Glioblastoma (GBM) is characterized by significant
genetic heterogeneity among tumor cells. This heterogeneity is
referred to as ‘polymorphism’, as tumor cells rapidly undergo
mitosis, resulting in the formation of numerous subclones and
uncertainty regarding the state of the genome. Understanding the
heterogeneity of GBM depends on the location of sampling and
analysis of subclonal fractionation or indeterminate genomic
status. Tumor cells are influenced by both genetic factors and
environmental elements in the microenvironment, resulting in a
complex regulation process (1). In
addition, intrinsic differences in subclonal tumor cells that arise
from random mutations can create distinct niches within limited
lesions. These cells are then forced to compete for growth and
nutrients (2). Various subclonal
tumor cells have the ability to modify the tumor microenvironment
in order to obtain adaptations. These adaptations include inducing
angiogenesis to acquire nutrient supply, interfering with immune
stimulation/inhibition checkpoint pathways to promote immune
evasion and remodeling the extracellular matrix to promote
metastasis (3). However, rather
than actively modifying the environment, multiple mechanisms guide
the evolution of tumor cells through the selection of subclones
with the most adaptive phenotype by environmental factors (1). Multiregional whole-exome or genome
sequencing has revealed that there is significant variation in the
genetic makeup of tumor cells across different anatomical locations
and within the same tumor over time (4). Furthermore, tumor heterogeneity has a
significant impact on both the immune microenvironment and the
infiltration of various immune cells within tumors, such as
cytotoxic T lymphocytes (CTLs) (5),
myeloid antigen-presenting cells (6) and cancer-associated fibroblasts (CAFs)
(7). This heterogeneity can vary
greatly between different types of immune cells, leading to further
complexity in understanding the immune response to tumors. It has
been discovered that the genetic structure of tumor cells and the
components of the immune microenvironment interact with each other.
This interaction results in a more complex alteration of both the
heterogeneity of tumor cells and the heterogeneity of the tumor
microenvironment. Consequently, the heterogeneity of tumor cells is
constantly evolving. It has been discovered that the genetic
structure of tumor cells and the components of the immune
microenvironment interact with each other. This interaction results
in a more complex alteration of both the heterogeneity of tumor
cells and the heterogeneity of the tumor microenvironment.
Consequently, the heterogeneity of tumor cells is constantly
evolving (8). Thus, tumor
heterogeneity has an important role in tumor development.
High-throughput sequencing methods are utilized to
analyze the mutational spectrum and evolutionary trajectories of
tumor cells. These studies reveal a wide range of genetic tumor
heterogeneity in both spatial and temporal dimensions, encompassing
diverse single-nucleotide mutations, insertions, deletions and copy
number variations (9,10). In primary tumors, mutations in
driver genes frequently provide a survival advantage and give rise
to a dominant clonal population. By contrast, mutations in
noncoding regions do not provide significant growth advantages
during tumor evolution (11).
Therefore, tumor evolution and spatiotemporal heterogeneity are
driven by genetic instability originating from both clonal and
subclonal tumor cells. This genetic heterogeneity also shapes the
antigenic profile of the tumor, ultimately contributing to the
heterogeneity of the tumor immune microenvironment (12). Neoantigens, which are primarily
derived from non-synonymous mutations and insertions/deletions,
were found to be the primary drivers of CD8+T
cell-specific differences. This suggests that genetic instability
is the root cause of the heterogeneity observed in the immune
microenvironment. Neoantigens have a significant role in the
variation of CD8+T cell specificity, highlighting the
importance of genetic instability in the diversity of the immune
microenvironment. Studies have shown that neutrophils, macrophage
M2 polarization and the inflammatory index are associated with
worse prognosis and overall survival in glioblastoma (13–15).
These findings indirectly suggest the presence of an inflammatory
response in tumor cells, as well as a deficiency in the mechanisms
that can trigger an immune response.
The formation of heterogeneity in the tumor immune
microenvironment is shaped by epigenetic remodeling of tumor cells,
as evidenced by various studies (16). This remodeling primarily occurs
through alterations in DNA modification, chromatin accessibility
and post-transcriptional regulation, such as gene expression
mediated by non-coding RNA interference. Epigenetic modifications
have a crucial role in accelerating the malignant transformation of
tumor cells and influencing the tumor immune microenvironment
(17). Various mechanisms,
including methylation, chromatin instabilities and epigenetic
remodeling, provide adaptive advantages to tumor cells in response
to their environment, such as in the progression of lung cancer
in situ (16). These
epigenetic modifications result in marked heterogeneity in both
spatial and longitudinal dimensions of genetic progeny, and also
impact tumor progression and immunogenicity by altering chromatin
accessibility and expression through immune-related elements
(18). Epigenetic modifications
have been linked to high levels of heterogeneity in various tumor
types, such as acute myeloid leukemia and glioblastoma, similar to
genetic instability.
Tumor cells and immune components in the
microenvironment are constantly exposed to radiation and
chemotherapy, leading to adaptive mechanisms in both tumor and
immune cells to establish a new balance (19). This balance is affected by the
intrinsic heterogeneity in tumor cell driver mutations or molecular
signatures, resulting in varying responses to therapy. In cases
where localized tumor clones fail to survive treatment, they
release large amounts of ATP through autophagy-mediated cell death
(20). ATPs have the ability to
promote chemotaxis and generate an inflammatory response in tumors.
However, overcoming immune heterogeneity has been shown to be
associated with therapeutic resistance and radio-resistance
(21). On the other hand, the
presence of extracellular nucleases can rapidly digest ATPs to
adenosine in the extracellular matrix, creating an inhibitory
immune microenvironment. This can hinder the immune response. In
addition, T-cell phenotypes can change significantly in response to
immune checkpoint blockade, affecting the immune cells. The immune
response to cancer treatment is characterized by a change of T-cell
subsets and cytokine production (22). For instance, certain patients with
glioblastoma who undergo chemotherapy exhibit a marked increase in
CD8+T cell proliferation post-treatment. The intricate
and ever-changing interplay between drug therapy, tumor cells and
immune cells has a crucial role in shaping the heterogeneous immune
microenvironment over time and space.
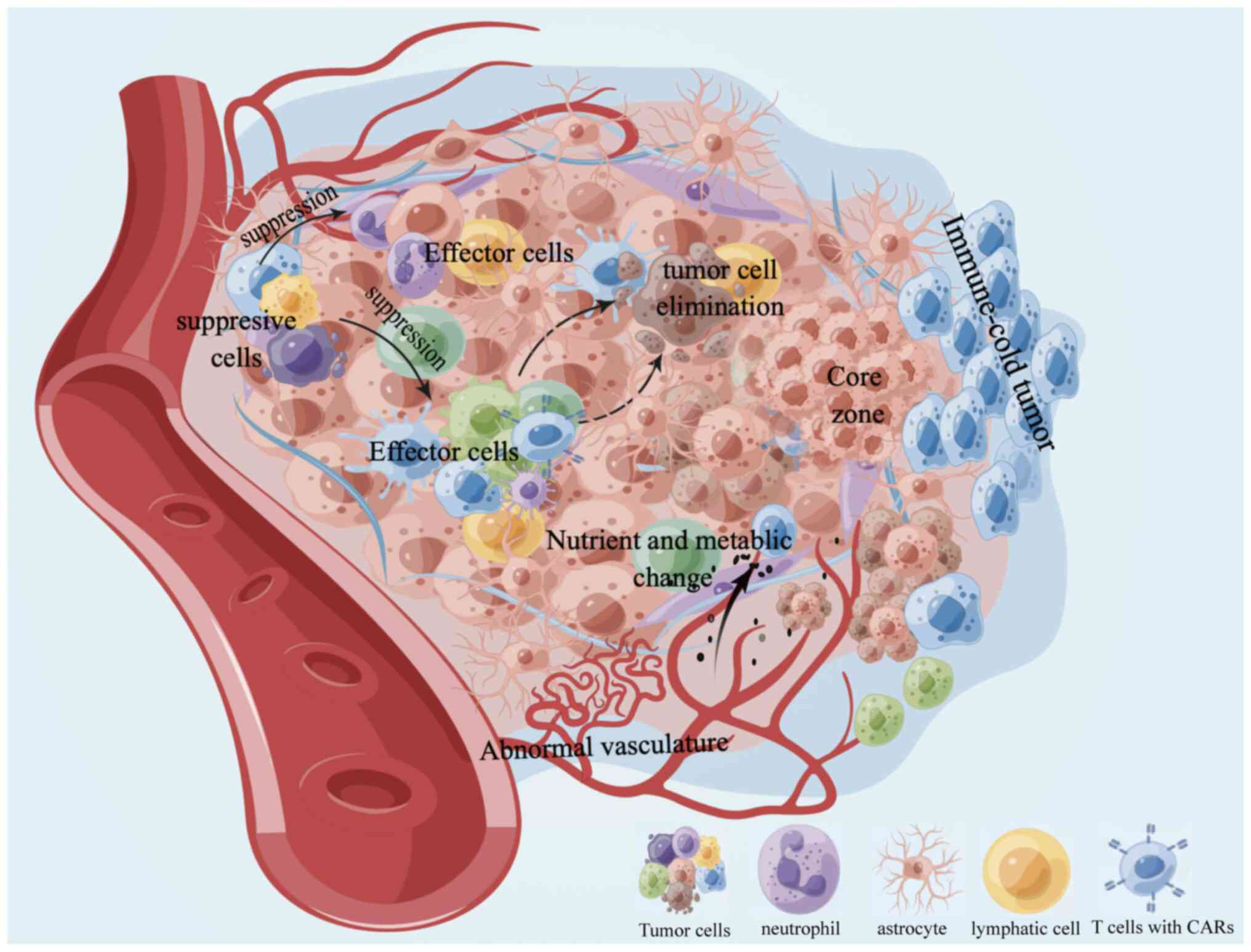
The tumor immune microenvironment is primarily
characterized by two components: Tumor and non-tumor. These
components are spatially distinct and have different localization
and abundance/activity. For instance, the inhibitory immune
checkpoint programmed cell death 1 (PD-1) ligand 1 is expressed on
the surface of tumor cells (23),
while immunosuppressive or pro-inflammatory cytokines are secreted
by both tumor and non-tumor cells (24,25).
Inhibitory or effector cells can infiltrate the microenvironment
and the state of the vasculature can also impact the immune
response (26). In addition, the
spatial distance of the marginal region and the distribution of
metabolized nutrients has a role in the microenvironment (27,28).
The spatial variations discussed in a subject paper have a
significant impact on both clinical prognosis and treatment
response (29). Various tumor types
have been found to exhibit intertumoral heterogeneity of immune
cells beyond T-cell subsets (30).
For instance, in glioma, macrophages with CD44 and CD109 phenotypes
were predominantly situated in the stroma, while the expression of
soluble CD10 was abnormally elevated in the core area compared to
the edge area (31).
Across different phenotypes, there is heterogeneity
in T-cell clonality, proliferative potential, differentiation
stage, functional polarization, cytokine secretion profile and
metabolic environment. In terms of T-cell repertoire propensity,
expanded/proliferated T-cell receptors (TCRs) are further
categorized as common TCR clones (which are detected in all regions
within the tumor) or regional clones (which have a heterogeneous
distribution) (30). The burden of
common and regional nonsynonymous mutations was found to be
positively correlated with the number of common and regional TCR
clones, suggesting regional heterogeneity and antigen-driven T-cell
proliferation. In addition, the metabolic profile has been
identified as a crucial regulator of the immune microenvironment,
potentially influencing the proliferation potential and
adaptability of cancer cells to their surroundings (32). The proliferation potential and
adaptability of cancer cells in the surrounding environment are
affected by metabolic characteristics, resulting in the production
of immunosuppressive mediators such as lactate and adenosine. These
mediators reduce the effectiveness of cytotoxic immune
surveillance. Cells with high glycolytic activity and malignant
cells can switch their metabolic pathways to anabolic reactions,
which can impact their behavior (33).
Tumor and immune cells can be influenced by both
genetic and non-genetic environmental factors, which can ultimately
impact the progression of the disease and the response to antitumor
therapy. In addition, these factors can also affect the dynamics of
the tumor cells themselves (34,35).
For instance, RNA-Seq analysis conducted on patients with
pancreatic ductal adenocarcinoma demonstrated significant
alterations in the composition of immune cell infiltration as the
disease progressed from a non-invasive lesion to an invasive
phenotype (36). Disease
progression across multiple tumor types is typically characterized
by a decrease in infiltration of CD8+T cells and dendritic cells,
as well as an abnormal accumulation or expansion of
immunosuppressive cells such as T-regulatory cells, myeloid-derived
suppressor cells or CAFs. In addition, impaired cytolytic activity,
expansion of the cell repertoire, clonality restriction and
progressive T- and B-cell exhaustion are also commonly observed.
Furthermore, spatiotemporal heterogeneity has been observed in
different tumor types, such as lung cancer (37), melanoma (38) and patients with intracranial
metastatic disease (39). The
significance of this heterogeneity for disease prognosis is
reinforced by the inverse relationship between the presence of
immune-favorable regions or lesions in individual patients and
disease control and survival outcomes (40). The present review focuses on new
theragnostic and therapeutic approaches that can combat the
heterogeneity of the tumor microenvironment (Fig. 1; Table
I).
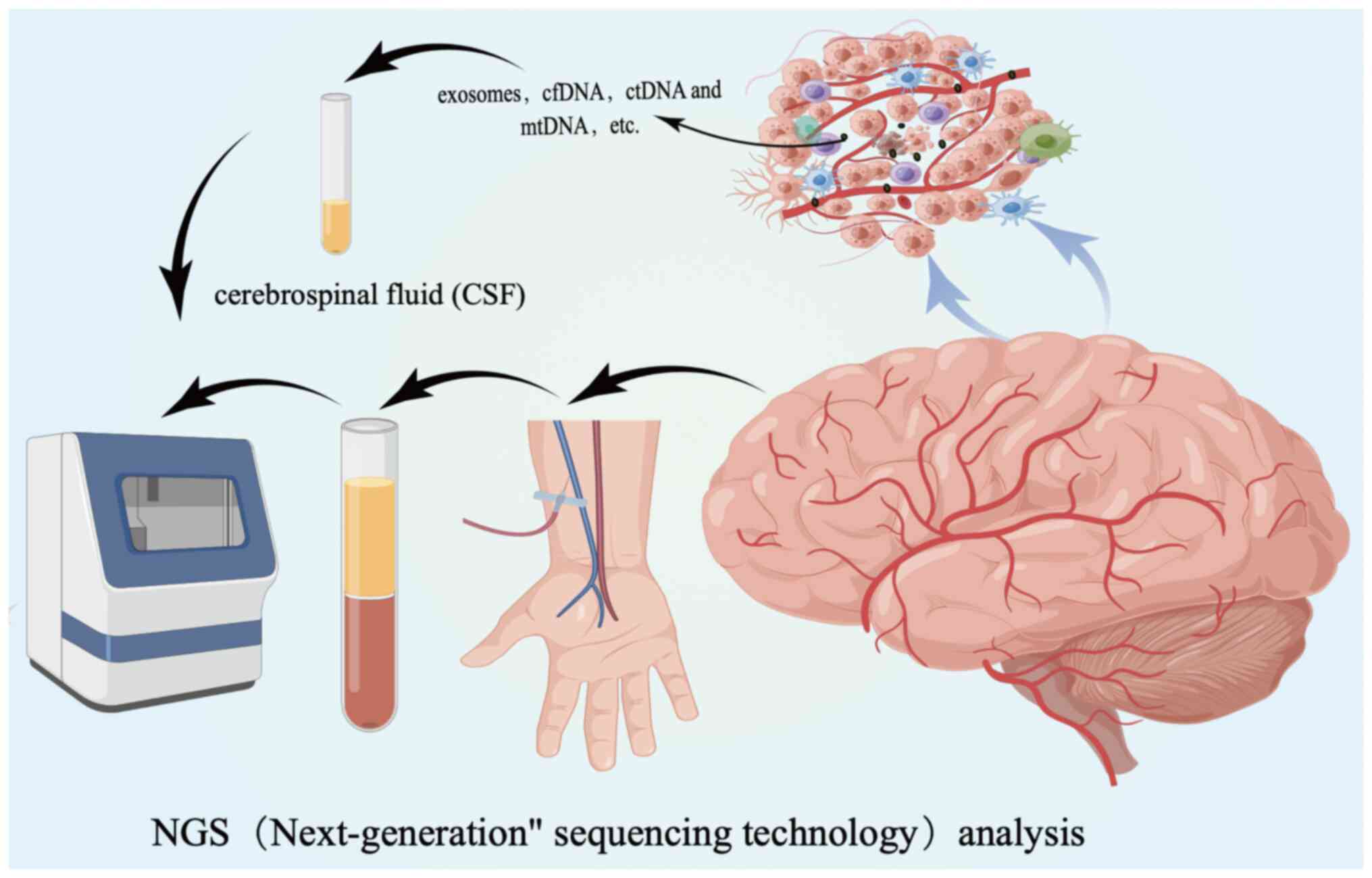
Recent studies have demonstrated the potential of
identifying extracellular vesicles, DNA and RNA particles, which
are collectively known as ‘liquid biopsies’, in monitoring disease
progression. This approach offers a glimpse into the heterogeneity
of the entire tumor cell population. Liquid biopsy is a quick and
cost-effective method for obtaining tumor-related information.
Integrating the analysis of extracellular vesicles and related
circulating nucleic acids into clinical practice can aid in
establishing a non-invasive diagnosis, conducting complex tumor
therapy and monitoring disease progression throughout the clinical
course (41). There is currently no
agreement among researchers regarding the optimal nucleic acid
types, biological fluids or pre-analysis/analysis techniques for
obtaining the most accurate results (42,43).
However, high-throughput sequencing technology, nanopore sequencing
technology and digital drop PCR have shown promise in analyzing the
genome and detecting various circulating DNA types, such as
cell-free DNA (cfDNA), circulating tumor DNA (ctDNA) and
mitochondrial DNA (mtDNA). These technologies can potentially
enhance the application of liquid biopsy technology. cfDNA is a
double-stranded structure that is usually 150–200 base pairs long
and has a concentration of ~10–15 ng/l. In patients with cancer,
the level of cfDNA increases significantly and the composition of
cfDNA changes over time. It is important to note that ctDNA, which
originates from tumor cells, is the major fraction of cfDNA
(44). The detection of
mutation-carrying ctDNA in plasma provides valuable information
about genetic changes in tumor tissue. However, ctDNA levels are
typically very low and have a short half-life in the case of GBM.
However, with the aid of highly sensitive and tumor-specific NGS,
mutations in B-RAF proto-oncogene, serine/threonine protein kinase,
isocitrate dehydrogenase (IDH1) and IDH2, and amplification of
tyrosine kinase receptor 2, mesenchymal-epithelial transition
factor, EGFR and platelet-derived growth factor receptor (PDGFR)α,
have been identified. In cases of primary brain malignancies, high
levels of ctDNA can be detected in cerebrospinal fluid (45). Changes in mtDNA can occur at an
early precancerous stage and elevated levels of mtDNA are
indicative of a poor prognosis. In addition, mtDNA may be present
in the future and is crucial in the early detection of new-onset
and recurrent GBM (46) (Fig. 2).
The objective of identifying new therapeutic targets
is to hinder various signaling pathways and stop the growth of
drug-resistant subclones, as well as immunosuppressive effects
(47). To date, the Food and Drug
Administration has approved two antineoplastic agents, temozolomide
and bevacizumab, and Tumor Treating Fields therapy for GBM after
postoperative radiotherapy and chemotherapy for glioblastoma
(48). In the BELOB study, the
combination of bevacizumab and lomustine was proven to
significantly enhance the overall survival rate of patients who
suffer from recurrent GBM (49).
Furthermore, the effectiveness of neoadjuvant pembrolizumab therapy
has been demonstrated in cases of recurrent and operable
glioblastoma (50). Glioblastoma is
characterized by an immunosuppressive tumor microenvironment,
minimal antigen presentation, unique antigen escape mechanisms and
direct immunosuppression (51).
Overall patient survival may be increased through an individualized
approach that combines drugs and various treatments (Table I).
Tumor cells can evade cellular immune responses by
binding specific antigens expressed on the surface of T cells, such
as CTL-associated protein (CTLA)-4 and PD-1, to their corresponding
ligands on the tumor surface. This results in reduced activation of
cytotoxic T cells, which may lead to the tumor cells avoiding
elimination. However, monoclonal antibodies that selectively block
these antigens have been found to stimulate an immune response.
According to clinical trials, anti-CTLA4 (nivolumab, pilimumab) and
anti-PD1 (pembrolizumab) have shown promising results (51). In addition, the enzyme indoleamine
2,3-dioxygenase, which has a crucial role in T-cell activity, has
exhibited checkpoint activity inhibition through the use of
methylated tryptophan indole oxime.
Vaccine therapy is an innovative approach to
treating the immune system, and it has the potential to be a
valuable tool in the battle against tumors. When tumor-associated
and tumor-specific antigens are introduced, they stimulate an
immune response. For instance, a protein vaccine called
lindopipate, which targets EGFR variant III, has shown promising
early results (52). One potential
target for cancer treatment is survivin, a member of the inhibitor
of apoptosis protein family, which can be targeted through the use
of a protein-based vaccine called SurVaxM (53). Another effective approach is the use
of activated autologous dendritic cells, such as DCVax. The most
prominent personalized cancer vaccine is heat shock protein (HSP)
peptide complex-96, which works by training the patient's immune
system to recognize the gp96 HSP and its associated proteins. This
protein is extracted from the patient's own tumor tissue (54).
The use of viral vectors that can integrate into the
host tumor genome is a form of immunotherapy. These vectors carry
genes that code for enzymes or other proteins that can be lethal to
tumor cells. Toca FC is a prodrug of the cytosine deaminase gene
and 5-fluorouracil, which can cross the blood-brain barrier and be
converted to 5-fluorouracil locally (55). Oncolytic viruses are viral particles
that have been genetically engineered to target and destroy tumor
cells, while leaving surrounding brain tissue unharmed.
Delta-24-RGD has been shown to have a strong antitumor effect on
its own and has also been effective when used in combination with
pembrolizumab (56). Due to the
variable infectivity of the virus, differential changes in
treatment may occur. Consequently, viral therapy should be utilized
as a complementary treatment rather than as a standalone
option.
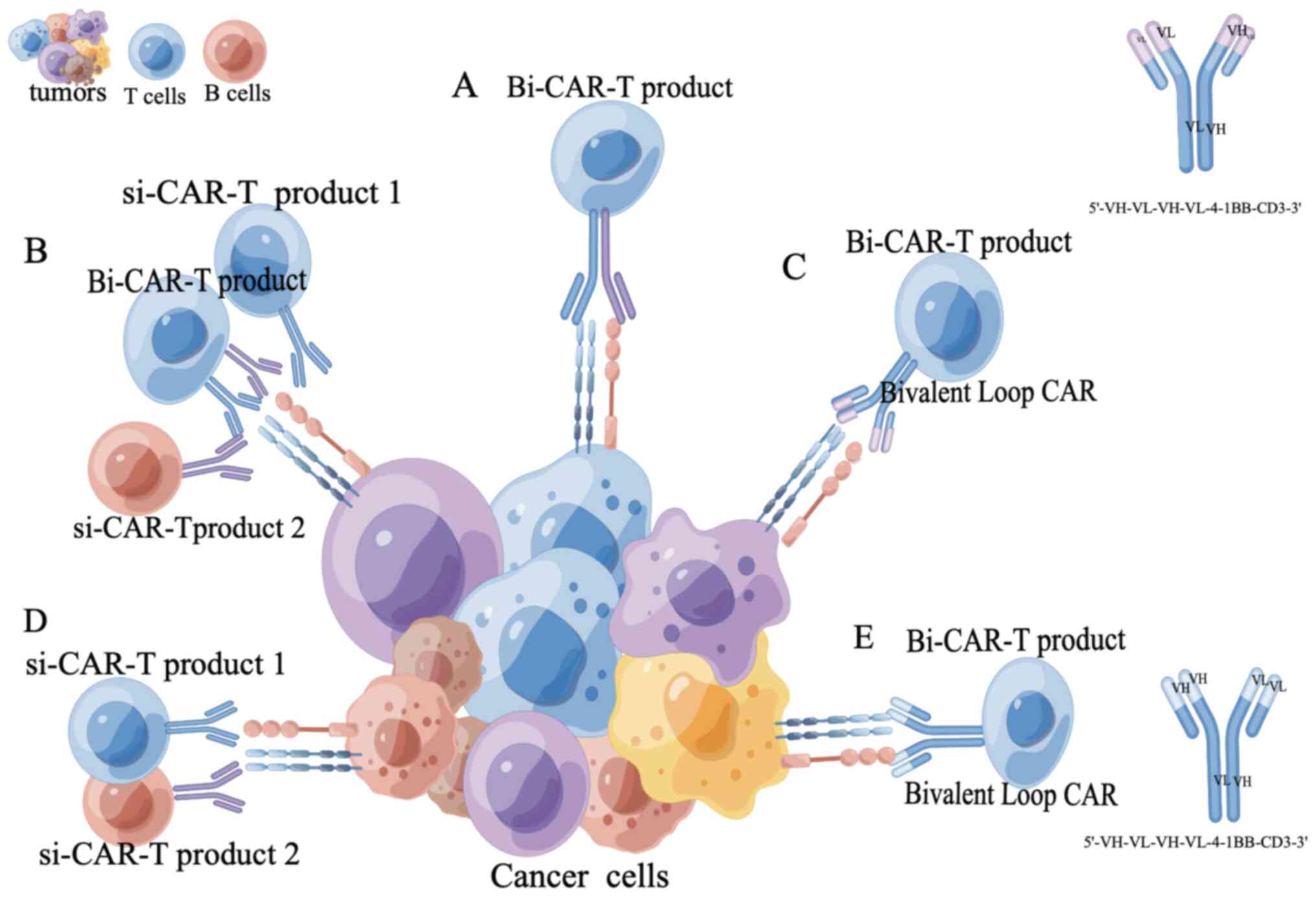
CAR-T cells targeting CD19 have been successful in
treating recurrent hematological malignancies, with an objective
response rate of >70%. Promising conversion rates have also been
achieved with B-cell maturation antigen's chlorpyrifos and
sitagliptin's self-microspheres, reaching 73 and 97%, respectively
(57). CAR-T cell therapy was shown
to have limitations, such as recurrence due to limited persistence
or functional inhibition of CAR-T cells or antigen escape (58). The use of dual-target CAR-T cell
therapy was shown to be more effective than single-target CAR-T
cells, and to also have a lower incidence of severe cytokine
release syndrome and no neurotoxicity (59). However, to achieve optimal results
in dual target CAR-T cell therapy, further optimization of CAR
target selection and CAR structure is necessary (60). Multi-target CAR-T cell therapy is a
crucial area of research with the potential to revolutionize tumor
immunotherapy in the future (Fig.
3).
Signaling cascades are crucial for various cellular
processes, such as proliferation, differentiation, migration,
intercellular communication and survival. Tumor heterogeneity in
GBM can lead to various outcomes, such as increased proliferation,
abnormal angiogenesis and evasion of cell apoptosis pathways. The
RTK (RAS-PI3K mTOR) pathway is of particular interest in GBM
research due to mutations in EGFR, VEGFR and PDGFR. Small-molecule
tyrosine kinase (TK) inhibitors, such as Nivozumab and Rigofenib,
selectively block single or multiple TK receptors (61). In addition, PI3K inhibitors, such as
Bupanib and GDC-0084, dual PI3K/mTOR inhibitors such as Dacoxib,
and mTOR inhibitors such as AZD8055, have been identified (62). As Myc is highly expressed in
glioblastoma, the combination of the CDK9 inhibitor zoteracib and
temozolomide have been shown to reduce the level of Myc (63,64).
Cell surface antigens, such as TK receptors found on glioma cells,
have the potential to be therapeutic targets for antibody-drug
conjugates. Clinical trials involving L19TNF have demonstrated low
toxicity and the combination of L19TNF with other chemotherapy
drugs, such as ICIs, may improve efficacy (65).
The search for new interventions at the genomic
level has resulted in the development of epigenetic treatments.
These treatments involve modifications of gene expression,
transcription and translation, which have the potential to suppress
oncogenes and support suppressor genes. By preventing
proliferation, causing cell cycle arrest and promoting apoptosis,
these interventions can effectively combat cancer. In addition,
miRNAs with epigenetic effects and histone deacetylase inhibitors
are potential drug candidates that can prevent glioma cell
proliferation. The drugs Vorinostat and Panobinostat were
investigated in conjunction with another medication for the
treatment of glioblastoma (66).
Glioblastoma is known for its anti-radiation
properties, which are attributed to various mechanisms, such as
microRNA, tumor heterogeneity, glioma stem cells, tumor
microenvironment, hypoxia, metabolic changes and DNA damage/repair.
One crucial player in DNA repair and prevention of cell apoptosis
is poly(ADP ribose) polymerase PARP. The PARP inhibitor Olaparib
has shown promising results in the treatment of breast cancer.
Research is currently being conducted on the use of Veliparib and
Fluzopanib in combination for the treatment of GBM (67). The detection and repair of DNA
double-strand breaks caused by radiation-induced DNA damage is
crucial and can be facilitated by DNA-dependent protein kinases
(DNA-PK). Inhibitors of DNA-PK, such as AZD7648 and NU7441, have
been studied (68). ATM and ATR
kinases have a crucial role in regulating DNA damage repair and
maintaining genomic stability. A promising development in this
field is the testing of AZD1390, an ATM molecular inhibitor, in
phase I trials for patients with GBM (69).
The optimization of GBM radiation therapy has a
crucial role in reducing side effects on normal brain tissue. Focal
brain radiation therapy has been implemented due to progress in
accuracy. Three-dimensional techniques such as intensity-modulated
radiation therapy and volume arc therapy have led to a decrease in
non-tumor tissue dose (70). The
particle therapy approach, including techniques such as gamma knife
and Zap-X, aims to minimize the dose to the edge of the tumor. This
can help to reduce the amount of radiation received by adjacent
tissues and organs at risk of the tumor. Proton beam therapy and
carbon ion radiotherapy are currently being extensively researched
for their potential to achieve this goal (71).
Tumor development is influenced by both genetic and
non-genetic factors, along with an imbalanced immune metabolism. It
is essential to identify potential targets for new therapeutic
regimens to shape the metabolic characteristics and immune
heterogeneity of the tumor microenvironment. By utilizing liquid
biopsy techniques to detect heterogeneous vesicles, valuable
insight into the presence of immune components in the tumor
microenvironment may be obtained. The present review proposes the
combination of radiotherapy and chemotherapy, as well as CAR-T
single-target and multi-target therapy, to address the
heterogeneity of tumor patients. Furthermore, liquid biopsy
technology is suggested as a means to provide an accurate treatment
basis and propose personalized treatment plans. Further research is
needed to determine the applicability of these strategies in
patients with different types of solid cancers and develop
effective treatment approaches. Precise and personalized treatment
plans are crucial, as they not only improve the prognosis and
maximize the treatment effect for tumor patients but also enhance
overall survival time and quality of life.
Not applicable.
This work was supported by grants from the National Natural
Science Foundation of China (grant no. 81960541/82060455).
Not applicable.
Conceptualization, WK; investigation, ZXM and WSL;
writing-original draft preparation, WSL, HFM and WK; writing-review
and editing, QZ. All authors have read and agreed to the published
version of the manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Junttila MR and de Sauvage FJ: Influence
of tumor micro-environment heterogeneity on therapeutic response.
Nature. 501:346–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nawaz S and Yuan Y: Computational
pathology: Exploring the spatial dimension of tumor ecology. Cancer
Lett. 380:296–303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greaves M and Maley CC: Clonal evolution
in cancer. Nature. 481:306–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Cazzato E, Ladewig E, Frattini V,
Rosenbloom DI, Zairis S, Abate F, Liu Z, Elliott O, Shin YJ, et al:
Clonal evolution of glioblastoma under therapy. Nat Genet.
48:768–476. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Azizi E, Carr AJ, Plitas G, Cornish AE,
Konopacki C, Prabhakaran S, Nainys J, Wu K, Kiseliovas V, Setty M,
et al: Single-cell map of diverse immune phenotypes in the breast
tumor microenvironment. Cell. 174:1293–1308.e36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chevrier S, Levine JH, Zanotelli VRT,
Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H,
et al: An immune atlas of clear cell renal cell carcinoma. Cell.
169:736–749.e18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Costa A, Kieffer Y, Scholer-Dahirel A,
Pelon F, Bourachot B, Cardon M, Sirven P, Magagna I, Fuhrmann L,
Bernard C, et al: Fibroblast heterogeneity and immunosuppressive
environment in human breast cancer. Cancer Cell. 33:463–479.e10.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jia Q, Wu W, Wang Y, Alexander PB, Sun C,
Gong Z, Cheng JN, Sun H, Guan Y, Xia X, et al: Local mutational
diversity drives intratumoral immune heterogeneity in non-small
cell lung cancer. Nat Commun. 9:53612018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Salmon H, Remark R, Gnjatic S and Merad M:
Host tissue determinants of tumor immunity. Nat Rev Cancer.
19:215–227. 2019.PubMed/NCBI
|
|
10
|
Dentro SC, Leshchiner I, Haase K,
Tarabichi M, Wintersinger J, Deshwar AG, Yu K, Rubanova Y,
Macintyre G, Demeulemeester J, et al: Characterizing genetic
intratumor heterogeneity across 2,658 human cancer genomes. Cell.
184:2239–2254.e39. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kumar S, Warrell J, Li S, McGillivray PD,
Meyerson W, Salichos L, Harmanci A, Martinez-Fundichely A, Chan
CWY, Nielsen MM, et al: Passenger mutations in more than 2,500
cancer genomes: Overall molecular functional impact and
consequences. Cell. 180:915–927.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rosenthal R, Cadieux EL, Salgado R, Bakir
MA, Moore DA, Hiley CT, Lund T, Tanić M, Reading JL, Joshi K, et
al: Neoantigen-directed immune escape in lung cancer evolution.
Nature. 567:479–485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv Y, Zhang S, Liu Z, Tian Y, Liang N and
Zhang J: Prognostic value of preoperative neutrophil to lymphocyte
ratio is superior to systemic immune inflammation index for
survival in patients with glioblastoma. Clin Neurol Neurosurg.
181:24–27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pasqualetti F, Giampietro C, Montemurro N,
Giannini N, Gadducci G, Orlandi P, Natali E, Chiarugi P, Gonnelli
A, Cantarella M, et al: Old and new systemic immune-inflammation
indexes are associated with overall survival of glioblastoma
patients treated with radio-chemotherapy. Genes (Basel).
13:10542022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Montemurro N, Pahwa B, Tayal A, Shukla A,
De Jesus Encarnacion M, Ramirez I, Nurmukhametov R, Chavda V and De
Carlo A: Macrophages in recurrent glioblastoma as a prognostic
factor in the synergistic system of the tumor microenvironment.
Neurol Int. 15:595–608. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Flavahan WA, Gaskell E and Bernstein BE:
Epigenetic plasticity and the hallmarks of cancer. Science.
357:eaal23802017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marks DL, Olson RL and Fernandez-Zapico
ME: Epigenetic control of the tumor microenvironment. Epigenomics.
8:1671–1687. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ishak CA, Classon M and De Carvalho DD:
Deregulation of retroelements as an emerging therapeutic
opportunity in cancer. Trends Cancer. 4:583–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dagogo-Jack I and Shaw AT: Tumour
heterogeneity and resistance to cancer therapies. Nat Rev Clin
Oncol. 15:81–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Martins I, Ma Y, Kepp O, Galluzzi
L and Kroemer G: Autophagy-dependent ATP release from dying cells
via lysosomal exocytosis. Autophagy. 9:1624–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stagg J and Smyth MJ: Extracellular
adenosine triphosphate and adenosine in cancer. Oncogene.
29:5346–5358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yost KE, Satpathy AT, Wells DK, Qi Y, Wang
C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, et al:
Clonal replacement of tumor-specific T cells following PD-1
blockade. Nat Med. 25:1251–1259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zou W, Wolchok JD and Chen L: PD-L1
(B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms,
response biomarkers, and combinations. Sci Transl Med.
8:328rv42016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Batlle E and Massagué J: Transforming
growth factor-β signaling in immunity and cancer. Immunity.
50:924–940. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding R, Liu S, Wang S, Chen H, Wang F, Xu
Q, Zhu L, Dong X, Gu Y, Zhang X, et al: Single-cell transcriptome
analysis of the heterogeneous effects of differential expression of
tumor PD-L1 on responding TCR-T cells. Theranostics. 11:4957–4974.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lamplugh Z and Fan Y: Vascular
microenvironment, tumor Immunity and Immunotherapy. Front Immunol.
12:8114852021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu T, Dai LJ, Wu SY, Xiao Y, Ma D, Jiang
YZ and Shao ZM: Spatial architecture of the immune microenvironment
orchestrates tumor immunity and therapeutic response. J Hematol
Oncol. 14:982021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Montenegro F and Indraccolo S: Metabolism
in the tumor microenvironment. Adv Exp Med Biol. 1263:1–11. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Binnewies M, Roberts EW, Kersten K, Chan
V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI,
Ostrand-Rosenberg S, Hedrick CC, et al: Understanding the tumor
immune microenvironment (TIME) for effective therapy. Nat Med.
24:541–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Joshi K, de Massy MR, Ismail M, Reading
JL, Uddin I, Woolston A, Hatipoglu E, Oakes T, Rosenthal R, Peacock
T, et al: Spatial heterogeneity of the T cell receptor repertoire
reflects the mutational landscape in lung cancer. Nat Med.
25:1549–1559. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bastola S, Pavlyukov MS, Yamashita D,
Ghosh S, Cho H, Kagaya N, Zhang Z, Minata M, Lee Y, Sadahiro H, et
al: Glioma-initiating cells at tumor edge gain signals from tumor
core cells to promote their malignancy. Nat Commun. 11:46602020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lambrechts D, Wauters E, Boeckx B, Aibar
S, Nittner D, Burton O, Bassez A, Decaluwé H, Pircher A, Van den
Eynde K, et al: Phenotype molding of stromal cells in the lung
tumor microenvironment. Nat Med. 24:1277–1289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu J and Thompson CB: Metabolic
regulation of cell growth and proliferation. Nat Rev Mol Cell Biol.
20:436–450. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sayaman RW, Saad M, Thorsson V, Hu D,
Hendrickx W, Roelands J, Porta-Pardo E, Mokrab Y, Farshidfar F,
Kirchhoff T, et al: Germline genetic contribution to the immune
landscape of cancer. Immunity. 54:367–386.e8. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maynard A, McCoach CE, Rotow JK, Harris L,
Haderk F, Kerr DL, Yu EA, Schenk EL, Tan W, Zee A, et al:
Therapy-induced evolution of human lung cancer revealed by
single-cell RNA sequencing. Cell. 182:1232–1251.e22. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bernard V, Semaan A, Huang J, San Lucas
FA, Mulu FC, Stephens BM, Guerrero PA, Huang Y, Zhao J, Kamyabi N,
et al: Single-cell transcriptomics of pancreatic cancer precursors
demonstrates epithelial and microenvironmental heterogeneity as an
early event in neoplastic progression. Clin Cancer Res.
25:2194–2205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mascaux C, Angelova M, Vasaturo A, Beane
J, Hijazi K, Anthoine G, Buttard B, Rothe F, Willard-Gallo K,
Haller A, et al: Immune evasion before tumor invasion in early lung
squamous carcinogenesis. Nature. 571:570–575. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Riaz N, Havel JJ, Makarov V, Desrichard A,
Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH,
et al: Tumor and microenvironment evolution during immunotherapy
with nivolumab. Cell. 171:934–949.e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang T, Yan Y, Zhou K, Su C, Ren S, Li N,
Hou L, Guo X, Zhu W, Zhang H, et al: Characterization of evolution
trajectory and immune profiling of brain metastasis in lung
adenocarcinoma. NPJ Precis Oncol. 5:62021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
AbdulJabbar K, Raza SEA, Rosenthal R,
Jamal-Hanjani M, Veeriah S, Akarca A, Lund T, Moore DA, Salgado R,
Al Bakir M, et al: Geospatial immune variability illuminates
differential evolution of lung adenocarcinoma. Nat Med.
26:1054–1062. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gatto L, Franceschi E, Di Nunno V, Tosoni
A, Lodi R and Brandes AA: Liquid biopsy in glioblastoma management:
From current research to future perspectives. Oncologist.
26:865–878. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Postel M, Roosen A, Laurent-Puig P, Taly V
and Wang-Renault SF: Droplet-based digital PCR and next generation
sequencing for monitoring circulating tumor DNA: A cancer
diagnostic perspective. Expert Rev Mol Diagn. 18:7–17. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Karlin-Neumann G: Improved liquid biopsies
with combined digital PCR and next-generation sequencing. Am Lab
Mag. 48:17–19. 2016.
|
|
44
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Piccioni DE, Achrol AS, Kiedrowski LA,
Banks KC, Boucher N, Barkhoudarian G, Kelly DF, Juarez T, Lanman
RB, Raymond VM, et al: Analysis of cell-free circulating tumor DNA
in 419 patients with glioblastoma and other primary brain tumors.
CNS Oncol. 8:CNS342019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mair R, Mouliere F, Smith CG, Chandrananda
D, Gale D, Marass F, Tsui DWY, Massie CE, Wright AJ, Watts C, et
al: Measurement of plasma cell-free mitochondrial tumor DNA
improves detection of glioblastoma in patient-derived orthotopic
xenograft models. Cancer Res. 79:220–230. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Janjua TI, Rewatkar P, Ahmed-Cox A, Saeed
I, Mansfeld FM, Kulshreshtha R, Kumeria T, Ziegler DS, Kavallaris
M, Mazzieri R and Popat A: Frontiers in the treatment of
glioblastoma: Past, present and emerging. Adv Drug Deliv Rev.
171:108–138. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wong ET, Lok E and Swanson KD: Clinical
benefit in recurrent glioblastoma from adjuvant NovoTTF-100A and
TCCC after temozolomide and bevacizumab failure: A preliminary
observation. Cancer Med. 4:383–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Taal W, Oosterkamp HM, Walenkamp AM,
Dubbink HJ, Beerepoot LV, Hanse MC, Buter J, Honkoop AH, Boerman D,
de Vos FY, et al: Single-agent bevacizumab or lomustine versus a
combination of bevacizumab plus lomustine in patients with
recurrent glioblastoma (BELOB trial): A randomised controlled phase
2 trial. Lancet Oncol. 15:943–953. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cloughesy TF, Mochizuki AY, Orpilla JR,
Hugo W, Lee AH, Davidson TB, Wang AC, Ellingson BM, Rytlewski JA,
Sanders CM, et al: Neoadjuvant anti-PD-1 immunotherapy promotes a
survival benefit with intratumoral and systemic immune responses in
recurrent glioblastoma. Nat Med. 25:477–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Di Cintio F, Dal Bo M, Baboci L, De Mattia
E, Polano M and Toffoli G: The molecular and microenvironmental
landscape of glioblastomas: Implications for the novel treatment
choices. Front Neurosci. 14:6036472020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Reardon DA, Desjardins A, Vredenburgh JJ,
O'Rourke DM, Tran DD, Fink KL, Nabors LB, Li G, Bota DA, Lukas RV,
et al: Rindopepimut with bevacizumab for patients with relapsed
EGFRvIII-expressing glioblastoma (ReACT): Results of a double-blind
randomized phase II trial. Clin Cancer Res. 26:1586–1594. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fenstermaker RA, Ciesielski MJ, Qiu J,
Yang N, Frank CL, Lee KP, Mechtler LR, Belal A, Ahluwalia MS and
Hutson AD: Clinical study of a survivin long peptide vaccine
(SurVaxM) in patients with recurrent malignant glioma. Cancer
Immunol Immunother. 65:1339–1352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Y, Mudgal P, Wang L, Wu H, Huang N,
Alexander PB, Gao Z, Ji N and Li QJ: T cell receptor repertoire as
a prognosis marker for heat shock protein peptide complex-96
vaccine trial against newly diagnosed glioblastoma. Oncoimmunology.
9:17494762020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cloughesy TF, Landolfi J, Vogelbaum MA,
Ostertag D, Elder JB, Bloomfield S, Carter B, Chen CC, Kalkanis SN,
Kesari S, et al: Durable complete responses in some recurrent
high-grade glioma patients treated with Toca 511 + Toca FC. Neuro
Oncol. 20:1383–1392. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ene CI, Fueyo J and Lang FF: Delta-24
adenoviral therapy for glioblastoma: Evolution from the bench to
bedside and future considerations. Neurosurg Focus. 50:E62021.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Raje N, Berdeja J, Lin Y, Siegel D,
Jagannath S, Madduri D, Liedtke M, Rosenblatt J, Maus MV, Turka A,
et al: Anti-BCMA CAR T-Cell therapy bb2121 in relapsed or
refractory multiple myeloma. N Engl J Med. 380:1726–1737. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lemoine J, Ruella M and Houot R: Born to
survive: How cancer cells resist CAR T cell therapy. J Hematol
Oncol. 14:1992021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang H, Gao L, Liu L, Wang J, Wang S, Gao
L, Zhang C, Liu Y, Kong P, Liu J, et al: A Bcma and CD19 bispecific
CAR-T for relapsed and refractory multiple myeloma. Blood.
134:31472019. View Article : Google Scholar
|
|
60
|
Xie B, Li Z, Zhou J and Wang W: Current
status and perspectives of dual-targeting chimeric antigen receptor
T-cell therapy for the treatment of hematological malignancies.
Cancers (Basel). 14:32302022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen H, Kuhn J, Lamborn KR, Abrey LE,
DeAngelis LM, Lieberman F, Robins HI, Chang SM, Yung WKA, Drappatz
J, et al: Phase I/II study of sorafenib in combination with
erlotinib for recurrent glioblastoma as part of a 3-arm sequential
accrual clinical trial: NABTC 05-02. Neurooncol. 2:vdaa1242020.
|
|
62
|
Jia Q, Wang A, Yuan Y, Zhu B and Long H:
Heterogeneity of the tumor immune microenvironment and its clinical
relevance. Exp Hematol Oncol. 11:242022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang H, Tao Z, Feng M, Li X, Deng Z, Zhao
G, Yin H, Pan T, Chen G, Feng Z, et al: Dual PLK1 and STAT3
inhibition promotes glioblastoma cells apoptosis through MYC.
Biochem Biophys Res Commun. 533:368–375. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu J, Yuan Y, Long Priel DA, Fink D, Peer
CJ, Sissung TM, Su YT, Pang Y, Yu G, Butler MK, et al: Phase I
study of zotiraciclib in combination with temozolomide for patients
with recurrent high-grade astrocytomas. Clin Cancer Res.
27:3298–3306. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Weiss T, Puca E, Silginer M, Hemmerle T,
Pazahr S, Bink A, Weller M, Neri D and Roth P: Immunocytokines are
a promising immunotherapeutic approach against glioblastoma. Sci
Transl Med. 12:eabb23112020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zang L, Kondengaden SM, Che F, Wang L and
Heng X: Potential epigenetic-based therapeutic targets for glioma.
Front Mol Neurosci. 11:4082018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Higuchi F, Nagashima H, Ning J, Koerner
MVA, Wakimoto H and Cahill DP: Restoration of temozolomide
sensitivity by PARP inhibitors in mismatch repair deficient
glioblastoma is independent of base excision repair. Clin Cancer
Res. 26:1690–1699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kopa P, Macieja A, Gulbas I, Pastwa E and
Poplawski T: Inhibition of DNA-PK potentiates the synergistic
effect of NK314 and etoposide combination on human glioblastoma
cells. Mol Biol Rep. 47:67–76. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lesueur P, Chevalier F, El-Habr EA, Junier
MP, Chneiweiss H, Castera L, Müller E, Stefan D and Saintigny Y:
Radiosensitization effect of talazoparib, a PARP inhibitor, on
glioblastoma stem cells exposed to low and high linear energy
transfer radiation. Sci Rep. 8:36642018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shaffer R, Nichol AM, Vollans E, Fong M,
Nakano S, Moiseenko V, Schmuland M, Ma R, McKenzie M and Otto K A:
A comparison of volumetric modulated arc therapy and conventional
intensity-modulated radiotherapy for frontal and temporal
high-grade gliomas. Int J Radiat Oncol Biol Phys. 76:1177–1184.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bunevicius A and Sheehan JP: Radiosurgery
for glioblastoma. Neurosurg Clin N Am. 32:117–128. 2021. View Article : Google Scholar : PubMed/NCBI
|