Introduction
Gliomas are the most prevalent primary brain tumors,
representing 81% of malignant brain tumors. According to the
classification of the World Health Organization (WHO), gliomas can
be divided into four grades: Grades I and II corresponding to
low-grade gliomas, and grades III and IV corresponding to
high-grade gliomas (1). Generally,
relatively high grades are associated with a poor prognosis. The
median overall survival (OS) of patients with grade III glioma is
~3 years, whereas that of patients with grade IV glioma is only 15
months (2,3). Elucidating the molecular mechanisms
underlying the tumorigenesis of gliomas is critical in order to
improve the therapeutic efficacy of glioma treatment and prolong
the OS of patients.
High mobility group box 1 (HMGB1) is a non-histone
DNA-binding protein mainly located in the nucleus of eukaryotic
cells under homeostatic conditions. In response to stress, it
translocates from the nucleus to the cytoplasm and is released from
the cell (4). HMGB1 participates in
various cellular functions, mainly depending on its subcellular
localization (4). In the cytoplasm,
HMGB1 induces autophagy by binding to Beclin-1 and regulates
mitochondrial quality (5). In the
extracellular matrix, HMGB1 functions as a damage-associated
molecule that participates in multiple cellular activities,
including cell-cell interactions, pro-inflammatory cytokine
production, cell proliferation, differentiation, invasion and
autophagy (4,6). The dysregulation of HMGB1 has been
shown to be associated with a number of diseases, particularly
cancer. In solid tumors, HMGB1 usually translocates from the
nucleus to the cytoplasm (6). High
levels of HMGB1 have been observed in various types of
malignancies, including lung cancer, breast cancer, head and neck
squamous cell carcinoma, colon cancer, nasopharyngeal carcinoma and
glioma (7–10). Extracellular HMGB1 can function as a
paracrine or autocrine factor to drive tumor growth, proliferation,
migration and angiogenesis (11).
HMGB1 expression is frequently upregulated in gliomas. Its
overexpression is associated with glioma progression and a poor
prognosis (12,13). The transcription and translocation
of HMGB1 from the nucleus to the cytoplasm mediate autophagy and
glioma growth (12). It has been
demonstrated that extracellular HMGB1 released by patient-derived
glioma cells following temozolomide (TMZ) treatment increases the
formation of glioma stem cells, which further induce resistance to
TMZ (14). Controversially, Li
et al (15) revealed that
the unconventional autophagy-based secretion of HMGB1 in
glioblastoma promoted chemosensitivity to TMZ through macrophage
polarization. Thus, the translocation and release of HMGB1 from
glioma cells are crucial steps in glioma progression. However, the
exact mechanisms by which HMGB1 is translocated and the precise
subcellular pathway of HMGB1 in gliomas during its release are
poorly understood.
The microenvironment in solid tumors is poor in
nutrients and glucose-deprived, owing to the high rate of tumor
cell consumption (16). Cancer cell
proliferation is dependent on extracellular nutrient acquisition;
however, glucose, amino acids and lipids are usually in short
supply, due to inadequate tumor perfusion (17). Nutrient deficiency is closely
related to metabolic changes in tumor cells and promotes
tumorigenesis. Therefore, in the present study, the subcellular
localization and secretion of HMGB1 in starved glioma cells were
investigated, in order to clarify the translocation and release
pathways of HMGB1 in glioma cells under nutrient-poor conditions.
It was revealed that HMGB1 translocated from the nucleus to the
cytoplasm and was then secreted into the exterior of starved glioma
cells. HMGB1 in the cytoplasm was distributed within or around the
mitochondria, endoplasmic reticulum (ER), peroxisomes,
autophagosomes, early endosomes, late endosomes, lysosomes,
cytoskeleton and mitochondria-associated ER membranes (MAMs). The
Manders' overlap coefficient of HMGB1 in these compartments was
markedly altered upon its release. In addition, autophagy mediated
the release of HMGB1 in starved glioma cells. The findings of the
present study provided a novel perspective to further clarify the
mechanisms by which HMGB1 affects gliomas under starvation.
Materials and methods
Cases
A total of six glioma and three normal brain tissue
sections (human samples isolated during decompression operations)
were obtained from the Department of Histology at the 988th
Hospital of the Joint Logistic Support Force (Zhengzhou, China).
Additionally, glioblastoma tissue (male, 66 years old) obtained
from the Department of Neurosurgery of the same hospital was used
for transmission electron microscopy. The age range and median age
of the patients were 36 to 66 years and 52.7 years, respectively.
The WHO grade, sex and age of all seven glioma cases are listed in
Table SI. The isolated tissues
were first fixed in 4% paraformaldehyde (PFA) at 4°C for 24 h
before the subsequent treatments. The entire storage process, from
isolation to storage, was completed within 30 min. Glioma samples
were validated by experienced clinical pathologists in the hospital
in accordance with the WHO classification (2016) (18). Written informed consent was obtained
from all patients with glioma and the families of the three
patients with traumatic brain injury who underwent decompression
surgery. The present study was conducted in accordance with the
Declaration of Helsinki, and the protocol was approved by the
Ethics Committee of the 988th Hospital of the Joint Logistic
Support Force (Zhengzhou, China).
Cells and reagents
HA1800 astrocytes were purchased from Shanghai Ji
Ning Industrial Co., Ltd., and three human glioma cell lines [U251,
U87-MG (ATCC version, glioblastoma of unknown origin and U118-MG)]
were purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Mycoplasma testing was performed on
the cell lines (data not shown), and the cells were authenticated
using STR profiling. The cells were sub-cultured in Dulbecco's
modified Eagle's medium (Biological Industries; cat. no.
C3110-0500) supplemented with 10% fetal bovine serum (BSA;
Biological Industries; cat. no. 04-001-1ACS) and 1%
penicillin-streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.; cat. no. P1400) and maintained in 5% CO2 at
37°C in a humidified incubator. The glioma cells were treated with
Hank's balanced salt solution (HBSS; Beijing Solarbio Science &
Technology Co., Ltd.; cat. no. H1025) at the indicated time
intervals (0, 0.5, 1, 2, 3 and 4 h) for starvation stress induction
or H2O2 (300 µM; Laiyang City Shuangshuang
Chemical Co., Ltd.) for 4 h for oxidative stress. Wortmannin (WOR;
Shanghai Selleck Chemicals Co., Ltd.; cat. no. S2758; 0.5 µM) and
chloroquine (CQ; Shanghai Selleck Chemicals Co., Ltd.; cat. no.
S6999; 20 µM) were used to inhibit early and late autophagy,
respectively. U251 glioma cells were pretreated with WOR or CQ for
2 h and then stimulated with or without HBSS for 3 h in the
presence of WOR or CQ at 37°C in a humidified incubator.
Cell transfection
The cells were cultured in six-well culture plates
until they reached 80% confluency, after which they were
transfected with 2 µg pEGFP-C1 or the HMGB1 recombinant
overexpression plasmid pEGFP-HMGB1-C1 (Shanghai Sangon Biotech Co.,
Ltd.) using Simple-Fect Transfection Reagent (Zhengzhou Kebang
Biological Technology Co., Ltd.) for 16 h at 37°C in a humidified
incubator in accordance with the manufacturer's protocol. The
culture medium was then switched to a normal medium. After ~24 h,
the experiments described in the following sections were
conducted.
Immunohistochemistry (IHC)
IHC was performed as previously described (19). Briefly, deparaffinized glioma slices
(5-µm-thick) were permeabilized with 0.3% Triton X-100 (Beijing
Biotopped Co., Ltd.; cat. no. T6200G) in PBS for 20 min at room
temperature (RT). Subsequently, the cells were blocked with
blocking solution [5% BSA (Beijing Solarbio Science &
Technology Co., Ltd.; cat. no. A8020)] plus 0.3% Triton X-100 in
phosphate-buffered saline (PBS) for 1 h at RT. Subsequently, the
cells were incubated with anti-HMGB1 (Abcam; cat. no. ab18256;
1:500), anti-ATP synthase F1 subunit alpha (ATP5A; Abcam; cat. no.
ab14748; 1:200), anti-Lysosomal-associated membrane protein 1
(LAMP1; CST Biological Reagents Co., Ltd.; cat. no. 9091#; 1:100),
anti-Ras-related protein 5 (Rab-5; Abcam; cat. no. ab18211; 1:200),
anti-glial fibrillary acidic protein [(GFAP; Abcam; cat. no.
ab279290; 1:500) and anti-calnexin (CANX) antibodies
sMilliporeSigma; cat. no. SAB2501291; 1:200] overnight at 4°C. All
sections were rinsed with PBS and then incubated with the following
fluorescein-conjugated secondary antibodies for 2 h at RT:
Fluorescein-conjugated goat anti-rabbit IgG (H+L) (Beijing ZSGB-BIO
Co. Ltd.; cat. no. ZF-0311; 1:100), Alexa Fluor 488 donkey
anti-rabbit IgG (H+L) (Abcam; cat. no. ab150073; 1:500),
rhodamine-conjugated goat anti-mouse IgG (H+L) (Beijing ZSGB-BIO
Co. Ltd; cat. no. ZF-0313; 1:100) and Alexa Fluor 633 donkey
anti-goat IgG (H+L) (Invitrogen Trading Shanghai Co. Ltd; cat. no.
A-21082; 1:500). Nuclei were stained with
4′6-diamidino-2-phenylindole (DAPI; Beijing Solarbio Science &
Technology Co., Ltd.; cat. no. C0060; 1:5,000) for 15 min at RT.
Images were acquired using an Olympus confocal microscope (LSM;
cat. no. FV1000; Olympus Corporation) and analyzed using Adobe
Photoshop CS6 (Adobe Systems, Inc.; version 13.0.1×64) and ImageJ
software (National Institutes of Health; version 1.4.3.67).
Immunocytochemistry (ICC)
ICC was performed as previously described (19). Following appropriate treatment with
HBSS, the cells were washed twice with ice-cold PBS, fixed with
freshly prepared 4% PFA in PBS for 15 min at RT, and then
permeabilized with 0.3% Triton X-100 (Beijing Biotopped Co., Ltd.;
cat. no. T6200G) in PBS for 20 min at RT. Subsequently, the cells
were blocked with blocking solution (as mentioned for IHC) for 1 h
at RT and then incubated with the following primary antibodies
overnight at 4°C: Anti-HMGB1 (Abcam; cat. no. ab18256; 1:1,000),
anti-LAMP1 (China-based branch, CST Biological Reagents Co., Ltd.;
cat. no. 9091; 1:500), anti-microtubule-associated proteins 1A/1B
light chain 3B (LC3B; CST Biological Reagents Co., Ltd.; cat. no.
3868; 1:500), anti-catalase (Abcam; cat. no. ab16731; 1:200),
anti-CANX (MilliporeSigma; cat. no. SAB2501291; 1:500), anti-Rab5
(Abcam; cat. no. ab18211; 1:500), anti-Rab7 (Abcam; cat. no.
ab137029; 1:500), anti-GFAP (Abcam; cat. no. ab279290; 1:500) and
anti-Sigma 1 receptor (Sigma1-R) antibodies (Abcam; cat. no.
ab53852; 1:100). The cells were incubated with the secondary
antibodies used in IHC for 2 h at RT and then counterstained with
DAPI at RT for 15 min. In several experiments, cells were preloaded
with MitoTracker Red CMXRos for 25 min at 37°C in a humidified
incubator before fixation to label the mitochondria (Thermo Fisher
Scientific, Inc.; cat. no. M7512). Images were obtained by Olympus
confocal microscope and analyzed using Adobe Photoshop CS6 (Adobe
Systems, Inc.; version 13.0.1×64) and ImageJ software (National
Institutes of Health; version 1.4.3.67).
Enzyme-linked immunosorbent assay
(ELISA)
Cell supernatants were collected and assayed using
an HMGB1 ELISA kit (Cusabio Technology, LLC; cat. no. CSB-E08223h)
in accordance with the manufacturer's instructions. Briefly, 100 µl
standards or samples were added to each well for 2 h at 37°C.
Following the removal of the liquid, 100 µl biotin-conjugated HMGB1
antibody (Cusabio Technology, LLC; cat. no. CSB-E08223h) was added
with for 1 h at 37°C and then washed three times with wash buffer.
Horseradish peroxidase-conjugated goat anti-rabbit IgG (Cusabio
Technology, LLC; cat. no. CSB-E08223h) was added for 1 h at 37°C.
After being washed five times, 90 µl of 3,3,5,5′
tetramethylbenzidine substrate were added to each well and the
plate was incubated for 30 min at 37°C. The reaction was terminated
by adding 50 µl of stop solution, and the optical density was then
read at 450 nm using a microplate tester (Multskan Mk3, Thermo
Fisher Scientific, Inc.) within 5 min.
Immunoelectron microscopy
Glioma cells were harvested and immediately fixed in
4% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) and incubated
for 2 h at 4°C. Following centrifugation at 100 × g for 5 min at
4°C, samples immersed in 4% glutaraldehyde in 0.1 M phosphate
buffer (pH 7.4) were sent to the biomedical testing center of
Tsinghua University, and the remaining steps were performed. The
samples were coated with 4 nm gold and viewed in a JEM-1400
electron microscope (JEOL, Ltd.).
Western blotting (WB)
Glioma cells were lysed in RIPA lysis buffer
(Beijing Solarbio Science & Technology Co., Ltd.; cat. no.
R0010). The protein amounts in the whole-cell lysates were
determined using the BCA Protein Assay kit (Beijing Solarbio
Science & Technology Co., Ltd.; cat. no. PC0020). In total,
20–50 µgs protein extracts were separated on a 12% SDS-PAGE and
then transferred to polyvinylidene fluoride membranes
(MilliporeSigma; cat. no. IPVH00010). Following blocking with 5%
non-fat dry milk solution (in Tris-buffered saline wash buffer with
1‰ Tween-20) for 1 h at RT, the membranes were probed overnight at
4°C with the following primary antibodies: Anti-HMGB1 (Abcam; cat.
no. ab18256; 1:1,000), anti-CANX (MilliporeSigma; cat. no.
SAB2501291; 1:500), anti-ATP5A (Abcam; cat. no. ab14748; 1:1,000),
anti-β-actin (OriGene Technologies, Inc.; cat. no. TA-09; 1:2,000),
anti-LaminB1 (China-based branch, Cell Signaling Technology, Inc.;
cat. no. 13435; 1:1,000), anti-LC3B (Cell Signaling Technology,
Inc. 3868; 1:1,000), anti-GAPDH (Hangzhou Xianzhi Biological
Technology Co., Ltd.; cat. no. AB-P-R 001; 1:1,000), and anti-p62
(China-based branch, Cell Signaling Technology, Inc.; cat. no.
23214, 1:1,000). Subsequently, the membranes were incubated with
peroxidase-conjugated goat anti-mouse or anti-rabbit IgG secondary
antibodies (Beijing ZSGB-BIO Co. Ltd; cat. no. ZB-2305 or ZB-2301,
respectively; 1:5,000) for 2 h at RT. An enhanced chemiluminescent
substrate (Dalian Meilun Biology Technology Co., Ltd.; cat. no.
MA0186) was used to visualize the protein bands. Relative band
intensities were quantified using ImageJ software (National
Institutes of Health; version 1.4.3.67).
Subcellular fractionation
U87-MG glioma cells in 10 dishes, 10 cm each, were
treated with HBSS (Beijing Solarbio Science & Technology Co.,
Ltd.; cat. no. H1025) for 1 h at 37°C and then harvested in lysis
buffer [20 mM HEPES-KOH (pH 7.2, Sigma-Aldrich (Shanghai) Trading
Co.Ltd., cat. no. V900477), 400 mM sucrose (Beijing Solarbio
Science & Technology Co., Ltd.; cat. no. S8271), and 1 mM EDTA
(Beijing Solarbio Science & Technology Co., Ltd.; cat. no.
E8030)] supplemented with protease and phosphatase inhibitor
(Dalian Meilunbio Co. Ltd., cat. no. MB12707-1) and 0.3 mM
dithiothreitol (Beijing Solarbio Science & Technology Co.,
Ltd.; cat. no. D8220). The cell lysates were then homogenized by
using a 22G needle. The homogenates were centrifuged at 800 × g for
10 min at RT to pellet the nuclei and cytoskeleton. The ER, crude
mitochondria, and pure mitochondria in the cytoplasm were
fractionated following previously described protocols, and analyzed
using WB (20). Anti-CANX
(MilliporeSigma; cat. no. SAB2501291; 1:500) antibody and ATP5A
(Abcam; cat. no. ab14748; 1:200) were used as markers for the ER
and pure mitochondria proteins, respectively. Autophagosomes were
fractionated following the method described by Zhang et al
(21). The homogenates were
sequentially centrifuged at 3,000 × g for 10 min, 25,000 × g for 20
min, and 100,000 × g for 30 min at RT (TLA100.3 rotor; Beckman
Coulter, Inc.), and the pelleted membranes were collected at each
speed, and analyzed using WB. Anti-LC3B antibody (CST Biological
Reagents Co., Ltd.; cat. no. 3868; 1:500) was used as marker for
the autophagosomes.
Protease K protection assay
The pelleted membranes that were previously
centrifuged at 25,000 × g and 100,000 × g were resuspended in
buffer containing 25 µg/ml protease K [Sigma-Aldrich (Shanghai)
Trading Co. Ltd., cat. no. 3115828001], with or without 1% Triton
X-100 (Beijing Biotopped Co., Ltd.; cat. no. T6200G) and then
incubated on ice for 30 min. The reaction was terminated by adding
5X SDS loading buffer (Beijing Dingguo Changsheng Biotechnology
Co., Ltd., cat. no. WB-0091), heated in boiling water for 10 min,
and analyzed using WB.
Lactate dehydrogenase (LDH) release
assay
LDH release was measured as a physiological index of
cell membrane damage. U87-MG glioma cells were treated with HBSS to
induce starvation stress at the indicated time intervals (0, 0.5,
1, 2, 3 and 4 h). U251 glioma cells were stimulated with or without
HBSS (3 h), WOR (3 h, 0.5 µM), or CQ (3 h, 20 µM). The release of
LDH from these cells was measured using an LDH cytotoxicity assay
kit (Beyotime Institute of Biotechnology; cat. no. C0016) in
accordance with the manufacturer's instructions. Briefly, the
supernatant of the treated glioma cells was collected and added
with LDH reagent. The mixture was incubated in the dark for 30 min
at RT, and the OD490 value was measured using a microplate tester
(Multskan Mk3, Thermo Fisher Scientific, Inc.).
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using the CCK-8 assay
(Dalian Meilun Biotech Co., Ltd.; cat. no. MA0218-5) following the
manufacturer's protocol. Briefly, cells were seeded in 96-well
plates and grown to 80% confluency. In total, a quantity of 10 µl
CCK-8 reagent was added to the treated glioma cells and incubated
for 2 h at 37°C in a humidified incubator with 5% CO2.
The absorbance was obtained at 450 nm using a microplate tester
(Multskan Mk3, Thermo Fisher Scientific, Inc.).
Statistical analysis
All assays were performed independently and at least
in triplicate. Statistical results are expressed as mean ± SD. An
unpaired Student's t-test and one-way analysis of variance (ANOVA)
followed by Tukey's post-hoc test were used for statistical
analyses using GraphPad Prism software (Dotmaticx; Version
8.3.0.538). Manders' overlap coefficient was calculated using the
JACoP ImageJ plugin (National Institutes of Health; version
1.4.3.67) as previously described (22). Line fluorescence tracing from the
images was performed using OriginPro software (OriginLab; version
8.5). P<0.05 was considered to indicate a statistically
significant difference.
Results
Translocation of HMGB1 in glioma
cells
To examine the translocation of HMGB1, its
distribution was first analyzed in normal and glioma tissues. As
presented in Fig. 1, HMGB1 was
mainly distributed in the nucleus of the normal brain tissues
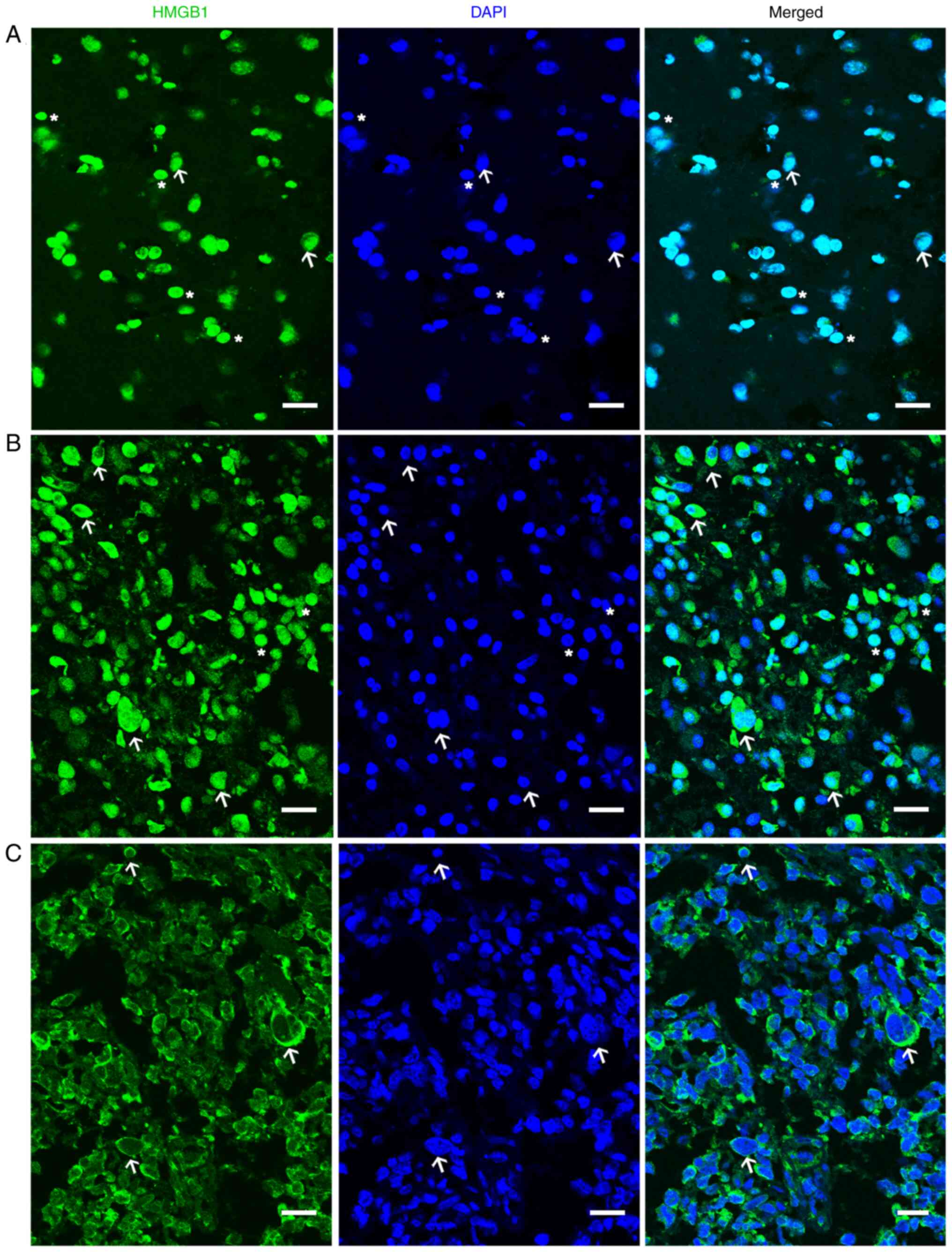
(Fig. 1A), whereas in the glioma
tissues, it was localized in the nucleus and cytoplasm (Fig. 1B and C). In certain cells, the
distribution of HMGB1 solely inside the cytoplasm was observed
(Fig. 1C). The difference in the
localization of HMGB1 demonstrated in Fig. 1B and C was potentially due to glioma
heterogeneity. The cytoplasmic HMGB1 localization index of HMGB1
has been shown to be moderately associated with tumor stage in a
previously published study on cholangiocarcinoma (23). HMGB1 is mainly localized to the
nucleus under homeostatic conditions (24). In the present study, when the cells
were treated with HBSS (starvation), HMGB1 was released from the
nucleus into the cytoplasm and extracellular medium (Fig. 2A-C). The cytoplasmic expression of
HMGB1 reached its maximum 1 h after HBSS treatment (Fig. 2B), whereas its extracellular
secretion reached its maximum at 2 h (Fig. 2C). LDH release assay revealed that
the increase in HMGB1 secretion was not attributed to non-specific
membrane permeability (Fig.
2D).
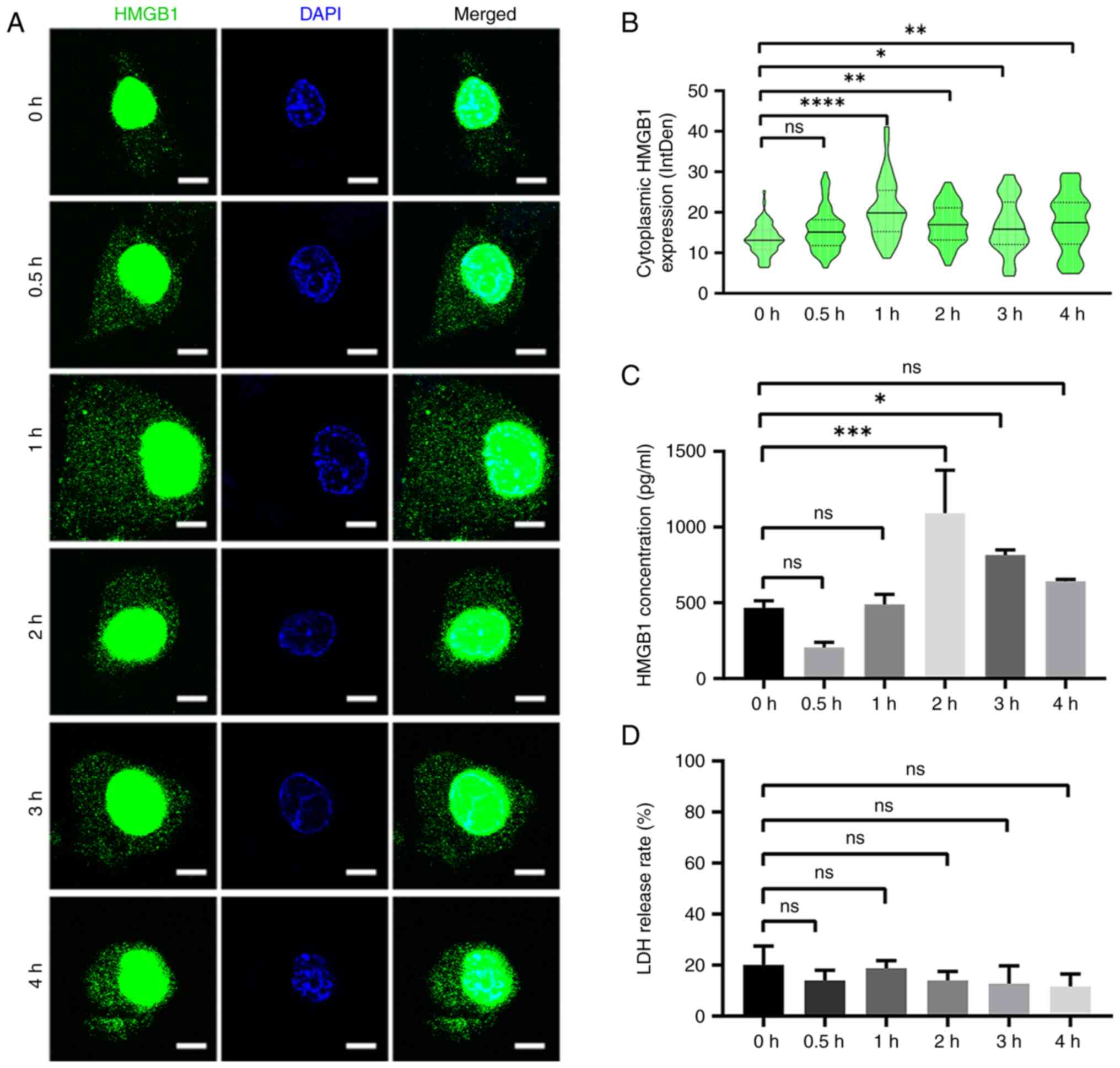 | Figure 2.HMGB1 translocation at the indicated
time intervals. (A) HMGB1 staining in U87-MG glioma cells at 0,
0.5, 1, 2, 3 and 4 h following starvation with HBSS. Nuclei were
stained with 4′6-diamidino-2-phenylindole. Scale bar, 5 µm. (B)
Cytoplasmic immunoreactivity of HMGB1 at the indicated time
intervals was determined (n=3, 16–26 cells per replicate). (C)
Supernatants of U87-MG glioma cells treated with HBSS at indicated
time intervals were collected, and HMGB1 was detected using ELISA
(n=3). (D) The cytotoxicity of HBSS was measured using an LDH
release assay (n=3). Data represent the mean ± SD; Statistical
significance was assessed using one-way ANOVA followed by Tukey's
post hoc test. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. ns, not significant; HMGB1, high mobility group
box 1; HBSS, Hank's balanced salt solution; LDH, lactate
dehydrogenase. |
Co-localization of HMGB1 and specific
marker proteins of cellular compartments in glioma
The association between HMGB1 and the cytosolic
compartments was then investigated and the subcellular localization
of HMGB1 was determined using ICC. The results revealed that HMGB1
co-localized with Mitotracker Red, CANX, catalase, LC3B, LAMP1,
Rab5 and Rab7, indicating that HMGB1 was imported into the
mitochondria, ER, peroxisomes, autophagosomes, lysosomes, early
endosomes and late lysosomes of glioma cells. Oxidative stress with
H2O2 was also induced the HMGB1 translocation
to the cytoplasm and colocalization with Mitotracker Red in glioma
cells (Fig. S1A and B).
Furthermore, merged images of HMGB1 with GFAP were obtained,
indicating that HMGB1 was associated with the cytoskeleton in
glioma cells (Figs. 3A and B, and
4A and B). Notably, the staining of
GFAP in the glioma cells did not reveal the cytoskeleton-like
structure, as was expected. A similar result has been confirmedsin
previous studies (25,26). This finding may be attributed to the
moderate immune response of glioma cells to GFAP (27).
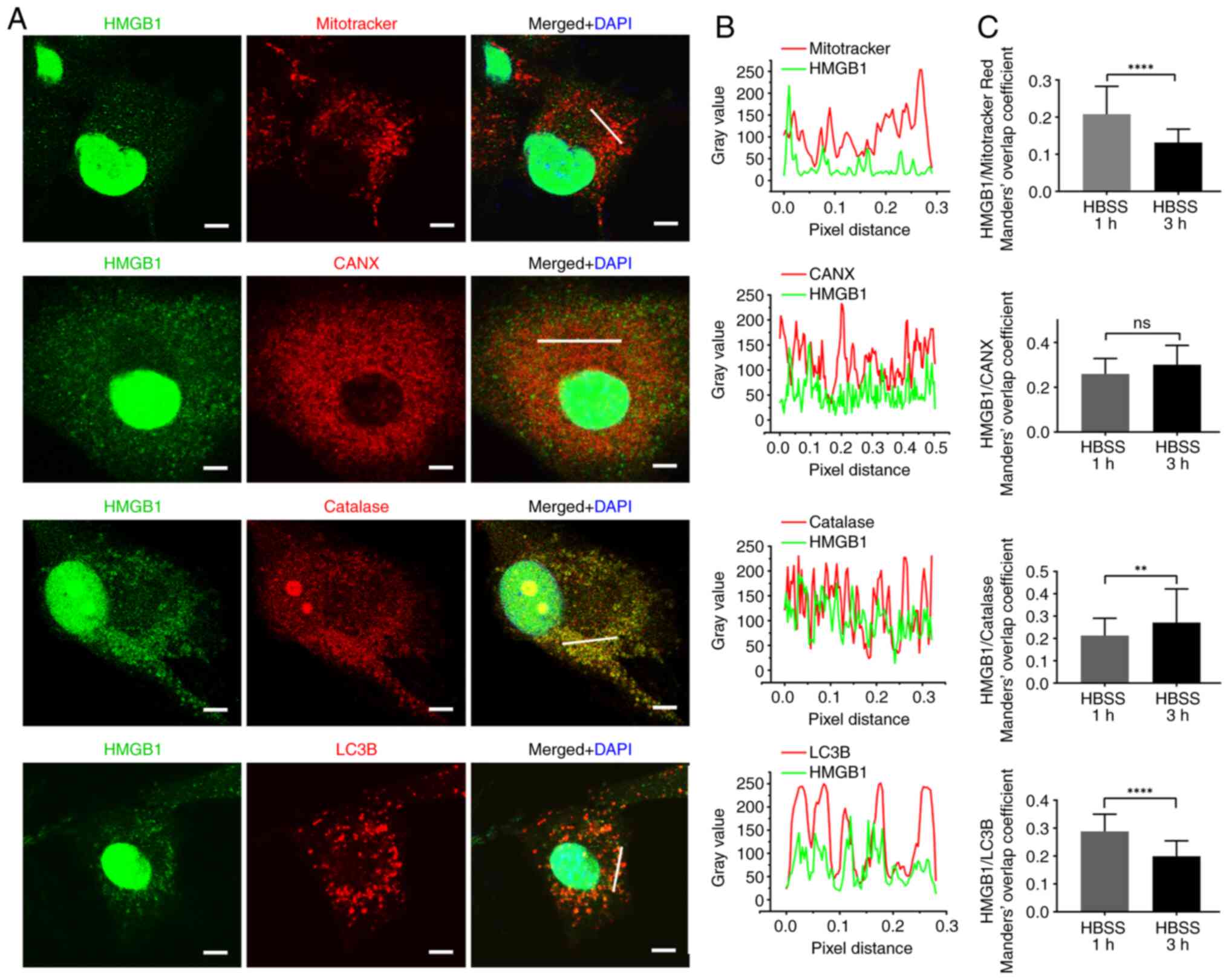 | Figure 3.Immunocytochemistry HMGB1 with
mitrotracker red, CANX, catalase and LC3B. (A) Staining for HMGB1
with mitrotracker red probe, CANX, catalase and LC3B antibodies in
U87-MG glioma cells at 3 h following HBSS stimulation. Nuclei were
stained with 4′6-diamidino-2-phenylindole. Scale bars, 5 µm. (B)
Line fluorescence tracing from images in (A). (C) Manders' overlap
coefficient analysis of HMGB1 and each marker protein 1 and 3 h
following HBSS stimulation (n=3, 20–33 cells per replicate). Data
represent the mean ± SD; an unpaired Student's t-test was used to
evaluate statistical significance. **P<0.01 and ****P<0.0001.
ns, not significants HMGB1, high mobility group box 1; CANX,
calnexin; LC3B, microtubule-associated proteins 1A/1B light chain
3B; HBSS, Hank's balanced salt solution. |
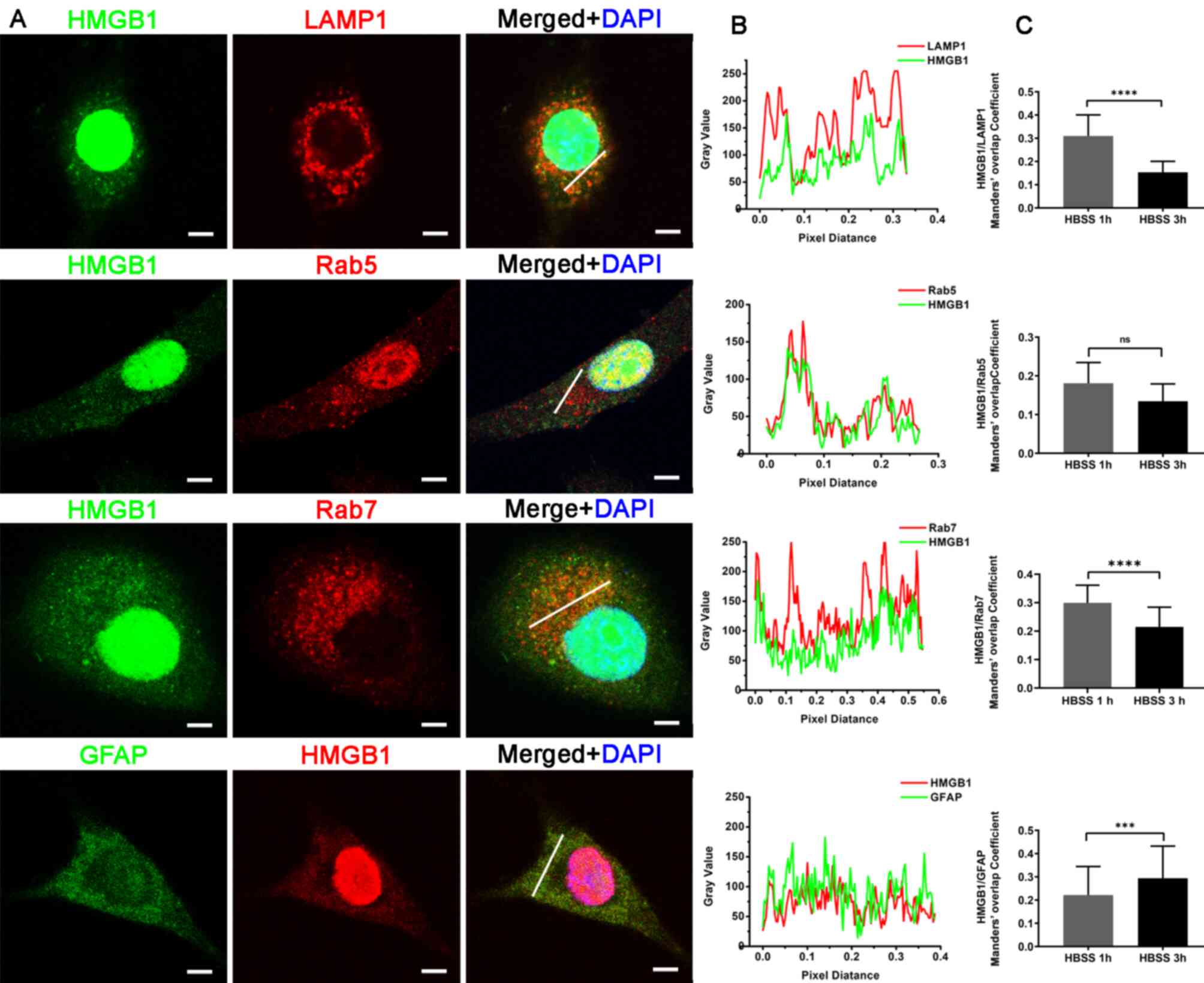 | Figure 4.HMGB1 with LAMP1, Rab5, Rab7 and
GFAP. (A) Staining for HMGB1 with LAMP1, Rab5, Rab7 and GFAP
antibodies in U87-MG glioma cells at 3 h following HBSS
stimulation. Nuclei were stained with 4′6-diamidino-2-phenylindole.
Scale bars, 5 µm. (B) Line fluorescence tracing from images in (A).
(C) Manders' overlap coefficient analysis of HMGB1 and each marker
protein 1 and 3 h after HBSS stimulation (n=3, 20–33 cells per
replicate). Data represent the mean ± SD; an unpaired Student's
t-test was used to evaluate the statistical significance.
***P<0.001 and ****P<0.0001. ns, not significant; HMGB1, high
mobility group box 1; LAMP1, lysosomal-associated membrane protein
1; GFAP, glial fibrillary acidic protein; HBSS, Hank's balanced
salt solution. |
In order to explore the association of HMGB1 with
these compartments during secretion, Manders' overlap coefficient
between HMGB1 and the aforementioned marker proteins were analyzed,
and compared at 1 and 3 h following starvation. Manders' overlap
coefficient, which is commonly used to examine protein
co-localization (28,29), ranged between 0 and 1 in the present
study. The higher the Manders' overlap rate, the higher the
fluorescence overlap rate. The results demonstrated that the
Manders' overlap coefficient between HMGB1 and MitoTracker Red,
LC3B, LAMP1 and Rab7 was significantly lower at 3 h than at 1 h
following starvation, whereas that between HMGB1 and catalase or
GFAP increased (Figs. 3C and
4C). The change in the Manders'
overlap coefficient suggests that HMGB1 is dynamically associated
with cytoplasmic compartments in starved glioma cells. In addition,
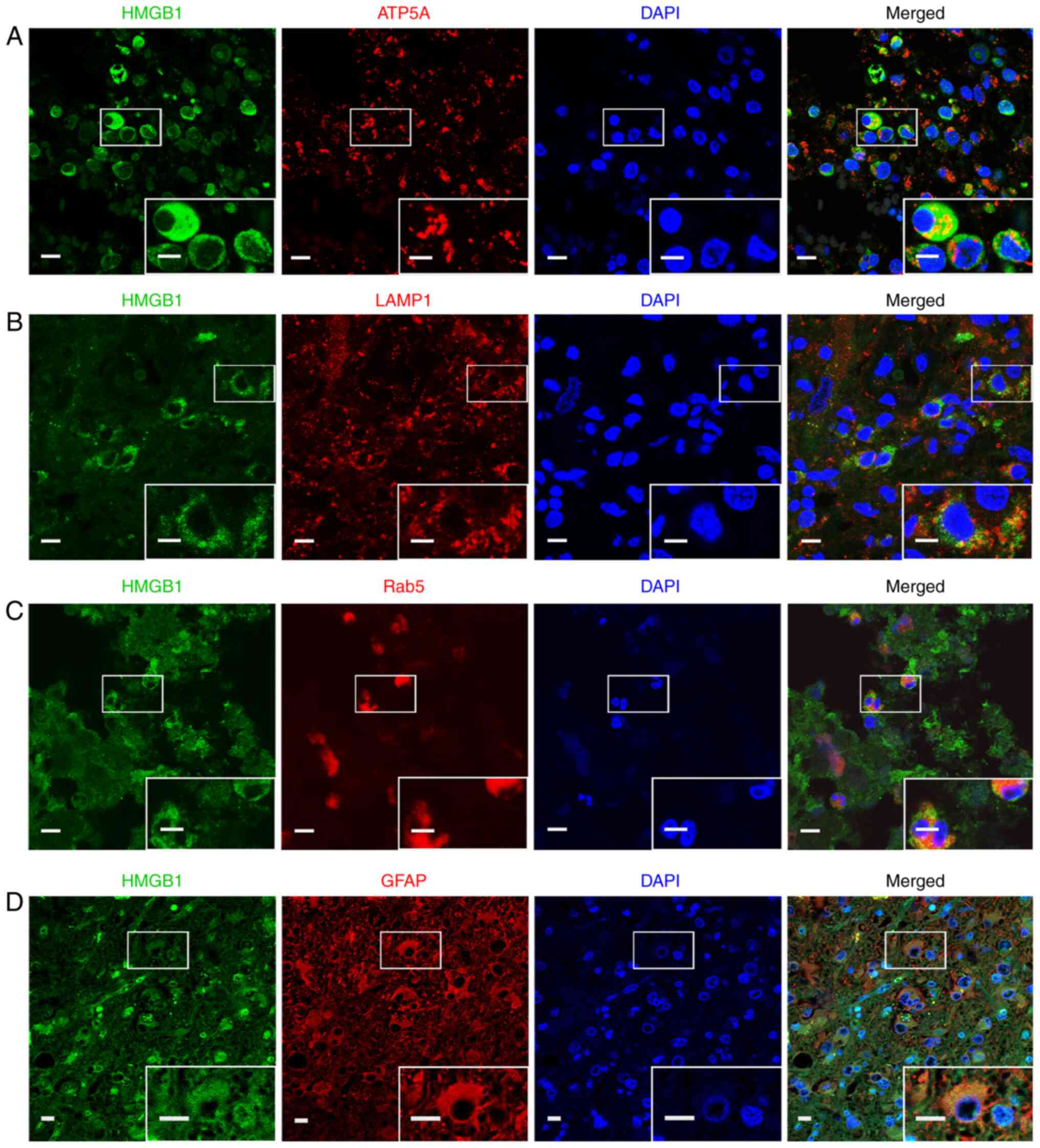
the subcellular localization of HMGB1 was examined in glioma
tissues. Considering the limitations in antibody species and
reactivity, only the co-localization of HMGB1 with ATP5A
(mitochondria), LAMP1 (lysosome), Rab5 (endosome) and GFAP
(cytoskeleton) was detected (Fig.
5). The results revealed that HMGB1 and these marker proteins
co-localized in glioma tissues.
Ultrastructural characterization of
subcellular localization of HMGB1 in glioma cells
To gain further insight into the translocation of
HMGB1 in glioma cells, HMGB1 localization was examined at the
ultrastructural level by performing immunogold electron microscopy
on HBSS-treated glioma cells. The results revealed that gold
particles were positioned in the nucleus, cytoplasm and
extracellular matrix of the starved glioma cells (Fig. 6A-C), indicating that HMGB1 was
released from the nucleus into the cytoplasm and outside of the
cell, following starvation stress. In the cytoplasm, gold particles
were present within or around the membrane structures. According to
the morphological criteria, gold particles were localized within or
around the mitochondria (Fig. 6D),
ER (Fig. 6E and J), small vesicles
(Fig. 6F), endosomes (Fig. 6G), coated vesicles (Fig. 6H) and autolysosomes (Fig. 6I). Furthermore, gold particles were
associated with the cytoskeleton (Fig.
6J). Notably, some gold particles were free and did not bind to
specific structures in the cytoplasm (Fig. 6K). Collectively, these experiments
revealed that HMGB1 translocated within or around membrane
organelles and the cytoskeleton of the starved glioma cells.
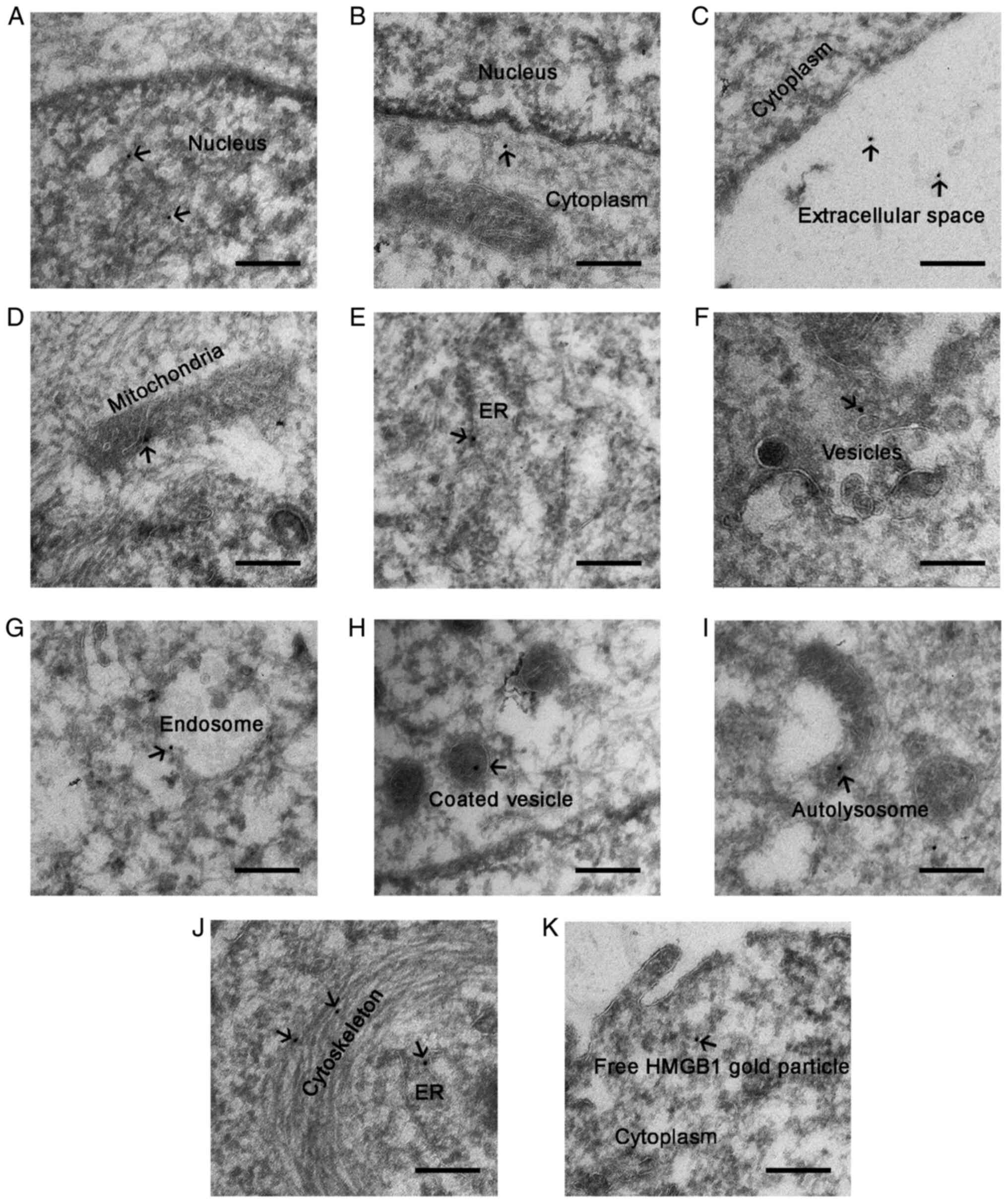 | Figure 6.Electron microscopy images of HMGB1
in U87-MG glioma cells treated with HBSS for 3 h. HMGB1
localization was visualized using 10 nm gold particles (indicated
by arrows). HMGB1 was localized in the (A) nucleus, (B) cytoplasm,
and (C) extracellular space. In the cytoplasm, gold particles were
found within or around (D) the mitochondria, (E and J) ER, (F)
small vesicles, (G) endosomes, (H) coated vesicles, (I)
autolysosomes, and (J) the cytoskeleton. (K) Free gold particles of
HMGB1 in the cytoplasm. Scale bar, 200 nm. HMGB1, high mobility
group box 1; HBSS, Hank's balanced salt solution; ER, endoplasmic
reticulum. |
HMGB1 is enriched in membrane-bound
compartments
Subsequently, it was tested whether cytosolic HMGB1
in the starved glioma cells is enclosed in membrane-bound
compartments through subcellular fractionation (20,30).
Cell lysates of HBSS-treated U87-MG cells were sequentially
centrifuged to pellet the nuclei (precipitated along with the
cytoskeleton), ER, crude mitochondria and pure mitochondrial
pellets, respectively. The pellets were then immunoblotted for the
expression of HMGB1 and organelle markers. Lamin B1, β-actin, CANX
and ATP5A were used to label nuclear proteins, cytosolic proteins,
ER and mitochondria, respectively. β-actin expression was observed
in the nuclei fraction (Fig. 7A).
This may be attributed to the nuclei and cytoskeleton precipitating
together, and β-actin belonging to the cytoskeletal protein. The
results revealed the expression of HMGB1 in both the nuclei and the
cytoplasm (Fig. 7A). Moreover,
HMGB1 expression was also detected in the ER, crude mitochondria
and pure mitochondrial fractions (Fig.
7B), indicating that HMGB1 was enriched in the membrane-bound
compartments after starvation.
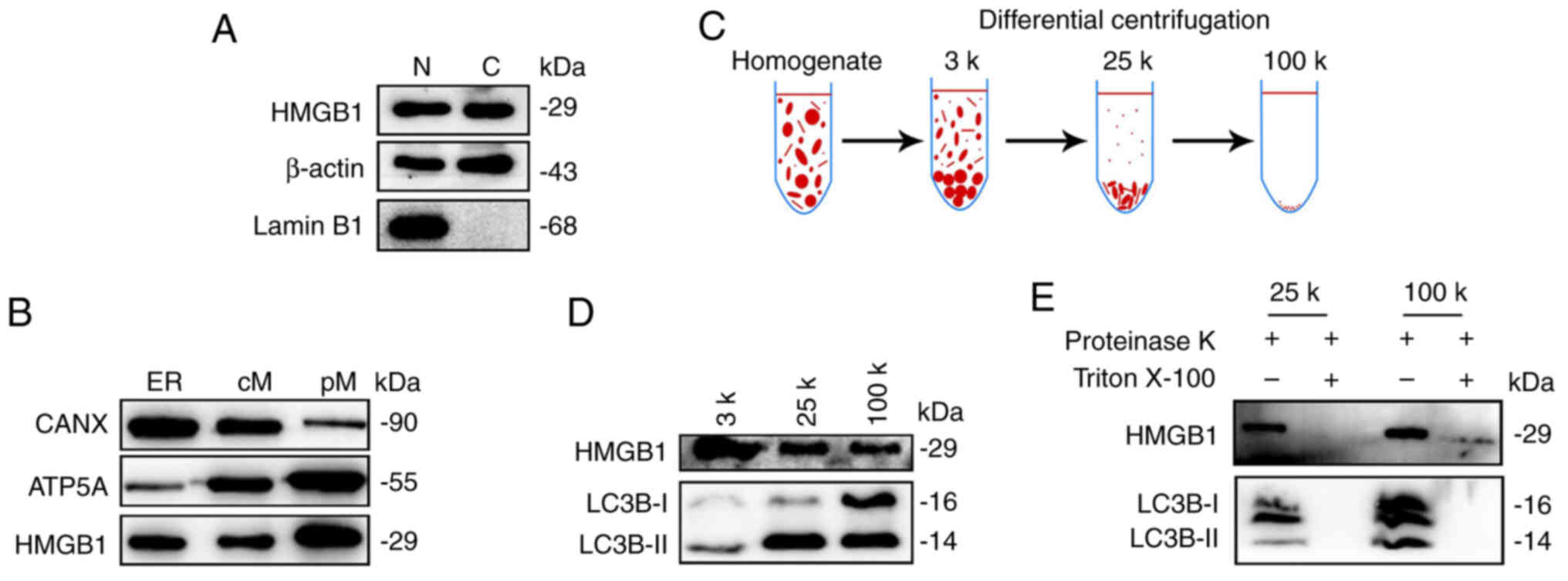 | Figure 7.HMGB1 is enriched in membrane-bound
compartments. (A) Pellets containing nuclei and cytoskeletons (N)
and cytoplasmic proteins (C) were collected from the HBSS-treated
U87-MG glioma cells and immunoprobed with Lamin B1 (marker for the
nuclear proteins), β-actin (marker for the cytosolic proteins) and
HMGB1. (B) Pellets containing ER, cM and pM were fractionated and
immunoprobed with HMGB1, the ER marker, CANX, and the mitochondria
marker, ATP5A. HMGB1 was detected in the membrane-bound
compartments. (C) Membrane fractionation scheme. Briefly, U87-MG
cells were starved in HBSS for 1 h, collected and then homogenized.
Cell lysates were differentially centrifuged at 3,000 × g, 25,000 ×
g, and 100,000 × g at RT. (D) The expression levels of LC3B and
HMGB1 in different fractionations were measured using western
blotting. (E) Proteinase K digestion was performed using the 25-k
and 100-k membrane fractions. HMGB1, high mobility group box 1;
HBSS, Hank's balanced salt solution; ER, endoplasmic reticulum; cM,
crude mitochondria; pM, pure mitochondria; CANX, calnexin; ATP5A,
ATP synthase F1 subunit alpha; LC3B, microtubule-associated
proteins 1A/1B light chain 3B. |
To further demonstrate the translocation of HMGB1
into membrane-bound compartments, autophagosomes were isolated
according to a membrane fractionation procedure as previously
described (21) (Fig. 7C). Overall, 3, 25 and 100 k membrane
pellet fractions were first obtained by applying differential
centrifugation to the HBSS-treated U87-MG glioma cells. LC3B-II is
enriched in the 25-k fraction (21). WB demonstrated that HMGB1 was
enriched in all three fractions, with the 25- and 100-k fractions
co-localizing with LC3B-II (Fig.
7D). However, LC3B-I was strongly expressed in the 100-k
fraction (Fig. 7D). In general, the
ratio of LC3B-II to LC3B-I represents the level of autophagy
(31). Therefore, it was considered
that autophagosomes were mainly enriched in the 25-k fraction in
this experiment. Protease K protection experiments demonstrated
that HMGB1 was sequestered in the membrane vesicular structures in
the 25- and 100-k fractions (Fig.
7E). Accordingly, these data suggested that HMGB1 translocated
to the autophagosomes (25-k fraction) and other types of membrane
vesicular structures (100-k fraction) of the starved glioma
cells.
HMGB1 localizes at the MAMs in
glioma
Considering that HMGB1 is dually localized to the
mitochondria and ER, the present study then wished to investigate
whether HMGB1 localizes to the ER and mitochondria contact sites,
termed MAMs. MAMs are highly specialized subcellular compartments
located between the mitochondria and ER. They are ER membranes that
are in close proximity to the mitochondria, although being
biochemically distinct from pure ER and pure mitochondria (32). In the cytoplasm, HMGB1 is
distributed non-uniformly and adopts a punctate or patchy
distribution, a staining pattern similar to that of MAM-localized
proteins (33,34). Herein, firstly, MAMs were examined
in glioma cells and tissues. The ER and mitochondria were labeled
with CANX and Mitotracker or ATP5A, respectively. The MAMs, the
double labeled sites, were calculated through Manders' overlap
coefficient. Compared with the HA1800 astrocytes or the normal
brain tissues, MAMs were significantly increased in both the glioma
cells (Fig. S2A and B) and glioma
tissues (Fig. S2C and D). In
addition, it was observed that the mitochondria with partial or
total cristolysis were in close association with ER profiles
establishing MAMs in the transmission electron microscopy images of
a case of glioblastoma (Fig. S2E).
These results indicated that MAMs were increased in glioma.
Subsequently, it was examined whether HMGB1 localized at MAMs
through triple ICC staining. The results demonstrated that some
HMGB1 puncta were associated with ER and mitochondrial markers in
the glioma tissues and HBSS-treated glioma cells (Figs. 8A and B, and S3, as indicated by the arrows).
Furthermore, it was examined whether HMGB1 localizes at MAMs by
imaging glioma cells (expressing HMGB1-EGFP) in which the
mitochondria and ER were labeled with their respective fluorescent
probes. The transfection control for the HMGB1 transfection
efficiency evaluation was performed through ICC and WB (Figs. S4A and B). The results revealed
that HMGB1-EGFP puncta were generally co-localized with both ER and
mitochondrial markers (Fig. 8C, as
indicated by the arrows). In addition, it was observed that HMGB1
puncta were associated with the MAM marker Sigma1-R (Fig. 8D, as indicated by the arrows).
Immunoelectron microscopy further indicated that HMGB1 gold
particles were localized in the narrow cytoplasmic cleft outlining
both the mitochondria and expanded ER, thereby confirming that
HMGB1 puncta localized at MAMs (Fig. 8E
and F). Taken together, these results identified HMGB1 as a
novel, to the best of our knowledge, MAM protein in glioma cells
following starvation.
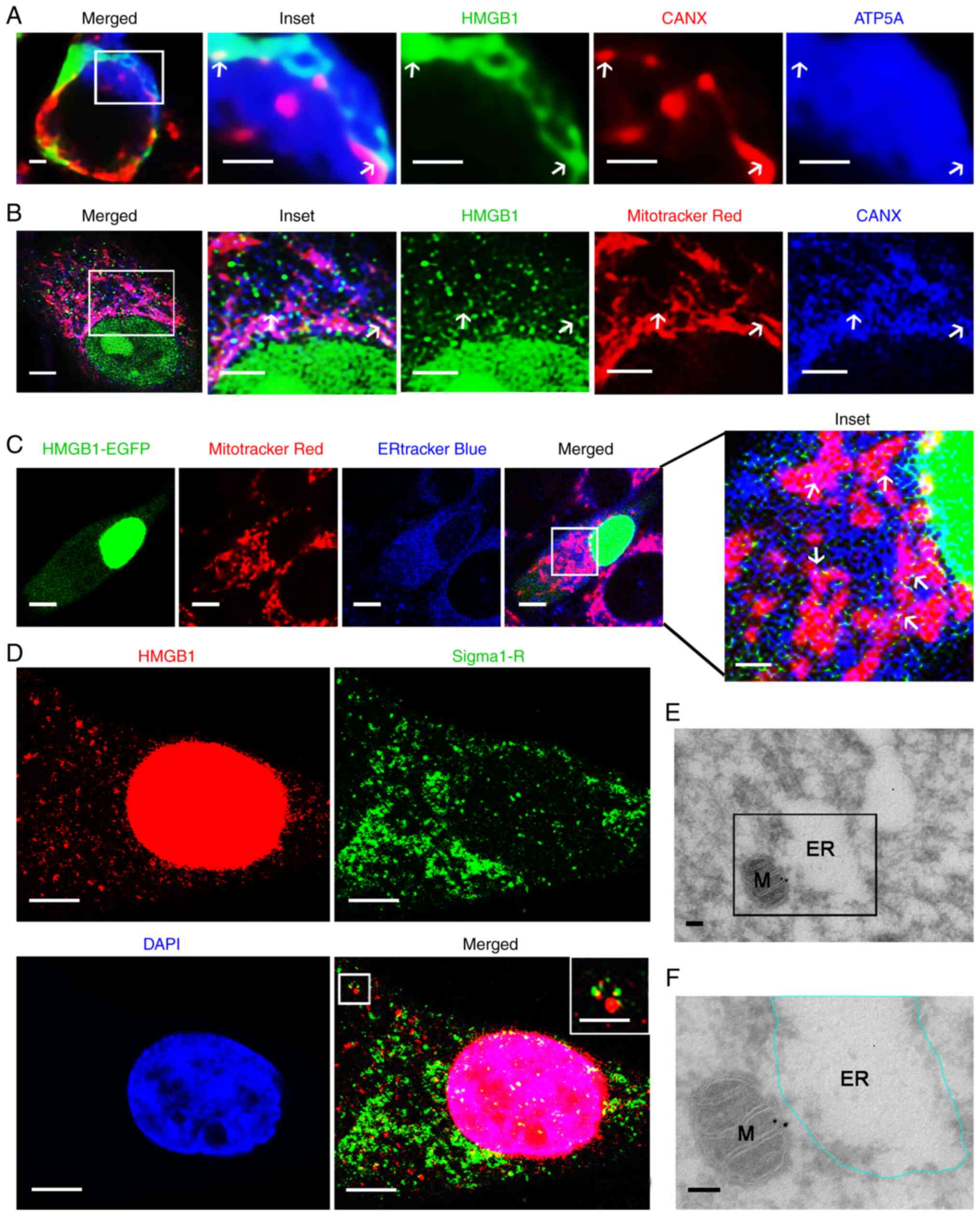 | Figure 8.HMGB1 is localized at MAMs in glioma.
(A) Paraffin-embedded glioma sections were co-stained for HMGB1 and
the ER marker, CANX, as well as the mitochondrial protein, ATP5A.
HMGB1 was detected at MAMs, as indicated by arrows. The ‘inset’
image is an enlarged image of the boxed region in the merged image.
(B) HBSS-treated U87-MG cells were co-stained for HMGB1, CANX and
Mitotracker Red. Arrows indicate the localization of HMGB1 at MAMs.
The ‘inset’ image is an enlarged image of the boxed region in the
merged image. (C) U87-MG cells were transfected with the HMGB1-EGFP
plasmid and then treated with HBSS. ER and mitochondria were
labeled with ERtracker Blue and Mitotracker Red, respectively. The
‘inset’ image is an enlarged image of the boxed region in the
merged image. White arrows indicate the localization of HMGB1 in
MAMs. (D) HBSS-treated U87-MG cells were co-stained for HMGB1 and
Sigma1-R. The boxed arean the upper right corner is an enlarge
image of the boxed area on the upper left corner. (E)
Representative images of HBSS-treated U87-MG cells showing the
localization of HMGB1 by immune gold. The boxed area and expanded
ER are amplified and outlined, respectively, in (F). M,
mitochondria. Scale bars: (A) 1 µm, (B) lower magnified image 3 µm
and higher magnified images 2 µm, (C) lower magnified images 5 µm
and higher magnified image 1 µm, (D) 2 µm, and inset image 1 µm, (E
and F) 100 nm. HMGB1, high mobility group box 1; MAMs,
mitochondria-associated endoplasmic reticulum membranes; ER,
endoplasmic reticulum; CANX, calnexin; ATP5A, ATP synthase F1
subunit alpha; HBSS, Hank's balanced salt solution; Sigma1-R, sigma
1 receptor. |
Autophagy contributes to the release
of HMGB1 in glioma cells following starvation
Autophagy positively contributes to HMGB1 secretion
via an export pathway in non-tumor cells (35,36).
In the present study, HMGB1 translocated to the autophagosomes,
lysosomes, endosomes and MAMs [the assembly origin site of
autophagosomes (37)], all of which
are implicated in autophagy. These results suggest that autophagy
mediates the release of HMGB1 from starved glioma cells. As
demonstrated in Figs. 9A and B, and
S5, the expression of the
autophagy marker, LC3B-II, was significantly increased, while that
of the autophagy-related protein sequestosome 1 (p62; this protein
is incorporated into the autophagosome and then degraded during
autophagic process (38–40), was significantly decreased,
indicating an increase in autophagy levels following HBSS
treatment. Co-treatment with the early autophagy inhibitor, WOR,
decreased the expression of the autophagy marker, LC3B-II. WOR
inhibits the initiation of autophagy (41). Considering that CQ inhibits the
autophagic flux by decreasing autophagosome-lysosome fusion
(42), the accumulation of LC3B-II
was observed upon CQ treatment. The results of ELISA demonstrated
that HMGB1 secretion was promoted following HBSS treatment;
however, the secretion was inhibited after WOR and CQ were applied
(Fig. 9C). The results of LDH
release and CCK-8 assays revealed that the increased secretion of
HMGB1 was not due to increased non-specific membrane permeability
or cell death (Fig. 9D and E).
Furthermore, we observed accumulation of HMGB1 containing
autophagosomes after the autophagic flux was inhibit by CQ in
glioma cells (Fig. 9F-H).
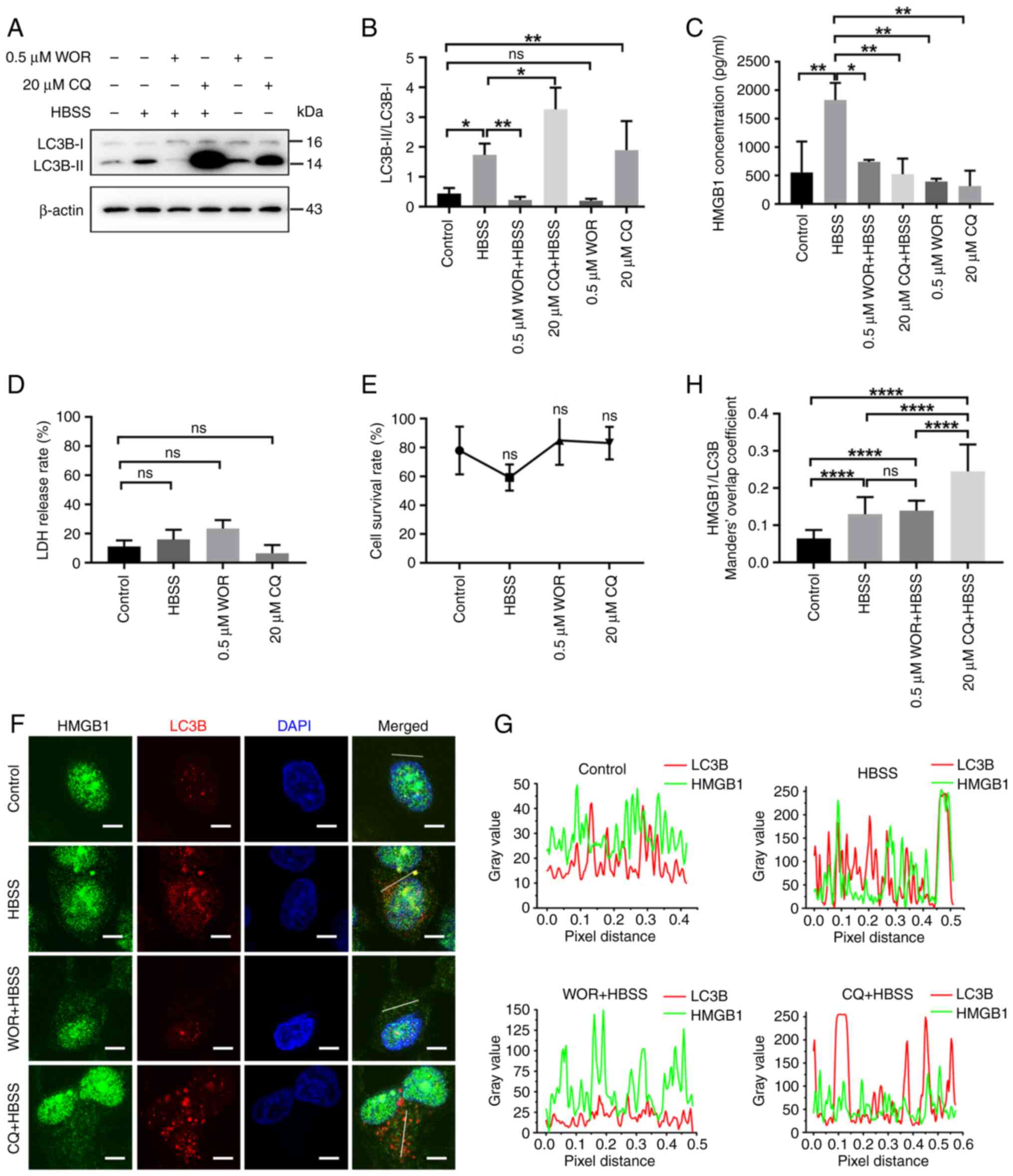 | Figure 9.HMGB1 secretion is regulated by
autophagy-mediated secretion in glioma cells. (A) U251 glioma cells
were pretreated with 0.5 µM WOR and 20 µM CQ for 2 h and then
treated with HBSS for 3 h. Whole cell lysates immunoblotted with
anti-LC3B and anti-β-actin antibodies. (B) Statistical analysis of
LC3B-II/LC3B-I ratio (n=3). (C) U251 glioma cells were treated with
0.5 µM WOR and 20 µM CQ for 2 h and then treated with HBSS for 3 h.
The secreted HMGB1 levels were measured using ELISA (n=3). (D) LDH
release assay determined the cytotoxicity of HBSS, 0.5 µM WOR and
20 µM CQ (n=3). (E) Treatment with HBSS (3 h), WOR (3 h, 0.5 µM),
or CQ (3 h, 20 µM) did not induce the apoptosis of U251 glioma
cells, as determined using CCK-8 assay (n=3). (F) U251 glioma cells
were treated with 0.5 µM WOR and 20 µM CQ for 2 h and then treated
with HBSS for 3 h. Immunostaining of HMGB1 and the autophagosome
marker, LC3B, in U251 glioma cells subjected to different
treatments. (G) Line fluorescence tracing from images in (F). (H)
Manders' overlap coefficient analysis of HMGB1 and LC3B (n=3, 10–15
cells per replicate). Data represent the mean ± SD; one-way ANOVA
followed by Tukey's post-hoc test was used to evaluate statistical
significance. *P<0.05, **P<0.01 and ****P<0.0001. ns, not
significant. HMGB1, high mobility group box 1; WOR, wortmannin; CQ,
chloroquine; HBSS, Hank's balanced salt solution; LC3B,
microtubule-associated proteins 1A/1B light chain 3B; LDH, Lactate
dehydrogenase; CCK-8, Cell Counting Kit-8. |
Discussion
Previous studies have investigated the translocation
of HMGB1 to neurons (43),
immunocytes (5) and glioma cells
(44). These studies clearly
revealed the translocation of HMGB1 from the nucleus to the
extracellular space through cytosolic compartments. Although HMGB1
in the cytoplasm generally exhibits a characteristic granular
staining pattern (43,45), information regarding its subcellular
localization of HMGB1 in the cytoplasm is limited, particularly in
cancer cells (46). In the present
study, the translocation of HMGB1 in glioma cells under starvation
stress was confirmed. It was observed that HMGB1 was expressed in
the nucleus, cytoplasm and extracellular space of the glioma cells.
This result is consistent with the tissue array results in a
previous study by the authors (44). Similarly, in cultured glioma cells,
HMGB1 was released from the nucleus into the cytoplasm and was
eventually secreted into the exterior under conditions of
starvation. In addition to starvation, it was also observed that
H2O2-induced stress caused HMGB1 to
translocate from the nucleus to the cytoplasm and locate on the
mitochondria (Fig. S1). This
result indicates that HMGB1 translocation is common in glioma cells
under stress. ICC and immunoelectron microscopy revealed that HMGB1
was imported into the mitochondria, ER, peroxisomes,
autophagosomes, lysosomes, early endosomes, late endosomes and the
cytoskeleton. In addition, the localization of HMGB1 in these
compartments changed with prolonged starvation, indicating a
dynamic association between HMGB1 and cytosolic compartments during
its release. The release of HMGB1 has been shown to promote tumor
growth and metastases through its cytokine, chemokine and growth
factor activities, whereas cytoplasmic HMGB1 increases
chemoresistance due to its pro-autophagic activity (47,48).
HMGB1 cannot enter the ER for processing as there is no signaling
peptide enabling this action; that is, HMGB1 is rather secreted via
an endosome-lysosome or autophagy-mediated non-canonical pathway
and not extracellularly via the classical ER-Golgi secretion
pathway (36,49). HMGB1 co-localizes with the lysosomal
marker protein, LAMP1, suggesting that HMGB1 may be secreted into
the extracellular matrix from lysosomes or endosome-lysosomes
(50). Different cell types release
HMGB1 via different mechanisms. In monocytes, HMGB1 is first
translocated from the nucleus to the cytoplasm and is then
encapsulated in secretory lysosomes or other membrane organelles.
These membrane structures then fuse with the cell membrane to
release HMGB1 (50). Kim et
al (36) reported that early
autophagy and late endosomes mediate the exocrine pathway of HMGB1
in 293T cells, human monocyte THP-1 cells, and mouse embryonic
fibroblasts. In psoriatic keratinocytes, early and late autophagy
play pivotal roles in the extracellular secretion of HMGB1
(35).
Understanding the HMGB1 secretory pathway is
crucial, since extracellular HMGB1 has pro-inflammatory functions
and serves as a multifunctional alarm to regulate cell
proliferation, tissue remodeling and tumor progression (36). In the present study, treatment with
WOR, which inhibits the initiation of autophagy (41) and CQ, which reduces
autophagosome-lysosome fusion (42), prevent the extracellular secretion
of HMGB1, supporting that HMGB1 secretion was mediated by early and
late autophagy in glioma cells under starvation stress. Elmaci
et al (51) reported that
HMGB1 was secreted into the extracellular space via autophagy in
dying glioma cells. The data of the present study demonstrated the
translocation of HMGB1 to subcellular compartments during its
release; cytosolic HMGB1 in glioma cells can be imported into or
around autophagosomes, endosomes, lysosomes and MAMs. Endosomes,
particularly late endosomes, are destined to evolve into lysosomes
(52). MAMs mark the initiation
sites of autophagosome formation (37). Thus, all these cellular compartments
are involved in autophagy. Additionally, the autophagy-lysosomal
pathway may mediate the selective release of HMGB1 in glioma cells
under nutrient-poor conditions, which do not induce cell death or
non-specific membrane permeability.
In addition to the autophagy-related cellular
compartments, HMGB1 translocation inside the lumen of the
mitochondria and ER was observed, as well as in the peroxisomes and
cytoskeleton. Previous studies have examined the localization of
HMGB1 in the mitochondria, demonstrating also that HMGB1
translocation to the mitochondria may affect mitochondrial
morphology, energy metabolism, and autophagy (53). In addition, HMGB1 has a high
affinity to mitochondrial DNA (mtDNA) released from injured
hepatocellular carcinoma cells (54). Thus, HMGB1 localization in the
mitochondria may be associated with remodeling of morphology and
energy metabolism, as well as mtDNA release following starvation.
Moreover, HMGB1 was localized in decomposed mitochondria engulfed
by autolysosomes (Fig. 6I),
suggesting that HMGB1 may mediate mitophagy in starved glioma
cells. In addition, hyperoxic conditions increase the generation of
mitochondrial reactive oxygen species and the overexpression and
secretion of HMGB1 in the brain and lungs, indicating a potential
association between mitochondrial dysfunction and HMGB1
translocation (53). HMGB1 lacks
leader peptides and thus cannot enter the conventional ER-to-Golgi
secretory pathway (36). However,
herein, immunoelectron microscopy demonstrated that HMGB1 particles
were localized in the ER lumen (Fig. 6E
and J). Previous studies have revealed that HMGB1 plays an
essential role in ER stress (55,56).
However, the association between HMGB1 and the ER in gliomas
requires further elucidation in future studies. Wang et al
(43) demonstrated that HMGB1 can
be imported into the peroxisomes of ischemic neurons. In the
present study, HMGB1 and the peroxisome marker protein catalase
co-localized in glioma cells. Catalase is an
H2O2-degrading enzyme. In addition, other
redox-related enzymes can also be detected in peroxisomes.
Therefore, it was hypothesized that peroxisomes are involved in the
regulation of the redox state and related intracellular signaling
pathways (57,58). In addition, HMGB1 co-localized with
GFAP, and the Manders' overlap coefficient between them increased
with the release of HMGB1. GFAP is a well-known marker protein for
astrocytes and an intermediate filament. Previous studies have
reported the co-localization of HMGB1 and GFAP (25,59).
The cytoskeleton tracks vesicular trafficking, and aberrant
vesicular trafficking and cytoskeletal interactions have been
observed in cancer cells (60).
Accordingly, it was hypothesized that the cytoskeleton is required
for vesicular trafficking and extracellular secretion of HMGB1 in
glioma cells under conditions of starvation.
The present study has some limitations. For example,
it was not investigated whether HMGB1 is transported to the Golgi
apparatus. Immunoelectron microscopy with gold particles of
different sizes is suitable for precisely distinguishing between
organelles. In addition, ascorbate peroxidase-based proximity
labeling technique can be used to image HMGB1 profiles in
subcellular compartments using electron microscopy (61,62).
Several functional factors, including Golgi reassembly stacking
protein 2, ADP ribosylation factor 1 and secretion-associated
Ras-related GTPase 1A, are required for secretory autophagy
(36). The Ras-related proteins
RAB8a, RAB11a and RAB27a regulate polarized membrane trafficking
and plasma membrane fusion (63–65).
Furthermore, vesicle-associated membrane protein 7,
synaptotagmin-7, or altering the dynamics of lysosomes by
inhibiting ADP ribosylation factor like GTPase 8B, could prevent
lysosome exocytosis (66–68). Therefore, knockdown or
overexpression experiments are necessary to verify the functions of
these factors in HMGB1 secretion. The present study was conducted
primarily in vitro. In vivo experiments also need to be
conducted to complete the entire study design.
In conclusion, the present study demonstrated that
starvation-induced stress induced the translocation of HMGB1 from
the nucleus to the cytoplasm and extracellular space in glioma
cells. In the cytoplasm, HMGB1 was transported to various cellular
compartments, including the mitochondria, ER, MAMs, peroxisomes,
autophagosomes, early endosomes, late endosomes, lysosomes and
cytoskeleton. Early and late autophagy may be involved in the
extracellular secretion of HMGB1 from glioma cells under the same
conditions (Fig. 10).
Extracellular HMGB1 is a risk factor for several malignancies. The
aforementioned results contribute to the further understanding of
the translocation and secretion pathways of HMGB1, which may
provide a novel perspective for designing inhibitors of HMGB1
secretion for the treatment of glioma.
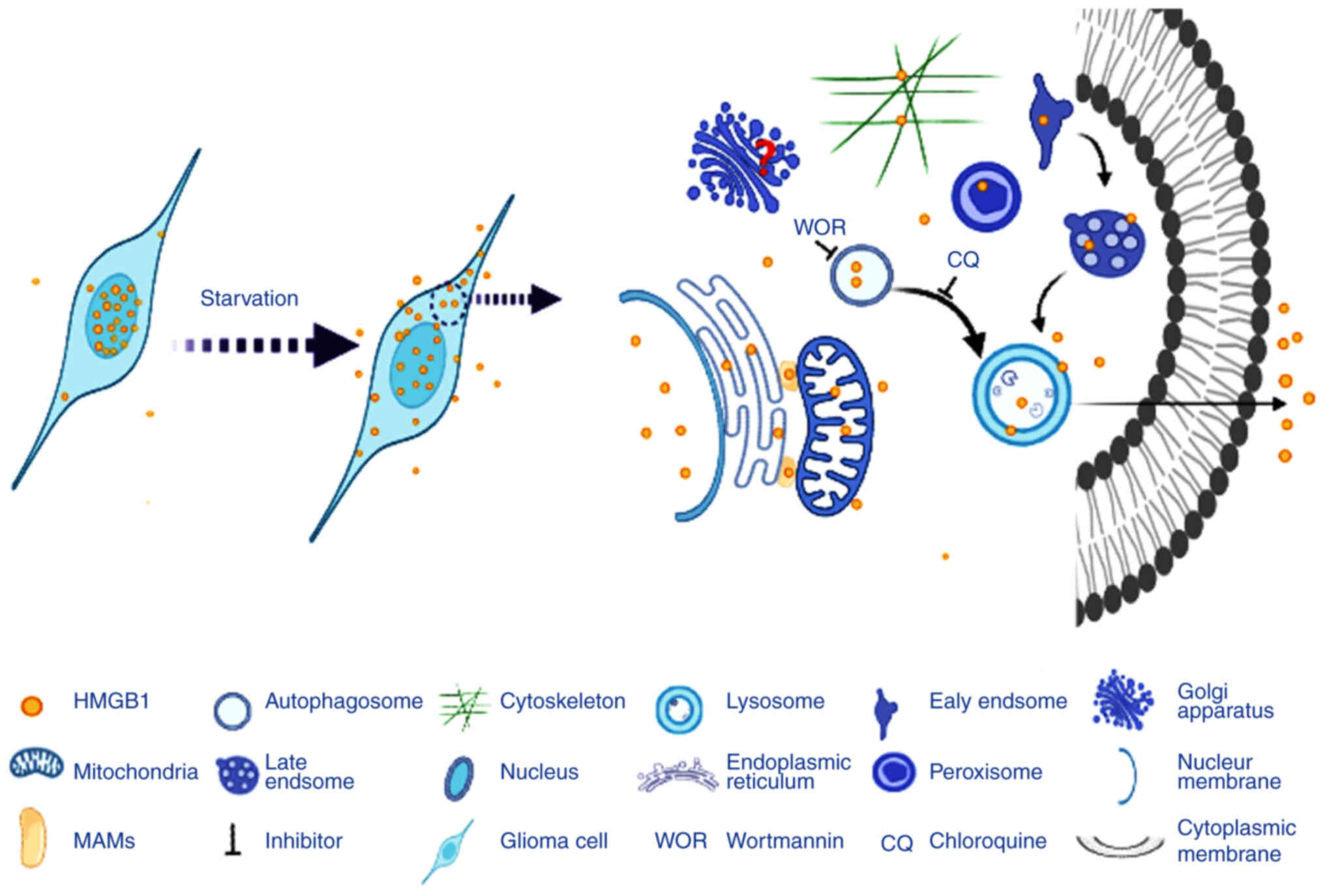 | Figure 10.Schematic diagram demonstrating the
subcellular localization of HMGB1 in glioma cells. Starvation
stress triggers the translocation of HMGB1 from the nucleus to the
cytoplasm and the extracellular milieu. In the cytoplasm, HMGB1 can
be imported into the mitochondria, ER, MAMs, peroxisomes,
autophagosomes, lysosomes, early endosomes, late endosomes and
cytoskeleton. Early (inhibited by WOR) and late autophagy
(inhibited by CQ) mediated the extracellular secretion of HMGB1.
HMGB1, high mobility group box 1; ER, endoplasmic reticulum; MAMs,
mitochondria-associated endoplasmic reticulum membranes; WOR,
wortmannin; CQ, chloroquine. |
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Xia Chu from the
Department of Histology at the 988th Hospital of the Joint Logistic
Support Force (Zhengzhou, China) for providing the tissue sections,
and Dr Ying Li from the Biomedical Testing Center of Tsinghua
University (Beijing, China) for providing technical assistance.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81402455), China Postdoctoral
Science Foundation (grant no. 2019M653946) the Cultivation Fund of
Zhengzhou University in 2021 (grant no. JC21835040) and the opening
project of State Key Laboratory of Explosion Science and Technology
(Beijing Institute of Technology) (grant no. KFJJ23-09M).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
All authors (XC, AY and LJ) contributed to the
conception and design of this study. Material preparation, data
collection and analyses were performed by XC and AY. The first
draft of the manuscript was written by LJ, and all authors
commented on previous versions of the manuscript. All authors
confirm the authenticity of the raw data and have read and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the principles of the Declaration of Helsinki. Approval was granted
by the Ethics Committees of 988th Hospital of the Joint Logistic
Support Force (Zhengzhou, China). Written informed consent was
obtained from all the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Perry A and Wesseling P: Histologic
classification of gliomas. Handb Clin Neurol. 134:71–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu S, Tang L, Li X, Fan F and Liu Z:
Immunotherapy for glioma: Current management and future
application. Cancer Lett. 476:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mendonça Gorgulho C, Murthy P, Liotta L,
Espina V and Lotze MT: Different measures of HMGB1 location in
cancer immunology. Methods Enzymol. 629:195–217. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Le Y, Wang Y, Zhou L, Xiong J, Tian J,
Yang X, Gai X and Sun Y: Cigarette smoke-induced HMGB1
translocation and release contribute to migration and NF-κB
activation through inducing autophagy in lung macrophages. J Cell
Mol Med. 24:1319–1331. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang S and Zhang Y: HMGB1 in inflammation
and cancer. J Hematol Oncol. 13:1162020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Völp K, Brezniceanu ML, Bösser S, Brabletz
T, Kirchner T, Göttel D, Joos S and Zörnig M: Increased expression
of high mobility group box 1 (HMGB1) is associated with an elevated
level of the antiapoptotic c-IAP2 protein in human colon
carcinomas. Gut. 55:234–242. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akaike H, Kono K, Sugai H, Takahashi A,
Mimura K, Kawaguchi Y and Fujii H: Expression of high mobility
group box chromosomal protein-1 (HMGB-1) in gastric cancer.
Anticancer Res. 27:449–457. 2007.PubMed/NCBI
|
|
9
|
Ranganathan A, Gunnarsson O and Casarett
D: Palliative care and advance care planning for patients with
advanced malignancies. Ann Palliat Med. 3:144–149. 2014.PubMed/NCBI
|
|
10
|
Seidu RA, Wu M, Su Z and Xu H: Paradoxical
role of high mobility group box 1 in glioma: A suppressor or a
promoter? Oncol Rev. 11:3252017.PubMed/NCBI
|
|
11
|
Xue J, Suarez JS, Minaai M, Li S, Gaudino
G, Pass HI, Carbone M and Yang H: HMGB1 as a therapeutic target in
disease. J Cell Physiol. 236:3406–3419. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao M, Zhang Y, Jiang Y, Wang K, Wang X,
Zhou D, Wang Y, Yu R and Zhou X: YAP promotes autophagy and
progression of gliomas via upregulating HMGB1. J Exp Clin Cancer
Res. 40:992021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang XJ, Zhou SL, Fu XD, Zhang YY, Liang
B, Shou JX, Wang JY and Ma J: Clinical and prognostic significance
of high-mobility group box-1 in human gliomas. Exp Ther Med.
9:513–518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao XY, Zang J, Zheng MH, Zhang YF, Yue
KY, Cao XL, Cao Y, Li XX, Han H, Jiang XF and Liang L: Temozolomide
treatment induces HMGB1 to promote the formation of glioma stem
cells via the TLR2/NEAT1/Wnt pathway in glioblastoma. Front Cell
Dev Biol. 9:6208832021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Fu WJ, Chen XQ, Wang S, Deng RS,
Tang XP, Yang KD, Niu Q, Zhou H, Li QR, et al: Autophagy-based
unconventional secretion of HMGB1 in glioblastoma promotes
chemosensitivity to temozolomide through macrophage M1-like
polarization. J Exp Clin Cancer Res. 41:742022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grasmann G, Smolle E, Olschewski H and
Leithner K: Gluconeogenesis in cancer cells-repurposing of a
starvation-induced metabolic pathway? Biochim Biophys Acta Rev
Cancer. 1872:24–36. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Finicle BT, Jayashankar V and Edinger AL:
Nutrient scavenging in cancer. Nat Rev Cancer. 18:619–633. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Komori T: The 2016 WHO classification of
tumours of the central nervous system: The major points of
revision. Neurol Med Chir (Tokyo). 57:301–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan W, Song Y, Ren Z, Cheng X, Li P, Song
H and Jia L: Glioma cells are resistant to inflammation-induced
alterations of mitochondrial dynamics. Int J Oncol. 57:1293–1306.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prajapati P, Wang WX, Nelson PT and
Springer JE: Methodology for subcellular fractionation and MicroRNA
examination of mitochondria, mitochondria associated ER membrane
(MAM), ER, and cytosol from human brain. Methods Mol Biol.
2063:139–154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang M, Kenny SJ, Ge L, Xu K and Schekman
R: Translocation of interleukin-1β into a vesicle intermediate in
autophagy-mediated secretion. Elife. 4:e112052015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Penjweini R, Deville S, Haji Maghsoudi O,
Notelaers K, Ethirajan A and Ameloot M: Investigating the effect of
poly-l-lactic acid nanoparticles carrying hypericin on the
flow-biased diffusive motion of HeLa cell organelles. J Pharm
Pharmacol. 71:104–116. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suwannakul N, Midorikawa K, Du C, Qi YP,
Zhang J, Xiang BD, Murata M and Ma N: Subcellular localization of
HMGB1 in human cholangiocarcinoma: Correlation with tumor stage.
Discov Oncol. 12:492021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Satoh TK: The role of HMGB1 in
inflammatory skin diseases. J Dermatol Sci. 107:58–64. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang X, Zhou W, Li X, Ren J, Ji G, Du J,
Tian W, Liu Q and Hao A: Graphene oxide suppresses the growth and
malignancy of glioblastoma stem cell-like spheroids via epigenetic
mechanisms. J Transl Med. 18:2002020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Asperen JV, Robe PAJT and Hol EM: GFAP
alternative splicing and the relevance for disease-a focus on
diffuse gliomas. ASN Neuro. 14:175909142211020652022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uceda-Castro R, van Asperen JV, Vennin C,
Sluijs JA, van Bodegraven EJ, Margarido AS, Robe PAJ, van Rheenen J
and Hol EM: GFAP splice variants fine-tune glioma cell invasion and
tumour dynamics by modulating migration persistence. Sci Rep.
12:4242022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pastorek L, Sobol M and Hozák P:
Colocalization coefficients evaluating the distribution of
molecular targets in microscopy methods based on pointed patterns.
Histochem Cell Biol. 146:391–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khushi M, Napier CE, Smyth CM, Reddel RR
and Arthur JW: MatCol: A tool to measure fluorescence signal
colocalisation in biological systems. Sci Rep. 7:88792017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Le Vasseur M, Chen VC, Huang K, Vogl WA
and Naus CC: Pannexin 2 localizes at ER-mitochondria contact sites.
Cancers (Basel). 11:3432019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Klionsky DJ, Abdel-Aziz AK, Abdelfatah S,
Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu
YP, Acevedo-Arozena A, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy (4th edition).
Autophagy. 17:1–382. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Morciano G, Marchi S, Morganti C, Sbano L,
Bittremieux M, Kerkhofs M, Corricelli M, Danese A,
Karkucinska-Wieckowska A, Wieckowski MR, et al: Role of
mitochondria-associated ER membranes in calcium regulation in
cancer-specific settings. Neoplasia. 20:510–523. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Murley A, Sarsam RD, Toulmay A, Yamada J,
Prinz WA and Nunnari J: Ltc1 is an ER-localized sterol transporter
and a component of ER-mitochondria and ER-vacuole contacts. J Cell
Biol. 209:539–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eisenberg-Bord M, Shai N, Schuldiner M and
Bohnert M: A tether is a tether is a tether: Tethering at membrane
contact sites. Dev Cell. 39:395–409. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Z, Zhou H, Zheng H, Zhou X, Shen G,
Teng X, Liu X, Zhang J, Wei X, Hu Z, et al: Autophagy-based
unconventional secretion of HMGB1 by keratinocytes plays a pivotal
role in psoriatic skin inflammation. Autophagy. 17:529–552. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim YH, Kwak MS, Lee B, Shin JM, Aum S,
Park IH, Lee MG and Shin JS: Secretory autophagy machinery and
vesicular trafficking are involved in HMGB1 secretion. Autophagy.
17:2345–2362. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu Y, Chen H, Zhang L, Lin X, Li X, Zhuang
H, Fan H, Meng T, He Z, Huang H, et al: The AMPK-MFN2 axis
regulates MAM dynamics and autophagy induced by energy stresses.
Autophagy. 17:1142–1156. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bjørkøy G, Lamark T, Brech A, Outzen H,
Perander M, Overvatn A, Stenmark H and Johansen T: p62/SQSTM1 forms
protein aggregates degraded by autophagy and has a protective
effect on huntingtin-induced cell death. J Cell Biol. 171:603–614.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Komatsu M, Waguri S, Koike M, Sou YS, Ueno
T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al:
Homeostatic levels of p62 control cytoplasmic inclusion body
formation in autophagy-deficient mice. Cell. 131:1149–1163. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pankiv S, Clausen TH, Lamark T, Brech A,
Bruun JA, Outzen H, Øvervatn A, Bjørkøy G and Johansen T:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–24145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Turkoz Uluer E, Kilicaslan Sonmez P,
Akogullari D, Onal M, Tanriover G and Inan S: Do wortmannin and
thalidomide induce apoptosis by autophagy inhibition in 4T1 breast
cancer cells in vitro and in vivo? Am J Transl Res. 13:6236–6247.
2021.PubMed/NCBI
|
|
42
|
Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr
M, Hijlkema KJ, Coppes RP, Engedal N, Mari M and Reggiori F:
Chloroquine inhibits autophagic flux by decreasing
autophagosome-lysosome fusion. Autophagy. 14:1435–1455. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang D, Liu K, Fukuyasu Y, Teshigawara K,
Fu L, Wake H, Ohtsuka A and Nishibori M: HMGB1 translocation in
neurons after ischemic insult: Subcellular localization in
mitochondria and peroxisomes. Cells. 9:6432020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jia L, Song Y, Song H, Wang G, Fan W, Li
X, Zheng H and Yao A: Overexpression of high mobility group box 1
(HMGB1) has no correlation with the prognosis in glioma. Biomark
Med. 13:851–863. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fu L, Liu K, Wake H, Teshigawara K,
Yoshino T, Takahashi H, Mori S and Nishibori M: Therapeutic effects
of anti-HMGB1 monoclonal antibody on pilocarpine-induced status
epilepticus in mice. Sci Rep. 7:11792017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kostova N, Zlateva S, Ugrinova I and
Pasheva E: The expression of HMGB1 protein and its receptor RAGE in
human malignant tumors. Mol Cell Biochem. 337:251–258. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Taguchi A, Blood DC, del Toro G, Canet A,
Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, et al: Blockade of
RAGE-amphoterin signalling suppresses tumour growth and metastases.
Nature. 405:354–360. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bald T, Quast T, Landsberg J, Rogava M,
Glodde N, Lopez-Ramos D, Kohlmeyer J, Riesenberg S, van den
Boorn-Konijnenberg D, Hömig-Hölzel C, et al:
Ultraviolet-radiation-induced inflammation promotes angiotropism
and metastasis in melanoma. Nature. 507:109–113. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
New J and Thomas SM: Autophagy-dependent
secretion: Mechanism, factors secreted, and disease implications.
Autophagy. 15:1682–1693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Andersson U, Yang H and Harris H:
High-mobility group box 1 protein (HMGB1) operates as an alarmin
outside as well as inside cells. Semin Immunol. 38:40–48. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Elmaci I, Alturfan EE, Cengiz S, Ozpinar A
and Altinoz MA: Neuroprotective and tumoricidal activities of
cardiac glycosides. Could oleandrin be a new weapon against stroke
and glioblastoma? Int J Neurosci. 128:865–877. 2018.PubMed/NCBI
|
|
52
|
Fraser J, Simpson J, Fontana R,
Kishi-Itakura C, Ktistakis NT and Gammoh N: Targeting of early
endosomes by autophagy facilitates EGFR recycling and signalling.
EMBO Rep. 20:e477342019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Okuma Y, Becker LB, Hayashida K, Aoki T,
Saeki K, Nishikimi M, Shoaib M, Miyara SJ, Yin T and Shinozaki K:
Effects of post-resuscitation normoxic therapy on oxygen-sensitive
oxidative stress in a rat model of cardiac arrest. J Am Heart
Assoc. 10:e0187732021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Y, Yan W, Tohme S, Chen M, Fu Y, Tian
D, Lotze M, Tang D and Tsung A: Hypoxia induced HMGB1 and
mitochondrial DNA interactions mediate tumor growth in
hepatocellular carcinoma through Toll-like receptor 9. J Hepatol.
63:114–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang GZ, Zhang K, Yang SQ, Zhang Z, Chen
S, Hou BJ and Yuan JY: VASPIN reduces inflammation and endoplasmic
reticulum stress of renal tubular epithelial cells by inhibiting
HMGB1 and relieves renal ischemia-reperfusion injury. Eur Rev Med
Pharmacol Sci. 24:8968–8977. 2020.PubMed/NCBI
|
|
56
|
Tu L, Long X, Song W, Lv Z, Zeng H, Wang
T, Liu X, Dong J and Xu P: MiR-34c acts as a tumor suppressor in
non-small cell lung cancer by inducing endoplasmic reticulum stress
through targeting HMGB1. Onco Targets Ther. 12:5729–5739. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Di Cara F, Andreoletti P, Trompier D,
Vejux A, Bülow MH, Sellin J, Lizard G, Cherkaoui-Malki M and Savary
S: Peroxisomes in immune response and inflammation. Int J Mol Sci.
20:38772019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lismont C, Revenco I and Fransen M:
Peroxisomal hydrogen peroxide metabolism and signaling in health
and disease. Int J Mol Sci. 20:36732019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fan H, Tang HB, Chen Z, Wang HQ, Zhang L,
Jiang Y, Li T, Yang CF, Wang XY, Li X, et al: Inhibiting HMGB1-RAGE
axis prevents pro-inflammatory macrophages/microglia polarization
and affords neuroprotection after spinal cord injury. J
Neuroinflammation. 17:2952020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Muro S: Alterations in cellular processes
involving vesicular trafficking and implications in drug delivery.
Biomimetics (Basel). 3:192018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Nguyen TMT, Kim J, Doan TT, Lee MW and Lee
M: APEX proximity labeling as a versatile tool for biological
research. Biochemistry. 59:260–269. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lam SS, Martell JD, Kamer KJ, Deerinck TJ,
Ellisman MH, Mootha VK and Ting AY: Directed evolution of APEX2 for
electron microscopy and proximity labeling. Nat Methods. 12:51–54.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang J, Zhang L, Chen H, Lei Y, Zhang T,
Wang Y, Jin P, Lan J, Zhou L, Huang Z, et al: Regorafenib induces
lethal autophagy arrest by stabilizing PSAT1 in glioblastoma.
Autophagy. 16:106–122. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Song L, Tang S, Han X, Jiang Z, Dong L,
Liu C, Liang X, Dong J, Qiu C, Wang Y and Du Y: KIBRA controls
exosome secretion via inhibiting the proteasomal degradation of
Rab27a. Nat Commun. 10:16392019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wall AA, Condon ND, Luo L and Stow JL:
Rab8a localisation and activation by Toll-like receptors on
macrophage macropinosomes. Philos Trans R Soc Lond B Biol Sci.
374:201801512019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wu PH, Onodera Y, Giaccia AJ, Le QT,
Shimizu S, Shirato H and Nam JM: Lysosomal trafficking mediated by
Arl8b and BORC promotes invasion of cancer cells that survive
radiation. Commun Biol. 3:6202020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tian X, Zheng P, Zhou C, Wang X, Ma H, Ma
W, Zhou X, Teng J and Chen J: DIPK2A promotes STX17- and
VAMP7-mediated autophagosome-lysosome fusion by binding to VAMP7B.
Autophagy. 16:797–810. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sleiman M, Stevens DR, Chitirala P and
Rettig J: Cytotoxic granule trafficking and fusion in
synaptotagmin7-deficient cytotoxic T lymphocytes. Front Immunol.
11:10802020. View Article : Google Scholar : PubMed/NCBI
|