhibit gastric cancer
metastasis in vivo. However, further research is required in
order to elucidate the mechanisms through which GC-MSC-CM affect
the circ_0024107 levels in gastric cancer cells.
A previous study by the authors reported that CPT1A
was an unfavorable prognostic factor for all patients with gastric
cancer (19). However, when
considering MSC abundance in gastric cancer tissues, CPT1A became a
favorable prognostic factor for patients with a decreased abundance
of MSCs in gastric cancer tissues, but not for patients with an
enriched MSC abundance status. These findings indicate a complex
role of CPT1A in gastric cancer and the underlying mechanisms need
to be further elucidated. High levels of the two miRNAs (miR-5572
and miR-6855-5p) were shown to be associated with the favorable
prognosis of patients with a decreased number of MSCs, which
suggests a potential link between the two miRNAs and MSCs in
gastric cancer. Furthermore, apart from miR-5572, the other
regulatory axis molecules were consistently altered in GC-MSCs and
their expression exhibited a good correlation with each other in
MSCs. The overall trend of miR-5572 expression changes was
consistent with the prior findings (miR-5572 expression was
downregulated in GC-MSCs), but without a significant difference.
This discrepancy may be caused by their scattered expression in
these selected MSCs. To further analyze the association between the
regulatory axis molecules and to comprehensively evaluate the
clinical implication of circ_024017, more clinical gastric cancer
samples and GC-MSCs need to be included for further
assessments.
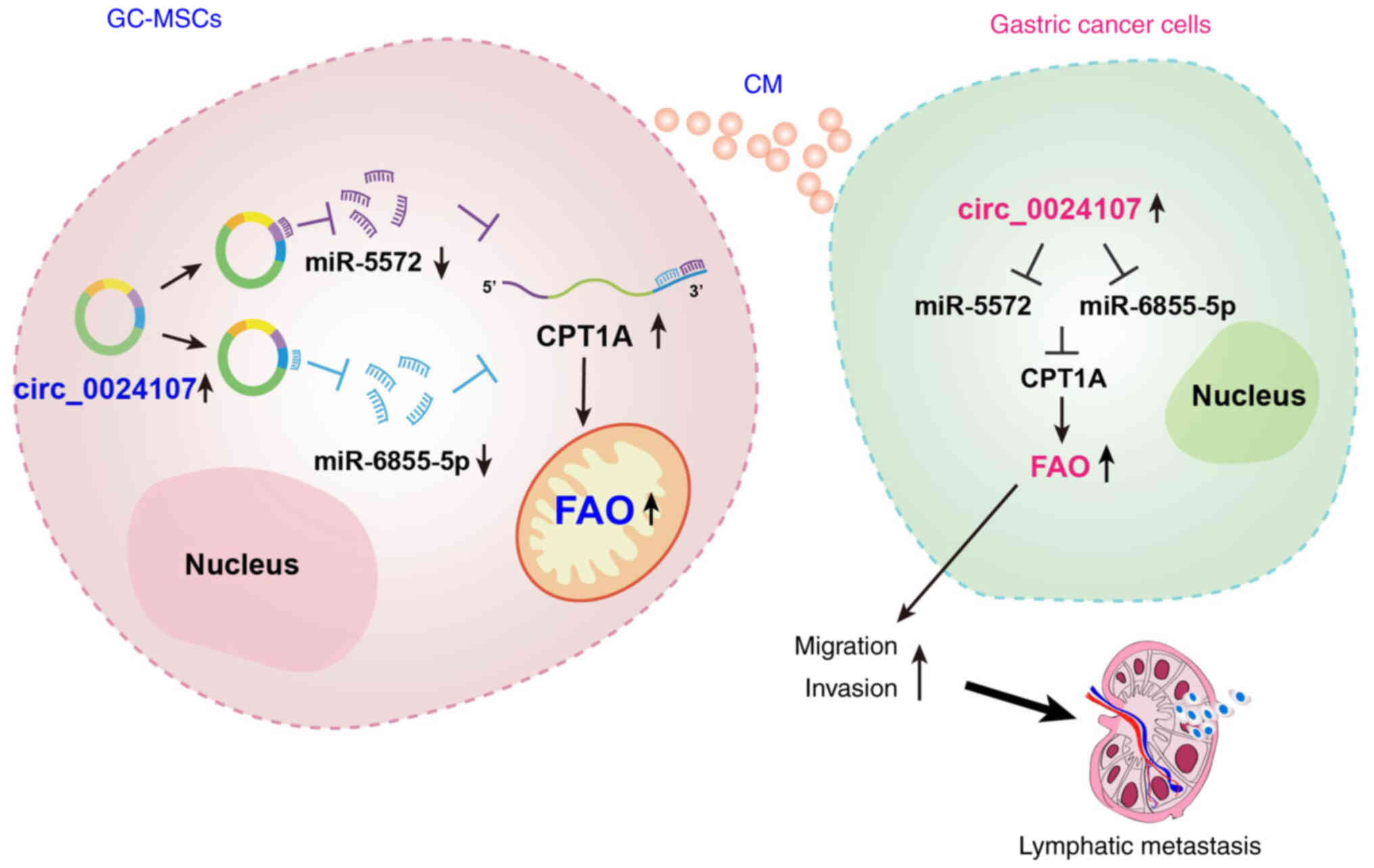
In conclusion, the present study identified that
circ_0024107, as a GC-MSC-derived novel circRNA, induced the FAO
metabolic reprogramming of GC-MSCs via the miR-5572/6855-5p-CPT1A
axis, which enables GC-MSC-CM to upregulate the circ_0024107 level
in gastric cancer cells, thus increasing FAO activity through the
same axis to enhance gastric cancer cell migration and invasion to
form lymphatic metastasis (Fig. 8).
These findings present novel insight into gastric cancer malignant
progression and may lead to the development of potential
therapeutic targets for gastric cancer.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81902510, 81772641 and 81972313),
the Medical Scientific Research Project of Jiangsu Provincial
Health Commission (grant no. Z2022022), the Suzhou Health Youth
Backbone Talent of National Mentor System (grant no. ngg2021043)
and the Postgraduate Research and Practice Innovation Program of
Jiangsu Province (grant no. KYCX22_3719).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW, CW and JX performed the research and wrote the
draft of the manuscript. ZG, XC and JH performed the statistical
analyses and interpreted the data. XC and HD provided technical and
material support, and contributed to the design of the study. WZ
and FH performed critical revisions of the manuscript and were
involved in design of the study. MW and CZ were involved in the
conception and design of the study
Ethics approval and consent to
participate
The collection of tissues from gastric cancer
patients was approved by the Affiliated Tumor Hospital of Nantong
University (approval no. 2021-017). The animal experiment was
approved by the Committee on Use and Care of Animals of Jiangsu
University [approval no. SYXK(Su) 2018-0053].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quail DF and Joyce JA: Microenvironmental
regulation of tumor progression and metastasis. Nat Med.
19:1423–1437. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cao H, Xu W, Qian H, Zhu W, Yan Y, Zhou H,
Zhang X and Xu X, Li J, Chen Z and Xu X: Mesenchymal stem cell-like
cells derived from human gastric cancer tissues. Cancer Lett.
274:61–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang M, Zhao C, Shi H, Zhang B, Zhang L,
Zhang X, Wang S, Wu X, Yang T, Huang F, et al: Deregulated
microRNAs in gastric cancer tissue-derived mesenchymal stem cells:
Novel biomarkers and a mechanism for gastric cancer. Br J Cancer.
110:1199–1210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang F, Wang M, Yang T, Cai J, Zhang Q,
Sun Z, Wu X, Zhang X, Zhu W, Qian H and Xu W: Gastric
cancer-derived MSC-secreted PDGF-DD promotes gastric cancer
progression. J Cancer Res Clin Oncol. 140:1835–1848. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu M, Wang M, Yang F, Tian Y, Cai J, Yang
H, Fu H, Mao F, Zhu W, Qian H and Xu W: miR-155-5p inhibition
promotes the transition of bone marrow mesenchymal stem cells to
gastric cancer tissue derived MSC-like cells via NF-κB p65
activation. Oncotarget. 7:16567–16580. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun L, Huang C, Zhu M, Guo S, Gao Q, Wang
Q, Chen B, Li R, Zhao Y, Wang M, et al: Gastric cancer mesenchymal
stem cells regulate PD-L1-CTCF enhancing cancer stem cell-like
properties and tumorigenesis. Theranostics. 10:11950–11962. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang M, Chen B, Sun XX, Zhao XD, Zhao YY,
Sun L, Xu CG, Shen B, Su ZL, Xu WR and Zhu W: Gastric cancer
tissue-derived mesenchymal stem cells impact peripheral blood
mononuclear cells via disruption of Treg/Th17 balance to promote
gastric cancer progression. Exp Cell Res. 361:19–29. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li W, Zhang X, Wu F, Zhou Y, Bao Z, Li H,
Zheng P and Zhao S: Gastric cancer-derived mesenchymal stromal
cells trigger M2 macrophage polarization that promotes metastasis
and EMT in gastric cancer. Cell Death Dis. 10:9182019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun L, Wang Q, Chen B, Zhao Y, Shen B,
Wang H, Xu J, Zhu M, Zhao X, Xu C, et al: Gastric cancer
mesenchymal stem cells derived IL-8 induces PD-L1 expression in
gastric cancer cells via STAT3/mTOR-c-Myc signal axis. Cell Death
Dis. 9:9282018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang M, Gong Z, Zhao X, Yu W, Huang F and
Dong H: Circular RNAs emerge as important regulators with great
potential for clinical application in gastric cancer. Biomark Med.
15:69–82. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shan C, Zhang Y, Hao X, Gao J, Chen X and
Wang K: Biogenesis, functions and clinical significance of circRNAs
in gastric cancer. Mol Cancer. 18:1362019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao L, Liu Y, Zhang S, Wei L, Cheng H and
Wang J and Wang J: Impacts and mechanisms of metabolic
reprogramming of tumor microenvironment for immunotherapy in
gastric cancer. Cell Death Dis. 13:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui MY, Yi X, Zhu DX and Wu J: Aberrant
lipid metabolism reprogramming and immune microenvironment for
gastric cancer: A literature review. Transl Cancer Res.
10:3829–342. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huo J, Guan J and Li Y: Metabolism
reprogramming signature associated with stromal cells abundance in
tumor microenvironment improve prognostic risk classification for
gastric cancer. BMC Gastroenterol. 22:3642022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang M, Zhao X, Qiu R, Gong Z, Huang F, Yu
W, Shen B, Sha X, Dong H, Huang J, et al: Lymph node
metastasis-derived gastric cancer cells educate bone marrow-derived
mesenchymal stem cells via YAP signaling activation by exosomal
Wnt5a. Oncogene. 40:2296–2308. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma Y, Temkin SM, Hawkridge AM, Guo C, Wang
W, Wang XY and Fang X: Fatty acid oxidation: An emerging facet of
metabolic transformation in cancer. Cancer Lett. 435:92–100. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang M, Yu W, Cao X, Gu H, Huang J, Wu C,
Huang J, Wu C, Wang L, Sha X, et al: Exosomal CD44 transmits lymph
node metastatic capacity between gastric cancer cells via
YAP-CPT1A-mediated FAO reprogramming. Front Oncol. 12:8601752022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim SJ, Khadka D and Seo JH: Interplay
between solid tumors and tumor microenvironment. Front Immunol.
13:8827182022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pavlova NN and Thompson CB: The emerging
hallmarks of cancer metabolism. Cell Metab. 23:27–47. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dey P, Kimmelman AC and DePinho RA:
Metabolic codependencies in the tumor microenvironment. Cancer
Discov. 11:1067–1081. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang W, Fu K, Sun H, Rong D, Wang H and
Cao H: CircRNA microarray profiling identifies a novel circulating
biomarker for detection of gastric cancer. Mol Cancer. 17:1372018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Feng Z, Xu Y, Jiang S, Zhang Q,
Zhang Z, Wang K, Li X, Xu L, Yuan M, et al: Novel roles of LSECtin
in gastric cancer cell adhesion, migration, invasion, and lymphatic
metastasis. Cell Death Dis. 13:5932022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng S, Hu C, Lin H, Li G, Xia R, Zhang
X, Su D, Li Z, Zhou Q, Chen R, et al: circCUL2 induces an
inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via
the activation of the MyD88-dependent NF-κB signaling pathway. J
Exp Clin Cancer Res. 41:712022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ba L, Xue C, Li X, Zhang M, Yang Y, Han Q,
Sun Z and Zhao RC: Gastric cancer cell-derived exosomes can
regulate the biological functions of mesenchymal stem cells by
inducing the expression of circular RNA circ_0004303. Stem Cells
Dev. 30:830–842. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu H, Liu B, Chen Z, Li G and Zhang Z:
MSC-induced lncRNA HCP5 drove fatty acid oxidation through
miR-3619-5p/ AMPK/PGC1alpha/CEBPB axis to promote stemness and
chemo-resistance of gastric cancer. Cell Death Dis. 11:2332020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He W, Liang B, Wang C, Li S, Zhao Y, Huang
Q, Liu Z, Yao Z, Wu Q, Liao W, et al: MSC-regulated lncRNA
MACC1-AS1 promotes stemness and chemoresistance through fatty acid
oxidation in gastric cancer. Oncogene. 38:4637–4654. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen L and Shan G: CircRNA in cancer:
Fundamental mechanism and clinical potential. Cancer Lett.
505:49–57. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T,
Xu T and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu H, Xu J, Gong G, Zhang Y and Wu S:
CircARL8B contributes to the development of breast cancer via
regulating miR-653-5p/HMGA2 axis. Biochem Genet. 59:1648–1665.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Q, Yang Z, Ding H, Li H, Wang W and
Pan Z: CircWHSC1 promotes breast cancer progression by regulating
the FASN/AMPK/mTOR axis through sponging miR-195-5p. Front Oncol.
11:6492422021. View Article : Google Scholar : PubMed/NCBI
|