Introduction
Radiation therapy is an effective method for reducing tumor size and surgical range before resection (1). Radiation therapy can also improve the quality of life of patients by relieving symptoms, including pain and nausea (2). However, each patient experiences a different response to radiation therapy and a number of patients with colorectal cancer are resistant to radiation therapy (3). In addition, patient prognosis can deteriorate in cases with low or no response to treatment due to cancer progression (4). Patients with a good response to radiation therapy can be successfully treated with this type of therapy alone, without requiring additional treatments that may be associated with adverse side effects (5).
Neuromedin U (NMU) is a secreted neuropeptide encoded by the NMU gene. NMU is synthesized as a 174-amino acid precursor peptide that is subsequently processed into a 25-amino acid mature peptide through post-translational modifications (6). NMU is expressed in the central nervous system, intestines, thyroid gland, lymphocytes, spleen, adipose tissue, keratinocytes, endothelial cells and mast cells. In addition, NMU has a variety of functions in different organs, including cell development regulation, pain recognition, blood pressure regulation, muscle contraction, energy homeostasis and sleep regulation. These functions of NMU are mediated through binding with the G protein-coupled receptors (GPCRs) NMU receptor (NMUR)1 and NMUR2 (7,8). NMU contains a C-terminal pentapeptide repeat, which enables binding with its receptors, and N-terminal signaling peptides that are associated with NMU secretion (6). NMUR1 is primarily expressed in the gastrointestinal tract, kidneys, lungs, male genitourinary system, cardiovascular system and immune system, whereas NMUR2 is primarily expressed in the central nervous system (9). These GPCRs can couple with Gq/11 and Gi, but not with Gs (10). Recent studies have reported that NMU is upregulated in various types of cancer, such as lung, skin, thyroid, endometrial and pancreatic cancer, and affects cancer growth and development (11,12). However, the function of NMU in colorectal cancer is not well understood.
The Hippo pathway serves an essential role in tumor development and progression (13). Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) are key factors in the Hippo pathway, and are regulated through phosphorylation by serine-threonine kinases, such as mammalian ste2-like kinase 1/2 and large tumor suppression kinase 1/2 (LATS1/2). Phosphorylation causes YAP and TAZ to bind to 14-3-3 or induce ubiquitination of themselves, resulting in impaired YAP and TAZ nuclear translocation (14,15). YAP acts as a co-transcription factor that combines with transcription factors in the transcriptional enhanced associate domain family to activate target genes, such as connective tissue growth factor (CTGF), cysteine-rich angiogenic inducer 61 (CYR61) and AXL (16). Similarly, TAZ interacts with the peroxisome proliferator-activated receptor, T-box transcription factor 5, runt-related transcription factor 1 and paired box 3 transcription factor families (17). YAP and TAZ can also be activated by LATS1/2 inhibition, which is mediated by Gq/11- and G12/13-coupled receptors, whereas Gs-coupled receptors promote LATS1/2 activity and inhibit YAP and TAZ (18). Furthermore, YAP and TAZ activation has been shown to induce resistance to anticancer therapies, including radiation therapy, in various tumors (19).
In the present study, biomarkers able to predict patient response to radiation therapy were explored to improve the selection of appropriate therapies for patients with colorectal cancer. Among the various candidate genes, NMU was selected as a potential biomarker for radiation resistance in colorectal cancer. Subsequently, the molecular mechanism underlying the effects of NMU on colorectal cancer was identified.
Materials and methods
Cell culture
HCT-8, HT-29 and HCT 116 cells were cultured in Roswell Park Memorial Institute medium (Welgene, Inc.), supplemented with 10% fetal bovine serum (Corning, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Welgene, Inc.). These cells (5×105) were plated in 100-mm culture dishes and were cultured at 37°C in a 5% CO2 humidified incubator. When the cells achieved 70–80% confluence, subculture was performed. For 2-Gy ionizing radiation (IR) treatment, cells were γ-irradiated for 34 sec from a 137 Cs γ-ray source (GC-3000 Elan; Atomic Energy of Canada, Ltd.) administered at a dose of 3.5 Gy/min.
Analysis of The Cancer Genome Atlas (TCGA) data analysis
Gene expression levels (n=478) were obtained from the TCGA data portal (http://tcga-data.nci.nih.gov/tcga/) and cBioPortal (http://www.cbioportal.org/public-portal/), to analyze the expression profile and prognostic significance of the NMU gene in patients with colorectal cancer. To calculate the effect of NMU expression on patient survival, UALCAN (ualcan.path.uab.edu) was used to create a Kaplan-Meier plot with TCGA data. The fragments per kilobase of transcripts per million (FPKM) for each gene were used for quantification of expression and patients were divided based on the expression level into one of the two groups: NMU low (<3.0 FPKM) or NMU high (>3.0 FPKM).
Cell counting assay
HCT-8, HT-29 and HCT 116 cells were detached by treatment with trypsin-EDTA and stained with 0.1% trypan blue for 5 min at room temperature. Cell numbers were measured using a LUNA-FX7 Automated Cell Counter (Logos Biosystems; Aligned Genetics).
Clonogenic cell survival assay
Cells were seeded into 6-well cell culture plates at 100, 200 or 500 cells/well. After seeding, cells were irradiated with 0, 2 or 3 Gy IR for 0, 34 or 51 sec using a GC-3000 Elan (Atomic Energy of Canada, Ltd.). After irradiation, cells were incubated in a 5% CO2 humidified incubator at 37°C for 2 weeks. When a single cell grew into a colony (>50 cells), the culture medium was removed and cells were fixed in 4% paraformaldehyde at 4°C for 20 min. After removing the fixation buffer, colonies were stained with 0.5% crystal violet solution at room temperature for 30 min. The crystal violet solution was gently removed and the number of colonies in the entire well was counted, and relative survival fraction was calculated by plating efficiency (PE) and surviving fraction (SF). PE=number of colonies formed/number of cells seeded ×100. SF=number of colonies formed after IR treatment/number of cells seeded × PE.
Wound-healing assay
Cells were seeded at 3×105 cells/well in 24-well plates. After cells became 100% confluent, scratch wounds were made using a 1-mm pipette tip, and the cells were incubated at 37°C in RPMI media containing 2% serum for 24 h. Wound healing was observed using an optical microscope (Eclipse Ts2; Nikon Corporation) and images were obtained using an MShot Digital Imaging System V1.1.6 (Guangzhou Micro-shot Technology Co., Ltd.) and analyzed using ImageJ software V1.53 (National Institutes of Health).
TUNEL staining assay
Cells were seeded at 1×104 cells/well in 8-well slides and incubated at 37°C for 24 h. Each slide was exposed to 2 Gy radiation at room temperature. After 24 h, the culture medium was removed, and the cells were fixed using 4% paraformaldehyde for 25 min at 4°C, followed by permeabilization with 0.2% Triton X-100 at room temperature for 5 min. TUNEL staining was performed using a DeadEND™ Fluorometric TUNEL system kit (Promega Corporation). The cells were stained in a Coplin jar by immersing the slides in 40 ml propidium iodide solution freshly diluted to 1 µg/ml in PBS for 10 min at room temperature in the dark and mounted by 50 µl VECTASHIELD with DAPI (cat. no. H-1200; Vector Laboratories, Inc.) at room temperature. The fluorescence intensity was measured using a confocal microscope (LSM 900; Carl Zeiss AG).
Total RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
RNA was extracted from cells using Tri-RNA Reagent (Favorgen), according to the manufacturer's protocol. Subsequently, 1 µg extracted RNA was subjected to complementary cDNA synthesis using the All-In-One cDNA Master Mix (CellScript™, CellSafe) at 42°C for 1 h. qPCR amplification was performed using the qPCR SYBR 2X Master Kit (cat. no. 18102; Mbiotech) and gene-specific oligonucleotide primer pairs (Table I) from Bioneer Corporation. Amplification was conducted with the CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories, Inc.). The thermocycling conditions consisted of initial denaturation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec and annealing/extension at 60°C for 30 sec. GAPDH expression levels were used to normalize the mRNA expression levels in each sample. The 2−ΔΔCq method was used to calculate the relative gene expression (20).
 |
Table I.
Oligonucleotide primers used for reverse transcription-quantitative PCR.
|
Table I.
Oligonucleotide primers used for reverse transcription-quantitative PCR.
| Gene |
Sequence |
| NMU |
F: 5′-CACAGAAGTTGGGCAAGTCA-3′ |
| |
R: 5′-TGCTGACCTTCTTCCATTCC-3′ |
| NMUR1 |
F: 5′-CAATCTGTGCCACATACCTG-3′ |
| |
R: 5′-AAACAGTAGCGTGCGGAAAT-3′ |
| NMUR2 |
F: 5′-ACTTCTTCCTCCCCGTGTCT-3′ |
| |
R: 5′-GCGCCACATCTCATAGACCT-3′ |
| AXL |
F: 5′-ACACCCCAGAGGTGCTAATG-3′ |
| |
R: 5′-ACGAGAAGGCAGGAGTTGAA-3′ |
| CTGF |
F: 5′-CTTGCGAAGCTGACCTGGAAGA-3′ |
| |
R: 5′-CCGTCGGTACATACTCCACAGA-3′ |
| CYR61 |
F: 5′-GGAAAAGGCAGCTCACTGAAGC-3′ |
| |
R: 5′-GGAGATACCAGTTCCACAGGTC-3′ |
| GAPDH |
F: 5′-CGACCACTTTGTCAAGCTCA-3′ |
| |
R: 5′-AGGGGTCTACATGGCAACTG-3′ |
Transfection
HCT-8, HT-29 and HCT 116 cells were cultured to ~70% confluence in 60-mm plates. For NMU and NMUR1 knockdown, 20 nM small interfering RNAs (siRNAs) targeting NMU or NMUR1, or control siRNA (Table II) were transfected using G-fectin transfection reagent (Genolution, Inc.) overnight at 37°C. After changing the media, the cells were incubated at 37°C for 24 h and subsequent experiments for proliferation, migration and TUNEL assays were performed. To assess the exogenous effect of NMU on NMUR1 knockdown, the cells were treated with 1 µM NMU recombinant protein (NMU-25; cat. no. ab141007; Abcam) at 37°C for 24 h. For NMU overexpression, 1 µg NMU-expression pCMV3 vector (Sino Biological, Inc.) was transfected using Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. An empty vector was used as the control.
 |
Table II.
Duplex siRNA sequences used for transfection.
|
Table II.
Duplex siRNA sequences used for transfection.
| siRNA |
Sequence |
| NMU |
Sense: 5′-CUUUGCAAGUCAAAGUCGAUU-3′ |
| |
Antisense: 5′-UCGACUUUGACUUGCAAAGUU-3′ |
| NMUR1 |
Sense: 5′-GCACGCUACUGUUUGAGAU-3′ |
| |
Antisense: 5′-GCACGCCUACCAACUACUA-3′ |
| Negative control |
Sense: 5′-CCUCGUGCCGUUCCAUCAGGUAGUU-3′ |
| |
Antisense: 5′-CUACCUGAUGGAACGGCACGAGGUU-3′ |
Total protein extraction and nuclear fractionation
Cells were washed twice with phosphate-buffered saline and harvested with protein extraction solution (Elpis Biotech, Inc.) containing protease-phosphatase inhibitor (Cell Signaling Technology, Inc.). The nuclear fraction was prepared using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Protein concentrations were quantified with the bicinchoninic acid method (Pierce Biotechnology, Inc.) using 1 µg/ml bovine serum albumin (Pierce; Thermo Fisher Scientific, Inc.) as a standard.
Western blot analysis
Proteins (25 µg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10% gels and were transferred to a nitrocellulose membrane. The membranes were blocked using 5% skimmed milk (MilliporeSigma) for 1 h at room temperature, and soaked in primary antibody solutions (diluted to 1:1,000) against GAPDH (ca. no. sc47724), β-actin (cat. no. sc81178; Santa Cruz Biotechnology, Inc.), lamin A/C (cat. no. 4777), YAP (cat. no. 4912), TAZ (cat. no. 70148), LATS1 (cat. no. 9153), phosphorylated (p)-YAP (cat. no. 4911), p-TAZ (cat. no. 70148) and p-LATS1 (cat. no. 8654) (all Cell Signaling Technology, Inc.) overnight at 4°C. Membranes were then washed five times (10 min/wash) with Tris-buffer containing 1% Tween 20 and were incubated with horseradish peroxidase (HRP)-conjugated IgG goat anti-rabbit (cat. no. 7074) or HRP-conjugated IgG goat anti-mouse antibodies (cat. no. 7076) (diluted to 1:10,000; Cell Signaling Technology, Inc.) for 1 h at room temperature. Protein expression was detected using Western Lighting Plus-ECL (PerkinElmer, Inc.).
Clinical data and heatmap generation
A total of 6 patients with pathologically proven locally advanced rectal cancer, tested between April 2018 and May 2019, whose neoadjuvant chemo-radiation therapy outcomes were categorized as complete response (n=3) or poor response (n=3) based on total mesorectal excision specimens, were selected for inclusion. Clinical data from patients with colorectal cancer were provided by the Korea Institute of Radiological and Medical Sciences (Seoul, South Korea; Table III). For the genetic information of patients with colorectal cancer, microarray data obtained from our previous study were used (5). Heatmaps were constructed using MeV software V4.7 (http://mev.tm4.org/) based on the provided clinical data.
 |
Table III.
Clinical patient information.
|
Table III.
Clinical patient information.
| Characteristic |
RR-1 |
RR-2 |
RR-3 |
RS-1 |
RS-2 |
RS-3 |
| Sex |
Male |
Male |
Female |
Male |
Male |
Female |
| Age, years |
77 |
71 |
76 |
49 |
64 |
62 |
| Pre-RT MRI tumor length, cm |
4.2 |
5.4 |
6.5 |
6.5 |
3.8 |
3.5 |
| Post-RT MRI tumor length, cm |
2.3 |
5.3 |
2.5 |
4 |
1.6 |
2.5 |
| Tumor length on pathologya, cm |
2.5 |
7.5 |
3.5 |
3.8 |
1 |
1.2 |
| Regression grade after RT |
Minimal |
Minimal |
Minimal |
Complete |
Complete |
Complete |
Statistical analysis
Data are presented as the mean ± standard deviation from three independent experiments and were analyzed using GraphPad Prism 9.0 software (GraphPad Software, Inc.). The statistical significance of differences between groups was determined using one-way ANOVA and subsequent Tukey's multiple comparison post hoc test, or using Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of NMU is increased in radiation-resistant colorectal cancer
Microarray analysis was performed in our previous study (5) to confirm the gene transcriptional characteristics of radiation-resistant (RR) colorectal cancer. Six patients with colorectal cancer were classified as RR or radiation-sensitive (RS) according to tumor regression grade following radiation therapy. The patients were assessed by pelvis magnetic resonance imaging and 18-fluoro-2-deoxy-glucose positron emission tomography scanning before and after radiotherapy (5). The clinical information of these patients is summarized in Table III. Normal tissue was not used as a control for patients with cancer, but cancer tissues were compared according to the radiosensitivity of patients with cancer. The microarray analysis showed that 325 genes were expressed at higher levels in RR patients than in RS patients (cutoff of fold-change >2). The differential expression of the top 50 genes is presented in the heatmap in Fig. 1A. RT-qPCR was performed on the top 50 differentially expressed genes to verify the microarray results. Our previous study (5) showed that 40 genes were expressed at higher levels in RR colorectal cancer than in RS colorectal cancer (cutoff of fold-change >2). The expression levels of 20 genes are presented in Fig. 1B. By analyzing these data, NMU was selected as a candidate biomarker for RR colorectal cancer. The relationship between NMU expression and prognosis among patients with colorectal cancer was examined using data from The Cancer Genome Atlas, which revealed a significant association between NMU expression (Fig. 1C) and prognosis (Fig. 1D). Based on these data, NMU may affect the development of radiation resistance and the progression of colorectal cancer.
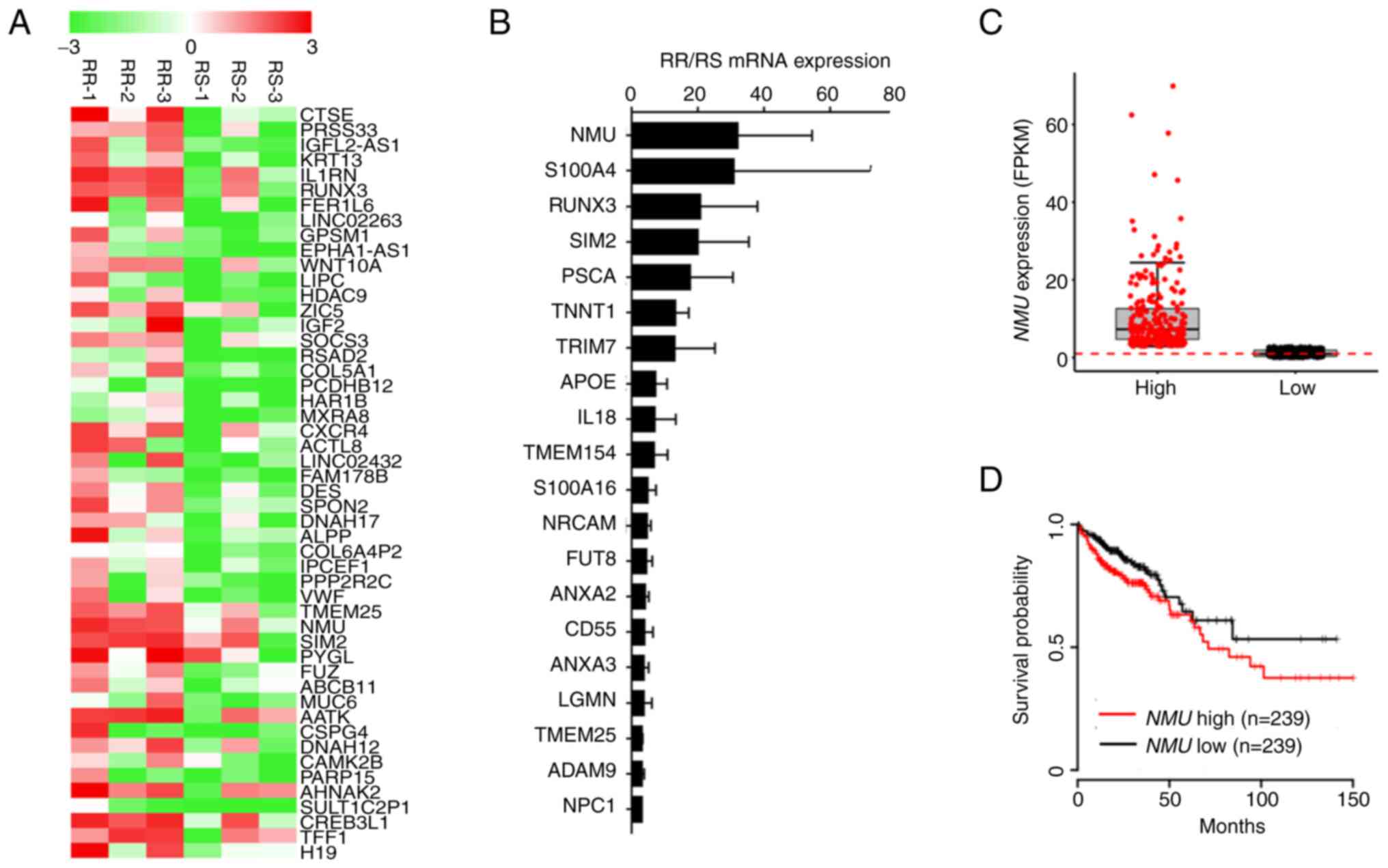 |
Figure 1.
Expression of NMU is increased in RR colorectal cancer tissues. (A) Heatmap showing 50 genes that were highly expressed in RR colorectal cancer organoids (RR-1, −2 and −3) compared with RS colorectal cancer organoids (RS-1, −2 and −3). (B) Reverse transcription-quantitative PCR data showing 20 genes that were highly expressed in RR colorectal cancer organoids (RR-1, −2 and −3) compared with RS colorectal cancer organoids (RS-1, −2 and −3). (C) NMU expression levels for each patient group. (D) Kaplan-Meier plot of survival among 478 patients with colorectal cancer based on NMU expression levels. Patients were classified as having high (n=239) or low (n=239) NMU expression levels. These data were obtained from The Cancer Genome Atlas. FPKM, fragments per kilobase of transcript per million; NMU, neuromedin U; RR radiation-resistant; RS, radiation-sensitive.
|
NMU knockdown inhibits colorectal cancer cell proliferation, colony formation and migration
The present study first confirmed the expression levels of NMU and its receptors, NMUR1 and NMUR2, in HCT-8, HT-29 and HCT 116 colorectal cancer cells. NMU expression was high in HCT-8 cells, and relatively low in HT-29 and HCT 116 cells. Conversely, the expression levels of NMUR1 and NMUR2 were highest in HCT 116 cells (Fig. S1). Given the observed relationship between NMU upregulation and poor patient survival, the present study investigated whether NMU knockdown affected colorectal cancer cell proliferation in the presence and absence of radiation. siRNA targeting NMU was used to transfect HCT-8 and HCT 116 colorectal cancer cells, which resulted in an NMU depletion efficiency of ~40% (Fig. 2A). Colorectal cancer cells transfected with NMU siRNA exhibited reduced cell proliferation and this inhibition was further enhanced when these cells were irradiated (Fig. 2B). HCT-8 and HCT 116 cells transfected with NMU siRNA showed 17 and 34% reductions in colony-forming ability following 3 Gy irradiation in the clonogenic assay compared with that in cells transfected with control siRNA (Fig. 2C). Subsequently, the effect of NMU knockdown on the migration of colorectal cancer cells was assessed using the monolayer wound-healing assay. NMU siRNA reduced the migration of HCT-8 and HCT 116 cells, and migration was further reduced when these cells were exposed to radiation (Fig. 2D). TUNEL staining confirmed that NMU depletion increased the irradiation-induced apoptosis of colorectal cancer cells (Fig. 2E). These data indicated that NMU knockdown may affect cell survival, cell migration and radiation resistance in colorectal cancer cells.
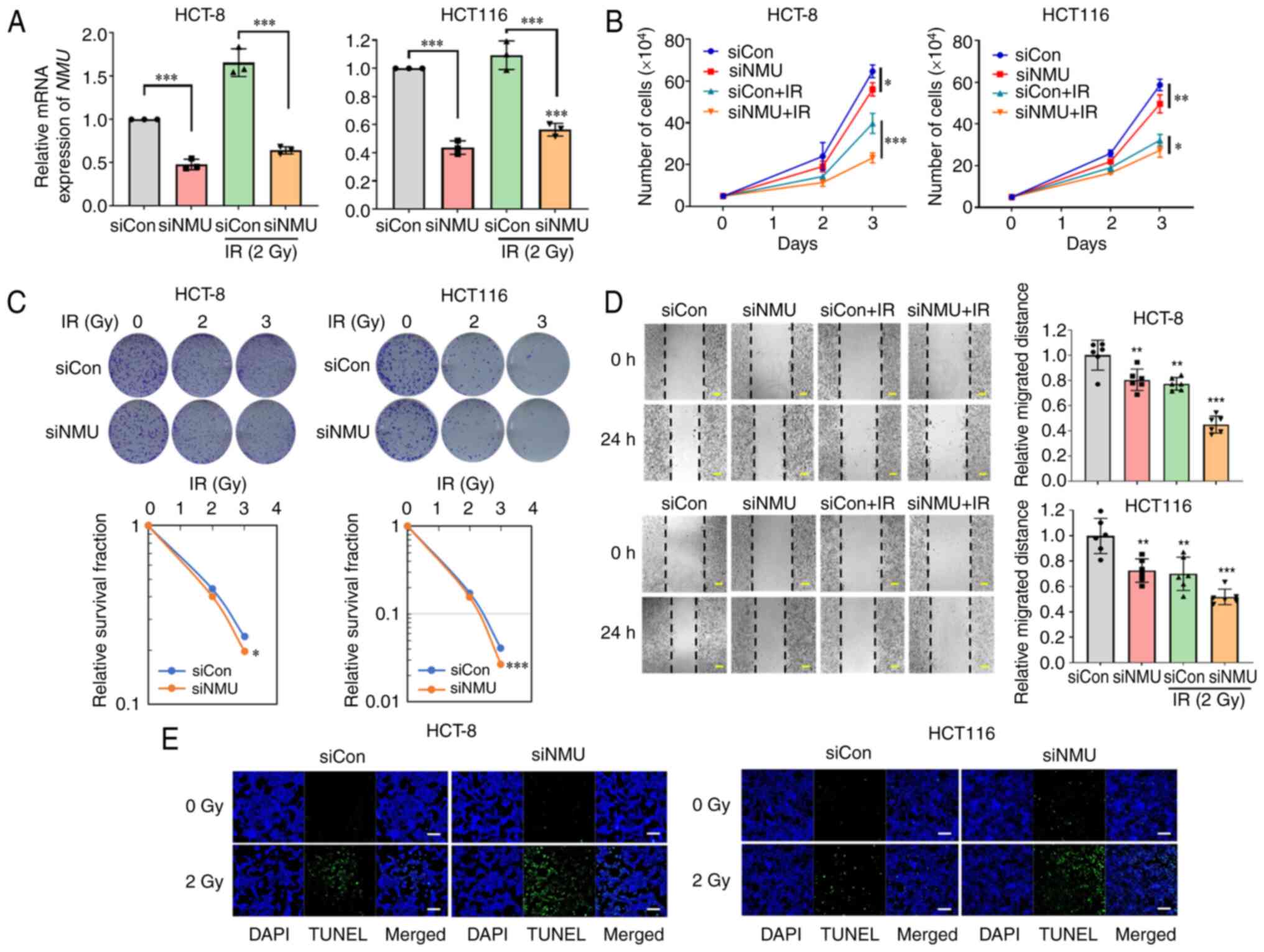 |
Figure 2.
NMU knockdown inhibits colorectal cancer cell proliferation, colony formation and migration. (A) HCT-8 and HCT 116 cells were transfected with 20 nM siNMU or siCon. Cells were harvested 6 h after exposure to 2 Gy radiation. **P<0.01 and ***P<0.001 compared with siCon-transfected group with or without IR. (B) siNMU-transfected cells were incubated for 3 days after treatment with 2 Gy radiation. The numbers of live cells were measured via Cell Counting Kit-8 assay. *P<0.05, **P<0.01 and ***P<0.001 compared with siCon-transfected group with or without IR. (C) HCT-8 and HCT 116 cells were transfected with siNMU or siCon. The cells were incubated for 2 weeks after exposure to the indicated radiation doses. The number of colonies was measured using ImageJ software. *P<0.05 and ***P<0.001 compared with siCon-transfected group. (D) Confluent siNMU-transfected cells were plated and exposed to 2 Gy radiation when the cells reached 100% confluence. Images were captured for 24 h, starting immediately after the cells were wounded. The widths of wounds were measured using ImageJ software. Scale bars, 200 µm. *P<0.05, **P<0.01 and ***P<0.001 compared with siCon-transfected group. (E) siNMU-transfected cells were exposed to 2 Gy radiation for 24 h and were stained with TUNEL (green) and DAPI (blue). Scale bars, 50 µm. Con, control; IR, irradiation; NMU, neuromedin U; si, small interfering.
|
NMU overexpression induces cell proliferation and radiation resistance in colorectal cancer cells
To evaluate the effect of NMU overexpression on colorectal cancer cells, HT-29 and HCT 116 cells were transfected with a DNA vector containing the NMU open reading frame, resulting in the successful overexpression of NMU (Fig. 3A). Following exposure to 3 Gy irradiation, NMU-overexpressing HT-29 and HCT 116 cells showed 22 and 200% increases in colony formation compared with that in control vector-transfected cells (Fig. 3C). The proliferation and migration of colorectal cancer cells were increased following NMU overexpression. In addition, NMU overexpression restored or offset reductions in proliferation and migration induced by irradiation (Fig. 3B and D). Irradiation-induced apoptosis was also reduced in cells with NMU overexpression (Fig. 3E). These data indicated that NMU overexpression may promote malignant phenotypes and protect against the irradiation-induced apoptosis of colorectal cancer cells.
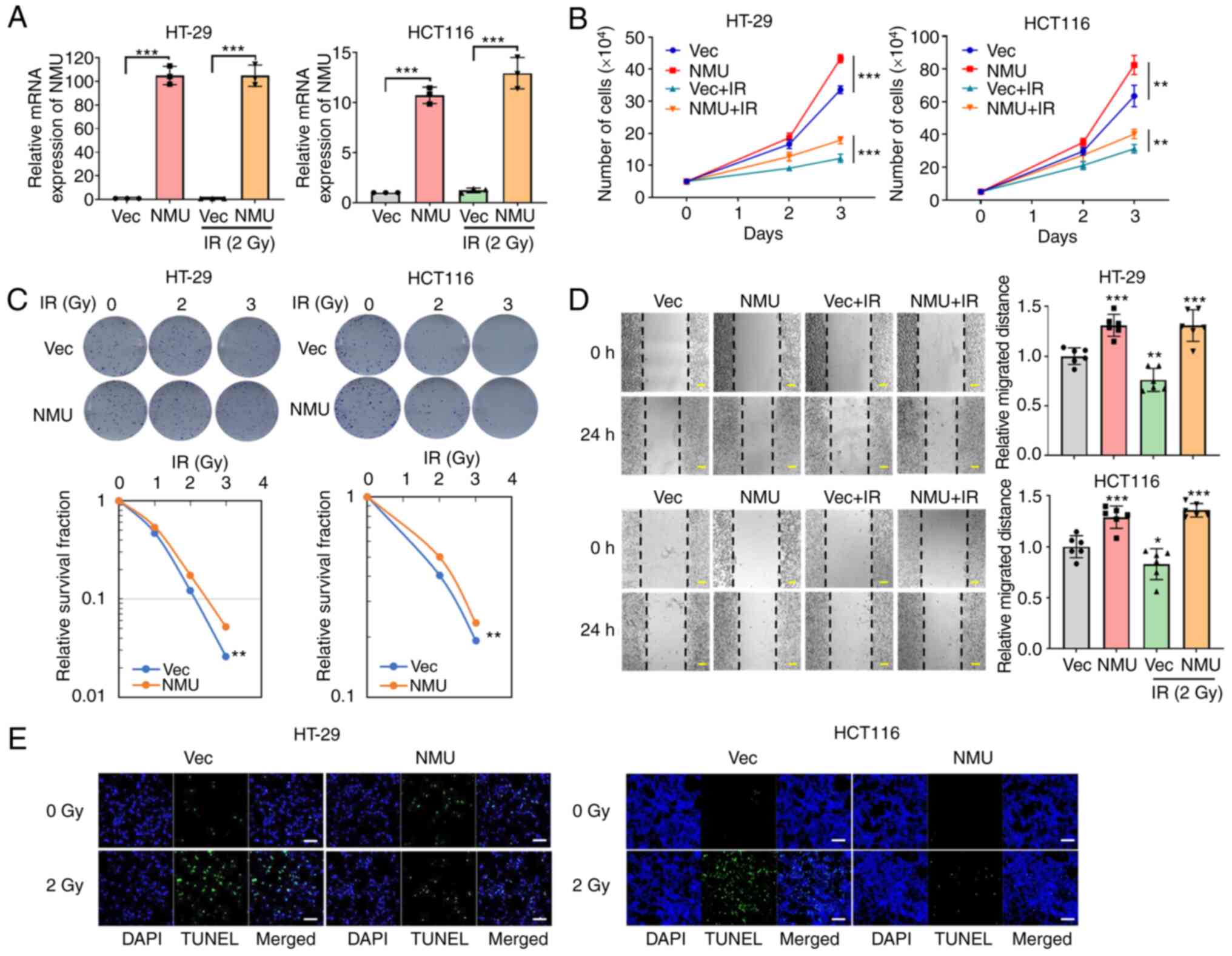 |
Figure 3.
NMU overexpression induces cell proliferation and radiation resistance in colorectal cancer cells. (A) HT-29 and HCT 116 cells were transfected with NMU or control DNA vectors. The cells were harvested 6 h after exposure to 2 Gy radiation. ***P<0.001 for comparison between Vec-transfected group and NMU-transfected group. (B) Numbers of live cells were measured via Cell Counting Kit-8 assay after irradiating cells with 2 Gy. **P<0.01 compared with Vec-transfected group in HCT 116 and ***P<0.001 compared with Vec-transfected group in HT-29. (C) NMU-overexpressing cells were exposed to the indicated radiation doses for 2 weeks. The number of colonies was measured using ImageJ software. **P<0.01 compared with Vec-transfected group. (D) Cells were plated and grown to 100% confluence in a 60-mm culture dish. Images were captured 24 h after transfection with NMU vector and treatment with 2 Gy radiation. The widths of wounds were measured using ImageJ software. Scale bars, 200 µm. **P<0.01 and ***P<0.001 compared with Vec-transfected group. (E) NMU-overexpressing cells were exposed to 2 Gy radiation for 24 h and were stained with TUNEL (green) and DAPI (blue). Scale bars, 50 µm. Con, control; IR, irradiation; NMU, neuromedin U; Vec, empty vector.
|
NMU contributes to YAP and TAZ activity in colorectal cancer cells
YAP and TAZ are associated with radiation resistance in various types of cancer (21,22). To investigate whether NMU is involved in the Hippo pathway, which regulates YAP and TAZ activity, changes in YAP and TAZ expression following NMU knockdown or overexpression were determined by western blotting. In HT-29 cells, NMU depletion increased the expression levels of p-LATS1, p-YAP and p-TAZ, and similar results were observed under irradiated conditions (Fig. 4A). To confirm that NMU enhances the nuclear translocation of YAP and TAZ, which are known to induce cancer cell proliferation and tumorigenesis, nuclear fractionation and western blotting were performed. siRNA-mediated NMU depletion decreased the nuclear translocation of YAP and TAZ (Fig. 4B). By contrast, NMU overexpression reduced the expression levels of p-LATS1, p-YAP and p-TAZ (Fig. 4C), and increased the nuclear translocation of YAP and TAZ (Fig. 4D). Similar outcomes were observed under irradiated conditions. Taken together, these results suggested that NMU may be involved in regulating YAP and TAZ activity in colorectal cancer cells.
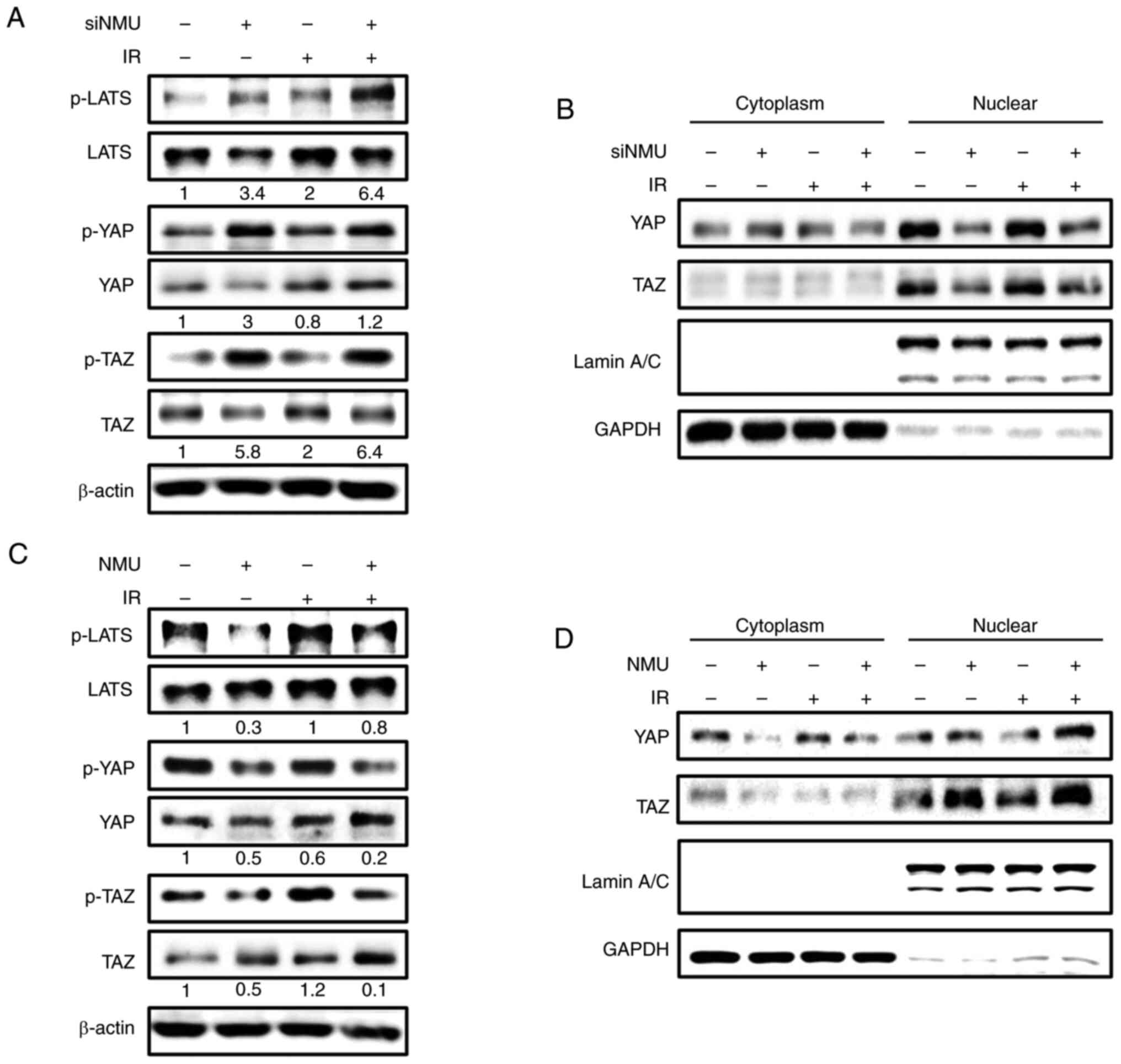 |
Figure 4.
NMU contributes to YAP and TAZ activity in colorectal cancer cells. (A) HT-29 cells were transfected with siNMU (+) or control siRNA (−). Cells were harvested 6 h after exposure to 2 Gy radiation. Phosphorylation levels of LATS1, YAP and TAZ were measured by western blotting and normalized to the total amount of each protein. β-actin was used as the loading control. (B) siNMU-transfected cells were harvested 6 h after exposure to 2 Gy radiation, and the cytoplasm and nucleus were isolated. The presence of the nuclear marker lamin A/C and the cytoplasmic marker GAPDH in the appropriate fractions was confirmed by western blotting. (C) HT-29 cells were transfected with NMU cDNA and exposed to 2 Gy radiation. Phosphorylation levels of LATS1, YAP and TAZ were measured by western blotting and normalized to the total amount of each protein. (D) Cytoplasm and nucleus were isolated from NMU-overexpressing cells. Protein expression levels of YAP and TAZ in the cytoplasmic and nuclear fractions were measured by western blotting. A representative image from three independent experiments is shown. IR, irradiation; LATS, large tumor suppression kinase; NMU, neuromedin U; p-, phosphorylated; si, small interfering; TAZ, transcriptional co-activator with PDZ-binding motif; YAP, Yes-associated protein.
|
NMU activates YAP and TAZ, and induces colorectal cancer cell proliferation and migration in an NMUR1-dependent manner
The present study investigated whether agonist activation of NMUR1 by NMU affects colorectal cancer cell proliferation and migration. In HCT 116 cells, NMUR1 was depleted using siRNA (Fig. 5A), followed by treatment with NMU-25, which is the functional form of NMU in humans. Treatment with NMU-25 induced colorectal cancer cell proliferation and migration, similar to the effects observed with NMU overexpression; however, NMU-25 had no effect on cells transfected with NMUR1 siRNA (Fig. 5B and C). Nuclear fractionation and western blotting revealed increased nuclear translocation of YAP and TAZ in colorectal cancer cells following treatment with NMU-25, but the effects of NMU-25 treatment were attenuated by NMUR1 siRNA (Fig. 5D). As well as the increased nuclear translocation of YAP and TAZ in cells treated with NMU-25, the expression levels of the YAP and TAZ transcriptional target genes AXL, CTGF and CYR61 were increased, and these effects were abolished in NMUR1-depleted cells (Fig. 5E). These results suggested that the NMU-indued migratory phenotypes observed in colorectal cancer cells involve YAP and TAZ activity and are mediated through NMUR1 signaling.
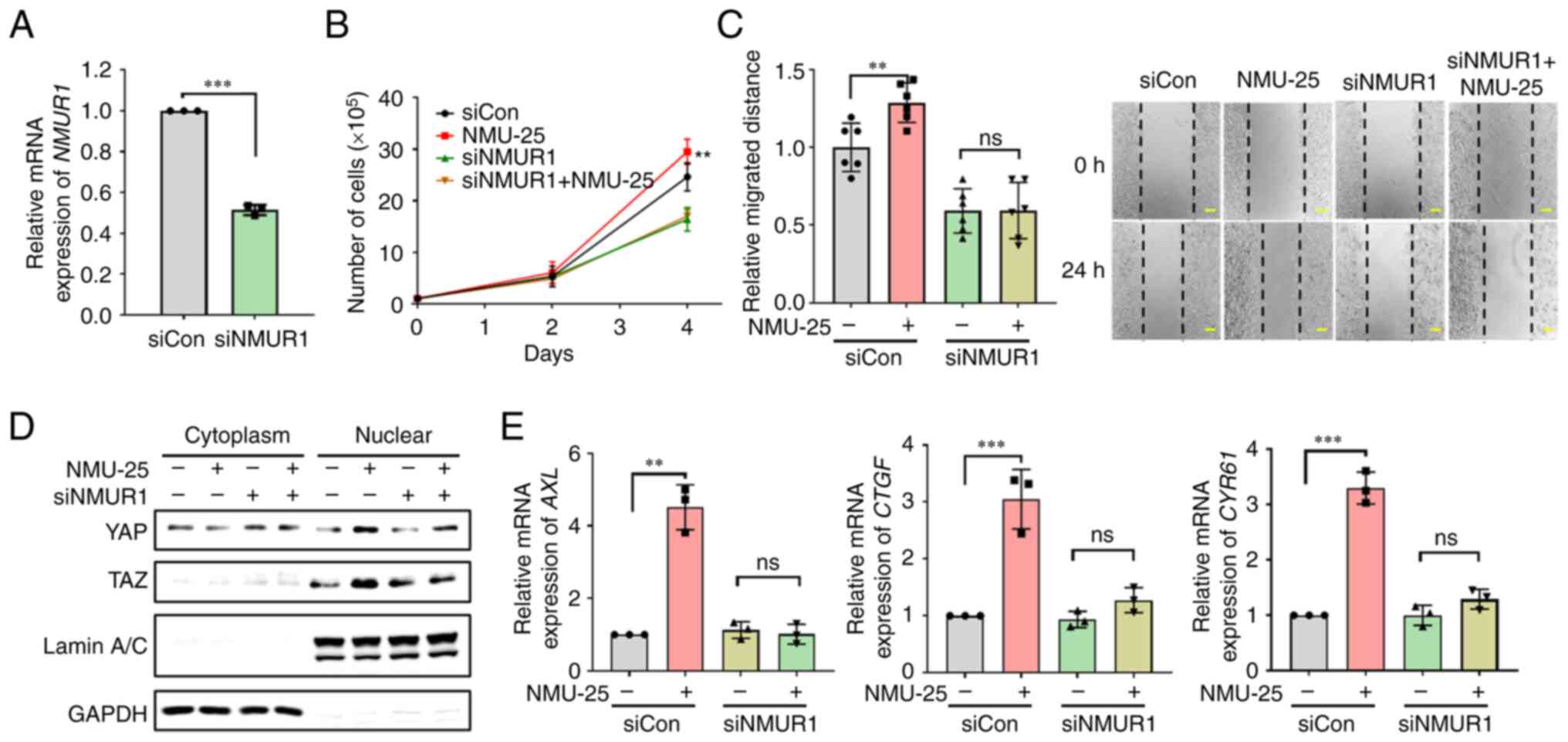 |
Figure 5.
NMU activates YAP and TAZ, and induces colorectal cancer proliferation and migration in an NMUR1-dependent manner. (A) HCT 116 cells were transfected with 20 nM siNMUR1. ***P<0.001 compared with siCon-transfected group. (B) siNMUR1-transfected cells were treated with 1 µM NMU-25 and incubated for 4 days. The numbers of live cells were measured via Cell Counting Kit-8 assay after seeding the cells for 2 or 4 days. **P<0.01 compared with siCon-transfected group. (C) siNMUR1-transfected cells were grown to 100% confluence and treated with 1 µM NMU-25. Images were captured for 24 h, starting immediately after the cells were wounded. The widths of wounds were measured using ImageJ software. Scale bars, 200 µm. **P<0.01 compared with siCon-transfected group. (D) siNMUR1-transfected cells were harvested 24 h after treatment with 1 µM NMU-25, and the cytoplasmic and nuclear fractions were isolated from cells. The presence of the nuclear marker lamin A/C and the cytoplasmic marker GAPDH in the appropriate fractions was confirmed by western blotting. (E) Relative expression levels of YAP transcriptional target genes was measured in HCT 116 cells. The cells were transfected with 20 nM siNMUR1 and incubated 24 h after treatment with 1 µM NMU-25. Expression levels of AXL, CTGF and CYR61 were measured by reverse transcription-quantitative PCR analysis. **P<0.01 and ***P<0.001 compared with siCon-transfected group. Con, control; CTGF, connective tissue growth factor; CYR61, cysteine-rich angiogenic inducer 61; NMU, neuromedin U; NMUR1, NMU receptor 1; si, small interfering; TAZ, transcriptional co-activator with PDZ-binding motif; YAP, Yes-associated protein.
|
Discussion
Different therapies should be considered for individual patients to maximize effectiveness and minimize side effects in cancer (1). Differences in the reactivity to radiation therapy among patients with colorectal cancer highlight the need to predict reactivity to ensure successful treatment (4,5). The accurate prediction of the reactivity of patients to radiation therapy can facilitate the selection of a suitable treatment and minimize potential side effects. Clinical biomarkers, such as epidermal growth factor receptor (EGFR), p53, Bcl-2, Bax and p21, have been suggested to serve as RR biomarkers; however, the clinical application of RR biomarkers in colorectal cancer remains controversial, and a number of classical biomarkers cannot be used to distinguish patient reactivity to radiation therapy (23,24). In addition, the mechanisms underlying the response to radiation therapy in colorectal cancer have not been fully established. Therefore, it is important to identify biomarkers that are able to predict radiation resistance in colorectal cancer, and to define the functions and mechanisms of potential biomarkers (25).
To identify genes associated with radiation resistance in colorectal cancer, gene expression was analyzed in organoids derived from the biopsy tissues of patients with different reactivities to radiation treatments for colorectal cancer. NMU, which has previously been identified in various types of cancer (11,26) but has not been well studied in colorectal cancer, was selected from the possible candidates for further study. To determine whether NMU could serve as a suitable biomarker candidate for RR colorectal cancer, the functions of NMU in colorectal cancer cells were evaluated through phenotypic observations.
To evaluate whether NMU expression levels were associated with radiation resistance in colorectal cancer cells, the survival rates of cells exposed to various radiation doses were calculated using a clonogenic assay. NMU knockdown significantly reduced the survival rate, with larger effects observed under irradiated conditions. Conversely, NMU overexpression increased the survival rate in response to radiation. NMU knockdown reduced colorectal cancer cell proliferation and migration, and enhanced the effects of radiation on these phenotypes. In addition, NMU knockdown enhanced the irradiation-induced apoptosis of colorectal cancer cells. NMU overexpression produced the opposite outcomes, increasing colorectal cancer cell proliferation and migration, and reducing the impacts of radiation on these phenotypes. NMU overexpression also inhibited radiation-induced apoptosis. These results revealed that NMU expression could regulate the response of colorectal cancer cells to radiation.
NMU has been reported to serve roles in various cancer types (11). In non-small cell lung cancer, NMU knockdown can affect the expression of forkhead box protein M1, which has been suggested to serve as a therapeutic target due to its effects on cell proliferation (27). In clear cell renal cell carcinoma, increased NMU expression due to inactivation of the Von Hippel-Lindau tumor-suppressing gene has been shown to increase cell migration and invasion (28). In endometrial cancer, NMU promotes proliferation, migration and the epithelial-mesenchymal transition (EMT) through Src signaling (29). In pancreatic adenocarcinoma, NMU induces c-Met expression, which increases cell motility and invasiveness (26). In bladder cancer, NMU has been reported to be negatively regulated by Rho GDP dissociation inhibitor 2 (RhoGDI2), and decreased RhoGDI2 expression results in the upregulation of NMU, increased tumor growth and metastasis (30). In human EGFR2-positive breast cancer, NMU upregulation is associated with increased expression of interleukin-6 and EMT markers, accompanied by treatment resistance, aggression, migration and invasion (31). In neuroblastoma, NMU is involved in the development of the tumor microenvironment (32). In acute myeloid leukemia, NMU was selected as a target gene of c-Myb, and changes in cell growth were observed with changes in NMU expression (33). In colorectal cancer, NMU has been shown to be upregulated by Snail, an EMT biomarker, and is associated with early-stage EMT (34) and invasion (35). These reports supported the phenotypic changes observed in colorectal cancer cells in the present study. Various intracellular molecular changes induced by changes in NMU expression likely contributed to the identified RR phenotypes in the present study.
NMU signal transmission generally involves coupling with the GPCRs NMUR1 and NMUR2, which bind to Gq/11 and Gi (10). In addition, the non-canonical NMU receptor neurotensin receptor 1 also binds with Gq/11, Gi/o and G12/13 to transmit signaling (36). Signaling by combinations of GPCRs and G proteins has been reported to affect the Hippo pathway (18), which is involved in growth, drug resistance and radiation resistance in various types of cancer (19). The Hippo pathway was identified as a mediator of the mechanisms underlying NMU-mediated changes leading to RR phenotypes in colorectal cancer cells in the present study.
The activities of the transcriptional regulators YAP and TAZ have been associated with radiation resistance in various types of cancer (37). In urothelial cell carcinoma, YAP depletion promotes radiation-induced DNA damage and apoptosis (38). In endometrial cancer, the depletion of YAP reduces the radiation dose required to induce apoptosis (39). By contrast, YAP activation in medulloblastoma prevents radiation-induced apoptosis and promotes proliferation through IGF2/AKT pathway activation (21). In addition, high YAP expression has been reported in RR head and neck squamous cell carcinoma (40). Moreover, YAP and TAZ activation has been reported to have positive effects on the growth, invasion, migration and cancer stem cell-like attributes in various cancer types, including colorectal cancer (41,42). In the present study, NMU knockdown inhibited the nuclear translocation of YAP and TAZ through LATS1 phosphorylation, whereas NMU overexpression decreased LATS1 phosphorylation and increased YAP and TAZ translocation.
Based on these reports and the experimental results of the present study, NMU appears to activate YAP and TAZ, leading to the induction of RR phenotypes, including enhanced proliferation, migration and survival of colorectal cancer cells. Therefore, NMU may serve as a biomarker of radiation resistance and a potential therapeutic target in colorectal cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from the Korea Institute of Radiological and Medical Sciences (grant no. 50531-2022), which is funded by the Ministry of Science and Information and Communications Technology of the Korean government.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The raw data of RNA sequencing generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE226034).
Authors' contributions
MKS and SRK wrote the original draft of the manuscript. MKS, JEP and SRK investigated the mechanism of NMU. YK and USS analyzed clinical data. JL, EJK, YK and USS were involved in data validation and curation. HK and KSK confirmed the authenticity of all the raw data, and were involved in study supervision. KSK contributed to manuscript editing and funding acquisition. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was conducted according to the guidelines of The Declaration of Helsinki and was approved by the Ethics Committee of Korea Cancer Center Hospital (approval no. KIRAMS-2017-07-001).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Kim YB and Suh CO: Evolution of radiotherapy: High-precision radiotherapy. J Korean Med Assoc. 51:604–611. 2008. View Article : Google Scholar
|
|
2
|
Gunderson LL and Tepper JE: Clinical radiation oncology. Elsevier Health Sciences; 2015
|
|
3
|
Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG, Ru G, Xu HT and Cao JP: Role of miR-100 in the radioresistance of colorectal cancer cells. Am J Cancer Res. 5:545–559. 2015.PubMed/NCBI
|
|
4
|
Jin H, Gao S, Guo H, Ren S, Ji F, Liu Z and Chen X: Re-sensitization of radiation resistant colorectal cancer cells to radiation through inhibition of AMPK pathway. Oncol Lett. 11:3197–3201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee J, Kwon J, Kim D, Park M, Kim K, Bae I, Kim H, Kong J, Kim Y, Shin U and Kim E: Gene expression profiles associated with radio-responsiveness in locally advanced rectal cancer. Biology (Basel). 10:5002021.PubMed/NCBI
|
|
6
|
Austin C, Lo G, Nandha K, Meleagros L and Bloom S: Cloning and characterization of the cDNA encoding the human neuromedin U (NmU) precursor: NmU expression in the human gastrointestinal tract. J Mol Endocrinol. 14:157–169. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Howard AD, Wang R, Pong SS, Mellin TN, Strack A, Guan XM, Zeng Z, Williams DL Jr, Feighner SD, Nunes CN, et al: Identification of receptors for neuromedin U and its role in feeding. Nature. 406:70–74. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martinez VG and O'Driscoll L: Neuromedin U: A multifunctional neuropeptide with pleiotropic roles. Clin Chem. 61:471–482. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shan L, Qiao X, Crona JH, Behan J, Wang S, Laz T, Bayne M, Gustafson EL, Monsma FJ Jr and Hedrick JA: Identification of a novel neuromedin U receptor subtype expressed in the central nervous system. J Biol Chem. 275:39482–39486. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brighton PJ, Szekeres PG and Willars GB: Neuromedin U and its receptors: Structure, function, and physiological roles. Pharmacol Rev. 56:231–248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Przygodzka P, Soboska K, Sochacka E and Boncela J: Neuromedin U: A small peptide in the big world of cancer. Cancers (Basel). 11:13122019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Q, Han L, Ruan S, Shen S, Cao Q, Cai X, Yan Y, Peng B and Hua Y: The prognostic value of neuromedin U in patients with hepatocellular carcinoma. BMC Cancer. 20:952020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan J, Li S, Chi P, Xu Z, Lu X and Huang Y: Lentivirus-mediated RNA interference targeting WWTR1 in human colorectal cancer cells inhibits cell proliferation in vitro and tumor growth in vivo. Oncol Rep. 28:179–185. 2012.PubMed/NCBI
|
|
14
|
Edgar BA: From cell structure to transcription: Hippo forges a new path. Cell. 124:267–273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al: Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21:2747–2761. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sturbaut M, Bailly F, Coevoet M, Sileo P, Pugniere M, Liberelle M, Magnez R, Thuru X, Chartier-Harlin MC, Melnyk P, et al: Discovery of a cryptic site at the interface 2 of TEAD-towards a new family of YAP/TAZ-TEAD inhibitors. Eur J Med Chem. 226:1138352021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu C, Huang W and Lei Q: Regulation and function of the TAZ transcription co-activator. Int J Biochem Mol Biol. 2:247–256. 2011.PubMed/NCBI
|
|
18
|
Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al: Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 150:780–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim MH and Kim J: Role of YAP/TAZ transcriptional regulators in resistance to anti-cancer therapies. Cell Mol Life Sci. 74:1457–1474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fernandez-L A, Squatrito M, Northcott P, Awan A, Holland EC, Taylor MD, Nahlé Z and Kenney AM: Oncogenic YAP promotes radioresistance and genomic instability in medulloblastoma through IGF2-mediated Akt activation. Oncogene. 31:1923–1937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Wang Y, Zhou D, Wang K, Wang X, Wang X, Jiang Y, Zhao M, Yu R and Zhou X: Radiation-induced YAP activation confers glioma radioresistance via promoting FGF2 transcription and DNA damage repair. Oncogene. 40:4580–4591. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Das P, Skibber JM, Rodriguez-Bigas MA, Feig BW, Chang GJ, Wolff RA, Eng C, Krishnan S, Janjan NA and Crane CH: Predictors of tumor response and downstaging in patients who receive preoperative chemoradiation for rectal cancer. Cancer. 109:1750–1755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kremser C, Trieb T, Rudisch A, Judmaier W and de Vries A: Dynamic T(1) mapping predicts outcome of chemoradiation therapy in primary rectal carcinoma: Sequence implementation and data analysis. J Magn Reson Imaging. 26:662–671. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Passiglia F, Bronte G, Castiglia M, Listì A, Calò V, Toia F, Cicero G, Fanale D, Rizzo S, Bazan V and Russo A: Prognostic and predictive biomarkers for targeted therapy in NSCLC: For whom the bell tolls? Expert Opin Biol Ther. 15:1553–1566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ketterer K, Kong B, Frank D, Giese NA, Bauer A, Hoheisel J, Korc M, Kleeff J, Michalski CW and Friess H: Neuromedin U is overexpressed in pancreatic cancer and increases invasiveness via the hepatocyte growth factor c-Met pathway. Cancer Lett. 277:72–81. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Takahashi K, Furukawa C, Takano A, Ishikawa N, Kato T, Hayama S, Suzuki C, Yasui W, Inai K, Sone S, et al: The neuromedin U-growth hormone secretagogue receptor 1b/neurotensin receptor 1 oncogenic signaling pathway as a therapeutic target for lung cancer. Cancer Res. 66:9408–9419. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Harten SK, Esteban MA, Shukla D, Ashcroft M and Maxwell PH: Inactivation of the von Hippel-Lindau tumour suppressor gene induces neuromedin U expression in renal cancer cells. Mol Cancer. 10:892011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin TY, Wu FJ, Chang CL, Li Z and Luo CW: NMU signaling promotes endometrial cancer cell progression by modulating adhesion signaling. Oncotarget. 7:10228–10242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Y, McRoberts K, Berr SS, Frierson HF Jr, Conaway M and Theodorescu D: Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene. 26:765–773. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Martinez VG, Crown J, Porter RK and O'Driscoll L: Neuromedin U alters bioenergetics and expands the cancer stem cell phenotype in HER2-positive breast cancer. Int J Cancer. 140:2771–2784. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang D, Zhang X, Li Z, Xu F, Tang C and Chen H: Neuromedin U and neurotensin may promote the development of the tumour microenvironment in neuroblastoma. PeerJ. 9:e115122021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shetzline SE, Rallapalli R, Dowd KJ, Zou S, Nakata Y, Swider CR, Kalota A, Choi JK and Gewirtz AM: Neuromedin U: A Myb-regulated autocrine growth factor for human myeloid leukemias. Blood. 104:1833–1840. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Przygodzka P, Papiewska-Pajak I, Bogusz H, Kryczka J, Sobierajska K, Kowalska MA and Boncela J: Neuromedin U is upregulated by Snail at early stages of EMT in HT29 colon cancer cells. Biochim Biophys Acta. 1860:2445–2453. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Przygodzka P, Sochacka E, Soboska K, Pacholczyk M, Papiewska-Pająk I, Przygodzki T, Płociński P, Ballet S, De Prins A and Boncela J: Neuromedin U induces an invasive phenotype in CRC cells expressing the NMUR2 receptor. J Exp Clin Cancer Res. 40:2832021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Besserer-Offroy É, Brouillette RL, Lavenus S, Froehlich U, Brumwell A, Murza A, Longpré JM, Marsault É, Grandbois M, Sarret P and Leduc R: The signaling signature of the neurotensin type 1 receptor with endogenous ligands. Eur J Pharmacol. 805:1–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo Y, Luo J, Zou H, Liu C, Deng L and Li P: Context-dependent transcriptional regulations of YAP/TAZ in cancer. Cancer Lett. 527:164–173. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ciamporcero E, Shen H, Ramakrishnan S, Yu Ku S, Chintala S, Shen L, Adelaiye R, Miles KM, Ullio C, Pizzimenti S, et al: YAP activation protects urothelial cell carcinoma from treatment-induced DNA damage. Oncogene. 35:1541–1553. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tsujiura M, Mazack V, Sudol M, Kaspar HG, Nash J, Carey DJ and Gogoi R: Yes-associated protein (YAP) modulates oncogenic features and radiation sensitivity in endometrial cancer. PLoS One. 9:e1009742014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Akervall J, Nandalur S, Zhang J, Qian CN, Goldstein N, Gyllerup P, Gardinger Y, Alm J, Lorenc K, Nilsson K, et al: A novel panel of biomarkers predicts radioresistance in patients with squamous cell carcinoma of the head and neck. Eur J Cancer. 50:570–581. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vigneron AM, Ludwig RL and Vousden KH: Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev. 24:2430–2439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee WY, Kuo CC, Lin BX, Cheng CH, Chen KC and Lin CW: Podocalyxin-like protein 1 regulates TAZ signaling and stemness properties in colon cancer. Int J Mol Sci. 18:20472017. View Article : Google Scholar : PubMed/NCBI
|



















